More than 10,000 enhancers located in intergenic regions in Arabidopsis, which regulate gene expression, were predicted using genomic data based on hypersensitivity to DNase I digestion.
Abstract
Enhancers are important regulators of gene expression in eukaryotes. Enhancers function independently of their distance and orientation to the promoters of target genes. Thus, enhancers have been difficult to identify. Only a few enhancers, especially distant intergenic enhancers, have been identified in plants. We developed an enhancer prediction system based exclusively on the DNase I hypersensitive sites (DHSs) in the Arabidopsis thaliana genome. A set of 10,044 DHSs located in intergenic regions, which are away from any gene promoters, were predicted to be putative enhancers. We examined the functions of 14 predicted enhancers using the β-glucuronidase gene reporter. Ten of the 14 (71%) candidates were validated by the reporter assay. We also designed 10 constructs using intergenic sequences that are not associated with DHSs, and none of these constructs showed enhancer activities in reporter assays. In addition, the tissue specificity of the putative enhancers can be precisely predicted based on DNase I hypersensitivity data sets developed from different plant tissues. These results suggest that the open chromatin signature-based enhancer prediction system developed in Arabidopsis may serve as a universal system for enhancer identification in plants.
INTRODUCTION
Gene expression in eukaryotes is regulated by the orchestrated binding of regulatory proteins to promoters, enhancers, and other cis-regulatory DNA elements (CREs). Promoters and enhancers are the most common and best understood CREs. Most eukaryotic genes contain a single promoter located close to the transcription start site (TSS). Enhancers recruit transcription factors (TFs) to regulate the transcription of target genes, often in a cell type- or tissue-specific manner (Shen et al., 2012). The expression of a gene can be regulated by multiple enhancers at different developmental stages and/or in different tissues. In addition, enhancers act independently of their distance and orientation to the promoters of target genes (Bulger and Groudine, 2011). Thus, enhancers can be located in the front or at the ends of genes, within or far away from genes, which makes enhancers significantly more difficult to identify than promoters. Remarkably, only a handful of enhancers, especially those that are located in intergenic regions, have been identified in plants (Yang et al., 2005; Clark et al., 2006; McGarry and Ayre, 2008; Schauer et al., 2009; Raatz et al., 2011).
Enhancers have been extensively investigated in humans and model animal species. Genome-wide identification of enhancers became possible due to the advent of next-generation sequencing techniques. Several transcriptional cofactors have been well characterized in mammalian species. These cofactors connect TFs to the basal transcription machinery (such as the RNA Polymerase II complex) or remodel the chromatin status to help the transcription apparatus and TFs to access the chromosomal DNA (Näär et al., 2001). Genome-wide mapping of the binding sites of these cofactors, including p300/CBP and MED1, has provided the most important information of the genomic locations of enhancers (Heintzman et al., 2009; Visel et al., 2009; Kim et al., 2010). Genes encoding homologs of mammalian p300/CBP and MED1 (the Arabidopsis thaliana homologs of the Mediator complex) were identified in Arabidopsis, and their roles in regulation of plant gene expression have been documented (Han et al., 2007; Kidd et al., 2011). However, the Arabidopsis genome encodes five CPB/p300-like proteins and the Arabidopsis Mediator complex contains 27 subunits (Han et al., 2007; Kidd et al., 2011). These transcriptional cofactors have not been fully characterized in Arabidopsis and other plant species and their potential in enhancer identification has not been exploited. Several histone modification marks have also been used for enhancer prediction in model animal species (Heintzman et al., 2007, 2009; Creyghton et al., 2010; Zentner et al., 2011; Hnisz et al., 2013).
Genomic regions encompassing an active CRE, which would be bound by regulatory protein(s), are depleted of nucleosomes or under dynamic nucleosome modification or displacement (Henikoff et al., 2009; Jin et al., 2009). A common characteristic associated with such genomic regions is a pronounced sensitivity to DNase I cleavage (Gross and Garrard, 1988). These genomic regions, named DNase I hypersensitive sites (DHSs), can be identified by DNase I digestion followed by high-throughput sequencing (DNase-seq). DHS has been proven to be a reliable chromatin signature for genome-wide mapping of various CREs, including enhancers, in model animal species (Guertin et al., 2012; Shen et al., 2012; The_ENCODE_Project_Consortium, 2012). We recently developed DHS maps in Arabidopsis using both leaf and flower tissues (Zhang et al., 2012a). Here, we exploited the potential of DHS as a sole chromatin marker for enhancer prediction. A set of 10,044 intergenic enhancers were predicted based on the DHS data sets. We selected 14 predicted enhancers for validation using the Escherichia coli β-glucuronidase (GUS) reporter gene. Remarkably, 10 of the 14 putative enhancers were confirmed by the reporter assays, including five of the six leaf- or flower-specific enhancers. Thus, DHS-based prediction serves as a promising enhancer identification system that can potentially be applied in any plant species with a sequenced genome.
RESULTS
Enhancer Prediction Based on DHSs Located in Intergenic Regions
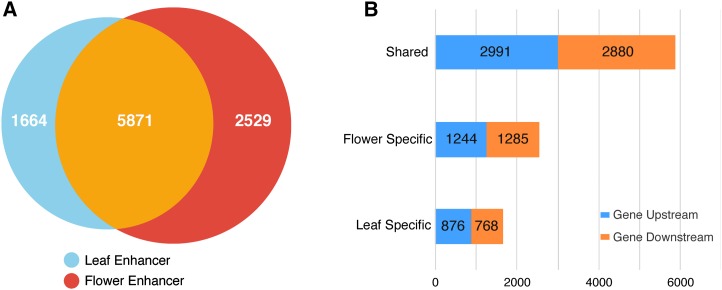
The immense success of enhancer prediction based on chromatin features in mammalian species prompted us to test the possibility of predicting plant enhancers using the DHS data sets developed in Arabidopsis. We previously identified 38,290 and 41,193 DHSs in leaf and flower tissues, respectively (Zhang et al., 2012a). To distinguish putative enhancers from any promoters, we excluded all DHSs located within 1.5 kb upstream of the TSS of each gene. Only DHSs located in intergenic regions (downstream of transcription terminal site and >1.5 kb upstream of TSS) were considered to be putative enhancers. Based on these criteria, we identified a total of 10,044 DHSs as putative intergenic enhancers in the Arabidopsis genome (Supplemental Data Set 1), including 5871 DHSs shared by leaf and flower tissues (common enhancers) and 1644 and 2529 DHSs specific to leaf and flower tissue, respectively (tissue-specific enhancers) (Figure 1).
Figure 1.
Tissue Specificity and Genomic Locations of Predicted Enhancers in Arabidopsis.
(A) The total numbers of three types of predicted enhancers: leaf-specific enhancers, flower-specific enhancers, and the enhancers common to both tissues.
(B) Genomic locations (Gene Upstream, located upstream of the TSS of a gene; Gene Downstream, located downstream of the transcription terminal site of a gene) of the three types of predicted enhancers.
Most enhancers regulate their most proximal genes (Mendenhall et al., 2013; Ghavi-Helm et al., 2014; Kvon et al., 2014). Chromatin conformation capture (3C) data revealed that local intrachromosome interaction represents a dominant structural feature of genome conformation in Arabidopsis (Feng et al., 2014; Wang et al., 2015). Thus, we projected that each predicted enhancer will most likely regulate the expression of its most adjacent gene(s). We identified the most proximal genes flanking each of the predicted intergenic enhancer (see Methods). We then analyzed the expression patterns of these genes using RNA-seq data developed from the same tissues used in DNase-seq (Zhang et al., 2012a). We identified 1651 most proximal genes (fragments per kilobase of transcript per million mapped reads [FPKM] > 1 in leaf or flower tissue) associated with the 2529 putative flower-specific enhancers. The expression levels of these genes were significantly higher in flower tissue (mean of FPKM, 58.21; median of FPKM, 15.19) than in leaf tissue (mean of FPKM, 54.13; median of FPKM, 10.78) (P value < 8.8 × 10−9, K-S test) (Figure 2A). Similarly, we identified 1362 most proximal genes (FPKM > 1) associated with the 1644 putative leaf-specific enhancers. The expression levels of these genes were higher in leaf tissue (mean of FPKM, 63.63; median of FPKM, 14.88) than in flower tissue (mean of FPKM, 40.85; median of FPKM, 13.82) (P value: 0.0081, K-S test). These results support the impact of tissue-specific enhancers on the tissue-specific expression of the proximal genes.
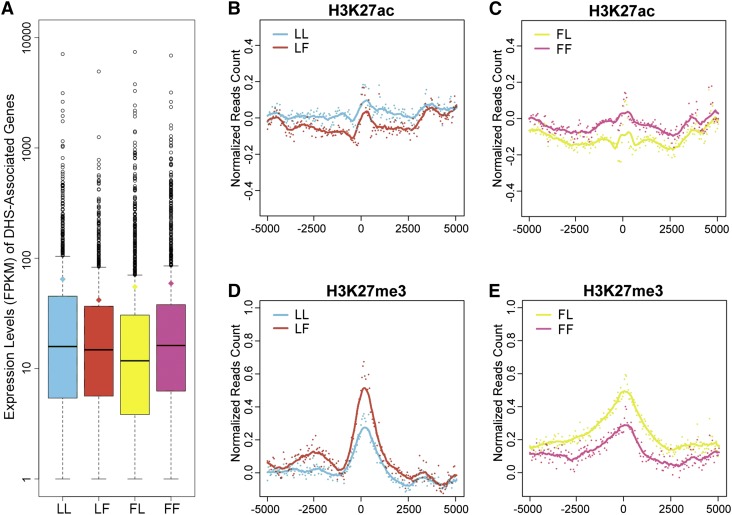
Figure 2.
Gene Expression and Histone Modifications Associated with the Predicted Tissue-Specific Enhancers.
(A) Box plots of the expression levels of genes most proximal to the predicted tissue-specific enhancers. The y axis represents the expression level (FPKM), where the whiskers refer to the statistical quartiles of data. Only genes with FPKM > 1 were included in the analysis.
(B) to (E) Histone modifications associated with the predicted tissue-specific enhancers and the flanking ±5-kb regions. The middle point (nucleotide position) of the tissue-specific enhancers was aligned as the center (0 point). Each dot represents the ChIP-seq read account of a tissue-specific enhancer. The read accounts were normalized using shared DHSs as the background in 50-bp windows. Solid lines represent smoothed score based on dot plot. ChIP-seq data from leaf and flower tissues were labeled in green and red, respectively.
(B) Leaf-specific enhancers and their association with leaf tissue-derived H3K27ac (sky blue) and flower tissue-derived H3K27ac (vermilion).
(C) Flower-specific enhancers and their association with flower tissue-derived H3K27ac (reddish purple) and leaf tissue-derived H3K27ac (yellow).
(D) Leaf-specific enhancers and their association with leaf tissue-derived H3K27me3 (sky blue) and flower tissue-derived H3K27me3 (vermilion).
(E) Flower-specific enhancers and their association with flower tissue-derived H3K27me3 (reddish purple) and leaf tissue-derived H3K27me3 (yellow).
LL, leaf-specific enhancers in leaf tissue; LF, leaf-specific enhancers in flower tissue; FL, flower-specific enhancers in leaf tissue; FF, flower-specific enhancers in flower tissue.
Histone Modification Patterns Associated with the Predicted Enhancers
Active mammalian enhancers are distinctly associated with the histone modification mark H3K27ac (Heintzman et al., 2007, 2009; Creyghton et al., 2010; Zentner et al., 2011). In contrast, poised mammalian enhancers are distinguished by the absence of H3K27ac and the enrichment of H3K27me3 (Rada-Iglesias et al., 2011). To investigate the possible association of these histone modification marks with enhancers in A. thaliana, we developed chromatin immunoprecipitation followed by sequencing (ChIP-seq) data sets for H3K27ac and H3K27me3 using both leaf and flower tissues that were at the same developmental stages as the tissues used for DNase-seq (see Methods). We examined ChIP-seq profiles along ±5-kb regions flanking each predicted enhancer. These surrounding regions were clearly enriched in both histone modification marks (Supplemental Figure 1A). In contrast, the same regions were depleted of H3K27me1 and H3K9me2, two classical marks associated with heterochromatin in Arabidopsis (Luo et al., 2013) (Supplemental Figure 1A).
We then examined if the predicted tissue-specific enhancers were associated with unique histone modification patterns. We used common enhancers (predicted by both leaf and flower DHS data) as a control to normalize the H3K27ac and H3K27me3 ChIP-seq read count along ±5-kb regions flanking the predicted enhancers. We then examined the histone modification patterns around tissue-specific enhancers. The predicted leaf-specific enhancers and flanking regions were more enriched in leaf tissue-derived H3K27ac than in flower tissue-derived H3K27ac (Figure 2B). Similarly, the predicted flower-specific enhancers and flanking regions were more enriched in flower tissue-derived H3K27ac than leaf-tissue-derived H3K27ac (Figure 2D). Thus, H3K27ac was positively correlated with tissue-specific enhancers (Supplemental Figure 1B). By contrast, the H3K27me3 enrichment flanking the enhancers showed opposite patterns to those of H3K27ac. Specifically, leaf-specific enhancers were less enriched in leaf tissue-derived H3K27me3 than flower tissue-derived H3K27me3 (Figure 2C). Similarly, flower-specific enhancers were less enriched in flower tissue-derived H3K27me3 than leaf tissue-derived H3K27me3 (Figure 2E). Thus, H3K27me3 was negatively correlated with tissue-specific enhancers (Supplemental Figure 1B), suggesting that H3K27me3 is likely a repressor mark and is associated with poised enhancers.
Noncoding RNAs Associated with the Predicted Enhancers
Several recent studies in mammalian species revealed that enhancers are associated with noncoding RNA (De Santa et al., 2010; Kim et al., 2010; Ørom et al., 2010). RNA Polymerase II (RNAPII) was bound to 25% of the neuronal activity-regulated enhancers in mouse. Short and nonpolyadenylated enhancer RNAs were detected from almost all of the RNAPII-bound enhancers and were correlated with mRNA synthesis from nearby genes (Kim et al., 2010; Andersson et al., 2014). Long intergenic noncoding RNAs (lincRNAs) in the human genome have properties reminiscent of enhancers (Ørom et al., 2010). Depletion of a number of lincRNAs led to decreased expression of their neighboring protein-coding genes. In addition, these lincRNA loci were also occupied by RNAPII and coactivator p300/CBP (Ørom et al., 2010).
The Arabidopsis genome contains 13,230 intergenic transcripts or noncoding RNAs (ncRNAs), including 6480 lincRNAs (Liu et al., 2012). We found that a total of 1991 (19.8%) of the intergenic DHSs have a minimum of 1 bp overlap with a ncRNA transcript, and 1338 of the 1991 DHSs overlap with lincRNA transcripts. We randomly selected 10,044 intergenic sequences (300 bp each, the length of the average-sized DHS, no overlap between any two selected sequences) and examined their overlap (a minimum of 1 bp) with ncRNAs. This simulation was performed 10,000 times. The mean overlapping rate was 3.52% with a sd of 0.18%. Thus, the overlapping rate between predicted enhancers with ncRNAs is significantly higher than the overlapping rates using random data sets (empirical, P value < 1 × 10−4).
Validation of the Predicted Enhancers Using Reporter Assay
A GUS-based reporter system was developed to validate the putative enhancers (Supplemental Figure 2). The predicted enhancers, ranging from 384 to 806 bp, were amplified from Arabidopsis genomic DNA and cloned into a reporter vector consisting of a minimal 35S promoter (−50 to −2 bp) and the GUS reporter gene (Jefferson, 1987). The constructs were then transformed into wild-type Arabidopsis (Col-0) plants and the transgenic plants were assayed for GUS expression. A predicted enhancer would be validated if GUS expression was consistently observed in the transgenic plants.
A total of 14 predicted intergenic enhancers were selected for validation (Supplemental Table 1), including three circadian-regulated enhancers that were identified previously by the traditional enhancer trapping method (Michael and McClung, 2003). Each of these three enhancers overlapped with an intergenic DHS and thus was also predicted to be an enhancer in our data set. Two positive and two negative reporter constructs were also used in validation. The two positive constructs included a known enhancer from the 35S promoter (−200 to −39 bp) (Benfey et al., 1989) that was placed in opposite directions in the reporter construct (−200 to −39 bp and −39 to −200 bp, respectively, constructs 35sEn and 35sEnR in Figure 3E). An empty vector, pKGWSF7.0, that contained the backbone of the reporter construct (Karimi et al., 2007) and a construct with only the minimal 35S promoter (construct mini35s in Figure 3E) were used as negative controls.
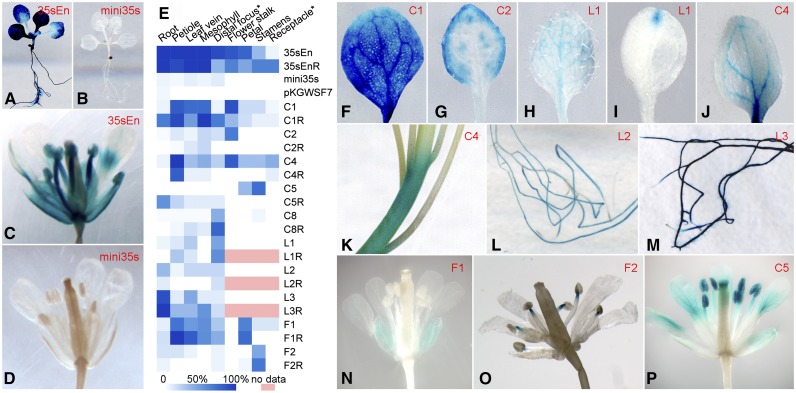
Figure 3.
Validation of Predicted Intergenic Enhancers and Representative GUS Expression Patterns from Transgenic Plants Transformed with Different Enhancer Constructs.
Construct names are the same as those in Table 1.
(A) A complete transgenic seedling transformed with the positive control construct 35sEn.
(B) A complete transgenic seedling transformed with the negative control construct mini35s.
(C) A complete flower of a transgenic plant harboring the construct 35sEn.
(D) A complete flower of a transgenic plant harboring the construct mini35s.
(E) A heat map of GUS expression in different tissues of transgenic plants transformed with different enhancer constructs. White indicates no GUS expression was detected in any plants. Blue indicates that GUS expression was detected in 100% of the transgenic plants. *Distal focus, distal focus of leaf; *Receptacle, floral receptacle.
(F) A leaf of a transgenic plant transformed with construct C1, showing almost uniform GUS staining throughout the entire leaf.
(G) A leaf of a transgenic plant transformed with construct C2, showing enhanced signals along the edge of the leaf. Similarly stained leaves were also observed from transgenic plants harboring constructs C1 and F1.
(H) A leaf of a transgenic plant transformed with construct L1, showing uniform GUS staining in most veins.
(I) A leaf of a transgenic plant transformed with construct L1, showing GUS signals at the distal focus of the leaf. Similarly stained leaves were also observed from transgenic plants harboring constructs L2 and C8.
(J) A leaf of a transgenic plant transformed with construct C4, showing strong GUS staining in the main veins and the base of the leaf. Similarly stained leaves were also observed from transgenic plants harboring construct L3.
(K) GUS-stained stem from a transgenic plant harboring construct C4. Similarly stained stems were also observed from transgenic plants harboring constructs C1 and C2.
(L) GUS-stained roots from a transgenic plant transformed with construct L2.
(M) GUS-stained roots from a transgenic plant harboring construct L3.
(N) A complete flower of a transgenic plant harboring construct F1, showing specific signals in sepals. A similar GUS staining pattern in flowers was also observed in transgenic plants harboring construct C1.
(O) A complete flower of a transgenic plant harboring construct F2, showing specific signals at the tips of filaments attached to the anthers.
(P) A complete flower of a transgenic plant harboring construct C5.
Both positive constructs generated strong GUS signals in different tissues (Figures 3A and 3C). In contrast, the two negative constructs produced inconsistent and faint GUS signals in a few transgenic plants (Figures 3B and 3D; Supplemental Table 2). Ten of the 14 enhancer candidates (71%) consistently generated GUS signals in different tissues with different signal intensities (Figure 3E; Supplemental Table 2). GUS signals were detected in various tissues/organs, including leaf (Figures 3F to 3J), stem (Figure 3K), root (Figures 3L and 3M), and different parts of flower (Figures 3N to 3P). Thus, these 10 candidates were validated to have enhancer function. We also randomly selected 10 intergenic regions (N1 to N10) that were not associated with DHSs (Supplemental Table 1). These regions, ranging from 293 to 713 bp, were cloned into the reporter vector as non-enhancer controls. The transgenic plants derived from these nonenhancer constructs showed similar GUS expression patterns to those of the two negative control constructs (Supplemental Figure 3 and Supplemental Table 2).
Tissue-Specific Function of the Predicted Enhancers
Among the 14 predicted enhancers selected for validation, three were leaf-specific DHSs (leaf-specific enhancers, L1 to L3) and three were flower-specific DHSs (flower-specific enhancers, F1 to F3). All three leaf-specific enhancers generated distinct GUS expression patterns in leaves, but not in flowers, in the transgenic plants (Table 1). In addition, L2 and L3 also generated GUS signals in roots (Figures 3L and 3M). This was not unexpected because we used the entire seedling, including root tissue, in DNase-seq experiments.
Table 1. General GUS Expression Patterns in Transgenic Lines from Predicted Enhancers.
| Predicted Enhancer (Short Name) | Size (bp) | Tissues with GUS Expression |
||
|---|---|---|---|---|
| Root (F/R)a | Leaf (F/R) | Flower (F/R) | ||
| Common DHS 1 (C1)b | 568 | −/− | +++/+++ | +++/+ |
| Common DHS 2 (C2)b | 684 | −/− | +/+ | +++/− |
| Common DHS 3 (C3)b | 393 | −/NA | −/NA | −/NA |
| Common DHS 4 (C4) | 580 | −/− | +++/+++ | +++/− |
| Common DHS 5 (C5) | 580 | −/++ | −/− | +++/− |
| Common DHS 6 (C6) | 384 | −/NA | −/NA | −/NA |
| Common DHS 7 (C7) | 403 | −/NA | −/NA | −/NA |
| Common DHS 8 (C8) | 562 | −/− | +/+++ | −/− |
| Leaf DHS 1 (L1) | 588 | −/− | +/++ | −/NA |
| Leaf DHS 2 (L2) | 444 | +/+ | +/+ | −/NA |
| Leaf DHS 3 (L3) | 625 | +++/+++ | +/+++ | −/NA |
| Flower DHS 1 (F1) | 622 | −/− | +++/+++ | +++/+++ |
| Flower DHS 2 (F2) | 533 | −/− | −/− | +/++ |
| Flower DHS 3 (F3) | 806 | −/NA | −/NA | −/NA |
Forward/reverse direction of each predicted enhancer within the report construct. +++, More than 75% positive transgenic plants or 50 to 75% positive transgenic plants with consistently strong signals in the same tissue(s); ++, 50 to 75% positive transgenic plants with consistently weak signals in the same tissue(s); +, <50% positive transgenic plants with consistently strong signals in the same tissue(s); −, <50% positive transgenic plants with inconsistent signals in different tissue(s) or no signals in any tissues; NA, not available.
C1, C2, and C3 are three putative enhancers identified previously by the enhancer trapping method (Michael and McClung, 2003).
Two of three flower-specific enhancers, F1 and F2, generated GUS signals in flowers in the transgenic plants. F2 generated distinct GUS signals that were highly specific to the tips of filaments attached to the anthers (Figure 3O). Unambiguous GUS signals were not observed in the rest of the transgenic plants. F1 generated specific signals in the sepals of the flowers (Figure 3N). Interestingly, F1 also generated signals in various cell types in leaf tissue (Figure 3E). Since sepals are modified leaves, we hypothesize that the sepal specificity of the endogenous F1 is regulated by additional flanking elements. The transgenic F1 lost the tissue specificity and became active in other organs related to sepals. In addition, neither F1 nor F2 generated signals in roots.
Eight of 14 predicted enhancers were associated with DHSs in both leaf and flower tissues (common enhancers, C1 to C8). Several of them generated signals in both leaf and flower tissues (Figure 3E, Table 1). However, these common enhancers often generated more consistent and/or stronger GUS signals in one specific tissue in transgenic plants. For example, C4 generated consistent (12/12 transgenic plants) and strong GUS signals in petiole and flower stalk but much weaker and less frequent signals in other tissues.
Three enhancers, C8, L1, and L2, generated relatively weak GUS signals in leaves. However, these weak signals were consistently observed in different transgenic plants, which were clearly different from the inconsistent weak signals observed from the negative control constructs. Thus, these results suggest that these three DNA fragments have relatively low levels of enhancer activity in leaf tissue (Supplemental Table 2).
The Impact of Orientation of the Predicted Enhancers in Reporter Constructs
Enhancers are known to function in an orientation and distance independent manner (Timko et al., 1985; Fluhr et al., 1986; Nagy et al., 1987; Fang et al., 1989). We developed reverse-orientated constructs from all 10 validated enhancers (Figure 3E, Table 1). Most reverse constructs generated similar GUS signal patterns in transgenic plants compared with the corresponding forward constructs. However, the forward and reverse constructs of C2, C4, and C5 produced different GUS expression patterns. Both C2 and C2R (the reverse construct of C2) generated strong GUS signals in leaves, but only C2, not C2R, generated strong signals in flower stalks (Supplemental Table 2). Similarly, both C4 and C4R generated GUS signals in leaves, but only C4, not C4R, generated signals in flower stalks. C5, but not C5R, generated strong signals in stamens (Supplemental Table 2). In contrast, C5R, but not C5, generated weak signals at the branching points of the lateral roots (Table 1).
Open Chromatin Structure Associated with the Transgenic Enhancer L3
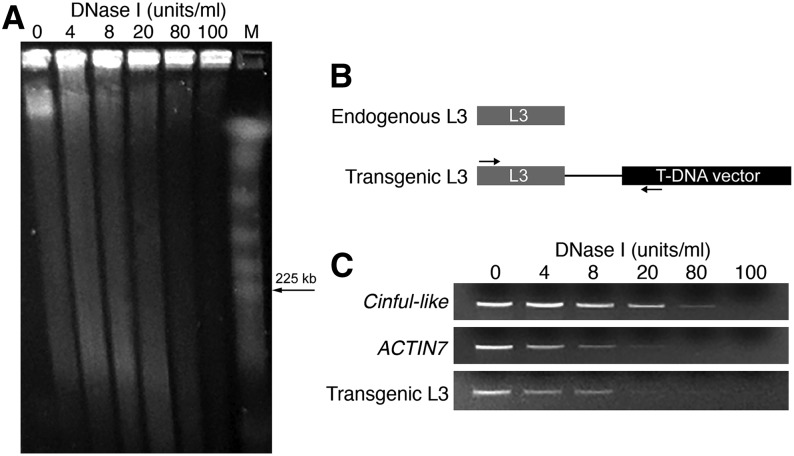
We developed a DNase I-PCR procedure (see Methods) to examine the open chromatin status of a transgenic enhancer. We used a transgenic L3 as a target because the L3 enhancer consistently generated strong GUS signals in both leaf and root tissues (see below). Nuclei were isolated from the leaf tissue of a L3 transgenic line in the T3 generation. The nuclei were digested by a gradient amount of DNase I and the levels of digestion were examined by pulsed-field gel electrophoresis (PFGE) (Figure 4A). DNAs were then isolated from the digested nuclei for PCR analysis. A specific primer pair was designed to distinguish the transgenic L3 from the endogenous L3 (Figure 4B). The silent transposable element gene Cinful-like and active gene ACTIN7 were previously demonstrated to be associated with closed and open chromatin, respectively, using DNase I-PCR (Shu et al., 2013). The transgenic L3 locus showed a similar sensitivity to DNase I digestion to that of the ACTIN7 gene based on the DNase I-PCR assay (Figure 4C), which confirmed that the transgenic L3 locus maintains an open chromatin status.
Figure 4.
DNase I-PCR Assay of the DNase I Sensitivity of a Transgenic Enhancer Sequence.
(A) Gradient DNase I digestion of the same amount of chromatin. The degree of DNase I digestion was assessed by PFGE.
(B) A diagram illustrating the design of a PCR primer that allows the transgenic L3 locus, but not the endogenous L3, to be specifically amplified.
(C) DNase I-PCR shows differential DNase I sensitivity of chromatin in selected genomic regions. The DNase I sensitivity of the transgenic L3 locus was similar to that of the open chromatin control ACTIN7.
Dissection of the Compound Enhancer L3
It was interesting to note that L3, which spans 625 bp, consistently generated strong GUS signals in roots in 16/18 (89%) of the transgenic plants (Figure 5A). In contrast, GUS activity in leaves was detected in only 10/18 (56%) of the transgenic plants. In addition, the GUS signals were more variable in leaves than those observed in root tissues. We suspected that this enhancer may span two or more independent enhancers. To test this hypothesis, we divided the 625-bp L3 fragment into six different subfragments (#1 to #6, respectively; Table 2, Figure 5B). These subfragments were separately cloned into the reporter vector. Construct #5 (198 bp) generated strong GUS signals in roots in 10/12 (83%) of the transgenic plants, but in leaves only in 3/12 (25%) of the transgenic plants (Figure 5C, Table 2). Thus, construct #5 showed similar enhancer activity to that of the entire L3 construct. Interestingly, construct #3 (122 bp) generated GUS signals in leaves in 9/12 (75%) of the transgenic plants, but in roots only in 2/12 (17%) of the transgenic plants. In addition, construct #3 generated strong leaf signals but weak root signals compared with the L3 construct (Figure 5C). These results indicate that L3 is indeed a fused DHS consisting of at least two different enhancers (#5 and #3). The root (#5) enhancer displays a dominant function in reporter assays when both the root and leaf (#3) enhancers are included in a single construct (L3 and #1). Independent #3 construct, which is likely alleviated or freed from the suppression effect from #5, showed a much stronger enhancer activity in leaves.
Figure 5.
Dissection of the Compound Enhancer L3.
(A) Two representative transgenic plants showing GUS expression patterns. The plant on the left shows weak GUS staining in its lower leaves. The plant on the right shows almost no GUS staining in leaves.
(B) A diagram showing the sizes and relative positions of the six subconstructs derived from L3.
(C) GUS expression patterns of three independent and representative transgenic plants derived from each of the six subconstructs.
Table 2. GUS Expression in Transgenic Plants Harboring Constructs Derived from Enhancer L3.
| Enhancer Construct (bp) | GUS Expression in Different Tissues of Transgenic Plants |
||||
|---|---|---|---|---|---|
| Root | Leaf | Petiole | Leaf Vein | Mesophyll | |
| L3 (625) | 16/18a | 10/18b | 2/18 | 8/18 | 2/18 |
| #1 (477) | 12/18 | 6/18 | 2/18 | 5/18 | 1/18 |
| #2 (298) | 7/18 | 6/18 | 3/18 | 4/18 | 1/18 |
| #3 (122) | 2/12 | 9/12 | 5/12 | 7/12 | 6/12 |
| #4 (172) | 1/12 | 2/12 | 2/12 | 1/12 | 1/12 |
| #5 (198) | 10/12 | 3/12 | 3/12 | 2/12 | 2/12 |
| #6 (198) | 4/12 | 4/12 | 3/12 | 0/12 | 1/12 |
Sixteen of the 18 transgenic plants showed GUS expression in roots.
Ten of the 18 transgenic plants showed signals in at least one of the three cell types (petiole, leaf vein, and mesophyll).
A T-DNA Insertion Disrupted the Integral Function of the L3 Enhancer
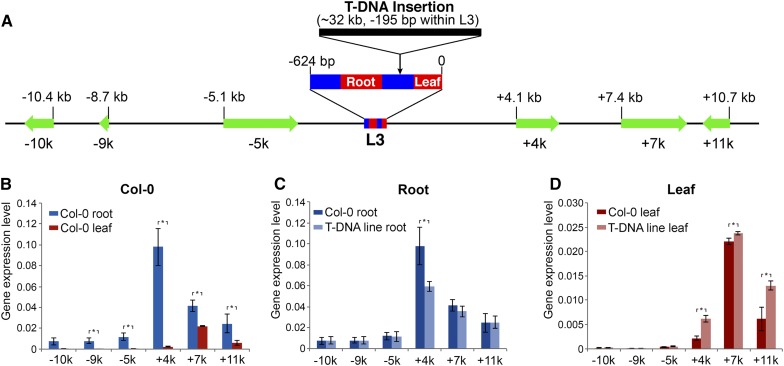
The strong GUS expression in roots driven by the L3 enhancer prompted us to conduct further analysis of the impact of this enhancer on its proximal genes. We identified a T-DNA line (GABI-Kat 909A07) that was inserted in the middle of the L3 enhancer. The T-DNA was inserted in the −195-bp position in L3, which separated the leaf and root enhancers and moved the root-specific enhancer away from its downstream genes (Figure 6A). Our initial PCR-based analysis suggested that the T-DNA insert in this line was not a single 5.7-kb insertion but was either rearranged and/or contained multiple T-DNA copies. Quantitative PCR using primers targeting different regions of the T-DNA indicated that the insert contained approximately four copies of the T-DNA (Supplemental Figure 4A). Genomic DNA from the T-DNA line was digested with XhoI, whose restriction site is absent in the T-DNA sequence, and hybridized with a probe derived from the T-DNA. A single large band (>20 kb) was detected in the T-DNA line (Supplemental Figure 4B). Finally, we conducted DNA fiber-based fluorescence in situ hybridization (fiber-FISH) analysis (Jackson et al., 1998) using the T-DNA as a probe. The fiber-FISH signals averaged 10.0 ± 2.4 μm (n = 30), representing 32.1 ± 7.7 kb (Supplemental Figure 4C). These results showed that this T-DNA line contains a complex insertion consisting of four to five copies of the T-DNA.
Figure 6.
Expression of Six Genes Flanking Enhancer L3 in Wild-Type Col-0 and T-DNA Line GABI-Kat 909A07.
(A) A diagram displaying the relative positions of L3 enhancer and the six flanking gene. The sizes (bp) of the enhancer and six genes are drawn according to scale. The L3 enhancer is enlarged to show the relative positions of the leaf enhancer and root enhancer within L3. The T-DNA insertion, which is nearly 32 kb long, is also enlarged and is not drawn according to scale.
(B) Relative expression levels of six flanking genes in roots vs. leaves in wild-type Col-0. Five genes showed a significantly higher level of expression in roots than in leaves.
(C) The relative expression levels of the six genes in roots in the two lines. Only gene +4k showed a higher expression level in wild-type Col-0 than in the T-DNA line.
(D) The relative expression levels of the six genes in leaves in the two lines. Three genes, +4k, +7k, and +11k, showed higher expression levels in the T-DNA line than in wild-type Col-0. The error bars represent sd based on qRT-PCR data from three biological replicates. All asterisks indicate significant difference (P < 0.05) from pairwise comparison using unpaired t test.
We examined the expression of three genes flanking both sides of the L3 enhancer in wild-type Arabidopsis (Col-0 ecotype) and the T-DNA line. These six genes were named −10k, −9k, −5k, +4k, +7k, and +11k, respectively, based on their distance from the L3 enhancer (Figure 6A). We conducted qRT-PCR analysis of the six flanking genes in both leaves and roots. Interestingly, all six genes, except for −10k, showed higher expression levels in roots than in leaves (Figure 6B; Supplemental Table 3). The +4k gene showed a particularly high level of expression in roots, suggesting that this gene is positively regulated by the root-specific enhancer within L3 (Figure 5). We then compared the expression level of each gene in the same tissue in Col-0 versus the T-DNA line. Surprisingly, the +4k gene showed a significantly higher expression level in Col-0 than in the T-DNA line in root tissue (Figure 6C), suggesting that the insertion of the T-DNA had a negative impact on the expression of this gene in roots. In contrast, the expression levels of the five other genes were similar in Col-0 and the T-DNA line (Figure 6C). In addition, the three downstream genes (+4k, +7k, and +11k) showed higher expression levels in the T-DNA line than in Col-0 in leaves (Figure 6D). We propose that the separation of the leaf enhancer from the root enhancer released the dominant effect of the root enhancer, resulting in an enhanced effect of the leaf enhancer on the three downstream genes. Thus, this T-DNA insert functioned as a enhancer insulator, which is similar to a classical example that a gypsy retrotransposon blocks the function of an upstream enhancer of the Drosophila melanogaster yellow gene (Geyer and Corces, 1992).
DISCUSSION
Efficiency of DHS-Based Enhancer Prediction in Arabidopsis
Enhancer trapping, a technique developed in Drosophila (O’Kane and Gehring, 1987), has been used as a general method for enhancer discovery in plants. An enhancer trap construct, combining a minimal promoter with a reporter gene, can be randomly integrated into the genome of a target plant by transformation. The reporter gene will be activated if the construct is integrated into a position adjacent to an enhancer (or a promoter). A large number of enhancer/promoter trapping lines were developed in several plant species, including Arabidopsis (Sundaresan et al., 1995; Campisi et al., 1999), rice (Oryza sativa) (Wu et al., 2003; Yang et al., 2004; Johnson et al., 2005), and poplar (Populus trichocarpa) (Groover et al., 2004). Unfortunately, the enhancer trapping approach has a major drawback. A positive trapping line would only indicate a potential enhancer located close to the reporter gene. However, this potential enhancer could be located either 5′ or 3′ of the reporter gene at variable distances. Thus, isolation of the enhancer sequence from a trapping line often involves tedious cloning and transformation assays (Yang et al., 2005). Very few enhancers have been isolated using the enhancer trapping approach in plants.
Enhancers in mammalian species can be effectively predicted based on data sets associated with transcriptional cofactors, histone modification marks, and DHSs (Heintzman et al., 2009; Lee et al., 2011; Hnisz et al., 2013). This bioinformatics-based method can correctly locate enhancers that were previously characterized based on experimental data (Heintzman et al., 2009). Computationally predicted enhancers were also validated by luciferase reporter assays. For example, seven out of nine (78%) predicted enhancers in human HeLa cells were confirmed by reporter assays (Heintzman et al., 2009). Similarly, six of seven (86%) predicted mouse neuronal enhancers were confirmed by luciferase reporter assays (Kim et al., 2010). In this study, we validated 10 of the 14 (71%) putative enhancers predicted solely based on DHS data sets. The success rate of our prediction is only slightly below the rates reported in mammalian species. Three of these 14 enhancers, C1-C3, were also identified by the enhancer trapping method (Michael and McClung, 2003). However, only C1 and C2 were validated by our reporter assays. These results show that the DHS-based approach represents a promising enhancer prediction system in plants. This system has two major advantages compared with the traditional enhancer trapping method: (1) It does not require the generation and maintenance of a large number of transgenic lines, which can only be performed in a few plant species. (2) The predicted enhancers are positioned within small genomic regions (<1 kb) and can be readily cloned for validation. In addition, DHS data sets can be readily developed in any plant species with a sequenced genome (Zhang et al., 2012a, 2012b). We should also be able to add additional data sets, such as transcriptional cofactor binding sites, to further refine the prediction parameters and increase the prediction accuracy.
Tissue Specificity of Predicted Enhancers Based on DHS Data Sets
Enhancers often show tissue/organ-specific function. For example, 2543, 561, and 2105 putative enhancers were predicted to be associated with mouse forebrain, midbrain, and limb, respectively. Only 21 of these were found in all three tissues (Visel et al., 2009). A similar specificity was observed for mammalian enhancers developed in several other tissues (Blow et al., 2010; Visel et al., 2013).
We used the relative DNase-seq read enrichment of a DHS in leaves versus flowers to predict the tissue specificity of each putative enhancer. This prediction was remarkably accurate. All three predicted leaf-specific enhancers (L1-L3) showed activities in leaves but not in flowers (Table 1). The two predicted flower-specific enhancers (F1 and F2) generated GUS signals in highly restricted tissues in flowers (Figures 3M and 3N). In addition, only leaf-specific enhancers, not flower-specific enhancers, generated GUS signals in roots, which agrees with the fact that complete seedlings, including roots, were used to generate the “leaf DHS data set.” Therefore, it will be valuable to develop additional DHS data sets from various tissue types in Arabidopsis, similar to the large number of DHS data sets developed from various types of human cell lines (The_ENCODE_Project_Consortium, 2012), which will be critical for developing a complete collection of all enhancers in the Arabidopsis genome.
Number of Enhancers in the Arabidopsis Genome
The exact number of enhancers in the human genome is unclear. However, epigenomic profiling of different tissues and diverse cell lines has revealed that the human genome contains hundreds of thousands of enhancers, which vastly outnumbers the ∼20,000 protein-encoding genes (The_ENCODE_Project_Consortium, 2012; Pennacchio et al., 2013). It is an open question whether the expression of plant genes is regulated by as many enhancers as is that of human genes. Nevertheless, the 10,044 intergenic enhancers predicted in this study is clearly an underestimate of the total number of enhancers in Arabidopsis. First, we only selected DHSs that are 1.5 kb away from a TSS as putative enhancers. However, many plant enhancers reside very close to the promoters of genes, often merely a few hundred base pairs away (Timko et al., 1985; Fluhr et al., 1986; Nagy et al., 1987). Thus, our prediction mainly includes distant intergenic enhancers, but excludes those located very close to the promoters. Second, both leaf and flower are complex tissues consisting of many different cell types. If a genomic region (enhancer) is only associated with regulatory protein(s) in a small percentage of cells in such complex tissues, its corresponding DHS may be masked by the majority of the cells in which the same region is associated with nucleosomes. Thus, our current DHS data sets developed from leaf and flower tissues may not include DHSs associated with less dominant cell types in the tissues. Third, both DHS mapping and GUS reporter-based validation procedures may not detect some weak enhancers. Therefore, obtaining a complete collection of all enhancers in Arabidopsis is a highly desirable goal, yet a daunting challenge for the plant science community.
METHODS
Plant Materials
Seeds of Arabidopsis thaliana ecotype Col-0 and T-DNA line GABI-Kat 909A07 were germinated in one-half-strength Murashige and Skoog medium. The seedlings were grown under 16-h light (white fluorescent lamps with 150 μmol m−2 s−1 light)/8-h dark cycles at 23°C for collecting tissues or were transferred into potting soil and grown under the same light-dark conditions until flowering. The T-DNA line GABI-Kat 909A07 was ordered from Bielefeld University. A homozygote insertion line was isolated by genotyping using PCR primers for enhancer construct L3 (Supplemental Table 1) and primers for T-DNA insertion (gene-specific primer 5′-GTATTATCTCGAAGCTCAGCGTTT-3′, T-DNA primer for PCR 5′-ATAATAACGCTGCGGACATCTACATTTT-3′, and T-DNA primer for sequencing 5′-ATATTGACCATCATACTCATTGC-3′). PCR products were sequenced to confirm the genomic position of the T-DNA insert.
ChIP-seq and Analysis
ChIP was performed following previously published protocols (Nagaki et al., 2003). Both leaf and flower tissues were used in the ChIP experiments. The tissues collected from Arabidopsis Col-0 plants were at the same developmental stages as those used in the original DNase-seq experiments (Zhang et al., 2012a). Thus, the DNase-seq and ChIP-seq data sets are directly comparable. Commercial antibodies against H3K27ac (07-360; Millipore) and H4K27me3 (07-499; Millipore) were used in ChIP experiments. ChIP-seq libraries were developed following previously published protocols (Zhang et al., 2012b) and were sequenced using the Illumina HiSequation 2000 platform. The 100-bp paired-end ChIP-seq sequence reads were mapped to the TAIR10 reference genome using bowtie2 (Langmead and Salzberg, 2012). ChIP-seq data of H3K27me1, H3K27me3, and H3K9me3 developed by Luo et al. (2013) were downloaded and mapped to the reference genome. We aligned the middle site of all predicted enhancers and plotted the ChIP-seq and DNase-seq reads along ±5 kb flanking both sides of each enhancer using a heat map. We calculated normalized ChIP-seq read counts of H3K27ac and H3K27me3 data sets from leaf and flower tissue, respectively. We then used normalized reads from leaf and subtracted normalized reads from flower to determine occupancy change along the ±5-kb regions flanking the middle site of the predicted enhancers.
Enhancer Prediction and Analysis
We previously identified 38,290 and 41,193 DHSs in leaf and flower tissues, respectively, in Arabidopsis (Zhang et al., 2012a). Intergenic DHSs were considered to be potential enhancers. We measured the distance from the middle point of each intergenic DHS to the TSS of its flanking gene(s). Only the DHSs that are at least 1500 bp away from TSSs were selected as putative enhancers. For each predicted enhancer, we selected a gene that has the shortest distance between its TSS and the enhancer. This gene is considered to be the “most proximal gene” of the enhancer. The expression of these most proximal genes were analyzed using the RNA-seq data sets developed from tissues from Arabidopsis plants at the same developmental stages as those used in DNase-seq (Zhang et al., 2012a).
Enhancer Validation
A set of 14 DHSs located in intergenic regions were selected for validation. A GUS reporter system was developed for validation (Supplemental Figure 2). The sequence of each predicted enhancer was amplified from the genomic DNA of Col-0 (Supplemental Table 1) and was ligated with the minimal 35S promoter (−50 to −2 bp) from CaMV. The ligated product was cloned into an entry vector pENTR (Invitrogen catalog number K240020), followed by recombination reaction between the entry vector and pKGWFS 7.0 vector (Karimi et al., 2007). The reporter constructs were transferred into Agrobacterium tumefaciens strain GV3101 and then introduced into Arabidopsis ecotype Col-0 using the floral dip method (Clough and Bent, 1998). For selection of transformants, seeds were selected on solid medium containing kanamycin (50 µg/mL). Plants were grown in soil under normal greenhouse conditions (18 to 22°C, 16/8 h light/dark).
Transgenic plants were examined for GUS activity according to the published procedure (Jefferson, 1987) with only minor modifications. The samples were immersed in GUS-staining solution (100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.1% [v/v] Triton, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, and 0.05% [w/v] X-Gluc), vacuum infiltrated for 10 to 15 min, and incubated at 37°C overnight. The GUS-stained tissues were then cleared in 80% ethanol three times for more than 5 h or until the green pigmentation dissipated. The cleared tissues were observed directly under a microscope. The images were collected using an EPSON Perfection 4180 scanner and a LEICA MZ16F microscope.
DNase I-PCR Assay
Nuclei were isolated from 2-week-old leaf tissues using published protocols (Nagaki et al., 2003). The DNase I-PCR protocol was developed by following a published procedure (Shu et al., 2013) with several modifications. Briefly, aliquots of nuclei in digestion buffer (Roche Applied Science; catalog number 04716728001) were transferred to six PCR tubes and kept on ice. A DNase I (RNase-free; 10 U/μL; Roche Applied Science; catalog number 04716728001) dilution series was prepared by step-wise dilution using digestion buffer. The final DNase I concentrations in the digestion mixtures were 0, 4, 8, 20, 50, and 100 units mL−1, and the final digestion volume was 100 μL. The digestion mixtures were incubated at 37°C for 2 min and then stopped by adding 57 μL digestion buffer and 17 μL 0.5 M EDTA. Equal volume of 2% low melt agarose gel was added to the tube and mixed by pipetting. Eighty-five microliters of nuclei and agarose gel mixture from last step was transferred into Bio-Rad plug modes (catalog number 170-3713), using four plugs for each digestion. One of the four plugs was used for PFGE (20 to 60 switch time, 17.5 h, 6 V/cm), and the remaining three plugs were used for the DNA recovery. DNA was recovered by processing in order with an equal volume of phenol, phenol:chloroform, and chloroform, respectively. The precipitated DNA was dissolved in 90 μL of nuclease-free water, 1 μL (10 ng) of which was used for each PCR reaction. PCR was performed with 20 μL total reaction volume using PrimeSTAR GXL DNA polymerase (Takara R050A) according to the manufacturer’s protocol. The promoter region (considered to be open chromatin) of ACTIN7 (1646 bp) was selected as a positive control, while a transposable element gene, Cinful-like (1651 bp) (considered to be closed chromatin), was used as a negative control (Shu et al., 2013). The DNase I sensitivity of the transgenic L3 locus (1512 bp) in a transgenic line was examined and compared with the controls. Primers for amplifying the transgenic L3 locus are: forward, 5′-caccAATCGGGATGGTCCGCATAA-3′; reverse: 5′-TCGGCCATGATATAGACGTT-3′.
qRT-PCR, DNA Gel Blot Hybridization, and Fiber-FISH
Total RNA was extracted using Trizol reagent (Invitrogen). Approximately 5 µg of total RNA was isolated from triplicate samples of leaf and root tissues from Col-0 and T-DNA insertion line GABI-Kat 909A07, and cDNA was synthesized using the Superscript first-strand synthesis kit (Invitrogen) according to the manufacturer’s instructions. PCR amplification was performed with SYBR Green Real-time PCR Master Mix reagent (Toyobo) on a StepOne Plus Real-Time PCR apparatus (Applied Biosystems), using the following program: 95°C for 1 min, 40 cycles of 95°C for 5 s, and 60°C for 1 min. Primers for all analyzed genes were designed using the Primer 3 and BLAST programs. Gene ACT2 (At3g18780) (Czechowski et al., 2005) was used to normalize the expression data. The relative expression level of each gene in different tissues (leaf and flower) of Col-0 or in the same tissues of different lines (Col-0 and T-DNA line) was calculated using the 2−△CT method (Schmittgen and Livak, 2008). DNA gel blot hybridization and fiber-FISH were performed according to previously published protocols (Jackson et al., 1998; Miller et al., 1998)
Accession Numbers
The histone modification ChIP-seq data sets have been submitted to the National Center for Biotechnology Information databases under BioProject ID PRJNA252965.
Supplemental Data
Supplemental Figure 1. Histone modification patterns associated with predicted enhancers in Arabidopsis.
Supplemental Figure 2. Diagram of the development of enhancer validation constructs.
Supplemental Figure 3. Heat map of GUS expression in different tissues of transgenic plants transformed with 10 nonenhancer constructs and control constructs.
Supplemental Figure 4. Characterization of the T-DNA inserted in L3 enhancer.
Supplemental Table 1. Genomic locations and primers for all predicted enhancers and nonenhancer controls.
Supplemental Table 2. GUS expression in transgenic plants derived from all enhancer constructs and nonenhancer control constructs.
Supplemental Table 3. Expression of six genes flanking enhancer L3 in wild-type Col-0 and T-DNA line GABI-Kat 909A07.
Supplemental Data Set 1. Chromosomal locations, positions relative to adjacent genes of all predicted enhancers.
Supplementary Material
Acknowledgments
We thank Patrick Krysan for valuable comments on the article and Yufeng Wu, Qunyan He, and Yonghua Han for technical assistance. This work was supported by the Program for Introducing Talents to Universities (B07017) to B.L. and Grants MCB-0923640 and MCB-1412948 from the National Science Foundation and a Vilas Associate Fellowship from the University of Wisconsin-Madison to J.J.
AUTHOR CONTRIBUTIONS
J.J. conceived the research. B.Z. and W.Z. performed the experiments. B.Z., T.Z., and J.J. analyzed the data. J.J., B.L., B.Z., W.Z., and T.Z. wrote the article.
Glossary
- CRE
cis-regulatory DNA element
- TF
transcription factor
- TSS
transcription start site
- DHS
DNase I hypersensitive site
- DNase-seq
DNase I digestion followed by high-throughput sequencing
- FPKM
fragments per kilobase of transcript per million mapped reads
- ChIP-seq
chromatin immunoprecipitation followed by sequencing
- lincRNA
long intergenic noncoding RNA
- ncRNA
noncoding RNA
- PFGE
pulsed-field gel electrophoresis
- fiber-FISH
fiber-based fluorescence in situ hybridization
Footnotes
Articles can be viewed online without a subscription.
References
- Andersson R., et al. ; FANTOM Consortium (2014). An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey P.N., Ren L., Chua N.H. (1989). The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 8: 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow M.J., et al. (2010). ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 42: 806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., Groudine M. (2011). Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi L., Yang Y., Yi Y., Heilig E., Herman B., Cassista A.J., Allen D.W., Xiang H., Jack T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17: 699–707. [DOI] [PubMed] [Google Scholar]
- Clark R.M., Wagler T.N., Quijada P., Doebley J. (2006). A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38: 594–597. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Creyghton M.P., et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107: 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., Tusi B.K., Muller H., Ragoussis J., Wei C.L., Natoli G. (2010). A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 8: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R.X., Nagy F., Sivasubramaniam S., Chua N.H. (1989). Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell 1: 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Cokus S.J., Schubert V., Zhai J., Pellegrini M., Jacobsen S.E. (2014). Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell 55: 694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr R., Kuhlemeier C., Nagy F., Chua N.H. (1986). Organ-specific and light-induced expression of plant genes. Science 232: 1106–1112. [DOI] [PubMed] [Google Scholar]
- Geyer P.K., Corces V.G. (1992). DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6: 1865–1873. [DOI] [PubMed] [Google Scholar]
- Ghavi-Helm Y., Klein F.A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E.E.M. (2014). Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512: 96–100. [DOI] [PubMed] [Google Scholar]
- Groover A., Fontana J.R., Dupper G., Ma C., Martienssen R., Strauss S., Meilan R. (2004). Gene and enhancer trap tagging of vascular-expressed genes in poplar trees. Plant Physiol. 134: 1742–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D.S., Garrard W.T. (1988). Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57: 159–197. [DOI] [PubMed] [Google Scholar]
- Guertin M.J., Martins A.L., Siepel A., Lis J.T. (2012). Accurate prediction of inducible transcription factor binding intensities in vivo. PLoS Genet. 8: e1002610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.K., Song J.D., Noh Y.S., Noh B. (2007). Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J. 49: 103–114. [DOI] [PubMed] [Google Scholar]
- Heintzman N.D., et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39: 311–318. [DOI] [PubMed] [Google Scholar]
- Heintzman N.D., et al. (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J.G., Sakai A., Loeb G.B., Ahmad K. (2009). Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 19: 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. (2013). Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.A., Wang M.L., Goodman H.M., Jiang J. (1998). Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41: 566–572. [PubMed] [Google Scholar]
- Jefferson R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5: 387–405. [Google Scholar]
- Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G. (2009). H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 41: 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.A.T., Hibberd J.M., Gay C., Essah P.A., Haseloff J., Tester M., Guiderdoni E. (2005). Spatial control of transgene expression in rice (Oryza sativa L.) using the GAL4 enhancer trapping system. Plant J. 41: 779–789. [DOI] [PubMed] [Google Scholar]
- Karimi M., Depicker A., Hilson P. (2007). Recombinational cloning with plant gateway vectors. Plant Physiol. 145: 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd B.N., Cahill D.M., Manners J.M., Schenk P.M., Kazan K. (2011). Diverse roles of the Mediator complex in plants. Semin. Cell Dev. Biol. 22: 741–748. [DOI] [PubMed] [Google Scholar]
- Kim T.K., et al. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon E.Z., Kazmar T., Stampfel G., Yáñez-Cuna J.O., Pagani M., Schernhuber K., Dickson B.J., Stark A. (2014). Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512: 91–95. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Karchin R., Beer M.A. (2011). Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res. 21: 2167–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L., Arenas-Huertero C., Chua N.H. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Sidote D.J., Zhang Y., Kerstetter R.A., Michael T.P., Lam E. (2013). Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. 73: 77–90. [DOI] [PubMed] [Google Scholar]
- McGarry R.C., Ayre B.G. (2008). A DNA element between At4g28630 and At4g28640 confers companion-cell specific expression following the sink-to-source transition in mature minor vein phloem. Planta 228: 839–849. [DOI] [PubMed] [Google Scholar]
- Mendenhall E.M., Williamson K.E., Reyon D., Zou J.Y., Ram O., Joung J.K., Bernstein B.E. (2013). Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 31: 1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., McClung C.R. (2003). Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 132: 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.T., Dong F., Jackson S.A., Song J., Jiang J. (1998). Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics 150: 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näär A.M., Lemon B.D., Tjian R. (2001). Transcriptional coactivator complexes. Annu. Rev. Biochem. 70: 475–501. [DOI] [PubMed] [Google Scholar]
- Nagaki K., Talbert P.B., Zhong C.X., Dawe R.K., Henikoff S., Jiang J. (2003). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163: 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F., Boutry M., Hsu M.Y., Wong M., Chua N.H. (1987). The 5′-proximal region of the wheat Cab-1 gene contains a 268-bp enhancer-like sequence for phytochrome response. EMBO J. 6: 2537–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kane C.J., Gehring W.J. (1987). Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. USA 84: 9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom U.A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q., Guigo R., Shiekhattar R. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A., Bickmore W., Dean A., Nobrega M.A., Bejerano G. (2013). Enhancers: five essential questions. Nat. Rev. Genet. 14: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raatz B., Eicker A., Schmitz G., Fuss E., Müller D., Rossmann S., Theres K. (2011). Specific expression of LATERAL SUPPRESSOR is controlled by an evolutionarily conserved 3′ enhancer. Plant J. 68: 400–412. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer S.E., Schlüter P.M., Baskar R., Gheyselinck J., Bolaños A., Curtis M.D., Grossniklaus U. (2009). Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. Plant J. 59: 987–1000. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V., Ren B. (2012). A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H., Gruissem W., Hennig L. (2013). Measuring Arabidopsis chromatin accessibility using DNase I-polymerase chain reaction and DNase I-chip assays. Plant Physiol. 162: 1794–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V., Springer P., Volpe T., Haward S., Jones J.D.G., Dean C., Ma H., Martienssen R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810. [DOI] [PubMed] [Google Scholar]
- The_ENCODE_Project_Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed]
- Timko M.P., Kausch A.P., Castresana C., Fassler J., Herrera-Estrella L., Van den Broeck G., Van Montagu M., Schell J., Cashmore A.R. (1985). Light regulation of plant gene expression by an upstream enhancer-like element. Nature 318: 579–582. [DOI] [PubMed] [Google Scholar]
- Visel A., et al. (2009). ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., et al. (2013). A high-resolution enhancer atlas of the developing telencephalon. Cell 152: 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu C., Roqueiro D., Grimm D., Schwab R., Becker C., Lanz C., Weigel D. (2015). Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 25: 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Li X., Yuan W., Chen G., Kilian A., Li J., Xu C., Li X., Zhou D.X., Wang S., Zhang Q. (2003). Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 35: 418–427. [DOI] [PubMed] [Google Scholar]
- Yang W., Jefferson R.A., Huttner E., Moore J.M., Gagliano W.B., Grossniklaus U. (2005). An egg apparatus-specific enhancer of Arabidopsis, identified by enhancer detection. Plant Physiol. 139: 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.Z., Peng H., Huang H.M., Wu J.X., Ha S.R., Huang D.F., Lu T.G. (2004). Large-scale production of enhancer trapping lines for rice functional genomics. Plant Sci. 167: 281–288. [Google Scholar]
- Zentner G.E., Tesar P.J., Scacheri P.C. (2011). Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 21: 1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhang T., Wu Y., Jiang J. (2012a). Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell 24: 2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wu Y., Schnable J.C., Zeng Z., Freeling M., Crawford G.E., Jiang J. (2012b). High-resolution mapping of open chromatin in the rice genome. Genome Res. 22: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.