ALIX associates with the ESCRT-III complex to mediate trafficking to the vacuole of protein cargoes, including phosphate transporters and brassinosteroid receptor BRI1.
Abstract
Prior to the release of their cargoes into the vacuolar lumen, sorting endosomes mature into multivesicular bodies (MVBs) through the action of ENDOSOMAL COMPLEX REQUIRED FOR TRANSPORT (ESCRT) protein complexes. MVB-mediated sorting of high-affinity phosphate transporters (PHT1) to the vacuole limits their plasma membrane levels under phosphate-sufficient conditions, a process that allows plants to maintain phosphate homeostasis. Here, we describe ALIX, a cytosolic protein that associates with MVB by interacting with ESCRT-III subunit SNF7 and mediates PHT1;1 trafficking to the vacuole in Arabidopsis thaliana. We show that the partial loss-of-function mutant alix-1 displays reduced vacuolar degradation of PHT1;1. ALIX derivatives containing the alix-1 mutation showed reduced interaction with SNF7, providing a simple molecular explanation for impaired cargo trafficking in alix-1 mutants. In fact, the alix-1 mutation also hampered vacuolar sorting of the brassinosteroid receptor BRI1. We also show that alix-1 displays altered vacuole morphogenesis, implying a new role for ALIX proteins in vacuolar biogenesis, likely acting as part of ESCRT-III complexes. In line with a presumed broad target spectrum, the alix-1 mutation is pleiotropic, leading to reduced plant growth and late flowering, with stronger alix mutations being lethal, indicating that ALIX participates in diverse processes in plants essential for their life.
INTRODUCTION
Trafficking of cargo proteins coming from the plasma membrane (PM) or the Golgi apparatus (GA) to the vacuole occurs through multivesicular bodies (MVBs) (Winter and Hauser, 2006). These organelles, also termed late endosomes or prevacuolar compartments (PVCs), contain internal vesicles that will be delivered, together with their cargoes, into the lumen of vacuoles/lysosomes upon MVB fusion with the tonoplast (Winter and Hauser, 2006). This process plays a central role in controlling the reutilization, storage, or degradation of membrane components and thus regulates fundamental biological processes including membrane turnover, defense against pathogens, development, hormone transport, nutrient uptake, and cell signaling. In the case of membrane-associated regulatory proteins, the MVB route allows modulation of their function by regulating their abundance at the PM and in other vesicular compartments (e.g., endosomes).
The sorting of most integral membrane proteins into intraluminal vesicles (ILVs) is dependent on the attachment of ubiquitin to their cytosolic domains, although ubiquitin-independent sorting mechanisms also exist (McNatt et al., 2007). Selective packaging of protein cargoes into ILVs of MVB is mediated by ESCRT (ENDOSOMAL SORTING COMPLEXES REQUIRED FOR TRANSPORT) protein complexes (Conibear, 2002; Winter and Hauser, 2006; Nickerson et al., 2007; Henne et al., 2011). The latter consist of several cytosolic proteins of the VPS-E (class E Vacuolar Protein Sorting) class that are organized into five complexes: ESCRT-0-, -I, -II, -III, and ESCRT-III-associated SKD1/Vps4 complex (Winter and Hauser, 2006; Richardson et al., 2011). ESCRT complexes work sequentially and in concert to transfer ubiquitinated cargo proteins to the next complex until cargoes get into ILVs. Thus, targeted proteins are first recognized by subunits of the ESCRT-0 complex, which also recruit the ESCRT-I complex to the endosome surface. ESCRT-I, together with ESCRT-II, orchestrates cargo protein sorting and initiates membrane invagination, whereas ESCRT-III allows cargo concentration, cargo engulfment into ILVs, and ILV scission. During this stage, function of Doa4/UBPY (in yeast and humans) and AMSH (in animals and plants) deubiquitinases is required to remove ubiquitin tags from cargoes. In the last step, SKD1/Vps4 complex promotes disassembly and dissociation of the ESCRT machinery from the MVB surface (Katzmann et al., 2001; Babst et al., 2002a, 2002b; Martin-Serrano et al., 2003; Babst, 2005; Isono et al., 2010; Katsiarimpa et al., 2011; Wright et al., 2011).
Putative homologs for most canonical ESCRT components have been found in plants, except for ESCRT-0 subunits (Leung et al., 2008; Winter and Hauser, 2006; Reyes et al., 2011). However, only some of them have been characterized in detail. The latter include Arabidopsis thaliana ESCRT-I component ELCH (ortholog of Vps23p/TSG101 in yeast and mammals), ESCRT-III subunits VPS2 (Vps2p/CHMP2), VPS20 (Vps20p/CHMP6), VPS24 (Vps24p/CHMP3), SNF7 (Snf7/CHMP4), and ESCRT-III-associated SKD1 (Vps4p/SKD1), and CHMP1A and B (Did2p/CHMP1) proteins (Spitzer et al., 2006, 2009; Katsiarimpa et al., 2013; Cai et al., 2014). In addition, plant-specific ESCRT components have been identified, such as PROS, a positive regulator of SKD1, and FREE1, a FYVE domain-containing protein (also termed FYVE1) that interacts with ESCRT-I complexes at MVBs to allow sorting of ubiquitinated protein targets into ILVs (Gao et al., 2014, 2015; Reyes et al., 2014; Kolb et al., 2015). In most cases, complete loss of function of ESCRT components has deleterious effects in plants, leading to lethality at early developmental stages. In accord with their indispensability, plant ESCRT components have been shown to mediate essential processes, including cytokinesis and autophagy, besides their implication in MVB-mediated trafficking of protein cargoes (Spitzer et al., 2006, 2015; Gao et al., 2015). Known targets of the MVB route in plants include, among others, transmembrane auxin carriers PIN1, PIN2, and AUX1, iron transporter IRT1, boron transporter BOR1, flagellin receptor FLS2, brassinosteroid receptor BRI1, and members of the PHT1 family of high-affinity Pi transporters (Geldner et al., 2001, 2007; Kleine-Vehn et al., 2006, 2008; Spitzer et al., 2009; Barberon et al., 2011; Bayle et al., 2011; Kasai et al., 2011; Spallek et al., 2013).
The PHT1 family (nine members in Arabidopsis; PHT1;1 to PHT1;9) is conserved across plant species and allows Pi uptake into root cells and its distribution throughout different plant organs (Shin et al., 2004; Nussaume et al., 2011; Ayadi et al., 2015). Accumulation of PHT1 proteins in the PM is tightly regulated at both transcriptional and posttranslational levels (Bayle et al., 2011). Thus, under Pi deprivation conditions, expression of PHT1 genes is increased in a PHOSPHATE STARVATION RESPONSE1 (PHR1)-dependent manner (Rubio et al., 2001). Transcription factor PHR1 and homolog proteins play a fundamental role during plant adaptation to growth under low Pi, beyond regulation of PHT1 genes. PHR1 and PHL1 were shown to regulate up to 70% of Pi starvation-induced (PSI) genes in Arabidopsis (Bustos et al., 2010). Accordingly, mutants with reduced PHR1 function display impaired Pi starvation responses, including reduced root-to-shoot ratio, decreased accumulation of anthocyanins, and low Pi uptake (Bustos et al., 2010). Upon induction of PHT1 gene expression (Rubio et al., 2001; Bustos et al., 2010; Chiou and Lin, 2011), newly synthesized PHT1 proteins are then sorted from the endoplasmic reticulum (ER) to the PM into COPII-coated secretory vesicles. Correct ER-to-PM trafficking of PHT1 proteins requires the function of PHF1 (PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1), a Sec12-related protein that allows PHT1 exit from the ER (González et al., 2005; Bayle et al., 2011). By contrast, under Pi-rich conditions, a CK2 kinase phosphorylates PHT1 proteins inhibiting their exit from the ER; additionally, PHT1 proteins at the PM and the GA are packaged into endosomes and sorted to vacuoles for degradation to reduce their protein levels at the PM and, therefore, Pi uptake (Bayle et al., 2011; Chen et al., 2015). Endocytosis of PHT1 proteins also occurs under low-Pi conditions, although in this case most Pi transporters are returned to the PM via recycling endosomes (Bayle et al., 2011). In spite of our current knowledge about endosomal trafficking of PHT1 proteins, the molecular mechanisms, likely involving ESCRT complex function, that regulate their sorting into MVBs remain unclear.
The function of ESCRT complexes in the MVB route is aided by additional factors that include, among others, Bro1 domain-containing proteins (Odorizzi et al., 2003; Luhtala and Odorizzi, 2004; Kim et al., 2005). The latter correspond to a conserved multifunctional class of proteins extensively studied in yeast (represented by Bro1 and Rim20) and animals (ALIX), where they associate with ESCRT-III complex by interacting with their Snf7/Vps32/CHMP4 subunit and facilitate cargo sorting and ILV formation (Galindo et al., 2007; McCullough et al., 2008; Wemmer et al., 2011; Bissig and Gruenberg, 2014). Mutations impairing Bro1-Snf7 interaction severely inhibit ILV formation and cargo sorting in yeast, therefore altering trafficking and degradation of protein cargoes at the lytic vacuoles. An Arabidopsis Bro1/ALIX-related protein has been examined in yeast two-hybrid analysis of ESCRT protein interactions, but functional characterization of this protein in plants is lacking (Zhou et al., 2010; Richardson et al., 2011). Here, by means of a screen for mutations that suppress Pi starvation response defects of phr1 mutants, we identified ALIX, the Arabidopsis homolog of Bro1/ALIX proteins. Like its animal and yeast counterparts, Arabidopsis ALIX is a cytosolic protein that associates with MVB and enables protein cargo trafficking from the PM to the vacuole. Indeed, we show that ALIX binds to ESCRT-III component SNF7 in vivo and mediates sorting and vacuolar degradation of high-affinity transporter PHT1;1. Partial loss-of-function mutants for ALIX display defects in vacuole biogenesis that, together with altered Pi transporter trafficking, may underlie the altered Pi uptake and distribution in these mutants. ALIX seems to regulate additional plant biological processes possibly by mediating trafficking of other protein cargoes, such as brassinosteroid receptor BRI1. Together, our results establish ALIX as a major player in membrane protein processing through the plant endomembrane system and in vacuolar biogenesis.
RESULTS
Isolation of Mutants Suppressing phr1 Altered Responses to Pi Starvation
To obtain insights into the molecular mechanisms regulating PHR1 activity, we performed a screen for phr1 suppressors based on recovering the reduced accumulation of anthocyanins in phr1 seedlings in response to Pi starvation compared with wild-type plants (Rubio et al., 2001). M2 seedlings of an ethyl methanesulfonate (EMS)-mutagenized phr1-1 population (∼360,000) were grown in medium lacking Pi for 10 d, and plants displaying increased shoot pigmentation, compared with phr1 controls, were selected as candidates for further analysis. By sequencing the PHR1 locus in all candidates, we confirmed 44 cases in which reversion of the phr1 phenotype was not due to mutations in the PHR1 gene or to contamination with the wild-type PHR1 allele. These mutant lines were termed sphr (suppressor of phr1). Among them, the sphr1 mutant (hereafter termed alix-1 for the sake of simplicity; see next section) was selected for further analysis since, in addition to showing increased anthocyanin levels, other phenotypic defects of phr1 mutants were also partially suppressed. The alix-1 plants showed increased PSI (Pi-STARVATION INDUCED) gene expression and Pi levels compared with phr1 controls (Figure 1). However, alix-1 mutants displayed similar alterations in root morphology as phr1 mutants when grown in low-Pi medium (i.e., shorter primary and lateral roots compared with wild-type plants; Figure 1; Supplemental Figure 1), indicating that the alix-1 mutation could not recapitulate all PHR1 functions. Additionally, alix-1 mutant plants showed morphological and developmental alterations that did not depend on the phr1 mutation. Thus, alix-1 mutant lines both in the phr1 (alix-1 phr1) and wild-type (alix-1 PHR1) backgrounds displayed reduced growth when cultivated in complete medium and late flowering under long day conditions (Supplemental Figures 1 and 2). Reduced growth could be observed in petioles and rosette leaves of mutant plants (Supplemental Figure 1). The latter also showed highly serrated margins and curled surface compared with wild-type leaves. Similar phenotypes were found in cauline leaves where, in addition, leaf tips pointed upwards instead of downwards as in wild-type plants. These results indicate that the alix-1 mutation has pleiotropic effects in plant growth and development, including in plant Pi homeostasis. These analyses were performed using alix-1 mutants backcrossed four times with either phr1 or wild-type plants. Phenotypic characterization of the progeny obtained from heterozygous alix-1 plants indicated that the alix-1 mutation is recessive (Supplemental Table 1).
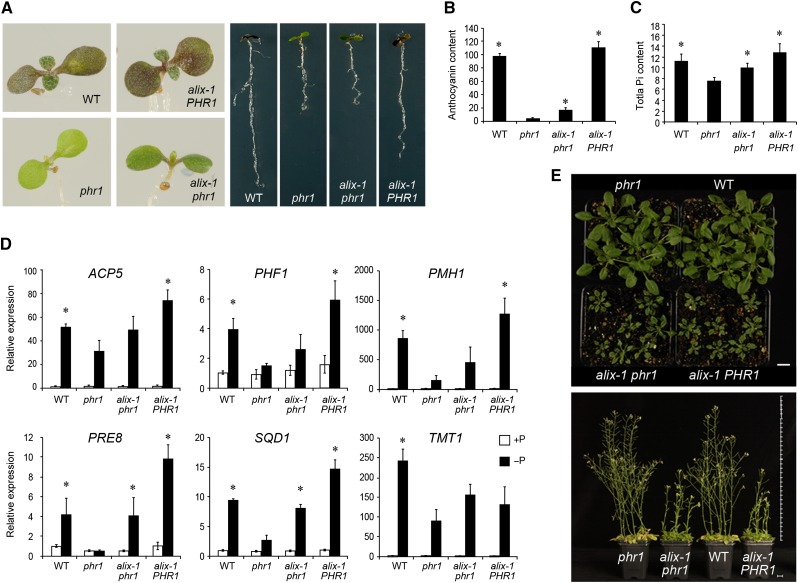
Figure 1.
Molecular and Physiological Characterization of alix-1 Mutants.
(A) Photographs of wild-type (WT), phr1, alix-1 phr1, and alix-1 PHR1 plants grown in Pi-deficient medium for 10 d.
(B) and (C) Histograms showing anthocyanin and Pi content in wild-type, phr1, alix-1 phr1, and alix-1 PHR1 plants. Seedlings were grown for 12 d in Pi-deficient medium for anthocyanin measurements (Abs 530 nm/g fresh weight; n = 8) and for 10 d in complete medium for Pi content analysis (μM/g fresh weight; n = 6). Error bars indicate standard deviations.
(D) RT-qPCR analysis of the expression of representative PSI genes in wild-type, phr1, alix-1 phr1, and alix-1 PHR1 seedlings grown under low Pi (−P; 30 µM Pi) and Pi-sufficient (+P; 500 µM Pi) conditions. ACTIN8 was used as a housekeeping reference gene. Expression levels are relative to Pi-rich-grown wild-type values, which were normalized to 1. Data represent the mean of three biological replicates with sd.
(E) Pictures of 28-d-old (up) and 49-d-old (down) wild-type, phr1, alix-1 phr1, and alix-1 PHR1 plants grown in soil. Bars = 1 cm.
*P < 0.05 (Student’s t test) with respect to the phr1 mutant in the same experimental conditions.
Positional Cloning of the ALIX Gene and the Characteristics of Its Protein
The ALIX gene was positionally cloned on the basis of a cross between the alix-1 phr1-1 mutant (Columbia ecotype; Rubio et al., 2001) and a phr1-1 mutant introgressed five times into the Landsberg erecta (Ler) background (see Methods). A single nucleotide change (G to A) was identified in the second exon of the At1g15130 locus that produced a missense mutation (Gly260-to-Asp) in the predicted protein sequence (Figure 2). Transformation of alix-1 phr1 mutant plants with a 4-kb genomic region spanning the whole At1g15130 locus (gALIX alix-1 phr1) rescued the phr1 phenotypes. Thus, similar to phr1 mutants, gALIX alix-1 phr1 plants showed low anthocyanin levels in shoots and reduced expression of PSI genes under low Pi conditions and decreased Pi content under Pi-sufficient supply, as well as normal growth and development at the adult stage when grown in complete media (Figure 2; Supplemental Figure 3).
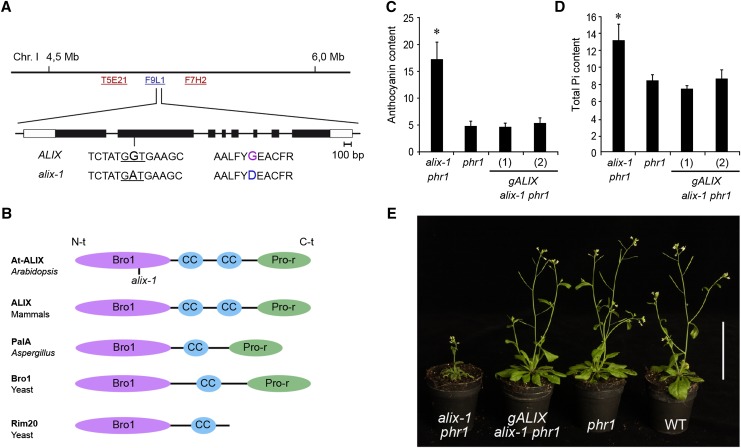
Figure 2.
Positional Cloning of the ALIX Gene and the Characteristics of Its Protein.
(A) Position of the ALIX locus on chromosome 1 of Arabidopsis (BAC F9L1). The sequence surrounding the alix-1 mutation (G-to-A transition), as well as the resulting amino acid change (Gly260-to-Asp), is shown. The exon structure of ALIX is represented with boxes (light, untranslated; dark, coding region).
(B) Diagrams depicting the domain organization of At-ALIX and homolog proteins in mammals (ALIX), A. nidulans (PalA), and yeast (Bro1 and Rim20). ALIX-related proteins comprise a Bro1 domain (Bro1) in the N-terminal region (N-t), followed by one or two coiled-coil (CC) domains and a proline-rich (Pro-r) motif in the C-terminal region (C-t). The position of the alix-1 mutation in the Bro1 domain of At-ALIX protein is indicated.
(C) and (D) Complementation of alix-1 mutant defects using a construct containing the ALIX genomic region. Plants corresponding to phr1, alix-1 phr1, and alix-1 phr1 transformed with a construct containing the ALIX genomic region (gALIX alix-1 phr1) were grown for 12 d in Pi-deficient medium for anthocyanin measurements (Abs 530 nm/g fresh weight; n = 8) and for 10 d in complete medium for Pi content analysis (μM/g fresh weight; n = 6). Error bars indicate standard deviations. *P < 0.05 (Student’s t test) with respect to the phr1 mutant in the same experimental conditions.
(E) Plants corresponding to the same genotypes as in (C), together with wild-type (WT) plants were grown in soil for 45 d under long-day conditions. Bar = 5 cm.
ALIX mRNA encodes a theoretical protein of 846 amino acids, with similar size and domain structure to mammalian ALIX (amino acid identity 21%) and related functional counterparts in Aspergillus nidulans, PalA (amino acid identity 22%), and Saccharomyces cerevisiae, Bro1 (amino acid identity 13%) and Rim20 (amino acid identity 19%). These proteins are composed of three main domains. A Bro1 domain in the N-terminal region, a proline-rich domain in the C-terminal region (except for Rim20), and connecting both domains, a relatively uncharacterized sequence containing one or two coiled-coil domains (Odorizzi, 2006) (Figure 2). Similar domain composition was found in At-ALIX, which is predicted to contain two coiled-coil domains. Sequence alignment of At-ALIX-related proteins showed greater conservation in their Bro1 domain region (Supplemental Figure 4).
At-ALIX Is an Essential Protein
The nature of the alix-1 mutation did not allow discerning whether it corresponded to a total loss of ALIX function. To characterize the effect of the ALIX null mutation, two Arabidopsis T-DNA mutants were obtained from public collections. These contained T-DNA insertions in the fifth intron (alix-2 mutant) and in the last exon (i.e., eight; alix-3), respectively (Figure 3). RT-PCR assays showed that the T-DNA insertion in the alix-2 allele abolished accumulation of its transcript. However, alix-3 mutant alleles still expressed truncated ALIX transcripts encoding an ALIX chimera that lacks 140 amino acids at the C terminus (Figure 3). After analyzing the genotype of seedlings from progeny of heterozygous alix-2 ALIX and alix-3 ALIX mutant plants, we were unable to find homozygous mutants for any of them, suggesting that ALIX is essential for plant viability (Supplemental Tables 2 and 3). Indeed, while analyzing the offspring of heterozygous alix-2 and alix-3 mutants growing in complete media, we observed a significant number of ungerminated seeds (∼15% for alix-2 and 7.5% for alix-3; Supplemental Table 4). The latter contained fully developed embryos, although they were not viable, as shown by viability assays based on tetrazolium staining (Figure 3B). Genotyping of embryos from nongerminating seeds showed they corresponded to homozygous mutants (Figure 3B). Together, these results indicate that alix-2 and alix-3 correspond to null recessive mutations that affect the seed-to-seedling developmental phase transition.
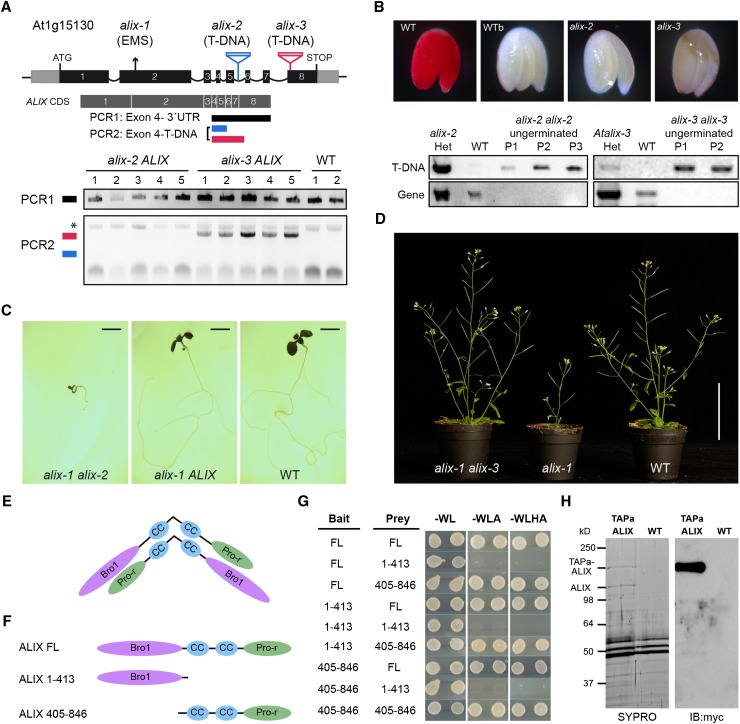
Figure 3.
ALIX Forms Dimers and Is Essential for Plant Life.
(A) A diagram of At-ALIX genomic region showing the position of the EMS-induced G-to-A mutation in alix-1 plants and that of T-DNA insertions in alix-2 and alix-3 mutants. In lower panels, PCR analysis was used to detect potential truncated ALIX transcripts in the cDNA of alix-2 and alix-3 heterozygous mutants (five lines per mutant). An asterisk indicates a nonspecific band amplified in all samples.
(B) Genotyping and tetrazolium staining shows that embryos from ungerminated seeds of the alix-2 and alix-3 progenies correspond to homozygous mutants that are not viable. Embryos from imbibed (WT) and boiled wild-type (WTb) seeds were used as a control. PCR analysis of wild-type and alix mutant embryos was performed as described in Methods to detect T-DNA insertions in the ALIX gene. P represents pools of 15 embryos.
(C) Photographs of 10-d-old nonviable transheterozygous alix-1 alix-2 and viable heterozygous alix-1 ALIX and wild-type seedlings. Bars = 0.2 cm.
(D) Photographs of trans-heterozygous alix-1 alix-3, homozygous alix-1, and wild-type seedlings grown in soil. Bar = 5 cm.
(E) Model for hypothetical ALIX dimerization following an antiparallel disposition.
(F) and (G) Yeast two-hybrid assays using full-length (FL) and truncated versions (comprising the Bro1 domain, amino acids 1 to 413; or the coiled coils plus the Pro-rich region, amino acids 405 to 846) of ALIX. Transformed yeast cells were grown in SD-WL medium as a transformation control and in SD-WLA and SD-WLHA media for interaction assays.
(H) Copurification of TAPa-ALIX and endogenous ALIX proteins. TAPa-purified proteins were separated in a 10% SDS-PAGE gel and subjected to immunoblot analysis using anti-myc. SYPRO staining was used to visualize differentially purified protein bands prior to their mass spectrometry analysis. Wild-type protein extract was used as a TAPa negative control.
Lower than expected (25%) ratios of ungerminated seeds suggested defects in the transmission of mutant alleles through either or both gametes. To test this, genetic transmission of alix-2 and alix-3 through the male and female gametes was determined by carrying out reciprocal test crosses in which heterozygous alix-2 and alix-3 mutants were crossed with the wild type. Reduced transmission efficiency for the alix-2 mutant allele through the female gametophyte was observed, whereas transmission of the alix-3 allele through both sexes was unaffected (Supplemental Table 5). We also tested whether interallelic interactions occur between the three alix mutations. For this, homozygous alix-1 mutants were crossed with either heterozygous alix-2 ALIX or alix-3 ALIX plants. When the progeny of alix-1 alix-1 × alix-2 ALIX crosses were grown in complete medium, two different plant populations were observed with a 1:1 segregation; one of them corresponded to viable plants that totally resembled wild-type controls, and the other consisted of seedlings that died soon after germination and cotyledon expansion (Figure 3C). Genotyping of these two populations showed that the latter corresponded to transheterozygous alix-1 alix-2 lines. By contrast, all individuals from the F1 population obtained after crossing alix-1 and alix-3 ALIX mutants were similar to wild-type plants (Figure 3D). Plant genotyping showed that the two possible genetic constitutions alix-1 alix-3 and alix-1 ALIX were equally represented. Since these two plant populations showed identical phenotypes, we concluded that interallelic complementation occurs between the alix-1 and alix-3 mutations.
ALIX Forms Dimers
Interallelic complementation is frequently associated with mutant alleles corresponding to dimeric/oligomeric proteins (Clifford and Schüpbach, 1994; Busch et al., 1996; Simin et al., 1998). In mammals, ALIX dimerizes in vitro and in vivo via the V domain, the central region of ALIX composed of two coiled coils. For ALIX dimerization, the V domain must be open, which allows antiparallel interaction of the ALIX V domain arms (Pires et al., 2009) (Figure 3E). To test whether At-ALIX also forms dimers, we performed yeast two-hybrid assays using full-length and truncated versions of At-ALIX comprising the Bro1 domain (amino acids 1 to 413) or the coiled coils plus the Pro-rich region (amino acids 405 to 846) (Figure 3F). The C-terminal portion of At-ALIX (amino acids 405 to 846) interacted with all protein versions in at least one bait/prey orientation (Figure 3G), pointing to a role for this domain in intramolecular interactions and/or dimerization of At-ALIX, as observed in mammals (Pires et al., 2009). To confirm At-ALIX dimerization in vivo, tandem affinity purification (TAP) assays were performed using soluble protein extracts from Arabidopsis lines expressing an N-terminal fusion of At-ALIX to the TAPa tag (Rubio et al., 2005). Mass spectrometry analysis and immunoblots of TAP-purified proteins showed that endogenous ALIX protein associated with the TAPa-ALIX fusion in planta (Figure 3H; Supplemental Table 6).
ALIX Localizes in the Cytosol and Is Associated to MVBs
To get insights on ALIX function in plants and how it functions in plant responses to Pi starvation and Pi homeostasis, we first analyzed the subcellular localization of ALIX under different Pi regimes. With this aim, we generated Arabidopsis alix-1 phr1 transgenic lines expressing a fully functional fusion of the ALIX genomic sequence to the GFP, under the control of its own promoter (ALIXpro:GFP-gALIX; Figure 4; Supplemental Figure 5). Immunoblots showed that the GFP-ALIX fusion remains intact in vivo, whereas overexpression of an ALIX-GFP fusion yields truncated fragments. Similar GFP-ALIX protein levels were found under Pi-low and -rich conditions. Lack of responsiveness of ALIX to variations in Pi supply was also found at the transcript level (Supplemental Figure 6). Microsomal fractionation experiments using ALIXpro:GFP-gALIX seedlings showed that GFP-ALIX was mainly found in the cytosolic fraction, although a small proportion could be also detected in membranous fractions (Figure 4D). Accordingly, association of ALIX to membranes could be disrupted by treatment with detergent (Triton X-100). High salt concentration and urea treatments also released ALIX from microsomal fractions, indicating that such association is weak and depends on ALIX conformation (Figure 4E). In agreement with microsomal fractionation results, confocal imaging of root cells from ALIXpro:GFP-gALIX seedlings showed that, independently of Pi supply, most GFP-ALIX was uniformly located in the cytosol, although a small fraction formed punctate structures likely representing vesicular compartments (Figure 4F). Mammalian ALIX and yeast Bro1 are mainly cytosolic proteins that associate transiently with MVBs/PVCs, allowing protein cargo delivery to the lysosome/lytic vacuole lumen (Vito et al., 1999; Odorizzi et al., 2003; Odorizzi, 2006; Morita et al., 2007). To test whether At-ALIX associates with MVBs in plant cells, we obtained Arabidopsis plants expressing GFP-ALIX and the MVB marker mCherry-RabF2b. In parallel, additional cell compartment markers, corresponding to the Golgi apparatus, trans-Golgi network (TGN)/early endosome (EE), and recycling endosome, were coexpressed with GFP-ALIX in Arabidopsis (Dettmer et al., 2006; Geldner et al., 2009; Supplemental Table 7). Confocal imaging and quantification of signals from the GFP and mCherry/mRFP channels from root epidermal cells of these lines showed significant overlap only in the case of GFP-ALIX and the MVB marker (Figure 4G).
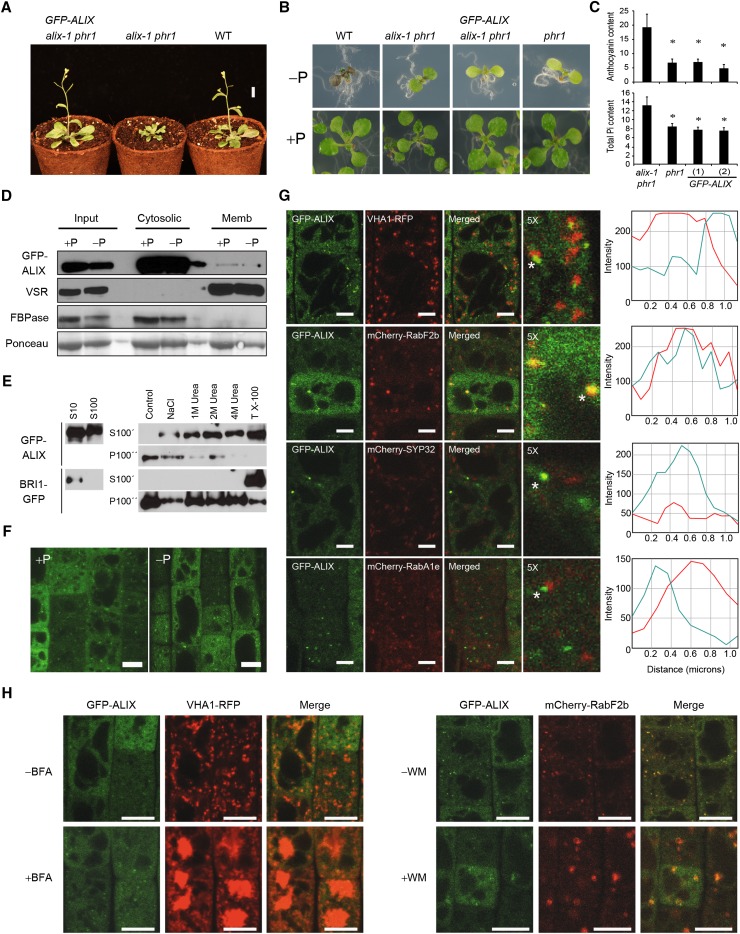
Figure 4.
Functional GFP-ALIX Fusion Protein Localizes in Both Cytosolic and Microsomal Fractions.
(A) Plants corresponding to the wild type (WT), alix-1 phr1, and alix-1 phr1 transformed with a construct containing the ALIX genomic region fused to GFP driven by the ALIX promoter (GFP-ALIX) were grown in soil for 3 weeks before photographs were taken. Bar = 1 cm.
(B) Plants as in (A) together with phr1 were grown in Pi-deficient and Pi-rich media for 12 d before photographs were taken.
(C) Histograms showing anthocyanin and Pi content in phr1, alix-1 phr1, and GFP-ALIX (two independent transgenic lines) plants. Error bars indicate standard deviations. *P < 0.05 (Student’s t test) with respect to the phr1 mutant in the same experimental conditions.
(D) Isolation of microsomes from postnuclear fractions (Input) of 10-d-old GFP-ALIX seedlings grown under Pi-rich (+P) and -deficient (−P) conditions. GFP-ALIX was found in the soluble fraction (Cytosolic), which corresponds to cytosol, as well as associated to microsomes (Memb).
(E) Membrane association of GFP-ALIX can be disrupted with chaotropic agents and detergents. Supernatant (S10) samples were ultracentrifuged to give soluble fractions (S100) and pellets. Pellets were resuspended in homogenization buffer without additives (control); with 1 M NaCl (NaCl); with 1, 2, or 4 M urea; or with 1% Triton X-100 (TX-100) and ultracentrifuged again, giving wash fractions (S100’) and pellets. This procedure was repeated to give washed pellets (P100’’). Same procedures were followed with BRI1-GFP as a detergent-solubilized control.
(F) Confocal images of root epidermal cells from 5-d-old GFP-ALIX seedlings grown under Pi-deficient (−P) and -rich (+P) conditions. Bars = 10 μm.
(G) Confocal images of root epidermal cells of 5-d-old seedlings expressing GFP-ALIX and different cell compartment markers; VHA1-RFP (early endosome), mCherry-RabF2b (MVB), mCherry-SYP32 (Golgi), mCherry-RabA1e (recycling endosome). The 5× enlarged images of merged color channels are shown. ImageJ quantification of green (turquoise lines) and red (red lines) signal intensities for spots indicated by asterisks. Bars = 5 μm.
(H) BFA (left panels) and WM (right panels) treatments of 5-d-old GFP-ALIX seedlings expressing VHA1-RFP (for BFA) or mCherry-Rabf2b (for WM). Bars = 10 μm.
To further characterize the association of ALIX with endosomal compartments, we analyzed the effects of brefeldin A (BFA) and wortmannin (WM) on the localization of GFP-ALIX. BFA disrupts the membrane recycling pathway between the ER and the GA, forming membranous aggregates termed brefeldin bodies that consist of aggregated TGN/EEs and recycling endosomes with the ER and the GA (Wee et al., 1998; Baldwin et al., 2001; Geldner et al., 2001; Ritzenthaler et al., 2002; Tse et al., 2004, 2007). WM blocks protein cargo trafficking to vacuoles and causes enlargement of MVBs, which facilitates their visualization (Fernandez-Borja et al., 1999; Geldner et al., 2001; Jaillais et al., 2006). In addition to GFP-ALIX, seedlings used for BFA treatments also expressed the TGN/EE marker VHA1-RFP, whereas for WM treatments, they expressed the MVB marker mCherry-RabF2b. WM treatment revealed GFP-ALIX association with swollen MVBs (Figure 4H). By contrast, BFA, which prompted relocalization in brefeldin bodies of VHA1-RFP, had no effect on GFP-ALIX. These results indicate that ALIX localizes to MVBs, as has been shown for ESCRT-III associated proteins (Spitzer et al., 2009).
ALIX Associates with Components of the ESCRT-III Complex
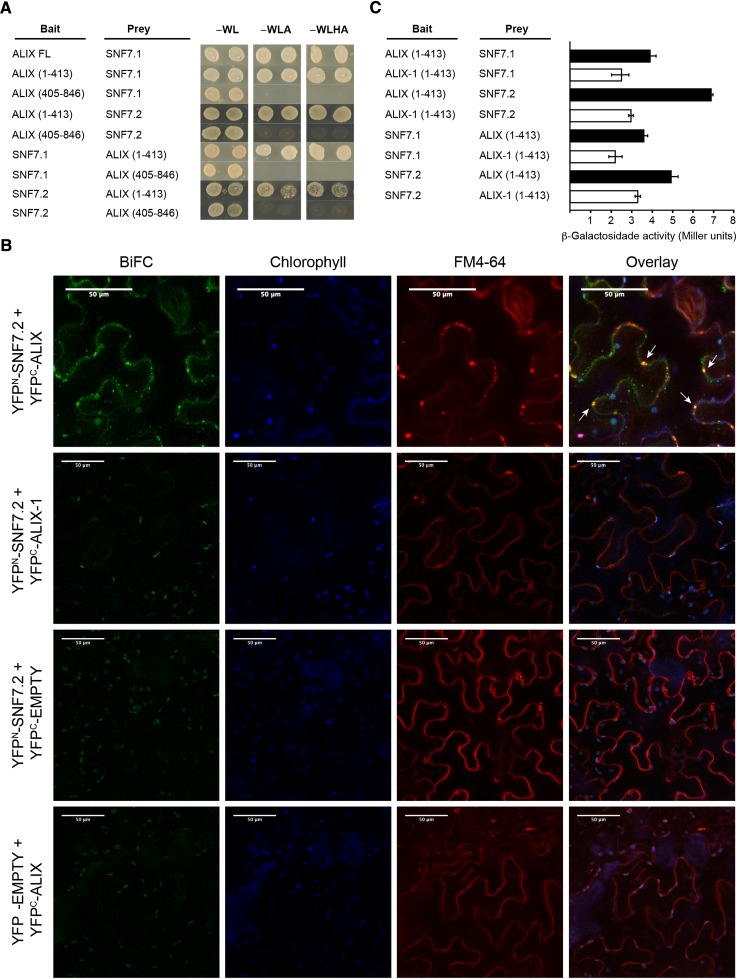
Studies in yeast indicate that Bro1 protein physically interacts through its Bro1 domain with Snf7/Vps32, a component of ESCRT-III complex that mediates protein cargo trafficking through the endomembrane system (Boysen and Mitchell, 2006; Wemmer et al., 2011). Given its localization in MVBs, we studied whether At-ALIX also associates with the ESCRT-III complex. We tested the ALIX-SNF7 interaction in yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays (Figure 5; Supplemental Figures 7 and 8). For these assays, we used the two Arabidopsis Vps32/Snf7 homologs SNF7.1 (At4g29160) and SNF7.2 (At2g19830) (Richardson et al., 2011; Ibl et al., 2012). ALIX directly interacted with both SNF7.1 and SNF7.2. As in the case of yeast Bro1, these interactions were mediated by the Bro1 domain of At-ALIX and took place in vesicle compartments, as shown by colocalization of reconstituted YFP fluorescence driven by ALIX-SNF7.2 interaction and the FM4-64 dye (Figure 5). Interestingly, versions of ALIX fusions containing the alix-1 mutation displayed reduced interaction with SNF7 proteins compared with wild-type versions that could not be explained by lower accumulation of the mutant protein fusions in these experiments (Figure 5; Supplemental Figures 7 to 9). Reduced physical interaction with SNF7 proteins provides a potential explanation at the molecular level for defects caused by alix-1 point mutation in planta.
Figure 5.
ALIX Interacts with ESCRT-III Complex Component VPS32/SNF7 through Its Bro1 Domain.
(A) Yeast two-hybrid assays showing interaction between the Bro1 domain of At-ALIX and SNF7.1 and SNF7.2. ALIX full-length (FL) and truncated versions (comprising the Bro1 domain, amino acids 1 to 413; or the coiled coils plus the Pro-rich region, amino acids 405 to 846) were used. Transformed yeast cells were grown in SD-WL medium as a transformation control and in SD-WLA and SD-WLHA media for interaction assays.
(B) BiFC assays show that ALIX, but not a version containing the alix-1 mutation, interacts with SNF7.2 in vivo. FM-4-64 (5 μM) was injected in Nicotiana benthamiana leaf epidermal cells expressing different construct combinations as indicated. Leaves were observed by confocal imaging after 60 min. Reconstitution of YFP fluorescence indicates that the corresponding ALIX and SNF7 constructs directly interact. White arrows show YFP fluorescence colocalization with FM4-64 signal (red channel). Plastid autofluorescence due to chlorophyll is shown in the blue channel. Bars = 50 μm.
(C) alix-1 mutation reduces the ability of the Bro1 domain to interact with SNF7 proteins. β-Galactosidase assays were performed on yeast cotransfected with plasmids expressing indicated recombinant proteins. Error bars indicate standard deviations. n = 6.
alix-1 Mutants Are Defective in Vacuolar Biogenesis
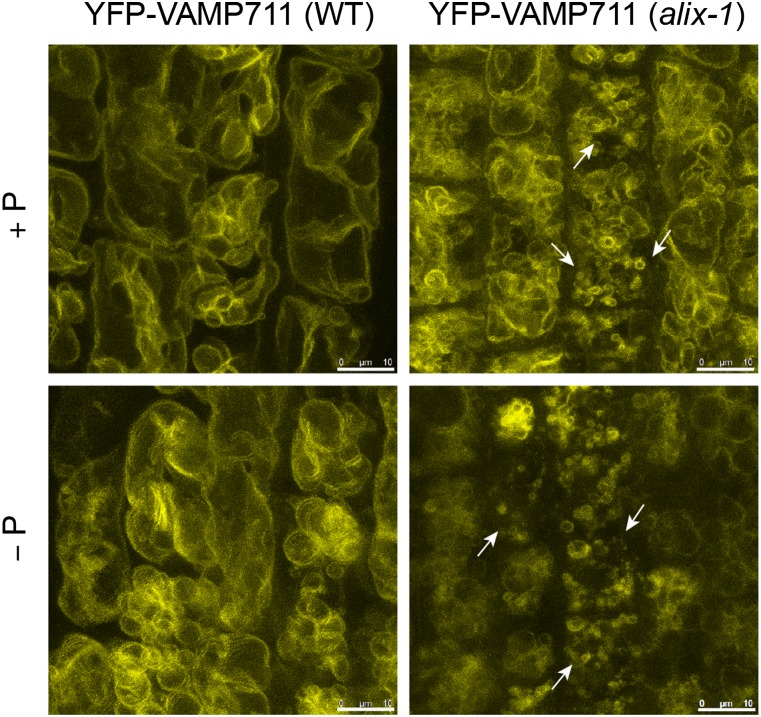
ALIX association with MVBs and interaction with ESCRT-III complex subunits SNF7.1 and 2 suggested that phenotypic defects in alix-1 mutants could be due to altered function of the endomembrane system. To test this hypothesis, we first used FM4-64 dye to check whether endocytosis and general vesicle trafficking were altered in alix-1 plants. Upon cell treatment, FM4-64 rapidly stains the PM by inserting into the outer leaflet of the PM lipid bilayer. As time passes, FM4-64 is internalized from the PM to endosomes and reaches the tonoplast (Bolte et al., 2004). FM4-64 staining of epidermal root cells at different time points did not show any difference detectable by confocal microscopy between alix-1 mutants and wild-type plants. However, a closer look at FM4-64-stained cells in alix-1 at late stages (180 min) suggested that vacuole morphology was altered in these mutants compared with the wild-type control (Supplemental Figure 10). To confirm this defect, alix-1 mutant plants expressing the tonoplast YFP-VAMP711 fusion were obtained. Confocal microscopy analysis showed increased number of vacuoles of smaller size than those in wild-type plants, independently of Pi supply (Figure 6). Vacuolar defects were even more obvious in transheterozygous alix-1 alix-2 plants expressing the YFP-VAMP711 marker, in which ALIX function should be severely compromised (Supplemental Figure 11). Vacuolar morphology in alix-1 could be rescued by expression of GFP-ALIX, as shown by BCECF-AM staining of vacuoles, providing additional proof of the functionality of this fusion protein (Supplemental Figure 12). These data reveal a role of ALIX in vacuolar biogenesis. Defective vacuolar morphology has been associated with mutations impairing ESCRT-III activity (Spitzer et al., 2009; Isono et al., 2010), which is congruent with the presumed functional interaction between ALIX and ESCRT-III.
Figure 6.
alix-1 Mutants Are Defective in Vacuolar Size and Morphology.
Confocal images of root cells from 5-d-old wild-type (WT) and alix-1 mutants overexpressing the tonoplast marker YFP-VAMP711 (Geldner et al., 2009). Increased number of vacuoles of smaller size (shown by arrows) than those in controls can be observed in alix-1 mutants independently of Pi status. Bars = 10 μm.
alix-1 Mutation Alters PHT1;1-GFP Trafficking
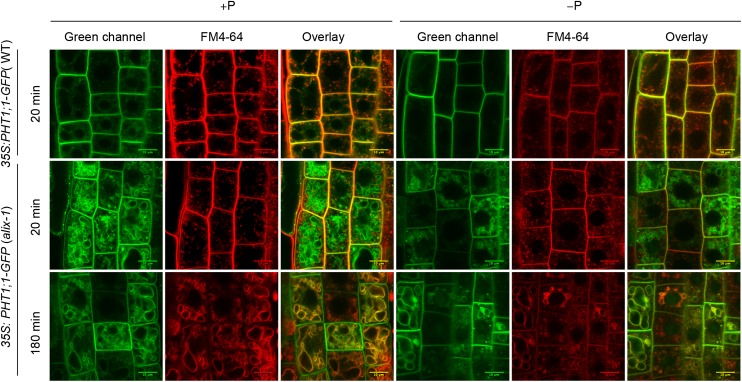
It has been previously shown that PHT1 levels are tightly regulated by the endomembrane system to allow proper maintenance of plant Pi homeostasis (Bayle et al., 2011; Chen et al., 2015). ALIX association to ESCRT-III complexes and the effect of alix-1 in Pi starvation responses led us to test the possibility that ALIX may participate in the trafficking of PM-located Pi transporters. We introgressed a 35S:PHT1;1-GFP marker (González et al., 2005) into the alix-1 mutant background. Interestingly, PHT1;1-GFP, which is located in the PM and in sorting endosomes in wild-type plants (Bayle et al., 2011), was additionally found in the tonoplast in alix-1 mutants (Figure 7). This effect was preferentially observed under Pi-rich conditions, when excess Pi transporters are readily endocytosed and sorted to the vacuolar lumen for degradation in wild-type plants (Bayle et al., 2011), suggesting that alix-1 mutation interferes with internalization of the transporters in MVB and results in their accumulation in the tonoplast.
Figure 7.
PHT1;1-GFP Is Mislocalized in alix-1 Mutants.
Confocal images of root epidermal cells from 5-d-old wild-type (WT) and alix-1 seedlings overexpressing a PHT1;1-GFP fusion grown in +P and −P conditions. Seedlings were treated with 2 μM FM4-64 for 5 min, washed, and visualized after 20 and 180 min. The green and red channels correspond to the PHT1;1-GFP and membrane-associated FM4-64 fluorescence, respectively. Overlay of both channels in images after 180 min shows PHT1;1-GFP localization in alix-1 tonoplasts. Bars = 10 μm.
ALIX-mediated control of Pi transporter trafficking most likely occurs at a later stage than that regulated by PHF1, which allows correct ER-to-PM trafficking of PHT1;1 (González et al., 2005; Bayle et al., 2011). This notion is supported by our results showing that PHT1;1 was mostly located in the ER in alix-1 phf1 double mutants, similar to what is observed in the single phf1 mutant, yet a small fraction of Pi transporter could exit the ER and reach the tonoplast (as shown by partial colocalization with FM4-64) (Supplemental Figure 13). These results indicate that PHF1 and ALIX regulate independent processes during PHT1 trafficking, with PHF1 acting in a stage prior to that of ALIX.
ALIX Regulates Vacuolar Degradation of PHT1;1
To further investigate the role of ALIX in PHT1;1 trafficking, we tested whether the alix-1 mutation alters PHT1;1-GFP targeting to sorting endosomes and their delivery and degradation at the lytic vacuole. For this, 35S:PHT1;1-GFP seedlings in wild-type and alix-1 backgrounds, grown in different Pi regimes, were treated with WM. Upon treatment with WM, confocal imaging showed that alix-1 mutation and Pi regime did not significantly alter PHT1;1-GFP localization in sorting endosomes (Supplemental Figure 14). Trafficking of PHT1;1 transporters to the vacuole involves their packaging into MVBs that mature from TGN/EE compartments (Bayle et al., 2011; Scheuring et al., 2011). To analyze whether alix-1 mutation alters this step in PHT1;1 trafficking, 35S:PHT1;1-GFP alix-1 seedlings were treated with BFA. Confocal imaging of root cells showed similar PHT1;1-GFP localization in brefeldin bodies in all conditions tested, indicating that alix-1 mutation and Pi regime did not alter PHT1;1 trafficking through these BFA-sensitive compartments (Supplemental Figure 14).
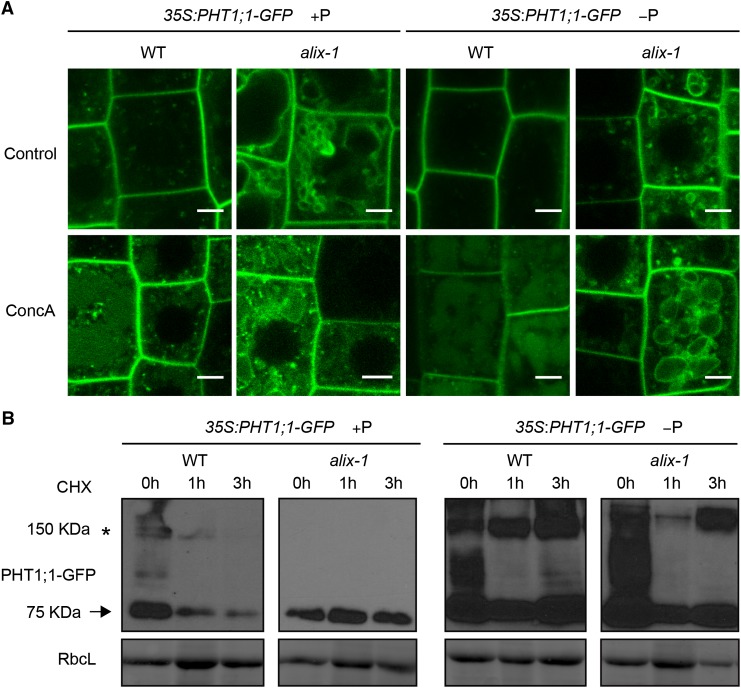
To analyze whether alix-1 mutation, and subsequent PHT1;1 mislocalization, alter PHT1;1 vacuolar degradation, 35S:PHT1;1-GFP seedlings in wild-type and alix-1 mutant backgrounds were treated with concanamycin A (ConcA), a specific inhibitor of vacuolar H+-ATPases that reduces acidification of lytic compartments and, as a consequence, blocks protein degradation at the vacuole (Páli et al., 2004). As expected, PHT1;1-GFP increasingly accumulated in the vacuole lumen of wild-type plants grown in Pi-sufficient medium compared with low-Pi medium (Figure 8). This accumulation in the lumen could be also observed in 35S:PHT1;1-GFP alix-1 plants, albeit to reduced levels, indicating that the alix-1 hypomorphic mutation does not fully impair internalization of Pi transporter at the MVB for degradation at the vacuole. To analyze more precisely whether the rate of PHT1;1 degradation is altered by alix-1 mutation, 35S:PHT1;1-GFP seedlings in both wild-type and alix-1 mutant backgrounds grown under different Pi supply conditions were treated with protein synthesis inhibitor cycloheximide. As previously reported, PHT1;1-GFP degradation mainly occurred under Pi-rich conditions in wild-type plants. Interestingly, alix-1 mutation clearly reduced PHT1;1-GFP degradation under Pi-sufficient conditions compared with wild-type controls (Figure 8).
Figure 8.
alix-1 Mutation Alters PHT1;1-GFP Degradation.
(A) Confocal images of root cells from 5-d-old 35S:PHT1;1-GFP seedlings in wild-type (WT) and alix-1 mutant backgrounds treated with 1 μΜ ConcA for 6 h. Bars = 5 μm.
(B) Immunoblots showing PHT1;1-GFP (arrow) degradation over time in 10-d-old 35S:PHT1;1-GFP seedlings in wild-type and alix-1 mutant backgrounds grown in +P or −P conditions, incubated or not during 1 and 3 h with 50 μM cycloheximide. Anti-GFP was used to detect PHT1;1-GFP. Ponceau staining of the large subunit of Rubisco (RbcL) was used as loading control. An asterisk indicates the position of PHT1;1-GFP aggregates as previously reported by Bayle et al. (2011).
Pi Uptake and Vacuolar Storage Are Altered in alix-1 Mutants
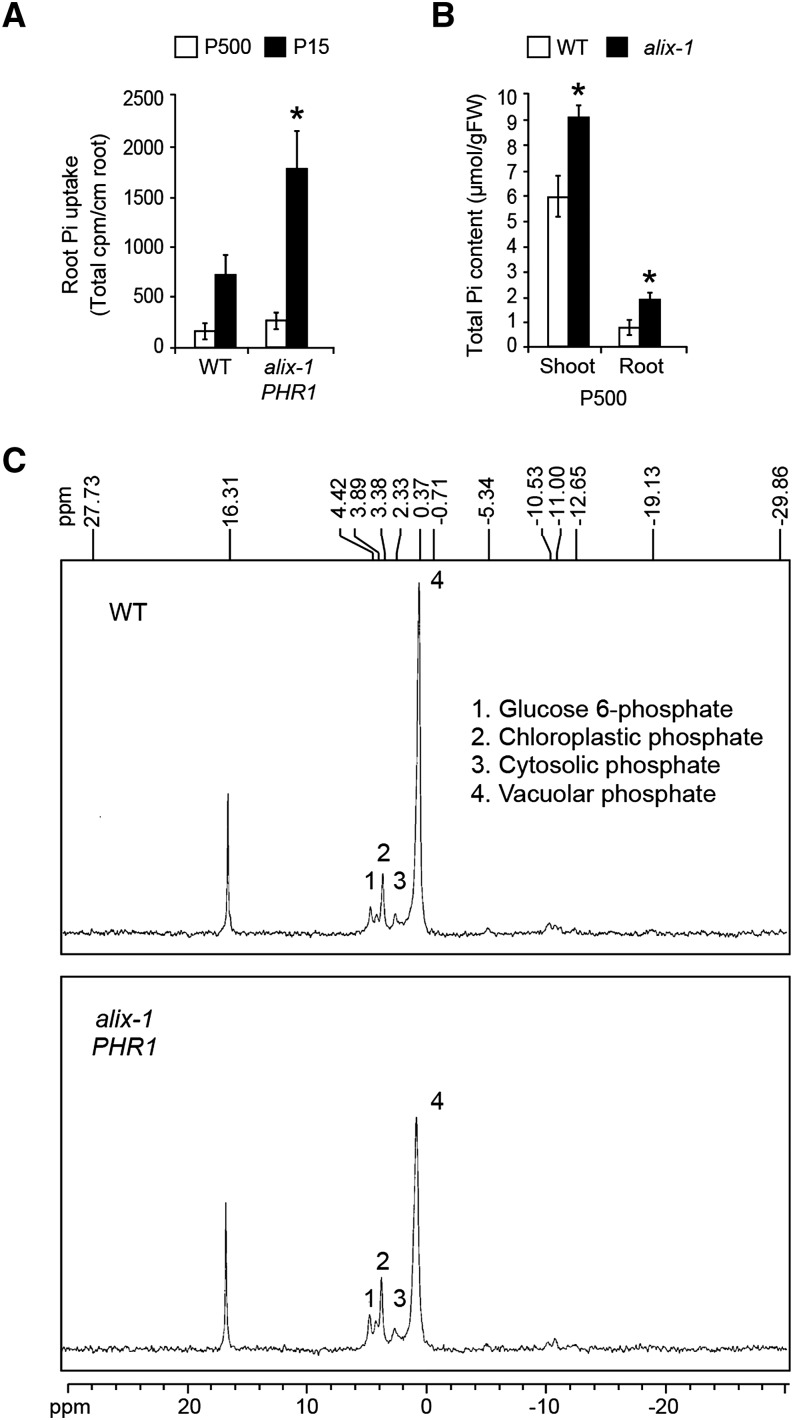
Results showing that alix-1 mutation provokes PHT1;1-GFP mislocalization in the tonoplast and reduced degradation at the vacuole led us to examine whether Pi uptake and storage are altered in alix-1 mutants. Toward this, 33PO4 absorption capacity was measured in alix-1 plants grown under Pi-sufficient or low-Pi conditions and compared with wild-type controls. Pi uptake was increased in alix-1 roots compared with the wild type (Figure 9A). Increased Pi uptake correlated with increased total Pi content in alix-1 plants, as shown by higher Pi levels in shoots and roots of alix-1 seedlings compared with wild-type plants (Figure 9B). We also studied whether the alix-1 mutation alters Pi subcellular distribution. For this, in vivo NMR experiments were conducted using alix-1 mutant and wild-type roots from plants grown in Pi-sufficient conditions. Interestingly, vacuolar Pi levels in alix-1 root cells decreased by ∼30% compared with wild-type levels. On the contrary, glucose-6-phosphate and chloroplast plus cytoplasmic Pi levels were increased by ∼18 and ∼37%, respectively, in alix-1 mutant plants (Figure 9C; Supplemental Table 8). Together, our findings indicate that alix-1 mutation leads to major alterations in Pi homeostasis.
Figure 9.
Pi Uptake and Vacuolar Storage Are Altered in alix-1 Mutants.
(A) Phosphate absorption capacity of alix-1 mutants compared with wild-type plants. 33PO4 uptake was measured in plants grown for 12 d in low Pi (15 μM) or Pi-sufficient (500 μM) medium. Data represent the means and standard deviations of results obtained for 15 plants.
(B) Pi content analysis in shoots and roots of alix-1 mutants and wild-type plants grown for 12 d in Pi-sufficient (500 μM) medium. Error bars indicate standard deviations. n = 10. *P < 0.05 (Student’s t test) with respect to the wild type in the same experimental conditions.
(C) In vivo 31P-NMR spectra from roots of alix-1 and wild-type seedlings grown in Pi-sufficient (500 μM) medium. Peaks from left to right are assigned to glucose-6-phosphate (1), chloroplastic Pi (2), (3) cytoplasmic Pi, and (4) vacuolar Pi.
ALIX Also Mediates Vacuolar Degradation of BRI1
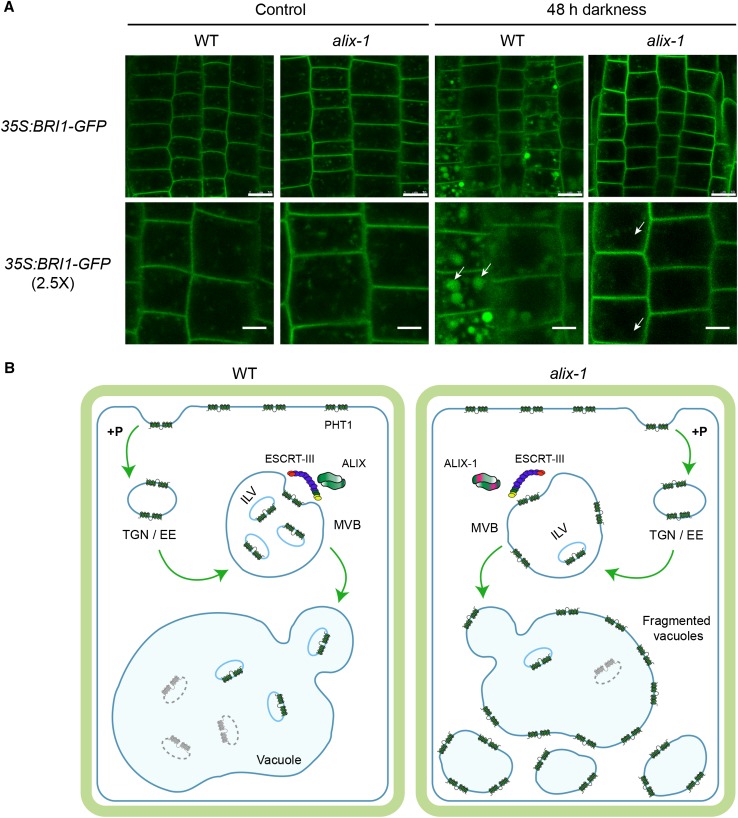
To test whether ALIX participates in the sorting and vacuolar degradation of additional protein cargoes rather than being specific for PHT1 proteins, we analyzed the subcellular localization of BRI1-GFP, a known target of the MVB route in Arabidopsis (Geldner et al., 2007), in the alix-1 mutant background. It was previously reported that degradation of GFP in the vacuole is light dependent (Tamura et al., 2003). Accordingly, we observed increased fluorescence in the vacuole lumen of BRI1-GFP wild-type plants incubated in the dark compared with that of light-grown seedlings. However, this effect was missing in alix-1, indicating that ALIX activity is involved in sorting of BRI1-GFP to the vacuole lumen (Figure 10A). Notably, BRI1-GFP did not accumulate in the tonoplast of alix-1 cells, which suggests that when not correctly internalized into ILV, it is recycled back to the plasma membrane. By contrast, PHT1;1 in the alix-1 background was sorted to the vacuole-limiting membrane. Differential accumulation of PHT1;1 and BRI1 in alix-1 mutants suggests that specific intracellular routes involving ALIX function regulate their trafficking through the plant endomembrane system.
Figure 10.
Vacuolar Degradation of BRI1 Is Defective in alix-1 Mutants.
(A) Root epidermal cells from 5-d-old seedlings overexpressing BRI1-GFP in wild-type (WT) and alix-1 backgrounds grown in Pi-rich conditions were kept in the dark or in light for 48 h before they were observed by confocal imaging. White arrows point to vacuoles. Bars = 10 μm except in 2.5× zoom images, where bars = 5 μm.
(B) Proposed model for defects in PHT1 localization in alix-1 mutants. Under Pi-rich conditions (+P), Pi transporters (PHT1) are readily endocytosed at the plasma membrane and transported to the TGN/EE. TGN/EE vesicles mature to MVBs, where cargo proteins are packaged into ILVs by the action of ESCRT complexes and associated proteins (i.e., ALIX). Finally, MVBs fuse with vacuoles and release ILVs to the vacuole lumen where they are degraded together with their cargoes. In alix-1 mutants, ALIX-1 association with ESCRT-III complexes is altered, and PHT1;1 proteins are not correctly internalized into ILVs, being retained in the tonoplast upon MVB fusion with vacuoles. This effect leads to reduced PHT1 degradation. Defects in cargo trafficking in alix-1 also provoke vacuolar fragmentation, which alters Pi storage and subcellular distribution, and, therefore, Pi homeostasis.
DISCUSSION
In this study, we describe At-ALIX as the founding plant representative of a conserved eukaryotic protein that associates with MVBs and enables protein cargo trafficking to the plant vacuole. We identified ALIX in a search for mutations that suppress Pi starvation response defects in phr1-1 mutants. Although phr1-1 likely corresponds to a null mutation (Rubio et al., 2001), a suppressor screen was possible because, in terms of PHR1 function, phr1-1 plants behave as leaky mutants due to functional redundancy between PHR1-related proteins (Bustos et al., 2010). The alix-1 mutants displayed increased Pi starvation responses compared with phr1-1, suggesting that the ALIX gene function affects Pi homeostasis and/or Pi starvation signaling in Arabidopsis. Accordingly, functional characterization of ALIX showed it has an effect in Pi uptake and vacuolar storage, very likely by facilitating high-affinity Pi transporter trafficking and vacuole morphogenesis. Further analysis of alix-1 mutants indicated that ALIX regulates diverse plant biological processes (i.e., flowering time, leaf development, and plant growth), possibly by mediating trafficking of other protein cargoes, such as BRI1.
Notably, characterization of null mutants, which could not germinate even though embryo development was apparently normal, showed that ALIX is essential for plant life, as has been reported for other ESCRT-III-associated components (Spitzer et al., 2009; Isono et al., 2010). These results are at variance with those observed for yeast bro1 and A. nidulans palC mutations, which do not affect cell viability under normal growth conditions, possibly because there are at least two partially redundant Bro1-domain containing proteins in fungi (Luhtala and Odorizzi, 2004; Tilburn et al., 2005). Interallelic complementation and RT-PCR assays showed that a putative truncated protein from the Arabidopsis alix-3 allele could complement alix-1 defects. Thus, alix-1 and alix-3 products, which affect different ALIX domains, might complement each other’s defects by forming heterodimers. In the case of alix-2, gene products could not be detected, indicating that a single of copy of alix-1 allele in transheterozygous alix-1 alix-2 mutants is insufficient to allow seedling growth progression.
Differences were also observed in gametophytic transmission efficiency rates of alix-2 and alix-3 alleles. Transmission efficiency of alix-2 through the female gametophyte was decreased by half, which could explain the reduced frequency of homozygous mutants observed in the progeny of heterozygous alix-2 plants. However, gametophytic transmission defects were not observed for alix-3. In this context, reduction in the homozygous alix-3 mutant class might be explained by reduced compatibility between male and female gametes harboring the alix-3 mutation. In this scenario, compatibility defects would be observed only during self-fertilization of heterozygous alix-3 mutants but not in reciprocal crosses with the wild type.
Previous analysis of the Arabidopsis cytosolic proteome identified ALIX as a component of this cell compartment (Ito et al., 2011). In line with this, using cell fractionation experiments and confocal microscopy, we found that ALIX mainly localizes in the cytosol, although it can be associated, to a much lesser extent, with specific membrane compartments. These results are in agreement with previous reports that mammalian ALIX and yeast Bro1 are cytosolic proteins that associate transiently with MVBs/PVCs (Vito et al., 1999; Odorizzi et al., 2003). Recruitment of ALIX/Bro1 to MVBs/PVCs occurs via interaction of their Bro1 domains with ESCRT-III complex components Vps32/Snf7 and CHMP4, in yeast and mammals, respectively (Odorizzi et al., 2003; Morita et al., 2007; Wemmer et al., 2011). Similarly, we demonstrated by yeast two-hybrid and BiFC experiments that At-ALIX interacts through its Bro1 domain with SNF7.1 and SNF7.2, the two Arabidopsis homologs of Vps32/Snf7/CHMP4 (Winter and Hauser, 2006). Colocalization of BiFC interactions and FM4-64 staining indicated that ALIX binding to SNF7 proteins takes place in endosomal compartments. The latter likely correspond to MVBs as shown by colocalization of a GFP-ALIX fusion with the MVB marker mCherry-RabF2b.
We also found that ALIX protein derivatives harboring the alix-1 mutation (Gly260-to-Asp) in the Bro1 domain showed reduced ability to interact with SNF7 proteins in both yeast two-hybrid and BiFC assays. Crystallographic studies have shown that the Bro1 domain structure resembles a boomerang containing 14 α-helices and three β-sheets that form a tetratricopeptide (TPR) pocket and two exposed hydrophobic patch substructures (Odorizzi et al., 2003; Kim et al., 2005; Boysen and Mitchell, 2006; Fisher et al., 2007; Boysen et al., 2010). Mutant and structural analyses showed that whereas the second hydrophobic patch is responsible for Bro1/ALIX binding to SNF7/CHMP4, the TPR pocket helps to stabilize intramolecular interactions between different parts of the Bro1 domain. Gly-260 lays in α-helix 9 that is part of the TPR pocket. This position is relatively conserved in other Bro1 domain-containing proteins, in which similar noncharged residues (e.g., Ala and Ser) can be found. It is likely that substitution of Gly-260 by Asp, a charged amino acid, affects the TPR structure to alter the Bro1 conformation, leading to reduced interaction with SNF7 proteins. It would be interesting to test whether a similar substitution in other Bro1 domain-containing proteins hampers their ability to associate with ESCRT-III complexes.
At-ALIX association with ESCRT-III complex subunits SNF7.1 and SNF7.2 at MVBs correlates with a function in general control of protein trafficking to the vacuole, as has been shown for its counterparts in fungi and animals (Bissig and Gruenberg, 2014). In agreement with this, we found alix-1 mutation alters trafficking of both PHT1;1 and BRI-GFP fusions, reducing their vacuolar degradation. During this process, ALIX likely acts at late stages during endosomal transport, according to our data using vesicle trafficking inhibitors and mutants altering PHT1;1 sorting. The fact that PHT1;1-GFP localizes in the tonoplast of alix-1 root cells strongly supports this notion and indicates that ALIX plays a role during packaging of cargo proteins into ILVs. Thus, loss of ALIX function would cause reduced internalization into ILVs of PHT1;1 proteins that are targeted for vacuolar degradation, being retained in the tonoplast when MVBs fuse with vacuoles (Figure 10B). Similar missorting phenomena have been reported for transmembrane auxin carriers PIN1, PIN2, and AUX1 in chmp1a chmp1b double mutants, which display altered ESCRT-III function and impaired ILV formation (Spitzer et al., 2009). Interestingly, although BRI1-GFP fusion failed to be delivered to the vacuole for degradation in alix-1 cells, it did not accumulate in the tonoplast, but rather it might have been recycled to the PM. Differential accumulation of PHT1;1 and BRI1 in alix-1 mutants might reflect that ALIX function concerns multiple intracellular routes, each used by a differential subset of cargo proteins. In this context, distinct types of endosomal compartments have been shown to mediate sorting of specific cargo proteins in Arabidopsis. This is the case of SORTING NEXIN1 (SNX1)- and GNOM-containing endosomes that specifically facilitate trafficking of PIN2 and PIN1 proteins, respectively (Jaillais et al., 2006). In accordance with our hypothesis, it has been previously reported that whereas PHT1;1 trafficking is likely mediated by SNX1-containing endosomes, BRI1 sorting is facilitated by GNOM-labeled endosomes (Bayle et al., 2011; Irani et al., 2012). The use of alix-1 mutants may help to unveil the mechanistic differences between these two types of endosomal carriers.
One question raised by this study is how alix-1 mutation suppresses phr1 phenotypes. We speculate that altered subcellular distribution of Pi in alix-1 mutants affects Pi homeostasis, leading to increased Pi starvation responses. Similarly, it has been recently reported that Arabidopsis pht4;6 mutants, which lack a Golgi-localized Pi carrier belonging to the major facilitator superfamily of permeases, also display altered intracellular Pi compartmentation and enhanced PSI gene expression, in spite of their total Pi content remaining unchanged (Hassler et al., 2012). Together, these findings support the notion that proper sensing of Pi levels from all subcellular pools governs general plant responses to Pi deprivation. In the case of alix-1, lower vacuolar Pi levels and missorting of Pi transporters to the tonoplast may suggest that PHT1;1 proteins are exporting Pi out of the vacuole in mutant cells. In this scenario, acidification of the cytosol should be expected, as PHT1 proteins are Pi:H+ symporters that cotransport two to four protons along with each Pi ion that is absorbed (Ullrich-Eberius et al., 1981; Sakano, 1990; Nussaume et al., 2011). Such changes in intracellular pH can be registered as variations in NMR spectra. However, the fact that alix-1 and wild-type plants displayed similar NMR spectra profiles suggests that no significant alterations in intracellular pH are occurring and, therefore, that PHT1;1 proteins are not exporting Pi and H+ out of the vacuole. Nevertheless, this scenario cannot be definitively discarded and should be further investigated in greater depth. Alternatively, reduced vacuolar Pi levels could be explained by altered morphology of this cell compartment in alix-1 mutants. Although alix-1 root cells contained an increased number of vacuoles compared with wild-type plants, their average size was reduced, which is expected to reduce vacuolar volume and consequently may limit Pi storage capacity (Hassler et al., 2012).
Defective vacuolar morphology of alix-1 uncovers a role for members of the ALIX family in vacuolar biogenesis. Defects in vacuolar morphology have also been associated with mutations altering the last step of ILV formation. In fact, this is the case for mutations affecting ESCRT-III-associated proteins such as VPS4/SKD1 and AMSH deubiquitinases, which lead to an excess of membrane components that triggers vacuolar fragmentation. Fragmented vacuoles provide increased membrane surface to cope with these extra membrane components (Spitzer et al., 2009; Isono et al., 2010; Shahriari et al., 2010). Similar vacuolar defects in alix-1 mutants further support a role for ALIX, likely as part of ESCRT-III complexes, in ushering cargo proteins and other membrane components into ILVs that might be relevant for proper vacuole biogenesis.
Future studies should seek to better understand how ALIX contributes to the dynamics of the plant endomembrane system, including vacuolar morphogenesis, and to identify additional cargo proteins whose trafficking depends on ALIX. Additionally, future efforts should aim to determine whether ALIX mediates other biological processes, such as cytokinesis, apoptosis, and viral infection, as has been shown for other Bro1-related proteins in fungi and animals (Missotten et al., 1999; Fisher et al., 2007; Morita et al., 2007). The insights gained in these studies will be crucial for assessing ALIX and ESCRT-III roles in maintaining plant cell homeostasis. Based on the fact that total loss of function of ALIX or known ESCRT components is lethal in plants, the viability of the alix-1 mutant makes it highly valuable.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants used in this study were of the Col-0 ecotype. For the positional cloning of alix-1, phr1 plants in the Ler background were also used. The alix T-DNA insertion mutants were obtained from the GABI-Kat collection (https://www.gabi-kat.de/; alix-2, line 837H11; and alix-3, line 780B02). WAVE and BRI1-GFP lines were kindly provided by Niko Geldner (Geldner et al., 2007, 2009). Plants were grown in Johnson medium (Johnson et al., 1957) with modifications (Bates and Lynch, 1996) at 21°C under a 16-h-light/8-h-dark cycle using cool white fluorescent light conditions (100 μmol m−2 s−1). Specific treatments were performed as stated in each experiment (see below and figure legends). The effect of variation in Pi supply was analyzed by growing plants in Pi-rich (+P; 1 mM KH2PO4) and Pi-deficient (−P; no addition of KH2PO4) media unless a different Pi concentration is specified. For BiFC experiments, Nicotiana benthamiana plants were grown in soil in the greenhouse at 22°C under a 16-h-light/8-h-dark photoperiod prior to agroinfiltration of leaves with the corresponding constructs (Sparkes et al., 2006).
Constructs for Expression in Plants
For alix-1 phr1 complementation experiments, a 4-kb genomic fragment of DNA containing the ALIX gene was obtained by PCR from wild-type plants and cloned into the pBIB plasmid (Becker, 1990). To this end, an ALIX genomic fragment (containing the promoter plus the first 1765 bp of the coding region) was PCR amplified using primers with SacII and EagI restriction sites: SacII-5, 5′-CGAAAGTCCGCGGCAAGGCCTCTCTAATTCATAAAC-3′, and EagI-3, 5′-GAAGTACTTACCGGCCGTCGATCAAGAATAG-3′. Upon digestion of the PCR product, the resulting DNA fragment was cloned into SacII-EagI sites of the pBluescript SK+ plasmid (construct 1). Then, the rest of the ALIX coding region (from nucleotide 1759 to the STOP codon) plus the 3′ untranslated regions was amplified by PCR using specific primers with EagI and EcoRI restriction sites: EagI-5, 5′-CTATTCTTGATCGACGGCCGGTAAGTACTT-3′, and EcoRI-3, 5′-GGTCTGAATTCTGCAATACATAAGCAAGGATGTACC-3′. Upon digestion of the PCR product, the resulting DNA fragment was cloned into EagI-EcoRI sites in construct 1 to obtain construct 2. By partial digestion of construct 2 with SacI-EcoRI, the complete ALIX genomic region was obtained and cloned into the pBIB binary vector (Becker, 1990), which confers hygromycin resistance in plants.
For ALIX subcellular localization experiments, plants expressed under the control of the At-ALIX promoter, a posttranslational fusion of the GFP to the N terminus of ALIX, were obtained. To this end, the GFP cDNA without stop codon plus three codons encoding Gly residues (as a linker) was PCR amplified using specific primers with NcoI restriction sites: NcoI-5, 5′-GTGTCCATGGTGAGCAAGGGCGAGGAG-3′, and NcoI-3, 5′-CATAGCCATGGCTCCTCCTCCCTTGTACAGCTCGTCCATGCC-3′. Upon digestion of the PCR product, the resulting DNA fragment was cloned into the NcoI site in construct 2 to obtain construct 3. By partial digestion of construct 3 with SacI-EcoRI, the complete ALIX genomic region plus GFP (ALIXpro:GFPgALIX) region was obtained and cloned into the pBIB binary vector.
Plant transformation was performed by transferring the resulting constructs in binary vectors to Agrobacterium tumefaciens C58C1 (pGV2260) competent cells (Deblaere et al., 1985). Transformation of Arabidopsis plants was performed by the floral dip method (Clough and Bent, 1998). T1 transgenic seeds were selected based on their hygromycin resistance, and T3 homozygous progenies were used for further studies.
Isolation of sphr Mutants
Approximately 50,000 phr1-1 seeds were mutagenized with EMS by treating hydrated seeds (soaked 16 h at 4°C in distilled water) with 0.3% EMS for 13 h at room temperature. Then, seeds were washed 15 times with sterile water (10 min each time), stratified at 4°C for 2 d, and then sown directly onto soil in 240 pots (M1 families). The progenies of these pots (M2 families) were collected, stored, and labeled separately. Around 1500 seeds of each M2 family were sown directly on Pi-deficient medium for 10 d. Seedlings showing anthocyanin accumulation in cotyledons were recovered on fresh, Pi-rich medium and transferred to soil to obtain their M3 progeny. M3 seeds were analyzed for the inheritance of the observed phenotype and the presence of the phr1 mutation to verify anthocyanin accumulation was due to new mutations and not to a contamination with wild-type plants. Primers used to PCR amplify and sequence the phr1 mutation were phr1-5, 5′-TGCAGACACCAAGCAACAACGATAGTG-3′, and phr1-3, 5′-TCACTACCGCCAAGACTGTTGACAGCC-3′.
Genetic Analysis and Positional Cloning of alix-1
Prior to mutant characterization, alix-1 phr1 mutant plants were backcrossed four times to both phr1-1 (Rubio et al., 2001) and wild-type plants. The progeny of these crosses was used to test the linkage of the different phenotypes of alix-1 plants to a single recessive mutation. The ALIX gene was cloned by a map-based chromosome walking procedure on the basis of a cross between the alix-1 phr1-1 mutant (Col ecotype) and a phr1-1 mutant introgressed five times into the Ler background. By use of 1415 F2 seedlings showing the alix-1 phenotype, the ALIX gene was mapped to chromosome 1 (in BACs F9L1) with a series of simple sequence length polymorphism markers (Bell and Ecker, 1994) and cleaved amplified polymorphic sequences (Konieczny and Ausubel, 1993) available in the Cereon collection of nucleotide polymorphisms between Col and Ler ecotypes (http://www.arabidopsis.org/cereon/index.html). Therefore, the ALIX locus was defined to a region of ∼140 kb that contained 32 genes. Among them, first candidate genes to be sequenced were selected based on their differential expression in response to Pi starvation (according to transcriptomic data in Bustos et al., 2010; At1g14870, At1g15100, and At1g15260). Since no mutations were found in these Pi starvation-responsive genes, all other genes in this region were sequenced.
Computer Programs for Protein and Nucleic Acid Analysis
Gene sequences were obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and The Arabidopsis Information Resource (http://www.arabidopsis.org) databases. Sequence alignment was performed using the T-COFFEE (Notredame et al., 2000; http://www.ch.embnet.org/software/TCoffee.html) and GENOMATIX (http://www.genomatix.de/cgi-bin/dialign/dialign.pl) programs. Protein domain predictions were performed using the SMART (Letunic et al., 2004) and COILS (Lupas, 1996; www.ch.EMBnet.org) programs.
Physiological Measurements
The method of Ames (1966) was used to determine the total Pi content in roots and shoots of 12-d-old seedlings grown on Pi-sufficient (500 μM) and low-Pi (15 μM) media. Anthocyanins were extracted from aerial parts of plants grown on Pi-lacking medium for 12 d and measured as described (Swain and Hillis, 1959). For flowering time measurement, plants were grown in the greenhouse at 22°C under continuous long-day photoperiods (16 h light/8 h darkness) in a mixture of soil and vermiculite (3:1). Flowering initiation was measured as the number of days to flowering and the leaf number at flowering. The first estimation corresponds to the number of days from the planting date until the opening of the first flower. Leaf number was calculated as the total number of rosette and cauline leaves in the main inflorescence at flowering time. Mean values were compared using Student’s t test. All experiments were repeated at least three times with similar results.
Analysis of Embryo Viability
To analyze embryo viability from mature seeds, seeds were scarified using a 20% bleach solution (containing 0.1% Triton X-100), stained with 1% tetrazolium (Sigma-Aldrich), and cleared with a lactophenol solution. Stained embryos were observed under a Leica LMD-6000 microscope (Verma et al., 2013). To genotype ungerminated seeds, genomic DNA was isolated from pools of 15 embryos released from their integuments using needles.
RT-qPCR
RNA used for RT-qPCR experiments was isolated from seedlings using the RNeasy Plant Mini kit (Qiagen) and DNase digestion to remove genomic DNA contamination. Three biological replicates, each consisting of tissue pooled from 15 to 20 plants from different plates, were taken for each extraction. cDNA was synthesized from 2 μg total RNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems). RT-qPCR reactions were performed in an Applied Biosystems 7300 real-time PCR system using the FastStart Taqman Probe Master-Rox (Roche); CT values were obtained with a 7300 Systems SDS software v.1.3 (Applied Biosystems). Relative expression changes were calculated by the comparative CT method; x-fold change is calculated as 2–ΔΔCt. ΔCt values were calculated as the difference between the CT value of each PSI gene analyzed and the CT value of ACTIN8. ΔΔCt was the difference between ΔCt of a given sample and the ΔCt value of the wild-type control grown under Pi-rich conditions (Livak and Schmittgen, 2001). Primers and probes used are described in Supplemental Table 9.
To amplify truncated ALIX transcripts from heterozygous null mutants, RNA and cDNA were prepared as above. For PCRs, the following primers were used: exon 4, 5′-GAGGCAACTGGAAAATCTTGG-3′; UTR3′, 5′- TAGACACACATCAAGTACTCACAGG-3′; and T-DNA, 5′-CCCATTTGGACGTGAATGTAGACAC-3′.
Yeast Two-Hybrid Experiments
To assay protein-protein interactions by yeast two-hybrid assays, plasmids were cotransformed into Saccharomyces cerevisiae AH109 cells, following standard heat shock protocols (Chini et al., 2007). Successfully transformed colonies were identified on yeast synthetic dropout lacking Leu and Trp. These colonies were resuspended in water and transferred to selective media lacking Ade, His, Leu, and Trp. Yeast cells were incubated at 30°C for 6 d. Empty vectors were cotransformed as negative controls. To test ALIX dimerization and its interaction with SNF7 proteins, different gene versions were PCR amplified and cloned into the pGADT7 and the pGBKT7 vectors using Gateway technology (Clontech), including coding sequences for full-length ALIX (ALIX, FL), ALIX Bro1 domain (N-terminal region; amino acids 1 to 413), ALIX C-terminal region (containing the coiled coils and the Pro-rich domain; amino acids 405 to 846), ALIX-1 (ALIX FL containing the alix-1 mutation), ALIX-1 1-413 (ALIX 1-413 version containing the alix-1 mutation), SNF7.1, and SNF7.2 (both full-length versions). β-Galactosidase assay was performed by monitoring the LacZ reporter gene expression directly upon addition of o-nitrophenyl-β-d-galactopyranoside to the liquid culture extracts (Clontech). For figure preparation, representative colonies for each bait:prey construct combination from independent plates were used.
Protein extracts from yeast cells were prepared using a trichloroacetic acid protein extraction technique (Foiani et al., 1994).
BiFC Experiments
Different combinations of Agrobacterium clones expressing fusion proteins as indicated were coinfiltrated into the abaxial surface of leaves from 3-week-old N. benthamiana plants (Sparkes et al., 2006). Leaves were also coinfiltrated with p19, which suppresses gene silencing (Voinnet, 2003). Empty vectors were used as negative controls. Fluorescence was visualized in epidermal cells of leaves 3 d after infiltration using a Leica TCS SP5 confocal microscope (Leica). Endosomes were observed 60 min after infiltration of leaves with a 5 μM FM4-64 water solution.
Protein extraction was performed using Laemmli 2× solution (also containing 4 M urea and 100 mM DTT).
Subcellular Fractionation, Protein Degradation, Tandem Affinity Purification, and Immunoblot Assays
For subcellular fractionation assays, fresh 10-d-old seedlings were homogenized in 2 volumes of cold extraction buffer (50 mM HEPES, pH 7.9, 300 mM sucrose, 150 mM NaCl, 10 mM K-Acetate, 5 mM EDTA, 1 mM PMSF, and 1× complete protease inhibitor [Roche]) and centrifuged at 278g for 5 min at 4°C. Samples of the first supernatant were kept as input control (S10). The rest of S10 supernatant was centrifuged for 60 min at 100,000g, and two fractions were obtained; supernatant S100 and pellets. The S100 supernatants, containing soluble cytosolic proteins, were also kept. Pellets were resuspended in 1 original volume of homogenization buffer without additives (control) or with 1 M NaCl (Na), 0.1 M Na2CO (pH 10.9), 1, 2, or 4 M urea, or 2% Triton X-100, and after 30 min centrifuged again at 100,000g for 90 min, giving wash fractions S100’ and pellets. This procedure was repeated to give washed pellets P100’’.
For protein degradation assays, 10-d-old seedlings were grown in Johnson solid media supplemented or not with 1 mM Pi for 10 d and then transferred to liquid Johnson media containing 50 μM cycloheximide (Sigma-Aldrich) in the presence or absence of 1 mM Pi. Whole-plant samples were harvested at specific time points as indicated and homogenized as in microsomal fractionation experiments (with no addition of Triton X-100). Then, samples were centrifuged at 278g at 4°C for 5 min. Protein concentration in the supernatant was determined by Bradford assay (Bio-Rad).
TAPa experiments were performed as described (Rubio et al., 2005).
For immunoblots, samples were denatured, separated on 7.5% SDS-PAGE gels, and transferred onto polyvinylidene difluoride membranes (Millipore). Membranes were probed with different antibodies. Antibodies used for immunodetections were anti-GFP-HRP (1:5000 dilution; Milteny Biotec), anti-HA-HRP (1:2000; Roche), anti-MYC-HRP (1:2000 dilution; Santa Cruz Biotechnology), anti-VSR (1:1000 dilution; Zouhar et al., 2010), and anti-cFBPase (1:5000 dilution; Agrisera). For BiFC protein detection, anti-GFP was used (1:1000 dilution; Clontech). The secondary antibody was anti-rabbit-HRP (1:10,000 dilution; GE Healthcare Life Sciences).
In-Gel Protein Digestion and Sample Preparation
Bands of interest from SYPRO-stained gels were excised manually, deposited in 96-well plates, and processed automatically in a Proteineer DP (Bruker Daltonics). The digestion protocol used was based on Shevchenko et al. (1996) with minor variations: Gel plugs were washed first with 50 mM ammonium bicarbonate and second with acetonitrile (ACN) prior to reduction with 10 mM DTT in 25 mM ammonium bicarbonate solution, and alkylation was performed with 55 mM indole-3-acetic acid in 50 mM ammonium bicarbonate solution. Gel pieces were then rinsed first with 50 mM ammonium bicarbonate and second with ACN and dried under a stream of nitrogen. Proteomics Grade Trypsin (Sigma-Aldrich) at a final concentration of 16 ng/μL in 25% ACN/50 mM ammonium bicarbonate solution was added and the digestion took place at 37°C for 4 h. The reaction was stopped by adding 50% ACN/0.5% trifluoroacetic acid for peptide extraction. The tryptic-eluted peptides were dried by speed vacuum centrifugation and were resuspended in 4 μL MALDI solution (30% ACN/15% isopropanol/0.5% trifluoroacetic acid). A 0.8-μL aliquot of each peptide mixture was deposited onto a 384-well OptiTOF Plate (AB SCIEX) and allowed to dry at room temperature. A 0.8-μL aliquot of matrix solution (3 mg/mL α-cyano-4-hydroxycinnamic acid in MALDI solution) was then deposited onto the dried digest and allowed to dry at room temperature.
MALDI Peptide Mass Fingerprinting, Tandem Mass Spectrometry Analysis, and Database Searching
For MALDI-TOF/TOF analysis, samples were automatically acquired in an ABi 4800 MALDI TOF/TOF mass spectrometer (AB SCIEX) in positive ion reflector mode (the ion acceleration voltage was 25 kV for MS acquisition and 2 kV for tandem mass spectrometry [MS/MS]), and the obtained spectra were stored in the ABi 4000 Series Explorer Spot Set Manager. Peptide mass fingerprinting (PMF) and MS/MS fragment ion spectra were smoothed and corrected to zero baseline using routines embedded in ABi 4000 Series Explorer Software v3.6. Each PMF spectrum was internally calibrated with the mass signals of trypsin autolysis ions to reach a typical mass measurement accuracy of <25 ppm. Known trypsin and keratin mass signals, as well as potential sodium and potassium adducts (+21 D and +39 D) were removed from the peak list. To submit the combined PMF and MS/MS data to MASCOT software v.2.5.10 (Matrix Science), GPS Explorer v4.9 was used, searching in the Viridiplantae protein database from Uniprot-SwissProt repository (UKBsp_20140703; 2,069,501 sequences; 705,582,891 residues). The following search parameters were used: enzyme, trypsin; allowed missed cleavages, 1; carbamidomethyl cysteine as fixed modification by the treatment with iodoacetamide; variable modifications, oxidation of methionine; mass tolerance for precursors was set to ± 50 ppm and for MS/MS fragment ions to ±0.3 D. The confidence interval for protein identification was set to ≥95% (P < 0.05), and only peptides with an individual ion score above the identity threshold were considered correctly identified.
Tracer and Drugs, Microscopy, and Image Processing
Seedlings visualized in live imaging experiments were grown vertically for 5 d and incubated in Johnson liquid medium supplemented or not with 1 mM KH2PO4 plus each specific drug at concentrations and periods described as follows: For FM4-64 (Invitrogen) internalization assays, seedlings were incubated for 5 min with 2 μM FM4-64 and rinsed with water two times. Visualization was performed every 20 min. FM4-64 stock was 4 mM in DMSO. For BFA experiments, seedlings were incubated for 90 min in darkness with 50 μM BFA (Sigma Aldrich). BFA stock was 100 mM in DMSO. For WM (Invitrogen) treatments, seedlings were incubated with 33 μM WM for 30 min under dark conditions. WM stock was 50 mM in DMSO. For ConcA (Sigma-Aldrich) assays, seedlings were incubated with 1 μM ConcA for 6 h. ConcA stock was 50 mM in ethanol. One hour incubation in darkness with 5 μM BCECF-AM [2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein acetoxymethyl ester; Sigma-Aldrich] was used to stain vacuoles. BCECF-AM stock was 1.6 mM in DMSO.
Confocal laser scanning microscopy was performed on three different inverted microscopes: Leica TCS SP2 (DBMV, Université de Lausanne, Switzerland), Leica TCS SP5 (CNB-CSIC, Madrid, Spain), and Zeiss LSM 780 (CEA-Cadarache, Saint-Paul-lès-Durance, France). Image processing was done with ImageJ from the National Institutes of Health (http://imagej.nih.gov/ij/).
Phosphate Uptake Experiments
Pi uptake experiments were performed according to Narang et al. (2000) and Misson et al. (2004). Twenty-four seedlings were used for each genotype and condition. Twelve-day-old seedlings grown in Pi-sufficient (500 μM) and low-Pi conditions (15 μM) were incubated separately in Pi incubation solution containing 33PO4 (5 mM MES, 0.1 mM CaCl2, 50 μM KH2PO4, and 0.15 μCi/mL 33PO4) for 2 h. After that, they were incubated in desorption medium (5 mM MES, 0.1 mM CaCl2, and 1 mM KH2PO4) for 2 h at 4°C. Scintillation cocktail (2 mL; InstaGel; Perkin-Elmer) was added to samples, and the radioactivity was measured using a liquid scintillation analyzer (TRI CARB, Packard Instrument Company).
NMR Analyses
NMR analyses were performed according to Roby et al. (1987). Ten-gram samples of roots from 3-week-old plants grown in Pi-sufficient conditions (500 μM) were cut into 4- to 5-mm2 pieces, vacuum-infiltrated in perfusion solution [5 mM glucose, 10 mM KNO3, 0.5 mM Ca(NO3)2, 1 mM KCl, 0.5 mM MgSO4, and 50 μM KH2PO4 at pH 6.0], and placed into a 25-mm glass tube under constant perfusion. In vivo 31P-NMR spectra were recorded on a spectrometer (AMX 400; Bruker) as described previously (Aubert et al., 1996). ImageJ v1.37 software (http://rsb.info.nih.gov/ij) was used to analyze NMR peak areas.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ALIX (At1g15130), PHR1 (At4g28610), PHT1;1 (At5g43350), SNF7.1 (At4g29160), and SNF7.2 (At2g19830). MS proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD002075 and 10.6019/PXD002075.
Supplemental Data
Supplemental Figure 1. Morphological comparisons between alix-1, phr1, and wild-type controls.
Supplemental Figure 2. Flowering time defects in alix-1 mutants.
Supplemental Figure 3. Complementation of alix-1 mutant defects.
Supplemental Figure 4. Multiple sequence alignment of ALIX-related proteins.
Supplemental Figure 5. Complementation of transcriptional defects in alix-1 mutants using a protein fusion of ALIX to GFP.
Supplemental Figure 6. ALIX gene expression and GFP-ALIX protein accumulation under different Pi supply conditions.
Supplemental Figure 7. Negative controls and expression analysis of bait and prey fusions used in yeast two-hybrid assays displayed in Figures 3 and 5.
Supplemental Figure 8. BiFC assays showing interaction between ALIX and SNF7.1 in vivo.
Supplemental Figure 9. Expression analysis of protein fusions used in BiFC assays displayed in Figure 5.
Supplemental Figure 10. Analysis of endocytosis and vesicle trafficking in alix-1 mutants.
Supplemental Figure 11. Mutants displaying reduced ALIX function are defective in vacuolar size and morphology.
Supplemental Figure 12. Vacuolar morphology defects in alix-1 mutants can be rescued by expression of a GFP-ALIX fusion.
Supplemental Figure 13. ALIX acts as a later stage than PHF1 during PHT1;1 trafficking.
Supplemental Figure 14. alix-1 mutation does not alter PHT1;1 localization in sorting endosomes and brefeldin bodies.
Supplemental Table 1. Phenotypic characterization of the progeny obtained from heterozygous alix-1 plants.
Supplemental Table 2. Segregation data from selfed alix-2 ALIX progeny.
Supplemental Table 3. Segregation data from selfed alix-3 ALIX progeny.
Supplemental Table 4. Germination analysis of the progeny of alix-2 ALIX and alix-3 ALIX heterozygous plants compared with wild-type (ALIX ALIX) plants.
Supplemental Table 5. Transmission efficiency of the alix-2 and alix-3 alleles through male and female gametes.
Supplemental Table 6. Protein identification details obtained with the Abi 4800 MALDI TOF/TOF mass spectrometer (AB SCIEX) and the GPS explorer v4.9 (AB SCIEX) software package combined with search engine Mascot version 2.5.10 (Matrix Science).
Supplemental Table 7. Reporter lines used in this study as described (Dettmer et al., 2006; Geldner et al., 2009).
Supplemental Table 8. Analysis of intracellular Pi pools in roots of wild-type and alix-1 PHR1 plants grown in Pi-sufficient media (500 μM).
Supplemental Table 9. List of primers and probes (Roche) used in qRT-PCR analyses
Supplementary Material
Acknowledgments
We thank Alfonso Muñoz, Enrique Rojo, Jan Zouhar, Maite Sanmartín, Michael Sauer, Miguel Angel López, Erika Isono, Claus Schwechheimer, Hélène Javot, Marie-Christine Thibaud, and Niko Geldner’s lab members for helpful discussion on the article. We thank Tsuyoshi Nakagawa and Sumie Ishiguro providing pGWB vectors, ABRC for cDNA clones, and GABI-KAT for T-DNA mutants. We also thank the confocal microscopy and in vitro plant culture facilities at the Centro Nacional de Biotecnología. This research was supported by the Spanish Ministry of Economy and Competitiveness (MINECO; National Research Program, Grants BIO2010-18820 and BIO2013-46539-R to V.R.; BIO2011-29085 to J.P.-A.; INNPACTO Program, Grant IPT-310000-2010-9 to J.P.-A.; CONSOLIDER Program, Grant 2007-28317 to J.P.-A.; and PLANT-KBBE Program, Grant EUI2008-03742 to L.N. and V.R.). X.C.-L. and L.C. were recipients of FPI fellowships from MINECO. C.R. was a recipient of a La Caixa PhD fellowship.
AUTHOR CONTRIBUTIONS
X.C.-L., L.C., N.G., L.N., J.P.-A., and V.R. conceived the study and designed the experiments. X.C.-L., L.C., E.M., C.R., M.L.I., E.G., M.I.P., R.B., and V.R. generated lines and constructs and performed the experiments. X.C.-L., L.C., E.M., R.B., N.G., L.N., J.P.-A., and V.R. analyzed the data. J.P.-A. and V.R. were co-senior authors of this study and wrote the article. All authors revised the article.
NOTE ADDED IN PROOF
While this article was under revision, an article by Kalinowska et al. was accepted for publication in PNAS. In this article, the authors also reported that ALIX is essential in Arabidopsis and is involved in intracellular trafficking and vacuole biogenesis.
Kalinowska, K., Nagel, M.K., Goodman, K., Cuyas, L., Anzenberger, F., Alkofer, A., Paz-Ares, J., Braun, P., Rubio, V., Otegui, M.S., and Isono, E. (2015). Arabidopsis ALIX is required for the endosomal localization of the deubiquitinating enzyme AMSH3. Proc. Natl. Acad. Sci. USA, http://dx.doi.org/10.1073/pnas.1510516112.
Glossary
- PM
plasma membrane
- MVB
multivesicular body
- GA
Golgi apparatus
- PVC
prevacuolar compartment
- ILV
intraluminal vesicle
- ER
endoplasmic reticulum
- EMS
ethyl methanesulfonate
- Ler
Landsberg erecta
- TGN
trans-Golgi network
- EE
early endosome
- BFA
brefeldin A
- WM
wortmannin
- BiFC
bimolecular fluorescence complementation
- ConcA
concanamycin A
- TPR
tetratricopeptide
- ACN
acetonitrile
- MS/MS
tandem mass spectrometry
- PMF
peptide mass fingerprinting
References
- Ames B.N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8: 115–118. [Google Scholar]
- Aubert S., Alban C., Bligny R., Douce R. (1996). Induction of beta-methylcrotonyl-coenzyme A carboxylase in higher plant cells during carbohydrate starvation: evidence for a role of MCCase in leucine catabolism. FEBS Lett. 383: 175–180. [DOI] [PubMed] [Google Scholar]
- Ayadi A., David P., Arrighi J.F., Chiarenza S., Thibaud M.C., Nussaume L., Marin E. (2015). Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol. 167: 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M. (2005). A protein’s final ESCRT. Traffic 6: 2–9. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D.J., Estepa-Sabal E.J., Meerloo T., Emr S.D. (2002a). Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3: 271–282. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D.J., Snyder W.B., Wendland B., Emr S.D. (2002b). Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3: 283–289. [DOI] [PubMed] [Google Scholar]
- Baldwin T.C., Handford M.G., Yuseff M.I., Orellana A., Dupree P. (2001). Identification and characterization of GONST1, a golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 13: 2283–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates T.R., Lynch J.P. (1996). Stimulation of root hair elongation in Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 19: 529–538. [Google Scholar]
- Bayle V., Arrighi J.F., Creff A., Nespoulous C., Vialaret J., Rossignol M., Gonzalez E., Paz-Ares J., Nussaume L. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.J., Ecker J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144. [DOI] [PubMed] [Google Scholar]
- Bissig C., Gruenberg J. (2014). ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol. 24: 19–25. [DOI] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173. [DOI] [PubMed] [Google Scholar]
- Boysen J.H., Mitchell A.P. (2006). Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol. Biol. Cell 17: 1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boysen J.H., Subramanian S., Mitchell A.P. (2010). Intervention of Bro1 in pH-responsive Rim20 localization in Saccharomyces cerevisiae. Eukaryot. Cell 9: 532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch M., Mayer U., Jürgens G. (1996). Molecular analysis of the Arabidopsis pattern formation of gene GNOM: gene structure and intragenic complementation. Mol. Gen. Genet. 250: 681–691. [DOI] [PubMed] [Google Scholar]
- Bustos R., Castrillo G., Linhares F., Puga M.I., Rubio V., Pérez-Pérez J., Solano R., Leyva A., Paz-Ares J. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhuang X., Gao C., Wang X., Jiang L. (2014). The Arabidopsis endosomal sorting complex required for transport III regulates internal vesicle formation of the prevacuolar compartment and is required for plant development. Plant Physiol. 165: 1328–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., et al. (2015). The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 27: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Chiou T.J., Lin S.I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62: 185–206. [DOI] [PubMed] [Google Scholar]
- Clifford R., Schüpbach T. (1994). Molecular analysis of the Drosophila EGF receptor homolog reveals that several genetically defined classes of alleles cluster in subdomains of the receptor protein. Genetics 137: 531–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Conibear E. (2002). An ESCRT into the endosome. Mol. Cell 10: 215–216. [DOI] [PubMed] [Google Scholar]
- Deblaere R., Bytebier B., De Greve H., Deboeck F., Schell J., Van Montagu M., Leemans J. (1985). Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 13: 4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Borja M., Wubbolts R., Calafat J., Janssen H., Divecha N., Dusseljee S., Neefjes J. (1999). Multivesicular body morphogenesis requires phosphatidyl-inositol 3-kinase activity. Curr. Biol. 9: 55–58. [DOI] [PubMed] [Google Scholar]
- Fisher R.D., Chung H.Y., Zhai Q., Robinson H., Sundquist W.I., Hill C.P. (2007). Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128: 841–852. [DOI] [PubMed] [Google Scholar]
- Foiani M., Marini F., Gamba D., Lucchini G., Plevani P. (1994). The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14: 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo A., Hervás-Aguilar A., Rodríguez-Galán O., Vincent O., Arst H.N. Jr., Tilburn J., Peñalva M.A. (2007). PalC, one of two Bro1 domain proteins in the fungal pH signalling pathway, localizes to cortical structures and binds Vps32. Traffic 8: 1346–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Luo M., Zhao Q., Yang R., Cui Y., Zeng Y., Xia J., Jiang L. (2014). A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr. Biol. 24: 2556–2563. [DOI] [PubMed] [Google Scholar]
- Gao C., Zhuang X., Cui Y., Fu X., He Y., Zhao Q., Zeng Y., Shen J., Luo M., Jiang L. (2015). Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. USA 112: 1886–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. [DOI] [PubMed] [Google Scholar]
- Geldner N., Hyman D.L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E., Solano R., Rubio V., Leyva A., Paz-Ares J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler S., Lemke L., Jung B., Möhlmann T., Krüger F., Schumacher K., Espen L., Martinoia E., Neuhaus H.E. (2012). Lack of the Golgi phosphate transporter PHT4;6 causes strong developmental defects, constitutively activated disease resistance mechanisms and altered intracellular phosphate compartmentation in Arabidopsis. Plant J. 72: 732–744. [DOI] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., Emr S.D. (2011). The ESCRT pathway. Dev. Cell 21: 77–91. [DOI] [PubMed] [Google Scholar]
- Ibl V., Csaszar E., Schlager N., Neubert S., Spitzer C., Hauser M.T. (2012). Interactome of the plant-specific ESCRT-III component AtVPS2.2 in Arabidopsis thaliana. J. Proteome Res. 11: 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani N.G., et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8: 583–589. [DOI] [PubMed] [Google Scholar]
- Isono E., Katsiarimpa A., Müller I.K., Anzenberger F., Stierhof Y.D., Geldner N., Chory J., Schwechheimer C. (2010). The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell 22: 1826–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Batth T.S., Petzold C.J., Redding-Johanson A.M., Mukhopadhyay A., Verboom R., Meyer E.H., Millar A.H., Heazlewood J.L. (2011). Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. J. Proteome Res. 10: 1571–1582. [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Fobis-Loisy I., Miège C., Rollin C., Gaude T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109. [DOI] [PubMed] [Google Scholar]
- Johnson C.M., Stout P.R., Broker T.C., Carlton A.B. (1957). Comparative chlorine requirements of different plants species. Plant Soil 8: 337–353. [Google Scholar]
- Kasai K., Takano J., Miwa K., Toyoda A., Fujiwara T. (2011). High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J. Biol. Chem. 286: 6175–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiarimpa A., Anzenberger F., Schlager N., Neubert S., Hauser M.T., Schwechheimer C., Isono E. (2011). The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization. Plant Cell 23: 3026–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiarimpa A., Kalinowska K., Anzenberger F., Weis C., Ostertag M., Tsutsumi C., Schwechheimer C., Brunner F., Hückelhoven R., Isono E. (2013). The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell 25: 2236–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D.J., Babst M., Emr S.D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155. [DOI] [PubMed] [Google Scholar]
- Kim J., Sitaraman S., Hierro A., Beach B.M., Odorizzi G., Hurley J.H. (2005). Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell 8: 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Swarup R., Bennett M., Friml J. (2006). Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN1. Plant Cell 18: 3171–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb C., Nagel M.K., Kalinowska K., Hagmann J., Ichikawa M., Anzenberger F., Alkofer A., Sato M.H., Braun P., Isono E. (2015). FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol. 167: 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410. [DOI] [PubMed] [Google Scholar]
- Leung K.F., Dacks J.B., Field M.C. (2008). Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9: 1698–1716. [DOI] [PubMed] [Google Scholar]
- Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. (2004). SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32: D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Luhtala N., Odorizzi G. (2004). Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166: 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. (1996). Prediction and analysis of coiled-coil structures. Methods Enzymol. 266: 513–525. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P.D. (2003). Role of ESCRT-I in retroviral budding. J. Virol. 77: 4794–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Fisher R.D., Whitby F.G., Sundquist W.I., Hill C.P. (2008). ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. USA 105: 7687–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt M.W., McKittrick I., West M., Odorizzi G. (2007). Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol. Biol. Cell 18: 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J., Thibaud M.C., Bechtold N., Raghothama K., Nussaume L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 55: 727–741. [DOI] [PubMed] [Google Scholar]
- Missotten M., Nichols A., Rieger K., Sadoul R. (1999). Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 6: 124–129. [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H.Y., Morham S.G., Gygi S.P., Rodesch C.K., Sundquist W.I. (2007). Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26: 4215–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang R.A., Bruene A., Altmann T. (2000). Analysis of phosphate acquisition efficiency in different Arabidopsis accessions. Plant Physiol. 124: 1786–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D.P., Russell M.R., Odorizzi G. (2007). A concentric circle model of multivesicular body cargo sorting. EMBO Rep. 8: 644–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217. [DOI] [PubMed] [Google Scholar]
- Nussaume L., Kanno S., Javot H., Marin E., Pochon N., Ayadi A., Nakanishi T.M., Thibaud M.C. (2011). Phosphate import in plants: Focus on the PHT1 transporters. Front. Plant Sci. 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G. (2006). The multiple personalities of Alix. J. Cell Sci. 119: 3025–3032. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Katzmann D.J., Babst M., Audhya A., Emr S.D. (2003). Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116: 1893–1903. [DOI] [PubMed] [Google Scholar]
- Páli T., Dixon N., Kee T.P., Marsh D. (2004). Incorporation of the V-ATPase inhibitors concanamycin and indole pentadiene in lipid membranes. Spin-label EPR studies. Biochim. Biophys. Acta 1663: 14–18. [DOI] [PubMed] [Google Scholar]
- Pires R., et al. (2009). A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure 17: 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F.C., Buono R., Otegui M.S. (2011). Plant endosomal trafficking pathways. Curr. Opin. Plant Biol. 14: 666–673. [DOI] [PubMed] [Google Scholar]
- Reyes F.C., Buono R.A., Roschzttardtz H., Di Rubbo S., Yeun L.H., Russinova E., Otegui M.S. (2014). A novel endosomal sorting complex required for transport (ESCRT) component in Arabidopsis thaliana controls cell expansion and development. J. Biol. Chem. 289: 4980–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson L.G., Howard A.S., Khuu N., Gidda S.K., McCartney A., Morphy B.J., Mullen R.T. (2011). Protein-protein interaction network and subcellular localization of the Arabidopsis rhaliana ESCRT machinery. Front. Plant Sci. 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler C., Nebenführ A., Movafeghi A., Stussi-Garaud C., Behnia L., Pimpl P., Staehelin L.A., Robinson D.G. (2002). Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14: 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roby C., Martin J.B., Bligny R., Douce R. (1987). Biochemical changes during sucrose deprivation in higher plant cells. Phosphorus-31 nuclear magnetic resonance studies. J. Biol. Chem. 262: 5000–5007. [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martín A.C., Iglesias J., Leyva A., Paz-Ares J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15: 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V., Shen Y., Saijo Y., Liu Y., Gusmaroli G., Dinesh-Kumar S.P., Deng X.W. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41: 767–778. [DOI] [PubMed] [Google Scholar]
- Sakano K. (1990). Proton/phosphate stoichiometry in uptake of inorganic phosphate by cultured cells of Catharanthus roseus (L.) G. Don. Plant Physiol. 93: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D., Viotti C., Krüger F., Künzl F., Sturm S., Bubeck J., Hillmer S., Frigerio L., Robinson D.G., Pimpl P., Schumacher K. (2011). Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858. [DOI] [PubMed] [Google Scholar]
- Shahriari M., Keshavaiah C., Scheuring D., Sabovljevic A., Pimpl P., Häusler R.E., Hülskamp M., Schellmann S. (2010). The AAA-type ATPase AtSKD1 contributes to vacuolar maintenance of Arabidopsis thaliana. Plant J. 64: 71–85. [DOI] [PubMed] [Google Scholar]
- Shin H., Shin H.S., Dewbre G.R., Harrison M.J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39: 629–642. [DOI] [PubMed] [Google Scholar]
- Simin K., Bates E.A., Horner M.A., Letsou A. (1998). Genetic analysis of punt, a type II Dpp receptor that functions throughout the Drosophila melanogaster life cycle. Genetics 148: 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallek T., Beck M., Ben Khaled S., Salomon S., Bourdais G., Schellmann S., Robatzek S. (2013). ESCRT-I mediates FLS2 endosomal sorting and plant immunity. PLoS Genet. 9: e1004035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Spitzer C., Li F., Buono R., Roschzttardtz H., Chung T., Zhang M., Osteryoung K.W., Vierstra R.D., Otegui M.S. (2015). The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 27: 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C., Reyes F.C., Buono R., Sliwinski M.K., Haas T.J., Otegui M.S. (2009). The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C., Schellmann S., Sabovljevic A., Shahriari M., Keshavaiah C., Bechtold N., Herzog M., Müller S., Hanisch F.G., Hülskamp M. (2006). The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133: 4679–4689. [DOI] [PubMed] [Google Scholar]
- Swain T., Hillis W. (1959). Phenolic constituents of Prunus domestica. I. Quantitative analysis of phenolic constituents. J. Sci. Food Agric. 10: 63–68. [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.I., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555. [DOI] [PubMed] [Google Scholar]
- Tilburn J., Sánchez-Ferrero J.C., Reoyo E., Arst H.N. Jr., Peñalva M.A. (2005). Mutational analysis of the pH signal transduction component PalC of Aspergillus nidulans supports distant similarity to BRO1 domain family members. Genetics 171: 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y.C., Lam S.K., Jiang L. (2007). Enigmatic brefeldin a. Plant Signal. Behav. 2: 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y.C., Mo B., Hillmer S., Zhao M., Lo S.W., Robinson D.G., Jiang L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich-Eberius C.I., Novacky A., Fischer E., Lüttge U. (1981). Relationship between energy-dependent phosphate uptake and the electrical membrane potential in Lemna gibba G1. Plant Physiol. 67: 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P., Kaur H., Petla B.P., Rao V., Saxena S.C., Majee M. (2013). PROTEIN L-ISOASPARTYL METHYLTRANSFERASE2 is differentially expressed in chickpea and enhances seed vigor and longevity by reducing abnormal isoaspartyl accumulation predominantly in seed nuclear proteins. Plant Physiol. 161: 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vito P., Pellegrini L., Guiet C., D’Adamio L. (1999). Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J. Biol. Chem. 274: 1533–1540. [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2003). RNA silencing bridging the gaps in wheat extracts. Trends Plant Sci. 8: 307–309. [DOI] [PubMed] [Google Scholar]
- Wee E.G., Sherrier D.J., Prime T.A., Dupree P. (1998). Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell 10: 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer M., Azmi I., West M., Davies B., Katzmann D., Odorizzi G. (2011). Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. J. Cell Biol. 192: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter V., Hauser M.T. (2006). Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 11: 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M.H., Berlin I., Nash P.D. (2011). Regulation of endocytic sorting by ESCRT-DUB-mediated deubiquitination. Cell Biochem. Biophys. 60: 39–46. [DOI] [PubMed] [Google Scholar]
- Zhou C., Yin Y., Dam P., Xu Y. (2010). Identification of novel proteins involved in plant cell-wall synthesis based on protein-protein interaction data. J. Proteome Res. 9: 5025–5037. [DOI] [PubMed] [Google Scholar]
- Zouhar J., Muñoz A., Rojo E. (2010). Functional specialization within the vacuolar sorting receptor family: VSR1, VSR3 and VSR4 sort vacuolar storage cargo in seeds and vegetative tissues. Plant J. 64: 577–588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.