Meiotic crossover in Arabidopsis is shaped by the combination of three major sequence motifs surrounded by a distinctive epigenetic landscape.
Abstract
The rate of crossover, the reciprocal exchanges of homologous chromosomal segments, is not uniform along chromosomes differing between male and female meiocytes. To better understand the factors regulating this variable landscape, we performed a detailed genetic and epigenetic analysis of 737 crossover events in Arabidopsis thaliana. Crossovers were more frequent than expected in promoters. Three DNA motifs enriched in crossover regions and less abundant in crossover-poor pericentric regions were identified. One of these motifs, the CCN repeat, was previously unknown in plants. The A-rich motif was preferentially associated with promoters, while the CCN repeat and the CTT repeat motifs were preferentially associated with genes. Analysis of epigenetic modifications around the motifs showed, in most cases, a specific epigenetic architecture. For example, we show that there is a peak of nucleosome occupancy and of H3K4me3 around the CCN and CTT repeat motifs while nucleosome occupancy was lowest around the A-rich motif. Cytosine methylation levels showed a gradual decrease within ∼2 kb of the three motifs, being lowest at sites where crossover occurred. This landscape was conserved in the decreased DNA methylation1 mutant. In summary, the crossover motifs are associated with epigenetic landscapes corresponding to open chromatin and contributing to the nonuniformity of crossovers in Arabidopsis.
INTRODUCTION
The process of meiotic recombination is initiated by DNA double-strand break (DSB) induction. When the ends of a broken DNA invade homologous sequences on the homologous chromosome, a heteroduplex intermediate is formed and its resolution gives rise to a crossover (CO) event, namely, the reciprocal exchange of large homologous chromosomal segments and/or to a non-crossover event (San Filippo et al., 2008), namely, a nonreciprocal exchange of short DNA sequences found in the heteroduplex intermediate. This can result in heterozygosity loss or gene conversion (San Filippo et al., 2008; Baudat et al., 2013). It should be noted that only a minority of the DSBs induced at the onset of meiosis turn into CO events (Youds and Boulton, 2011). Understanding the regulation and the landscape of CO and non-crossover events has been a major endeavor in genetic research, as early as a century ago with studies on genetic linkage in sweet pea (Lathyrus odoratus; Bateson et al., 1906), followed by Thomas Hunt Morgan’s (Morgan, 1911) work in Drosophila melanogaster, determining the rate of meiotic CO between linked loci.
Owing to the augmentation of mapped genetic markers, studies have benefited from an increasing degree of resolution in the mapping of CO events. One approach is high-throughput screening of many recombination events in a defined region. In this approach, two notable methods are pollen typing (Drouaud et al., 2013) and the use of fluorescent markers in seed (Melamed-Bessudo et al., 2005) and in pollen tetrads (Preuss et al., 1994; Francis et al., 2006, 2007; Berchowitz and Copenhaver, 2008; Sun et al., 2012; Yelina et al., 2013). With the recent advent of high-throughput sequencing technologies, it became possible to map historical recombination events using linkage disequilibrium, as has been done for human, mice, Arabidopsis thaliana, and maize (Zea mays) (Myers et al., 2008; Gore et al., 2009; Brunschwig et al., 2012; Choi et al., 2013). It is also possible to map recombination events at high resolution, by crossing strains with defined sequence polymorphisms, followed by whole-genome sequencing of the resulting progenies. High-resolution maps of genome-wide recombination events are now available for Saccharomyces cerevisiae, Drosophila, maize, and Theileria parva (Mancera et al., 2008; Comeron et al., 2012; Henson et al., 2012; Li et al., 2015; Rodgers-Melnick et al., 2015). Furthermore, high-throughput sequencing, combined with single-cell technology, contributed to the detection of meiotic recombination of single sperm (Lu et al., 2012b; Wang et al., 2012) and single oocyte (Hou et al., 2013) genotyping. Precise detection of the DSBs initiating meiotic recombination can be obtained through methods involving immunopurification of DSB binding proteins, such as Sporulation 11 (SPO11), Radiation sensitive 51, and Disrupted Meiotic cDNA1 (Smagulova et al., 2011; Choi and Henderson, 2015).
Recent studies were conducted to assess the recombination landscape of Arabidopsis by dense genome mapping (Giraut et al., 2011), as well as whole-genome sequencing (Lu et al., 2012a; Yang et al., 2012; Wijnker et al., 2013). The work of Giraut et al. (2011) and others (Vizir and Korol, 1990; Copenhaver et al., 1998; Melamed-Bessudo and Levy, 2012) showed a lack of uniformity and distinct CO landscapes in the male versus female lineage. Recently, CO events were mapped through whole-genome sequencing of F2 plants (Yang et al., 2012), meiotic tetrads (Lu et al., 2012a), and dihaploids (Wijnker et al., 2013) or through sequencing of ecotypes and linkage disequilibrium analysis (Choi et al., 2013). This enabled the identification of sequence motifs that are enriched at CO loci, such as A-rich motifs and CTT repeats (Horton et al., 2012; Choi et al., 2013; Wijnker et al., 2013).
We sequenced the whole genome of 24 F2 plants resulting from a cross between the Columbia (Col) and Landsberg erecta (Ler) accessions. Using these data, together with information from previous studies (Lu et al., 2012a; Yang et al., 2012; Wijnker et al., 2013), a data set of 737 different CO events was compiled, 424 of which were localized at very high resolution (<2000 bp). Using this data set, we verified the previously reported enrichment of A-rich and CTT repeat motifs at sites of CO (Horton et al., 2012; Choi et al., 2013; Wijnker et al., 2013). In addition, we identified the CCN repeat, a newly characterized motif that is associated with CO events and is enriched in subtelomeric regions that are characterized with a higher CO rate in the male compared with female cell lineage. A characteristic epigenetic landscape was found around CO motifs. We propose that the sequence motifs, together with the associated epigenetic landscapes, contribute to the nonrandom CO distribution in the Arabidopsis genome.
RESULTS
Defining CO Sites
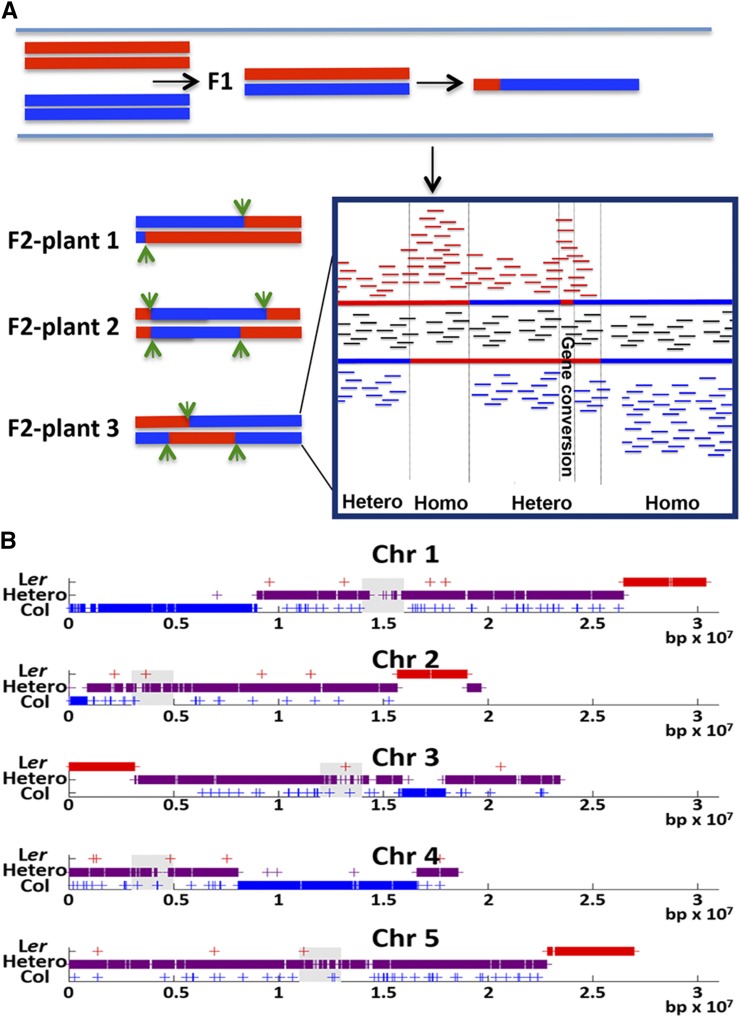
To study the control of the meiotic recombination landscape in Arabidopsis, whole-genome sequencing of 24 individual F2 progeny plants derived from a cross between two Arabidopsis ecotypes, Col and Ler, which differ every ∼300 bp on average, in sequence polymorphisms (Lu et al., 2012a), was performed. We used a meiotic tester line previously generated by transformation of the Col line with two seed-specific fluorescent markers on chromosome 3, i.e., GFP and RFP, mapped ∼15 centimorgans apart (Melamed-Bessudo et al., 2005). A CO event between these markers resulted in red only or green only seeds. F2 plants that underwent CO events between these markers were chosen for whole-genome sequencing, as they served as a positive control for our ability to identify CO events.
To ensure high-quality reads for each of the F2 individuals, the quality of the reads was verified with fastqc (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc) (for a detailed explanation and read depths, see Methods and Supplemental Table 1). Detection of recombination events was done by alignment of the sequence reads to the Arabidopsis reference genome TAIR10. For each individual F2, we determined the percentage of zygosity along a given chromosome section, according to the ratio of reads that supported each parental single nucleotide polymorphism (SNP). Regions with only one type of SNP (90 to 100%) were considered homozygous, while regions with two types of SNPs (50% ± 10%) were considered heterozygous. Crossover events that were consistent and were not followed by a reverse transition in the following 50 kb were manually selected as transitions of DNA segments from homozygosity to heterozygosity and vice versa (Figure 1A). The CO site was defined as the region between the closest homozygote and heterozygote SNPs (Figure 1B). In total, 201 CO sites were gathered from the 24 F2 plants analyzed. This is close to the expected number of CO events according the rate reported by Giraut et al. (2011) per male and female gametes (5.575 and 3.3, respectively), i.e., 8.875 COs per F2 plant, which amounts to 213 expected CO events in 24 plants (8.875 × 24 = 213). To generate a large data set of CO sites, the 201 CO events from this analysis were combined with previously published raw data (Lu et al., 2012a; Yang et al., 2012) and with the CO events listed in the Wijnker et al. (2013) study. Although the frequency of gene conversion events in the data of Yang et al. (2012) was not supported by other studies (Qi et al., 2014), the quality of the sequence data for CO detection was not challenged (Qi et al., 2014) and was supported by our quality control analysis. The combined data set included 737 CO events with fine resolution (median: 1534 bp). Their genomic distribution is shown in Supplemental Figure 1. Crossover events between the fluorescent seed markers on chromosome 3 (positive control) were detected in all the 24 sequenced plants. This subset of events represents only 24/737 (∼3%) of all CO events and has only a negligible effect on the genome-wide distribution of CO events. Overall, the 737 CO sites were dispersed throughout the genome (Supplemental Figure 1), with a distribution fairly similar to the recombination landscape previously published using molecular markers (Spearman correlation = 0.51) (Giraut et al., 2011).
Figure 1.
Identification of CO Events through Next-Generation Sequencing of F2 Plants.
(A) Overall design of detection of CO events. DNA segments are colored according to the supporting parent-specific SNP: Ler (red) and Col (blue). Samples with SNPs from both parents were considered heterozygotes (purple). Reads supporting parent-specific SNPs are shown as the horizontal lines in the frame. Green arrows and vertical bars (in frame) show transition between zygosity states.
(B) Recombination landscape in the five chromosomes (Chr) of one F2 plant. Centromeres are indicated as gray boxes. Plus signs indicate patches different from the surrounding zygotic level that have reverse transition to the surrounding zygotic level within a distance smaller than 50 kb.
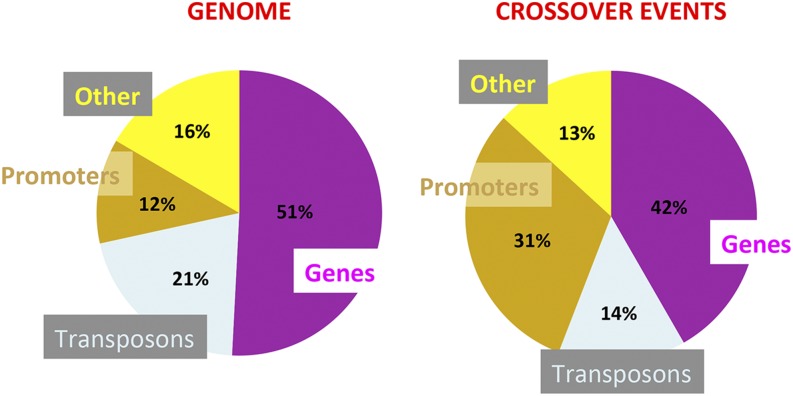
Owing to the high-resolution mapping of the CO events, 31% of the events were identified in promoters (defined as the 500 bp segments upstream of the transcription start site), although promoters represent only 12% of the entire genome (Figure 2), suggesting preferential occurrence of COs in promoter regions. Conversely, COs in transposable elements occurred less frequently than expected (14% compared with 21%). Crossovers occurred to a large extent (42%) in genes, as defined in TAIR10, namely, starting from the transcription start site up to the end of the transcript, although at a rate slightly lower than expected (51%) (Figure 2).
Figure 2.
Association of Motifs with Known Genomic Features.
Percentage of genomic features, such as promoters (defined as 500 bp upstream of transcription start site), genes (5′ and 3′ untranslated regions, exons, and introns), transposons, and others (all the remaining sequences, such as non-annotated repeats and tRNAs), within the whole genome (left panel) and within the data set of the 737 CO events (right panel). Genes are represented in purple, promoters in brown, transposons in light blue, and “others” in yellow. COs that overlapped with more than one feature were counted once for each feature.
Analysis of CO Motifs
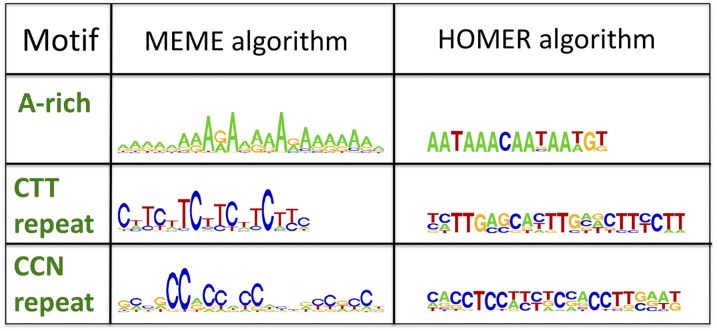
To search for motifs that are enriched in the CO regions, we compared the regions of the 424 CO events that mapped at high resolution (<2000 bp, 837 bp average length) to a set of similar size composed of 300 random 1000 bp sequences from the TAIR10 genome (henceforth, Rand genome). Using the MEME suite (Bailey et al., 2009) (discriminative analysis), a significant enrichment of sequence motifs at CO sites was detected. Two of these motifs were the A-rich motif (mean E-value = 1.67e-186) and the CTT repeat (mean E-value = 7.67e-65) (Figure 3), which were previously described as being associated with CO hot spots (Horton et al., 2012; Choi et al., 2013; Wijnker et al., 2013). In addition, this analysis revealed a novel motif composed of repeated cytosine pairs (CCN; mean E-value = 6.33e-22) (Figure 3). Notably, this CCN repeat motif resembled the human CO hot spot motif (Myers et al., 2008) (Figure 3). All three CO motifs, i.e., the A-rich, CTT, and CCN repeats, were abundant throughout the genome, reaching 85186, 50,977, and 58,803 copies, respectively. Interestingly, the abundance of the three motifs was significantly reduced in the ∼0.5- to 2-Mb region around the centromeres (Figure 4A; Supplemental Figure 2). To further validate the identified motifs, particularly the new CCN repeat, we used a second motif discovery tool called HOMER, an algorithm that accounts for sequence bias in the data set and provides a P value based on cumulative binomial distributions of motif occurrences in the target sequences versus flanking background sequences (target sequences<<background sequences) (Heinz et al., 2010). The same high-resolution CO data set used for the previous analysis was used as the input and the whole genome was used as a control. Surprisingly, the novel CCN motif was found to be the most significant of the three motifs (P = 1e-14) (Figure 3). The A-rich motif was found to be much less significant than the other motifs when using HOMER (P = 1e-10) than when using MEME (P = 1e-186) (Figure 3). We found that the regions where CO occurred were, on average, AT-rich (Supplemental Figures 3A to 3C); therefore, we tested the hypothesis that the highly abundant A-rich motif decreased in significance when using HOMER because it does not contrast well with its immediate background surrounding. When searching for a decay in motif signal in windows of 2 kb from the CO site, the A-rich motif was found to maintain a high rate of occurrence at long distances from the CO events (Supplemental Figure 3D). This finding suggests the possibility of a regional effect, rather than a motif-specific effect. By contrast, the CO rate decayed rapidly around the CCN repeat motif (Supplemental Figure 3D), suggesting a motif-specific effect. The CTT repeat motif as found by MEME (Figure 3) was not identified using HOMER; rather, a variation of this motif (Figure 3) was found to be significant (P = 1e-11).
Figure 3.
DNA CO Motifs.
Logo plots of DNA sequence showing motifs enriched in CO regions. Three motifs were identified using two different algorithms: MEME (left) and HOMER (right). An A-rich motif (E-value = 1.67e-186; P value = 1e-10), a CTT repeat motif (E-value = 7.67e-65; P value = 1e-11), and a CCN repeat motif (E-value = 6.33e-22; P value = 1e-14). The significance was expressed as E-value when using MEME or P value when using HOMER.
Figure 4.
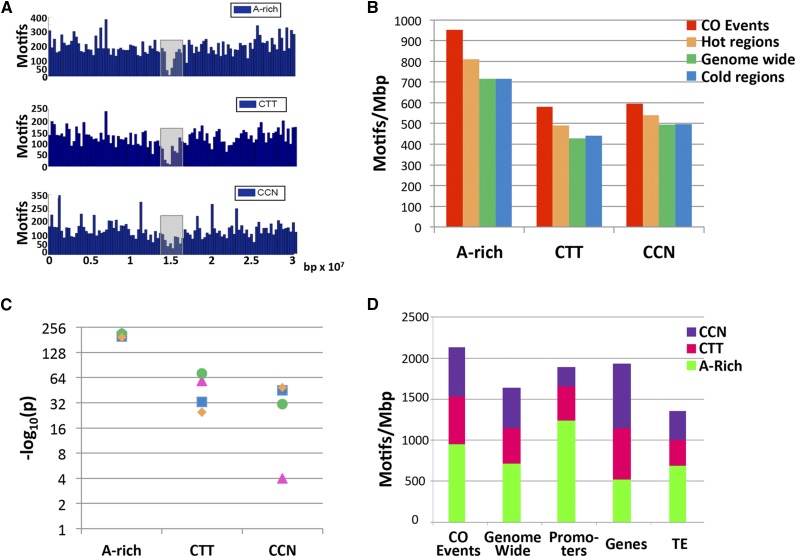
Abundance and Distribution of the Three CO Motifs.
(A) Distribution of the A-rich, CTT repeat, and CNN repeat motifs in chromosome 1 in the upper, middle, and lower panels, respectively. The pericentric region (∼2 Mb around the centromere) is shown as a gray box.
(B) Motif abundance (normalized as the number of motifs per Mb) is shown for the high-resolution CO data set (red), hot regions (orange), genome-wide (green), and cold regions (blue).
(C) Manhattan plot of the statistical significance (-log10 of P value) of the CO data set compared with each of the contrast groups: genome-wide (green circles), subtelomeric female-specific cold intervals (pink triangle), general cold intervals (blue squares), and hot regions (orange diamonds).
(D) Distribution of motifs within various data sets. CO events: data set of 737 CO events; genome wide: whole genome; promoters: 500 bp upstream of the transcription start site; genes: 5′ and 3′ untranslated regions, exons, introns, and transposable elements (TE).
The abundance of motifs is shown (Figure 4B) for various regions of the genome, namely, the CO data set including 424 CO events, the hot regions data set, the total genome, and data set of cold regions. The hot and cold regions and the regions showing differences between male and female, were defined based on the statistical analyses performed by Giraut et al. (2011). In brief, a male and a female CO data set were obtained using reciprocal backcrosses of a Col × Ler F1 and genotyping with SNP markers. A region was considered hot or cold if its CO rate was significantly higher or lower than the average rate in the same chromosome arm (Giraut et al., 2011). The regions where CO rates were significantly different between the male and female were also determined using appropriate statistical analyses (Giraut et al., 2011).
The A-rich motif was the most abundant in the CO events (952.8 motif/Mb), followed by the CTT repeat (581.4 motif/Mb) and the CCN repeat (595.5 motif/Mb) (Figure 4B). A closer examination demonstrated that motifs were not only enriched in CO regions (as expected) but were also enriched in hot versus cold regions (Figure 4B). The statistical significance of the difference between the CO data set (424 CO events mapped with the highest resolution <2000 bp) and the contrast groups (hot regions, random data set, and general cold regions) is presented in a Manhattan plot (Figure 4C). All the motifs showed a highly significant difference between the CO data set and the contrast groups (Figure 4C), with the exception of the CCN repeat motif, which was enriched in COs, when compared with general cold intervals (4.33e-22) or to a random genome (6.33e-22), but was not significantly enriched when compared with the female-specific cold regions (3.27e-03), which almost completely overlapped with the subtelomeric regions (Figure 4C). In other words, the CCN motif is abundant in subtelomeric regions, which are recombinogenic in the male cell lineage but cold in the female.
In addition, when assessing the abundance of motifs in genomic features, genes, promoters (500 bp upstream to the transcription start site), and transposable elements (Figure 4D), the A-rich motif was found to be mostly associated with promoters, while the CCN motif was found to be enriched in gene bodies (Figure 4D).
Epigenetic Architecture of COs around Motifs
Epigenetic imprints such as cytosine methylation (Melamed-Bessudo and Levy, 2012; Mirouze et al., 2012; Yelina et al., 2012; Colomé-Tatché et al., 2012) and histone occupancy and modifications were shown to be associated with CO rates (Yelina et al., 2012; Choi et al., 2013). Moreover, one of the motifs (CCN) is a target for H3K4 trimethylation in human (Hayashi et al., 2005; Baudat et al., 2010). Therefore, we analyzed the epigenetic architecture of CO regions and around motifs, using the epigenetic modification data set compiled by Zemach et al. (2013) from somatic tissues (see details in Methods). The analysis was performed around the three motifs throughout the genome, in cold or hot CO intervals, and around the 424 motifs where COs actually occurred.
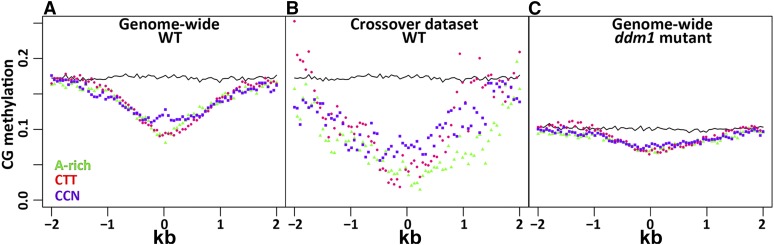
The abundance of the relevant modification was averaged for every 50 bp interval both 1 to 2 kb upstream and downstream of the motif (Figures 5 and 6; Supplemental Figures 4 and 5). The prominent characteristics of the motif-centric epigenetic landscape were the shape of the distribution and the degree of difference between hot and cold intervals. For example, a heterochromatic marker, such as CG methylation, showed hypomethylation at the ∼2-kb region around the three motifs (Figure 5A), with cold regions being more methylated than hot regions (Supplemental Figure 5A). We observed that the CG methylation levels were even lower when we analyzed the CO sites in our high-resolution data set (Figure 5B). The other types of cytosine methylation, CHH and CHG, showed a similar pattern to CG methylation (Supplemental Figures 4A to 4D, 5C, and 5D).
Figure 5.
The CG Methylation Landscape around the CO Motifs.
The ratio of methylated CG dinucleotides versus total CGs is shown in 50-bp bins around the CO motifs, namely, the CCN repeat (purple square), CTT repeat (red circle), and the A-rich motif (green triangle). As a control, CG methylation was analyzed around randomly picked sequences (continuous black line). Motifs are located at the center (0 bp) of the x axis. Each point is the average CG methylation for all the motifs in a given bin. The analysis was performed for all the motifs in the wild-type genome (A), for the data set of the actual CO events detected here (B), and for the whole genome in the ddm1 mutant (C).
Figure 6.
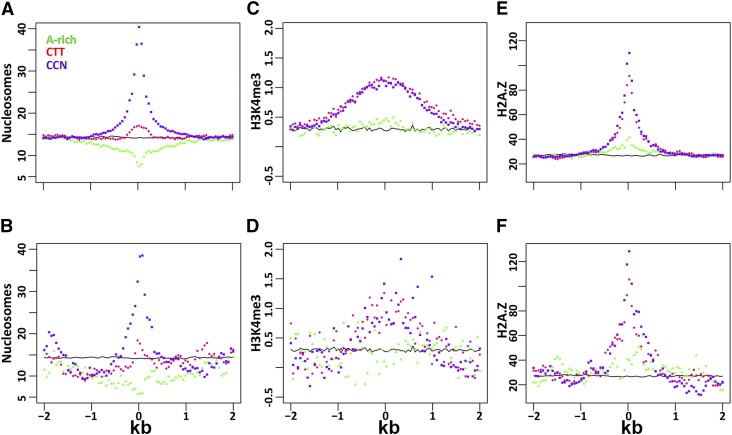
Epigenetic Landscape around the CO Motifs.
Epigenetic marks were analyzed around the CCN repeat (purple square), the CTT repeat (red circle), the A-rich CO motifs (green triangle), and the randomly picked control sequences (continuous black line). All the motifs are at the center of the x axis (0 bp). The average epigenetic mark value is shown on the y axis for 50-bp bins from the center of the x axis and up to 2000 bp upstream and downstream of the motifs. The epigenetic marks were analyzed in the whole wild-type genome ([A], [C], and [E]) and for the data set of CO events ([B], [D], and [F]). The epigenetic marks analyzed here are the nucleosome occupancy ([A] and [B]), H3K4me3 modifications ([C] and [D]), and variations in H2A.Z histone marks ([E] and [F]).
Previous experiments pointed to the DNA methylation maintenance gene DDM1 as a regulator of CO rates (Melamed-Bessudo and Levy, 2012). We tested whether hypomethylation at the motifs is DDM1 dependent. To this end, we analyzed the cytosine methylation landscape around the motifs in the ddm1 mutant (Figure 5C). As expected, the average degree of methylation was lower in the ddm1 mutant than in the wild type. However, the pattern of methylation was maintained, with the lowest degree of methylation found close to the motif itself. Interestingly, the methylation level around the motif was even lower in the data set of actual CO events (Figures 5B and 5C) than in the ddm1 mutant.
High nucleosome occupancy is also a heterochromatic marker (Chodavarapu et al., 2010). CO events are more likely to occur in euchromatic regions (Giraut et al., 2011). Moreover, A/T dinucleotides are known to have low nucleosome occupancy compared with C/G dinucleotides where the occupancy is high (Kaplan et al., 2009). As expected, based on sequence only, nucleosome occupancy was low at the A-rich motif, was slightly higher at the CTT repeat and was high at the CCN motif (Figure 6A). Occupancy was higher in cold versus hot regions for the CCN motif (Supplemental Figure 5G). A peak of occupancy was also observed around motifs found in the actual CO regions (Figure 6B). When analyzing H3 presence, another parameter of nucleosome occupancy, the general trends were similar to those of nucleosome occupancy, namely, depletion around the A-rich motif and enrichment around the CCN motif (Supplemental Figures 4E and 5H), supporting the functional significance of the linkage between motifs, nucleosome occupancy, and COs.
H3K4me3 and H2A.Z are marks for euchromatin and were shown to be associated with CO regions (Giraut et al., 2011; Choi et al., 2013). We show here that these marks form a peak around the CCN repeats motifs, with higher peaks in CO sites (Figures 6C to 6F), with differences between the hot and cold intervals (Supplemental Figures 5E and 5I). The CTT repeat showed patterns similar to those observed with the CCN repeat for H3K4me3 (Figures 6C and 6D) and H2A.Z (Figures 6E and 6F). No such peaks were noted around the A-rich motif, reflecting the low nucleosome occupancy (Figures 6C to 6F). Another euchromatic modification, H3K4me2, showed patterns similar to those of H3K4me3, in both cold and hot regions and in CO sites (Supplemental Figures 4G, 4H, 5E and 5F).
DISCUSSION
The three CO sequence motifs described here were shown to be associated with a nonrandom landscape of recombination; they are less abundant in centromeric regions (Figure 4A) and in cold regions (Figure 4B). In addition, the CCN repeat was associated with CO events throughout the genome, except for the female-specific cold regions that are mostly subtelomeric (Figure 4C). The A-rich motif was associated with promoters where CO events are enriched, while the CCN motif was enriched in genes where 42% of the COs occur (Figure 4D). This nonrandom association of motifs and COs could thus partly explain the nonrandom landscape of recombination in the Arabidopsis genome, including the low rates of CO in centromeric regions, in cold regions, and in subtelomeric regions of the female lineage.
Previous works have shown a correlation between epigenetic marks and meiotic recombination (Melamed-Bessudo and Levy, 2012; Mirouze et al., 2012; Colomé-Tatché et al., 2012; Yelina et al., 2012). We expanded on these findings by performing a motif-centric analysis of epigenetic marks throughout the genome. It should be noted that this study used public data on epigenetic marks that were obtained from somatic tissues at the seedling stage (as compiled by Zemach et al. [2013]). Thus, some reprogramming of epigenetic marks during meiosis, which might have affected our analysis, cannot be ruled out. However, some degree of conservation of the epigenetic patterns and especially of H3 methylations during plant development, including during meiosis, is expected (Feng et al., 2010; Oliver et al., 2013), in contrast to the extensive epigenetic reprogramming found in animals during meiosis (Sasaki and Matsui, 2008).
The presence of a motif per se, is not a strong predictor of CO. Indeed, motifs were enriched in all COs regions (Figure 4A); however, only by a small margin. On the other hand, the epigenetic landscape was strongly affected around motifs (Figures 5 and 6; Supplemental Figures 4 and 5). Thus, the combination of the sequence motif, together with the associated epigenetic changes, might be key in defining recombination sites. The type of motif epigenetic modification association differs between motifs. For example, the A-rich motif might act through its negative effect on nucleosome occupancy, while other motifs lead to increased occupancy (Figures 6A and 6B; Supplemental Figures 4E, 4F, 5G, and 5H). A-rich sequences are known to be stiff and not to fold around nucleosomes (Segal and Widom, 2009); therefore, the various core histone modifications cannot take place in the absence of nucleosomes (Figures 6C to 6F, A-rich motif). In this respect, the A-rich motif, which is associated with most CO events (Figure 3), seems to influence CO independently of the two other motifs. The A-rich signal proved persistent for large distances from the CO site (Supplemental Figure 3D). Nucleosome depletion extended for ∼2 kb around the A-rich motif in the associated CO sites; therefore, this effect must be mutually exclusive to that of the two other nucleosome-dependent motifs (Figures 6A and 6B). For the CTT and CCN motifs, nucleosome occupancy was increased, as could have been predicted based on sequence only (Kaplan et al., 2009). In that case, motifs and CO regions are characterized by peaks of modifications associated with euchromatin, such as H3K4me3 or H2A.Z. In the case of cytosine methylation, the association between the motifs and the epigenetic mark was similar for the three motifs. This suggests that the deep and wide cytosine methylation valley around the three motifs is established, regardless of histone presence (Figures 5A and 5B).
Taken together, while different motifs show different epigenetic associations, in all cases, recombinogenicity correlated with increased chromatin accessibility, either through nucleosome depletion or through euchromatic nucleosome modifications and through cytosine hypomethylation. The A-rich motif seems to be a mark of A/T-rich and recombinogenic regions and to a lesser extent of a local motif-specific effect.
The CCN motif, described here for the first time in plants, was associated with H3K4me3 modifications (Figure 6). These modifications were shown to peak around transcription start sites and spread through both promoters and genes (Choi et al., 2013), where most of the CO events took place (Figure 2). Interestingly, the CCN motif was associated with recombination within genes but not within promoters (Figure 4D). The mechanism that links the CCN repeat motif, H3K4me3 modifications, and COs is not known in plants. Interestingly, in human, most COs are associated with the CCNCCNTNNCCNC motif, which is recognized by the zinc finger domain of PRDM9, a protein that was shown to trimethylate H3K4 in germ cells (Hayashi et al., 2005) and to initiate meiotic recombination through recruitment of DSB-inducing SPO11 (Baudat et al., 2010). Plants do not have a PRDM9 homolog and the CCN repeat motif only partially overlaps with the human motif; therefore, a different and yet unknown mode of action must be linking the CCN repeat motif to H3K4me3 modifications and COs in plants. In Drosophila, high-resolution mapping of 106,964 recombination events led to the discovery of 18 motifs, two of which partially overlap the CCN repeat and four of which resemble the A-rich motif (Comeron et al., 2012). In summary, while there are substantial differences in the regulation of CO between species, there seems to be also some similarities that are probably related to chromatin accessibility.
The cytosine methylation data, together with the availability of the methylation-deficient mutant ddm1, provided valuable information. Remarkably, cytosine methylation was lowest around the CO motifs (Figure 5B). DDM1 is a chromatin remodeling protein from the SWI2/SNF2 family (Brzeski and Jerzmanowski, 2003), which, in principle, can directly interact with the motifs, promoting chromatin remodeling, as well as the demethylation gradient around the motif. To investigate this possibility, we checked the degree of CG methylation around the motif in the ddm1 mutant (Figure 5C; Supplemental Figure 5B). The same valley shape was observed in the ddm1 mutant and the DDM1 wild type, but with the lowest point being lower than in the wild type, consistent with the findings of enhanced CO in the ddm1 and met1 mutants (Melamed-Bessudo and Levy, 2012; Yelina et al., 2012). Thus, the methylation gradient around the motifs is DDM1 independent. Additional insight can be gained by the analysis of the CG methylation of the subset of motifs where CO events were actually observed (Figure 5B). While this subset showed the same shape as motifs from all around the genome, the bottom of the valley was even lower than in the ddm1 mutant (Figures 5B and 5C), further supporting the tight association between motifs and epigenetic marks and its functional significance. Moreover, it suggests a threshold model for the effect of cytosine methylation on CO, i.e., lower methylation levels will provide a more favorable context for CO (Figure 5).
In summary, the data presented here suggest that sequence motifs together with associated epigenetic modifications define recombinogenic regions and shape the nonuniform recombination landscape in the genome. While the different motifs seem to have different modes of action, overall, modifications around motifs show features of increased chromatin accessibility through low nucleosome occupancy, histone modifications, or decreased cytosine methylation; these features were most prominent in the actual CO regions. Moreover, the CCN repeat, a novel CO motif, provides potential clues into the male-female CO differences in subtelomeric regions. This work raises several questions, for example, with regards to how CO motifs interact with the chromatin and cytosine modification factors to promote CO. Likewise, it is intriguing to weigh whether the meiotic recombination landscape is just the reflection of epigenetic modifications established already in somatic tissues for some other purpose (e.g., promoters or genes) or, alternatively, is shaped by meiotic-specific factors.
METHODS
Sample Preparation
The Col meiotic tester line (Melamed-Bessudo et al., 2005) was crossed with Ler to obtain F1 plants. All plants were grown in a climatic control room with 18 h light and 6 h darkness. Fluorescence of F2 seeds was used to identify recombination events, namely, seeds that were green only or red only. These seeds were grown to produce F2 plants. DNA was extracted from F2 leaves using NucleoSpin Plant II (Macherey-Nagel) according to the manufacturer's protocol.
Sequencing Procedure
Sequencing was performed at the Weizmann Institute of Science, High Throughput Sequencing Unit. Genomic DNA was sheared by sonication to 200- to 600-bp fragments. A total of 10 ng fragmented DNA was used to prepare the libraries, as described by Blecher-Gonen et al. (2013). Sequence quality was assessed using fastqc (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). Low-quality sequences were trimmed to ensure high quality reads. Sequencing data are detailed in Supplemental Table 1.
CO Event Detection
Reads from our samples, as well as from publicly available resources (Lu et al., 2012a; Yang et al., 2012), were aligned to the TAIR10 Arabidopsis thaliana reference genome using Bowtie2. Ler SNPs (Lu et al., 2012a) were introduced to the reference genome and the reads were realigned. The SAMtools (view and Mpileup) and Varscan2 (pileup2snp) packages were employed for SNP, calling for reads with high map quality score (>30) and without PCR duplicates. An SNP was considered homozygous if it was supported by at least 90 to 100% of the reads with a criterion of a minimum of eight reads, while regions with two types of SNPs (50% ± 10%) were considered heterozygous. Crossover events were determined according to transition of zygosity levels of large DNA segments. For each F2 individual, determination of the percentage of zygosity along a given chromosome section was according to the ratio of reads that support each parental-specific SNP from the SNP calling analysis. Crossover events were determined as transitions of DNA segments from the state of homozygosity to heterozygosity and vice versa, not followed by a reverse transition in the following 50 kb. A CO data set was compiled from this analysis and was added to published CO events (Wijnker et al., 2013). From this data set, a subset of 424 events with the highest resolution <2000 bp was chosen for a comparison of different control groups.
Association with Genomic Features
The presence of the high-resolution data set was assessed in transposable elements, genes, and promoters (500 bp upstream to the transcription start site). If a CO event overlapped more than one feature, it was counted for all the features it overlapped. The expectation for a genomic feature was calculated according to the base pair content of the feature.
Motif Discovery
The high-resolution 424 CO event data set was checked for enrichment of motifs using MEME (Bailey et al., 2009). First, the CO data set was checked for motif enrichment without assuming any background bias without discriminative analysis. Next, a discriminative analysis was used for motif enrichment. As background sequences, three sets from the following control groups were randomly selected: genome wide, female-specific cold intervals (Giraut et al., 2011), and general cold intervals (Giraut et al., 2011). The female-specific cold intervals (Giraut et al., 2011) are regions that were described to have a significantly different CO rate when compared with that of males in the same region. General cold intervals (Giraut et al., 2011) were described as male cold intervals that overlapped or did not have a significantly different CO rate than females in the same region. Each of the background sets was composed of 300 sequences, and each sequence in a set was 1000 bp long. The CO subset was independently compared with each control group. An independent analysis was performed to validate the results using HOMER (Heinz et al., 2010). The CO data set was tested versus the whole genome for motif lengths of 8, 10, 12, 15, and 22.
Motif Centric Epigenetic Data Analysis
Epigenetic data were provided by Assaf Zemach and were described by Zemach et al. (2013). Motif locations were determined genome-wide by FIMO from MEME suite (Bailey and Elkan, 1994; Bailey et al., 2009). The epigenetic data were binned every 50 bp. The mean values 1000 to 2000 bp up- and downstream of each of the motifs were calculated for all the occurrences in a certain region.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF143940 for DDM1.
Supplemental Data
Supplemental Figure 1. Distribution of CO events.
Supplemental Figure 2. Distribution of the three crossover motifs.
Supplemental Figure 3. AT/GC content and motif signal.
Supplemental Figure 4. Average epigenetic modifications 2000 bp around all the motifs.
Supplemental Figure 5. Average epigenetic modifications 2000 bp around all the motifs in hot and cold intervals.
Supplemental Table 1. Number of reads yield from a lane of Hi-Seq Illumina machine of two different library preparation protocols.
Supplementary Material
Acknowledgments
We thank Assaf Zemach for the data on epigenetic modifications in Arabidopsis, Shai Lubliner, Ephraim Kenigsberg, and Amos Tanay for help with the motifs analysis and for advice, members of the Levy and Barkai laboratories for useful comments, Yehudit Posen for language editing of the article, and the European Research Council for partial funding of the genome data analysis (EU-FP7 TRACTAR grant to A.A.L.).
AUTHOR CONTRIBUTIONS
S.S. performed the bioinformatics analysis and initiated some of the analyses. C.M.-B. performed most of the experimental work. Y.D. validated some of the recombination events. N.B. cosupervised the work. A.A.L. planned and cosupervised the work and wrote parts of the article.
Glossary
- CO
crossover
- DSB
double-strand break
- Ler
Landsberg erecta
- SNP
single nucleotide polymorphism
- Col
Columbia
Footnotes
Articles can be viewed online without a subscription.
References
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- Bateson W., Saunders E.R., Punnett R.C. (1906). Further experiments on inheritance in sweet peas and stocks: Preliminary account. Proc. R. Soc. Lond. B 77: 236–238. [Google Scholar]
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., Przeworski M., Coop G., de Massy B. (2010). PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Imai Y., de Massy B. (2013). Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 14: 794–806. [DOI] [PubMed] [Google Scholar]
- Berchowitz L.E., Copenhaver G.P. (2008). Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat. Protoc. 3: 41–50. [DOI] [PubMed] [Google Scholar]
- Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., Amit I. (2013). High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 8: 539–554. [DOI] [PubMed] [Google Scholar]
- Brunschwig H., Levi L., Ben-David E., Williams R.W., Yakir B., Shifman S. (2012). Fine-scale maps of recombination rates and hotspots in the mouse genome. Genetics 191: 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeski J., Jerzmanowski A. (2003). Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J. Biol. Chem. 278: 823–828. [DOI] [PubMed] [Google Scholar]
- Chodavarapu R.K., et al. (2010). Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Henderson I.R. (2015). Meiotic recombination hotspots - a comparative view. Plant J. 83: 52–61. [DOI] [PubMed] [Google Scholar]
- Choi, K., Zhao, X., Kelly, K.A., Venn, O., Higgins, J.D., Yelina, N.E., Hardcastle, T.J., Ziolkowski, P.A., Copenhaver, G.P., Franklin, F.C.H., McVean, G., and Henderson, I.R. (2013). Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45: 1327–1336. [DOI] [PMC free article] [PubMed]
- Colomé-Tatché M., et al. (2012). Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc. Natl. Acad. Sci. USA 109: 16240–16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J.M., Ratnappan R., Bailin S. (2012). The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver G.P., Browne W.E., Preuss D. (1998). Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl. Acad. Sci. USA 95: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J., Khademian H., Giraut L., Zanni V., Bellalou S., Henderson I.R., Falque M., Mézard C. (2013). Contrasted patterns of crossover and non-crossover at Arabidopsis thaliana meiotic recombination hotspots. PLoS Genet. 9: e1003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Jacobsen S.E., Reik W. (2010). Epigenetic reprogramming in plant and animal development. Science 330: 622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis K.E., Lam S.Y., Copenhaver G.P. (2006). Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol. 142: 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis K.E., Lam S.Y., Harrison B.D., Bey A.L., Berchowitz L.E., Copenhaver G.P. (2007). Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3913–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraut L., Falque M., Drouaud J., Pereira L., Martin O.C., Mézard C. (2011). Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7: e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore M.A., Chia J.M., Elshire R.J., Sun Q., Ersoz E.S., Hurwitz B.L., Peiffer J.A., McMullen M.D., Grills G.S., Ross-Ibarra J., Ware D.H., Buckler E.S. (2009). A first-generation haplotype map of maize. Science 326: 1115–1117. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Yoshida K., Matsui Y. (2005). A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438: 374–378. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson S., Bishop R.P., Morzaria S., Spooner P.R., Pelle R., Poveda L., Ebeling M., Küng E., Certa U., Daubenberger C.A., Qi W. (2012). High-resolution genotyping and mapping of recombination and gene conversion in the protozoan Theileria parva using whole genome sequencing. BMC Genomics 13: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton M.W., et al. (2012). Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Fan W., Yan L., Li R., Lian Y., Huang J., Li J., Xu L., Tang F., Xie X.S., Qiao J. (2013). Genome analyses of single human oocytes. Cell 155: 1492–1506. [DOI] [PubMed] [Google Scholar]
- Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J., Segal E. (2009). The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Li L., Yan J. (2015). Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat. Commun. 6: 6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Han X., Qi J., Yang J., Wijeratne A.J., Li T., Ma H. (2012a). Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 22: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., et al. (2012b). Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science 338: 1627–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L.M. (2008). High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C., Levy A.A. (2012). Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: E981–E988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C., Yehuda E., Stuitje A.R., Levy A.A. (2005). A new seed-based assay for meiotic recombination in Arabidopsis thaliana. Plant J. 43: 458–466. [DOI] [PubMed] [Google Scholar]
- Mirouze M., Lieberman-Lazarovich M., Aversano R., Bucher E., Nicolet J., Reinders J., Paszkowski J. (2012). Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 5880–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T.H. (1911). Random segregation versus coupling in Mendelian inheritance. Science 34: 384. [DOI] [PubMed] [Google Scholar]
- Myers S., Freeman C., Auton A., Donnelly P., McVean G. (2008). A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 40: 1124–1129. [DOI] [PubMed] [Google Scholar]
- Oliver C., Pradillo M., Corredor E., Cuñado N. (2013). The dynamics of histone H3 modifications is species-specific in plant meiosis. Planta 238: 23–33. [DOI] [PubMed] [Google Scholar]
- Preuss D., Rhee S.Y., Davis R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460. [DOI] [PubMed] [Google Scholar]
- Qi J., Chen Y., Copenhaver G.P., Ma H. (2014). Detection of genomic variations and DNA polymorphisms and impact on analysis of meiotic recombination and genetic mapping. Proc. Natl. Acad. Sci. USA 111: 10007–10012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E., Bradbury P.J., Elshire R.J., Glaubitz J.C., Acharya C.B., Mitchell S.E., Li C., Li Y., Buckler E.S. (2015). Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc. Natl. Acad. Sci. USA 112: 3823–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H. (2008). Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77: 229–257. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Matsui Y. (2008). Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 9: 129–140. [DOI] [PubMed] [Google Scholar]
- Segal E., Widom J. (2009). Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 19: 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F., Gregoretti I.V., Brick K., Khil P., Camerini-Otero R.D., Petukhova G.V. (2011). Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Ambrose J.H., Haughey B.S., Webster T.D., Pierrie S.N., Muñoz D.F., Wellman E.C., Cherian S., Lewis S.M., Berchowitz L.E., Copenhaver G.P. (2012). Deep genome-wide measurement of meiotic gene conversion using tetrad analysis in Arabidopsis thaliana. PLoS Genet. 8: e1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizir I.Y., Korol A.B. (1990). Sex difference in recombination frequency in Arabidopsis. Heredity (Edinb.) 65: 379–383. [Google Scholar]
- Wang J., Fan H.C., Behr B., Quake S.R. (2012). Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150: 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker E., et al. (2013). The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2: e01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Yuan Y., Wang L., Li J., Wang W., Liu H., Chen J.-Q., Hurst L.D., Tian D. (2012). Great majority of recombination events in Arabidopsis are gene conversion events. Proc. Natl. Acad. Sci. USA 109: 20992–20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina N.E., et al. (2012). Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 8: e1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina N.E., Ziolkowski P.A., Miller N., Zhao X., Kelly K.A., Muñoz D.F., Mann D.J., Copenhaver G.P., Henderson I.R. (2013). High-throughput analysis of meiotic crossover frequency and interference via flow cytometry of fluorescent pollen in Arabidopsis thaliana. Nat. Protoc. 8: 2119–2134. [DOI] [PubMed] [Google Scholar]
- Youds J.L., Boulton S.J. (2011). The choice in meiosis - defining the factors that influence crossover or non-crossover formation. J. Cell Sci. 124: 501–513. [DOI] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.-H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.