Microscopic dissection and RNA-seq in leaves and apices of barley identify genes important for floral transition, seed set and fertility in barley in response to photoperiod and variation at Ppd-H1.
Abstract
Timing of the floral transition and inflorescence development strongly affect yield in barley (Hordeum vulgare). Therefore, we examined the effects of daylength and the photoperiod response gene PHOTOPERIOD1 (Ppd-H1) on barley development and analyzed gene expression changes in the developing leaves and main shoot apices (MSAs) of barley by RNA sequencing. The daylength sensitivity of MSA development had two phases, floret primordia initiated under long and short days, whereas successful inflorescence development occurred only under long days. The transcripts associated with floral transition were largely regulated independently of photoperiod and allelic variation at Ppd-H1. The photoperiod- and Ppd-H1-dependent differences in inflorescence development and flower fertility were associated with the induction of barley FLOWERING LOCUS T orthologs: FT1 in leaves and FT2 in MSAs. FT1 expression was coregulated with transcripts involved in nutrient transport, carbohydrate metabolism, and cell cycle regulation, suggesting that FT1 might alter source-sink relationships. Successful inflorescence development correlated with upregulation of FT2 and transcripts related to floral organ development, phytohormones, and cell cycle regulation. Identification of photoperiod and stage-specific transcripts gives insights into the regulation of reproductive development in barley and provides a resource for investigation of the complexities of development and yield in temperate grasses.
INTRODUCTION
The timing of reproductive development has a major effect on yield in cereal crops such as barley (Hordeum vulgare). A better understanding of the developmental processes that determine potential seed number could enhance the efficiency of breeding programs aimed at improving grain yield. Reproductive development in temperate cereals is divided into three phases based on morphological changes of the shoot apex: leaf initiation (vegetative phase), floret initiation (early reproductive phase), and spike growth (late reproductive phase) (Appleyard et al., 1982; Slafer and Rawson, 1994; González et al., 2002). Waddington et al. (1983) developed a quantitative scale for barley and wheat (Triticum aestivum) development based on the morphogenesis of the shoot apex and carpels. This scale is based on the progression of the most advanced floret primordium and carpel of the inflorescence. The enlargement of the apical dome at Waddington stage (W) 1.0 represents an apex that is transitioning to a reproductive state and indicates the end of the vegetative phase. The emergence of the first floret primordia on the shoot apex at the double ridge stage (W1.5-W2.0) specifies a reproductive shoot apical meristem (SAM). At the stamen primordium stage (W3.5), the first floral organ primordia differentiate and stem elongation initiates. In barley, the induction of floret primordia on the inflorescence continues until the awn primordium stage (W5.0). Anthesis and pollination of the most advanced floret occurs at the last stage of the Waddington scale (W10.0).
The timing of developmental events before anthesis affects yield components, such as spike number per plant or seed number per spike (Fischer, 1985; Slafer, 2003; Arisnabarreta and Miralles, 2008). Several studies in wheat and barley have shown that the durations of different phenological phases of preanthesis development are genetically controlled and vary in their sensitivity to vernalization and photoperiod (Slafer and Rawson, 1994; González et al., 2002; Borrás-Gelonch et al., 2012a). Vernalization has a strong effect on vegetative phase duration and stem elongation (Purvis, 1934; Chen et al., 2009; Sasani et al., 2009). The vernalization requirement in winter barley is determined by the interaction between two genes: VERNALIZATION-H2 (Vrn-H2), a strong inhibitor of flowering under long-day (LD) conditions, and Vrn-H1. During vernalization, Vrn-H1 is upregulated and represses Vrn-H2 (Yan et al., 2003, 2004). Deletions of the Vrn-H2 locus or in the regulatory region of Vrn-H1 are characteristic of spring barley, which does not require vernalization (Hemming et al., 2009). The induction of Vrn-H1 parallels the floral transition in barley (von Zitzewitz et al., 2005), and the expression of Vrn1 is required for the floral transition in wheat (Pearce et al., 2013). In contrast to vernalization, variations in photoperiod have minor effects on the duration of the vegetative phase but strongly accelerate the early and late reproductive phases of inflorescence development in wheat and barley (Miralles and Richards, 2000; Slafer et al., 2001). The duration of the spike growth phase has been identified as a major determinant of seed number (Fischer, 1985; Miralles et al., 2000; Slafer, 2003; Reynolds et al., 2009; Alqudah and Schnurbusch, 2014) because of competition between the spike and stem for limited assimilates (González et al., 2003, 2011; Ghiglione et al., 2008). Quantitative trait loci with strong effects on the durations of individual preanthesis phases have been mapped close to PHOTOPERIOD1, Ppd-B1/Ppd-D1 and Ppd-H1 in wheat and barley, respectively (González et al., 2005; Borrás-Gelonch et al., 2010, 2012a, 2012b; Sanna et al., 2014). In barley, Ppd-H1 (also known as PRR37), a homolog of the Arabidopsis thaliana genes PSEUDO-RESPONSE REGULATOR3 (PRR3) and PRR7, is a key regulator that accelerates flowering under LDs. A recessive mutation in the conserved CCT domain of Ppd-H1 is associated with late flowering under inductive LDs (Turner et al., 2005). The mutated ppd-h1 allele has been selected in Northern European spring barley genotypes, whereas the dominant wild-type Ppd-H1 allele is prevalent in wild and Mediterranean winter barley (Jones et al., 2008). A latitudinal cline in the distribution of the functional variation at PPD1 in barley and wheat indicates that this gene has a strong adaptive effect on yield (Laurie et al., 1994, 1995; Worland et al., 1998; Cockram et al., 2007). Under LDs, PPD1 induces the expression of VRN3, a homolog of Arabidopsis FLOWERING LOCUS T (FT), and rice (Oryza sativa) Hd3a. FT and Hd3a proteins translocate from the leaves through the phloem to the SAM, where these proteins induce the switch from vegetative to reproductive growth (Corbesier et al., 2007; Tamaki et al., 2007). Faure et al. (2007) identified five different FT-like genes in barley: FT1 (Vrn-H3), FT2, FT3, FT4, and FT5. Similar to Arabidopsis, FT-like genes in cereals have been described as central regulators of the transition from vegetative to reproductive growth (Kojima et al., 2002; Lv et al., 2014). However, most studies have only examined the effects of PPD1 and downstream FT-like genes on the duration of the entire process from sowing to flowering (Shitsukawa et al., 2007; Chen et al., 2009; González et al., 2005; Borrás-Gelonch 2012a, 2012b). Therefore, little is known regarding the effects of these genes on the timing of individual preanthesis phases and on the formation of individual yield components.
In this study, we aimed to dissect the complexity of the morphological and transcriptional changes in barley leaves and shoot apices during the three distinct preanthesis phases. To this end, we characterized the effects of photoperiod and functional variation at Ppd-H1 on the timing of these phases and on concomitant transcriptional changes that occur before and after floral transition, at the transition apex and double ridge stage, and toward the end of the early reproductive phase, at the stamen primordium stage. This approach helped us to identify putative central regulators of floral development, independent of photoperiod, and to identify candidate genes acting in the photoperiod- and Ppd-H1-dependent flowering pathway in both the leaves and shoot apices. The parallel measurements of plant development and of leaf and main shoot apex (MSA) transcriptome changes provide a valuable resource for future investigations into the complex regulation of reproductive development and yield in barley.
RESULTS
Effects of Photoperiod and Allelic Variation at Ppd-H1 on MSA Development and Plant Architecture
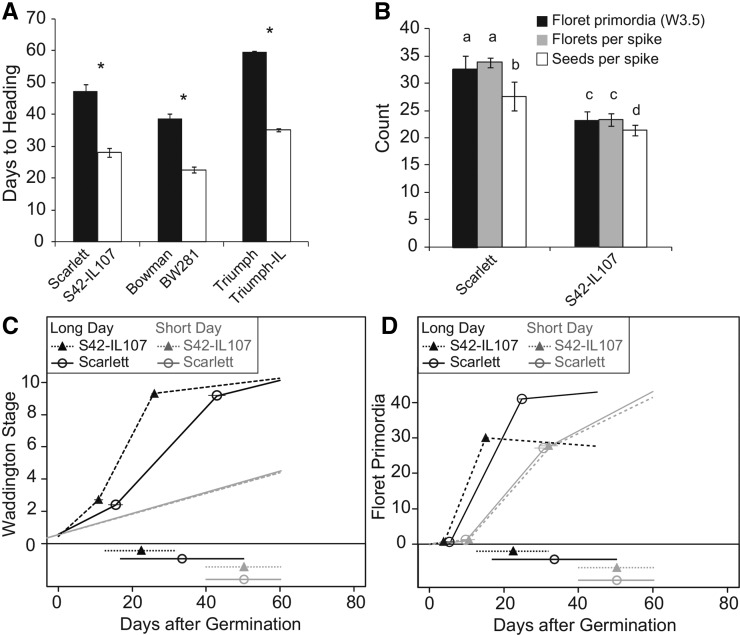
Our objective was to dissect alterations in preanthesis development in barley in response to variations in photoperiod and at Ppd-H1. We analyzed morphological changes of the MSA during development under LD and short-day (SD) conditions in three pairs of spring barley cultivars and introgression lines (ILs). The spring barley cultivars Scarlett, Bowman, and Triumph carried the mutant allele ppd-H1, and the ILs carried the dominant wild-type Ppd-H1 allele introgressed from either wild barley or a winter cultivar into the respective spring barley genetic background. We were primarily interested in identifying morphological and molecular differences resulting from allelic variation at Ppd-H1. Therefore, we focused our analyses on differences consistently detected between the spring barley cultivars and their respective ILs and did not further investigate the background variation between Scarlett, Bowman, and Triumph. The macroscopic development of all six genotypes was scored as heading as soon as the first awns became visible on the main culm. Under LDs, the main shoot spikes of Bowman, Scarlett, and Triumph headed at 39, 46, and 60 d after germination, respectively (Figure 1A). The ILs with the dominant Ppd-H1 allele flowered significantly earlier than their recurrent parents; BW281, S42-IL107, and Triumph-IL headed at 23, 27, and 35 d after germination, respectively. Consequently, the introgression of a dominant Ppd-H1 allele into the spring barley lines consistently accelerated reproductive development in all three spring barley backgrounds. Under LDs, the dissection of Scarlett and S42-IL107 plants revealed a biphasic pattern of MSA development with strong acceleration of development from the beginning of stem elongation until anthesis (Figure 1C). S42-IL107 exhibited only a moderate acceleration of the vegetative phase and early reproductive development until the beginning of internode elongation; however, a strong acceleration of inflorescence development from the beginning of stem elongation until heading was observed compared with Scarlett (Figure 1C; Supplemental Table 1). As for S42-IL107, the acceleration of early plant development in the other ILs resulted in a lower final number of leaves emerging from the main culm (Supplemental Figure 1A). Under SDs, MSA development was not affected by genetic variation at Ppd-H1 and progressed at a constant rate until abortion between W4.0 and W6.0 during stem elongation (Figures 1C and 2G; Supplemental Table 1). Consequently, none of the investigated genotypes flowered from the main shoot under SDs.
Figure 1.
Developmental Phenotypes of Barley Main Shoot Apices.
(A) Heading date of spring barley genotypes and Ppd-H1 introgression lines grown under LD conditions.
(B) Phenotypes of the main shoot spike recorded at stamen primordium stage (W3.5, floret primordia per inflorescence) or at plant maturity (florets per spike and seeds per spike) in Scarlett and S42-IL107 under LDs. The bars represent the means ± sd of 5 to 15 plants. Significant differences (P < 0.05) between spring barley and derived Ppd-H1 introgression lines and between spike-related phenotypes are indicated as asterisks and small letters on top of the charts, respectively.
(C) and (D) Broken-line regression analyses are shown for shoot apex development (C) and floret primordia appearance (D) on main shoot inflorescences of plants germinated and grown under SD or LD conditions. The positions of regression line breakpoints and their 95% confidence intervals are indicated above each chart. The slopes of individual segments of the composite regression lines representing the rate of apex development and floret primordia induction with 95% CIs are presented in Supplemental Table 11. The lines and symbols at the bottom of the plots indicate the timing of internode elongation.
Figure 2.
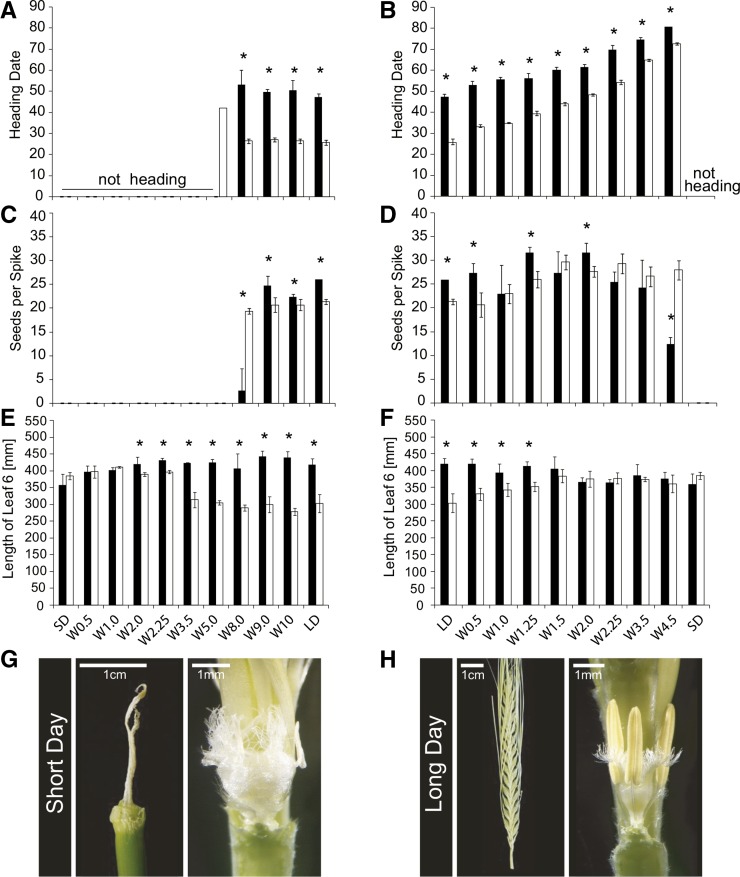
Effects of the Photoperiod on Plant Development and Spike-Related Traits of Scarlett and S42-IL107.
Plants of Scarlett (black) and S42-IL107 (white) were grown under SD or LD conditions. At different stages of MSA development (W0.5-W10), plants were transferred from LDs to SDs (left column) or from SDs to LDs (right column) and remained in the respective photoperiod until plant senescence. The following phenotypes of the main shoot were recorded: heading date ([A] and [B]), seed number per spike ([C] and [D]), and leaf blade length of leaf 6 ([E] and [F]). The bars represent the means ± sd over three plants. Significant differences (P < 0.05) between genotypes transferred from SDs to LDs and vice versa at the same developmental stage are indicated by asterisks above bar graphs. Aborted and developed main shoot spikes of Scarlett grown under SDs ([G], left) or LDs ([H], left), respectively. The carpels and stamen of main shoot spikes of Scarlett grown under LDs ([H], right) or transferred from LDs to SDs ([G], right) at Waddington stage 8.0 (see also [C]).
Furthermore, we tested whether the faster maturation of the MSAs in the ILs correlated with increased SAM activity. For this purpose, we determined the rate of floret primordia emergence at the MSAs in Scarlett and S42-IL107 (Figure 1D). The rate of floret primordia emergence increased in both genotypes under LDs compared with SDs and in S42-IL107 compared with Scarlett under LDs. S42-IL107 showed earlier induction of the first floret primordium under LDs and generated 2.7 floret primordia per day compared with 2.1 floret primordia per day for Scarlett (Supplemental Table 1). Allelic variation at Ppd-H1 affected the duration of inflorescence meristem activity, as floret primordia were induced until W4.0 and W6.0 in S42-IL107 and Scarlett, respectively. However, the number of floret primordia at the stamen primordium stage (W3.5) corresponded to the number of florets and seeds per spike, suggesting that floret primordia that emerged after W3.5 did not develop into fertile florets in both genotypes (Figure 1B; Supplemental Figure 1B). Under LDs, the duration of the early reproductive phase until W3.5 was longer in all spring barley lines that carried the mutated ppd-H1 allele compared with the ILs. Therefore, the number of floret primordia at the stamen primordium stage and, consequently, the number of seeds per spike were higher in Scarlett, Bowman, and Triumph than in the corresponding ILs (Figure 1B; Supplemental Figure 1B).
To further investigate the effects of the photoperiod on different phases of MSA development, we conducted a photoperiod shift experiment. Scarlett and S42-IL107 plants were transferred from SDs to LDs and vice versa at different stages of MSA development. Scarlett and S42-IL107 produced seeds only when shifted from LDs to SDs at W8.0 or thereafter, suggesting that exposure to LDs was necessary for normal inflorescence development, flowering, and seed set (Figures 2A, 2C, and 2G). When plants were shifted from SDs to LDs, the number of florets and seeds increased with the delay in floral transition, whereas a SD-LD shift after W1.5-W2.0 did not further increase the floret and seed numbers (Figure 2D; Supplemental Figure 2H). In addition, Scarlett had a higher number of florets and seeds than S42-IL107 when plants were shifted to LDs before W2.25, suggesting that the duration of the vegetative and possibly early reproductive phases determined the number of initiated florets. When plants were shifted from SDs to LDs at W4.5, S42-IL107, which has a stronger photoperiod response than Scarlett, produced more seeds (Figure 2D). S42-IL107 thus showed higher floret fertility under LDs. Plants that were shifted to SDs before W8.0 had strongly reduced plant heights and spike lengths (Supplemental Figures 2C and 2E). Consequently, LDs were required for internode elongation of the shoots and spikes in both genotypes. The effect of LDs on flowering time was quantitative; the delay in flowering increased with increasing delay in transfer from SDs to LDs (Figure 2B). The difference in the heading date between the two genotypes gradually decreased with increasing delay in the shift to LDs. The final number of leaves emerging from the main shoot gradually increased with prolonged SD treatment, whereas the difference in the final leaf number between genotypes was minimized (Supplemental Figure 2B). Consequently, the time of floral transition correlated with the final number of leaves. Additionally, the two genotypes differed in leaf size starting from leaves 5 and 6 (Figures 2E and 2F). S42-IL107 had shorter leaves than did Scarlett, and this difference increased with increasing leaf number and time under LDs.
Taken together, these results show that the initiation of floret primordia occurred under LDs and SDs, whereas spike growth and seed development, i.e., floret fertility, were strongly dependent on LDs and allelic variation at Ppd-H1. In addition, variation in photoperiod and genetic variation at Ppd-H1 had profound effects on plant architecture and determined internode elongation, leaf number, and leaf shape. Moreover, the number of floret primordia initiated until stem elongation (W3.5) corresponded to the final number of florets and seeds per spike, thus suggesting an important role for the early reproductive phase in determining yield potential.
Characterization of Transcriptional Changes in Leaves and at the Shoot Apex during the Vegetative and Early Reproductive Phases
To identify candidate genes for modulating the observed phenotypes, we investigated transcriptional changes during the course of MSA development, with a focus on the floral transition and floret initiation phases. Therefore, we conducted whole-transcriptome expression profiling of developing shoot apices during the vegetative (W0.5-W1.0) and early reproductive phases (W2.0-W3.5) of Scarlett and S42-IL107 plants grown under LDs. Under SDs, we examined changes in expression only in S42-IL107 because in this photoperiod, Scarlett and S42-IL107 did not differ in MSA development (Figures 1C and 1D; Supplemental Figure 3). MSA and leaf samples of the two genotypes grown under different photoperiods were harvested at the same developmental stages, but not at the same time after sowing. This approach allowed us to identify candidate genes for the regulation of shoot apex development both dependent on and independent of variations in photoperiod and Ppd-H1 in leaves and apices. We identified transcripts differentially regulated during development under both photoperiods and in both genotypes. Therefore, the regulation of these differentially expressed transcripts (DETs) was independent of photoperiod and genotype. On the other hand, we discovered Ppd-H1-dependent changes in gene expression by identifying transcripts that were differentially expressed between both photoperiods in S42-IL107 and between the genotypes under LDs, as the function of Ppd-H1 is LD specific. To corroborate that the observed effects were dependent on Ppd-H1 but not on the other genes in the introgression, we confirmed selected expression patterns by RT-qPCR in two additional ILs, carrying independent Ppd-H1 introgressions from different donor genotypes (Supplemental Figures 4 and 5).
To estimate whether a reference bias might influence our estimations of differential gene expression, we compared the DET density along the Scarlett chromosomes and in the introgressed regions. A reference bias may arise when RNA-seq data sets obtained from different genotypes are mapped onto a single reference and allelic variation becomes indistinguishable from the bona fide differential transcript expression (Degner et al., 2009). The DET density along the S42-IL107 introgressions on chromosome 2H did not noticeably deviate from the average values (Supplemental Figure 6). This finding suggested the absence of or only a minor effect of the reference bias in this study.
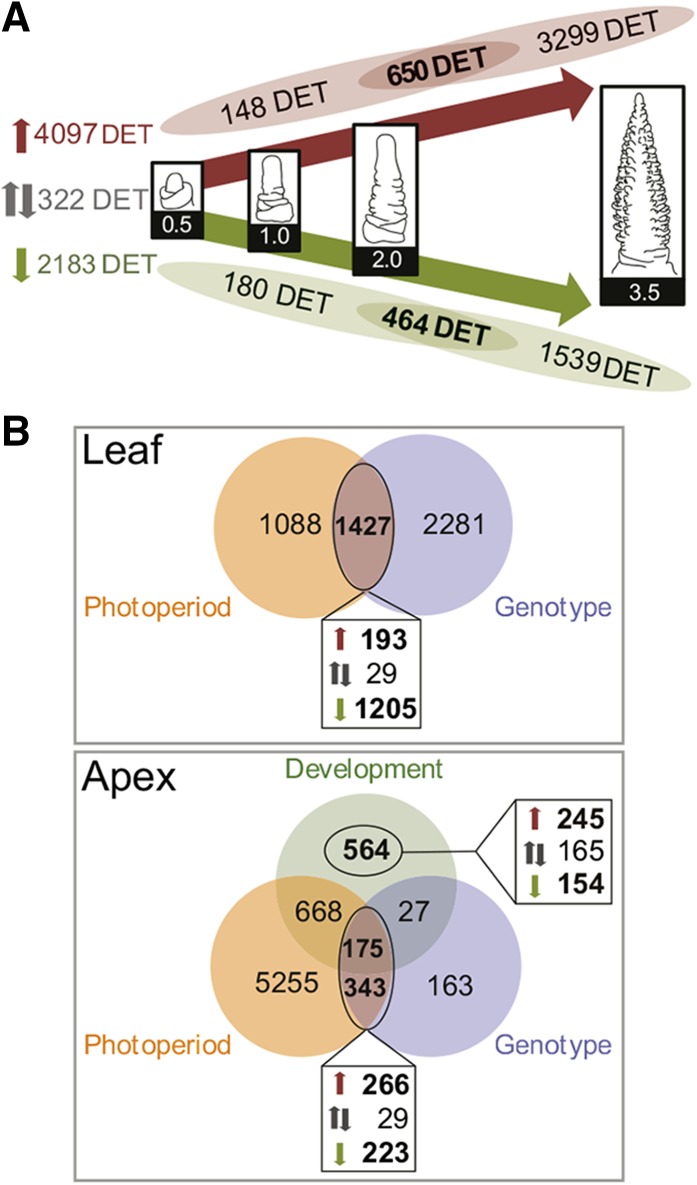
Transcriptome analysis revealed the expression of 25,152 transcripts at levels greater than 5 cpm in at least two libraries. Of these transcripts, 4044 and 2340 were exclusively expressed in leaf and MSA tissues, respectively. The tissue specificity of the detected transcripts was further supported by Gene Ontology (GO) term enrichment analysis. We identified the overrepresentation of transcripts related to metabolic processes in chloroplasts, such as amino acid and flavonoid biosynthesis, in the leaf-specific transcripts and processes related to cell cycle and meiosis among the MSA-specific transcripts. We identified 7604 DETs in leaf and MSA samples. Among these DETs, 6602 were differentially regulated during development in the MSAs in one or both photoperiods or genotypes (Figure 3A). Furthermore, 1427 DETs in the leaves and 518 DETs in the shoot apices were differentially regulated between photoperiods in S42-IL107 and between genotypes under LDs. These genes included transcripts that were differentially expressed during development and genes with stable expression levels across different developmental stages (Figure 3B). The 7604 DETs were characterized by 31 distinct expression profiles and three tissue-specific clusters, I-III (Figure 4; Supplemental Figure 7 and Supplemental Table 2). The DETs in cluster I, which were predominantly expressed in leaves, were enriched for photosynthesis- and light response-related GO terms (Supplemental Figure 8A). The DETs in cluster III, which had higher transcript levels at the shoot apex, were enriched for genes related to cell cycle regulation and meristem and flower development (Supplemental Figure 8C). The DETs in cluster II had similar expression levels in both the leaves and shoot apices and were enriched for genes involved in metabolic and developmental processes (Supplemental Figure 8B).
Figure 3.
Differential Gene Expression Analysis in Leaves and Developing Shoot Apices.
(A) DETs during shoot apex development of Scarlett under LD conditions or of S42-IL107 under both SD and LD conditions. Venn diagrams illustrate the intersections of DETs between shoot apices during floral transition (W0.5-W2.0) and DETs between the stamen primordium stage (W3.5) and prior developmental stages. Up- and downregulated transcripts during MSA development are highlighted in red and green, respectively. Transcripts with expression patterns that differ from continuous up- or downregulation during MSA development are shown in gray.
(B) Venn diagrams depict transcripts that are differentially regulated throughout MSA development (Development), DETs between LD- and SD-grown S42-IL107 plants (Photoperiod), and DETs between LD-grown S42-IL107 and Scarlett plants (Genotype) in leaves or in shoot apices. The boxes below each Venn diagram indicate DETs coregulated by LDs, and in S42-IL107, upregulated (red), downregulated (green), or DETs with opposing regulation by LDs and in S42-IL107 (black) are shown. DETs regulated during MSA development, independent of the photoperiod and the genotype, are depicted in the box on the right side of the Venn diagram. W, Waddington stage.
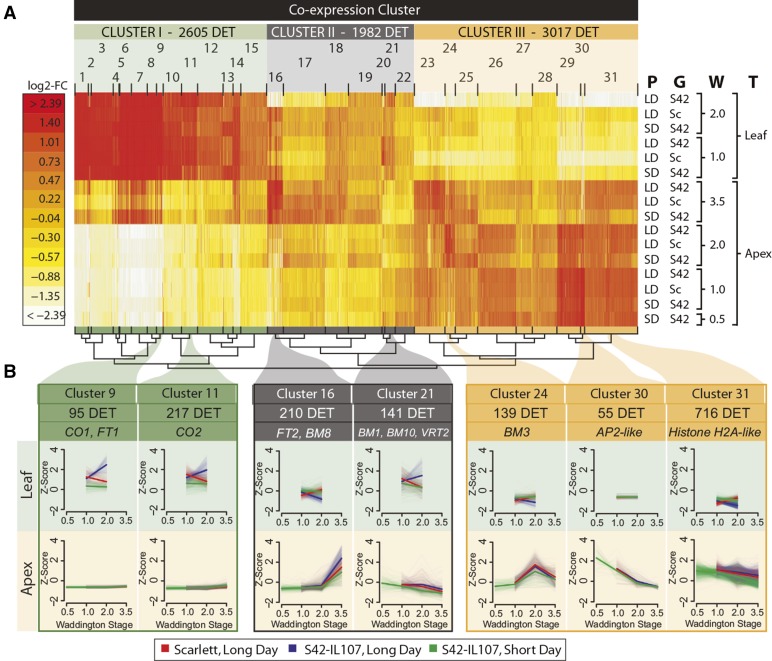
Figure 4.
Coexpression Clustering of DETs.
Coexpression clustering of DETs regulated during shoot apex development and DETs regulated between genotypes and between photoperiods in leaves and shoot apices.
(A) Heat map of coexpression clusters for 7604 DETs. Colors represent log2-fold changes (log2-FC) in expression levels relative to the mean transcript abundance across the tested conditions, i.e., leaf and apex samples of Scarlett (Sc) and S42-IL107 (S42), when plants were grown under SD and LD conditions and harvested at different developmental stages (Waddington stage 0.5-3.5). P, photoperiod; G, genotype; W, Waddington stage; T, tissue. Coexpression clusters (1-31) were assigned to three higher-level clusters (I-III) with tissue-specific expression patterns (Supplemental Table 2). The number and assignment of DETs to higher and lower level coexpression clusters are shown above the heat map. The similarity of coexpression clusters is indicated in the hierarchical tree structure below the heat map.
(B) Selected coexpression clusters representative of DETs during shoot apex development and DETs coregulated by LDs and in S42-IL107. The cluster sizes and coexpressed flowering time genes are indicated above the coexpression plots. The expression levels for individual transcripts (light colors) and the mean expression level across all transcripts within each cluster (bright color) were plotted. The coexpression plots are shown as the mean centered and scaled transcript levels (Z-score).
Of the 6602 DETs that were regulated during development, 4097 transcripts showed upregulation and 2183 transcripts showed downregulation during either the vegetative or early reproductive phase (Figure 3A). The expression patterns of 322 transcripts were distinct from consistent up- or downregulation during early or later stages of MSA development. A majority of the 6602 DETs were either induced (3299 DETs) or downregulated (1539 DETs) specifically at the stamen primordium stage (W2.0-3.5) and were not differentially regulated during floral transition (W0.5-W2.0). Photosynthesis- and light response-related genes were overrepresented among the 3299 DETs that were upregulated in shoot apices specifically at the stamen primordium stage (Figure 3A; Supplemental Tables 3 and 4). Additionally, unsupervised clustering clearly distinguished the apex- and leaf-derived expression data sets. The data sets for MSA samples at W3.5 clustered at an intermediate position between the leaf and MSA samples harvested before and immediately after floral transition (W0.5-W2.0; Supplemental Figure 9C). Consequently, the gene expression in shoot apices at the stamen primordium stage partially resembled the gene expression observed in the leaves. Accordingly, the inflorescences were greener at the stamen primordium stage (W3.5) than at the double ridge stage (W2.0; Supplemental Figure 3), suggesting that the synthesis of the photosynthetic apparatus in the MSAs followed a developmental program, as synthesis gradually increased in the absence of light signals inside the leaf sheath.
Since we were interested in identifying transcripts differentially regulated throughout MSA development, we excluded transcripts that were regulated only at the stamen primordium stage, e.g., photosynthesis-related genes. We focused our further analyses on a set of 1434 DETs that were differentially regulated in the MSAs during the vegetative and early reproductive phases, i.e., 650, 464, and 320 DETs that were upregulated, downregulated, or showed different expression patterns throughout MSA development, respectively (Figure 3A). Within this developmental set of 1434 DETs, 245 DETs were consistently upregulated and 154 DETs were downregulated during development, independent of genotype and photoperiod (Figure 3B). The GO enrichment analysis among the 1434 DETs highlighted the genes involved in the regulation of metabolic processes and inflorescence and floral meristem development (Supplemental Table 5).
The detailed description of the expression of transcripts in the reference set in different genotypes, tissues, and developmental stages; their functional annotations; their homology to Arabidopsis, rice, Brachypodium, and Sorghum genes; and their assignment to coexpression clusters can be found in Supplemental Data Sets 1 to 4.
Transcriptional Changes during Floral Transition Independent of Photoperiod and Allelic Variation at Ppd-H1
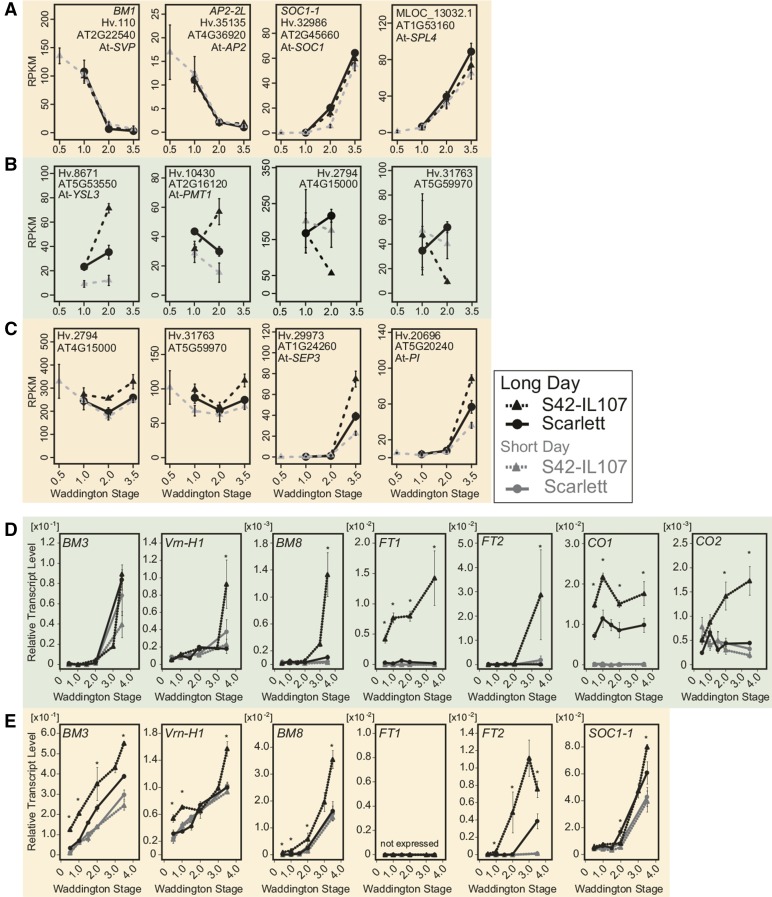
Within the 154 DETs that were downregulated independent of photoperiod and genotype in the MSAs, we identified genes with high expression levels at the vegetative stage and a strong reduction in expression upon floral transition (W2.0) (Supplemental Table 6). For example, the three MADS box transcription factors VEGETATIVE TO REPRODUCTIVE TRANSITION2 (VRT2; Hv.15491), BARLEY MADS BOX1 (BM1; Hv.110), and BM10 (Hv.19680), homologous to SHORT VEGETATIVE PHASE (SVP) in Arabidopsis, exhibited high expression levels in vegetative apices and were gradually downregulated during MSA development (Figures 3B and 5A, cluster 21; Supplemental Tables 6 and 7 and Supplemental Figure 10A). BM1 was completely downregulated at floral transition (W2.0), whereas reductions in BM10 and VRT2 transcript levels were less pronounced, and the complete repression of VRT2 occurred only at the stamen primordium stage (W3.5). Similar to BM1, transcripts in cluster 30 were repressed during MSA development, independent of the photoperiod and Ppd-H1 genotype (Figures 3B and 5A, cluster 30; Supplemental Table 7). Whereas BM1, BM10, and VRT2 were expressed in both the MSAs and leaves, the expression of transcripts in cluster 30 was restricted to the MSAs. Among these transcripts, we identified Hv.35135, encoding a homolog of the Arabidopsis floral homeotic gene AP2/EREB (AP2); Hv.11559, a homolog of GIBBERELLIN-2-OXIDASE (GA2ox), which is a major gibberellin catabolic enzyme in plants; and Hv.12609, a homolog of REDUCED VERNALIZATION RESPONSE1 (VRN1), which is involved in epigenetic gene silencing in Arabidopsis (Figure 5A; Supplemental Figure 10A and Supplemental Table 6). The complete or near-complete downregulation of these genes marked the transition from vegetative to reproductive MSAs, independent of the photoperiod and genotype.
Figure 5.
Expression Patterns of Selected Transcripts in Leaves and Shoot Apices of Scarlett and S42-IL107 as Determined by RNA Sequencing and RT-qPCR.
RNA sequencing-derived expression data of selected transcripts (A) differentially expressed during MSA development independent of the photoperiod and the genotype or differentially regulated between photoperiods and between genotypes in the leaves (B) or shoot apices (C). The normalized expression values are reported in reads per kilo base per million (RPKM). The error bars indicate sd across two to three independent RNA samples. The quantification of transcript levels by RT-qPCR in leaf samples (D) and samples enriched for apex tissue (E) at different stages of plant development. The transcript levels are shown relative to the transcript abundance of Actin. The error bars indicate the sd over three biological and two technical replicates. The asterisks highlight significant differences (P < 0.05) between the transcript levels of S42-IL107 and Scarlett when the plants were at the same developmental stage and grown under LD conditions.
Among genes with no or extremely low expression at the vegetative stage (W0.5-W1.0) and upregulation at floral transition (W2.0) under both LDs and SDs, we identified genes involved in the regulation of meristem development, such as KNOTTED1 (KN1; Hv.12878) and the transcript MLOC_13032.1, a homolog of the transcription factor SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE4 (SPL4) of Arabidopsis (Figure 5A; Supplemental Figure 10B and Supplemental Table 8). We also identified the induction of SOC1-1 (Hv.32986), a homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) in Arabidopsis, which was upregulated at floral transition under LDs and at the stamen primordium stage under SDs, independent of the genotype (Figures 5A and 5E; Supplemental Table 8). In addition, homologs of Arabidopsis genes involved in carbohydrate transport (Hv.10624; Arabidopsis SWEET15), nitrate transport (Hv.16527; NITRATE TRANSPORTER1), and hormone signaling and transport (Hv.15702; CBL-INTERACTING PROTEIN KINASE6) were induced in shoot apices after the induction of the first floret primordia, independent of the genotype and photoperiod (Supplemental Figure 10B and Supplemental Tables 8 and 9).
In summary, the expression of the barley MADS box gene BM1, of AP2-2L, and of KN1, as well as the barley homologs of SPL4, GA2ox, and VRN1, correlated with the floral transition, independent of both genotype and photoperiod, and might serve as molecular markers for the staging of floral transition.
Photoperiod- and Ppd-H1-Dependent Regulation of Transcript Expression in Leaves and Shoot Apices
To identify the candidate genes acting downstream of Ppd-H1 in the photoperiod-dependent regulation of preanthesis development, we focused on transcripts differentially regulated between photoperiods in S42-IL107 and between genotypes under LDs. In the leaves, we identified 1427 transcripts that were differentially regulated between photoperiods and between genotypes at the time of the floral transition (Figure 3B). The majority of 193 transcripts that were upregulated in the leaves in a Ppd-H1-dependent manner (Supplemental Table 10) showed high expression levels in the leaves and low or no expression in the MSAs (Figure 4, clusters 9, 11, and 12; Supplemental Figure 7). Among these transcripts, we identified CO1, CO2, and FT1, which were described previously as putative downstream targets of Ppd-H1 in barley (Supplemental Table 11; Turner et al., 2005). FT1 expression levels in all three tested spring barley genotypes were close to the detection limit as verified by RT-qPCR assays. In contrast, the ILs exhibited a >10-fold increase in FT1 expression compared with the spring barley genotypes at any stage of development (Figure 5D; Supplemental Figures 5A, 5B, and 10C). Similarly, the transcript levels of CO1 and CO2 increased under LDs in the ILs, while in BW281 and Triumph-IL, CO1 expression decreased to the levels of the recurrent parents after floral transition (Figure 5D; Supplemental Figures 5A and 5B). Among the transcripts in clusters 9, 11, and 12, FT1 was coregulated with genes involved in transmembrane transport (Supplemental Table 12), e.g., iron transport genes, such as Hv.8671, Hv.8787, and Hv.23326, homologs of the YELLOW STRIPE LIKE gene family of iron-phytosiderophore transporters; a potassium transport gene (Hv.4917); a sugar transport gene (Hv.10430; POLYOL/MONOSACCHARIDE TRANSPORTER1); and an amino acid transport gene (Hv.19083; a protein transporter from the ENTH/VHS/GAT family; Figures 4 and 5B; Supplemental Table 11 and Supplemental Figure 10C).
In the leaves, the majority of genes were downregulated in S42-IL107 grown under LDs, specifically at W2.0, when floret primordia emerged from the MSAs (1205 DETs; Figure 3B). GO enrichment among the downregulated genes highlighted biological processes related to cell proliferation, nucleosome assembly, and carbohydrate metabolism (Supplemental Table 13). The downregulation of transcripts encoding ribosomal and histone proteins, e.g., Hv.2794 and Hv.31763, in S42-IL107 (LD; Figure 5B), indicated that large-scale de novo protein synthesis and leaf development ceased earlier in the fast-developing S42-IL107 plants compared with Scarlett. Interestingly, ribosomal and histone proteins with reduced expression in the leaves of S42-IL107 were upregulated in the MSAs of S42-IL107 under LDs, suggesting that a reduction in biosynthetic activity in the leaves was associated with increased metabolic activity in the MSAs (Figure 5C; Supplemental Table 14). We also observed the downregulation of genes involved in leaf growth and patterning and auxin signaling in S42-IL107, such as homologs of KNOTTED1-LIKE/BREVIPEDICELLUS (Hv.105, Hv. 12878, and Hv.33953) and ASYMMETRIC LEAVES1 (Hv.12064) (Supplemental Figure 10C). The downregulation of transcripts involved in DNA packaging, cell division, and leaf development was associated with the smaller leaf size observed in the introgression line compared with Scarlett under LDs (Figures 2E and 2F).
In the MSAs, 518 DETs were coregulated between the photoperiods and genotypes (Figure 3B). In total, 266 DETs, including genes involved in the regulation floral organ development, hormone signaling, metabolism, disease resistance, cell cycle regulation, and nucleosome assembly, were upregulated under LD and in S42-IL107 (Supplemental Table 14). Within this group, we identified FT2, a member of the barley FLOWERING LOCUS T-LIKE gene family (Faure et al., 2007). In enriched MSA tissue, we detected the early induction of FT2 at or before floral transition (<W2.0) in BW281 and S42-IL107, respectively, and at lemma primordium stage (W3.0) in Triumph-IL (Figure 5E; Supplemental Figures 5A and 5B). The induction of FT2 in shoot apices preceded its induction in leaf tissue, where FT2 was upregulated at the stamen primordium stage (W3.5) in S42-IL107 and BW281 under LDs (Figure 3D; Supplemental Figure 5A). FT2 was coexpressed with 209 other transcripts in cluster 16 (Figure 4B; Supplemental Tables 15 and 16), which were associated with floral organ development. Among these transcripts, we identified MLOC_57803.1, Hv.29973, Hv.20696, and Hv.20726, representing barley genes that are homologous to the Arabidopsis floral homeotic genes SEPALLATA1 (SEP1), SEP3, PISTILATA (PI), and APETALA3 (AP3), respectively (Figures 4 and 5C). In addition, BM3, Vrn-H1 (BM5a), and BM8, which are barley homologs of the APETALA1/FRUITFUL (AP1/FUL) gene family of MADS box transcription factors in Arabidopsis (Schmitz et al., 2000; Trevaskis et al., 2007), were upregulated in the leaves and shoot apices during preanthesis development under both LDs and SDs (Figures 5D and 5E; Supplemental Figures 5A, 5B and 10D). The transcript levels of Vrn-H1 were first induced in the MSAs, followed by BM3 and BM8. We observed the Ppd-H1-dependent upregulation of Vrn-H1 before floral transition and at the beginning of stem elongation, of BM3 at all developmental phases, and of BM8 after floral transition. The differential regulation of the three BM genes indicated the subfunctionalization of the AP1/FUL-like genes during the photoperiod-dependent regulation of preanthesis development in barley.
To estimate the degree of coregulation between transcripts in the leaves and shoot apices with floret primordia formation in the MSAs, we correlated the RT-qPCR expression profiles obtained from the three Ppd-H1 ILs and their recurrent parents during shoot apex development (Supplemental Figure 11). We identified a positive correlation (r = 0.62) between FT1 expression in the leaves and FT2 transcript levels in the shoot apices. FT2 expression in the shoot apices also correlated positively with SOC1-1, BM3, and BM8. SOC1-1 expression levels showed the highest number of significant correlations (q < 0.05, |r| > 0.4) with transcripts expressed in the shoot apices and leaves. FT2 expression also predominantly correlated with the expression of AP1-/FUL-like transcripts in the leaves and shoot apices. Furthermore, the expression of SOC1-1, Vrn-H1, and BM3 correlated positively with the number of floret primordia formed at the shoot apex throughout development, indicating the potential contribution of these genes to the regulation of floret primordia induction and to the further development of floral organs.
In summary, the flowering time regulator FT1 was upregulated in the leaves of the ILs and coexpressed with genes involved in carbon, amino acid, and metal ion transport. The downregulation of the leaf transcripts involved in nucleosome assembly, cell proliferation, carbohydrate metabolism, and leaf growth at floral transition indicated a reduction in cellular biosynthetic activity and early leaf maturation in the fast-developing S42-IL107 plants compared with Scarlett, which was also reflected by the reduced leaf size of the ILs. In the MSA, the expression of SOC1-1, Vrn-H1, and BM3 correlated positively with floret primordia induction, whereas FT2 expression was associated with floret maturation and flowering. FT2 expression and fast inflorescence development correlated with the upregulation of genes involved in floral organ development and cell cycle regulation.
DISCUSSION
Previously, microarray hybridization was used to analyze gene expression changes during barley and wheat development. These studies resolved spatio-temporal gene expression changes during grain development (Drea et al., 2005; Wan et al., 2008) or provided an overview of the gene expression profiles in different barley tissues without exact developmental staging of the plant material (Druka et al., 2006; Winfield et al., 2009, Greenup et al., 2011, Hemming et al., 2012). Here, we used whole-transcriptome sequencing in the leaves and dissected MSAs at defined developmental stages to identify molecular changes during plant development, both dependent on and independent of the photoperiod and allelic variation at Ppd-H1. These analyses revealed the expression patterns of 25,152 transcripts, including 2683 additional transcript clusters not represented in previous transcriptome assemblies (Mayer et al., 2012; UniGene build #59 at NCBI). In comparison, RNA sequencing of eight different tissues of the barley cultivar Morex revealed 79,379 transcript clusters and resulted in an estimate of 30,400 genes for barley (Mayer et al., 2012). The use of a custom reference sequence for mapping the RNA sequencing data increased the efficiency of mapping and enhanced the detection of additional tissue-specific transcripts. Furthermore, the dissection and enrichment of MSA tissue likely increased the sensitivity of the detection of shoot apex-specific transcripts. Accordingly, the dissection of MSA samples resulted in a clear separation of leaf and apex samples in the unsupervised clustering of gene expression data and in the identification of transcripts with tissue-specific expression profiles. We propose that the information regarding transcript abundance and coexpression clusters at defined developmental stages of the MSAs will facilitate the formulation of biological hypotheses concerning the regulation of spike development in barley and guide the design of further experiments required for functional validation. Consequently, we provide a valuable data set for future investigations into the environmental and genetic control of shoot apex development and yield components in barley.
Floral Transition and Floret Initiation Do Not Require LDs
The analysis of the effects of photoperiod on MSA development in spring barley revealed that floral transition and early reproductive development occurred under both LDs and SDs, whereas later stages of inflorescence development and flower maturation required LDs (Figure 6). Accordingly, the transcripts associated with floral transition were largely regulated independently of the photoperiod and allelic variation at Ppd-H1. One prominent example was BM1 of the SVP-like gene family, which was completely downregulated during floral transition under LDs and SDs. In Arabidopsis, SVP encodes a MADS box transcription factor, which delays floral transition by repressing floral integrator genes and gibberellin biosynthesis (Andrés et al., 2014). In barley, ectopic overexpression of BM1 induced floral reversion; however, a specific role for barley SVP-like genes in floral transition has not yet been demonstrated (Trevaskis et al., 2007). At the shoot apex, BM1 was coregulated with AP2-2L (Hv.35135; 7HL), an AP2 domain transcription factor homologous to Arabidopsis AP2, which is involved in a wide variety of developmental processes, including stem cell niche regulation (Würschum et al., 2006), floral organ determination (Krogan et al., 2012), and seed mass control (Ohto et al., 2005). In barley, the AP2 gene family consists of four members, of which AP2 (2HL) has been implicated in the regulation of spike density by determining the duration of rachis internode elongation (Houston et al., 2013). In this study, another Arabidopsis AP2 homolog, AP2-2L (7HL), was downregulated in the MSA specifically before floral transition. In Arabidopsis, AP2 represses floral transition under LDs and SDs through the direct transcriptional repression of SOC1 and AP1 (Yant et al., 2010). The repression of AP2-2L at the shoot apex correlated negatively with the upregulation of SOC1-1 during floral transition under both photoperiods, suggesting that AP2-2L acts as a floral repressor in barley. BM1 and AP2-2L expression correlated negatively with that of the barley homolog of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE4 (SPL4), which was upregulated at floral transition, independent of the photoperiod. In Arabidopsis, SPL genes control floral transition by binding directly to and activating the transcription of SOC1, AP1, FUL, and LEAFY (Wang et al., 2009; Yamaguchi et al., 2009). In addition, SPL4 is repressed directly or indirectly by SVP (Torti et al., 2012), suggesting that the negative relationship between SVP and SPL4 is conserved between Arabidopsis and barley. Interestingly, AP2 and SPL are regulated by the microRNAs miR156 and miR172 (Yant et al., 2010), which control developmental phase changes, such as the juvenile-to-adult and vegetative-to-reproductive transition, in Arabidopsis and other higher plant species (Wu and Poethig, 2006; Chuck et al., 2007; Mathieu et al., 2009; Wang et al., 2011). Therefore, future studies are required to elucidate the role of these two microRNAs in regulating floral transition in barley.
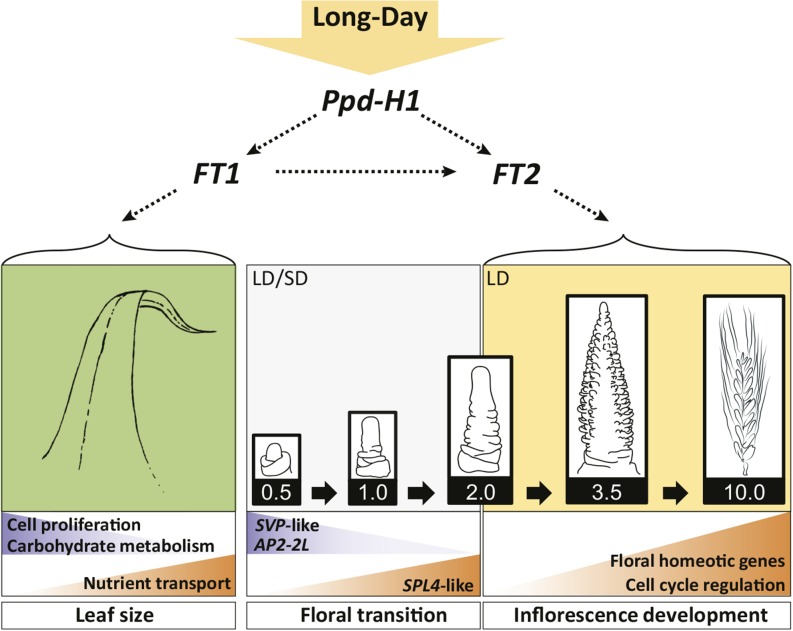
Figure 6.
Model for the Effects of the Photoperiod and Allelic Variation at Ppd-H1 on Morphological and Transcriptional Changes during Leaf and Shoot Apex Development in Spring Barley.
The floral transition and induction of floret primordia during the early reproductive phase of shoot apex development (Waddington stages 0.5-2.0) occurred under SD and LD conditions. LD conditions and the photoperiod-responsive Ppd-H1 allele predominantly affected the inflorescence development during the spike growth and stem elongation phase. Accordingly, differentially expressed transcripts at early developmental stages were mainly regulated independently of the photoperiod and allelic variation in Ppd-H1, as reflected by the repression of three SVP-like genes and the AP2-domain transcription factor AP2-2L or the induction of a SPL4-like gene. The photoperiod- and Ppd-H1-dependent promotion of leaf and inflorescence maturation and flower fertility correlated with the induction of the FT-like genes FT1 in leaves and FT2 in shoot apices. Accordingly, FT1 was positively coregulated with transcripts involved in carbon, amino acid, and metal ion transport and negatively coregulated with transcripts associated with leaf growth, cell proliferation, and carbohydrate metabolism in the leaves. In the shoot apices, FT2 was coregulated with floral homeotic genes and transcripts related to cell cycle regulation. Arrows in the figure indicate genetic and expression correlations between genes. Backgrounds of genes and processes up- and downregulated are colored in orange and blue, respectively.
In summary, phenotypic and molecular analyses suggested that floral transition and floret primordia induction in spring barley were not strongly controlled by the photoperiod or by the variation at Ppd-H1. Accordingly, the photoperiod- and Ppd-H1-independent downregulation of BM1 and AP2-2L and the upregulation of SPL4-like correlated with the appearance of floret initials and, thus, floral transition of the MSAs under SDs and LDs.
The Induction of the Photoperiod Pathway in the Leaves and MSAs Correlates with Floret Fertility
The late reproductive phase of stem elongation has been described as the most important phase for yield because competition between the spike and stem for limited assimilates during this phase induces the abortion of floret primordia (Ghiglione et al., 2008; González et al., 2011; Alqudah and Schnurbusch, 2014). This conclusion was based on the positive correlation between the duration of stem elongation and the number of fertile florets and final seeds, obtained from macroscopic observations of barley and wheat development (Miralles and Richards, 2000; González et al., 2003; Slafer, 2003). By contrast, we showed that the number of floret primordia that were initiated before the beginning of stem elongation (W3.5) corresponded to the final number of seeds per spike (Figure 1B; Supplemental Figure 1B). Therefore, these findings support the idea that the early reproductive phase is critical for yield, as this phase determines the maximum number of floret primordia that may develop into seeds (Appleyard et al., 1982; Kitchen and Rasmusson, 1983). Accordingly, a shortening of the early reproductive phase in S42-IL107 with a dominant Ppd-H1 allele reduced the number of floret primordia compared with Scarlett. By contrast, we observed that S42-IL107, with fast stem elongation, exhibited higher floret fertility and seed set (Figure 2D). Consequently, LDs and the dominant Ppd-H1 allele decreased the number of initiated floret primordia and of subsequently formed florets (Figures 1B and 1D; Supplemental Figures 1B and 2H); however, they increased floret fertility during the late reproductive phase of stem elongation (Figure 2D).
This finding is consistent with previously reported effects of Ppd-D1 on reducing the number of floret primordia per spike but increasing floret fertility in wheat (Worland et al., 1998). Two recent studies have shown that the application of gibberellin under SDs accelerated floral transition in wheat and barley, but both species failed to produce seeds under SDs, suggesting that in addition to gibberellin, a signal generated only under LDs is necessary for floret fertility in these temperate crops (Pearce et al., 2013; Boden et al., 2014). The failure to set seeds did not reflect a shortage of photoassimilates under SDs in this study, as day-neutral barley genotypes, which activate the LD photoperiod pathway under SD conditions, produced fertile flowers and seeds (Faure et al., 2012; Campoli et al., 2013; Pankin et al., 2014). These observations suggested that the LD photoperiod response pathway is crucial for inflorescence development.
The photoperiod-dependent differences in inflorescence development were associated with 1427 and 518 transcripts in the leaves and MSAs, respectively, which were differentially regulated by photoperiod and Ppd-H1. In the leaf, FT1, which acts downstream of Ppd-H1 (Turner et al., 2005), was upregulated under LD and in the ILs compared with the spring barley cultivars, whereas no expression was detected under SDs. Interestingly, LD-dependent FT1 expression in the leaf was coregulated with genes involved in transmembrane transport, including genes involved in carbon, amino acid, and iron transport at floral transition, well before the development of strong sink organs and the onset of inflorescence and plant growth. Consequently, FT1 and a concomitant upregulation of sugar, amino acid, and metal ion transporters might prime the plant for fast inflorescence and plant growth under LDs. The failure to induce these processes correlated with the abortion of the main shoot spike under SDs. Therefore, the expression of FT1 may induce changes in source-sink relationships and consequently lead to higher floret fertility. Further studies are needed to investigate how flowering time genes participate in the remobilization and transport of nutrients and assimilates from source to the sink organs and the partition of dry matter in plants.
In the MSAs, the 518 transcripts regulated by photoperiod and Ppd-H1 included genes involved in floral organ development, hormone signaling, metabolism, chromatin modification, and nucleosome assembly. Among these genes, we identified the expression of FT2, a close homolog of FT1, in the MSAs. In MSA-enriched tissue, FT2 was expressed before floral transition under LDs in S42-IL107 and BW281, which carried the photoperiod-responsive Ppd-H1 allele, and the expression of FT2 in the MSAs correlated positively with the expression of FT1 in the leaf. Homologs of FT2 in Brachypodium and wheat have recently been suggested to act downstream of FT1 in the leaf (Lv et al., 2014), but a role for FT-like genes in the MSA has not yet been described in temperate cereals. However, two recent studies in Arabidopsis have shown that FT is expressed in a photoperiod- and CONSTANS-independent manner in inflorescences and siliques, where this gene is important for the maintenance of inflorescence and floral meristem identity (Liu et al., 2014; Müller-Xing et al., 2014). In Arabidopsis, a transient shift for a few days to LD growth conditions is sufficient to irreversibly commit the plants to flower (Corbesier et al., 2007). By contrast, continuous exposure to LD conditions was required for successful inflorescence development in barley. Future studies should investigate whether successful inflorescence development is dependent on the stable expression of FT2 in the MSA, similar to the proposed role of silique-expressed FT in maintaining floral development in Arabidopsis.
The transcripts of genes involved in auxin response and transport, homologous to DWARF IN LIGHT1, AUXIN RESPONSE FACTOR6 (ARF6), ARF8, and WALLS ARE THIN1 of Arabidopsis, were upregulated in MSAs under LDs. Changes in auxin homeostasis may be important for floral organ initiation in barley, as demonstrated in Arabidopsis, where auxin distribution within the periphery of the inflorescence meristems specifies the site of floral meristem initiation and mediates organ growth and patterning (Krizek, 2011). We also observed the photoperiod- and Ppd-H1-dependent upregulation of barley homologs of the Arabidopsis floral homeotic genes AP1/FUL, SEP1, SEP3, PI, and AP3, which are involved in floral organ formation (Figure 5C; Supplemental Figures 5A, 5B, and 10D). In parallel to transcripts involved in floral organ initiation and formation, transcripts implicated in cell cycle, chromatin modification, and nucleosome assembly showed higher expression under LDs, particularly in S42-IL107 compared with Scarlett (Figure 4, clusters 16 and 31; Supplemental Tables 16 and 17). This finding suggested that MSAs at the same developmental stage are characterized by higher cellular biosynthetic and mitotic activities under LDs than SDs and in S42-IL107 compared with Scarlett, consistent with the observed differences in MSA development at later stages.
Our results indicated that FT1 and FT2 play important roles in reproductive development beyond floral transition in temperate grasses. Future studies should dissect the roles of FT1 and FT2 in inflorescence development and identify downstream targets associated with successful flowering and seed set in barley.
Genetic Variation at Ppd-H1 Correlates with the Development of MSAs, Stems, and Leaves
Photoperiod and Ppd-H1 affected inflorescence development, internode elongation, leaf number, and leaf size, suggesting that these processes are tightly coordinated (Figure 2; Supplemental Figure 2). Early growth termination of the leaves in the fast-developing S42-IL107 was consistent with the downregulation of a large number of genes involved in DNA packaging, cell proliferation, and leaf development during floral transition.
Similar pleiotropism of flowering time regulators has been observed in other species, e.g., SINGLE FLOWER TRUSS, the tomato (Solanum lycopersicum) ortholog of FT, affected flowering time, stem growth, leaf maturation, and compound leaf complexity (Lifschitz et al., 2006; Shalit et al., 2009). In addition, major regulators of flowering time in rice, such as GRAIN NUMBER, PLANT HEIGHT AND HEADING DATE7 (GHD7) and GHD8, apart from flowering time, also controlled plant height and seeds per panicle (Xue et al., 2008; Yan et al., 2011). Pleiotropic effects of flowering time regulators might be a consequence of changes in source-sink relationships triggered by the transition from vegetative to reproductive growth. However, studies in Arabidopsis and tomato also suggested that FT-like genes and the downstream targets SOC1 and AP1/FUL may control the activity of leaf meristems (Teper-Bamnolker and Samach, 2005; Melzer et al., 2008; Shalit et al., 2009; Burko et al., 2013). For example, Melzer et al. (2008) showed that FT and its downstream targets SOC1 and FUL controlled determinacy of leaf and axillary meristems independent of flowering time in Arabidopsis. Arabidopsis 35S:FT soc1-3 and 35S:FT ful-2 plants flowered early but displayed a marked increase in longevity and rosette size. Similarly, the strong upregulation of FT1 and the AP1/FUL-like homologs Vrn-H1, BM3, and BM8 in the leaves of S42-IL107 might have affected determinacy of leaf meristems as suggested by the reduced leaf size in the IL compared with Scarlett. Future studies should investigate the mechanisms of how flowering time regulators influence leaf growth, which is an important agronomic trait that determines photosynthetic capacity and vegetative biomass yield.
METHODS
Plant Materials
In this study, we used three spring barley (Hordeum vulgare) genotypes carrying a natural mutation in the CCT domain of Ppd-H1 (reduced photoperiod response; Turner et al., 2005) and three derived backcross lines carrying introgressions of the photoperiod-responsive dominant Ppd-H1 allele. The spring barley genotypes were Scarlett, Bowman, and Triumph, and the derived ILs were S42-IL107, BW281, and Triumph-IL. S42-IL107 and BW281 carry introgressions of the dominant Ppd-H1 allele from wild barley (Druka et al., 2011; Schmalenbach et al., 2011). Triumph-IL is a BC4F2-selected IL derived from the doubled haploid population of a cross between Triumph and the winter cultivar Igri (Laurie et al., 1995). This line was kindly provided by David Laurie (John Innes Centre, Norwich, UK). The sizes of the introgressions in S42-IL107 and BW281 were reported previously by Schmalenbach et al. (2011) and Druka et al. (2011), respectively. The 9K Infinium iSelect barley array was used to genotype the Triumph-IL line (Supplemental Figure 4).
Plant Cultivation and Phenotyping
For all experiments, the plants were sown in the soil Mini Tray (Einheitserde) in 96-cell growing trays (100 mL/cell). Plants were maintained at 4°C for 3 d, followed by germination under SD conditions (8 h, 22°C day; 16 h, 18°C night; PAR 270 µM/m2 s). Subsequent to germination, the plants were transferred to LD conditions (16 h, 22°C day; 8 h, 18°C night) or cultivation was continued under SDs. For the RNA sequencing experiment, Scarlett and S42-IL107 were germinated and maintained under SDs for 7 d before separation into LD and SD conditions.
Three representative plants per genotype and photoperiod were dissected every 3 to 4 d from germination to seed set in two independent experiments. At each time point, the developmental stage of the MSA was determined according to the quantitative scale of Waddington et al. (1983), in the text referred to as Waddington stage (W), reflecting the development of the most advanced floret primordium on the MSA. In addition, morphological phenotypes of the main shoot, i.e., the number of emerged leaves and the number of floret primordia, were recorded for each genotype during development. The heading date (at Z49; Zadoks et al., 1974), number of florets per spike, and number of grains per spike were recorded at plant maturity for 10 plants per genotype. Minor modifications of the Waddington scale were performed, i.e., Waddington stage 0.5 (W0.5) was assigned to shoot apices before the elongation of the apical dome present in transition apices at W1.0. Images of apices were obtained using Diskus imaging software (version 4.8.0.4562; Hilgers Technisches Büro) and a stereomicroscope (model MZ FLIII; Leica) equipped with a digital camera (model KY-F70B; Leica).
Broken-line regressions were calculated for MSA development and for floret primordia emergence using the “segmented” package (version 0.2-9.5; Muggeo, 2003, 2008) in the statistical software R (version 3.0.1; R Development Core Team, 2008). Regression models were fitted for the presence of none to four breakpoints, and the model with the highest Bayesian information criterion score was selected. Slopes of the individual linear segments and their 95% confidence intervals were extracted from the broken-line regression models.
Significant genetic differences in morphological phenotypes recorded at plant maturity were identified by Student’s t tests.
Photoperiod Shift Experiment
Seeds of Scarlett and S42-IL107 were germinated in 96-well planting trays under SDs. Subsequently, plants of both genotypes were transferred to 3-liter pots, and cultivation was continued under SDs or LDs, respectively. At eight stages of MSA development (W0.5-4.5) under SDs and nine stages (W0.5-10) under LDs, three plants per genotype were dissected to determine the developmental stage of the MSA before the transfer of another three plants from SDs to LDs or vice versa. The cultivation of three plants per genotype was continued constantly under either SD or LD conditions. The heading date (at Z49; Zadoks et al., 1974) and the final leaf number of the main shoot were recorded before plant maturity for each plant. At plant maturity, the height, spike length, floret number, and seeds per spike were recorded for the main shoot of each plant. The experiment was terminated at 150 d after germination, as many plants grown under SD conditions did not flower.
Library Preparation and RNA Sequencing
For RNA sequencing, leaf and shoot apex tissues were harvested from the main shoots of Scarlett and S42-IL107 plants grown under SDs at W0.5, W1.0, W2.0, and W3.5 and under LDs at W1.0, W2.0, and W3.5. The samples were harvested at 3 h before the end of the light period. Before each sampling time point, three plants per genotype and per condition were dissected, and the developmental stage was recorded. For apex sampling, the leaves surrounding the MSA were removed manually, and the apex was cut with a microsurgical stab knife (5-mm blade at 15° [SSC#72-1551]; Sharpoint, Surgical Specialties) under a stereomicroscope to confirm the developmental stage of each harvested MSA. Pools of apices were collected within a total harvest period of 2 h by a team of six people. The MSA samples included tissues of young leaf and floret primordia as indicated in Supplemental Figure 12A. The samples collected during the vegetative phase (W0.5 and W1.0) consisted of 25 to 30 pooled apices. At the double ridge stage (W2.0) and stamen primordium stage (W3.5), 15 and 7 shoot apices were pooled, respectively. The leaf samples were harvested from a subset of seven plants, of which apex tissue was collected at the time of floral transition (W1.0 and W2.0). The harvested leaf tissue was restricted to the distal part of the leaf at ∼2 to 4 cm before the leaf tip. The leaf and apex samples designated for RNA sequencing were harvested in three replicate pools. For gene expression analysis by RT-qPCR, the leaf and shoot apex tissues were harvested at four to six stages between W0.5 to W5.0 from the main shoot of all six genotypes grown under SDs and LDs. Each leaf and shoot apex sample was composed of pooled tissues of five plants. The harvested shoot apex samples were enriched for shoot apex tissue, i.e., parts of the crown and young leaf primordia surrounding the inflorescence were included (Supplemental Figure 12B).
The samples harvested for RNA extraction were frozen immediately in liquid nitrogen and stored at −80°C. Total RNA, excluding miRNAs, was extracted from ground tissue using an RNeasy Micro Kit (Qiagen) and TRIzol (Life Technologies) for RNA sequencing and qRT-PCR, respectively. Residual DNA was removed using a DNA-free kit (Ambion). RNA extraction and DNase treatment were performed according to the manufacturer’s instructions. The RNA concentration and integrity were determined using a 2100 Bioanalyzer (Agilent) before RNA library preparation for RNA sequencing. Owing to the column-based RNA purification method of the RNeasy Micro Kit, only transcript species larger than 200 nucleotides were further processed for RNA sequencing.
cDNA libraries were prepared according to the TruSeq RNA sample preparation protocol (version 2; Illumina). Clonal sequence amplification and generation of sequence clusters were conducted using a cBot (Illumina). Single-end sequencing was performed using a HiSequation 2000 (Illumina) platform by multiplexing 12 libraries (libraries A-X, 1st set) and 24 libraries (libraries A1-AE1, 2nd set). In total, 47 libraries were sequenced, generating 672,463,624 1 × 100 bp single-end reads. Detailed information regarding the sequencing results is presented in Supplemental Table 18.
The sequencing data quality was verified using FastQC software (version 0.10.1; http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) before further processing using the CLC Genomics workbench (version 6.0.4; CLCbio). PCR duplicates were removed from the raw sequencing data using the Duplicate Read Removal plug-in of CLC. The reads were trimmed with an error probability limit calculated from the Phred scores of 0.05, allowing for a maximum of two ambiguously called nucleotides per read. Reads shorter than 60 bp, subsequent to the quality-based trimming, were removed from the data set. After PCR duplicate removal and quality-based filtering, 391,047,834 reads were retained, corresponding to an average of 59% of the raw sequencing data per library (Supplemental Figure 9A).
Differential Gene Expression Analysis
For transcriptome analysis, we used a custom reference sequence consisting of 68,739 transcripts (see Supplemental Methods). Quality filtered reads of each library were mapped against the reference sequence using the RNA-Seq Analysis tool of the CLC Genomics workbench with default parameters (Supplemental Table 18). Counts of uniquely mapped reads were extracted and used for downstream analyses. Differentially expressed transcripts were identified with the R/Bioconductor package “edgeR” (version 3.2.3; Robinson et al., 2010) using the generalized linear model with the factors genotype, photoperiod, and developmental stage of the MSAs. Five separate models were specified because under SDs, only S42-IL107 MSA and leaf samples were subjected to RNA sequencing. For DET calling, individual contrasts were defined to extract DETs between individual developmental stages in MSA samples, between photoperiods, and between genotypes in MSA and leaf samples. DETs were called at a false discovery rate (FDR) < 10−4. Additionally, DETs detected between individual developmental stages at the MSA required an absolute log2-fold change > 1.
For DET calling, only transcripts with expression levels greater than 5 cpm in at least two libraries were retained. Tables with raw counts for all transcripts and with normalized expression levels for the expressed transcripts are provided as supplemental annotation tables (Supplemental Data Sets 3 and 4). The expression values of the filtered transcripts were correlated between individual libraries in order to verify the quality of biological replication (Supplemental Table 19). One library was identified for S42-IL107 (MSA library under SD at W3.5, biological replicate 1), with a correlation coefficient of r ≤ 0.86 compared with its biological replicates. However, transcripts with low correlations between the biological replicates in this set of libraries were also among the transcripts with the highest variation between biological replicates in other library sets. Thus, we decided to retain the library with a low correlation coefficient for differential gene expression analysis, as biological rather than technical reasons were causative for the differences in expression values in this library.
To estimate the effect of the reference bias on estimates of differential gene expression between different genotypes, the introgressions of S42-IL107 and the identified DETs were located on the barley chromosomes using the PopSeq map (Mascher et al., 2013). Density of the DETs was defined as the number of the DETs divided by the number of all genes in 5-centimorgan sliding windows with a 2-centimorgan step and was plotted along the barley chromosomes using R.
Coexpression Analysis
Coexpression analysis was performed on reads per kilo base per million normalized expression levels of 7604 DETs, which were composed of 6602 transcripts differentially regulated during shoot apex development or transcripts coregulated between photoperiods and between genotypes in either leaves (1427 DETs) or shoot apices (518 DETs; Figures 3A and 3B). To optimize the number of coexpression clusters, negative binomial models were fitted for different numbers of coexpression clusters ranging from 5 to 120 using the R package “MBCluster.Seq” (version 1.0; Si et al., 2014). Convergence of the EM algorithm for estimation of cluster centers was called in a maximum of 103 iterations. The final number of clusters was determined based on visual inspection of the hybrid tree supported by high average probability of clustered transcripts (Supplemental Table 20).
The overrepresentation of identified DET subsets within coexpression clusters was examined using Pearson’s χ2 test in R (Monte Carlo simulation, 2000 replicates).
The overrepresentation of particular GO terms within the coexpression clusters and identified subsets of DETs was estimated against the GO-annotated reference (18,890 of 25,152 expressed transcripts) using Fisher’s exact tests (FDR, q < 0.05) implemented in the Blast2GO software (version 2.5.0; Conesa et al., 2005).
Verification of Gene Expression by RT-qPCR Assays and Correlation Network Analysis
DNase-treated total RNA (1 µg) was reverse-transcribed using SuperScript II reverse transcriptase (Life Technologies) according to the manufacturer’s instructions. The expression levels of the target genes were quantified by RT-qPCR. The reactions were set up, and RT-qPCRs were conducted as reported by Campoli et al. (2012a, 2012b) using gene-specific primers (Supplemental Table 21). Actin, GAPDH, and Ubiquitin showed stable expression across tissues, developmental stages, and photoperiods in the RNA sequencing experiments, and Actin was selected for the relative quantification of the target gene expression levels in the qRT-PCR assays. The RT-qPCR data for each target gene are presented as average expression levels over three biological replicates, with two technical replicates per reaction, relative to the expression levels of the Actin reference gene. Pearson’s correlation coefficients were calculated between RT-qPCR-derived transcripts levels of genes expressed in leaves and shoot apices and between transcript levels and number of floret primordia emerged from the MSA using the R package “Hmsic” (version 3.14-4). Correlations with |r| > 0.4 and an FDR of q < 0.05 were plotted using the R package “qgraph” (version 1.2.3; Epskamp et al., 2012).
Accession Numbers
Illumina data are in the European Short Read Archive (PRJEB8748). Accession numbers of transcripts probed by RT-qPCR are provided in Supplemental Table 21.
Supplemental Data
Supplemental Figure 1. Development related phenotypes of Scarlett/S42-IL107, Bowman/BW281, and Triumph/Triumph-IL.
Supplemental Figure 2. Effects of photoperiod during plant development on leaf emergence, stem elongation, and spike traits of Scarlett and S42-IL107.
Supplemental Figure 3. Development of the main shoot apex of Scarlett and S42-IL107.
Supplemental Figure 4. Size and flanking markers of Ppd-H1 introgressions on chromosome 2H.
Supplemental Figure 5. Validation of transcript levels in leaves and shoot apices of Bowman/BW281 and Triumph/Triumph-IL.
Supplemental Figure 6. Distribution and density of differentially expressed transcripts along the barley chromosomes.
Supplemental Figure 7. Expression profiles of 7604 DETs in 31 coexpression clusters
Supplemental Figure 8. Overrepresentation of Gene Ontology terms among transcripts in coexpression clusters I-III.
Supplemental Figure 9. Overview of RNA sequencing statistics.
Supplemental Figure 10. Expression patterns of selected transcripts in leaves and shoot apices of Scarlett and S42-IL107 determined by RNA sequencing.
Supplemental Figure 11. Correlation network for gene expression data in leaves and shoot apices.
Supplemental Figure 12. Representative main shoot apices dissected for RNA extraction.
Supplemental Figure 13. Schematic representation of the RNA sequencing pipeline.
Supplemental Table 1. Slopes of broken-line regressions performed on MSA development and floret primordia induction in Scarlett and S42-IL107 under SD and LD.
Supplemental Table 2. Average differences in expression levels (RPKM) between transcripts in coexpression clusters I-III.
Supplemental Table 3. GO term enrichment for 3299 transcripts specifically upregulated at stamen primordium stage.
Supplemental Table 4. GO term enrichment for 798 transcripts upregulated during floral transition.
Supplemental Table 5. GO term enrichment for 1434 DETs commonly expressed during floral transition and at stamen primordium stage.
Supplemental Table 6. Selected transcripts downregulated during MSA development independent of the photoperiod and genotype.
Supplemental Table 7. Coexpression clusters enriched for transcripts downregulated during MSA development independent of the photoperiod and genotype.
Supplemental Table 8. Selected transcripts upregulated during MSA development independent of the photoperiod and genotype.
Supplemental Table 9. Coexpression clusters enriched for transcripts upregulated during MSA development independent of the photoperiod and genotype.
Supplemental Table 10. Coexpression clusters enriched for transcripts upregulated in leaves by long photoperiods and in S42-IL107.
Supplemental Table 11. Selected transcripts upregulated in leaves by long photoperiods and in S42-IL107.
Supplemental Table 12. GO term enrichment of transcripts in coexpression clusters 9, 11, and 12.
Supplemental Table 13. GO term enrichment for 1205 transcripts downregulated in leaves by long photoperiods and in S42-IL107.
Supplemental Table 14. Selected transcripts upregulated in shoot apices by long photoperiods and in S42-IL107.
Supplemental Table 15. Coexpression clusters enriched for transcripts upregulated in shoot apices by long photoperiods and in S42-IL107.
Supplemental Table 16. GO term enrichment for transcripts in coexpression cluster 16.
Supplemental Table 17. GO term enrichment for transcripts in coexpression cluster 31.
Supplemental Table 18. Overview of RNA samples and sequencing statistics.
Supplemental Table 19. Correlation analysis for quality control of biological replicates used for RNA sequencing.
Supplemental Table 20. Statistics of coexpression clustering.
Supplemental Table 21. Oligonucleotide sequences used in qRT-PCR assays.
Supplemental Data Set 1. Annotation of differentially expressed transcripts.
Supplemental Data Set 2. Blast2GO annotation of transcripts in reference sequence.
Supplemental Data Set 3. RPKM transcript levels of expressed transcripts.
Supplemental Data Set 4. Read counts of all transcripts in the reference sequence.
Supplemental Methods. Design of reference sequence.
Supplementary Material
Acknowledgments
We thank Klaus Pillen (Martin Luther University Halle, Germany), Robbie Waugh (James Hutton Institute, UK), and David Laurie (John Innes Center, Norwich, UK) for seeds of barley lines. We also thank Kerstin Luxa and Andrea Lossow for excellent technical assistance. This work was supported by the Max Planck Society, by International Max Planck Research School fellowships to B.D. and A.P., and by the Deutsche Forschungsgemeinschaft under Grants SPP1530 (“Flowering time control: from natural variation to crop improvement”) and Excellence Cluster EXC1028.
AUTHOR CONTRIBUTIONS
B.D. and M.v.K. designed the research. B.D. performed the research. B.D. and A.P. analyzed data. B.D., A.P., and M.v.K. wrote the article.
Glossary
- SAM
shoot apical meristem
- LD
long day
- MSA
main shoot apex
- SD
short day
- IL
introgression line
- DET
differentially expressed transcript
- GO
Gene Ontology
- FDR
false discovery rate
Footnotes
Articles can be viewed without a subscription.
References
- Alqudah A.M., Schnurbusch T. (2014). Awn primordium to tipping is the most decisive developmental phase for spikelet survival in barley. Funct. Plant Biol. 41: 424–436. [DOI] [PubMed] [Google Scholar]
- Andrés F., Porri A., Torti S., Mateos J., Romera-Branchat M., García-Martínez J.L., Fornara F., Gregis V., Kater M.M., Coupland G. (2014). SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. USA 111: E2760–E2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard M., Kirby E.J.M., Fellowes G. (1982). Relationships between duration of phases in the pre-anthesis life cycle of spring barley. Aust. J. Agric. Res. 33: 917–925. [Google Scholar]
- Arisnabarreta S., Miralles D.J. (2008). Critical period for grain number establishment of near isogenic lines of two- and six-rowed barley. Field Crops Res. 107: 196–202. [Google Scholar]
- Boden S.A., Weiss D., Ross J.J., Davies N.W., Trevaskis B., Chandler P.M., Swain S.M. (2014). EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell 26: 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrás-Gelonch G., Denti M.B., Thomas W., Romagosa I. (2012a). Genetic control of pre-heading phases in the Steptoe x Morex barley population under different conditions of photoperiod and temperature. Euphytica 183: 303–321. [Google Scholar]
- Borrás-Gelonch G., Rebetzke G.J., Richards R.A., Romagosa I. (2012b). Genetic control of duration of pre-anthesis phases in wheat (Triticum aestivum L.) and relationships to leaf appearance, tillering, and dry matter accumulation. J. Exp. Bot. 63: 69–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrás-Gelonch G., Slafer G.A., Casas A.M., van Eeuwijk F., Romagosa I. (2010). Genetic control of pre-heading phases and other traits related to development in a double-haploid barley (Hordeum vulgare L.) population. Field Crops Res. 119: 36–47. [Google Scholar]
- Burko Y., Shleizer-Burko S., Yanai O., Shwartz I., Zelnik I.D., Jacob-Hirsch J., Kela I., Eshed-Williams L., Ori N. (2013). A role for APETALA1/fruitfull transcription factors in tomato leaf development. Plant Cell 25: 2070–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C., Drosse B., Searle I., Coupland G., von Korff M. (2012a). Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J. 69: 868–880. [DOI] [PubMed] [Google Scholar]
- Campoli C., Shtaya M., Davis S.J., von Korff M. (2012b). Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 12: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C., Pankin A., Drosse B., Casao C.M., Davis S.J., von Korff M. (2013). HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol. 199: 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Carver B.F., Wang S., Zhang F., Yan L. (2009). Genetic loci associated with stem elongation and winter dormancy release in wheat. Theor. Appl. Genet. 118: 881–889. [DOI] [PubMed] [Google Scholar]
- Chuck G., Cigan A.M., Saeteurn K., Hake S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39: 544–549. [DOI] [PubMed] [Google Scholar]
- Cockram J., Jones H., Leigh F.J., O’Sullivan D., Powell W., Laurie D.A., Greenland A.J. (2007). Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J. Exp. Bot. 58: 1231–1244. [DOI] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Degner J.F., Marioni J.C., Pai A.A., Pickrell J.K., Nkadori E., Gilad Y., Pritchard J.K. (2009). Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25: 3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea S., Leader D.J., Arnold B.C., Shaw P., Dolan L., Doonan J.H. (2005). Systematic spatial analysis of gene expression during wheat caryopsis development. Plant Cell 17: 2172–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A., et al. (2006). An atlas of gene expression from seed to seed through barley development. Funct. Integr. Genomics 6: 202–211. [DOI] [PubMed] [Google Scholar]
- Druka A., Franckowiak J., Lundqvist U., Bonar N., Alexander J., Houston K., Radovic S., Shahinnia F., Vendramin V., Morgante M., Stein N., Waugh R. (2011). Genetic dissection of barley morphology and development. Plant Physiol. 155: 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epskamp S., Cramer A.O.J., Waldorp L.J., Schmittmann V.D., Borsboom D. (2012). {qgraph}: Network visualizations of relationships in psychometric data. J. Stat. Softw. 48: 1–18. [Google Scholar]
- Faure S., Higgins J., Turner A., Laurie D.A. (2007). The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S., Turner A.S., Gruszka D., Christodoulou V., Davis S.J., von Korff M., Laurie D.A. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. USA 109: 8328–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R.A. (1985). Number of kernels in wheat crops and the influence of solar radiation and temperature. J. Agric. Sci. Camb. 105: 447–461. [Google Scholar]
- Ghiglione H.O., Gonzalez F.G., Serrago R., Maldonado S.B., Chilcott C., Curá J.A., Miralles D.J., Zhu T., Casal J.J. (2008). Autophagy regulated by day length determines the number of fertile florets in wheat. Plant J. 55: 1010–1024. [DOI] [PubMed] [Google Scholar]
- González F., Slafer G., Miralles D. (2005). Pre-anthesis development and number of fertile florets in wheat as affected by photoperiod sensitivity genes Ppd-D1 and Ppd-B1. Euphytica 146: 253–269. [Google Scholar]
- González F.G., Miralles D.J., Slafer G.A. (2011). Wheat floret survival as related to pre-anthesis spike growth. J. Exp. Bot. 62: 4889–4901. [DOI] [PubMed] [Google Scholar]
- González F.G., Slafer G.A., Miralles D.J. (2002). Vernalization and photoperiod responses in wheat pre-flowering reproductive phases. Field Crops Res. 74: 183–195. [Google Scholar]
- González F.G., Slafer G.A., Miralles D.J. (2003). Floret development and spike growth as affected by photoperiod during stem elongation in wheat. Field Crops Res. 81: 29–38. [Google Scholar]
- Greenup A.G., Sasani S., Oliver S.N., Walford S.A., Millar A.A., Trevaskis B. (2011). Transcriptome analysis of the vernalization response in barley (Hordeum vulgare) seedlings. PLoS One 6: e17900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming M.N., Fieg S., Peacock W.J., Dennis E.S., Trevaskis B. (2009). Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol. Genet. Genomics 282: 107–117. [DOI] [PubMed] [Google Scholar]
- Hemming M.N., Walford S.A., Fieg S., Dennis E.S., Trevaskis B. (2012). Identification of high-temperature-responsive genes in cereals. Plant Physiol. 158: 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston K., et al. (2013). Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc. Natl. Acad. Sci. USA 110: 16675–16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H., Leigh F.J., Mackay I., Bower M.A., Smith L.M., Charles M.P., Jones G., Jones M.K., Brown T.A., Powell W. (2008). Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol. Biol. Evol. 25: 2211–2219. [DOI] [PubMed] [Google Scholar]
- Kitchen B.M., Rasmusson D.C. (1983). Duration and inheritance of leaf initiation, spike initiation, and spike growth in barley. Crop Sci. 23: 939. [Google Scholar]
- Kojima S., Takahashi Y., Kobayashi Y., Monna L., Sasaki T., Araki T., Yano M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43: 1096–1105. [DOI] [PubMed] [Google Scholar]
- Krizek B.A. (2011). Auxin regulation of Arabidopsis flower development involves members of the AINTEGUMENTA-LIKE/PLETHORA (AIL/PLT) family. J. Exp. Bot. 62: 3311–3319. [DOI] [PubMed] [Google Scholar]
- Krogan N.T., Hogan K., Long J.A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139: 4180–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie D.A., Pratchett N., Bezant J.H., Snape J.W. (1994). Genetic analysis of a photoperiod response gene on the short arm of chromosome 2(2H) of Hordeum vulgare (barley). Heredity 72: 619–627. [Google Scholar]
- Laurie D.A., Pratchett N., Snape J.W., Bezant J.H. (1995). RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter x spring barley (Hordeum vulgare L.) cross. Genome 38: 575–585. [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Farrona S., Klemme S., Turck F.K. (2014). Post-fertilization expression of FLOWERING LOCUS T suppresses reproductive reversion. Front. Plant Sci. 5: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B., Nitcher R., Han X., Wang S., Ni F., Li K., Pearce S., Wu J., Dubcovsky J., Fu D. (2014). Characterization of FLOWERING LOCUS T1 (FT1) gene in Brachypodium and wheat. PLoS One 9: e94171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., et al. (2013). Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J. 76: 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K.F., Waugh R., Brown J.W., Schulman A., Langridge P., Platzer M., Fincher G.B., Muehlbauer G.J., Sato K., Close T.J., Wise R.P., Stein N.; International Barley Genome Sequencing Consortium (2012). A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716. [DOI] [PubMed] [Google Scholar]
- Melzer S., Lens F., Gennen J., Vanneste S., Rohde A., Beeckman T. (2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40: 1489–1492. [DOI] [PubMed] [Google Scholar]
- Miralles D.J., Richards R.A. (2000). Responses of leaf and tiller emergence and primordium initiation in wheat and barley to interchanged photoperiod. Ann. Bot. (Lond.) 85: 655–663. [Google Scholar]
- Miralles D.J., Richards R.A., Slafer G.A. (2000). Duration of the stem elongation period influences the number of fertile florets in wheat and barley. Funct. Plant Biol. 27: 931–940. [Google Scholar]
- Muggeo V.M.R. (2003). Estimating regression models with unknown break-points. Stat. Med. 22: 3055–3071. [DOI] [PubMed] [Google Scholar]
- Muggeo V.M.R. (2008). Segmented: an R package to fit regression models with broken-line relationships. R News 8: 20–25. [Google Scholar]
- Müller-Xing R., Clarenz O., Pokorny L., Goodrich J., Schubert D. (2014). Polycomb-group proteins and FLOWERING LOCUS T maintain commitment to flowering in Arabidopsis thaliana. Plant Cell 26: 2457–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M.A., Fischer R.L., Goldberg R.B., Nakamura K., Harada J.J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102: 3123–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankin A., Campoli C., Dong X., Kilian B., Sharma R., Himmelbach A., Saini R., Davis S.J., Stein N., Schneeberger K., von Korff M. (2014). Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 198: 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce S., Vanzetti L.S., Dubcovsky J. (2013). Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 163: 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis O.N. (1934). An analysis of the influence of temperature during germination on the subsequent development of certain winter cereals and its relation to the effect of length of day. Ann. Bot. (Lond.) 48: 919–955. [Google Scholar]
- R Development Core Team (2008). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing).
- Reynolds M., Foulkes M.J., Slafer G.A., Berry P., Parry M.A.J., Snape J.W., Angus W.J. (2009). Raising yield potential in wheat. J. Exp. Bot. 60: 1899–1918. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna G., Giunta F., Motzo R., Mastrangelo A.M., De Vita P. (2014). Genetic variation for the duration of pre-anthesis development in durum wheat and its interaction with vernalization treatment and photoperiod. J. Exp. Bot. 65: 3177–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasani S., Hemming M.N., Oliver S.N., Greenup A., Tavakkol-Afshari R., Mahfoozi S., Poustini K., Sharifi H.-R., Dennis E.S., Peacock W.J., Trevaskis B. (2009). The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare). J. Exp. Bot. 60: 2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalenbach I., March T.J., Bringezu T., Waugh R., Pillen K. (2011). High-resolution genotyping of wild barley introgression lines and fine-mapping of the threshability locus thresh-1 using the Illumina GoldenGate assay. G3 (Bethesda) 1: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Franzen R., Ngyuen T.H., Garcia-Maroto F., Pozzi C., Salamini F., Rohde W. (2000). Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol. Biol. 42: 899–913. [DOI] [PubMed] [Google Scholar]
- Shalit A., Rozman A., Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y., Lifschitz E. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 106: 8392–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitsukawa N., Ikari C., Shimada S., Kitagawa S., Sakamoto K., Saito H., Ryuto H., Fukunishi N., Abe T., Takumi S., Nasuda S., Murai K. (2007). The einkorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes Genet. Syst. 82: 167–170. [DOI] [PubMed] [Google Scholar]
- Si Y., Liu P., Li P., Brutnell T.P. (2014). Model-based clustering for RNA-seq data. Bioinformatics 30: 197–205. [DOI] [PubMed] [Google Scholar]
- Slafer G.A. (2003). Genetic basis of yield as viewed from a crop physiologist’s perspective. Ann. Appl. Biol. 142: 117–128. [Google Scholar]
- Slafer G.A., Rawson H.M. (1994). Sensitivity of wheat phasic development to major environmental factors: a re-examination of some assumptions made by physiologists and modellers. Funct. Plant Biol. 21: 393–426. [Google Scholar]
- Slafer G.A., Abeledo L.G., Miralles D.J., Gonzalez F.G., Whitechurch E.M. (2001). Photoperiod sensitivity during stem elongation as an avenue to raise potential yield in wheat. Euphytica 119: 191–197. [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker P., Samach A. (2005). The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S., Fornara F., Vincent C., Andrés F., Nordström K., Göbel U., Knoll D., Schoof H., Coupland G. (2012). Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B., Tadege M., Hemming M.N., Peacock W.J., Dennis E.S., Sheldon C. (2007). Short vegetative phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol. 143: 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R.P., Laurie D.A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034. [DOI] [PubMed] [Google Scholar]
- von Zitzewitz J., Szűcs P., Dubcovsky J., Yan L., Francia E., Pecchioni N., Casas A., Chen T.H.H., Hayes P.M., Skinner J.S. (2005). Molecular and structural characterization of barley vernalization genes. Plant Mol. Biol. 59: 449–467. [DOI] [PubMed] [Google Scholar]
- Waddington S.R., Cartwright P.M., Wall P.C. (1983). A quantitative scale of spike initial and pistil development in barley and wheat. Ann. Bot. (Lond.) 51: 119–130. [Google Scholar]
- Wan Y., Poole R.L., Huttly A.K., Toscano-Underwood C., Feeney K., Welham S., Gooding M.J., Mills C., Edwards K.J., Shewry P.R., Mitchell R.A. (2008). Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics 9: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Park M.Y., Wang L.J., Koo Y., Chen X.Y., Weigel D., Poethig R.S. (2011). miRNA control of vegetative phase change in trees. PLoS Genet. 7: e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield M.O., Lu C., Wilson I.D., Coghill J.A., Edwards K.J. (2009). Cold- and light-induced changes in the transcriptome of wheat leading to phase transition from vegetative to reproductive growth. BMC Plant Biol. 9: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland A.J., Börner A., Korzun V., Li W.M., Petrovíc S., Sayers E.J. (1998). The influence of photoperiod genes on the adaptability of European winter wheats. Euphytica 100: 385–394. [Google Scholar]
- Wu G., Poethig R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T., Gross-Hardt R., Laux T. (2006). APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18: 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Wu M.F., Yang L., Wu G., Poethig R.S., Wagner D. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17: 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T., Dubcovsky J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100: 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., Bennetzen J.L., Echenique V., Dubcovsky J. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W.H., Wang P., Chen H.X., Zhou H.J., Li Q.P., Wang C.R., Ding Z.H., Zhang Y.S., Yu S.B., Xing Y.Z., Zhang Q.F. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4: 319–330. [DOI] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T.T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., Zhang Q. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Zadoks J.C., Chang T.T., Konzak C.F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14: 415–421. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.