Genomic resequencing of laboratory strains of Chlamydomonas reveals a mosaic pattern of high (∼2%) genetic variation, which affects phenotype scoring and allows accurate strain identification.
Abstract
Chlamydomonas reinhardtii is a widely used reference organism in studies of photosynthesis, cilia, and biofuels. Most research in this field uses a few dozen standard laboratory strains that are reported to share a common ancestry, but exhibit substantial phenotypic differences. In order to facilitate ongoing Chlamydomonas research and explain the phenotypic variation, we mapped the genetic diversity within these strains using whole-genome resequencing. We identified 524,640 single nucleotide variants and 4812 structural variants among 39 commonly used laboratory strains. Nearly all (98.2%) of the total observed genetic diversity was attributable to the presence of two, previously unrecognized, alternate haplotypes that are distributed in a mosaic pattern among the extant laboratory strains. We propose that these two haplotypes are the remnants of an ancestral cross between two strains with ∼2% relative divergence. These haplotype patterns create a fingerprint for each strain that facilitates the positive identification of that strain and reveals its relatedness to other strains. The presence of these alternate haplotype regions affects phenotype scoring and gene expression measurements. Here, we present a rich set of genetic differences as a community resource to allow researchers to more accurately conduct and interpret their experiments with Chlamydomonas.
INTRODUCTION
Chlamydomonas reinhardtii is a unicellular green alga from the Chlorophyte lineage (Harris, 2009). For decades, this species has been at the forefront of research in photosynthesis and the function of the chloroplast, in the structure and function of cilia, and in elucidation of DNA methylation processes. Recently, it has proven to be quite useful in studies of algae to produce biofuels (Rochaix, 1995; Li et al., 2004; Merchant et al., 2012). A number of traits make Chlamydomonas a particularly useful reference organism. It can grow autotrophically or heterotrophically, making it ideal for studying photosynthesis mutants (Spreitzer and Mets, 1981; Grossman et al., 2010). Chlamydomonas is a powerful model for genetic studies because it has a well characterized and sequenced haploid genome and is capable of sexual recombination (Merchant et al., 2007). Lastly, it can be induced to produce neutral lipids or molecular hydrogen under certain conditions, which makes it an attractive model for biofuel research (Esquível et al., 2011; Goodenough et al., 2014).
Chlamydomonas has two distinct mating types, dubbed mt+ and mt–, with one of each required for sexual recombination (Harris, 2009). The mating type is conferred by an ∼200- to 400-kb region on chromosome 6 known as the mating locus (De Hoff et al., 2013). Under certain stresses, such as nitrogen deprivation, Chlamydomonas cells will become gametes. Opposite mating type gametes fuse during fertilization to form a diploid zygospore. Upon germination, the zygospore undergoes meiosis and releases two haploid mt+ and two haploid mt– zoospores, or occasionally four and four, which then resume vegetative growth (Smith and Regnery, 1950; Harris, 2009).
Most work on Chlamydomonas to date uses a limited number of interrelated strains we will refer to here as the standard laboratory strains. It has been reported that these strains trace their lineage to the work of Gilbert Smith in the 1940s and 50s (Smith, 1946; Smith and Regnery, 1950). Although these early studies with Chlamydomonas are somewhat poorly documented, it has been proposed that the standard laboratory strains all descend from a single zygospore isolated by Smith from a soil sample collected in a potato field in Massachusetts in 1945 (Harris, 2009). Smith studied Chlamydomonas isolates with a particular interest in sexual recombination (Smith and Regnery, 1950), and in subsequent years, he began supplying strains as matched mating pairs to interested researchers at other institutions (Sager, 1955; Ebersold, 1956).
The generally accepted model, as summarized in the Chlamydomonas Source Book (Kubo et al., 2002; Pröschold et al., 2005; Harris, 2009), groups the standard laboratory strains into three sublineages, each with an mt+ and an mt– member. Each mating type-matched pair of these is considered a principal strain, and they are referred to as the Sager lineage, the Cambridge lineage, and the Ebersold-Levine lineage. Perhaps because of this, it is commonly assumed that the two mating types within a lineage are essentially isogenic except at the mating locus and that each lineage is distinct from the other two (Harris, 2009; Cakmak et al., 2012).
In addition to these principal strains, researchers have crossed various Chlamydomonas lineages in pursuit of desired phenotypes. For example, CC-3269 (also known as 2137) is the result of a cross between a mt– strain in the Ebersold-Levine lineage and a mt+ strain from the Sager lineage (Spreitzer and Mets, 1981). It was selected for vigorous growth in the dark to use as the background strain in a study of photosensitive mutants. Similarly, strain g1 was selected for strong negative phototaxis and high transformation efficiency to be used as the background strain in a study of phototaxis (Pazour et al., 1995).
The standard laboratory strains are maintained in a common repository known as the Chlamydomonas Resource Center, which is presently hosted at the University of Minnesota, St. Paul. In addition to making thousands of strains available to researchers for a nominal fee, the Chlamydomonas Resource Center’s website (http://chlamycollection.org) provides the invaluable service of documenting the histories and known mutations for each of the strains it maintains. It is common in the Chlamydomonas community to refer to strains by a relevant mutant phenotype, such as cw15 for strains with a particular defect in cell wall production (Davies and Plaskitt, 1971; Loppes and Deltour, 1975; Scholz et al., 2011). However, this can create confusion when different unrelated strains share a common phenotype. The Chlamydomonas Resource Center gives each strain a unique designation with “CC-” followed by a serial number. For the purposes of clarity, we will refer to strains in this article primarily by their CC number, except for the few strains that are not presently part of the Chlamydomonas Resource Center’s collection or when we wish to draw a distinction between strains from different sources.
Despite their recent common ancestry, there are many readily observable phenotypic differences between the standard laboratory strains. For example, strains in the Ebersold-Levine lineage are unable to utilize nitrate as a nitrogen source, while those in the Sager lineage can (Harris, 2009). Strains differ in their ability to utilize micronutrients, in their responses to light, and in the production of a cell wall (Davies and Plaskitt, 1971; Pazour et al., 1995; Merchant et al., 2006). Some of these phenotypes have been traced to specific mutations, such as the nit1 and nit2 mutations that prevent nitrate utilization (Fernández et al., 1989). CC-1690, the mt+ strain in the Sager lineage, remains green when grown in the dark, whereas CC-1691, the mt– Sager strain, turns yellow due to a mutation at the y1 locus (Sager, 1955). Many other phenotypes are due to the interplay of multiple genetic loci. A good example of this is metal homeostasis. The intracellular concentration of iron is tightly regulated by Chlamydomonas via the action of a host of transporters and scavenging proteins (Glaesener et al., 2013). As the available iron becomes limited, these proteins engage in a coordinated process to import needed iron. When strains of Chlamydomonas are grown in limiting concentrations of iron, the effectiveness of these iron-harvesting mechanisms becomes readily observable as reduced growth and, in extreme cases, as chlorosis.
Given the recent advent of whole-genome sequencing (WGS) technologies, we resequenced a wide range of standard laboratory strains. In total, we compared 39 accessions, including mt+ and mt– representatives from all three lineages (listed in Table 1; described in detail in Supplemental Data Set 1). Each of these was aligned to the Chlamydomonas reference genome, which was generated from a mt+ member of the Ebersold-Levine lineage known as CC-503 (Merchant et al., 2007). By comparing these strains to CC-503 and to each other, we were able to examine the full range of genetic diversity. Unexpectedly, we noted that this genetic diversity was distributed unevenly throughout the genome and was in distinct, heritable patterns. Furthermore, the distribution of these patterns created a unique fingerprint for each strain that could be used to identify unknown or mislabeled strains and could be used to demonstrate interstrain relatedness.
Table 1. Sequenced Strains.
| Strain Name | Alternate Names | Mating Locus | Source | Reference | Mean Coverage | Total Nonduplicate Reads | Read Length |
|---|---|---|---|---|---|---|---|
| CC-503 | reference strain | mt+ | Chlamydomonas Resource Center 2012 | Merchant et al. (2007) | 140 | 199,339,380 | 100+100 |
| CC-2290 | S1 D2 | mt– | Chlamydomonas Resource Center 2011 | Gross et al. (1988) | 50 | 180,632,350 | 50+50 |
| CC-124 | 137c mt– | mt– | Chlamydomonas Resource Center 2012 | 73 | 187,721,897 | 100+100 | |
| CC-125 | 137c mt+ | mt+ | Chlamydomonas Resource Center 2012 | 113 | 277,120,089 | 100+100 | |
| 21gr | mt+ | Merchant group prior to 2011 | 41 | 114,184,946 | 50+50 | ||
| CC-1690 | 21gr | mt+ | Chlamydomonas Resource Center 2011 | 43 | 122,314,084 | 50+50 | |
| CC-1691 | 6145c | mt– | Chlamydomonas Resource Center 2013 | 71 | 95,422,478 | 100+100 | |
| CC-1009 | UTEX 89 | mt– | Chlamydomonas Resource Center 2012 | 54 | 76,043,571 | 100+100 | |
| CC-1010 | UTEX 90 | mt+ | Chlamydomonas Resource Center 2012 | 63 | 89,383,819 | 100+100 | |
| CC-425 | mt+ | Merchant group prior to 2011 | 9 | 15,044,693 | 100+100 | ||
| CC-620 | R3+ | mt+ | Chlamydomonas Resource Center 2013 | 57 | 76,312,279 | 100+100 | |
| CC-621 | NO– | mt– | Chlamydomonas Resource Center 2013 | 64 | 95,483,751 | 100+100 | |
| CC-4286a | 1A mt– | mt– | Chlamydomonas Resource Center 2011 | 16 | 20,123,761 | 100+100 | |
| CC-4287a | 3D mt+ | mt+ | Chlamydomonas Resource Center 2011 | 54 | 147,761,285 | 50+50 | |
| CC-4402b | isoloP | mt+ | Chlamydomonas Resource Center 2011 | 77 | 206,121,659 | 50+50 | |
| CC-4403b | isoloM | mt– | Chlamydomonas Resource Center 2011 | 41 | 112,179,184 | 50+50 | |
| 2137A+ | 2137 | mt+ | Robert Spreitzer 2012 | Spreitzer and Mets (1981) | 9 | 15,321,881 | 100+100 |
| CC-1021 | 2137 | mt+ | Merchant group prior to 2011 | Spreitzer and Mets (1981) | 6 | 10,288,676 | 100+100 |
| CC-3269 | 2137 | mt+ | Merchant group prior to 2011 | Spreitzer and Mets (1981) | 78 | 99,477,102 | 100+100 |
| CC-4532 | mt– | Laurens Mets 1981 | 261 | 414,042,238 | 76+76 | ||
| IAM C-9 | NIES-2235 | mt– | Hideya Fukuzawa 2012 | 93 | 120,110,144 | 100+100 | |
| CC-4425 | D66+ | mt+ | Arthur Grossman 2010 | Schnell and Lefebvre (1993) | 35 | 87,936,394 | 50+50 |
| SAG 73.72 | C-8 | mt+ | Maria Mittag 2012 | 68 | 99,517,895 | 100+100 | |
| CC-4051 | 4A+ | mt+ | Krishna Niyogi prior to 2011 | Soupene et al. (2004) | 53 | 138,309,332 | 50+50 |
| CC-4603c | 4Ax5.2– | mt– | Krishna Niyogi 2012 | Dent et al. (2005) | 17 | 27,416,092 | 100+100 |
| CJU10– | mt– | James Umen 2012 | 67 | 99,485,786 | 100+100 | ||
| g1 | mt+ | George Witman 2012 | 97 | 123,203,137 | 100+100 | ||
| S24– | mt– | Francis-Andre Wollman 2012 | 94 | 123,642,884 | 100+100 | ||
| T222+ | mt– | Francis-Andre Wollman 2012 | 75 | 101,789,719 | 100+100 | ||
| cw15 arg– | mt+ | Ralph Bock 2012 | Neupert et al. (2009) | 73 | 89,944,895 | 100+100 | |
| 302 | cw15 | mt+ | Peter Hegemann 2012 | 47 | 57,800,731 | 100+100 | |
| CC-4350 | 302 | mt+ | Chlamydomonas Resource Center 2012 | 67 | 84,406,501 | 100+100 | |
| CC-4351 | 325 | mt+ | Chlamydomonas Resource Center 2012 | 52 | 73,762,721 | 100+100 | |
| CC-4568 | 330 or cw15 | mt+ | David Dauvillée 2012 | Dauvillée et al. (2001) | 10 | 20,811,685 | 100+100 |
| CC-4348 | BAFJ5 or sta6 | mt+ | Ursula Goodenough 2012 | Zabawinski et al. (2001) | 51 | 134,558,203 | 50+50 |
| CC-4349 | Goodenough cw15 | mt– | Ursula Goodenough 2012 | 84 | 110,405,526 | 100+100 | |
| CC-4567 | STA6-C6 | mt+ | Ursula Goodenough 2012 | 11 | 15,536,158 | 100+100 | |
| CC-4504 | nrr1-1 | mt+ | Arthur Grossman 2010 | Gonzalez-Ballester et al. (2011) | 12 | 15,549,216 | 100+100 |
| pcc1-1 | mt+ | Arthur Grossman 2010 | Gonzalez-Ballester et al. (2011) | 31 | 44,027,724 | 100+100 | |
| crd2-1 | mt– | Merchant group 2004 | Eriksson et al. (2004) | 12 | 22,968,931 | 76+76 |
CC-4286 and CC-4287 are the result of 10 backcrosses to an unknown strain.
CC-4402 and CC-4403 are the result of 10 backcrosses to CC-124.
CC-4603 is the result of five backcrosses to CC-4051.
RESULTS
Divergent Phenotypes Are Evident in Wild-Type Strains
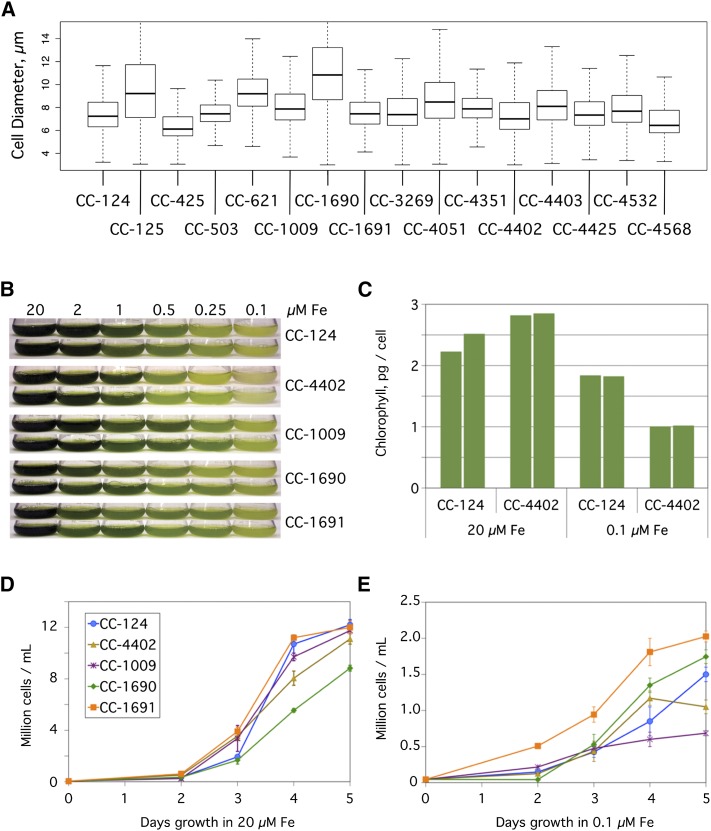
Despite the recent common ancestry of the standard laboratory strains, we observed that different wild-type laboratory strains often have significant phenotypic differences, including cell size. To examine this, duplicate samples of 1000 cells each from 16 commonly used laboratory strains were assayed for cell size by cellometer (Figure 1A). Despite being grown under ideal conditions, cells of the standard laboratory strains differed from each other considerably in both median size and size distribution. For example, CC-1690 cells were both 85% larger and had a 240% broader size distribution than those of CC-425 (mean diameter ± sd for CC-1690 is 11.3 ± 4.0 µm versus 6.1 ± 1.7 µm for CC-425). These differences in size did not correspond with differences in ploidy, as there was no significant difference in the DNA content per cell between the largest and smallest strains (P = 0.094). In an average of six samples of various cell densities, the largest strain, CC-1690, had 210 ± 60 fg DNA per cell versus 150 ± 40 fg DNA per cell for the smallest strain, CC-425 (mean ± sd).
Figure 1.
Phenotypic Diversity in Wild-Type Strains.
(A) Diversity of cell size. The size of Chlamydomonas cells from the indicated 16 strains were assayed by a cellometer. Results are plotted as a box plot indicating the median cell size (bold horizontal line), the upper and lower quartiles (ends of boxes), and the range (thin horizontal lines) from 1000 cells per sample.
(B) Growth of Chlamydomonas cells in a range of iron concentrations. The indicated strains were inoculated to a density of104 cells/mL in 100-mL cultures of TAP media containing iron concentrations ranging from 0.1 to 20 µM. Duplicate cultures were photographed after 5 d of growth.
(C) Chlorophyll content in Chlamydomonas cells. Chlorophyll content was measured on a per cell basis for duplicate cultures of the closely related CC-124 and CC-4402 strains in TAP medium plus 20 µM or 0.1 µM iron after 5 d of growth.
(D) and (E) Growth rates. Quantification of the number of cells in TAP medium supplemented with 20 µM iron (D) or 0.1 µM iron (E). Cells were counted by a hemocytometer daily for 5 d and plotted. Each point represents the mean (± range) of the cell count for duplicate cultures.
Another readily observable phenotypic difference is in iron homeostasis, which manifests as chlorosis and reduced growth when the available iron is insufficient. To examine this, we used several common wild-type laboratory strains of Chlamydomonas: CC-124, CC-1009, CC-1690, and CC-1691. All of these strains are directly descended from those originally distributed by Smith in the 1940s and 50s, and this group includes examples from all three lineages. In addition, we included CC-4402 (also known as isoloM) that was generated from 10 backcrosses to CC-124 and would therefore be expected to have a nearly identical phenotype to CC-124 (Lin et al., 2013). These strains were grown in iron concentrations ranging from 0.1 to 20 µM for 5 d to compare their relative abilities to tolerate limiting iron (Figure 1B). CC-1691 had a notable growth advantage over the other strains at 0.1 µM iron (Figure 1E). CC-1690 lagged behind the other strains in iron-replete media (Figure 1D), but grew relatively well when iron concentrations were limiting. Unexpectedly, CC-4402 was markedly more sensitive to limiting iron than CC-124. Not only were there fewer cells of CC-4402 after 5 d of growth in 0.1 µM iron (1.1 × 107 cells/mL versus 1.5 × 107 cells/mL, respectively), but the cells also had a lower chlorophyll content (1.0 pg/cell versus 1.8 for CC-124) (Figure 1C). Similar studies with additional strains revealed an even wider range of phenotypes (Supplemental Figure 1). For example, strains CC-4051 and CC-4532 were relatively tolerant of low iron, while strains CC-425 and CC-4351 were relatively sensitive.
Genome Resequencing Reveals Genetic Diversity of Laboratory Stains
In order to evaluate the genetic diversity among the standard laboratory strains of Chlamydomonas, 39 strains were chosen for WGS (Table 1). Many of these strains are ones that were contributed by members of the Chlamydomonas research community as their most frequently used wild-type strains (see Supplemental Data Set 1 for detailed strain histories). In addition to the 39 standard laboratory strains, one unrelated, but interfertile, isolate, CC-2290 (also known as S1 D2), was included for comparison (Gross et al., 1988). DNA was collected from the strains and sequenced on the Illumina platform. The resulting sequence data were aligned to version 5 of the reference Chlamydomonas genome (http://phytozome.jgi.doe.gov/pz/portal.html; 2014). After extensive quality filtering, ∼100,000,000 nonduplicate reads per strain were aligned to the reference genome, for an average coverage of 60× (Table 1; Supplemental Figure 2).
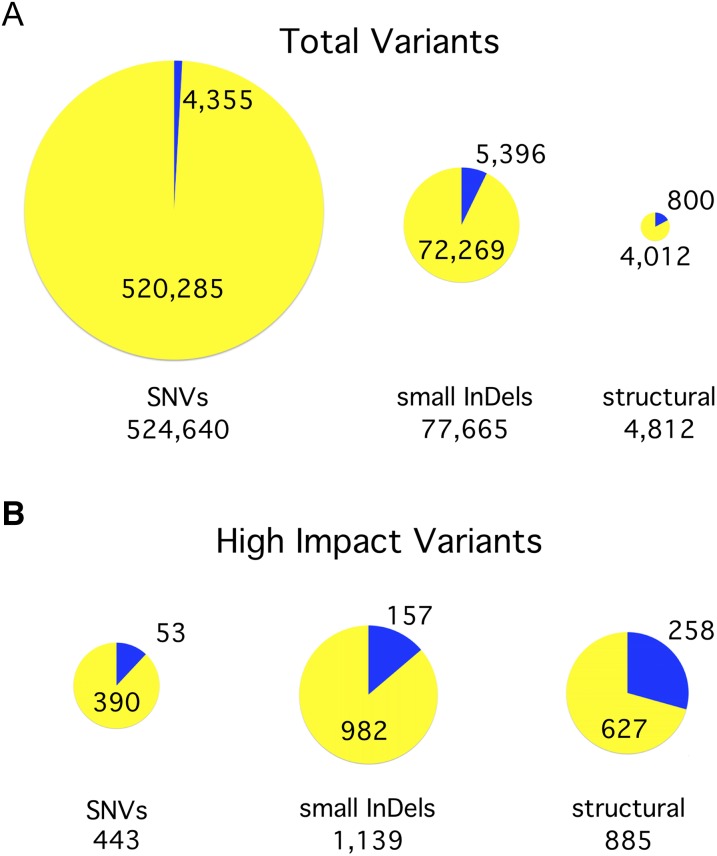
Next, we determined the number of variants relative to the reference genome, which were grouped into single nucleotide variants (SNVs), small insertions or deletions (InDels; ≤40 bp), and structural variants (Supplemental Data Sets 2 and 3). In the complete set of 39 standard laboratory strains, we observed 607,117 total variants (Figure 2A). This would average out to 1 variant per 180 bp in the Chlamydomonas 109-Mb genome, except that the actual distribution of variants is far more complex (see below). For the SNVs, there was a 1.431 ratio of transitions to transversions (Supplemental Table 1). A-to-T and T-to-A transversions were underrepresented, possibly due to the high (64%) GC content of the Chlamydomonas genome (Merchant et al., 2007).
Figure 2.
Summary of Variants by Type.
(A) Identification of variants. We identified 607,117 variants in total, including SNVs, InDels of 40 bp or less (small InDels), and structural variants (insertions, deletions, and inversions >40 bp). The size of each pie chart is proportional to the number of variants in the indicated class. The yellow portion of each pie indicates the percentage of variants in that class that are attributable to a second haplotype inherited from a divergent ancestral parent, referred to in this work as haplotype 2. The blue portion indicates those variants that arose in the laboratory since the original cross.
(B) Effect of variants on gene models. Each variant was graded based on what effect it was predicted to have on nearby gene models. Those variants that were rated to have a high impact on at least one gene are included here in proportionately sized pie charts.
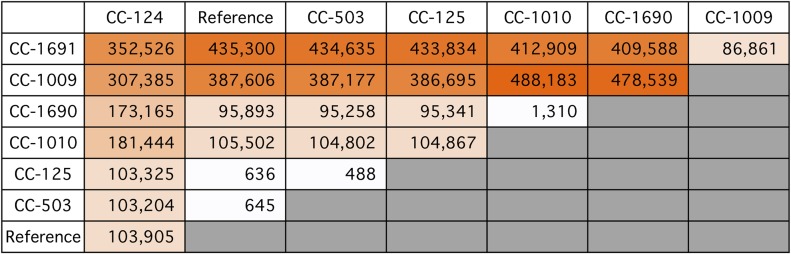
To determine how similar each strain is to its siblings, we calculated the number of pairwise SNVs for every pair of strains (Supplemental Data Set 4). The number of pairwise SNVs between any two strains ranged from 162 to 505,396, which represents 0.4% of the Chlamydomonas genome. A smaller subset of strains (Figure 3) that includes the reference strain and all of the original strains distributed by Smith showed a similar wide distribution of pairwise SNVs (from 488 to 488,183).
Figure 3.
1000-Fold Range of Pairwise SNVs between Representative Strains.
The number of pairwise SNVs for each pair of indicated strains is presented. A gradient from white to dark orange highlights the increasing numbers of SNVs. This figure includes exemplars of the original strains distributed by Smith, as well as the reference strain. A similar comparison of all strains can be found in Supplemental Data Set 4.
In the classic model of the standard laboratory strains, the strains within the three lineages should be very similar, but strains from different lineages should be more divergent. In contrast to that hypothesis, strains CC-1690 and CC-1691 (both from the Sager lineage) had 409,588 SNVs between them (Figure 3). This high number of pairwise SNVs makes those two strains some of the most divergent in the set. By contrast, CC-1690 has a relatively low 1310 SNVs when compared with CC-1010 (Cambridge lineage), despite the fact that these two strains come from different lineages.
The Genomes of the Chlamydomonas Standard Laboratory Strains Consist of Two Haplotypes
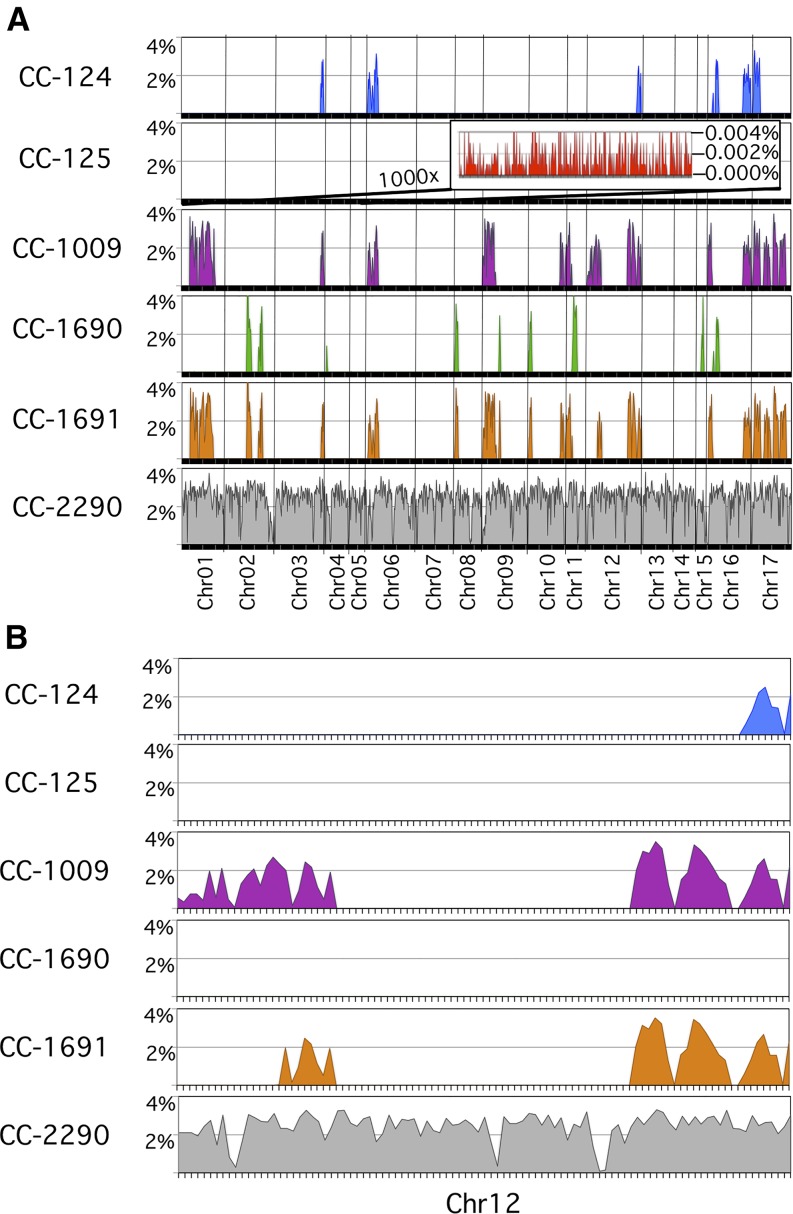
Given the observation that there were huge differences in the number of variants between any two strains in this set, we sought to determine if there was any pattern in how those variants are distributed. We plotted the number of small variants (SNVs and small InDels) as a percentage variant rate within nonoverlapping 100,000-bp windows (excluding Ns in the reference) over the length of the genome. A representative set of six strains is presented in Figure 4A, which includes examples from all three lineages and the unrelated isolate CC-2290, which had a 2.4% ± 0.7% variant rate that was fairly constant over the length of the genome. In contrast, CC-125 had a 1000-fold lower variant rate of 0.002% ± 0.002% over the length of its genome. Interestingly, the other standard laboratory strains that we examined exhibited a similar basal rate of 0.002% that sporadically jumped 1000-fold to 2.0% ± 1.0%. These regions of high-variation rate relative to the reference genome were observed to occur at several specific locations throughout the genome and in multiple strains (Figure 4A).
Figure 4.
Uneven Distribution of Variants across the Genome in Representative Strains.
(A) Discrete regions with a high variant rate that were found throughout the genome. The percentage of variant nucleotides was plotted for 100,000-bp windows over the length of each of the 17 chromosomes for the indicated strains. CC-2290 is an interfertile, but independent, isolate of Chlamydomonas. It has an average of 2.4% variant rate that is consistent across the genome. The other five strains included are direct descendants of the original strains distributed by Smith and are representative of all of the strains included in this study. Each of these standard laboratory strains has a biphasic variant rate that jumps 1000-fold from a 0.002% basal variant rate up to 2.0% in discrete regions throughout the genome. A representative portion of the basal variant rate for CC-125 is shown in the inset at 1000× magnification.
(B) An expanded view of chromosome 12. High variant rate regions are shared in different combinations between the standard laboratory strains.
These discrete blocks of 2% variation were clearly not regions of hypermutation because only one alternate nucleotide was observed for each SNV, and any two strains that shared the same high variant block shared the same complement of alternate nucleotides. For example, CC-124, CC-1009, and CC-1691 all shared the same alternate nucleotides within the 2% variant region at the end of chromosome 12 (Figure 4B). This region of alternate nucleotides extended further to the left for CC-1009 and CC-1691, but not for CC-124. These regions appeared to be highly stable since the same variant nucleotides were observed in multiple strains despite the fact that these strains have been propagated independently for many decades since leaving Smith’s laboratory.
The unrelated strain that we included in this analysis, CC-2290, had a mostly different set of variants than those that were shared between the standard laboratory strains within their high variant rate blocks. However, it is interesting to note that the rate of variants within these blocks, 2.0%, was comparable to the 2.4% rate of variation that we observed in CC-2290, as well as the 2.83% average variant rate for wild isolates of Chlamydomonas as reported by Flowers et al. (2015).
Given these results, we hypothesized that the regions of high variation represent genetic contributions from two ancestral strains that were ∼2% divergent from each other. In this model, the two divergent parents mated, and the descendants of that cross, now known as the standard laboratory strains, each carry different proportions of those two ancestral parents in a mosaic pattern. The observation that a strain carries a given block of 2% variation suggests that that strain has inherited that region from the opposite ancestral parent as the reference strain. For the purposes of distinguishing these regions in this work, we will arbitrarily refer to the reference strain as haplotype 1 and regions with the alternate nucleotides as haplotype 2. Collectively, we found that the regions of the genome within this population of strains that have two alternate haplotypes covered 25.2% of the total genome. All laboratory strains shared a single, common haplotype for the remaining 74.8% of the genome. For any one strain in this group, the proportion of the genome that is haplotype 2 ranged from 0% for CC-125 and CC-620 to a maximum of 21.4% for CC-1691.
Strikingly, of the 524,640 SNVs that we observed in this population, nearly all (520,285 = 99.2%) were due to the 25.2% of the genome with the two alternate haplotypes (Figure 2A). We observed a number of cases in which a daughter strain carried a smaller contiguous region of haplotype 2 than did its parent strain. This observation is consistent with a model in which the boundaries of haplotype 2 regions represent sites of meiotic recombination between strains with different haplotypes. For example, strain CC-3269 is known to be a cross of strains equivalent to CC-1690 and CC-124. The fact that CC-3269 had a smaller haplotype 2 region on chromosome 10 than CC-1690 is readily explained by a meiotic recombination event somewhere in the 52-nucleotide range between numbers 364,671 and 364,723 on chromosome 10. As shown in Supplemental Figure 3, CC-3269 had a number of haplotype 2-specific SNVs to the left of this locus, but only haplotype 1 nucleotides to the right. In contrast, its parents were either all haplotype 2 (CC-1690) or all haplotype 1 (CC-124) on either side of this locus.
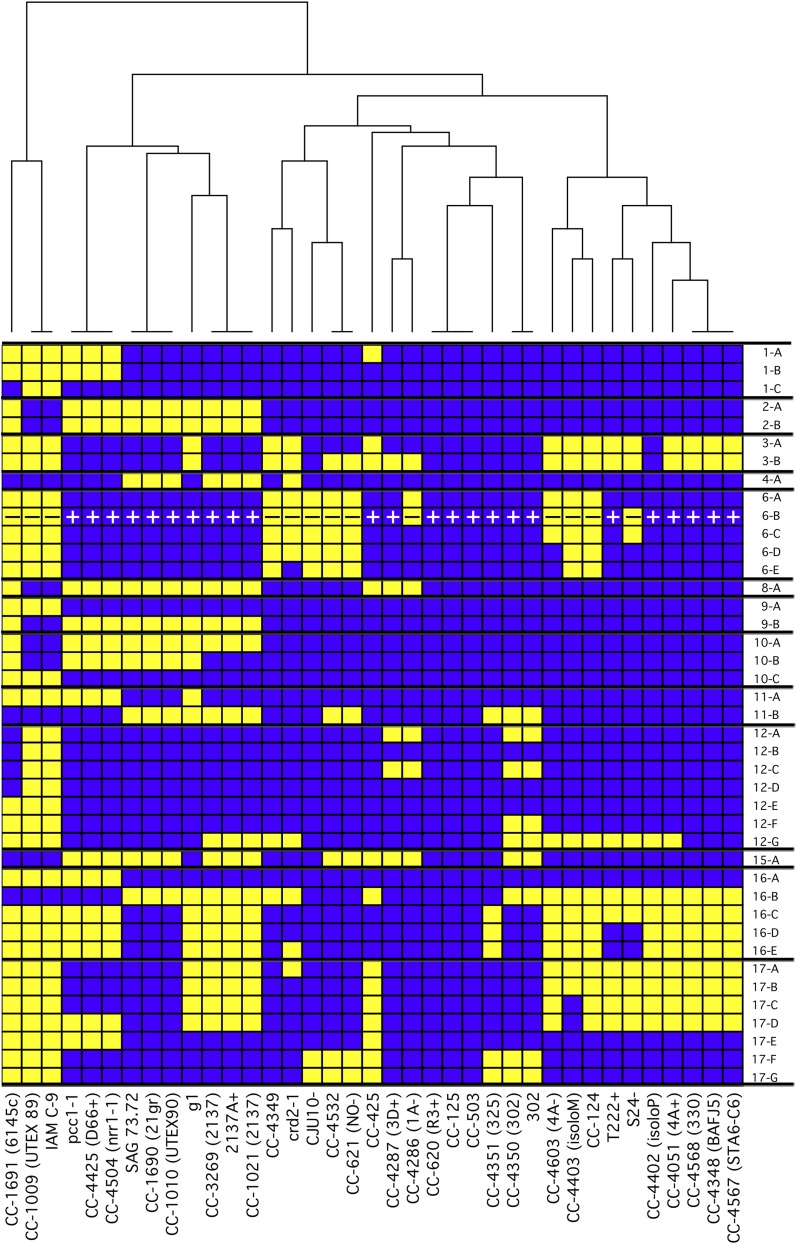
Our observations on the distribution of haplotype 2 regions suggested a convenient way to evaluate the relatedness of the different strains. We divided the haplotype 2 regions algorithmically into the minimum number of blocks such that any given strain in this set is either all haplotype 1 or all haplotype 2 within the bounds of the given block. This produced 41 distinct regions, the coordinates of which are indicated in Table 2. The sequenced strains were then graded in a binary fashion for their haplotype in each block, effectively creating a unique fingerprint for each strain (Figure 5). In order to examine the relatedness of the strains, a dendrogram was generated from these patterns and used to cluster the strains.
Table 2. Coordinates of Haplotype 2 Blocks in Version 5 of the Chlamydomonas Reference Genome.
| Block Name | Chromosome | Start | End |
|---|---|---|---|
| 1-A | chr01 | 1,717,427 | 3,188,098 |
| 1-B | chr01 | 3,543,098 | 5,920,858 |
| 1-C | chr01 | 6,115,512 | 6,320,512 |
| 2-A | chr02 | 4,066,433 | 4,888,063 |
| 2-B | chr02 | 6,165,360 | 6,925,459 |
| 3-A | chr03 | 8,412,815 | 8,861,169 |
| 3-B | chr03 | 8,861,169 | 9,028,966 |
| 4-A | chr04 | 151,353 | 238,452 |
| 6-A | chr06 | 1 | 281,650 |
| 6-B | chr06 | 288,650 | 1,360,343 |
| 6-C | chr06 | 1,360,343 | 1,423,425 |
| 6-D | chr06 | 1,423,425 | 1,761,623 |
| 6-E | chr06 | 1,761,623 | 1,967,222 |
| 8-A | chr08 | 118,000 | 713,465 |
| 9-A | chr09 | 1 | 2,366,573 |
| 9-B | chr09 | 2,926,229 | 3,118,228 |
| 10-A | chr10 | 62,001 | 364,697 |
| 10-B | chr10 | 364,697 | 644,298 |
| 10-C | chr10 | 5,882,189 | 6,570,585 |
| 11-A | chr11 | 16,001 | 1,280,878 |
| 11-B | chr11 | 1,280,878 | 2,193,206 |
| 12-A | chr12 | 1 | 437,166 |
| 12-B | chr12 | 505,166 | 630,166 |
| 12-C | chr12 | 697,496 | 843,594 |
| 12-D | chr12 | 974,762 | 1,719,321 |
| 12-E | chr12 | 1,719,321 | 2,483,019 |
| 12-F | chr12 | 7,219,514 | 8,662,505 |
| 12-G | chr12 | 8,971,075 | 9,725,409 |
| 15-A | chr15 | 842,837 | 1,276,864 |
| 16-A | chr16 | 9,001 | 736,752 |
| 16-B | chr16 | 868,455 | 1,952,379 |
| 16-C | chr16 | 6,342,055 | 6,386,055 |
| 16-D | chr16 | 6,398,154 | 6,974,942 |
| 16-E | chr16 | 7,015,856 | 7,777,706 |
| 17-A | chr17 | 270,199 | 352,132 |
| 17-B | chr17 | 352,132 | 475,231 |
| 17-C | chr17 | 475,231 | 1,134,081 |
| 17-D | chr17 | 1,134,081 | 1,442,180 |
| 17-E | chr17 | 2,153,378 | 3,333,368 |
| 17-F | chr17 | 3,813,427 | 5,983,702 |
| 17-G | chr17 | 5,983,702 | 6,075,633 |
Figure 5.
Distribution of Two Haplotypes in Laboratory Strains.
Within the population of standard laboratory strains, 25.2% of the genome was found to have either of two haplotypes with 2.0% sequence divergence between them. The haplotype of the reference strain, CC-503, was arbitrarily designated as haplotype 1 and the alternate regions as haplotype 2. We algorithmically determined the boundaries of the regions with two haplotypes and combined them into the fewest number of contiguous regions in which all strains are entirely one or the other haplotype. These are plotted for the indicated strains with blue representing haplotype 1 and yellow representing haplotype 2. The mating locus is indicated by + or –. A dendrogram was constructed to arrange the strains based on the similarity of their haplotypes. The coordinates of each block in version 5 of the Chlamydomonas reference genome are presented in Table 2.
Within this group of 39 strains, there were a few examples of strains that are nominally the same, only from different sources. For example, CC-3269 and 2137A+ are the same strain provided to us by the Chlamydomonas Resource Center and Robert Spreitzer, respectively. As expected for these strains, they had an identical haplotype fingerprint and clustered together in the dendrogram. Other strains, such as CC-4286 and CC-4287, were not the same, but closely related. As such, it is not surprising that these strains had similar haplotype fingerprints and clustered together. However, there were a number of surprises. Strains that had been believed to be closely related, such as CC-124 and CC-125, belonged to remote clades. In contrast, other strains not previously known to be related, such as CC-1690 and CC-1010, clustered to the same clade. Taken together, this approach revealed a number of unexpected groupings that warranted further examination.
Mislabeled Strains and Inaccurate Histories Were Identified
The strain identified here as CC-4532 was provided to this group by Laurens Mets in 1981 as strain 2137, which was the progeny of a cross between strains equivalent to CC-1690 and CC-124 (Spreitzer and Mets, 1981). Since that time, we have published numerous studies based on work with that strain (Yu et al., 1988; Long et al., 2008; Castruita et al., 2011). In this study, we resequenced this strain, along with two examples of 2137 from the Chlamydomonas collection (CC-3269 and CC-1021) and one from Robert Spreitzer (2137A+). In comparing their sequences, these last three examples of 2137 clustered tightly together in the same clade, but CC-4532 clustered to a different clade (Figure 5). Looking at the haplotype distribution, three of the four strains (2137A+, CC-1021, and CC-3269) shared the same distribution of haplotype 2 regions (Supplemental Figure 4A), and they had fewer than 2000 pairwise SNVs between them (Supplemental Data Set 4). The observed haplotype pattern was consistent with these strains being the F1 progeny of a cross of strains CC-1690 and CC-124. In contrast, strain CC-4532 was found to be completely distinct from the other strains. The presence of haplotype 1 in block 16-B and haplotype 2 in blocks 17-F and 17-G means that CC-4532 could not have arisen from a cross between those parental strains, as neither parent carries those haplotype regions (Supplemental Figure 4B). Based on its haplotype pattern, it instead appeared to be the same as strain CC-621 (also known as NO–), as both had the same haplotype and only 953 pairwise SNVs.
CC-4348, a sta6 mutant, was generated by insertional mutagenesis in a cw15 mutant strain known as strain 330 (Zabawinski et al., 2001). We acquired CC-4348 and its supposed parental strain for use in a study of TAG production (Blaby et al., 2013). A number of observations, such as incorrect mating type and the lack of the expected arginine auxotrophy, caused us to question whether the cw15 mutant strain we received, now called CC-4349, was the true parent of CC-4348. To test this, we obtained the parental strain directly from the group that produced CC-4348, and we sequenced all three. The second instance of the parental strain, now called CC-4568, did in fact have the same haplotype as CC-4348 (Supplemental Figure 4C) and only 669 SNVs relative to CC-4348 (Supplemental Data Set 4). In contrast, CC-4349 had a dramatically different haplotype and 80,934 SNVs relative to CC-4348. The absence of haplotype 2 blocks 16-C through 16-E, and 17-A through 17-D in CC-4349 means that strain cannot be the parental strain of CC-4348 (Supplemental Figure 4C).
Having identified a number of examples of incorrectly identified strains, we devised an amplification-based assay for the genotyping of individual strains. A set of 82 allele-specific (AS-PCR) primer pairs were designed for use in determining the haplotype of any of the standard laboratory strains using either qPCR or PCR amplification followed by analytical gel electrophoresis. When tested against DNA from strains CC-1009 and CC-1010, which had different haplotypes at each of the 41 regions (Supplemental Figure 5A), we were able to unambiguously score the haplotype of each region for both strains (Supplemental Figure 5B).
Isogenic Strain Pairs Were Compared to Evaluate Their Similarity
Within the 39 laboratory strains that we examined, there were three pairs of strains that have undergone multiple backcrossings in an attempt to produce isogenic pairs with both mating types.
CC-4286 (also known as 1A mt–) and CC-4287 (also known as 3D mt+) are reported by the Chlamydomonas Resource Center to have been made by Paul Lefebvre by crossing CC-124 and CC-125 and backcrossing the progeny to CC-125 10 times (http://chlamycollection.org). In personal communication stimulated by this work, Lefebvre subsequently corrected the history of strains CC-4286 and CC-4287, noting that they are the result of a cross between CC-620 (also known as R3+) and CC-621 (also known as NO–). However, the presence of haplotype 2 blocks in chromosome 12 that are absent in all of those strains suggested that the actual parents are none of these (Supplemental Figure 4E). Despite the confusion about their parentage, strains CC-4286 and CC-4287 shared the same distribution of haplotype 2, except at the mating locus. There were 12,609 SNVs between them, and virtually all of those were accounted for by the mating locus-proximal blocks on chromosome 6 (Supplemental Data Set 4). This makes CC-4286 and CC-4287 highly suitable for use as a near-isogenic mating pair.
In contrast, CC-4402 (also known as isoloP) and CC-4403 (also known as isoloM), which were made by a similar approach (Lin et al., 2013), were far less isogenic. These two strains were generated by Susan Dutcher’s group by crossing CC-124 and CC-125 and backcrossing the progeny to CC-124 10 times. The distribution of haplotype 2 regions was consistent with this history (Supplemental Figure 4D). However, the two strains were not yet isogenic despite the 10 backcrosses. In addition to the expected differences at the mating locus on chromosome 6, the two strains had different haplotype blocks on chromosomes 3 and 17. The difference in the haplotype 2 regions on chromosome 17 between CC-4403 and its parent suggested that a recombination event took place between nucleotides 475,219 and 475,322 during one of the 10 crosses, and this novel pattern was maintained for the remainder of the 10 backcrossings. Additionally, a region of haplotype 1 on chromosome 3 in CC-4402 persisted through 10 backcrossings to a strain with haplotype 2 at that locus. Given the differences between CC-4402 and CC-4403, it is not surprising that there were 52,065 SNVs between them (Supplemental Data Set 4). Even when excluding the mating locus-proximal regions on chromosome 6, there were still 25,982 pairwise SNVs between them.
Strain CC-4603 (also known as 4Ax5.2–) was created by Brian Chin in Krishna Niyogi's group to be an mt– version of CC-4051 (also known as 4A+) (Dent et al., 2005). It was produced by crossing CC-4051 with strain 17D– and then backcrossing the progeny to CC-4051 five times. As intended, CC-4603 had an identical haplotype to CC-4051 except at the mating locus (Supplemental Figure 4F). There were 14,228 SNVs between the two strains, and almost all of those are proximal to the mating locus (Supplemental Data Set 4). A recombination event between nucleotides 1,419,584 and 1,426,667 on chromosome 6 during the backcrossing most likely caused CC-4603 to have a reduced set of haplotype 2 alleles at the mating locus relative to most other mt– strains.
Transposon Position Jumping Was Widespread in Chlamydomonas
Chlamydomonas is known to harbor many class I and class II transposons. Given that we had observed over 4000 putative structural variants within this population (Figure 2), we wondered whether any of those could be attributed to transposon position jumping. We used the BLAST algorithm to identify the locations of MRC1, TOC1, TOC2, REM1, Bill, Gulliver, Pioneer, Tcr1, and Tcr3 transposons within the reference genome (Supplemental Data Set 5). Next, we compared those loci with the positions of the structural variants that we identified. Throughout the genome, there was widespread evidence for transposon position jumping between the strains. For example, in chromosome 16, we identified three sites of MRC1, four sites of Gulliver, one site of Bill, and two sites of TOC1 whose coordinates in the reference genome were tightly correlated with the coordinates of large deletions in our sequenced strains (Supplemental Figure 6). In total, we found 84 examples of transposon jumping between the strains (Supplemental Table 2). There were numerous examples of this for most transposons, including 25 examples of MRC1 and 27 examples of Gulliver. We found no evidence for transposon jumping for Pioneer. Interestingly, there were two examples of MRC1 that were present in the reference genome, but absent in all others, including the newly resequenced CC-503. This suggested that those MRC1 transposons had jumped between the time that the reference genome was first sequenced and the current study. In 20 of the 84 examples, the position of the transposon sequence matched exactly with the position of the large deletion. However, in the majority (58 of 84), the deletion was slightly larger than the transposon sequence (median = 4% larger).
The Effect of Haplotype 2 Variants on Gene Models Was Determined
We identified 592,554 small variants that were due to the alternate haplotype (Figure 2A). What effect do all of these variants have on the gene models? To examine this, we used SnpEff (Cingolani et al., 2012) to predict the effect of each variant on the corresponding coding sequence. Of the haplotype 2-specific small variants, 27.7% fell within known gene models in the Chlamydomonas version 5.5 gene annotations (Supplemental Figure 7A). Those 164,606 variants were further classified by the effect each would have on the coding region (Supplemental Figure 7B). The majority, 54.8%, were synonymous codon changes that should have no effect on the resulting protein. An additional 9389 variants (5.7%) were due to small InDels (average size 3.9 bp). These were disproportionately (8507 versus 882) InDels that preserved the reading frame. Only 277 (0.2%) nonsense variants were identified. The remaining 64,685 variants (39.3%) caused nonsynonymous codon changes.
For the nonsynonymous codon changes, we further classified them based on the change to the corresponding amino acid (Supplemental Figure 7C). A plurality of the nonsynonymous codon changes (44.2%) encoded an amino acid of the same class (hydrophobic, hydrophilic-charged, or hydrophilic-neutral).
Of the 4012 structural variants in the haplotype 2 regions, 627 (15.6%) were predicted to have a high impact on one or more genes (Figure 2B). The majority of these (537) were deletions with a mean size of 5 kb. The other structural variants predicted to affect genes included 15 duplications, 23 insertions, and 52 inversions (Supplemental Data Set 3). However, since structural variant predictions become less precise for regions that are highly divergent from the reference genome, some of these variants may leave the gene function intact.
Some examples of genes that were predicted to be affected by the haplotype 2 variants are highlighted in Supplemental Table 3 and Supplemental Figure 8. Many of these, such as the structural variant in chromosome 12 that removes the CGL49 locus, were found to be examples of large deletions that remove part or all of a gene. An inversion in chromosome 16 was predicted to disrupt the FAL18 locus. Smaller InDels that preserved the reading frame, such as DUR1, or caused a frameshift, such as HSP90A, are also indicated. Lastly, a few genes with high numbers of missense mutations, such as RSEP1 with its 28 nonsynonymous codons, are also listed.
Laboratory-Originated Mutations Affect Gene Models
While the great majority (99.1%) of SNVs were attributable to haplotype 2, an additional 4355 SNVs were identified that are likely due to mutations that have accumulated in the strains in the laboratory since the original zygospore was isolated (Figure 2A). Each strain, including our own clone of the reference strain, had between 450 and 1077 SNVs relative to the published reference genome sequence, with a mean of 770 laboratory-derived SNVs. It is impossible to know how many cell divisions each strain has undergone since the initial germination in 1945. Under ideal growth conditions, the doubling time of Chlamydomonas can be as high as four to five doublings per day (Harris, 2009). However, laboratory strains are routinely grown on solid agar where cells reach stationary phase and can be maintained this way for several weeks. As a rough estimate, if a newly plated cell undergoes 1 week of continuous division at four doublings per day, followed by three more weeks in stationary phase, this would average one doubling per day. From this estimate, the 770 laboratory-originated variants we observe would be due to a mutation accumulation rate of 0.03 variants division−1 genome−1. This rough estimation is in good agreement with a previously published estimate of mutation accumulation in laboratory grown cultures of Chlamydomonas: 0.0362 variants division−1 genome−1 (Ness et al., 2012).
What effect do these variants have on the gene models? A total of 206 of the postzygospore variants were predicted by SnpEff to cause a loss-of-function mutation in a gene model. When we examined this group of genes for their Gene Ontology descriptors, there was a significant enrichment of cell communication/signal transduction genes (P value = 0.026265) (Supplemental Table 4).
One noteworthy example of a laboratory mutation affecting a gene is NIT2. It has been generally understood that all strains in the Ebersold-Levine lineage are nit2 mutants (Harris, 2009). As expected, CC-125 and its descendants have a C-to-A transversion at chr03:4,696,755 that produces a nonsense mutation at amino acid 755 of NIT2 (Supplemental Figure 9). Unexpectedly, CC-124, the other strain from the Ebersold-Levine lineage, is wild-type at this locus and throughout the NIT2 gene. This phenotype is obscured by the fact that all of the Ebersold-Levine lineage strains carry a known histidine-to-glutamine missense mutation in the NIT1 gene at chr09:7,003,590 (Supplemental Figure 9). Perhaps because NIT2 is dispensable when laboratory cultures are grown with ammonium as a nitrogen source (Harris, 2009), we identified several novel mutations at the NIT2 locus. Strain CC-4425 (also known as D66+) and its descendants have a C-to-T transition at chr03:4,696,396 that creates a premature stop codon. CC-4286 has a private G-to-T transversion at chr03:4,695,629 that creates a serine-to-isoleucine mutation in NIT2, and strain g1 has a private frameshift InDel in NIT2 at chr03:4,696,234 (Supplemental Figure 9).
RNA-Seq Gene Expression Estimates Were Affected by Haplotype
In RNA-seq analyses, mRNA transcripts are converted to cDNA, sequenced, and aligned to the reference genome for the purposes of quantifying transcription. Given the relatively high 2% variant nucleotide rate in the haplotype 2 regions of the Chlamydomonas genome, we wondered whether aligning sequence reads from haplotype 2-encoded genes to an exclusively haplotype 1 reference genome would lead to artificially low estimates of RNA abundance. To test this, we made use of an RNA-seq data set from a previous comparison of strains CC-4348 and CC-4349 (Blaby et al., 2013). Having identified the haplotype 2 regions of these two strains by WGS (Supplemental Figure 4C), we generated a haplotype-accurate, strain-specific genome for both strains. For this analysis, we limited our study to either the 4.3 Mb of CC-4348 or the 4.4 Mb of CC-4349 that corresponds to haplotype 2 in those strains.
RNA-seq reads were mapped to both haplotypes for these regions using default settings with a commonly used read alignment tool, RNA-star (Dobin et al., 2013). When reads were mapped to the correct haplotype, the mismatch rate decreased by 31% ± 3% (n = 32). With this decreased numbers of mismatches, there was a corresponding 2.5% ± 0.5% (n = 32) increase in the number of mappable reads.
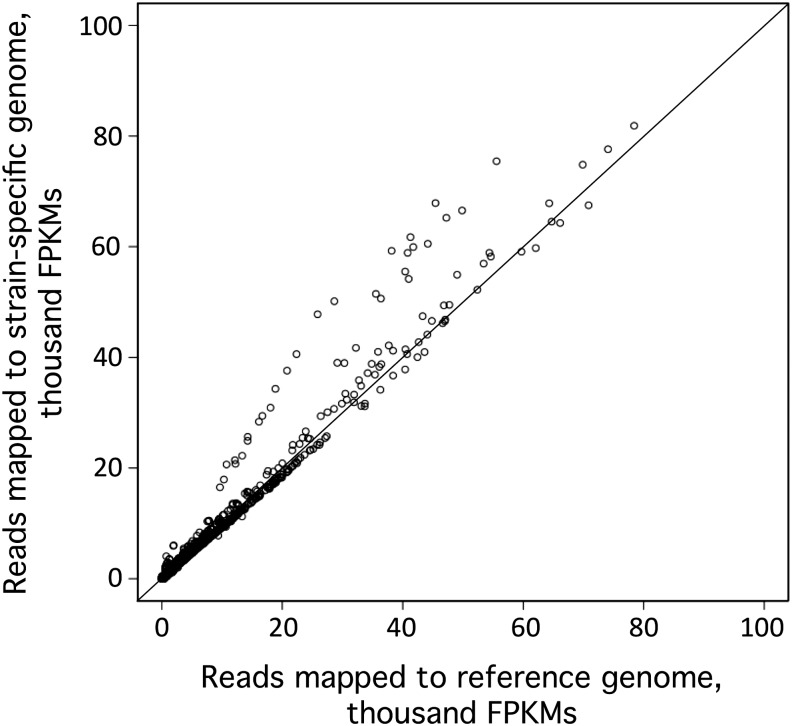
Next, we wanted to determine if the increase in the number of mappable reads affected gene expression estimates. We quantified the level of expression, in terms of fragments per kilobase of transcript per million mapped reads (FPKMs) for each gene, in each library, for both the true haplotype 2 genome and the false haplotype 1 genome (Figure 6). Not surprisingly, most genes were relatively unaffected, as evidenced by the data points that fell along the diagonal. However, the additional 2.5% of reads that became mappable when using a genome with the correct haplotype appeared to cluster to a small subset of genes, and this greatly affected the estimation of their expression levels. The expression estimates for these genes, which appear above the diagonal in Figure 6, were increased by as much as 16-fold.
Figure 6.
Impact of a Strain-Specific Genome on RNA-Seq Expression Estimates.
Reads from 16 independent CC-4348 RNA-seq libraries and 16 independent CC-4349 RNA-seq libraries were aligned in parallel to both the reference genome and a strain-specific genome for those regions identified by WGS to be haplotype 2 in the respective strains. The mRNA abundance for each gene in these regions was determined in terms of FPKMs for each library for both sets of alignments. The results are shown as a scatterplot of FPKMs as determined by alignment to the haplotype 1 reference genome (x axis) versus the same reads aligned to the haplotype 2 strain-specific genome (y axis).
RNA-Seq Data Were Used to Identify Strains
Given the high density of SNVs within the haplotype 2 regions, we speculated that RNA-seq data could also be used to determine the haplotype of a strain and therefore to identify the strain. To test this, we reexamined data from previous RNA-seq experiments. In one study, transcription from a strain that was believed to be 2137 was assayed by RNA-seq during growth in limiting iron (Urzica et al., 2012). As described above, we have since determined by genomic resequencing that this strain is distinct from the other true 2137 strains, and it has since been renamed CC-4532. A similar but independent study was conducted in the same strain grown in limiting copper (Castruita et al., 2011). The copper study also examined a mutant strain, crr1-2, that was generated in a background of CC-425 and then crossed. After aligning the RNA-seq reads to the present version of the genome, we manually scanned highly expressed genes in order to identify SNVs in the transcripts relative to the reference genome. Many of these SNVs corresponded to ones that we had identified as haplotype 2-specific variants. An example of one such region in haplotype block 17-F is presented in Supplemental Figure 10A. Reads from both studies from strain “2137” that aligned to the SDR28 locus (Cre17.g731350) carried a consistent pattern of SNVs relative to the reference. In contrast, reads from the crr1-2 strain lacked those variants. When the pattern of SNVs in the “2137” strain was compared with the two genomic haplotypes, the pattern of SNVs clearly matched haplotype 2 (Supplemental Figure 10B). In this manner, each haplotype block was scored for the presence or absence of haplotype 2-specific SNVs in highly expressed genes. The strain of Chlamydomonas identified as 2137 had the same haplotype as CC-4532, a pattern that is distinctly different from the confirmed 2137 strains 2137A+ and CC-1021.
DISCUSSION
Gilbert Smith's Original Lineages
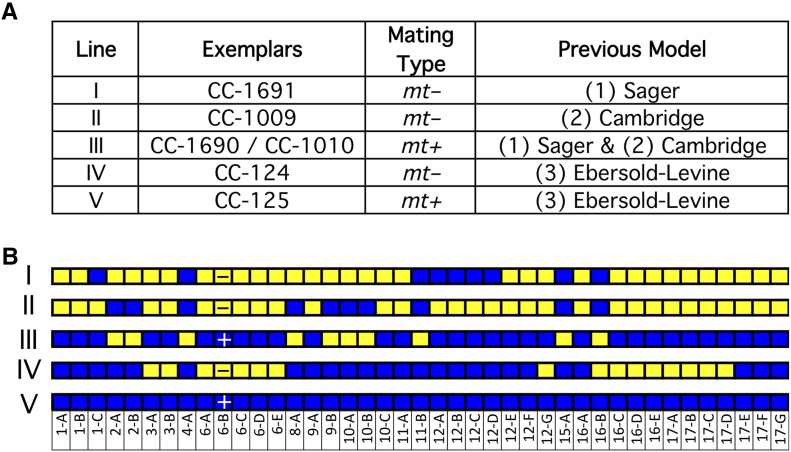
It is generally understood that Smith distributed matched pairs (mt+ and mt–) of Chlamydomonas strains as three different lineages and that each of these lineages is distinct from the others. To evaluate this, we included in our analysis three pairs of strains that are the direct descendants of those original strains: CC-1690 and CC-1691 for Sager; CC-1009 and CC-1010 for Cambridge; and CC-124 and CC-125 for Ebersold-Levine. The two strains in each pair were no more similar to each other than any two strains in any lineage. CC-124 has 103,325 SNVs relative to CC-125, due mostly to the 16 blocks of haplotype 2 for CC-124 versus none in CC-125. The distribution of haplotype 2 blocks in CC-1009 and CC-1010 are exactly opposite, suggesting that they may both be daughter cells from the same cross (Figure 7B). Unexpectedly, the mt+ strain from the Sager lineage, CC-1690, and the mt+ strain from the Cambridge lineage, CC-1010, appear to be the same strain. They have the same distribution of haplotype 2 regions and only 1310 pairwise SNVs between them.
Figure 7.
Improved Model for Gilbert Smith’s Original Strains.
(A) Five-strain model. WGS of the strains originally distributed by Smith supports a model with five distinct strains, labeled I to V. The relationship of these strains to the previous three-strain model is shown.
(B) The haplotype patterns of the five lineages. The + or – in block 6-B represents the mating type.
Given these results, we propose replacing the three-lineage model with a more accurate one in which there are five different lineages of Smith’s Chlamydomonas strains (Figure 7A). All of the strains that we have examined to date are consistent with the idea that Smith distributed these five strains, and all the standard laboratory strains could result from crosses of those five.
Collectively, 75% of all the genomes in all of the strains we examined come from only one of the parents that produced the original zygospore. If the strains that Smith distributed were F1 progeny from that cross, the contributions of the two parents would be expected to be closer to 50/50. Additionally, several chromosomes in the original five strains show evidence of multiple meiotic recombination events. These strains have been maintained clonally as haploids for many decades since leaving Smith’s laboratory, and Chlamydomonas requires cells of both mating types in order to mate. It therefore seems likely that the five original lineages were the result of an indeterminate number of crosses in Smith’s laboratory prior to distribution (Figure 8). The loss of much of the genetic diversity from the original two parents suggests two nonexclusive possibilities. First, it may be that Smith performed a number of backcrosses with his strains that diluted out the contribution of one of the parents. Second, it may be that Smith intentionally or unintentionally selected for certain traits that favored the alleles of one parent over the other. In support of this idea, strain CC-4402 retained haplotype 1 regions on chromosome 3 despite being backcrossed 10 times to a strain with the alternate haplotype at that locus. It is far more likely that this is due to a selective advantage for certain alleles than the 1 in 1024 chance that this occurred by random chance.
Figure 8.
Model for the Distribution of the Two Haplotypes.
The original zygospore that all standard laboratory strains are derived from is hypothesized to be the product of a mating between two strains that were 2% divergent in sequence, depicted here as blue versus yellow. In subsequent crosses, the resulting progeny lost much of this genetic variation so that in extant strains today there is only one haplotype for 74.8% of the genome. The other 25.2% of the genome may have one of two possible haplotypes in each strain, depending on which ancestral parent donated a given locus to that strain. Here, we arbitrarily designated the haplotype of the reference strain as haplotype 1 and the alternate as haplotype 2. Haplotype 2 regions are recognized by the relatively high (2%) frequency of variants relative to the reference strain.
Correcting Misidentified Strains
We have presented examples of strains, such as CC-4349 and CC-4532, which were found to be misidentified, here, as strains cw15-330 and 2137, respectively. In other instances, such as the parental strains of CC-4286 and CC-4287, we demonstrated that the purported lineage cannot be correct. Unfortunately, incorrectly identified strains such as these can sometimes confound the interpretation of experimental results. CC-4349 was provided to us as the parental strain of the sta6 mutant strain, CC-4348, and was used as a control for that strain in RNA-seq experiments (Blaby et al., 2013). These RNA-seq experiments were designed to isolate one variable: namely, the presence or absence of the STA6 protein. Instead, these two strains differ at 13 different haplotype blocks (Supplemental Figure 4C), and this helps to explain the phenotypic differences in mating type, cell size, and arginine auxotrophy that we observed (Blaby et al., 2013; Goodenough et al., 2014). Considering only the gene coding regions within those haplotype blocks, we confirmed the presence of over 24,000 SNVs and small InDels, affecting 619 genes, that distinguish these two strains. Collectively, this high degree of divergence serves to obscure the phenotypic effects that could otherwise be attributed to the sta6 mutation.
Another ongoing source of confusion is that strains are often identified only by a particular phenotypic characteristic. For example, four of the strains included in this study (CC-4349, CC-4568, CC-4350, and CC-4351) are frequently designated simply by the strain name cw15 due to their shared cell wall-deficient phenotype (Kondo et al., 1991; Pfannenschmid et al., 2003; Neupert et al., 2009; Goodson et al., 2011; Siaut et al., 2011). Upon sequencing, we observed that each of these four strains has a distinctly different haplotype and anywhere from 60,000 to 160,000 pairwise SNVs (Supplemental Figure 4G). To avoid ambiguity, it is imperative that researchers in the Chlamydomonas community avoid the use of phenotypes as the primary identifier of their strains. Submitting strains of interest to the Chlamydomonas Resource Center for inclusion in their collection will trigger the generation of a unique and unambiguous “CC-” name, which can alleviate this concern.
Given the importance of correctly identifying strains that are used in research, the distribution of haplotype 2 regions that we described here can be a great boon to researchers in the Chlamydomonas community. The pattern of haplotype 2 blocks can be viewed as a fingerprint for each strain. These patterns can be used to identify a strain and to provide insight on its parentage. While WGS is the premier method for identifying strains, we recognize that this approach is not always practical in terms of time and expense. Therefore, we provided a set of AS-PCR primers (Supplemental Data Set 6) that can be used to determine the haplotype of any standard laboratory strain in a matter of hours using inexpensive reagents (Supplemental Figure 5). Alternatively, we demonstrate here that RNA-seq data can also be repurposed for strain identification (Supplemental Figure 10).
Effects of Haplotype 2 on Genes
We employed SnpEff to predict the effect of haplotype 2 variants on the gene models. To validate these results, we performed extensive manual inspections of the supporting Illumina reads. For the most part, the predictions of missense and nonsense mutations due to SNVs were just as the SnpEff algorithm predicted. However, SnpEff had difficulty with other types of predictions, especially splice site variants. Most of the potential splice site mutants that were identified by SnpEff were due to small InDels that fell on exon-intron boundaries. However, on inspection, the majority of these InDels were ones in which both the reference and alternate alleles had identical nucleotides at the exon-intron border. Many of these were due to low complexity sequences, such as runs of TG dinucleotides, which are unusually abundant in the Chlamydomonas genome (Morris et al., 1986). When RNA-seq data were available, we confirmed that these potential splice site variants did not translate into alternatively spliced transcripts. Given this, we chose to omit the splice site variants from the SnpEff-predicted affected genes presented in Supplemental Figure 7B. This demonstrates the importance of critically reviewing the output of bioinformatics software.
Within the haplotype 2 regions, we identified many variants that are predicted to have some effect on gene coding sequences. However, given that both haplotypes are present in fully functional, wild-type strains, we did not expect to see many variants that cause a complete loss of function. Consistent with this idea, we observed a bias in haplotype-specific variants toward ones that are likely to preserve normal gene functioning. For example, there was a 10:1 ratio of in-frame versus out-of-frame InDels (Supplemental Figure 7B). If the number of nucleotides gained or lost were under no selective pressure, this ratio would be expected to be 1:2 in favor of out-of-frame InDels. Of the SNVs that fall within open reading frames, a significant majority (90,255 versus 64,685) cause silent rather than missense codon changes. Only 277 (0.2%) of the haplotype 2-specific small variants were predicted to cause nonsense mutations.
In work cosubmitted with this article, Flowers et al. (2015) also observed ways in which mutations present in the sequenced strains of Chlamydomonas were biased toward those that are less deleterious to gene expression. For example, they observed that 28% of genes with premature stop codons encode proteins that are truncated by 5% or less. Additionally, the ratio of nonsynonymous to synonymous codon changes is low in Chlamydomonas relative to land plants.
Identifying Mutations
Often, a primary goal of WGS is to identify the causative mutation of a particular phenotype (Schneeberger et al., 2009; Lin et al., 2013). For known mutations, such as the mutation in NIT1 common to many of the standard laboratory strains, we were able to correctly identify the expected mutant gene locus. However, the high number of variants between any two strains, coupled with the high percentage of gene models that still lack user-curated annotation (74% as of Phytozome version 10), make it extremely difficult to identify previously unknown causative mutations for specific phenotypes. Clearly, as has been observed previously (Lin et al., 2013), backcrossing to a wild-type strain multiple times with selection for the mutant phenotype is key to performing these sorts of genetic analyses. Despite the fact that our data set includes many cw15 mutant strains, we were unable to positively identify a genetic locus that was likely to be the source of that phenotype.
Isogenic Strains
In this set of 39 laboratory strains, there are three examples of strain pairs that were backcrossed with the goal of making them isogenic. Curiously, five backcrosses were sufficient to make CC-4051 and CC-4603 nearly isogenic, whereas twice as many backcrosses were insufficient in the case of CC-4402 and CC-4403. The persistence of haplotype 2 regions proximal to the mating locus is entirely expected in both cases, and these are the only blocks that are different between CC-4051 and CC-4603 (6-A through 6-C) (Supplemental Figure 4F). However, CC-4402 retained haplotype 2 regions on chromosome 3 (3-A and 3-B), and CC-4403 retained them on chromosome 17 (17-C and 17-D) (Supplemental Figure 4D). Surprisingly, these regions persisted through 10 crossings. One likely explanation for this is that there was unintentional selective pressure for alternate alleles at those loci. The group that produced these strains reported that they selected for high efficiency mating, so perhaps there are key genes related to sexual functioning in those regions (Lin et al., 2013). Indeed, Cre03.g201552 and Cre17.g705200, which are located in the relevant blocks of chromosomes 3 and 17, respectively, both have gametolysin peptidase M11 domains and multiple nonsynonymous codons between the two alleles. If the haplotype 1 alleles of these two genes conferred a more efficient mating phenotype, it could explain the persistence of these haplotype blocks in CC-4402 and CC-4403 across 10 matings. In other words, it may be unintended selective pressure that caused the statistically unlikely persistence of those regions.
METHODS
Chlamydomonas reinhardtii Strains and Culture Conditions
Chlamydomonas strains were acquired from various sources as indicated in Table 1. After sequencing, clonal populations of the strains were submitted to the Chlamydomonas Resource Center, where they are available for order as sequence-verified clones. The relevant strain identifiers are presented in Supplemental Table 5.
Cultures of each strain were grown in Innova incubators (New Brunswick Scientific) in 250-mL flasks filled with 100 mL Tris acetate-phosphate (TAP) medium at 24°C with 180 rpm agitation (Harris, 2009). Cultures were provided with 50 to 100 µmol m−2 s−1 continuous illumination by six cool white fluorescent bulbs (4100K) and three warm white fluorescent bulbs (3000K) per incubator. The medium was supplemented with Kropat’s trace elements solution (Kropat et al., 2011), including 20 µM iron except where noted.
For the iron homeostasis study, precultures in late-log growth phase in TAP supplemented with 5 µM iron (supplied as Fe/EDTA) were counted and diluted to104 cells/mL in fresh media with the indicated iron concentrations. Cell density was quantified by a hemocytometer. Chlorophyll content was assayed as described previously (Glaesener et al., 2013).
For the cell size study, two cultures per strain were grown in TAP medium as described above, and samples were collected during the late-log growth phase for analysis by the Cellometer Auto M10 (Nexcelom Bioscience). Cell diameter was determined by the Cellometer software from 1000 cells per sample and plotted as a box plot.
Nucleic Acid Preparation
Genomic DNA was prepared as follows. Each strain was clonally isolated two to four times on solid TAP-agar plates before being used to inoculate 100 mL liquid TAP cultures as described above. Total cellular DNA was prepared from stationary phase cultures (∼1 × 107 cells/mL). Cells from 50 mL of each culture were collected by centrifugation (3700g, 5 min, 4°C) and resuspended in 2 mL of Milli-Q purified water. Exactly 2 mL of the resuspended cells was transferred to a fresh tube and combined with 2 mL of 2× lysis solution (10 mM Tris-Cl, pH 7.5, 10 mM EDTA, 10 mM NaCl, 0.5% SDS, and 200 μg/mL proteinase K). After incubation for 2 h at 50°C, DNA was extracted by addition of 4 mL of phenol/chloroform, followed by vigorous shaking and centrifugation to separate the two phases (13,800g, 15 min, 10°C). Four milliliters of the aqueous phase was transferred to a clean tube and treated with 5 μL of 5 mg/mL of RNaseA for 30 min at 37°C, followed by an additional phenol/chloroform extraction as before. Four milliliters of the resulting aqueous phase was transferred to a clean tube. Next, polysaccharides were selectively precipitated by the addition of 1.4 mL of room temperature 100% ethanol, incubation for 15 min on wet ice, and centrifugation (13,800g, 10 min, 10°C). The supernatant (5.4 mL) was transferred to a clean tube, and the DNA was precipitated by the addition of 5.4 mL isopropanol, incubation for 15 min at room temperature, and centrifugation (19,800g, 30 min, 10°C). After the supernatant was discarded, the DNA pellets were air dried for 15 min at room temperature, resuspended in 500 μL of purified water, and transferred to 1.5-mL microcentrifuge tubes. DNA was precipitated again by the addition of 125 μL of 4 M NaCl and 625 μL of 20% polyethylene glycol-8000. The mixture was incubated for 30 min on wet ice, after which the DNA was collected by centrifugation (13,400g, 20 min, 4°C). The supernatant was removed by decanting, and the pellet was washed with 70% ethanol and air-dried for 15 min at room temperature. The resulting DNA was resuspended in 50 μL of purified water and the concentration determined by optical absorbance on a NanoDrop 2000 spectrophotometer (Thermo Scientific).
Determination of DNA Content per Cell
Liquid cultures of strains CC-425 and CC-1690 were grown to mid-log phase in TAP medium (Harris, 2009). Two samples of a volume of culture equivalent to 1 × 107, 2 × 107, and 4 × 107 total cells were used for DNA purification (see above). The DNA content per sample was determined in triplicate using the Qubit dsDNA HS assay kit (Life Technologies).
Genomic Library Preparation and Sequencing
For each strain, 1 μg of genomic DNA was sheared by the S-220 Adaptive Focused Acoustics system (Covaris) using the following settings: 10% duty cycle, 5.0 intensity, 200 bursts s−1, 120 s, and 6°C. The resulting fragments were used to make sequencing libraries using the TruSeq DNA sample preparation kit, version 1 (Illumina), following the low-throughput protocol. The concentrations of the resulting libraries were determined by the Qubit double-stranded DNA Broad Range assay kit (Invitrogen). Sequencing flow cells were prepared using the TruSeq cBot PE cluster generation kit, version 3 (Illumina), and sequencing was performed on a HiSeq2000 sequencer (Illumina).
Read Alignment
The raw sequences were aligned to the Chlamydomonas reference sequence (strain CC-503) version 5 with BWA mem, version 0.7.5a-r405 (Li and Durbin, 2009), using default parameters. Duplicate read pairs were removed using Picard MarkDuplicates, version 1.85(1345) (http://broadinstitute.github.io/picard) with default parameters. Numbers of unique reads and coverage are shown in Table 1.
Small Variant Detection
The Genome Analysis Toolkit (GATK), version 2.6-5-gba531bd (McKenna et al., 2010; DePristo et al., 2011), was used to prepare and call variants on the aligned, deduplicated reads. Specifically, reads from all strains were realigned together using GATK’s RealignerTargetCreator and IndelRealigner with default parameters, except -maxReadsForRealignment was doubled to 40,000. Bases were recalibrated using GATK’s BaseRecalibrator. Variants from a subset of 27 strains, with a minimum quality score of 120, were used as the known variant input for BaseRecalibrator. Additionally, base quality scores were capped by the BAQ algorithm (Li, 2011) using PrintReads. Variants were called for all strains together, but separately for SNVs and InDels, using GATK’s UnifiedGenotyper with -downsample_to_coverage increased to 10000, -sample_ploidy 1, and -stand_call_conf 20.0. Queue was used to parallelize jobs for RealignerTargetCreator, IndelRealigner, and UnifiedGenotyper.
“HET” Labeling
Variants with greater than one base strongly represented were presumed due to collapsed imperfect repeats in the reference genome. They were identified by running GATK’s UnifiedGenotyper in diploid mode and selecting variants called as heterozygous for at least one strain (with a genotype quality greater than 98) and with no strains called as a homozygous variant (with a genotype quality greater than 98). These variants were labeled as “HET” in the variant call format (vcf) files and were excluded from counts of SNVs.
Haplotype Variation
Variants presumed to have been present in the original zygote, i.e., variation between the two joining gametes, were identified by their clustering into two distinct haplotypes, as follows. After excluding variants either found in our CC-503 data, labeled as HET (see above), or with a depth-adjusted quality score <2.0, variants were counted in 10,000 base windows (excluding reference Ns). Counts were additionally separated on the basis of the set of strains that carry the variant. Haplotype regions were defined by merging neighboring windows with counts of >20 variants for identical sets of variant strains. Additionally, resulting regions with identical strain patterns within 460,000 bases of each other were merged. For windows that overlap two neighboring strain patterns, the midpoint of the overlap was taken as the transition point. The resulting 41 genome regions with both haplotypes represented are shown in Figure 5. Strains were clustered by haplotype pattern and plotted with dendrogram using the heatmap2 function of the gplots package in R (https://cran.r-project.org/web/packages/gplots/index.html).
Specific variants that follow the alternate haplotype strain patterns were identified and labeled “set=orig” in the vcf files using the following rules: (1) strain pattern matches one of the defined alternate haplotype regions on the same chromosome; (2) no more than one strain with a high quality mismatch to the strain pattern (i.e., called variant when should be reference or vice versa); (3) if less than three strains have high quality matching variant calls, then a minimum of three lower quality matching variant calls and <10 mismatches (at any quality) are required; and (4) if less than three strains have matching variant calls (at any quality) there must be at least one fewer mismatch. These rules allow for sites with high uncertainty and error and for occasional reversion in a single strain and did not lead to excessive sporadic overidentification, as determined visually using Integrative Genomics Viewer (IGV) (Robinson et al., 2011; Thorvaldsdóttir et al., 2013).
Variant Quality Score Recalibration: Creating a Training Set
In order to use GATK’s Variant Quality Score Recalibration, it was necessary to produce a set of high confidence variants to be used for training. The high frequency of certain SNV patterns due to shared ancestry was exploited for this purpose since false positives would only rarely be expected to match these patterns. Genotype patterns were counted for all nonfiltered SNVs on the main chromosomes, e.g., 000001100100100.0010001100.0110010000000000 represents the pattern of variant (1), reference (0), or uncalled (.) for a single variant across an ordered set of strains. After excluding patterns with any uncalled strains or with only a single variant strain, the top 50 most frequent patterns were identified. They ranged in frequency from 53,145 variants to 58 variants and totaled 327,268 variants (61% of all nonfiltered SNVs). As expected, the set is enriched for interhaplotype (set=orig) variants with only 838 (0.3%) due to shared ancestry within the laboratory. Transition/transversion (ts/tv) ratios were calculated for the variants within each pattern, and variants for patterns with ts/tv < 1.4 were manually inspected using IGV to ensure their high confidence. Ts/tv for the combined top 50 patterns was 1.48, while ts/tv for the remaining variants was 1.32. The same strain patterns were used to create a set of 45,105 high confidence InDel variants, of which 244 (0.5%) were laboratory-derived variants.
Variant Quality Score Recalibration and Filtering: Only Applied to Laboratory Variation
GATK’s VariantRecalibrator was run separately for SNVs and InDels using the training sets described above, prior=20.0, and the following parameters: for SNVs, target_titv 1.5 -an MQRankSum -an ReadPosRankSum -an FS -an QD -an DP; for InDels, an MQRankSum -an ReadPosRankSum -an FS -an DP. Subsequent manual inspection of variants within various VQSLOD tranches for both interhaplotype (original) variants and laboratory-derived variants made clear that filtering was not beneficial for the interhaplotype variants, but the laboratory-derived variants were substantially improved by VQSLOD filtering. A cutoff of VQSLOD > 1.32 was implemented for laboratory-derived SNVs, filtering 70% of the 15,453 previously unfiltered SNVs. This cutoff was chosen to minimize the total number of false positives and false negatives, crudely estimated by manual inspection to be 6% for each. A cutoff of VQSLOD > −0.30 was implemented for laboratory-derived InDels, filtering 18% of the 6873 previously unfiltered InDel variants.
All variants called by UnifiedGenotyper are included in Supplemental Data Set 2 as a vcf file. A filter field has been set based on the above analyses. The filters, as described above, and their nonunique counts are LowQual (7900), HET (31,061), VQSRTrancheSNP99.90to100.00 (4780), VQSRTrancheSNP99.00to99.90 (5112), VQSRTrancheSNP98.00to99.00 (932), VQSRTrancheINDEL99.90to100.00 (1019), and VQSRTrancheINDEL99.80to99.90 (246).
Identification of Structural Variants
Larger variants were identified using the tools Pindel (Ye et al., 2009) and BreakDancer (Chen et al., 2009) followed by extensive additional filtering steps. These filtering steps were based on trial and error using repeated manual inspection of the results in IGV and would therefore not be appropriately applied to other data sets. Variants of length <20 bp predicted by Pindel or BreakDancer were excluded since the true variants would mostly be redundant with those predicted by the small variant caller, GATK’s UnifiedGenotyper (see above).
For variants identified by Pindel to be included, at least one sequenced strain must have five supporting reads. The total number of supporting reads across samples with less than five supporting reads must be <10 (GT0_SRcount) and less than one-third of all reads. Samples with at least five supporting reads were labeled GT=1, others were labeled GT=0. Variants that overlap with UnifiedGenotyper variants labeled HET were labeled HET and excluded; otherwise, variants that overlap with UnifiedGenotyper variants were labeled RED for redundant and excluded, unless they were deletions >40 bp with a percentage change in size (from reference bases to alternate bases) of >50% or deletions >80 bp, or if they are inversions, small insertions, or tandem duplications >40 bp in size. Additionally, variants were excluded based on ratios of reference supporting reads to variant supporting reads across all GT=1 samples (GT1ratio) and ratios of normalized read depths for GT=0 over GT=1 (RDPratio) as follows: Variants were excluded if they were deletions with GT1ratio > 10 and RDPratio < 2, or duplications with GT1ratio > 32 and RDPratio < 0.7, or any other type with GT1ratio > 10.
For variants identified by BreakDancer to be included, at least one sequenced strain must have five supporting reads. The total number of supporting reads across samples with less than five supporting reads must be <10 (GT0_SRcount) and less than one-third of all reads. Samples with at least five supporting reads were labeled GT=1, and others were labeled GT=0. Additionally, variants were excluded based on ratios of reference supporting reads to variant supporting reads across all GT=1 samples (GT1ratio) as follows: Variants were excluded if they were deletions or insertions with GT1ratio > 10 or any other type with GT1ratio > 6. Also all variants of type BND were filtered as LowQual, as they were found to be mostly false positives due to repeats.
Variant Impact Prediction
The functional impact of each small variant (SNV or small InDel) was predicted using SnpEff (Cingolani et al., 2012). The results are included in the INFO field of the corresponding vcf output (see below). These predictions were graded by SnpEff for severity of the impact on the encoded protein. Predictions rated as either MODERATE or HIGH impact were selected for additional review (Supplemental Figure 7) and were manually compared with RNA-seq data (see below). SnpEff’s predictions were generally validated by the RNA-seq data, with one exception—the splice site variants. These HIGH impact calls, due to SNVs and InDels at exon-intron boundaries, were almost entirely unsupported by the RNA-seq data. As such, we downgraded the splice site variant calls from HIGH impact to LOW impact.
SnpEff was also used to predict the effects of the structural variants on gene models. However, SnpEff is not designed for such data and only worked on a subset of the variant types. The impact fields were adjusted as follows: For inversions, the impact was set to MODERATE if either end was in a gene or to HIGH if either end was not in the 5′ or 3′ untranslated region and both ends were not in the same intron. Both impacts were kept if the two ends were in different genes. For duplications, the impact was set to HIGH if the duplication was within one gene and both ends were not in the same intron or MODERATE if one end was in a gene and the other was outside of that gene.
Excluded variants are included in the vcf file with exclusion categories listed in the FILTER field.
Transposons
The sequences of the following Chlamydomonas transposons were downloaded from NCBI GenBank: Bill (DQ446204.1), Gulliver (AF019750.1 and AF019751.1), MRC1 (DQ446210.1), Pioneer1 (U19367.1), REM1 (AY227352.1), Tcr1 (DQ446205.1), Tcr3 (Y14652.1 and Y14653.1), TOC1 (X56231.1), and TOC2 (X84663.1). The positions of these transposons in the reference genome were identified by comparing FASTA files of the transposon sequences to version 5 of the Chlamydomonas genome on Phytozome using the BLAST algorithm. The resulting coordinates were used to generate a bed file of likely transposons (included in Supplemental Data Set 5). In order to identify putative sites of transposon jumping, the coordinates of the various transposons were compared for significant overlap to sites of large deletions identified by Pindel and BreakDancer.
Haplotype Determination by Allele-Specific Amplification
DNA was prepared from strains CC-1009 and CC-1010 as described above. Five microliters of DNA was diluted into 495 μL of purified water (final concentration ∼10 ng/µL). Custom oligonucleotides (Eurofins MWG Operon) were dissolved in purified water to a concentration of 3 µM as a 10× working stock. The sequences of each are provided in Supplemental Data Set 6. AS-PCR was performed in 96-well PCR plates on a CFX96 Optical Thermocycler (Bio-Rad) using 10 μL of iTaq Universal SYBR Green Supermix (Bio-Rad), 1 μL of each primer stock (300 nM final concentration), 5 μL of diluted DNA template, and 3 μL purified water. The thermocycler was run for 30 s at 95°C, followed by 30 cycles of 5 s at 95°C and 30 s at 63°C. Ten microliters of the resulting amplicons was mixed with bromophenol blue/xylene cyanol DNA loading dye and loaded onto a 2% agarose gel in TAE buffer (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA) plus 0.2 µg/mL ethidium bromide. The DNA was separated for ∼20 min at 8 V/cm and photographed on a UV transilluminator.
Strain-Specific Reference Genome and Annotations
The Custom Chlamy Generator (CCG) is a software program we created to generate strain-specific reference genomes and gff3 gene annotation files using the SNVs and small InDel data in Supplemental Data Set 1. Alternatively, CCG can take a user-supplied haplotype (such as determined by the AS-PCR assay described above) and generate strain-specific files for any other standard laboratory strain. CCG is available at https://bitbucket.org/gallaher/custom-chlamy-generator. Detailed instructions for use of CCG are included there.
RNA-Seq
Sequencing data from an RNA-Seq study performed with strains CC-4348 and CC-4349 was described previously (Blaby et al., 2013). Here, the resulting data were realigned to the Chlamydomonas reference genome (v5.0; see above) or a strain-specific genome generated by CCG. To isolate the effect of haplotype, subgenomes with both the reference sequence and the strain-specific sequence were generated with only those regions identified to be haplotype 2 in CC-4348 (blocks 3-A, 3-B, 6-A, 6-B, 6-C, 6-D, 6-E, 12-G, and 16-B; 4.3 Mb total) or haplotype 2 in CC-4349 (blocks 3-A, 3-B, 16-B, 16-C, 16-D, 16-E, 17-A, 17-B, 17-C, and 17-D; 4.4 Mb total). Alignment of the reads to each subgenome was performed with RNA-Star v2.4.0j using default settings (Dobin et al., 2013). FPKM expression estimates were calculated by cuffdiff v2.2.1 using default settings (Trapnell et al., 2013).
Accession Numbers
Version 5.0 of the Chlamydomonas reference genome (Creinhardtii_281_v5.0.fa.gz) and the corresponding version 5.5 gene annotations (Creinhardtii_281_v5.5.gene.gff3.gz) are available at Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html). Sequence data from this article can be found in the NCBI Short Read Archive sequence database under accession number SRP053354. Chlamydomonas transposons are available from NCBI GenBank under the following accession numbers: Bill (DQ446204.1), Gulliver (AF019750.1 and AF019751.1), MRC1 (DQ446210.1), Pioneer1 (U19367.1), REM1 (AY227352.1), Tcr1 (DQ446205.1), Tcr3 (Y14652.1 and Y14653.1), TOC1 (X56231.1), and TOC2 (X84663.1).
Supplemental Data
Supplemental Figure 1. Growth Phenotype of Iron Limitation on Additional Wild-Type Strains.
Supplemental Figure 2. Callable Loci.
Supplemental Figure 3. Evidence for Recombination between Haplotypes.
Supplemental Figure 4. Comparison of Haplotype Patterns for Selected Strains.
Supplemental Figure 5. Allele-Specific Amplification to Identify Haplotype.
Supplemental Figure 6. Examples of Transposon Position Jumping in Chromosome 16.
Supplemental Figure 7. Predicted Effects of Haplotype 2 Variants on Gene Models.
Supplemental Figure 8. Variants in Genes Attributable to Haplotype 2.
Supplemental Figure 9. Laboratory-Originated Mutations in Commonly Studied Genes.
Supplemental Figure 10. Determining Haplotype from RNA-Seq Data Identifies Mislabeled 2137 Strain.
Supplemental Table 1. Transversions and Transitions for All SNVs.
Supplemental Table 2. Transposon Position Jumping at 84 Loci.
Supplemental Table 3. Examples of Genes with Predicted Haplotype 2-Specific Variants.
Supplemental Table 4. Enrichment of Gene Ontology Terms in Laboratory-Originated Loss-of-Function Mutations.
Supplemental Table 5. Sequence-Verified Clones Available from the Chlamydomonas Resource Center.
The following materials have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.q1t7v.
Supplemental Data Set 1. Detailed Strain Histories.
Supplemental Data Set 2. SNV and Small InDel VCF File.
Supplemental Data Set 3. Structural Variant VCF File.
Supplemental Data Set 4. Pairwise SNVs for All Strains.
Supplemental Data Set 5. Transposon Position BED File.
Supplemental Data Set.6. Allele-Specific PCR Primer Sequences.
Supplementary Material
Acknowledgments
We thank Matt Laudon and the staff at the Chlamydomonas Resource Center for their invaluable work curating thousands of strains of Chlamydomonas and for keeping track of their respective histories. We also thank the members of the Chlamydomonas community that contributed strains to this work (as named in Table 1). We thank Patrice Salome and Stefan Schmollinger for their insightful comments on this article. Funding was provided by the National Institutes of Health R24 GM092473 and by the Office of Science (Biological and Environmental Research), U.S. Department of Energy (Grants DE-FC02-02ER63421 and DE-FD02-04ER-15529). Lastly, we thank the UCLA Broad Stem Cell Research Center High-Throughput Sequencing Core Resource for sequence service.
AUTHOR CONTRIBUTIONS
S.D.G., A.G.G., S.T.F.-G., M.P., and S.S.M. designed the research. S.D.G., A.G.G., and S.T.F.-G. performed the research and analyzed data. S.D.G. and S.T.F.-G. wrote the article.
Glossary
- WGS
whole-genome sequencing
- SNV
single nucleotide variant
- InDel
insertions or deletion
- FPKM
fragments per kilobase of transcript per million mapped reads
- TAP
Tris acetate-phosphate
- GATK
Genome Analysis Toolkit
- IGV
Integrative Genomics Viewer
- CCG
Custom Chlamy Generator
References
- Blaby I.K., et al. (2013). Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell 25: 4305–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak T., Angun P., Demiray Y.E., Ozkan A.D., Elibol Z., Tekinay T. (2012). Differential effects of nitrogen and sulfur deprivation on growth and biodiesel feedstock production of Chlamydomonas reinhardtii. Biotechnol. Bioeng. 109: 1947–1957. [DOI] [PubMed] [Google Scholar]
- Castruita M., Casero D., Karpowicz S.J., Kropat J., Vieler A., Hsieh S.I., Yan W., Cokus S., Loo J.A., Benning C., Pellegrini M., Merchant S.S. (2011). Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., et al. (2009). BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D., Colleoni C., Mouille G., Buléon A., Gallant D.J., Bouchet B., Morell M.K., d'Hulst C., Myers A.M., Ball S.G. (2001). Two loci control phytoglycogen production in the monocellular green alga Chlamydomonas reinhardtii. Plant Physiol. 125: 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D.R., Plaskitt A. (1971). Genetical and structural analyses of cell-wall formation in Chlamydomonas reinhardi. Genet. Res. 17: 33. [Google Scholar]
- De Hoff P.L., Ferris P., Olson B.J.S.C., Miyagi A., Geng S., Umen J.G. (2013). Species and population level molecular profiling reveals cryptic recombination and emergent asymmetry in the dimorphic mating locus of C. reinhardtii. PLoS Genet. 9: e1003724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R.M., Haglund C.M., Chin B.L., Kobayashi M.C., Niyogi K.K. (2005). Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 137: 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersold W.T. (1956). Crossing over in Chlamydomonas reinhardi. Am. J. Bot. 43: 408. [Google Scholar]
- Eriksson M., Moseley J.L., Tottey S., del Campo J.A., Quinn J., Kim Y., Merchant S. (2004). Genetic dissection of nutritional copper signaling in Chlamydomonas distinguishes regulatory and target genes. Genetics 168: 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquível M.G., Amaro H.M., Pinto T.S., Fevereiro P.S., Malcata F.X. (2011). Efficient H2 production via Chlamydomonas reinhardtii. Trends Biotechnol. 29: 595–600. [DOI] [PubMed] [Google Scholar]
- Fernández E., Schnell R., Ranum L.P., Hussey S.C., Silflow C.D., Lefebvre P.A. (1989). Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 86: 6449–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers J.M., et al. (2015). Whole-genome resequencing reveals extensive natural variation in the model green alga Chlamydomonas reinhardtii. Plant Cell 27: 2353–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaesener A.G., Merchant S.S., Blaby-Haas C.E. (2013). Iron economy in Chlamydomonas reinhardtii. Front. Plant Sci. 4: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ballester D., Pootakham W., Mus F., Yang W., Catalanotti C., Magneschi L., de Montaigu A., Higuera J.J., Prior M., Galvan A., Fernandez E., Grossman A.R. (2011). Reverse genetics in Chlamydomonas: a platform for isolating insertional mutants. Plant Methods 7: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U., et al. (2014). The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot. Cell 13: 591–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson C., Roth R., Wang Z.T., Goodenough U. (2011). Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot. Cell 10: 1592–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C.H., Ranum L.P.W., Lefebvre P.A. (1988). Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr. Genet. 13: 503–508. [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Karpowicz S.J., Heinnickel M., Dewez D., Hamel B., Dent R., Niyogi K.K., Johnson X., Alric J., Wollman F.-A., Li H., Merchant S.S. (2010). Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth. Res. 106: 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. (2009). The Chlamydomonas Sourcebook, 2nd ed (San Diego, CA: Academic Press; ). [Google Scholar]
- Kondo T., Johnson C.H., Hastings J.W. (1991). Action spectrum for resetting the circadian phototaxis rhythm in the CW15 strain of Chlamydomonas: I. Cells in darkness. Plant Physiol. 95: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S.S., Malasarn D. (2011). A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66: 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Abe J., Saito T., Matsuda Y. (2002). Genealogical relationships among laboratory strains of Chlamydomonas reinhardtii as inferred from matrix metalloprotease genes. Curr. Genet. 41: 115–122. [DOI] [PubMed] [Google Scholar]
- Li H. (2011). Improving SNP discovery by base alignment quality. Bioinformatics 27: 1157–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.B., et al. (2004). Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117: 541–552. [DOI] [PubMed] [Google Scholar]
- Lin H., Miller M.L., Granas D.M., Dutcher S.K. (2013). Whole genome sequencing identifies a deletion in protein phosphatase 2A that affects its stability and localization in Chlamydomonas reinhardtii. PLoS Genet. 9: e1003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.C., Sommer F., Allen M.D., Lu S.F., Merchant S.S. (2008). FER1 and FER2 encoding two ferritin complexes in Chlamydomonas reinhardtii chloroplasts are regulated by iron. Genetics 179: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppes R., Deltour R. (1975). Changes in phosphatase activity associated with cell wall defects in Chlamydomonas reinhardi. Arch. Microbiol. 103: 247–250. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., Allen M.D., Kropat J., Moseley J.L., Long J.C., Tottey S., Terauchi A.M. (2006). Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763: 578–594. [DOI] [PubMed] [Google Scholar]
- Merchant S.S., Kropat J., Liu B., Shaw J., Warakanont J. (2012). TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 23: 352–363. [DOI] [PubMed] [Google Scholar]
- Morris J., Kushner S.R., Ivarie R. (1986). The simple repeat poly(dT-dG).poly(dC-dA) common to eukaryotes is absent from eubacteria and archaebacteria and rare in protozoans. Mol. Biol. Evol. 3: 343–355. [DOI] [PubMed] [Google Scholar]
- Ness R.W., Morgan A.D., Colegrave N., Keightley P.D. (2012). Estimate of the spontaneous mutation rate in Chlamydomonas reinhardtii. Genetics 192: 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J., Karcher D., Bock R. (2009). Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 57: 1140–1150. [DOI] [PubMed] [Google Scholar]
- Pazour G.J., Sineshchekov O.A., Witman G.B. (1995). Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J. Cell Biol. 131: 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannenschmid F., Wimmer V.C., Rios R.-M., Geimer S., Kröckel U., Leiherer A., Haller K., Nemcová Y., Mages W. (2003). Chlamydomonas DIP13 and human NA14: a new class of proteins associated with microtubule structures is involved in cell division. J. Cell Sci. 116: 1449–1462. [DOI] [PubMed] [Google Scholar]
- Pröschold T., Harris E.H., Coleman A.W. (2005). Portrait of a species: Chlamydomonas reinhardtii. Genetics 170: 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix J.D. (1995). Chlamydomonas reinhardtii as the photosynthetic yeast. Annu. Rev. Genet. 29: 209–230. [DOI] [PubMed] [Google Scholar]
- Sager R. (1955). Inheritance in the green alga Chlamydomonas reinhardi. Genetics 40: 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A.H., Nielsen K.L., Jørgensen J.-E., Weigel D., Andersen S.U. (2009). SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Schnell R.A., Lefebvre P.A. (1993). Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz M., Hoshino T., Johnson D., Riley M.R., Cuello J. (2011). Flocculation of wall-deficient cells of Chlamydomonas reinhardtii mutant cw15 by calcium and methanol. Biomass Bioenergy 35: 4835–4840. [Google Scholar]
- Siaut M., Cuiné S., Cagnon C., Fessler B., Nguyen M., Carrier P., Beyly A., Beisson F., Triantaphylidès C., Li-Beisson Y., Peltier G. (2011). Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.M. (1946). The nature of sexuality in Chlamydomonas. Am. J. Bot. 33: 625–630. [PubMed] [Google Scholar]
- Smith G.M., Regnery D.C. (1950). Inheritance of sexuality in Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 36: 246–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene E., Inwood W., Kustu S. (2004). Lack of the Rhesus protein Rh1 impairs growth of the green alga Chlamydomonas reinhardtii at high CO2. Proc. Natl. Acad. Sci. USA 101: 7787–7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer R.J., Mets L. (1981). Photosynthesis-deficient mutants of Chlamydomonas reinhardii with associated light-sensitive phenotypes. Plant Physiol. 67: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urzica E.I., Casero D., Yamasaki H., Hsieh S.I., Adler L.N., Karpowicz S.J., Blaby-Haas C.E., Clarke S.G., Loo J.A., Pellegrini M., Merchant S.S. (2012). Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell 24: 3921–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. (2009). Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.M., Merchant S., Theg S.M., Selman B.R. (1988). Isolation of a cDNA clone for the gamma subunit of the chloroplast ATP synthase of Chlamydomonas reinhardtii: import and cleavage of the precursor protein. Proc. Natl. Acad. Sci. USA 85: 1369–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabawinski C., Van Den Koornhuyse N., D’Hulst C., Schlichting R., Giersch C., Delrue B., Lacroix J.M., Preiss J., Ball S. (2001). Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-glucose pyrophosphorylase. J. Bacteriol. 183: 1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.