Cell cycle G1/S arrest regulated by Bub2/Bfa1 GAP during conidial germination of Colletotrichum orbiculare was adapted to fulfill specific plant-pathogenic roles in virulence-associated processes.
Abstract
Morphogenesis in filamentous fungi depends on appropriate cell cycle progression. Here, we report that cells of the cucumber anthracnose fungus Colletotrichum orbiculare regulate G1/S progression via a two-component GAP, consisting of Budding-uninhibited-by-benomyl-2 (Bub2) and Byr-four-alike-1 (Bfa1) as well as its GTPase Termination-of-M-phase-1 (Tem1) to establish successful infection. In a random insertional mutagenesis screen of infection-related morphogenesis, we isolated a homolog of Saccharomyces cerevisiae, BUB2, which encodes a two-component Rab GAP protein that forms a GAP complex with Bfa1p and negatively regulates mitotic exit. Interestingly, disruption of either Co BUB2 or Co BFA1 resulted in earlier onset of nuclear division and decreased the time of phase progression from G1 to S during appressorium development. S. cerevisiae GTPase Tem1p is the downstream target of the Bub2p/Bfa1p GAP complex. Introducing the dominant-negative form of Co Tem1 into Co bub2Δ or Co bfa1Δ complemented the defect in G1/S progression, indicating that Co Bub2/Co Bfa1 regulates G1/S progression via Co Tem1. Based on a pathogenicity assay, we found that Co bub2Δ and Co bfa1Δ reduced pathogenesis by attenuating infection-related morphogenesis and enhancing the plant defense response. Thus, during appressorium development, C. orbiculare Bub2/Bfa1 regulates G1/S progression via Co Tem1, and this regulation is essential to establish plant infection.
INTRODUCTION
Colletotrichum orbiculare is the causal agent of cucumber anthracnose disease. The infection process is initiated upon recognition of appropriate surface cues by aseptate conidia. After the conidia germinate, the resultant germ tubes differentiate into dome-shaped appressoria and undergo nuclear division. Successful appressorium function and, thus, infection depend on the proper sequence of metabolic events, including melanin synthesis, peroxisome metabolism, establishment of cellular polarity and cell wall integrity, and signal transduction, occurring in the fungus (Kubo and Takano, 2013; Harata and Kubo, 2014). Importantly, a microtubule dynamics analysis revealed that a precise nuclear distribution inside cells is also required for normal appressorial development (Takano et al., 2001). In the rice blast fungus Magnaporthe oryzae, appropriate regulation of the cell cycle is necessary to establish infection. The fungus must enter the S phase to initiate appressorium formation, whereas entry into mitosis and autophagic programmed cell death in conidia are essential for the germ tube to differentiate into a functional appressorium (Veneault-Fourrey et al., 2006; Saunders et al., 2010a). In addition to cell cycle regulation, pathogenesis in M. oryzae also requires that cells undergo the correct number of divisions at precise locations (Saunders et al., 2010b). Thus, the progression of the cell cycle and cytokinesis seem to regulate cellular differentiation processes. However, the details of cell cycle regulation that could affect infection-related morphogenesis and pathogenesis are poorly understood in Colletotrichum species.
In the budding yeast Saccharomyces cerevisiae and the multimorphic fungus Candida albicans, exit from mitosis and entry into cytokinesis are regulated by the mitotic exit network (MEN), a GTPase-regulated kinase cascade (Bardin and Amon, 2001; Milne et al., 2014). The MEN consists of the central GTPase Tem1p, which is regulated by the two-component GTPase-activating protein (GAP), consisting of Bub2p and Bfa1p. In S. cerevisiae cells, the Bub2p/Bfa1p complex constitutes multiple cell cycle checkpoints that prevent mitotic exit (Hu and Elledge, 2002). Such checkpoints include the spindle position checkpoint (SPOC), the surveillance mechanism for proper orientation of the mitotic spindle (Yeh et al., 1995); the spindle assembly checkpoint (SAC), for proper attachment of all kinetochores to the spindle (Musacchio and Salmon, 2007); and the DNA damage checkpoint (DDC), for the proper protection of chromosomal DNA (Harrison and Haber, 2006; Valerio-Santiago et al., 2013). The fission yeast Schizosaccharomyces pombe and the filamentous fungus Aspergillus nidulans have a signaling pathway homologous to MEN, termed the septation initiation network (SIN). The primary role of this network is to regulate septation rather than mitotic exit (Bruno et al., 2001; Krapp and Simanis, 2008). The SIN consists of the GTPase Spg1 (Septum-promoting GTP binding protein 1), an ortholog of S. cerevisiae Tem1p, which regulates a protein kinase pathway that triggers contraction of the actomyosin ring and positively regulates septum formation (Schmidt et al., 1997). The two-component GAP consists of Cdc16 and Byr4, orthologs of S. cerevisiae Bub2p and Bfa1p, that negatively regulate the SIN signaling pathway by inactivating the GTPase Spg1 and thus preventing septum formation during interphase (Furge et al., 1998; Li et al., 2000). Despite many of the MEN and SIN components being highly conserved in yeast and filamentous fungi, each of the homologous genes differs in its function. In plant pathogenic fungi, the functions of the GTPase Tem1 and two-component GAP consisting of Bub2 and Bfa1 have been reported only for the basidiomycete Ustilago maydis, indicating that the Tem1 homolog Ras3 regulates nuclear envelope breakdown rather than mitotic exit or septum formation (Straube et al., 2005), but other components or events are currently lacking in plant pathogenic fungi. Thus, the characterization of the two-component GAP and its GTPase in C. orbiculare could provide insight into the roles of MEN/SIN in fungal pathogenicity.
In this study, we present evidence that appropriate G1/S progression is regulated by BUB2 and BFA1 through TEM1 during appressorium development and is required for typical infection-related morphogenesis and pathogenesis of C. orbiculare.
RESULTS
Screening for Mutants with Defects in Pathogenesis and Infection-Related Morphogenesis and Identifying Co BUB2 in C. orbiculare
To identify genes involved in pathogenesis and infection-related morphogenesis, we performed random gene-insertional mutagenesis of the wild-type strain 104-T of C. orbiculare using Agrobacterium tumefaciens-mediated transformation (AtMT). We screened 10,021 mutants for defective pathogenesis and abnormal infection-related morphogenesis on cucumber (Cucumis sativus) cotyledons detached from plants. We obtained a mutant, named PDM1 (Pathogenicity Deficient Mutant 1), which formed only a few penetration hyphae on the surface of cotyledons and induced either no lesions or only a few, in contrast to the wild type, which formed penetration hyphae in the host cucumber leaves and induced distinct necrotic lesions on the leaves.
DNA segments adjacent to the inserted plasmid were isolated from the PDM1 mutant using a thermal asymmetric interlaced-PCR. A BLASTp search (NCBI BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) revealed that the sequence adjacent to the T-DNA exhibited high similarity to the amino acid sequence of Bub2p of S. cerevisiae and to those of the filamentous fungi (Supplemental Figure 1). This BUB2 homolog of C. orbiculare was designated Co BUB2. Comparison of the amplified cDNA with genomic DNA verified that Co BUB2 encodes a protein of 472 amino acids and contains the Rab GAP domain. The amino acid sequences of Co Bub2 homologs are well conserved among yeast and filamentous fungi, especially in the Rab GAP domain, which has an E-value 4.99 e-33, suggesting that Co Bub2 has potential GAP activity in C. orbiculare. In S. cerevisiae, Bub2p constitutes three surveillance cell cycle checkpoints (SPOC, SAC, and DDC), which ensure that the replicated genomic material is protected from damage and correctly distributed between the daughter and mother cells by preventing mitotic exit (Hu and Elledge, 2002).
Co Bub2/Co Bfa1 GAP Plays a Key Role in G1/S Progression during Appressorium Development
To understand the regulatory mechanism of Co BUB2, we focused on partner genes that interact with Co Bub2. In S. cerevisiae, Bub2p forms a two-component GAP complex with Bfa1p and requires Bfa1p to stimulate GTP hydrolysis of the downstream GTPase Tem1p to regulate the exit from mitosis (Geymonat et al., 2002). We identified a putative BFA1 homolog, Co BFA1, by BLAST search of the C. orbiculare genome (Gan et al., 2013), which putatively encodes a protein of 1020 amino acids with high identity to the sequence of S. cerevisiae Bfa1p, with an E-value of 2e-05 (Supplemental Figure 2).
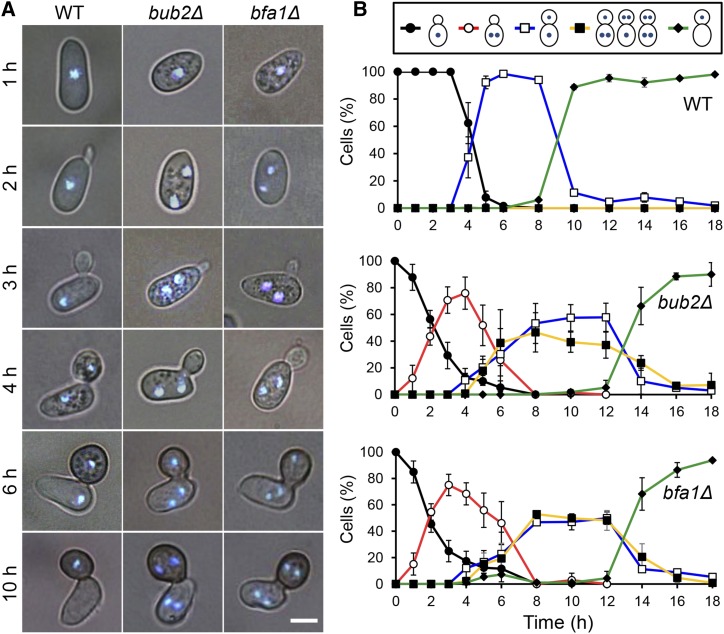
To investigate the function of Co BUB2 and Co BFA1, we generated the Co bub2Δ and Co bfa1Δ targeted disruption mutants (Supplemental Figure 3). Mutation of Co bub2 or Co bfa1 caused slightly delayed growth and a significant defect in conidiation (Supplemental Table 1). Furthermore, the mutation caused a defect in nuclear division during appressorium formation. The conidia of the wild type contained one nucleus and, following conidial germination, underwent one round of nuclear division during appressorium development at 4 h postincubation (hpi) (Figure 1). This resulted in the conidium and appressorium each having one nucleus. At 10 hpi, the nucleus that remained in the conidium had been broken down by autophagy, while the appressorium retained its one nucleus. Surprisingly, in Co bub2Δ and Co bfa1Δ, the nucleus in the conidium divided before the conidium germinated at 2 hpi, so that the pregermination conidium had two nuclei (Figure 1). After the development of the appressorium, in some germlings one nucleus migrated into the appressorium, resulting in one nucleus in the conidium and the other in the appressorium. The other germlings underwent a second round of mitosis, resulting in multiple nuclei in the conidium and appressorium. At 10 hpi, conidial autophagy in Co bub2Δ and Co bfa1Δ was significantly delayed compared with that in the wild type. Thus, in contrast to S. cerevisiae BUB2 and BFA1, Co BUB2 and Co BFA1 are involved in the timing of nuclear division and in the proper distribution of nuclei during appressorium development.
Figure 1.
Co BUB2 and Co BFA1 Are Involved in the Timing of Initiation of Nuclear Division during Appressorium Development.
(A) Time-course series of micrographs to show nuclei in conidia and infection structures of C. orbiculare wild type and Co bub2Δ and Co bfa1Δ mutants during appressorium development. Staining was performed using DAPI. Bar = 5 μm.
(B) Mean percentage (±se; n = 3) of cells with various patterns of nuclear distribution. Scoring: one nucleus retained in the conidium (black), two nuclei retained in the conidium (red), one nucleus in the conidium and the other in the appressorium through mitosis (blue), multiple nuclei in the conidium and appressorium through two or three rounds of mitosis (yellow), and one nucleus in the appressorium and degradation of the remaining nucleus in the conidium through autophagy (green). At least 200 conidia were scored at each time point.
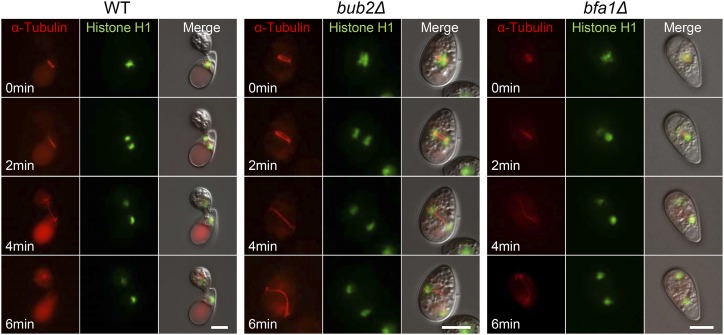
To investigate nuclear division and migration during appressorium development cytologically, we introduced the α-TUBULIN-mRFP fusion gene and HISTONE H1-GFP fusion gene into C. orbiculare to visualize microtubules and nuclei, respectively. In the wild type, microtubules were apparent at 4 to 5 hpi, that is, following appressorium development, and microtubules aligned parallel to the conidia-appressorium axis (Figure 2; Supplemental Movie 1). One spindle pole body remained in the conidium, and the other migrated into the appressorium within 6 min. In accordance with the movement of the microtubules, the mother nucleus divided; one mother nucleus remained in the conidium, while one daughter nucleus migrated into the appressorium. By contrast, in most conidia of Co bub2Δ and Co bfa1Δ, microtubules appeared at 2 hpi in the pregermination conidium, and the partner spindle pole body migrated to the edge of the conidium (Figure 2; Supplemental Movie 2). In the same way, the mother nucleus divided, and the two nuclei moved to the edge of the cell. These results confirm that mitosis of Co bub2Δ and Co bfa1Δ started ∼2 h earlier than that of the wild type.
Figure 2.
Earlier Mitosis of Co bub2Δ and Co bfa1Δ Visualized by Live-Cell Imaging.
C. orbiculare wild type, Co bub2Δ, and Co bfa1Δ showing microtubule dynamics and nuclear division. The αTUBULIN-mRFP and HISTONE H1-GFP gene fusion vectors were introduced into the three strains. Microtubules (red) and nuclei (green) were first observed in the appressorium-developing conidia of the wild type at 4 hpi, whereas they were first observed in the pregermination conidia in Co bub2Δ and Co bfa1Δ at 2 hpi. Bars = 5 μm.
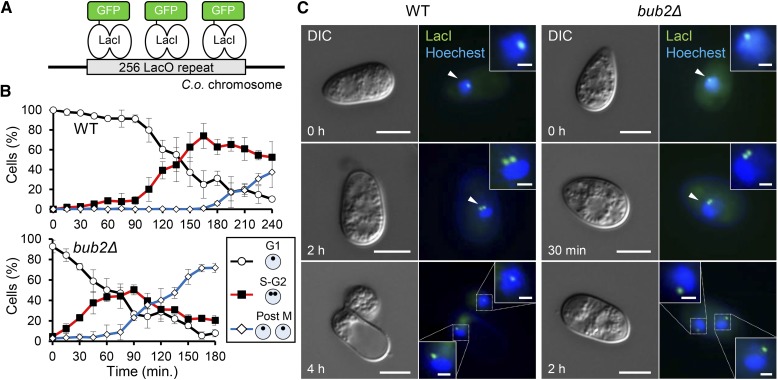
We therefore hypothesized that the cell cycle of Co bub2Δ and Co bfa1Δ might progress earlier than that of the wild type. To test this hypothesis, we evaluated the time of entry into S phase, with time 0 defined as the onset of conidial incubation, in Co bub2Δ as a representative strain during appressorium development. We used an established method in which a chromosomal locus is tagged with a fluorescent focus by recruiting a GFP-tagged Lac repressor fusion (GFP-LacI) to an integrated Lac operator array (LacO) (Robinett et al., 1996; Straight et al., 1996) (Figure 3A). In this chromosome tagging system, the time of DNA replication (S phase) is determined based on large-scale chromatin organization; that is, cells containing prereplicated DNA exhibit a single fluorescent spot, while cells that have undergone post-DNA replication harbor closely spaced double fluorescent spots, with one on each separated sister chromatid (Straight et al., 1996). To evaluate the timing of duplication of GFP spots in C. orbiculare nuclei, which refers to the timing of DNA replication (S phase), we constructed the S. pombe LacI-NLS-eGFP fusion protein (Straight et al., 1996; Nabeshima et al., 1998). Then, an array consisting of yeast vectors harboring 256 tandem copies of LacO (Yamamoto and Hiraoka, 2003) was ectopically introduced into the strain expressing the GFP-LacI reporter fusion. In the wild-type strain containing LacO/LacI-GFP, a single GFP spot was observed at a nucleus after the onset of conidial incubation (Figures 3B and 3C). After germination at 105 min, cells containing two GFP spots started to appear and such cells represented up to ∼80% of all cells by 165 min. At 240 min, mitosis was completed and each nucleus in the conidium and appressorium contained a single spot. On the other hand, in the Co bub2Δ mutant, a single GFP spot observed at the onset of conidial incubation decreased immediately after incubation, but double GFP spots in the nucleus were seen in up to ∼50% of Co bub2Δ cells by 60 min (Figures 3B and 3C). These results indicate that S phase starts after around 120 min of incubation in the wild type, but before 60 min in Co bub2Δ. Therefore, Co Bub2/Co Bfa1 regulates G1/S progression in the cell cycle during germination and appressorium formation.
Figure 3.
Co BUB2 and Co BFA1 Regulate G1/S Progression in C. orbiculare.
(A) Schematic representation of the C. orbiculare strain expressing GFP-LacI-NLS, which associates with the LacO array integrated into C. orbiculare chromosomes.
(B) Mean percentage (±se; n = 3) of wild-type (WT) and Co bub2Δ cells with three patterns of GFP-LacI spots. Scoring: single fluorescent spot at one nucleus, putative G1 phase (black); closely spaced double spots at one nucleus, putative post-S phase or G2 phase (red); and single spots at both nuclei, putative post-M phase (blue). At least 100 conidia were scored at each time point.
(C) Representative images of LacO/LacI-GFP introduced wild type and Co bub2Δ during appressorium development. Enlarged images of the nucleus with one or two LacI-GFP spots are shown in the boxed areas. Arrowheads indicate interphase nuclei. Nuclear staining was performed using Hoechst 33342. Bars in DIC panels = 5 μm; bars in enlarged image = 1 μm.
Next, we investigated whether regulation of G1/S progression by Co Bub2/Co Bfa1 is specific to morphogenesis from conidia. To test this idea, we monitored nuclear division of Co bub2Δ during hyphal growth in the presence of exogenous nutrients, which leads to the development of vegetative hyphae without appressorium formation (Takano et al., 1997). Under these conditions, before germination at 2 hpi, one round of nuclear division occurred in Co bub2Δ, but not in the wild type (Supplemental Figure 4). On the other hand, from 2 to 10 hpi in the vegetative hyphae, the increase in nuclear number in Co bub2Δ was similar to that in the wild type. Thus, Co BUB2 and Co BFA1 are involved in G1/S progression in pregermination conidia, but not in vegetative hyphal growth; their involvement seems to be specific for G1/S progression during appressorium development.
Co BUB2 and Co BFA1 Are Orthologs of BUB2 and BFA1 in S. cerevisiae
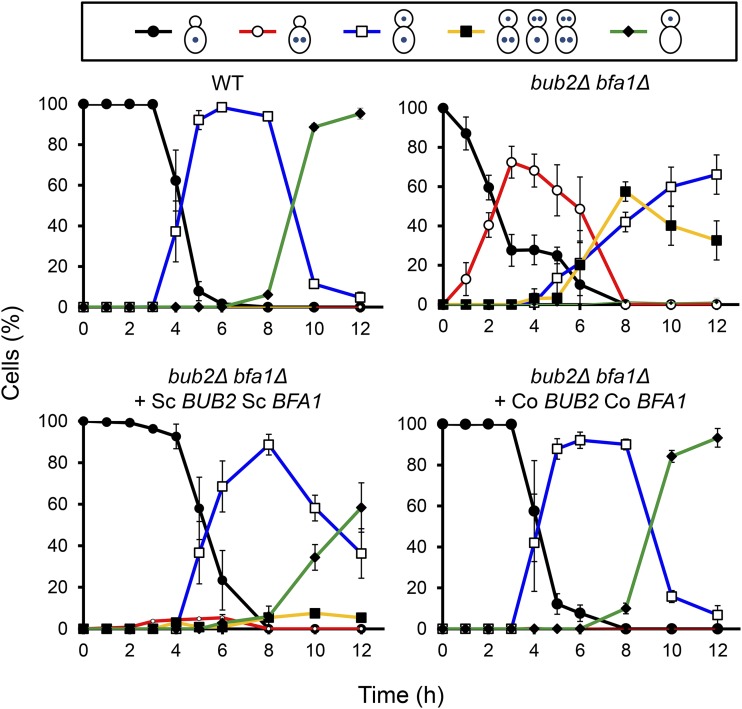
In S. cerevisiae, the Bub2p/Bfa1p GAP complex negatively regulates mitotic exit, while in C. orbiculare, Bub2/Bfa1 regulates proper G1/S progression. We therefore examined whether Co BUB2 and Co BFA1 were orthologs of BUB2 and BFA1 of S. cerevisiae. We conducted a complementation test that examined whether S. cerevisiae Bub2p/Bfa1p GAP activity regulated G1/S progression when introduced into C. orbiculare Co bub2Δ Co bfa1Δ double mutants. We constructed plasmids pBIBub2B and pBIBfa1Z that express open reading frames of S. cerevisiae BUB2 and BFA1 under the control of the Co BUB2 and Co BFA1 native promoters, respectively. We transformed Co bub2Δ Co bfa1Δ double mutants with a combination of S. cerevisiae BUB2 and BFA1. The wild-type phenotype was restored in transformants that expressed both S. cerevisiae BUB2 and BFA1. While the nuclear division of Co bub2Δ Co bfa1Δ started earlier in pregermination conidia, the nuclear division of the transformants that expressed both S. cerevisiae BUB2 and BFA1 started at a similar time point to the wild type in appressorium-forming conidia (Figure 4). This result indicates that the cell cycle defect of Co bub2Δ Co bfa1Δ was complemented by S. cerevisiae BUB2 and BFA1. Thus, we confirmed that S. cerevisiae BUB2 and BFA1, which regulate mitotic exit, are orthologs of C. orbiculare BUB2 and BFA1.
Figure 4.
Co BUB2 and Co BFA1 Genes Are Orthologs of BUB2 and BFA1 of S. cerevisiae.
Complementation of defects in nuclear behavior in Co bub2Δ Co bfa1Δ by introducing S. cerevisiae BUB2 and BFA1. Each nuclear distribution pattern was calculated (means ± se; n = 3). For the positive control, the Co bub2Δ Co bfa1Δ mutant expressing Co BUB2 and Co BFA1 was used. At least 200 conidia were scored at each time point.
Co Bub2 Forms a Two-Component GAP Complex with Co Bfa1, and Co Bub2/Co Bfa1 Interacts with GTPase Co Tem1
Since our results showed that the functions of Co Bub2 and Co Bfa1 differed from those of the respective homologs in yeast, we sought to determine the downstream target of Co Bub2/Co Bfa1 that regulates G1/S progression. In S. cerevisiae, the Bub2p/Bfa1p GAP complex negatively regulates Ras-like GTPase Tem1p, which triggers the exit from mitosis (Geymonat et al., 2002). Tem1p is activated in the GTP-bound form, and activity of Bub2p/Bfa1p GAP decreases the amount of the active GTP-bound form of Tem1p by stimulating GTP hydrolysis. We identified a gene, Co TEM1, from the C. orbiculare genome database (Gan et al., 2013), which showed 58% sequence identity with S. cerevisiae Tem1p (Supplemental Figure 5). The Co Tem1 sequence displays the highly conserved Rab family domain, suggesting that Co Tem1 potentially functions as a Rab GTPase in C. orbiculare.
To investigate whether Co Bub2, Co Bfa1, and Co Tem1 interact with each other, we performed yeast two-hybrid assays. First, the interaction of Co Bub2 and Co Bfa1 was examined. Co Bub2 contains the highly conserved Rab GAP domain and 124 amino acids of the N-terminal region that are not conserved in S. cerevisiae and S. pombe, showing only sequence similarity to proteins of filamentous fungi (Supplemental Figures 1 and 6). In Co Bfa1, 43 amino acids of two imperfect direct repeats (IDRs) at the C-terminal end have been reported to be required for interaction with the Co Bub2 homolog in S. pombe (Furge et al., 1998), and the two IDRs are conserved in filamentous fungi and yeast (Supplemental Figures 2 and 6). Yeast clones carrying Co Bfa1full grew only in the presence of CoBub2full and Co Bub21-124 on selective medium, but not in combination with CoBub2125-473, suggesting that Co Bfa1 interacts physically with the N-terminal region of Co Bub2 (Supplemental Figure 6). On the other hand, yeast clones carrying Co Bub2full grew only in the presence of Co Bfa1full and Co Bfa1784-1020, but not in combination with Co Bfa11-783, suggesting that Co Bub2 interacts physically with the IDR region of Co Bfa1. These results suggest that Co Bub2 forms the two-component GAP complex with Co Bfa1 and that the N-terminal region of Co Bub2 and the IDR region of Co Bfa1 are required for this interaction.
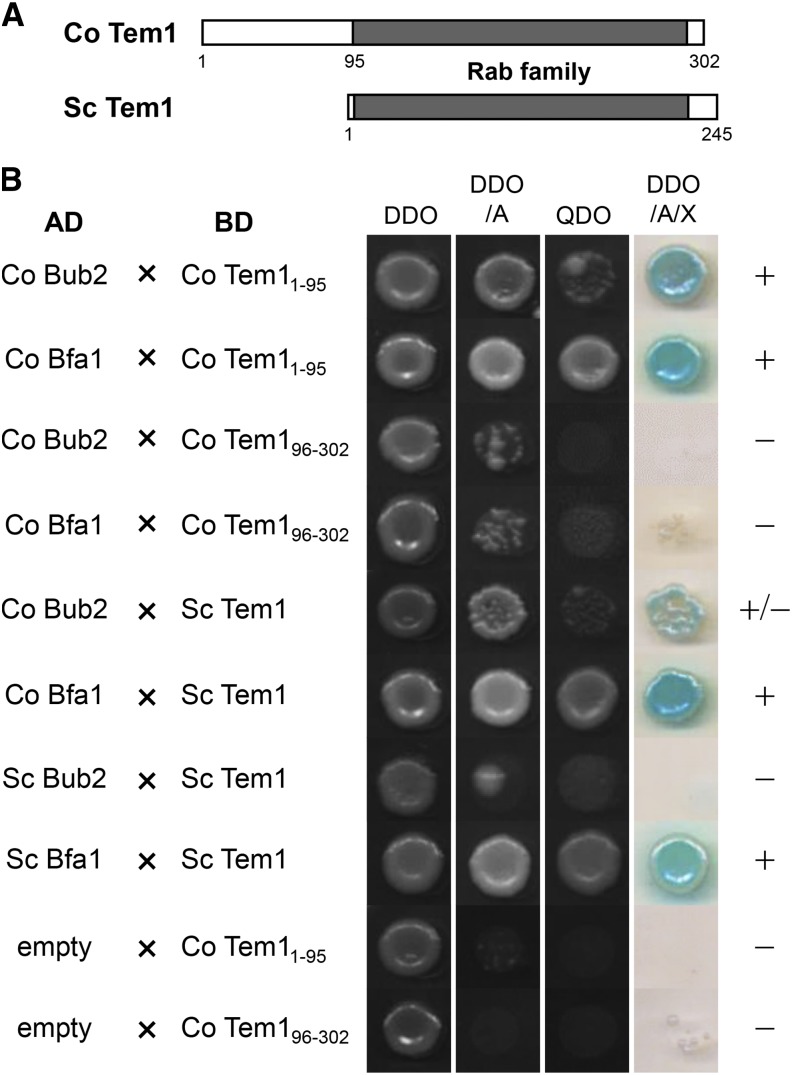
Next, we investigated whether the Co Bub2/Co Bfa1 GAP complex interacts with Co Tem1 in C. orbiculare, similar to the yeast homologs. Co Tem1 contains the highly conserved Rab family domain, and 95 amino acids of the N-terminal region that are not conserved in S. cerevisiae and S. pombe, but it displays similarity to the filamentous fungi proteins similar to the Co Bub2 sequence (Figure 5A; Supplemental Figure 5). Yeast clones carrying Co Bub2full or Co Bfa1full grew only in the presence of Co Tem11-95 on selective medium, but not in combination with Co Tem196-302 (Figure 5B), indicating that Co Tem1 interacts physically with Co Bub2 and Co Bfa1, and the N-terminal region of Co Tem1 is required for this interaction. We also tested the interaction of S. cerevisiae Tem1p with Co Bub2 and Co Bfa1. Yeast clones carrying Co Bfa1full grew in the presence of S. cerevisiae Tem1p on selective medium, suggesting that Co Bfa1 can interact with S. cerevisiae Tem1p, which lacks the conserved N terminus region of C. orbiculare Tem1. For control experiments, we also detected the interaction between Bfa1p and Tem1p of S. cerevisiae but did not detect an interaction between Bub2p and Tem1p of S. cerevisiae, consistent with published results in S. cerevisiae (Kim et al., 2008). Taken together, these results suggest that Co Bub2 forms a two-component GAP complex with Co Bfa1, and both Co Bub2 and Co Bfa1 interact with the GTPase Co Tem1 in C. orbiculare. Both Co Bub2 and Co Tem1 contain the N-terminal region that is conserved only in filamentous fungi, and these N-terminal regions of Co Bub2 and Co Tem1 are required for the interaction with Co Bfa1 and Co Bub2/Co Bfa1, respectively, suggesting that these N-terminal regions of Co Bub2 and Co Tem1 might have specific roles in filamentous fungi.
Figure 5.
Co Tem1 Interacts with Co Bub2 and Co Bfa1 in Yeast Two-Hybrid Assays.
(A) Schematic representation of C. orbiculare Tem1 and S. cerevisiae Tem1p. The 95-amino acid sequence of the N-terminal end of Co Tem1 is specific to filamentous fungi and missing in yeast. Rab family domains of Co Tem1 and Tem1p are labeled in gray.
(B) Interaction of Co Tem1 with Co Bub2 or Co Bfa1 and interaction of S. cerevisiae Tem1p with Co Bub2 or Co Bfa1, as determined by a yeast two-hybrid experiment. Interaction was assessed from yeast growth on SD media lacking –Trp –Leu (DDO), –Trp –Leu + AbA (DDO/A), –Trp –Leu –Ade –His (QDO), and –Trp –Leu + AbA + X-α-Gal (DDO/A/X). The empty-AD vector was used as a negative control. +, Interaction; –, no interaction.
Co Tem1 Is a Downstream Target of Co Bub2/Co Bfa1 and Regulates G1/S Progression and Septum Formation
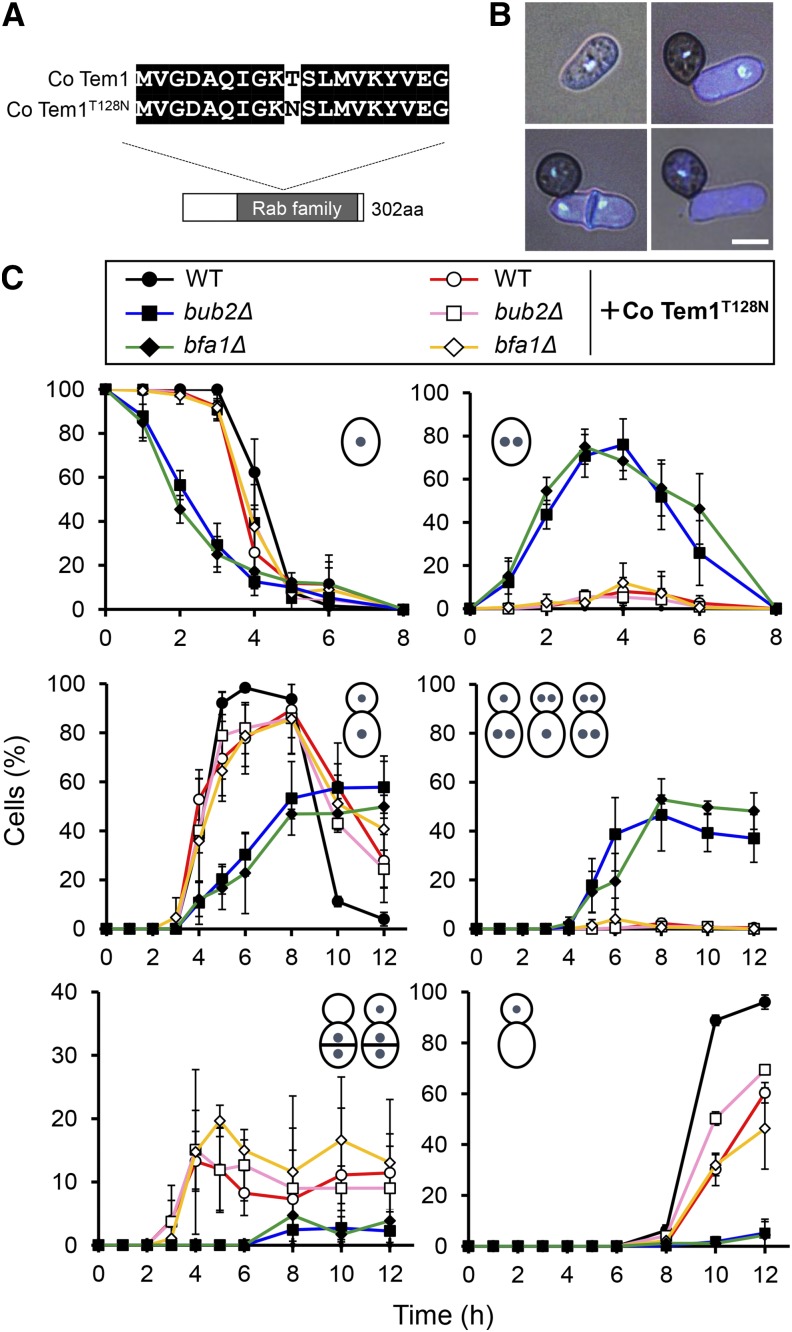
To investigate whether the two-component GAP Co Bub2/Co Bfa1 targets GTPase Co Tem1 to regulate G1/S progression in C. orbiculare, we generated a dominant-negative isoform of Co Tem1 analogous to the S. pombe homolog Spg1, a dominant-negative isoform, which has a Thr-to-Asn substitution at codon 24 (Schmidt et al., 1997). In S. pombe, the T24N mutant is predicted to titrate out the guanine nucleotide exchange factor and thus act as a dominant negative by inhibiting the signaling ability of the endogenous Spg1 (Schmidt et al., 1997). We then investigated whether Co bub2Δ and Co bfa1Δ expressing a dominant-negative form of Co Tem1 would restore the defective phenotype of G1/S arrest. A point mutation was generated in C. orbiculare Co Tem1 (Figure 6A), and the resulting allele was introduced by targeted allelic replacement into the C. orbiculare wild type and the Co bub2Δ and Co bfa1Δ mutants. The introduction of Co Tem1T128N restored the wild-type colony growth and restored conidia production, up to 40% over the very low conidia production in Co bub2Δ and Co bfa1Δ (Supplemental Table 1).
Figure 6.
Introduction of Co Tem1T128N Restored the Wild-Type Nuclear Behavior in Co bub2Δ and Co bfa1Δ.
(A) Schematic representation of the Co Tem1T128N allele, which was introduced into wild-type C. orbiculare and Co bub2Δ and Co bfa1Δ mutants.
(B) Representative images of nuclear distribution during appressorium morphogenesis in the Co Tem1T128N-introduced strain. Pregermination conidia and appressorium-forming conidia were fixed and nuclei stained with DAPI and calcofluor white at 3 and 10 hpi, respectively. Bar = 5 μm.
(C) Mean percentage (±se; n = 3) of cells with various patterns of nuclear distribution for the wild type, Co bub2Δ, and Co bfa1Δ, with and without the introduced Co Tem1T128N isoform. Lower left panel indicates conidia with two or three nuclei and an aberrant extra septum in the middle. For other scoring, see legend to Figure 1. At least 200 conidia were scored at each time point.
To investigate whether the nuclear behavior phenotype in Co bub2Δ and Co bfa1Δ is restored by introducing Co Tem1T128N, we analyzed the nuclear behavior of Co Tem1T128N-expressing transformants during appressorium differentiation. As expected, nuclear behavior was normal in most conidia from Co bub2Δ expressing Co Tem1T128N and from Co bfa1Δ expressing Co Tem1T128N (Figures 6B and 6C). While Co bub2Δ and Co bfa1Δ started nuclear division earlier and had more nuclei in the conidium and appressorium than in the wild type, most of the conidia from transformants expressing Co Tem1T128N had normal nuclear division and subsequent distribution to the conidium and appressorium. In addition to the restoration of nuclear division, autophagy also proceeded normally in most conidia from Co Tem1T128N-expressing transformants, while it was significantly delayed in Co bub2Δ and Co bfa1Δ. On the other hand, 20% of conidia from transformants expressing Co Tem1T128N, including the wild type expressing Co Tem1T128N, started nuclear division earlier and formed an aberrant extra septum at the middle of the germinating conidia. Since septa were rarely observed in germinating conidia of the wild type C. orbiculare, this result suggested that the Co TEM1T128N allele might have a dominant effect on regulating nuclear division and septation.
To further characterize the function of Co Tem1, we generated Co tem1 disruption mutants (Supplemental Figure 3). Although most Co tem1Δ conidia started nuclear division at 4 hpi without the extra septum that is typical of the wild type during appressorium differentiation, ∼20% of Co tem1Δ started nuclear division earlier in pregermination conidia and formed an aberrant extra septum at the middle of the germinating conidia (Supplemental Figure 7). This phenotype of Co tem1Δ was consistent with that of transformants expressing Co Tem1T128N. The S. pombe homolog Spg1, one of the SIN components, positively regulates septum formation (Schmidt et al., 1997). When the aseptate phenotype in S. pombe spg1 mutants and the aberrant extra septum in transformants expressing Co Tem1T128N and Co tem1Δ are considered together, Co TEM1 seems to be a negative regulator of septum formation, in contrast with the spg1 homolog in S. pombe. Therefore, these results indicate that Co Tem1 regulates two pathways, that is, G1/S progression under the control of Co Bub2/Co Bfa1 and negative regulation in septum formation.
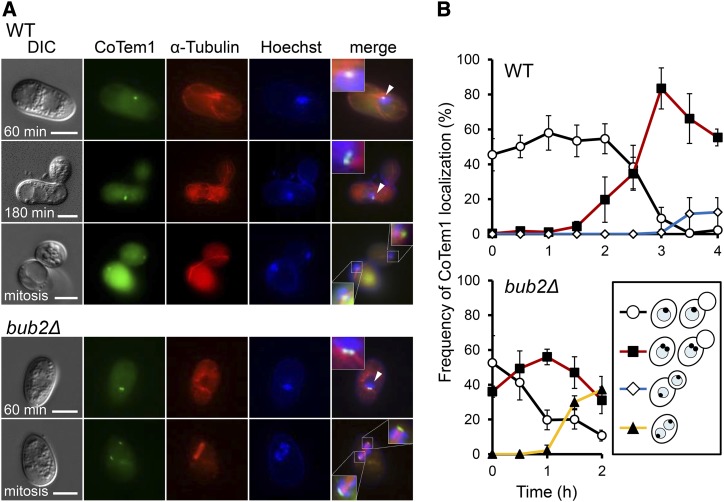
In S. cerevisiae and S. pombe, Co Tem1 homologs localize at spindle pole bodies (SPBs), and this localization is important for its function (Sohrmann et al., 1998; Bardin et al., 2000; Valerio-Santiago et al., 2013). We then analyzed the localization of Co Tem1 and investigated whether it associates with cell cycle progression during appressorium development in C. orbiculare. We constructed strains in which the Co TEM1 gene was C-terminally tagged with GFP under the control of its native promoter. In the pregerminated wild-type cells, one distinct Co Tem1-GFP signal was detected at the nuclear envelope (Figure 7A) in ∼50% of the cells (Figure 7B). In germinated conidia at 2 hpi, double Co Tem1-GFP signals appeared, and these signals were detected in ∼90% of the cells at 3 hpi. At 4 hpi, mitosis completed and double Co Tem1-GFP signals were symmetrically localized at both mother and daughter nuclei. To confirm that Co Tem1-GFP localizes to the SPBs, we observed Co Tem1-GFP in relation to α-Tubulin-mRFP signals. Microscopy observation demonstrated the colocalization of Co Tem1-GFP and α-Tubulin-mRFP during interphase and mitosis (Figure 7A). Especially during mitosis, Co Tem1-GFP signals were detected at both ends of mitotic spindle labeled by α-Tubulin-mRFP, suggesting that Co Tem1 localizes SPBs throughout the cell cycle. Therefore, these results indicate that Co Tem1 localizes putative SPBs in a cell cycle-dependent manner. To elucidate whether the Co Tem1 localization was affected by Co Bub2/Co Bfa1, we visualized the Co TEM1-GFP gene fusion in the Co bub2Δ mutant. From the onset of conidial incubation, single or double Co Tem1-GFP signals were detected at the nucleus of most cells (Figure 7A), and the frequency of double Co Tem1-GFP signals increased by 1 hpi (Figure 7B). During mitosis in pregerminating conidia, Co Tem1 localized at both ends of spindle. These results suggest that the dynamics of the Co Tem1-GFP signal is affected by the absence of Co Bub2. Thus, Co Tem1 localizes at SPBs in a cell cycle-dependent manner and regulates proper G1/S progression in C. orbiculare.
Figure 7.
Co Tem1 Localization throughout the Cell Cycle during Appressorium Development.
(A) Representative images of the wild type and Co bub2Δ expressing Co Tem1-GFP and α-Tubulin-mRFP during appressorium development. Enlargement of Co Tem1 localization is shown in the boxed areas. Arrowheads indicate interphase nuclei. Nuclei was stained with Hoechst 33342. Bars = 5 μm.
(B) Mean percentage (±se; n = 3) of cells expressing the Co TEM1-GFP gene fusion with four patterns of Co Tem1 localization. Scoring: single signal at one nucleus (black), double signals in one nucleus (red), and single signals at both nuclei after nuclear migration in appressorium developing conidia (blue) and in pregermination conidia (yellow). At least 100 conidia were scored at each time point.
Proper Cell Cycle Progression Is Required for Appressorium-Mediated Plant Invasion
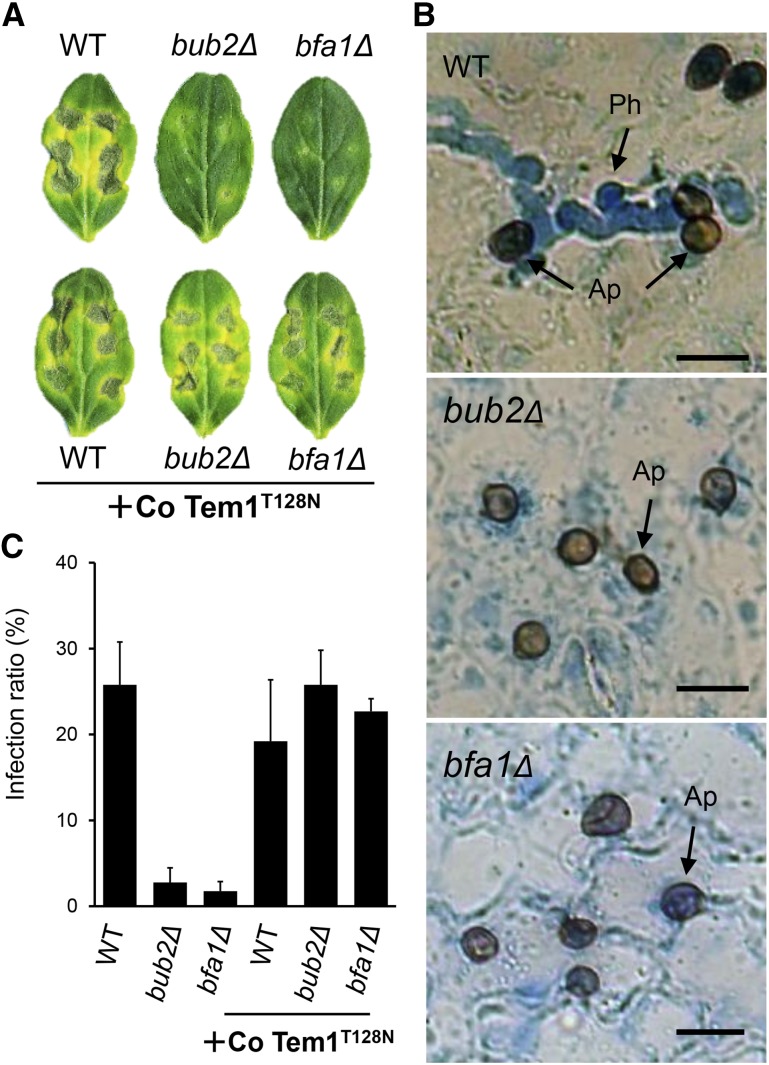
To assess the requirement of Co BUB2 and Co BFA1 for fungal pathogenicity, we inoculated cucumber cotyledons with a conidial suspension of the mutants. In contrast to the distinct necrotic lesions induced by the wild type, Co bub2Δ and Co bfa1Δ induced either no lesions or occasional small lesions (Figure 8A). Microscopy observation revealed that more than 25% of the appressoria of the wild-type strain formed penetration hyphae, which invaded the host plant tissue by 3 d after inoculation (Figures 8B and 8C). By contrast, Co bub2Δ and Co bfa1Δ formed very few penetration hyphae. To confirm that the Co Bub2/Co Bfa1 cascade is involved in appressorial function, pathogenicity and infection-related morphogenesis on the host plant surface were examined in Co bub2Δ and Co bfa1Δ expressing Co Tem1T128N. As expected, expression of Co Tem1T128N restored both infection-related morphogenesis and pathogenicity on the cucumber cotyledons (Figures 8A and 8C). This result indicates that Co Bub2/Co Bfa1 is involved in appressorial function through the Co Tem1 signal cascade. From these results, we hypothesized that the attenuated pathogenicity of Co bub2Δ and Co bfa1Δ resulted from the defect in infection-related morphogenesis.
Figure 8.
Defective Virulence of Co bub2Δ and Co bfa1Δ on Cucumber and Virulence Restored by Introduction of Co Tem1T128N.
(A) Disease symptoms with dark, yellowish sunken necrosis on cucumber cotyledon infected with C. orbiculare wild type, Co bub2Δ, Co bfa1Δ, and the introduced Co Tem1T128N isoform. Conidial suspensions were dropped onto a detached cucumber cotyledon and incubated at 24°C for 7 d.
(B) Micrographs showing penetration hyphae only in the wild type, not in Co bub2Δ, and Co bfa1Δ, on the lower surface of the cotyledon at 3 d after inoculation. Penetration hyphae were stained with lactophenol aniline blue. Ap, appressoria, Ph, penetration hyphae. Bars = 10 µm.
(C) Mean percentage (+se; n = 3) of appressoria forming penetration hyphae. At least 200 appressoria were scored.
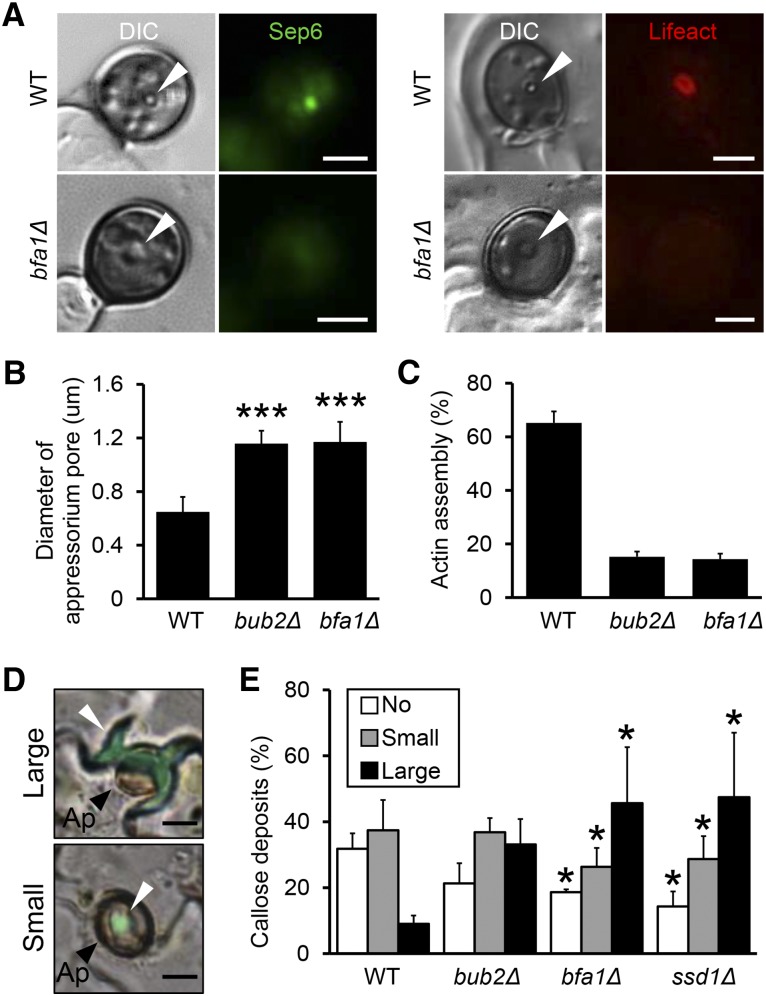
To investigate the relationship between cell cycle progression and appressorial penetration, we decided to focus on the septin and actin assemblies at the appressorium pore because they may provide rigidity to form the penetration peg. In M. oryzae, a septin assembly provides a scaffold for the toroidal F-actin network at the appressorium pore, a circular region from which the rigid penetration peg emerges (Dagdas et al., 2012). So we surmised that Co Bub2/Co Bfa1 affects the dynamics of the septins that might act as a scaffold for actin assembly at the appressorium pore, thereby causing the penetration defect in C. orbiculare mutants. To investigate this hypothesis, we visualized the septin cytoskeleton in Co bfa1Δ as a representative strain. A family of five septin genes has been identified in M. oryzae, and we expressed M. oryzae SEP6-GFP gene fusions in C. orbiculare. The Sep6-GFP signal showed dynamic localization during appressorium development (Supplemental Figure 8). We observed bright punctate structures in pregermination conidia, and after germination, the signal was observed at the tip of immature appressoria. In the mature appressorium, the Sep6-GFP signal was observed as an assembled spot at the neck of the appressorium. When we compared Sep6-GFP localization in Co bfa1Δ with that of the wild type, the frequency of localization of Sep6-GFP in Co bfa1Δ was greater, especially at the neck, the site of septation during cytokinesis. This result suggests that Co bfa1Δ is defective in completing cytokinesis in conidia and the processes for the development of the appressoria.
To assess the dynamics of the septins at the appressorium pore, we compared the localization of Sep6-GFP in Co bfa1Δ with that in the wild type. The wild type formed a distinct, narrow appressorium pore, where the Sep6-GFP signal was bright and punctate (Figures 9A and 9B). By contrast, Co bfa1Δ expressing Sep6-GFP formed a rather opaque and larger appressorium pore, where the Sep6-GFP signal was weaker than in the wild type. This result suggests that Co bfa1Δ has defects in assembling the septins at the appressorium pore.
Figure 9.
Co Bub2/Co Bfa1 Affects the Localization of Septin and Actin and the Defense Response of the Plant.
(A) Representative images showing septin and actin at the appressorium pore in the wild type and Co bfa1Δ during appressorium development. Strains harboring the SEP6-GFP gene were incubated on glass slides for 18 h, and strains harboring the LIFEACT-RFP gene were incubated on a cucumber cotyledon for 2 d. Arrowheads indicate appressorium pore. Bars = 2.5 μm.
(B) Diameter of the appressorium pore in the wild type, Co bub2Δ, and Co bfa1Δ with the LIFEACT-RFP gene (means + se; n = 4). At least 50 appressoria were scored. Each strain was incubated on a cucumber cotyledon for 2 d. Asterisk represents significant differences between the wild type and each mutant (Student’s t test; ***P < 0.005).
(C) Mean percentage (+se; n = 3) of conidia that formed an actin assembly at the appressorium pore in the Co bub2Δ and Co bfa1Δ strain with the LIFEACT-RFP gene. At least 50 appressoria were scored. Each strain was incubated on a cucumber cotyledon for 2 d.
(D) Representative images of papillae with callose deposits at sites of attempted penetration by C. orbiculare appressoria. Large: callose formation larger than boundaries of the appressorium; small: callose deposits smaller than boundaries of the appressorium. Arrowheads indicate papillae. Ap, appressoria. Bars = 5 μm.
(E) Mean percentage (+se; n = 4) of callose deposits at sites of attempted penetration by appressoria of the wild type, Co bub2Δ, Co bfa1Δ, and Δcossd1 at 3 dpi. At least 400 appressoria were scored. Asterisk represents significant differences between the wild type and each mutant (Tukey’s test; *P < 0.05).
To further investigate whether septin dynamics affect actin assembly at the appressorium pore, we visualized actin during appressorium development. As expected, the localization frequency of Lifeact-RFP at the appressorium pore was reduced, and the signal intensity was mostly fainter in Co bub2Δ and Co bfa1Δ than in the wild type (Figures 9A and 9C). These results indicate that the assembly of septin and actin at the appressorium pore was disturbed in Co bub2Δ and Co bfa1Δ.
Finally, we evaluated the possibility that the plant defense response is involved in the attenuated pathogenesis of Co bub2Δ and Co bfa1Δ. Unexpectedly, at sites of attempted penetration by Co bub2Δ and Co bfa1Δ, 80% of appressoria were accompanied by callose deposits, which were considerably larger than those formed by the wild type (Figures 9D and 9E). When inoculated leaves were stained with 3,3′-diaminobenzidine (DAB) to detect the accumulation of reactive oxygen species (ROS), 50% of plant cells attacked by Co bub2Δ or Co bfa1Δ appressoria showed positive DAB staining, as opposed to 0% of those attacked by the wild type (Supplemental Figure 9). To further evaluate the involvement of a plant defense reaction, we assayed infection after inoculation with Co bub2Δ or Co bfa1Δ on cucumber leaves that had been heat-shocked to impair resistance (Tanaka et al., 2007). The lesion formation by Co bub2Δ and Co bfa1Δ was partially restored on the leaves subjected to heat shock treatment (Supplemental Figure 9). Conclusively, these data suggest that Co Bub2/Co Bfa1 is required for pathogenesis via Co Tem1 and that the attenuated pathogenicity of Co bub2Δ and Co bfa1Δ resulted from a defect in septin- and actin-mediated penetration and in inducing a plant defense response.
DISCUSSION
The Novel Function of BUB2 and BFA1 in G1/S Progression in C. orbiculare
In S. cerevisiae, Bub2p constitutes a Rab GAP two-component complex with Bfa1p, and the Bub2p/Bfa1p complex regulates MEN as several cell cycle checkpoint components, such as SPOC, SAC, and DDC, in response to the misoriented spindle, unattached spindle to the kinetochore, and damaged DNA, respectively (Wang et al., 2000; Hu et al., 2001). By contrast, in S. pombe and A. nidulans, homologs of BUB2 and BFA1 act as negative regulators of the SIN, which is required for the onset of septum formation. SIN signaling activity is tightly regulated to ensure proper coordination of mitosis and cytokinesis (Furge et al., 1998; Li et al., 2000; Kim et al., 2006). In contrast to the results obtained so far for model yeast and fungi, we showed here that C. orbiculare homologs of BUB2 and BFA1, Co BUB2 and Co BFA1, play a major role in phase progression from G1 to S. This novel finding is supported by the following results. First, a mutation of Co bub2 and Co bfa1 initiated earlier nuclear division and caused binucleation in the pregermination conidia. Second, experiments using the LacO/LacI-GFP chromosome tagging system indicated that Co bub2Δ and Co bfa1Δ are defective in G1/S arrest. Thus, disruption of Co BUB2 and Co BFA1 did not significantly alter the phenotypes for mitotic exit or septum formation, while the primary functions of these homologs in S. cerevisiae, S. pombe, and A. nidulans are to inhibit the MEN or SIN signal cascade. From these phenotypic results, we wondered whether Co BUB2 and Co BFA1 are orthologs of S. cerevisiae BUB2 and BFA1. A complementation test with S. cerevisiae BUB2 and BFA1 showed the restoration of proper G1/S progression in the Co bub2Δ Co bfa1Δ double mutant. Furthermore, the physical association of Co Bub2 with Co Bfa1 in a yeast two-hybrid assay confirmed that Co Bub2 forms a complex with Co Bfa1 similar to the Bub2p/Bfa1p GAP complex of S. cerevisiae. Therefore, we conclude that Co BUB2 and Co BFA1 are orthologs of S. cerevisiae BUB2 and BFA1 but have distinct roles in G1/S progression in C. orbiculare.
Co Tem1 GTPase Is a Downstream Target of Co Bub2/Co Bfa1 Rab GAP
In S. cerevisiae and C. albicans, the Bub2p/Bfa1p Rab GAP complex negatively regulates the direct downstream GTPase Tem1p, a component of MEN (Bardin and Amon, 2001; Milne et al., 2014). In S. pombe and A. nidulans, an analogous cascade SIN is conserved, and Cdc16/BUBA and Byr4/BYRA, homologs of Bub2p and Bfa1p, negatively regulate the downstream GTPase Tem1p homolog Spg1/ASGA (Furge et al., 1998; Li et al., 2000; Kim et al., 2006). The tightly regulated signal cascade of the two-component GAP and GTPase prompted us to question whether Co Bub2/Co Bfa1 regulates G1/S progression mediated by the Tem1 homolog Co Tem1 or another GTPase specific to C. orbiculare.
Our yeast two-hybrid experiments suggested that the N-terminal region of Co Tem1 is important for its association with the Co Bub2/Co Bfa1 GAP complex in C. orbiculare. However, in the case of the interaction of C. orbiculare Bub2/Bfa1 with S. cerevisiae Tem1p, Co Bfa1 associated with S. cerevisiae Tem1p, which does not contain the N-terminal region found in C. orbiculare. Thus, this difference in the Tem1 sequence and the association with the Bub2/Bfa1 GAP complex could be the reason for the difference in the cellular response between C. orbiculare and S. cerevisiae.
We introduced the T128N dominant-negative form of Co Tem1 into Co bub2Δ or Co bfa1Δ to test for complementation. Introduction of Co Tem1T128N successfully complemented the defects of Co bub2Δ and Co bfa1Δ in the phase progression from G1 to S and autophagy, suggesting that Co Tem1 regulates G1/S progression through negative regulation by the Co Bub2/Co Bfa1 GAP complex. Thus, the two-component GAP Co Bub2/Co bfa1 seems to regulate G1/S progression through Co Tem1 in genetic and physical experiments.
Furthermore, we examined Co Tem1 localization during appressorium development. Co Tem1 localized to putative SPBs throughout cell cycle progression, and signal duplication of Co Tem1 increased from the onset of S phase, consistent with the duplication of SPBs reported in S. cerevisiae (Byers and Goetsch, 1975). In yeast, the homologs of Co Tem1 localize to SPBs and the localization affects MEN and SIN function (Sohrmann et al., 1998; Bardin et al., 2000). Therefore, Co Tem1 localization at SPBs was consistent with these reports, indicating that Co Tem1 regulates G1/S progression by localizing to SPBs.
A Role for Bub2/Bfa1 in Infection-Related Morphogenesis and Pathogenesis in C. orbiculare
The cell cycle is pivotal to cellular differentiation in multicellular eukaryotes, which must synchronize cell division to form specific tissues and organs effectively (Kipreos, 2005; Théry and Bornens, 2006; Cools and De Veylder, 2009). Most plant pathogenic fungi develop appressoria to rupture plant cuticle and invade plant tissue. Cell cycle progression in plant pathogenic fungi is coordinately regulated to form proper infection structures. In M. oryzae, S phase is necessary to initiate appressorium differentiation, while M phase and autophagy in conidia are essential for the development of functional appressoria that form the penetration peg (Veneault-Fourrey et al., 2006; Saunders et al., 2010a). By contrast, in U. maydis, cell cycle arrest in G2 phase is required for the induction of appressorium formation, and the arrest is held until the infective dikaryotic hyphae penetrate host plants (Castanheira et al., 2014). Thus, M. oryzae and U. maydis have distinctive mechanisms of appressorium development that are coordinated with proper cell cycle progression. It was hypothesized that differences in appressorium development depend on the type of infection strategy adopted by appressoria (Castanheira and Perez-Martín, 2015). In one strategy, high turgor pressure enables cuticle penetration and, in the other, maintenance of heterokaryosis is required for successful infection. An analysis of microtubules dynamics in C. orbiculare during appressorium development indicates that precise nuclear distribution is required for proper appressorial development (Takano et al., 2001). However, a precise molecular genetic analysis of cell cycle regulation affecting infection-related morphogenesis and pathogenesis had not been reported for any Colletotrichum species.
For the M. oryzae appressorium to rupture the leaf cuticle, septin assemblies and an extensive toroidal F-actin network must be established at the appressorium pore (Dagdas et al., 2012). In Co bub2Δ and Co bfa1Δ, the frequency of septin and actin assembly was lower than in the wild type and some of the appressorium pores in the mutants were larger in diameter than were those of the wild type. This result indicates that the attenuated assembly of septin and actin in Co bub2Δ and Co bfa1Δ decrease cortical rigidly at the appressorium pore enough that the fungus cannot penetrate the host cuticle.
In addition to this functional attenuation of appressorium vigor, these mutants also unexpectedly induced an increased plant defense response, such as callose deposit and ROS accumulation, where the appressoria attempted to penetrate the plant cells. Heat-treated leaves of the host plant were unaffected by the defect of penetration ability of the Co bub2Δ and Co bfa1Δ, also suggesting that the mutant defect in forming disease lesions is associated with a host defense response. In C. orbiculare, the pexophagy-related mutant coatg26 is defective not only in forming a penetration peg, but also in repressing the host defense response, thus leading to an inability to invade the host (Asakura et al., 2009). In M. oryzae, autophagy in the conidia was reported to have a pivotal role in the development of infection-related structures and for the avoidance of the plant immunity response (Talbot and Kershaw, 2009). Our analysis of Co bub2Δ and Co bfa1Δ showed that nuclei in the conidia were not degraded at the proper time, indicating a defect in autophagy.
Therefore, we propose that proper G1/S arrest by Co Bub2/Co Bfa1 during conidial germination is required to establish plant infection in C. orbiculare.
Global Regulation of Two-Component GAP and GTPase among Yeast and Filamentous and Dimorphic Fungal Species
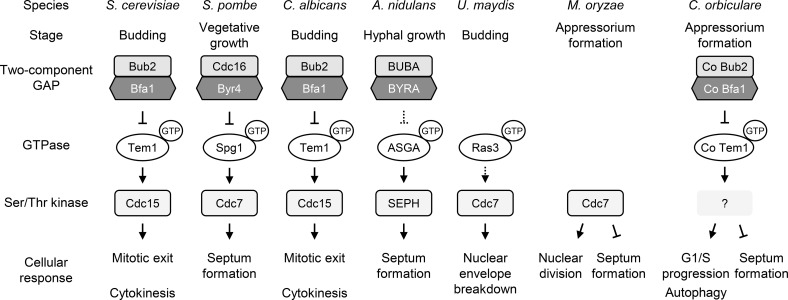
Eukaryotes possess a wide variety of conserved proteins related to the cell cycle, but the function of the respective proteins may not always be identical among various species. Our study showed that the Bub2/Bfa1-Tem1 cascade is conserved between yeast and C. orbiculare; however, this signal cascade has different functions in yeast and in other filamentous fungi. Our results suggest that C. orbiculare has plant pathogen-specific roles for the two-component GAP and its GTPase that are involved in functional development in various yeast and fungal species (Figure 10). In S. cerevisiae and C. albicans, Bub2p/Bfa1p negatively regulates GTPase Tem1p, thereby triggering the MEN pathway, which controls mitotic exit and cytokinesis via Ser/Thr kinase Cdc15p (Bardin and Amon, 2001; Finley et al., 2008; Milne et al., 2014). A similar cascade, the SIN, has been reported in S. pombe and A. nidulans, and its primary role appears to be in regulating septation rather than mitotic exit (Bruno et al., 2001; Krapp and Simanis, 2008; Kim et al., 2009). By contrast, the Tem1 homolog in the plant-pathogenic basidiomycete fungus U. maydis, GTPase Ras3, is required for nuclear envelope breakdown during open mitosis via Cdc7 kinase (Straube et al., 2005). In ascomycete plant pathogens, the Ser/Thr kinase gene SEP1, a homolog of S. cerevisiae CDC15, was identified in M. oryzae, and its encoded protein both coordinates nuclear division and negatively regulates cytokinesis, functions that are required for successful appressorial penetration (Saunders et al., 2010b). In this study, we found that two-component GAP Bub2/Bfa1 of C. orbiculare regulates G1/S progression and autophagy via GTPase Tem1. Unlike yeast and fungal species, a mutation of Co bub2 and Co bfa1 did not impair mitotic exit or septation, but instead affected G1/S progression during appressorium development and led to penetration failure on the host plant. The observation that Co Tem1T128N or Co tem1Δ mutants had an extra septum suggests that Co TEM1 negatively regulates septum formation, consistent with the report from M. oryzae sep1 mutants (Saunders et al., 2010b). However, this result is surprising because the homologs reported for yeast and other filamentous fungi positively regulate septum formation. Therefore, the signal cascade involving the two-component GAP and MEN/SIN components has been conserved, but the functions of the homologous genes in various species differ.
Figure 10.
Model for Bub2/Bfa1 Functions and the Tem1 Pathways.
Schematic comparison of signaling networks of two-component GAP, GTPase, and Ser/Thr kinase and the subsequent cellular response in yeast and filamentous fungi. Whereas the signal cascade is widely conserved, the cellular response is diverse in S. cerevisiae, S. pombe, C. albicans, A. nidulans, U. maydis, M. oryzae, and C. orbiculare, regulating mitotic exit, septum formation, nuclear envelope breakdown, and G1/S progression. In C. orbiculare, Co Bub2/Co Bfa1 two-component GAP has a novel function that regulates G1/S progression, and autophagy via GTPase Co Tem1 and Co Tem1 negatively regulates septum formation.
In conclusion, we consider that the Bub2/Bfa1 and Tem1 signaling cascade of C. orbiculare has been adapted for novel functional strategies to fulfill specific plant-pathogenic roles in different fungal species. Further investigation of Bub2/Bfa1 and Tem1 in other plant pathogens may provide insight into the diverse roles of these proteins in the cell cycle, which are likely to be necessary for virulence-associated processes in fungi.
METHODS
Strains and Growth Conditions
Strain 104-T (MAFF240422) of Colletotrichum orbiculare was used as the wild-type strain. All C. orbiculare strains were maintained at 24°C in darkness on 3.9% (w/v) potato dextrose agar (PDA) (Difco Laboratories) or SD medium (0.67% [w/v] yeast nitrogen base without amino acids, 2% [w/v] glucose, and 2% [w/v] agar). Escherichia coli DH5α-competent cells were used as a host for gene manipulation and maintained on Luria-Bertani medium at 37°C. Agrobacterium tumefaciens strain C58C1 was used as the T-DNA donor for fungal transformation and maintained on Luria-Bertani medium at 28°C.
Fungal Transformation
The AtMT protocol was applied with slight modifications of a previously described method (Tsuji et al., 2003). Hygromycin-resistant transformants were selected on PDA containing 100 µg/mL hygromycin B (Wako Chemicals) and 25 µg/mL meropenem hydrate (Sumitomo Dainippon Pharma). Bialaphos-resistant transformants were selected on SD medium containing 4 µg/mL bialaphos (Meiji Seika Kaisha) and 25 µg/mL meropenem hydrate. Sulfonylurea-resistant transformants were selected on SD medium containing 4 µg/mL chlorimuron ethyl (Maruwa Biochemical) and 25 µg/mL meropenem hydrate. For ectopic transformation of C. orbiculare with a fusion of Lac operator repeats and M. oryzae SEP6-GFP (Dagdas et al., 2012), polyethylene glycol-mediated protoplast transformation was performed as described previously (Kubo and Furusawa, 1991). All C. orbiculare strains generated in this study are listed in Supplemental Table 2.
Mutant Screening and Identification of C. orbiculare PDM1
Mutants of C. orbiculare PDM1 were screened using the AtMT protocol as described previously (Sakaguchi et al., 2010). The T-DNA inserted gene in the PDM1 mutant was identified using a thermal asymmetric interlaced-PCR protocol (Liu et al., 1995). Fungal genomic DNA flanking the T-DNA insert was analyzed as previously described (Tsuji et al., 2003). Amplified PCR products were sequenced with the Big-Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and an ABI PRISM 310 automated DNA sequencer (Applied Biosystems).
Gene Manipulations
For sequencing the entire region of the open reading frame of Co BUB2, Co BFA1, and Co TEM1, total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen), and cDNA synthesis and subsequent PCR amplification were performed using ReverTra Ace (Toyobo). The oligo(dT)12-20 primer was used for cDNA synthesis. Co BUB2, Co BFA1, and Co TEM1 were amplified using the primers shown in Supplemental Table 3. The resulting cDNA was sequenced by Eurofins Genomics.
Genomic DNA of C. orbiculare was isolated from mycelia, and DNA gel blot analysis was done by standard methods. DNA probes were labeled with DIG-dUTP using the BcaBEST DIG labeling kit (Takara Bio). Hybridized DNA was detected using Anti-Digoxygenin-AP Fab fragments (Roche Diagnostics), and light emission generated by enzymatic dephosphorylation of CDP-Star Detection Reagent (GE Healthcare) and by alkaline phosphatase was detected using the FujiFilm LAS1000 plus gel documentation system.
Plasmid Construction
Plasmids were derived from the following binary vectors: pBIG4MRHrev, carrying the hygromycin resistance gene cassette; pBIG4MRBrev, carrying the bialaphos resistance gene cassette; pBIG4MRSrev and pCAMBIA-Sur-RfA, carrying the sulfonylurea resistance gene cassette; and pBIG4MRNrev, carrying the neomycin resistance gene cassette. For plasmid constructions, the In-Fusion HD cloning kit (Clontech) or GENEART Seamless cloning and assembly kit (Life Technologies) was used. All primers used in this study are listed in Supplemental Table 3.
To generate gene disruption mutants, plasmids in which cloned genes were replaced with the hygromycin resistance gene cassette were constructed. For generating the Co bub2 disruption vector, a 1.0-kb fragment of the 5′ upstream region and a 1.0-kb fragment of the 3′ downstream region were amplified from C. orbiculare genomic DNA, and a 1.4-kb fragment of the hygromycin resistance gene was amplified from pBIG4MRHrev. These three fragments were inserted into a linearized pBIG4MRSrev. The same procedures were used to generate the Co bfa1 disruption vector and the Co tem1 disruption vector.
For chromosome tagging using lac operator/lac repressor system, two plasmids that contained the GFP-lac repressor fusions and lac operator repeats were constructed. To generate GFP-lac repressor fusion plasmids, the 3.4-kp lacI-NLS-SV40 poly(A) fragment was amplified from the yeast vector pYN15 (Straight et al., 1996; Nabeshima et al., 1998), and the fragment was inserted at the C-terminal of GFP driven by Aureobasidium pullulans TEF promoter (Fujihara et al., 2010). To construct Lac operator repeat plasmids, the 2.8-kb sulfonylurea resistance gene cassette was digested with SalI from pBIG4MRSrev and introduced into the SalI-digested yeast vector pCT31, which carries the 10.2-kb 256 Lac operator repeats (Yamamoto and Hiraoka, 2003).
For complementation assays of the mutants, plasmids that contained the complete gene, native promoter, and terminator were constructed. To generate the Co BUB2 complement vector, a Co BUB2 fragment that contained 1.0 kb of the 5′ upstream region, the open reading frame, and 1.0 kp of 3′ downstream region was amplified from C. orbiculare genomic DNA. The fragment was inserted into a linearized pBIG4MRSrev. The same procedures were used to construct the Co BFA1 complementation vector and the Co TEM1 complementation vector.
For visualizing of C. orbiculare Histone H1 (Co His1) and Co Tem1, a plasmid that carries the gene encoding GFP regulated by the native promoter of Co HIS1 or Co TEM1 was constructed. For constructing the Co HIS1-GFP fusion gene vector, the Co HIS1 complementation vector that carries the Co HIS1 ORF with 1.0 kb of the 5′ upstream region and 1.0 kb of the 3′ downstream region was constructed. The GFP fragment was amplified from pBIglyGFP and inserted at the C-terminal end of Co His1 in the complementation vector. For constructing the C. orbiculare Co TEM1-GFP fusion gene vector, the GFP fragment was amplified from pBICoHIS1GFP and inserted at the C-terminal end of Co Tem1 in the Co TEM1 complementation vector described above. For labeling microtubules, pBISCD1pmRFP1-α-TUB1S was used as previously described (Sakaguchi et al., 2008).
To complement the C. orbiculare mutants with S. cerevisiae BUB2 and BFA1, the open reading frame of Co BUB2 and Co BFA1 was replaced with S. cerevisiae BUB2 and BFA1, respectively. To construct the S. cerevisiae BUB2-replaced vector, the BUB2 open reading frame was amplified from the S. cerevisiae DNA and replaced with the Co BUB2 open reading frame in the Co BUB2 complementation vector. The same plasmid construction strategy was used to generate the S. cerevisiae BFA1-replaced vector.
To generate a dominant-negative form of Co TEM1, a 516-bp fragment containing the upstream region of the Co TEM1 open reading frame and a 522-bp fragment containing the downstream region of the Co TEM1 open reading frame were amplified from C. orbiculare genomic DNA to generate a point mutation, which has an ACG-to-AAT substitution at codon 128. These two fragments were inserted into the pBIG4MRBrev binary vector. The sulfonylurea-selectable marker gene was amplified and inserted downstream of the 3′ untranslated region of Co TEM1 in the Co TEM1 point mutation plasmid.
Yeast Two-Hybrid Interaction Assays
The yeast two-hybrid screen was performed using the instructions of the Matchmaker Gold Yeast Two-Hybrid System (Clontech). Full-length cDNAs of putative interaction partners were generated from 3-d-old C. orbiculare mycelia. The genes encoding the proteins tested for interaction were cloned into the pGBKT7 or pGADT7 vectors (Clontech) to express fusion proteins with the yeast GAL4 binding (BD) and activation domain (AD), respectively. All BD or AD constructs were used to transform the Gold or Y187 yeast strain, respectively (Clontech). After mating, diploid yeast was plated on double dropout synthetic selective medium lacking Trp and Leu (DDO) for mating control and on stringent medium supplemented with 100 ng/mL aureobasidin A and 20 μg/mL X-α-Gal (DDO/A/X) and incubated at 28°C for 5 d. Protein interactions were assessed by growth of diploid yeast on DDO, DDO/A, DDO/A/X, and stringent quadruple dropout synthetic selective medium lacking Trp, Leu, Ade, and His (QDO) compared with corresponding controls (empty vectors).
Chemical Treatments
For staining nuclei of living cells, 2 μL of 100 μg/mL Hoechst 33342 (Dojindo Laboratories) was added to cells on samples on the glass slide and incubated for 10 min. For staining nuclei of fixed cells, samples were fixed with 3 to 4% (v/v) formaldehyde in 0.1 M phosphate buffer (pH 7.0) and 0.2% Triton X-100 at room temperature for 1 h. Fixed cells were then washed twice with 0.1 M phosphate buffer, and 1-μL of 1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) was added directly to the cells on slides. For staining cell wall polysaccharides, 1-μL of 40 ng/mL Fluorescent Brightener 28 (Calcofluor white; MP Biomedicals) was added directly to the cells on slides. To examine penetration hyphae development in cucumber cotyledons, each sample was visualized using 0.1% lactophenol-aniline blue solution (Takano et al., 1997). For detecting callose deposits and ROS accumulation, each sample was stained with 0.01% (w/v) aniline blue in 0.15 M K2HPO4 and DAB (Yoshioka et al., 2003), respectively.
Pathogenicity Tests
C. orbiculare was tested for pathogenicity on detached cucumber cotyledons (Cucumis sativus) as described previously (Tsuji et al., 2003). Conidia of C. orbiculare were obtained from 7-d-old PDA cultures, and six drops of 10-μL of a conidial suspension (5 × 105 conidia/mL) were placed on the surface of cucumber cotyledons, and the tested strains were dropped onto five cucumber cotyledons. The cotyledons were incubated in a humid box at 24°C with a 16-h photoperiod for 7 d. For the heat shock treatment, detached cucumber cotyledons were dipped into distilled water at 50°C for 30 s and then inoculated with the test strains. For the control in all pathogenicity tests, distilled water was used instead of the conidial suspension.
Microscopy
For observation of appressorium formation, 20-μL of conidial suspension (105 conidia/mL 0.1% [w/v] yeast extract solution) was placed in the wells of multiwell glass slides (8-mm diameter; Matsunami Glass) and incubated in a humid environment at 24°C in the dark for 1 h. The yeast extract solution was then removed and replaced with distilled water. For observing penetration hyphae formation in vitro, a conidial suspension (105 conidia/mL distilled water) was placed on cellophane membranes (Wako Chemicals) and incubated at 24°C in the dark. For assessing penetration hyphae formation in planta, 10 μL of a conidial suspension (5 × 105 conidia/mL distilled water) was spotted onto the abaxial surface of cucumber cotyledons and incubated in a humid box at 24°C. After 3 d, we peeled off the lower epidermis of the cotyledons for observation on glass slides. For examining vegetative hyphae, the conidial suspension (105 conidia/mL 0.1% yeast extract solution) was placed on slides and incubated at 30°C.
Fluorescence was detected using a Zeiss Axio Imager M2 Upright microscope equipped with an AxioCam MRm digital camera and excitation/barrier filter set of 470 nm/509 nm for GFP and 595 nm/620 nm for RFP. Images were acquired with a 100× oil immersion lens (Plan Apochromat) using Axiovision 4.8 software. To observe the signal of GFP-lacI, 1.6× intermediate variable magnification was used. DAPI-stained and calcofluor white-stained samples were observed with a Nikon ECLIPSE E600 microscope equipped with a Keyence VB-7010 CCD color camera system. An excitation wavelength of 365/10 nm, dichroic mirror wavelength of 400 nm, and a barrier filter wavelength of 400 nm were used. Images were acquired with a 40× water immersion lens or a 100× oil immersion lens (Plan Fluor). To quantify the diameter of an appressorium pore, the images were analyzed using ImageJ (http://rsb.info.nih.gov/ij/).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: C. orbiculare Co Bub2 (ENH83696), Co Bfa1 (ENH87866), Co Tem1 (ENH82693), Co His1 (ENH76847); Saccharomyces cerevisiae Bub2 (NP_013771), Bfa1 (NP_012587), and Tem1 (NP_013647).
Supplemental Data
Supplemental Figure 1. Amino acid sequence alignment of Co Bub2 homologs.
Supplemental Figure 2. Amino acid sequence alignment of Co Bfa1 homologs.
Supplemental Figure 3. DNA gel blot analysis of targeted gene deletion mutants.
Supplemental Figure 4. Co BUB2 is not involved in G1/S progression during vegetative hyphal growth in C. orbiculare.
Supplemental Figure 5. Amino acid sequence alignment of Co Tem1 homologs.
Supplemental Figure 6. Co Bub2 interacts with Co Bfa1 in yeast two-hybrid assays.
Supplemental Figure 7. Co tem1Δ is impaired in septum formation but not in nuclear behavior during appressorium development.
Supplemental Figure 8. Co Bub2/Co Bfa1 affects the localization of septin during appressorium development in C. orbiculare.
Supplemental Figure 9. A defense response in plants was induced by attempted penetration by appressoria of C. orbiculare mutants Co bub2Δ and Co bfa1Δ.
Supplemental Table 1. Growth rate and conidiation of mutants on PDA medium.
Supplemental Table 2. Colletotrichum orbiculare strains used in this study.
Supplemental Table 3. Primers used in this study.
Supplemental Table 4. Saccharomyces cerevisiae strains used in this study.
Supplemental Movie 1. Time-lapse imaging of nuclear division during appressorium development in wild-type C. orbiculare.
Supplemental Movie 2. Time-lapse imaging of nuclear division in pregermination conidia of C. orbiculare bub2Δ.
Supplementary Material
Acknowledgments
We thank N.J. Talbot and O.R. Miriam (University of Exeter, UK) for the Sep6-GFP vector of M. oryzae, A. Yamamoto (Shizuoka University, Japan) and A. Belmont (University of Illinois) for lac operator and GFP-lacI plasmid, Y. Nishizawa and M. Nishimura (National Institute of Agrobiological Sciences, Japan) for pCAMBIA-Bar-RfA, and G. Tsuji (Kyoto Prefectural University, Japan) for the Lifeact-RFP vector. We thank B.E. Hazen for carefully reading the article and giving valuable suggestions. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (Grants 24248009, 15H05780, and KH20140023) and by the Mitsubishi Foundation (No. 24110).
AUTHOR CONTRIBUTIONS
F.F. designed and performed the research, analyzed the data, and wrote the article. Y.K. designed the research, analyzed the data, and wrote the article.
Glossary
- MEN
mitotic exit network
- SPOC
spindle position checkpoint
- SAC
spindle assembly checkpoint
- DDC
DNA damage checkpoint
- SIN
septation initiation network
- AtMT
Agrobacterium tumefaciens-mediated transformation
- hpi
hours postincubation
- IDR
imperfect direct repeat
- DAB
3,3′-diaminobenzidine
- ROS
reactive oxygen species
- PDA
potato dextrose agar
- DAPI
4′,6-diamidino-2-phenylindole
- SPB
spindle pole body
Footnotes
Articles can be viewed online without a subscription.
References
- Asakura M., Ninomiya S., Sugimoto M., Oku M., Yamashita S., Okuno T., Sakai Y., Takano Y. (2009). Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell 21: 1291–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A.J., Amon A. (2001). Men and sin: what’s the difference? Nat. Rev. Mol. Cell Biol. 2: 815–826. [DOI] [PubMed] [Google Scholar]
- Bardin A.J., Visintin R., Amon A. (2000). A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102: 21–31. [DOI] [PubMed] [Google Scholar]
- Bruno K.S., Morrell J.L., Hamer J.E., Staiger C.J. (2001). SEPH, a Cdc7p orthologue from Aspergillus nidulans, functions upstream of actin ring formation during cytokinesis. Mol. Microbiol. 42: 3–12. [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. (1975). Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124: 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira S., Mielnichuk N., Pérez-Martín J. (2014). Programmed cell cycle arrest is required for infection of corn plants by the fungus Ustilago maydis. Development 141: 4817–4826. [DOI] [PubMed] [Google Scholar]
- Castanheira S., Pérez-Martín J. (2015). Appressorium formation in the corn smut fungus Ustilago maydis requires a G2 cell cycle arrest. Plant Signal. Behav. 10: e1001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools T., De Veylder L. (2009). DNA stress checkpoint control and plant development. Curr. Opin. Plant Biol. 12: 23–28. [DOI] [PubMed] [Google Scholar]
- Dagdas Y.F., Yoshino K., Dagdas G., Ryder L.S., Bielska E., Steinberg G., Talbot N.J. (2012). Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 22: 1590–1595. [DOI] [PubMed] [Google Scholar]
- Finley K.R., Bouchonville K.J., Quick A., Berman J. (2008). Dynein-dependent nuclear dynamics affect morphogenesis in Candida albicans by means of the Bub2p spindle checkpoint. J. Cell Sci. 112: 466–476. [DOI] [PubMed] [Google Scholar]
- Fujihara N., Sakaguchi A., Tanaka S., Fujii S., Tsuji G., Shiraishi T., O’Connell R., Kubo Y. (2010). Peroxisome biogenesis factor PEX13 is required for appressorium-mediated plant infection by the anthracnose fungus Colletotrichum orbiculare. Mol. Plant Microbe Interact. 23: 436–445. [DOI] [PubMed] [Google Scholar]
- Furge K.A., Wong K., Armstrong J., Balasubramanian M., Albright C.F. (1998). Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol. 8: 947–954. [DOI] [PubMed] [Google Scholar]
- Gan P., Ikeda K., Irieda H., Narusaka M., O’Connell R.J., Narusaka Y., Takano Y., Kubo Y., Shirasu K. (2013). Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 197: 1236–1249. [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Smith S.J., Wheatley E., Rittinger K., Johnston L.H., Sedgwick S.G. (2002). Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 277: 28439–28445. [DOI] [PubMed] [Google Scholar]
- Harata K., Kubo Y. (2014). Ras GTPase activating protein CoIra1 is involved in infection-related morphogenesis by regulating cAMP and MAPK signaling pathways through CoRas2 in Colletotrichum orbiculare. PLoS One 9: e109045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J.C., Haber J.E. (2006). Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40: 209–235. [DOI] [PubMed] [Google Scholar]
- Hu F., Elledge S.J. (2002). Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints. Cell Cycle 1: 351–355. [PubMed] [Google Scholar]
- Hu F., Wang Y., Liu D., Li Y., Qin J., Elledge S.J. (2001). Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107: 655–665. [DOI] [PubMed] [Google Scholar]
- Kim J., Jang S.S., Song K. (2008). Different levels of Bfa1/Bub2 GAP activity are required to prevent mitotic exit of budding yeast depending on the type of perturbations. Mol. Biol. Cell 19: 4328–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Lu L., Shao R., Chin J., Liu B. (2006). Isolation of mutations that bypass the requirement of the septation initiation network for septum formation and conidiation in Aspergillus nidulans. Genetics 173: 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Zeng C.J., Nayak T., Shao R., Huang A.C., Oakley B.R., Liu B. (2009). Timely septation requires SNAD-dependent spindle pole body localization of the septation initiation network components in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 20: 2874–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos E.T. (2005). C. elegans cell cycles: invariance and stem cell divisions. Nat. Rev. Mol. Cell Biol. 6: 766–776. [DOI] [PubMed] [Google Scholar]
- Krapp A., Simanis V. (2008). An overview of the fission yeast septation initiation network (SIN). Biochem. Soc. Trans. 36: 411–415. [DOI] [PubMed] [Google Scholar]
- Kubo Y., Furusawa I. (1991). Melanin biosynthesis: Prerequisite for successful invasion of the plant host by appressoria of Colletotrichum and Pyricularia. In The Fungal Spore and Disease Initiation in Plants and Animals, Cole G.T., Hoch H.C., eds (New York: Plenum Publishing; ), pp. 205–217. [Google Scholar]
- Kubo Y., Takano Y. (2013). Dynamics of infection-related morphogenesis and pathogenesis in Colletotrichum orbiculare. J. Gen. Plant Pathol. 79: 233–242. [Google Scholar]
- Li C., Furge K.A., Cheng Q.C., Albright C.F. (2000). Byr4 localizes to spindle-pole bodies in a cell cycle-regulated manner to control Cdc7 localization and septation in fission yeast. J. Biol. Chem. 275: 14381–14387. [DOI] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463. [DOI] [PubMed] [Google Scholar]
- Milne S.W., Cheetham J., Lloyd D., Shaw S., Moore K., Paszkiewicz K.H., Aves S.J., Bates S. (2014). Role of Candida albicans Tem1 in mitotic exit and cytokinesis. Fungal Genet. Biol. 69: 84–95. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. (2007). The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8: 379–393. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa T., Straight A.F., Murray A., Chikashige Y., Yamashita Y.M., Hiraoka Y., Yanagida M. (1998). Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 9: 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S. (1996). In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135: 1685–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A., Miyaji T., Tsuji G., Kubo Y. (2008). Kelch repeat protein Clakel2p and calcium signaling control appressorium development in Colletotrichum lagenarium. Eukaryot. Cell 7: 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A., Tsuji G., Kubo Y. (2010). A yeast STE11 homologue CoMEKK1 is essential for pathogenesis-related morphogenesis in Colletotrichum orbiculare. Mol. Plant Microbe Interact. 23: 1563–1572. [DOI] [PubMed] [Google Scholar]
- Saunders D.G.O., Aves S.J., Talbot N.J. (2010a). Cell cycle-mediated regulation of plant infection by the rice blast fungus. Plant Cell 22: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders D.G.O., Dagdas Y.F., Talbot N.J. (2010b). Spatial uncoupling of mitosis and cytokinesis during appressorium-mediated plant infection by the rice blast fungus Magnaporthe oryzae. Plant Cell 22: 2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S., Sohrmann M., Hofmann K., Woollard A., Simanis V. (1997). The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 11: 1519–1534. [DOI] [PubMed] [Google Scholar]
- Sohrmann M., Schmidt S., Hagan I., Simanis V. (1998). Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 12: 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Belmont A.S., Robinett C.C., Murray A.W. (1996). GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Straube A., Weber I., Steinberg G. (2005). A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J. 24: 1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano Y., Kubo Y., Kuroda I., Furusawa I. (1997). Temporal transcriptional pattern of three melanin biosynthesis genes, PKS1, SCD1, and THR1, in appressorium-differentiating and nondifferentiating conidia of Colletotrichum lagenarium. Appl. Environ. Microbiol. 63: 351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano Y., Oshiro E., Okuno T. (2001). Microtubule dynamics during infection-related morphogenesis of Colletotrichum lagenarium. Fungal Genet. Biol. 34: 107–121. [DOI] [PubMed] [Google Scholar]
- Talbot N.J., Kershaw M.J. (2009). The emerging role of autophagy in plant pathogen attack and host defence. Curr. Opin. Plant Biol. 12: 444–450. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Yamada K., Yabumoto K., Fujii S., Huser A., Tsuji G., Koga H., Dohi K., Mori M., Shiraishi T., O’Connell R., Kubo Y. (2007). Saccharomyces cerevisiae SSD1 orthologues are essential for host infection by the ascomycete plant pathogens Colletotrichum lagenarium and Magnaporthe grisea. Mol. Microbiol. 64: 1332–1349. [DOI] [PubMed] [Google Scholar]
- Théry M., Bornens M. (2006). Cell shape and cell division. Curr. Opin. Cell Biol. 18: 648–657. [DOI] [PubMed] [Google Scholar]
- Tsuji G., Fujii S., Fujihara N., Hirose C., Tsuge S., Shiraishi T., Kubo Y. (2003). Agrobacterium tumefaciens-mediated transformation for random insertional mutagenesis in Colletotrichum lagenarium. J. Gen. Plant Pathol. 69: 230–239. [Google Scholar]
- Valerio-Santiago M., de Los Santos-Velázquez A.I., Monje-Casas F. (2013). Inhibition of the mitotic exit network in response to damaged telomeres. PLoS Genet. 9: e1003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneault-Fourrey C., Barooah M., Egan M., Wakley G., Talbot N.J. (2006). Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312: 580–583. [DOI] [PubMed] [Google Scholar]
- Wang Y., Hu F., Elledge S.J. (2000). The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr. Biol. 10: 1379–1382. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Hiraoka Y. (2003). Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 22: 2284–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Skibbens R.V., Cheng J.W., Salmon E.D., Bloom K. (1995). Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130: 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Numata N., Nakajima K., Katou S., Kawakita K., Rowland O., Jones J.D., Doke N. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15: 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.