A strategy for the design of novel enzymatic cores for bacterial microcompartments was devised and tested in cyanobacteria, which have potential uses as metabolic modules in plants.
Abstract
Bacterial microcompartments (BMCs) are self-assembling organelles that sequester segments of biochemical pathways within a protein shell. Given their functional diversity, BMCs constitute a rich source of metabolic modules for applications in synthetic biology. The carboxysome, the cyanobacterial BMC for CO2 fixation, has attracted significant attention as a target for installation into chloroplasts and serves as the foundation for introducing other types of BMCs into plants. Carboxysome assembly involves a series of protein-protein interactions among at least six gene products to form a metabolic core, around which the shell assembles. This complexity creates significant challenges for the transfer, regulation, and assembly of carboxysomes, or any of the myriad of functionally distinct BMCs, into heterologous systems. To overcome this bottleneck, we constructed a chimeric protein in the cyanobacterium Synechococcus elongatus that structurally and functionally replaces four gene products required for carboxysome formation. The protein was designed based on protein domain interactions in the carboxysome core. The resulting streamlined carboxysomes support photosynthesis. This strategy obviates the need to regulate multiple genes and decreases the genetic load required for carboxysome assembly in heterologous systems. More broadly, the reengineered carboxysomes represent a proof of concept for a domain fusion approach to building multifunctional enzymatic cores that should be generally applicable to the engineering of BMCs for new functions and cellular contexts.
INTRODUCTION
Bacterial microcompartments (BMCs) are a family of architecturally similar but functionally diverse self-assembling organelles composed entirely of protein (Axen et al., 2014; Kerfeld and Erbilgin, 2015). The first BMC identified was the carboxysome (Drews and Niklowitz, 1956). This ∼300-MD compartment (Cameron et al., 2013) is part of the cyanobacterial carbon concentrating mechanism (ccm) and enhances carbon fixation by sequestering Rubisco and carbonic anhydrase (CA) within a protein shell. In the carboxysome lumen, bicarbonate is converted into CO2 by CA; this effectively increases the proportion of CO2 to O2 in the vicinity of Rubisco. This favors Rubisco’s carboxylase activity, while the shell limits the loss of CO2 into the bulk cytosol (Cai et al., 2009). Two types of carboxysomes are found in cyanobacteria: α-carboxysomes, which contain form 1A Rubisco; and β-carboxysomes, which contain form 1B Rubisco (the type of Rubisco found in plants) (Price et al., 2008). The constituent core proteins also differ between the two types of carboxysome, and they appear to differ in mode of assembly. Recently it was proposed that a large, conserved multidomain protein (CsoS2) organizes the Rubisco in the α-carboxysome core (Cai et al., 2015). In contrast, assembly of the β-carboxysome is known to involve a sequence of protein domain interactions among multiple core proteins (Cameron et al., 2013).
Carboxysomes have captured the interest of researchers because of their proposed potential for enhancing CO2 fixation in plants (Price et al., 2013; Zarzycki et al., 2013). Recent efforts have led to important advances in expressing the β-carboxysome shell and cyanobacterial form 1B Rubisco in chloroplasts (Lin et al., 2014a, 2014b), which represent the first steps in assembling carboxysomes in plants. More broadly, these results bode well for the prospect of engineering novel types of metabolic nanoreactors based on BMC architectures in heterologous systems (Frank et al., 2013), including cyanobacteria and other microbial platforms for renewable chemical and biofuel production. However, for the realization of these goals a major hurdle remains: the assembly of the functional multiprotein metabolic core.
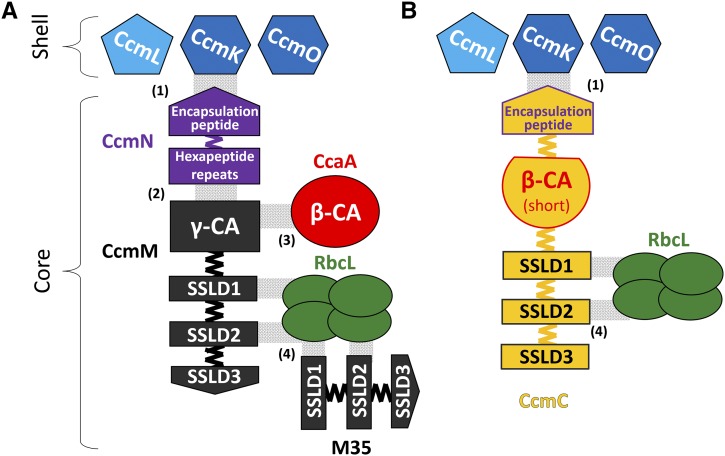
The cyanobacterial carboxysome offers an ideal system for testing strategies for developing engineered BMC cores because functional carboxysomes are essential for the survival of the cyanobacterial host. Furthermore, many of the subunits of the β-carboxysome have been structurally and functionally characterized. In Synechococcus elongatus PCC 7942 (hereafter Syn 7942), the β-carboxysome shell is formed by the structural proteins CcmK, CcmL, and CcmO; the core of the carboxysome is composed of CcmM, M35, and CcmN; the enzyme Rubisco (form 1B); and a β-CA, CcaA (Figure 1A). CcmM comprises an N-terminal γ-CA domain followed by three small subunit-like domains (SSLDs) with sequence homology to RbcS, the small subunit of Rubisco (Long et al., 2007). The ccmM gene encodes two essential carboxysome components: the full-length protein and a truncated form containing only the SSLDs (known as M35). CcmN contains multiple hexapeptide-repeats and, on its C terminus, an encapsulation peptide (EP), which is a short α-helical segment linked to the hexapeptide-repeat domains by a poorly conserved linker sequence (Kinney et al., 2012). While CcmM/M35 and CcmN are absolutely conserved and essential to carboxysome formation (Long et al., 2010; Kinney et al., 2012), CcaA deletion mutants still form carboxysomes (So et al., 2002b) but exhibit a high CO2-requiring (hcr) phenotype. Furthermore, many cyanobacteria lack CcaA (Zarzycki et al., 2013) and its function is presumably replaced by the γ-CA domain of CcmM (Peña et al., 2010). Given the challenges associated with introducing and regulating the expression of multiple genes necessary to assemble a BMC core in heterologous systems, including plants, a redesign to simplify the construction of the metabolic core is necessary.
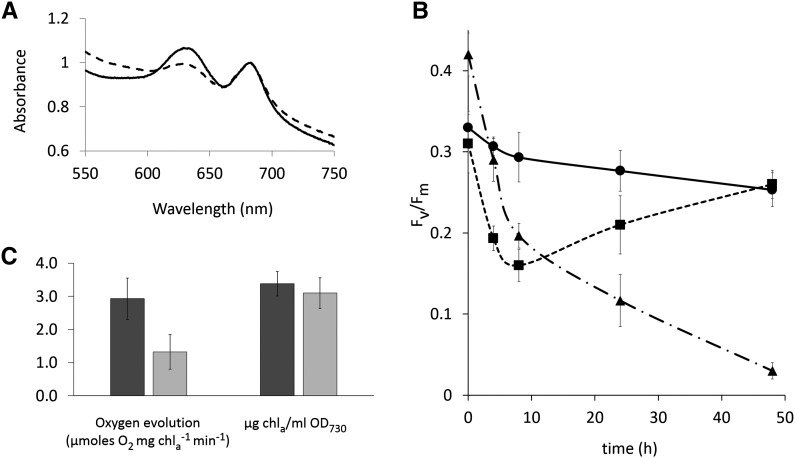
Figure 1.
Schematics of the Wild-Type and Streamlined Carboxysome Cores.
Assembly of the native β-carboxysome core (A) and assembly of the designed carboxysome core by the chimeric protein CcmC (yellow) (B). The SSLDs are numbered from the N (SSLD1) to C terminus (SSLD3). The specific details of their interactions with the large subunit of Rubisco are unknown, but it is assumed that they displace some of the RbcS subunits, which are not shown. Domains are colored as in Supplemental Figure 1, shell proteins are shown in blue, and four RbcL subunits of the L8S8 complex of Rubisco are shown in green. Gray shading denotes known noncovalent domain interactions with a numbers in parenthesis for the corresponding reference: (1) and (2) from Kinney et al. (2012), (3) from Long et al. (2007), and (4) from Long et al. (2010).
Here, we report the results of a protein domain-based approach to engineering a streamlined carboxysome core; we combined segments of four carboxysome components into a single chimeric protein. The synthetic core protein supports the assembly of functionally competent carboxysomes in cyanobacteria. These results not only provide an important advance for the installation of carboxysomes and other BMCs into chloroplasts, but they also represent a proof of concept of a design strategy based on protein domain interactions for constructing BMC cores with new functions.
RESULTS
Design of a Chimeric Protein That Supports Carboxysome Core Assembly and Growth in Air
Our approach took advantage of the observation that proteins evolve via domain fusions that are reflective of protein-protein interactions (Marsh et al., 2013). We predicted the domain boundaries in CcmM, CcmN, and CcaA of Syn 7942 using InterPro (Hunter et al., 2012) (Supplemental Figure 1A). We then constructed three chimeric genes whose protein products we surmised could potentially assemble into a carboxysome core: (1) ccaA-M35: the γ-CA domain (Pfam00132) of CcmM was replaced by β-CA (Pfam00484) (Supplemental Figure 2B); (2) M35-EP: the three SSLD domains (Pfam00101) and their native linkers were fused to the EP (Supplemental Figure 2C); and (3) M35-ccaA(short)-EP: contains three SSLDs and their native linkers, the β-CA, CcaA with a short segment of its C-terminal tail (So et al., 2002a) as a linker, and the EP from the C terminus of CcmN (Figure 1B; Supplemental Figures 1B and 2D).
A gene coding for a GFP-labeled large subunit of Rubisco (rbcL-GFP) was inserted into each strain for in vivo visualization of carboxysome formation by fluorescence microscopy (Savage et al., 2010). To test whether the chimeric proteins can assemble into a carboxysome core, Syn 7942 ccmM and ccmN were replaced with selectable marker genes (COREΔ2/RbcL-GFP strain; hcr phenotype). The chimeric genes (in the case of ccaA-M35, the ccmN gene was reintroduced in the same vector) were then transformed via double homologous recombination to replace the selectable markers of the COREΔ2/RbcL-GFP strain (placing the genes under the same regulation of the ccm operon genes) using growth in air as positive selection. Only M35-ccaA(short)-EP expression was able to rescue the hcr phenotype; we named this construct CcmC (“C” for chimeric) (Figure 1B). The resulting strain (COREΔ2/CcmC/RbcL-GFP) contains the original ccaA in its genome; therefore, to further substantiate the evident functional rescue by CcmC, we replaced ccaA with a gentamycin resistance gene (resulting in strain COREΔ3/CcmC/RbcL-GFP); this triple mutant strain was still able to grow in air.
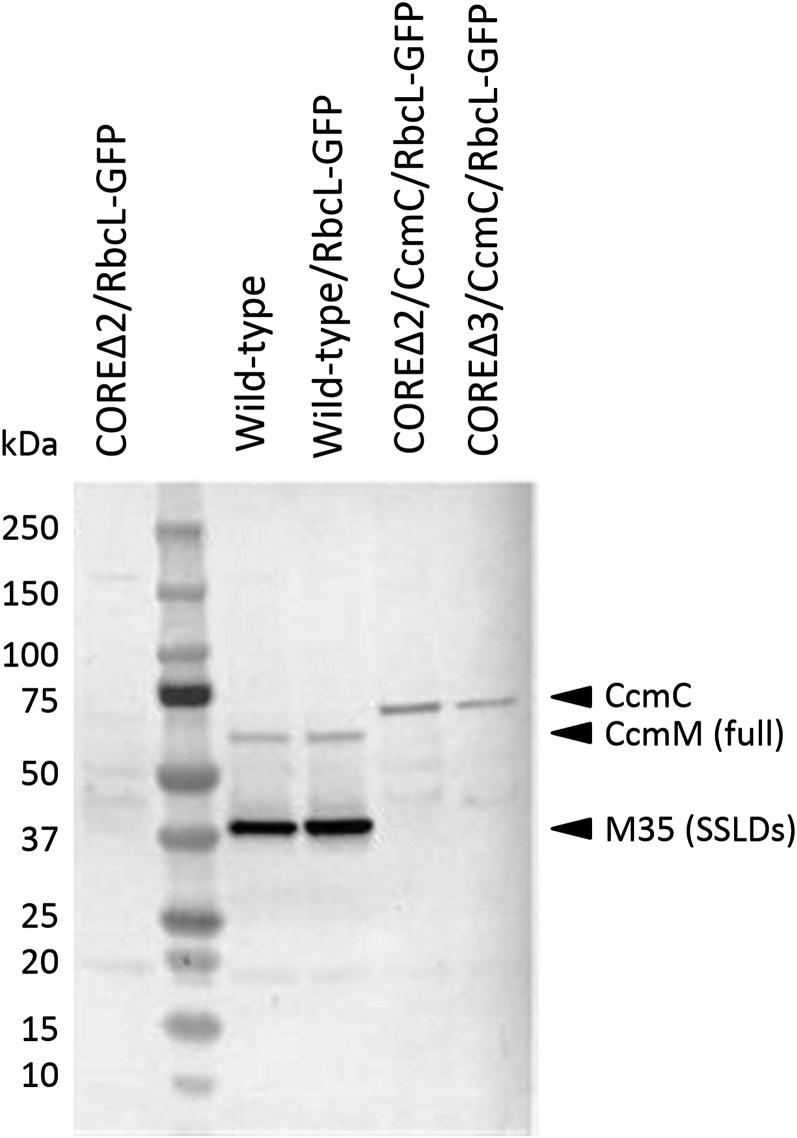
We confirmed the presence/absence of ccmM, ccmN, and ccaA by PCR (Supplemental Figure 3). Sequencing of the region between ccmL and ccmO further indicated that ccmC was integrated into the ccm operon (Supplemental Figure 4). Protein screening by immunoblot using polyclonal anti-CcmM antibodies showed no cross-reactivity with a total protein extract of the COREΔ2/RbcL-GFP strain, confirming the absence of those proteins (Figure 2). In contrast, signals at ∼37 kD (major) and at ∼63 kD (minor) were observed in the wild type and in the control strain expressing rbcL-GFP (hereafter wild-type/RbcL-GFP strain), corresponding to the two forms of CcmM required for carboxysome assembly in wild-type Syn 7942 carboxysomes (M35 and full-length CcmM; So et al., 2002b; Long et al., 2010). These two bands are absent in the COREΔ2/CcmC/RbcL-GFP and COREΔ3/CcmC/RbcL-GFP strains and replaced by cross-reactivity at ∼75 kD, corresponding to the fusion protein (predicted mass of 67 kD; Figure 2).
Figure 2.
Cross-Reactivity of the Chimeric Protein CcmC with Anti-CcmM Antibodies.
Whole cell lysates were blotted and probed using anti-CcmM antibodies. Protein extracts from controls (wild-type background) show two bands corresponding to the full-length and short form of CcmM, while the mutants (CcmC background) show one band due to the cross-reactivity of the antibody with the small subunit-like domains.
CcmC Structurally Replaces Four Proteins of the β-Carboxysome Core
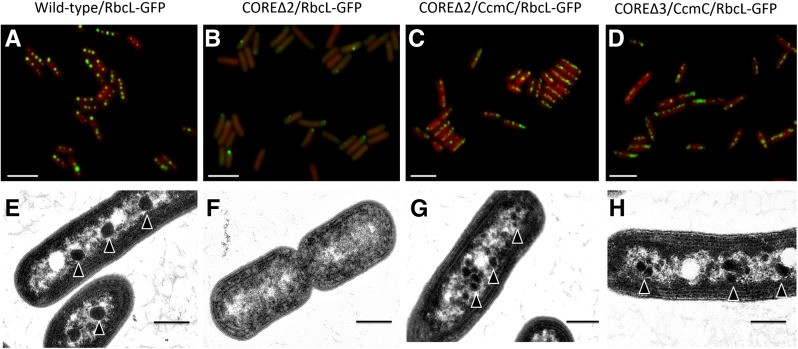
We used fluorescence and transmission electron microscopy to assay for formation of carboxysomes (Figure 3). In the wild-type/RbcL-GFP strain, the carboxysomes were in the typical arrangement, along the longitudinal axes of the cells (Figure 3A). RbcL-GFP in the COREΔ2/RbcL-GFP strain was diffuse throughout the cell, as expected for strains lacking carboxysomes (Cameron et al., 2013) (Figure 3B). Occasional polar foci (n = 150/556) were observed; these may be due to misfolded and aggregated labeled protein (polar localization of protein aggregates [Rokney et al., 2009] and false foci [Landgraf et al., 2012] have been observed in Escherichia coli) or due to interaction with the remaining gene products of the carboxysome operon. They are not indicative of carboxysome formation, as the COREΔ2/RbcL-GFP strain has an hcr phenotype. In contrast, abundant GFP-labeled carboxysomes were observed in the mutant strains COREΔ2/CcmC/RbcL-GFP and COREΔ3/CcmC/RbcL-GFP, and, although occasionally clustered, they still localized along the longitudinal axis of the cell (Figures 3C and 3D, respectively). The average carboxysome number (fluorescent puncta across the longitudinal plane) per cell in the wild-type/RbcL-GFP strain was 3.7 ± 1.1, while COREΔ2/CcmC/RbcL-GFP averaged 6.4 ± 1.8 and COREΔ3/CcmC/RbcL-GFP averaged 6.4 ± 2.0 (Supplemental Figure 5A).
Figure 3.
Structural Complementation of the Carboxysome Core Deletion Strains with the Chimeric Protein CcmC.
Top panel: Fluorescence microscopy of the strains expressing RbcL-GFP for carboxysome visualization of wild-type/RbcL-GFP (A), COREΔ2/RbcL-GFP (B), COREΔ2/CcmC/RbcL-GFP (C), and COREΔ3/CcmC/RbcL-GFP (D). Bottom panel: Electron micrographs of the same strains after incubation for at least 12 h in air in wild-type/RbcL-GFP (E), COREΔ2/RbcL-GFP (F), COREΔ2/CcmC/RbcL-GFP (G), and COREΔ3/CcmC/RbcL-GFP (H). Arrowheads indicate carboxysomes. Bars = 5 µm in (A) to (D) and 500 nm in (E) to (H).
We compared the amount of Rubisco per mg chlorophyll a by immunoblotting using antibodies against the large subunit RbcL. Both COREΔ2/CcmC/RbcL-GFP and COREΔ3/CcmC/RbcL-GFP strains contained more than a 2-fold increase in RbcL relative to the wild-type/RbcL strain (Supplemental Figure 5B). Analysis by transmission electron microscopy further confirmed carboxysome formation of native (Figure 3E) and streamlined carboxysomes (Figures 3G and 3H). The chimeric carboxysomes were smaller (average carboxysome diameter for wild-type/RbcL-GFP, 185 ± 28 nm; COREΔ2/CcmC, 103 ± 25 nm; and COREΔ3/CcmC, 95 ± 19 nm; Supplemental Figure 5C) and typically more numerous and clustered relative to wild-type carboxysomes. Abnormally shaped carboxysomes were occasionally observed (“rod carboxysomes”; Supplemental Figure 6); these have also been observed in the wild type (Gantt and Conti, 1969) and have been proposed to be a type of intermediate during carboxysome formation (Chen et al., 2013). Based on our data, these rod carboxysomes could also be indicative of a deficiency in CA activity, as carboxysome aggregation and morphological variation were also observed in the control strain ΔCcaA/RbcL-GFP (Supplemental Figure 7).
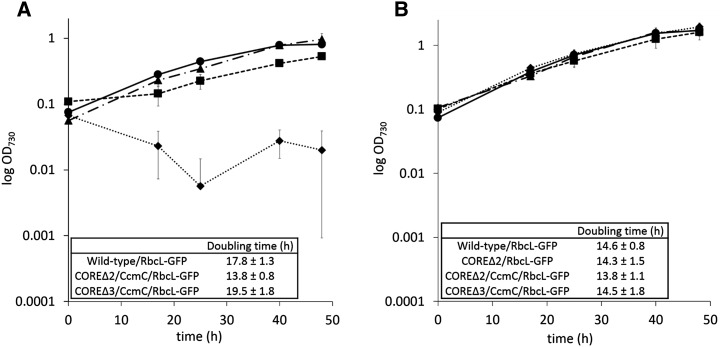
To determine if the reengineered carboxysomes function comparably to the wild-type/RbcL-GFP carboxysomes, we analyzed the growth of cells at the exponential phase under high CO2 (5%) and low CO2 (air) conditions. No growth difference was observed between the strains when incubated in high CO2 (Figure 4), as under these conditions, cyanobacterial CO2 fixation does not depend upon proper carboxysome formation. As expected, the COREΔ2/RbcL-GFP strain failed to grow in air, whereas the other strains were able to grow (Figure 4). The COREΔ2/CcmC/RbcL-GFP strain has the fastest doubling time among the reengineered strains tested, while the growth rates of COREΔ3/CcmC/RbcL-GFP and the wild-type/RbcL-GFP strain are comparable (Figure 4).
Figure 4.
Functional Complementation of the Carboxysome Core Deletion with the Chimeric Protein CcmC.
Changes in optical density (730 nm) over time of independent cultures grown in air (A) and 5% CO2 (B). n = 3; error bars = sd. Insets show doubling times calculated by exponential regression curve fitting (www.doubling-time.com/compute.php).
Physiology of a Cyanobacterial Strain with a Streamlined Carboxysome
We further examined the physiology of the triple deletion strain containing carboxysomes with synthetic cores (COREΔ3/CcmC/RbcL-GFP). The strain has an obvious pigmentation difference when compared with wild-type/RbcL-GFP, which can be attributed to decreased phycobilisome content (Figure 5A). We then measured the relative photosynthetic capacities of photosystem II (through quantification of chlorophyll fluorescence in dark adapted cells, Fv/Fm) upon transfer of the cultures from 3% CO2 to air (Figure 5B); Fv/Fm is widely used as a measure of the efficiency of the photosynthetic electron transport chain, which generates the ATP and reducing power that is consumed by the Calvin-Benson-Bassham (CBB) cycle (Baker, 2008). Accordingly, Fv/Fm has been used as a proxy for carboxysome function; carboxysome-deficient strains of Syn 7942 have an Fv/Fm approximating zero in 3% CO2 (Cameron et al., 2013). While the Fv/Fm of wild-type/RbcL-GFP remains relatively constant, a sharp decrease in Fv/Fm relative to the high-CO2 values is observed in both mutant core strains. The Fv/Fm in the COREΔ2/RbcL-GFP control strain declines toward zero and does not recover. The COREΔ3/CcmC/RbcL-GFP strain adapts within ∼10 h after the CO2 step-down and eventually reaches the same fluorescence levels as the wild-type/RbcL-GFP strain. As an additional complementary measure of photosynthetic activity, we compared oxygen evolution rates of air-grown cultures tested at high light intensity (950 µmoles photons m−2 s−1). Wild-type/RbcL-GFP produced more oxygen than the COREΔ3/CcmC/RbcL-GFP strain (2.9 ± 1.0 versus 1.3 ± 0.5 µmoles O2 µg Chla−1 min−1, respectively). These results demonstrate that the altered composition of the core has a net effect on the physiology of the cell relative to the wild-type/RbcL-GFP control (Figure 5); nevertheless, the reengineered core is immediately able to effectively support functional carboxysome assembly (Figures 3C and 3D) and photosynthesis (Figure 4).
Figure 5.
Comparison of Physiological Parameters in Wild-Type versus CcmC Backgrounds.
(A) Average absorbance spectra of whole cell suspensions normalized to chlorophyll a (663 nm).
(B) Change over time of Fv/Fm in cultures grown at 3% CO2 and transferred to air at time = 0 h.
(C) Oxygen evolution rates (normalized to chlorophyll a) at high light intensity of strains grown in air and supplemented with 10 mM bicarbonate and comparison of chlorophyll a per mL of OD730 culture. Solid line and dark-gray bar, wild-type/RbcL-GFP; dashed line and light-gray bar, COREΔ3/CcmC/RbcL-GFP; dash-dotted line, COREΔ2/ RbcL-GFP. n = 3 in (A) and (B); n ≤ 5 in (C). Error bars = sd.
DISCUSSION
The form and function of the carboxysome have been optimized by natural selection over millions of years. In this study, by considering the order of protein domain interactions in its assembly, it was possible, in a single step, to engineer a simplified carboxysome core by combining select portions of four gene products into a single chimeric protein (Figure 1). Our results support a domain-centric, as opposed to gene-centric, view of protein function (Kerfeld and Erbilgin, 2015; Lluch-Senar et al., 2015). Domain selection and ordering were key considerations in the design of chimeric cores. Two fusion designs failed to assemble functional carboxysome cores: the protein CcaA-M35 (which presumably fails to interact with CcmN, thus the formed core lacks an EP) and the M35-EP protein (which would require CcaA to have intrinsic potential for encapsulation). These results substantiate our current understanding of carboxysome assembly: the EP and an active CA are necessary for the formation of functional carboxysomes, while some domains (the γ-CA domain of CcmM and the hexapeptide repeats of CcmN) are expendable.
In CcmC, the SSLDs and the EP are fused to opposite ends of the β-CA domain; the SSLDs are available to interact with the large subunit of Rubisco and the EP can interact with the shell (Figure 1B; Supplemental Figure 1B). The resulting 67-kD chimeric protein replaces the 58-kD CcmM, the 35-kD M35, the 16-kD CcmN, and the 30-kD CcaA. The synthetic protein reduces the genomic load required to assemble a carboxysome by ∼1100 bp (18% of total message required for carboxysomes). Likewise, it reduces the number of proteins and, concomitantly, the need to balance the expression levels of four different genes. More broadly, these data demonstrate that functional domains from multiple core proteins of a BMC can be assembled on a single chimeric protein. CcmC contains scaffolding domains (the SSLDs are involved in nucleating Rubisco), an enzymatic domain (the CA domain), and an encapsulating domain (the EP). This strategy could be applied to the development of novel nanoreactors based on BMC architectures.
Specifically, the design and construction of a chimeric, simplified carboxysome core represents an advance in available approaches for introducing carboxysomes and other BMCs into plants, where the transfer of multiple genes presents a formidable challenge. However, the use of a fusion strategy does come at the expense of regulatory flexibility. The fixed stoichiometry of domains in CcmC (three SSLDs, one CA, and one EP) may account for the difference in efficiency between chimeric and native carboxysomes. For example, disruption of the CBB cycle increases the photoinactivation of photosystem II (Takahashi and Murata, 2005). The altered physiology of the strains with chimeric carboxysomes compared with the wild-type/RbcL-GFP strain may be a result of the effect of the chimeric carboxysome cores on the overall efficiency of the CBB cycle. The reduced phycobilisome content (Figure 5A) decreases photon capture, which otherwise would result in damage to photosystem II in strains with less efficient CBB cycles. The reduced oxygen evolution (Figure 5C) could also be due to oxidative damage of the oxygen evolving complex. The increased photoinhibition (as measured by Fv/Fm) upon a decrease in CO2 levels (Figure 5B) suggests less flexibility in adapting to environmental conditions.
Similarly, the reduced size of the streamlined carboxysomes (Figure 3; Supplemental Figure 5C) is likely attributable to the absence of some protein-protein interactions that occur in the wild-type carboxysome core; for example, the lack of free M35 protein, which aggregates Rubisco, may result in smaller carboxysome cores. Small carboxysome-like particles have been observed in Syn 7942 mutants with a decreased ratio of M35 to full-length CcmM (Long et al., 2010). The strains expressing the synthetic carboxysomes appear to compensate for their smaller size by increasing their number (Figure 3; Supplemental Figure 5A). Changes in carboxysome abundance, size, and shape (including rod carboxysomes) have been observed previously in strains defective in CA (Price and Badger, 1989). It is also possible that the CcmC core is somewhat deficient in CA activity, since the arrangement of the domains in CcmC may obstruct formation of the native CcaA oligomeric state (a dimer; So et al., 2002a).
The chimeric carboxysomes, although smaller, morphologically resemble wild-type carboxysomes (Figure 3); most importantly, they are able to support photosynthesis (Figure 4). This underscores the value of the carboxysome as a test system to evaluate approaches to engineering BMCs because cyanobacterial physiology and ultrastructure offer sensitive readouts of both structure and function. While the content of CcmC is stoichiometrically fixed (and therefore less likely to be able to respond flexibly to changes in metabolism), the results demonstrate that at least four protein domains combined into one non-native fusion protein can be enclosed in carboxysome shells by a single EP. The system can be further optimized for CO2 fixation, for example, at the molecular level by swapping in different CA modules with different kinetic properties or by altering domain stoichiometry within the fusion protein. At the cellular level, the system could be optimized using adaptive evolution approaches (Dragosits and Mattanovich, 2013).
More broadly, carboxysomes and other BMCs are the subject of intense research efforts for use as self-assembling organelles for compartmentalizing metabolism (Frank et al., 2013; Kerfeld and Erbilgin, 2015). Previous attempts to engineer BMCs have focused on associating heterologous proteins to shells using EPs. For example, through the addition of two different EPs to pyruvate decarboxylase and alcohol dehydrogenase, Lawrence et al. (2014) were able to repurpose a propanediol utilization compartment shell for ethanol production; Lin et al. (2014b) showed that the encapsulation peptide from CcmN targets yellow fluorescent protein into carboxysome-like structures formed in mutant wild tobacco (Nicotiana benthamiana) plants. In contrast, the approach reported here focuses on assembling a multifunctional BMC core: CcmC nucleates Rubisco, supplies CA activity, and recruits the shell. This approach allows the packaging of multiple protein domains within a BMC shell using only a single EP. Given that protein domains are the structural, functional, and evolutionary units of proteins, we propose that reengineering new functions in carboxysomes or designing metabolic cores for synthetic BMCs may be more readily tractable, as this technique focuses on domain structures and interactions rather than on targeting single gene products to BMC shells. The strategy reported here leverages the inherent modularity of proteins, by using domain fusions, for building new subcellular architectures. This approach should be especially valuable for plant synthetic biology.
METHODS
Cyanobacterial Strain and Growth Conditions
Synechococcus elongatus PCC 7942 (Syn 7942) cultures were grown in 250 mL baffled Erlenmeyer flasks with 60 mL BG-11 medium (Rippka et al., 1979) buffered with 10 mM HEPES, pH 8.0, under the following growth chamber settings: temperature of 30°C, light intensity of 40 µmoles photons m−2 s−1, shaking at 150 rpm, and CO2 concentrations of 5%, 3%, or air. Unless otherwise indicated, experiments were performed in cultures at exponential growth phase (OD730 = 0.4 to 0.7).
Mutant Generation
Syn 7942 cells were transformed as previously described (Kufryk et al., 2002). Cultures were grown to OD730 = 0.5 and concentrated to OD730 = 2.5 by centrifugation at 5000 rcf for 5 min. Five microliters of plasmids (∼1 µg of DNA) prepared from Escherichia coli DH5α cells were added to 400 μL of the cyanobacterial cell suspension and incubated for 6 h. The 400-µL aliquots were dried on Nuclepore track-etched polycarbonate membranes (GE Healthcare) on top of BG-11 plates and incubated for 12 to 24 h. The membranes were transferred to BG-11 plates with the proper selectable marker until resistant colonies were obtained. All mutant strains were transformed with pJCC008 plasmid (rbcL-GFP placed under the control of the ccmk2 promoter) (Cameron et al., 2013) for GFP labeling of the large subunit of Rubisco (RbcL) to enable carboxysome visualization by fluorescence microscopy. The carboxysome-minus strain COREΔ2/RbcL-GFP was generated by replacing synpcc7942_1423 and synpcc7942_1424 genes with a kanamycin resistance/sucrose sensitivity cassette obtained from the pPSBAII-KS plasmid (Lagarde et al., 2000) and using synpcc7942_1422 and synpcc7942_1425 sequences as flanking regions for double homologous recombination. Domains for the generation of chimeric proteins were assigned using the InterPro software (Hunter et al., 2012) and the HMM tool from the J. Craig Venter institute (http://blast.jcvi.org/web-hmm). DNA sequences were obtained from Cyanobase (http:// http://genome.microbedb.jp/cyanobase) and cloned by traditional methods (restriction digestion and ligation) (Sambrook and Russell, 2001) as follows. Plasmids with genes coding for the chimeric proteins (amino acids sequences found in Supplemental Figure 8; note that in the case of CcmC, the C-terminal extension of the β-CA was used as linker, and its terminal 14 amino acids were replaced by 18 amino acids comprising the EP) with synpcc7942_1422 and synpcc7942_1425 sequences as flanking regions were transformed into the COREΔ2/RbcL-GFP strain. Growth in air was used for positive selection and growth in 5% sucrose as confirmation. The COREΔ2/CcmC/RbcL-GFP strain is obtained after CcmC restores growth in air. CcaA (synpcc7942_1447) was interrupted in the COREΔ2/CcmC/RbcL-GFP strain and in wild-type/RbcL-GFP by insertion of a gentamycin resistance cassette and selection with 5 μg/mL gentamycin in solid BG-11 plates (resulting in COREΔ3/CcmC/RbcL-GFP strain and ΔCcaA/RbcL-GFP strain, respectively). Primers used are found in Supplemental Table 1.
Structural Modeling
The predicted domains obtained (Supplemental Figure 1A) were used as input for the automated mode of the SwissModel server (Biasini et al., 2014). The EP was manually added to the predicted structure of CcmC using Chimera software (Pettersen et al., 2004).
Spectrophotometric Measurements
Culture growth was monitored as the change in OD730. Chlorophyll a concentration was determined by absorbance measurements (at 663 nm) of methanol extracts from 1-mL culture aliquots and calculated according to Lichtenthaler (1987). Total cell spectra were obtained from 1-mL aliquots of cultures in exponential growth phase, which were diluted to OD730 = 0.3, and the obtained spectra were normalized to that of chlorophyll a (OD663). Doubling times were calculated using the exponential regression curve fitting online tool found at www.doubling-time.com/compute.php. All measurements were performed at least in triplicate from aliquots from different cultures (using the same inoculum from a BG-11 agar plate). All measurements were performed in a Nanodrop2000C spectrophotometer (Thermo Scientific).
PCR and Immunoblot Analysis
Standard PCR was performed as described in the manufacturer’s protocol using EconoTaq Plus Green 2X (Lucigen) and gene-specific primer pairs (Supplemental Table 1). For protein extraction, pellets from 50-mL culture aliquots were resuspended in 1 mL of lysis buffer (25 mM HEPES-NaOH, pH 7, 15 mM CaCl2, 5 mM MgCl2, 15% glycerol, 200 µM PMSF, and Complete, Mini protease inhibitor [Roche]) and broken in a BeadBug homogenizer (Biospec Products) by beating for six cycles of 30 s and 2 min of incubation in ice between each cycle. After 20 min of centrifugation at 20,000 rcf, 15-μL aliquots plus SDS loading dye were loaded onto an acrylamide gel (without boiling the sample) for SDS-PAGE. SDS-PAGE and immunoblot analysis were performed according to the manufacturer’s protocol (Bio-Rad’s bulletin 6376) using a polyclonal antibody from rabbit against Syn 7942 CcmM (dilution 1:5000; Rothamstead Research) as a primary antibody and goat anti-rabbit IgG-HRP (dilution 1:7000; Life Tech. 656120) as secondary antibody and 1-Step Ultra TMB-Blotting Solution as substrate (Thermo Scientific 37574). For densitometries, total protein extract samples from three independent cultures were normalized according to the peak absorbance at 663 nm, loaded at four decreasing serial dilutions, and blotted as described using Anti-RbcL antibody (Agrisera) at a dilution of 1:10,000. Densitometry measurements were performed on the different immunoblots using ImageJ software (Schneider et al., 2012).
Oxygen Evolution
Two-milliliter aliquots were harvested from exponential-phase cultures and supplemented with 10 mM bicarbonate prior to the measurement, and the steady state rate of oxygen evolution was determined at saturating light intensity (950 µmoles photons m−2 s−1) and 30°C using an LMI-6000 illuminator (Dolan-Jenner) and an Oxygraph Plus Clark-type electrode (Hansatech).
Fluorescence and Electron Microscopy
Cultures grown to OD730 = 0.5 in 3% CO2 were transferred to air and grown overnight. For fluorescence microscopy, 1-mL aliquots were concentrated by centrifugation (1500 rcf for 5 min and resuspended in 100 μL of BG11) and visualized (autofluorescence and GFP) using a Zeiss Axio Observer.D1 inverted microscope. For electron microscopy, pellets from 50-mL aliquots were chemically fixed with 2% glutaraldehyde in 50 mM phosphate buffer for 2 h at room temperature, followed by 1% osmium tetroxide for 2 h at room temperature, and block stained with 2% aqueous uranyl acetate overnight at 4°C. Cells were dehydrated in an increasing acetone series (2 min at 37°C; 20% acetone increments) and embedded in Spurr’s resin (15 min at 37°C; 25% increments) using an MS-9000 Laboratory Microwave Oven (Electron Microscopy Science). Sections (70-nm thick) were cut on a MYX ultramicrotome (RMC Products), positively stained with 6% uranyl acetate and Reynolds lead citrate (Reynolds, 1963), and visualized on a JEM 100CX II transmission electron microscope (JEOL) equipped with an Orius SC200-830 CCD camera (Gatan).
Quantum Efficiency of Photosystem II
Fv/Fm was determined in triplicate using 4-mL culture aliquots from biological replicates at exponential phase in cells dark adapted for three minutes as described previously (Cameron et al., 2013). Briefly, aliquots were diluted with BG-11 immediately before dark adaptation to a chlorophyll concentration of ∼1 to 2 µg/mL and measured using an Aquapen AP100 (Photon Systems Instruments). Measurement started at time = 0 h when the cultures were transferred from 3% CO2 to air.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers ABB57453 (CcmM: Synpcc7942_1423), ABB57454 (CcmN: Synpcc7942_1424), and ABB57477.1 (CcaA; Synpcc7942_1447).
Supplemental Data
Supplemental Figure 1. Schematic of domain architectures in CcmM, CcmN, CcaA, and CcmC.
Supplemental Figure 2. Schematics of native and designed chimeric carboxysome cores used in this study.
Supplemental Figure 3. Confirmation of genomic integration.
Supplemental Figure 4. DNA sequence of ccmC integrated into the ccm locus.
Supplemental Figure 5. Structural characterization of native and minimized carboxysomes.
Supplemental Figure 6. Additional transmission electron micrographs of carboxysomal core mutants.
Supplemental Figure 7. Carboxysome aggregation in the ΔCcaA/RbcL-GFP mutant.
Supplemental Figure 8. Amino acid sequences of the chimeric proteins.
Supplemental Table 1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank members of the Kerfeld laboratory for helpful discussions. We thank Jan Zarzycki for bioinformatics assistance, Tarryn Miller for assistance with fluorescence microscopy, Alicia Pastor for assistance with transmission electron microscopy, and Clara Cruet for assistance with strain sequencing. We thank Wim Vermaas for providing the PSBAII-KS plasmid. Support for this work was provided by the National Science Foundation (EF1105897) and the Office of Basic Energy Sciences, U.S. Department of Energy (Contract DE-FG02-91ER20021).
AUTHOR CONTRIBUTIONS
C.R.G.-E. and C.A.K. designed the research. C.R.G.-E. and T.B.S. conducted the research. C.R.G.-E. and C.A.K. wrote the article.
Glossary
- BMC
bacterial microcompartment
- CA
carbonic anhydrase
- SSLD
small subunit-like domain
- EP
encapsulation peptide
- CBB
Calvin-Benson-Bassham
References
- Axen S.D., Erbilgin O., Kerfeld C.A. (2014). A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLOS Comput. Biol. 10: e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59: 89–113. [DOI] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T.G., Bertoni M., Bordoli L., Schwede T. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42: W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F., Menon B.B., Cannon G.C., Curry K.J., Shively J.M., Heinhorst S. (2009). The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 4: e7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F., Dou Z., Bernstein S.L., Leverenz R., Williams E.B., Heinhorst S., Shively J., Cannon G.C., Kerfeld C.A. (2015). Advances in understanding carboxysome assembly in Prochlorococcus and Synechococcus implicate CsoS2 as a critical component. Life (Basel) 5: 1141–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron J.C., Wilson S.C., Bernstein S.L., Kerfeld C.A. (2013). Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155: 1131–1140. [DOI] [PubMed] [Google Scholar]
- Chen A.H., Robinson-Mosher A., Savage D.F., Silver P.A., Polka J.K. (2013). The bacterial carbon-fixing organelle is formed by shell envelopment of preassembled cargo. PLoS One 8: e76127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragosits M., Mattanovich D. (2013). Adaptive laboratory evolution—principles and applications for biotechnology. Microb. Cell Fact. 12: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G., Niklowitz W. (1956). [Cytology of Cyanophycea. II. Centroplasm and granular inclusions of Phormidium uncinatum]. Arch. Mikrobiol. 24: 147–162. [PubMed] [Google Scholar]
- Frank S., Lawrence A.D., Prentice M.B., Warren M.J. (2013). Bacterial microcompartments moving into a synthetic biological world. J. Biotechnol. 163: 273–279. [DOI] [PubMed] [Google Scholar]
- Gantt E., Conti S.F. (1969). Ultrastructure of blue-green algae. J. Bacteriol. 97: 1486–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S., et al. (2012). InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40: D306–D312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld C.A., Erbilgin O. (2015). Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol. 23: 22–34. [DOI] [PubMed] [Google Scholar]
- Kinney J.N., Salmeen A., Cai F., Kerfeld C.A. (2012). Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J. Biol. Chem. 287: 17729–17736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufryk G.I., Sachet M., Schmetterer G., Vermaas W.F. (2002). Transformation of the cyanobacterium Synechocystis sp. PCC 6803 as a tool for genetic mapping: optimization of efficiency. FEMS Microbiol. Lett. 206: 215–219. [DOI] [PubMed] [Google Scholar]
- Lagarde D., Beuf L., Vermaas W. (2000). Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 66: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf D., Okumus B., Chien P., Baker T.A., Paulsson J. (2012). Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9: 480–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence A.D., Frank S., Newnham S., Lee M.J., Brown I.R., Xue W.-F., Rowe M.L., Mulvihill D.P., Prentice M.B., Howard M.J., Warren M.J. (2014). Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth. Biol. 3: 454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382. [Google Scholar]
- Lin M.T., Occhialini A., Andralojc P.J., Parry M.A.J., Hanson M.R. (2014a). A faster Rubisco with potential to increase photosynthesis in crops. Nature 513: 547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.T., Occhialini A., Andralojc P.J., Devonshire J., Hines K.M., Parry M.A.J., Hanson M.R. (2014b). β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 79: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluch-Senar M., et al. (2015). Defining a minimal cell: essentiality of small ORFs and ncRNAs in a genome-reduced bacterium. Mol. Syst. Biol. 11: 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long B.M., Badger M.R., Whitney S.M., Price G.D. (2007). Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 282: 29323–29335. [DOI] [PubMed] [Google Scholar]
- Long B.M., Tucker L., Badger M.R., Price G.D. (2010). Functional cyanobacterial β-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol. 153: 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J.A., Hernández H., Hall Z., Ahnert S.E., Perica T., Robinson C.V., Teichmann S.A. (2013). Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell 153: 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña K.L., Castel S.E., de Araujo C., Espie G.S., Kimber M.S. (2010). Structural basis of the oxidative activation of the carboxysomal γ-carbonic anhydrase, CcmM. Proc. Natl. Acad. Sci. USA 107: 2455–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Price G.D., Badger M.R. (1989). Isolation and characterization of high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC7942: Two phenotypes that accumulate inorganic carbon but are apparently unable to generate CO2 within the carboxysome. Plant Physiol. 91: 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price G.D., Badger M.R., Woodger F.J., Long B.M. (2008). Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J. Exp. Bot. 59: 1441–1461. [DOI] [PubMed] [Google Scholar]
- Price G.D., Pengelly J.J.L., Forster B., Du J., Whitney S.M., von Caemmerer S., Badger M.R., Howitt S.M., Evans J.R. (2013). The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J. Exp. Bot. 64: 753–768. [DOI] [PubMed] [Google Scholar]
- Reynolds E.S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17: 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111: 1–61. [Google Scholar]
- Rokney A., Shagan M., Kessel M., Smith Y., Rosenshine I., Oppenheim A.B. (2009). E. coli transports aggregated proteins to the poles by a specific and energy-dependent process. J. Mol. Biol. 392: 589–601. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Savage D.F., Afonso B., Chen A.H., Silver P.A. (2010). Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327: 1258–1261. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A.K.-C., Cot S.S.-W., Espie G.S. (2002a). Characterization of the C-terminal extension of carboxysomal carbonic anhydrase from Synechocystis sp. PCC6803. Funct. Plant Biol. 29: 183–194. [DOI] [PubMed] [Google Scholar]
- So A.K.C., John-McKay M., Espie G.S. (2002b). Characterization of a mutant lacking carboxysomal carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Planta 214: 456–467. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Murata N. (2005). Interruption of the Calvin cycle inhibits the repair of photosystem II from photodamage. Biochim. Biophys. Acta 1708: 352–361. [DOI] [PubMed] [Google Scholar]
- Zarzycki J., Axen S.D., Kinney J.N., Kerfeld C.A. (2013). Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 64: 787–798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.