The rice AP2/ERF protein ERF3 interacts with the WUSCHEL-related homeobox protein WOX11 to regulate the cytokinin-responsive gene RR2 in development of crown roots.
Abstract
Crown roots are the main components of the fibrous root system in rice (Oryza sativa). WOX11, a WUSCHEL-related homeobox gene specifically expressed in the emerging crown root meristem, is a key regulator in crown root development. However, the nature of WOX11 function in crown root development has remained elusive. Here, we identified a rice AP2/ERF protein, ERF3, which interacts with WOX11 and was expressed in crown root initials and during crown root growth. Functional analysis revealed that ERF3 was essential for crown root development and acts in auxin- and cytokinin-responsive gene expression. Downregulation of ERF3 in wox11 mutants produced a more severe root phenotype. Also, increased expression of ERF3 could partially complement wox11, indicating that the two genes functioned cooperatively to regulate crown root development. ERF3 and WOX11 shared a common target, the cytokinin-responsive gene RR2. The expression of ERF3 and WOX11 only partially overlapped, underlining a spatio-temporal control of RR2 expression and crown root development. Furthermore, ERF3-regulated RR2 expression was involved in crown root initiation, while the ERF3/WOX11 interaction likely repressed RR2 during crown root elongation. These results define a mechanism regulating gene expression involved in cytokinin signaling during different stages of crown root development in rice.
INTRODUCTION
Cereals, such as rice (Oryza sativa) and maize (Zea mays), have a complex root system structure with several root types, including embryonic primary roots, lateral roots, and shoot-borne roots (also known as adventitious roots). The embryonic primary root develops shortly after germination. Shoot-borne roots initiate from stem nodes or coleoptile sections and are also called crown roots (Marcon et al., 2013). The fundamental difference between cereals and the dicot model plant Arabidopsis thaliana is that cereals have an extensive postembryonic crown-borne root system, which Arabidopsis lacks. To date, organization and cell differentiation processes in root development have been well characterized in Arabidopsis (reviewed in Scheres, 2002). During rice root morphogenesis, several developmental stages can be clearly distinguished, including crown root initiation, emergence, and elongation (Itoh et al., 2005; Coudert et al., 2010; Kitomi et al., 2011b; Wang et al., 2011). Although several key genes have been identified and characterized in the regulation of crown root development (Inukai et al., 2005; Liu et al., 2005; Kitomi et al., 2008; Liu et al., 2009; Zhao et al., 2009), the molecular mechanisms of crown root formation and the functional relationship between these genes are not known.
Available evidence indicates that regulation of crown root formation in rice and lateral root formation in Arabidopsis share several common characteristics. For instance, auxin biosynthesis or signaling-related mutants show morphological abnormalities in both rice crown roots and Arabidopsis lateral roots (De Smet and Jürgens, 2007; Kitomi et al., 2011b; Marcon et al., 2013). Exogenous treatment with auxin induces ectopic formation of lateral and adventitious roots in Arabidopsis (Schiefelbein, 2003; Verstraeten et al., 2013). Arabidopsis lateral root regulatory genes LATERAL ORGAN BOUNDARIES-DOMAIN16/ASYMMETRIC LEAVES2-LIKE18 (LBD16/ASL18) and LBD29/ASL16 are reported to function downstream of AUXIN RESPONSE FACTOR7 (ARF7)- and ARF19-dependent auxin signaling (Okushima et al., 2007; Lee et al., 2009; Goh et al., 2012). In rice, mutants or knockdown transgenic plants of genes involved in auxin signaling pathways and/or polar auxin transport (e.g., CROWN ROOTLESS1 [CRL1]/ADVENTITIOUS ROOTLESS1 [ARL1], CRL4/GNOM1, CAND1, PIN1, CRL5, Aux/IAA3, and MANNOSYL-OLIGOSACCHARIDE GLUCOSIDASE) display reduced or no crown root phenotypes (Inukai et al., 2005; Liu et al., 2005; Xu et al., 2005; Nakamura et al., 2006; Liu et al., 2009; Kitomi et al., 2011a; Wang et al., 2011; Zhu et al., 2012; Wang et al., 2014). In addition, a gain-of-function mutation of IAA11 affects lateral root development in rice (Nakamura et al., 2006; Zhu et al., 2012).
Cytokinin, which acts antagonistically to auxin, plays an important role in controlling root growth during postembryonic development. Cytokinin stimulates cell differentiation in the root proximal meristem, which leads to a decrease of meristem size and root growth (Dello Ioio et al., 2007, 2008; Bishopp et al., 2009; Ruzicka et al., 2009; Moubayidin et al., 2010). Mutations of Arabidopsis type-A cytokinin response factor genes ARR7 and ARR15 lead to misexpression of root development regulatory genes, such as SCARECROW, PLETHORA1 (PLT1), and WUSCHEL-RELATED HOMEOBOX5 (WOX5) (Lee et al., 2013; Müller and Sheen, 2008). Knockdown or overexpression of rice genes involved in cytokinin signaling pathway also affects root development. For instance, RR3 and RR5 overexpression plants produce more and longer lateral roots compared with the wild type when treated with exogenous cytokinin (Cheng et al., 2010). Overexpression of RR6 leads to dwarf phenotype with a poorly developed root system (Hirose et al., 2007).
Auxin-cytokinin crosstalk plays an important role in the regulation of root meristem activities (Müller and Sheen, 2008; Benková and Hejátko, 2009; Su et al., 2011; Durbak et al., 2012). Recent progress has revealed that the balance between cell differentiation and division, which is necessary for controlling root meristem size and root growth, is regulated by antagonistic action of cytokinin and auxin in Arabidopsis and rice (Dello Ioio et al., 2007; Kitomi et al., 2011b; Gao et al., 2014). In the root meristem, a primary cytokinin-response transcription factor, ARR1, activates SHORT HYPOCOTYL2/IAA3, a repressor of auxin signaling that negatively regulates the PIN auxin transport facilitator genes (Dello Ioio et al., 2008). Recent work reported that CYTOKININ OXIDASE4 mediates crown root development by integrating the interaction between cytokinin and auxin (Gao et al., 2014). We previously showed that rice WOX11, which belongs to the WOX transcription factor family, is involved in the activation of crown root development by regulating genes of both auxin and cytokinin signaling (Zhao et al., 2009). In this work, we identified the rice AP2/ETHYLENE-RESPONSIVE FACTOR (ERF) protein, ERF3, as a WOX11-interacting partner involved in rice crown root development. Our results showed that ERF3 was involved in crown root initiation and elongation. Further analysis indicated that ERF3 stimulated the expression of type-A RR gene RR2 in crown root initials. Overexpression of RR2 also promoted crown root initiation. Our data suggested that ERF3 regulation of crown root initiation may involve cytokinin signaling and that its interaction with WOX11 might enhance WOX11-mediated repression of RR2 or inhibit its function on RR2 activation during crown root elongation.
RESULTS
Identification of ERF3 as a WOX11-Interacting Protein
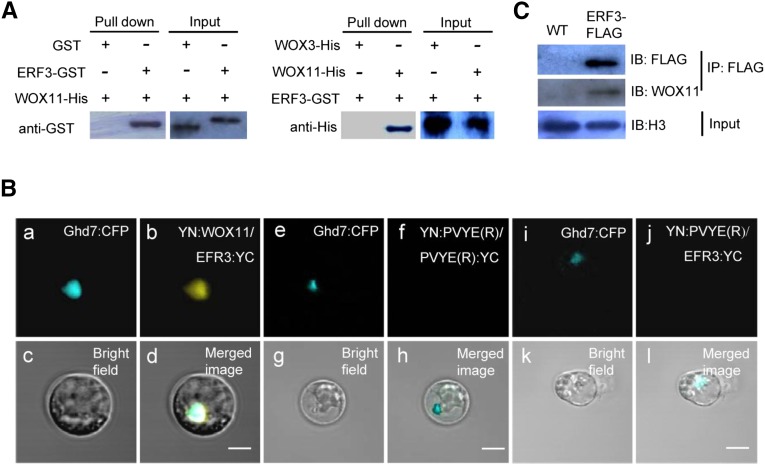
To elucidate the molecular basis by which WOX11 controls crown root development, yeast two-hybrid screening of WOX11-interacting proteins were performed using a rice root tip cDNA library. With the full-length WOX11 cDNA as bait, a total of 36 positive clones were isolated from the screening, which corresponded to 19 genes. One of the genes encoded ERF3 (Os01g58420), which accounted for nearly one-seventh (5 of 36) of all the positive clones. The clones isolated from the screening contained the whole coding region of ERF3. To validate the yeast two-hybrid data, both in vitro and in vivo experiments were performed to test the ERF3-WOX11 interaction. Recombinant full-length ERF3 and glutathione S-transferase (GST) fusion protein and the full-length WOX11 tagged with 6×His were produced in Escherichia coli and purified. His pull-down assays showed that ERF3-GST but not GST was retained by WOX11 (Figure 1A, left). Conversely, pull-down assays with GST revealed that WOX11-6×His but not WOX3-6×His was retained by ERF3 protein (Figure 1A, right) (Dai et al., 2007). To further confirm the interaction, bimolecular fluorescence complementation (BiFC) assays were performed in rice protoplasts. YFP was reconstituted when the coding sequences of ERF3 and WOX11 were coexpressed (Figure 1B, a to d). In contrast, coexpression of the ERF3-YFP C terminus and YFP-N terminus (Figure 1B, e to h), or the YFP protein C terminus and N terminus alone, did not show fluorescence (Figure 1B, i to l), which confirmed that the ERF3-WOX11 interaction is specific. Moreover, the BiFC experiments clearly revealed that ERF3-WOX11 interacted in the nucleus (Figure 1B, a to d), as YFP fluorescence fully overlapped with the nuclear protein Ghd7 (Xue et al., 2008) fused to CFP.
Figure 1.
ERF3 Directly Interacts with WOX11 in Vitro and in Vivo
(A) Pull-down assay of ERF3 interaction with WOX11. Left, ERF3-GST fusion protein or GST alone were incubated with WOX11-His in His beads. ERF3-GST but not GST was pulled down by the beads containing WOX11-His. Right, WOX11-His and WOX3-His were incubated with EFR3-GST in GST beads. WOX11-His but not WOX3-His was pulled down by the beads containing ERF3-GST.
(B) Interaction of ERF3 and WOX11 in rice protoplasts. Representative cells are shown, as imaged by confocal laser scanning microscopy. a, e, and i, Ghd7:CFP localization in the nucleus (blue fluorescence); b, detection in rice protoplasts of YN:WOX11 and ERF3:YC interaction, shown as yellow signal; f and j, as negative controls with YN:pVYNE(R)/pVYCE(R):YC and YN: pVYNE(R)/ERF3:YC; c, g, and k, bright field; d, h, and l, colocalization of three signals is indicated in merged images. Bars = 10 µm.
(C) In vivo coimmunoprecipitation assay of ERF3 and WOX11 interaction. Roots of 10-d-old seedling of ERF3-FLAG transgenic plants and the wild type (WT) were immunoprecipitated (IP) using an anti-FLAG polyclonal antibody and immunoblotted (IB) using anti-FLAG or anti-WOX11 antibodies as indicated.
Coimmunoprecipitation assays were performed to confirm the interaction between ERF3 and WOX11 in rice cells. Stable transgenic rice plants expressing ERF3-FLAG (under the maize ubiquitin promoter) were generated (Supplemental Figure 1). Crude protein extracts of roots from ERF3-FLAG and wild-type plants were immunoprecipitated by anti-FLAG antibody and then analyzed by immunoblotting with anti-FLAG and anti-WOX11 antibodies. As shown in Figure 1C, both ERF3-FLAG and WOX11 proteins were detected, further confirming the in vivo interaction between the two proteins.
ERF3 Displays a Partially Overlapping Expression Pattern with WOX11 in Developing Crown Roots and Is Responsive to Auxin and Cytokinin
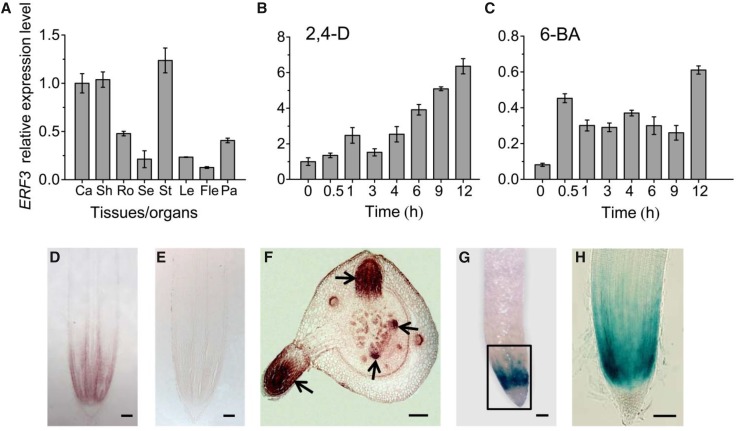
To study EFR3 expression, we first examined ERF3 mRNA accumulation in various organs/tissues at different developmental stages by RT-qPCR. The analysis showed that ERF3 was broadly expressed (Figure 2A). To gain insight into the functional significance of the interaction between ERF3 and WOX11, we further explored the expression pattern of ERF3 during rice crown root development by in situ hybridization and promoter-reporter lines. The results indicated that ERF3 was preferentially expressed in crown root initials, the crown root meristem (cell division) region, and in elongating crown roots (Figures 2D and 2F). This expression pattern overlapped with that of WOX11 but displayed some differences during crown root development, as WOX11 is expressed mainly after crown roots emergence and is hardly detectable in crown root initials (Zhao et al., 2009). In the promoter-reporter lines, GUS activity was mainly detected in the cell division zones of the primary root meristem (Figures 2G and 2H). This expression profile suggested that ERF3 and WOX11 might be involved in different stages of crown root development. The colocalization of WOX11 and ERF3 suggested that the ERF3-WOX11 pairs may function in rice crown root formation.
Figure 2.
ERF3 Expression in Rice Crown Root Development.
(A) Detection of ERF3 expression by RT-qPCR in callus (Ca), shoot (Sh), root (Ro), seedling (Se), stem (St), leaf (Le), flag leaf (Fle), and panicle (Pa). ACTIN1 was used as an internal control. Bars are means ± sd from three technical replicates.
(B) and (C) Kinetics of induction of ERF3 in response to plant hormones 2,4-D and 6-BA. The transcript level of ERF3 in roots of 10-d-old light-grown wild-type seedlings treated with 2,4-D or 6-BA for the indicated times were plotted as the relative expression (fold) of water-treated seedling during the same durations. The PCR signals were normalized with those of the ACTIN1 transcripts. Bars are means ± sd from three technical replicates.
(D) to (F) In situ hybridization detection of ERF3 transcripts in crown root tip (D) and primordia (F) with an antisense or a sense probe (E). Arrows indicate crown root initials at different stages. Bars = 25 μm.
(G) and (H) GUS activity in crown root tip. (H) is the boxed region in (G). Bars = 75 μm
As WOX11 expression is regulated by auxin and cytokinin, we wanted to know whether ERF3 was also regulated by these two hormones. Wild-type seedlings (10 d after germination) were transferred to liquid media containing 10−6 M 2,4-D or 10−5 M 6-benzylaminopurine (6-BA). The roots were harvested for RNA extraction at 0, 0.5, 1, 3, 4, 6, 9, and 12 h after hormone treatment. It is noteworthy that ERF3 transcript was induced by both 2,4-D and 6-BA after 1 or 0.5 h treatment, respectively (Figures 2B and 2C), suggesting that ERF3 responds to auxin and cytokinin signaling pathways.
Knockdown and Overexpression of ERF3 Affect Crown Root Formation in Rice
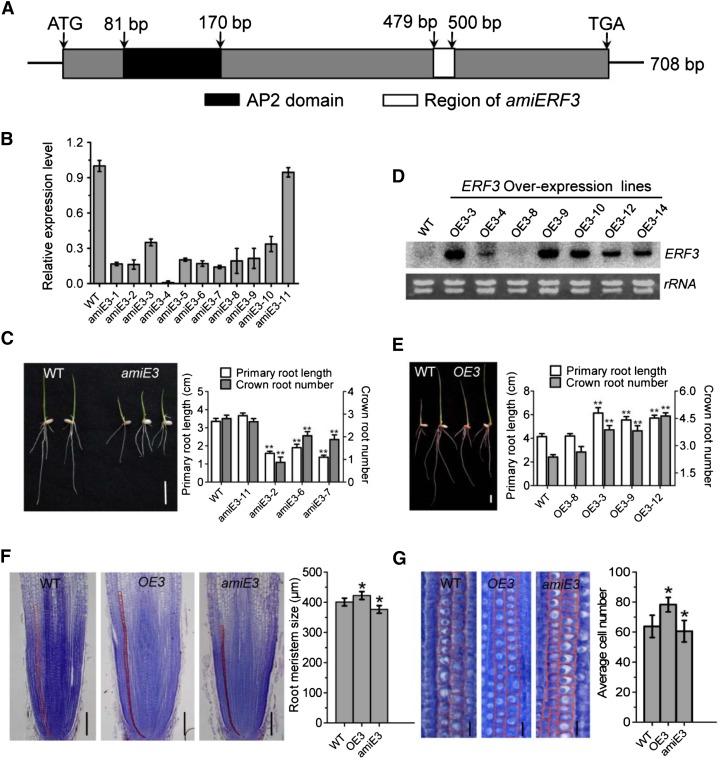
To study the function of ERF3 in rice root development, we generated transgenic plants expressing ERF3 artificial microRNAs (amiE3) (Figure 3A). Expression analysis of the transgenic population led to the identification of 10 lines (amiE3-1- amiE3-10) with reduced ERF3 mRNA levels (Figure 3B). Three of the artificial microRNA (amiRNA) lines (amiE3-2, amiE3-6, and amiE3-7) were selected for subsequent analysis. Line amiE3-11, which did not show any reduction of ERF3 transcripts, was used as a negative control in subsequent analysis (Figure 3B). Seven days after germination, the amiERF3 seedlings developed fewer crown roots than the wild type (Figure 3C). In addition, the primary root length and the plant height were also reduced (Figure 3C). Fourteen days after germination, the primary root length of the amiERF3 plants remained significantly shorter and the crown root number was reduced compared with the wild type (Table 1). To check if ERF3 regulated the initiation of crown root primordia, cross sections of the coleoptilar nodal region of 3- and 5-d-old seedlings were stained with toluidine blue. Crown roots were not initiated or retarded in amiERF3 transgenic lines compared with the wild type (Supplemental Figure 2B), indicating that ERF3 was essential for primary root elongation and crown root initiation.
Figure 3.
Analysis of ERF3 amiRNAs and Overexpression Transgenic Plants.
(A) Schematic representation of the ERF3 cDNA. The black box corresponds to the conserved domain (AP2 domain). The cDNA region used to construct the artificial microRNAs vector is indicated by the white box.
(B) RT-qPCR analysis of ERF3 transcripts in the wild type and 11 amiRNA (amiE3) transgenic lines. The PCR signals were normalized with ACTIN1 transcripts. Transcript level from the wild type was set at 1. Bars are means ± sd from three technical replicates.
(C) Comparison of primary root length and crown root number of 1-week-old seedlings between the wild type (WT; left) and the amiRNA line (amiE3; right). Picture and statistical data were taken from lines amiE3-2, amiE3-6, and amiE3-7, which showed similar phenotypes. amiE3-11 was used as a negative control. Bar = 1 cm.
(D) RNA gel blot analysis of ERF3 overexpression plants (OE3) in different transgenic lines compared with the wild type. The rRNA levels were revealed as controls. Statistical analyses of the data in (C) and (D) are presented in Table 1.
(E) Comparison of primary root length and crown root number of 1-week-old seedlings between wild-type (left) and overexpression plants (OE3; right). Picture and statistical data were taken from lines OE3-3, OE3-9, and OE3-12, which showed similar phenotypes. OE3-8 was used as a negative control. Bar = 1 cm.
(F) and (G) Histological analysis of crown root tip in the wild type, ERF3 amiRNAs (amiE3), and overexpression plants (OE3). Red lines delimit the meristem size (i.e., the distance between the quiescent center and the transition zone) (G). Cell number in meristem zone of wild-type, ERF3 overexpression (OE3), and amiRNA (amiE3) roots. Statistical analyses (t test) of data in (F) and (G) were performed with the wild type (n = 16), ERF3 overexpression (OE3; n = 20), and amiRNAs (amiE3; n = 17). Error bars in (F) and (G) represent sd. *P < 0.05.
Table 1. Comparison of Primary Root Length and Crown Root Number between the Wild Type, ERF3 Overexpressing (OE3), and RNAi Transgenic Lines (amiE3) 2 Weeks after Germination.
| Genotype (Plant Number) | Primary Root Length (cm) | Crown-Borne Root Number |
|---|---|---|
| Wild type (20) | 9.43 ± 0.38 | 4.96 ± 0.24 |
| amiE3-11 (16) | 9.02 ± 0.28 | 5.02 ± 0.23 |
| amiE3-2 (24) | 5.04 ± 0.25** | 2.71 ± 0.46** |
| amiE3-6 (17) | 5.78 ± 0.49** | 4.06 ± 0.39* |
| amiE3-7(21) | 5.65 ± 0.36** | 4.00 ± 0.27* |
| OE3-8 (14) | 9.61 ± 0.37 | 5.02 ± 0.15 |
| OE3-3 (17) | 13.97 ± 0.35** | 6.21 ± 0.11* |
| OE3-9 (13) | 14.55 ± 0.33** | 5.97 ± 0.29* |
| OE3-12 (29) | 13.82 ± 0.33** | 6.25 ± 0.21* |
Significant differences are indicated at the 5% (*) and 1% (**) probability levels (two-tailed t test). amiE3-11 and OE3-8 are negative controls.
To further confirm ERF3 function in root development, a vector containing the ERF3 cDNA under the control of the maize ubiquitin promoter was transformed into rice plants. RNA gel blot analysis revealed that several transgenic lines showed ERF3 overexpression (Figure 3D). Three overexpression lines (OE3-3, OE3-9, and OE3-12) and one negative transgenic line (OE3-8) were selected for further analysis. The overexpression plants produced a larger root system (with more and longer roots) 7 d after germination compared with the wild type and the transgenic negative control line (Figure 3E). Fourteen days after germination, the overexpression plants developed more crown roots and longer primary roots compared with the wild type and the control line (Table 1). These data suggested that elevated ERF3 expression level promoted crown root initiation and elongation in rice.
To study whether ERF3 regulated root meristem size, longitudinal sections of root tip of wild-type, amiERF3 (amiE3), and ERF3 overexpression (OE3) transgenic lines were stained with toluidine blue (Figure 3F). The root meristem of OE3 plants was longer than that of wild-type and amiERF3 plants (wild type, 400.71 ± 12.55; OE3, 422.29 ± 13.07*; amiE3, 376.38 ± 12.5*; data were obtained from three lines for each transgenic genotype with n > 15 for each line). Because root meristem size is correlated with the cell number in the meristem region and cell longitudinal length, examination of longitudinal section revealed that the root meristem of OE3 plants contained more cells (OE3, 78.77 ± 8.78**; wild type, 63.77 ± 12.4) and the cell longitudinal size was longer than that of the wild type (OE3, 11.25 ± 0.78*; wild type, 9.02 ± 0.40). While in amiERF3 plants root meristem, there were fewer cells (60.5 ± 9.16**) and shorter cell longitudinal length (8.03 ± 0.17*) compared with the wild type (Figure 3G; Supplemental Figure 2). These data suggested that ERF3 was likely to be involved in promoting both cell division and cell longitudinal elongation of the root meristem in rice.
ERF3 Functions in Regulating Expression of Crown Root Developmental and Hormone-Responsive Genes
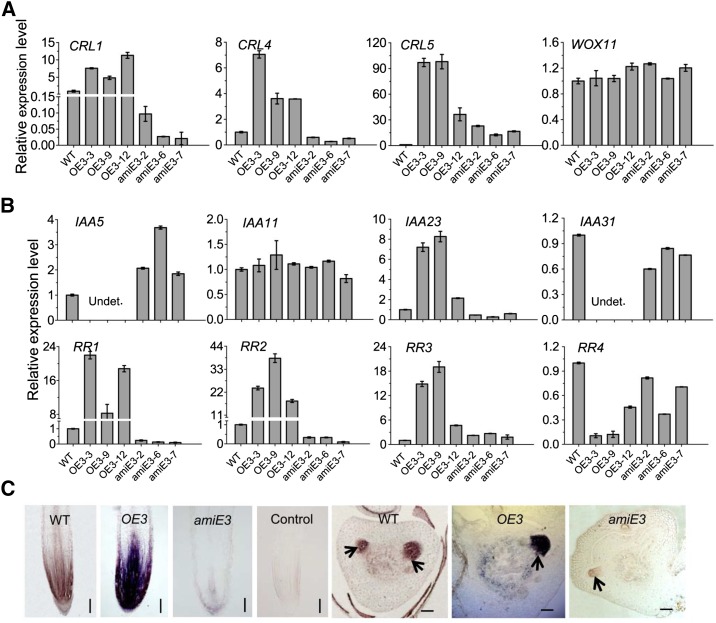
The CRL1, CRL4/GNOM1, CRL5, and WOX11 genes are involved in different regulatory pathways of crown root initiation and growth (Inukai et al., 2005; Liu et al., 2005; Kitomi et al., 2008, 2011a; Liu et al., 2009; Zhao et al., 2009). To determine whether ERF3 regulates these genes, we analyzed their mRNA levels in the roots of ERF3 transgenic plants. RT-qPCR results showed that CRL1, CRL4/GNOM1, and CRL5 were highly induced in ERF3 overexpression lines. In the amiERF3 lines, CRL1 and CRL4/GNOM1 were repressed but CRL5 was upregulated, suggesting that EFR3 might be directly or indirectly involved in regulation of these genes. However, WOX11 expression was not clearly affected in the transgenic plants (Figure 4A), suggesting that ERF3 was not involved in the regulation of WOX11.
Figure 4.
Expression of Root Development and Auxin/Cytokinin Response Genes in Crown Roots of ERF3 Transgenic Plants.
(A) Relative expression level of root developmental genes determined by RT-qPCR.
(B) Relative expression level of four rice Aux/IAA genes and four type-A RR genes determined by RT-qPCR.
The PCR signals in (A) and (B) were normalized with ACTIN1 transcripts. Transcript level from the wild type was set at 1. ACTIN1 was used as an internal control. Data represent the means ± sd of three independent biological replicates.
(C) In situ hybridization detection of RR2 transcripts in the crown root tip and crown root primordia in the wild type (WT), ERF3 overexpression (OE3), and ERF3 amiRNAs (amiE3) with an antisense or sense probe (control). Arrows indicate crown root initials. Bars= 75 μm.
To determine whether ERF3 was also involved in auxin and cytokinin signaling, we analyzed the expressions of four auxin-responsive Aux/IAA genes (IAA5, IAA11, IAA23, and IAA31) and four cytokinin-responsive type-A RR genes (RR1-RR4) in 7-d-old seedling root tips (∼8 to 10 mm long) of wild-type, three amiERF3 (amiE3-2, -6, and -7), and three ERF3 overexpression lines (OE3-3, -9, and -12) lines. These genes have been shown to be highly expressed in rice roots and to be regulated by WOX11 (Jain et al., 2006; Du et al., 2007; Zhao et al., 2009). We detected the IAA5 and IAA31 transcripts in the wild type and amiEFR3, but not in the overexpression plants. By contrast, IAA23 showed higher expression in the ERF3 overexpression and a lower expression in the amiEFR3 lines compared with the wild type. IAA11 displayed no difference in expression level between the transgenic and wild-type plants (Figure 4B). Therefore, up- or downregulation of ERF3 caused complex changes in the expression pattern of auxin-responsive genes.
Three of the four tested type-A RR genes (RR1, RR2, and RR3) were highly upregulated (10- to 40-fold) in the overexpression plants, while RR1 and RR2 were repressed in the amiERF3 lines. However, RR4 was repressed in both the overexpression and amiRNA lines, with more severe repression observed in the overexpression lines (Figure 4B). Furthermore, in situ hybridization experiments showed that RR2 had a higher expression in root meristem and crown root initials of ERF3 overexpression plants and a lower expression in amiERF3 lines compared with the wild type (Figure 4C). The results suggested that RR2 might be among immediate targets of ERF3 and RR genes might be subject to complex feedback control of cytokinin and auxin signaling possibly disturbed by up- or downregulation of ERF3.
ERF3 Directly Binds to RR2 and Positively Regulates RR2 Expression
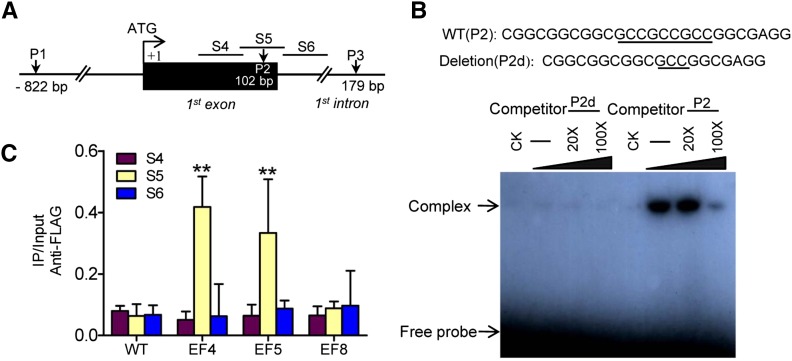
Our previous studies suggested that WOX11 could bind to the RR2 promoter region (1092 bp upstream of the ATG) in vitro and in vivo and directly suppresses RR2 expression in elongating crown roots (Zhao et al., 2009). The expression data shown in Figure 4 suggested that RR2 might be also targeted by ERF3. ERF proteins bind specifically to the GCC motif (GCCGCC), the core sequence of the ethylene-responsive element (ERE) (Hao et al., 1998). To investigate whether ERF3 directly binds to RR2, we analyzed RR2 genomic sequence and identified three GCC motifs in the RR2 locus: the first one (P1, 822 bp upstream to ATG) had two GCC repeats (GCCGCC) in the promoter region; the second (P2; 102 bp downstream to ATG) had three GCC repeats (GCCGCCGCC) in the first exon; and the third (P3; 179 bp downstream to ATG) had the two GCC repeats (GCCGCC) located in the first intron of RR2 (Figure 5A). To examine whether ERF3 protein directly bound to these element sequences, we performed electrophoresis mobility shift assays (EMSAs) using DNA fragments corresponding to P1, P2, and P3 regions and ERF3 protein produced in E. coli. As shown in Figure 5B and Supplemental Figure 3, the recombinant ERF3 only bound to P2, but not to P1 or P3. In addition, increasing molar excesses of unlabeled P2 fragment (competitor) inhibited the binding (Figure 5B). Furthermore, deletion of one GCC repeat from P2 also abolished the binding (Figure 5B). These results indicated that ERF3 bound specifically to the P2 site in the first exon of the gene in vitro.
Figure 5.
ERF3 Directly Binds to RR2 in Vitro and in Vivo.
(A) Schematic representation of three putative loci of ERF binding sites in the RR2 genomic sequence. P1 (−822 bp) including GCCGCC motif, P2 (within the first exon) including GCCGCCGCC motif, and P3 (within the first intron) including GCCGCC motif.
(B) Gel-shift assay of ERF3 protein binding to the first exon sequence (P2) of RR2 containing the ERF binding site (underlined) or a deletion version (P2d). E. coli-produced ERF3 protein was incubated with 32P-labeled P2 or P2d in the absence or presence of 20 or 100 M excess of the corresponding cold probes and analyzed by electrophoresis. The shifted band is indicated by the arrow. Three biological replicates were conducted.
(C) ChIP analysis of transgenic plants expressing ERF3-FLAG fusion protein. Nuclei from three ERF3-FLAG transgenic lines (EF4, EF5 and EF8, EF8 as a negative control) and the wild type (WT) were immunoprecipitated by anti-FLAG or without antibody. The precipitated chromatin fragments were analyzed by qPCR using three primer sets amplifying three RR2 regions (S4, S5, and S6), as indicated in (A). The relative nucleotide positions of the putative ERF binding site are indicated (with the initiation ATG codon being assessed as +1). One-tenth of the input chromatin was analyzed as control. Error bars represent means ± sd from three independent experiments.
To further confirm whether ERF3 bound to RR2 in vivo, chromatin fractions from wild-type and two ERF3-FLAG transgenic plants EF4 and EF5 together with a negative control EF8 (Supplemental Figure 1) were isolated and used for chromatin immunoprecipitation (ChIP) with anti-FLAG antibody. The precipitated products in the presence of the antibody as well as input (no antibody) were analyzed by qPCR using three primer sets (S4 to S6) corresponding to different regions around P2 (Figures 5A and 5C). As expected, only the S5 region covering the P2 sequence was clearly precipitated from the two ERF3-FLAG lines. The result indicated that ERF3 directly bound to RR2 in vivo.
ERF3 Promotes WOX11 Binding to RR2 in Vitro and in Vivo
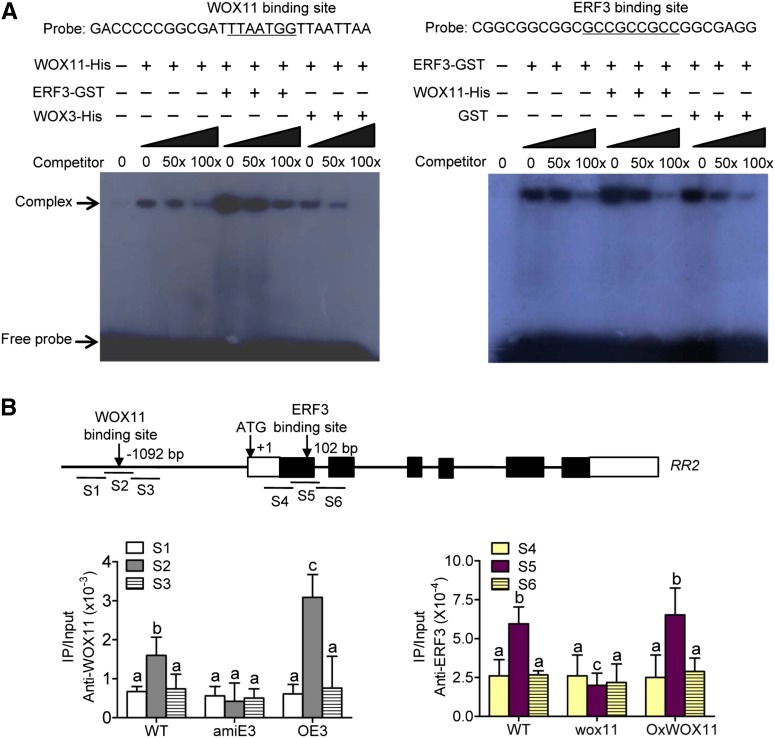
To check whether the ERF3/WOX11 interaction affected their binding to RR2, we performed EMSA using DNA fragments containing either the ERF3 (P2) or the WOX11 binding site in the RR2 genomic region as probes. The assays revealed that the binding of WOX11 to RR2 was enhanced by ERF3-GST, but not GST tag alone (Figure 6A, left). In contrast, the binding of ERF3-GST to RR2 was not apparently enhanced by inclusion of WOX11 (Figure 6A, right). These observations suggested that the physical interaction between ERF3 and WOX11 might promote WOX11 binding to the RR2 gene in vitro.
Figure 6.
ERF3 Promotes WOX11 Binding to RR2.
(A) Gel-shift assays of WOX11 binding to RR2 promoter containing the WOX11 binding site in the presence or absence of ERF3 (left) and ERF3 binding to RR2 first exon (P2) containing the ERF binding site in the presence or absence of WOX11 (right). E. coli-produced ERF3 and WOX11 proteins were incubated with 32P-labeled probes in the absence or presence of 50 or 100 M excess of the corresponding cold probes and analyzed by electrophoresis. The shifted band is indicated by the arrow. Three biological replicates were conducted.
(B) ChIP analysis of ERF3 and WOX11 binding to RR2 in different transgenic plants. Nuclei from OE3, amiE3, wox11, OxWOX11, and the wild type (WT) were immunoprecipitated by WOX11 (left) or ERF3 (right) antibodies. The precipitated chromatin fragments were analyzed by qPCR using six primer sets amplifying six RR2 regions (S1, S2, S3, S4, S5, and S6) as indicated. The relative nucleotide positions of the putative WOX11/ERF3 binding sites are indicated with arrows. One-tenth of the input (without antibody precipitation) chromatin was analyzed and used as control. Three biological replications were performed. Each value is the average ± sd from three independent experiments. Significant differences between samples (t test) are indicated by different letters.
To confirm the effect of the ERF3/WOX11 interaction on WOX11 binding to RR2 in vivo, we isolated chromatin fragments from roots of wild-type, amiERF3, and ERF3 overexpression plants and performed immunoprecipitation with the anti-WOX11 antibody. The precipitated products from the three genotypes as well as inputs were analyzed by qPCR using three primer sets (S1 to S3) corresponding to the WOX11 binding region in RR2. The results revealed that, compared with the S1 and S3 regions, the S2 region covering the WOX11 binding sequence was enriched in the wild type. The enrichment was greatly enhanced in ERF3 overexpression plants compared with wild-type plants (Figure 6B). These results suggested that ERF3 promoted WOX11 binding to the promoter region of RR2 in roots. Additionally, chromatin products from wild-type, wox11, and OxWOX11 (WOX11 overexpression) lines could also be precipitated by anti-ERF3. Further analysis revealed a clear enrichment of ERF3 binding to the S5 region in the first exon of RR2 compared with S4 and S6 regions in wild-type plants. However, the binding was reduced in wox11 mutant, but not clearly enriched in OxWOX11 plants (Figure 6B). The reduction may be due to downregulation of ERF3 in wox11 mutants (Supplemental Figure 4A). These data suggested that ERF3 promoted WOX11 binding to RR2 in rice roots.
Knockdown of ERF3 in wox11 Mutant Background Had a More Severe Effect on Crown Root Growth
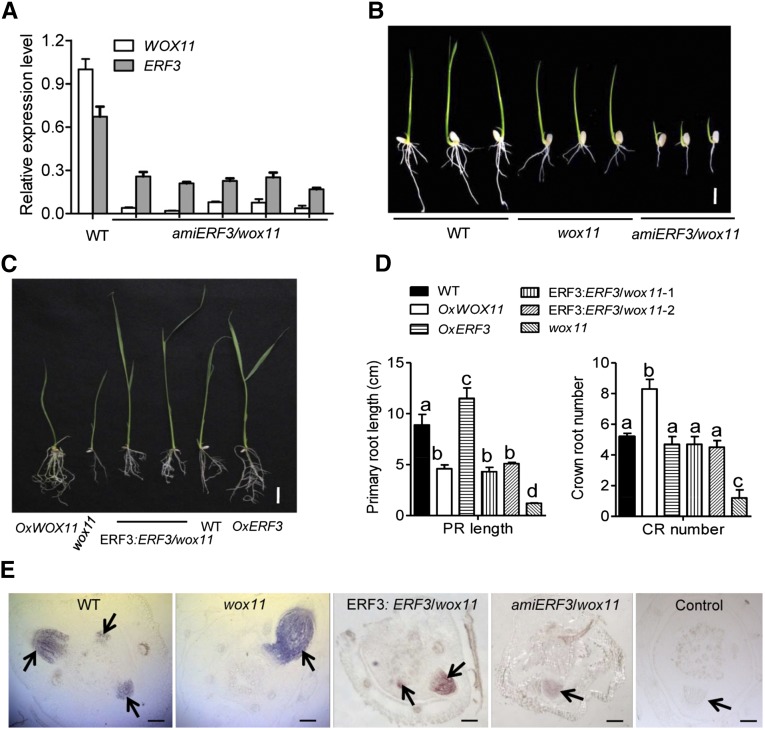
The phenotype of amiERF3, which showed fewer crown roots and shorter primary roots (Figure 3C), was similar to that of wox11 mutant (Zhao et al., 2009). Downregulation of ERF3 had no effect on the expression of WOX11 (Figure 4A). However, the level of ERF3 transcripts was decreased in wox11 (Supplemental Figure 4A), suggesting WOX11 might regulate ERF3. To further dissect the genetic relationship of ERF3 and WOX11 in controlling crown root development, we then generated ERF3 knockdown transgenic lines in the wox11 mutant background (amiERF3/wox11), in which ERF3 transcript was greatly reduced (Figure 7A). The amiERF3/wox11 lines displayed a more severe phenotype compared with the wox11 mutant. Some of the transgenic lines did not produce any primary or crown root during rooting stage (Supplemental Figure 4B), while others had much fewer and shorter crown roots in T1 lines compared with wox11 (Figure 7B, Table 2). Meanwhile, the transcript levels of RR2 clearly decreased in amiERF3/wox11 plants (Figure 7E), indicating an important role for the ERF3 and WOX11 interaction in regulating RR2 transcription. The more severe phenotype of amiERF3/wox11 suggested that WOX11 alone is not sufficient to fully maintain crown root formation in the absence of ERF3 and the two genes could act together during rice crown root development.
Figure 7.
Genetic Relationship between WOX11 and ERF3 in Controlling Rice Crown Root Development and RR2 Expression.
(A) Transcript levels of ERF3 and WOX11 in roots of wild-type (WT) and amiERF3/wox11 plants. The PCR signals were normalized with ACTIN1 transcripts. Transcript level from the wild type was set at 1. Bars are means ± sd from three technical replicates.
(B) to (D) Crown root phenotypes of indicated genotypes. OxWOX11, WOX11 overexpression lines; OxERF3, ERF3 overexpression lines; ERF3:ERF3/wox11, ERF3 cDNA under the ERF3 promoter in the wox11 background. Significant differences between samples (t test) are indicated by different letters. Bar = 1 cm in (B) and 5 cm in (C).
(E) RR2 transcripts detected by in situ hybridization in the wild type, wox11, ERF3:ERF3/wox11, and amiERF3/wox11. Arrows indicate crown root initials. Bar = 75 μm.
Table 2. Comparison of Primary Root Length and Crown Root Number between the Wild Type, wox11 Mutants, and amiERF3/wox11 Transgenic Plants 10 d after Germination.
| Genotype (Plant Number) | Primary Root Length (cm) | Crown-Borne Root Number |
|---|---|---|
| Wild type (16) | 2.94 ± 1.68 | 3.38 ± 0.58 |
| wox11 (14) | 1.83 ± 1.22* | 2.16 ± 0.27** |
| amiERF3/wox11 (34) | 1.87 ± 1.29* | 1.44 ± 0.85** |
Significant difference are indicated at the 5% (*) and 1% (**) probability levels (two-tailed t test).
Overexpression of ERF3 Partially Complemented the wox11 Root Phenotype
To further examine the above-mentioned interaction hypothesis, we introduced ERF3 under ERF3 promoter into wox11 background (ERF3:ERF3/wox11). ERF3:ERF3/wox11 plants produced more crown roots than wox11 mutants (Figures 7C and 7D), suggesting that increased EFR3 expression could partially complement the wox11 root phenotype. Previous results have shown that wox11 mutation induced RR2 expression in elongating crown roots (Zhao et al., 2009). To study whether EFR3 downregulation and overexpression affected RR2 expression in wox11 background, in situ hybridization and RT-qPCR analysis was performed. The analysis revealed that EFR3 downregulation reduced RR2 accumulation in elongating crown roots, whereas overexpression of ERF3 induced RR2 transcript in crown root initials in wox11 background (Figure 7E; Supplemental Figure 6A). These data confirmed the function of ERF3 in regulation of RR2 expression and suggested that ERF3 was a functional partner of WOX11. The two proteins might play different roles in temporal and spatial expression of RR2 during crown root initiation and emergence.
RR2 Modulated Root Development by Altered Cytokinin Signaling
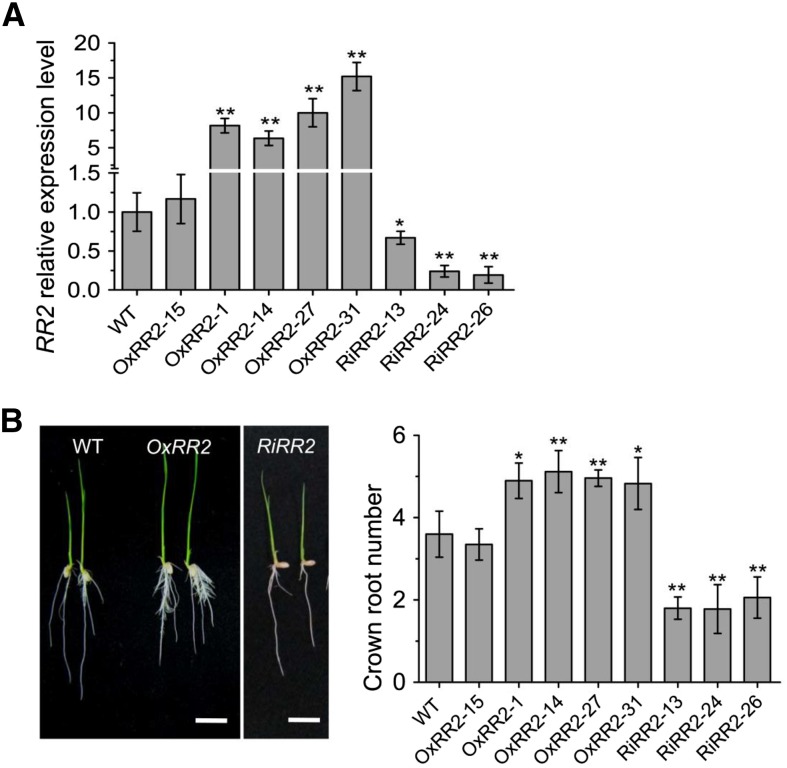
To study whether the crown root phenotypes in ERF3 and WOX11 transgenic and mutant plants were related to RR2 expression, we produced RR2 overexpression (OxRR2) and RNAi (RiRR2) transgenic plants (Figure 8A). Five overexpression lines (OxRR2-1, -14, -27, -31, and -15, among which line 15 is a negative control) and three RNAi lines (RiRR2-13, -24, and -26) were selected for detailed phenotypic analysis. The number of crown roots significantly increased in the OxRR2 lines and reduced in the RiRR2 lines compared with the wild type (Figure 8B), indicating that the RR2 expression level was positively correlated with rice crown root initiation. However, both OxRR2 and RiRR2 lines showed reduced root lengths compared with the wild type (Table 3), suggesting that the RR2 expression had a complex effect on root elongation. These results supported the hypothesis that RR2 expression might be involved in ERF3/WOX11-regulated crown root development.
Figure 8.
Root Phenotypes of RR2 Overexpression and RNAi Transgenic Plants.
(A) Relative RR2 transcript levels in the wild type (set as 1), five RR2 overexpression (OxRR2), and three RR2 RNAi lines (RiRR2). The PCR signals were normalized with ACTIN1 transcripts. Transcript level from the wild type was set at 1. Error bars represent sd. *P < 0.05 and **P < 0.01.
(B) Root phenotype of RR2 overexpression (OxRR2), three RNAi (RiRR2) transgenic plants, and the wild type (WT). Statistical analyses of the data are presented in Table 3. Bars = 1 cm.
Table 3. Comparison of Primary Root Length between the Wild Type, RR2 Overexpressing (OxRR2), and RNAi Transgenic Lines (RiRR2) 10 d after Germination.
| Genotype (Plant Number) | Primary Root Length (cm) |
|---|---|
| Wild type (20) | 6.01 ± 0.55 |
| OxRR2-15 (16) | 6.48 ± 0.71 |
| OxRR2-1 (16) | 4.66 ± 0.55** |
| OxRR2-14 (15) | 4.59 ± 0.78** |
| OxRR2-27 (15) | 4.36 ± 0.64** |
| OxRR2-31 (17) | 4.82 ± 0.81** |
| RiRR2-13 (15) | 3.48 ± 0.59** |
| RiRR2-24 (14) | 3.94 ± 0.63** |
| RiRR2-26 (15) | 3.41 ± 0.67** |
Significant differences are indicated at the 5% (*) and 1% (**) probability levels (two-tailed t test). OxRR2-15 is a negative control.
DISCUSSION
ERF3 Participates in Both the Initiation and Elongation Processes during Crown Root Development
Identification and characterization of genes affecting crown root initiation, emergence, and elongation process in rice have contributed to our increasing understanding of the genetic mechanisms underlying crown root development (Inukai et al., 2005; Liu et al., 2005; Kitomi et al., 2008, 2011b; Zhao et al., 2009; Wang et al., 2011). However, it is not clear what these crown root regulators interact with and how the interacting factors regulate crown root formation. In this study, we identified ERF3 as a WOX11-interacting partner in controlling crown root development. ERF3 belongs to the AP2/ERF transcription factor family (Nakano et al., 2006). Other rice AP2/ERF genes, such as CRL5, which contains two AP2 repeats, has been shown to regulate crown root initiation as well as other aspects of plant development in rice (Kitomi et al., 2011a). PLT1 to PLT6 are all expressed in the primordium of crown root (Li and Xue, 2011). Our data indicate that ERF3, which contains one AP2 repeat (Figure 3A), is involved in the control of crown root development likely acting at the initiation, emergence, and elongation steps (Figures 2F, 3C, and 3E, Table 1; Supplemental Figure 2B). Alteration of root meristem sizes in ERF3 overexpression and knockdown plants suggested that ERF3 may regulate cell division in root meristem (Figures 3F and 3G). It has been reported that overexpression of ERF3 (AP37) in rice under the control of the constitutive promoter Cc1 significantly increased grain yield by 16 to 57% over controls under severe drought conditions, yet exhibited no significant difference under normal growth conditions (Oh et al., 2009), which might be at least partially due to the well-developed root system in the plants overexpressing ERF3.
ERF3 Promoted Crown Root Development by Mediating Auxin-Cytokinin Signaling Gene Expression
Auxin signaling is required for crown root development, including crown root initiation and emergence. Exogenous auxin application increased crown roots number in rice seedlings (Inukai et al., 2005). Cytokinin also influences lateral root formation by disrupting lateral root initiation and patterning in Arabidopsis (Laplaze et al., 2007; Benková and Hejátko, 2009; Péret et al., 2009). Auxin-cytokinin crosstalk signaling plays key roles in root development and can coordinately regulate a series of genes (Dello Ioio et al., 2008; Müller and Sheen, 2008; Perilli et al., 2013). For example, transcription of root-specific putative homeobox genes ATHB53 and WOX11 are differentially regulated by auxin and cytokinin in Arabidopsis and rice, respectively (Son et al., 2005; Zhao et al., 2009). Our data show that auxin and cytokinin could rapidly but transiently induce ERF3 expression (Figures 2B and 2C) and that overexpression or knockdown of ERF3 either activated or repressed some auxin- and cytokinin-responsive genes (Figure 4B). Similarly, exogenously supplied auxin (indole3-acetic acid [IAA]) and cytokinin (6-BA) could rescue root phenotypes of amiERF3 plants (Supplemental Figure 5). These data suggested that ERF3 might be an auxin-cytokinin-responsive gene and the effect of exogenous hormones on crown root development might be partially mediated by elevation of ERF3 expression.
In Arabidopsis, disruption of eight of the 10 type-A ARR genes affected root development via altering the size of the apical meristem (Zhang et al., 2011). Mutation of a maize type-A RR gene, ABPHL1 (ABPH1), induced an increased root meristem size (Giulini et al., 2004). In rice, overexpression of RR3, RR5, and RR6 affected crown root development (Hirose et al., 2007; Cheng et al., 2010). RR2 is directly repressed by WOX11 during crown root development (Zhao et al., 2009). Recent work reported that RR1 regulates crown root initiation under the control of CRL5 (Kitomi et al., 2011a). These observations suggest that type-A RR genes, which negatively regulate cytokinin signaling, may play important roles in plant root development. The current study indicated that ERF3 directly targets RR2 and upregulates its expression during crown root initiation (Figures 4C, 5B, and 5C). Furthermore, the data showing that RR2 overexpression augmented crown root numbers and its knockdown had an opposite effect (Figure 8), which indicated that RR2 is a bona fide downstream target of ERF3 involved in crown root initiation. Taken together, these results suggested that activation of type-A RR2 gene by ERF3, which might repress cytokinin signaling in crown root initials, enhanced root meristem activity and promoted crown root formation.
The Gene Regulatory Pathway Controlling Crown Root Development in Rice
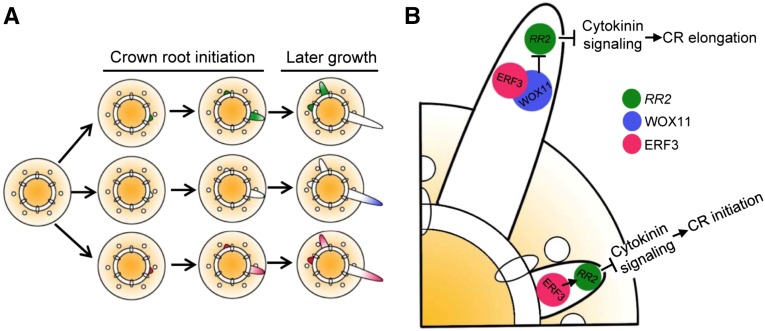
In rice, WOX11 is a key regulator in crown root development (Zhao et al., 2009). Thus far, the regulatory network by which WOX11 regulates downstream targets is unknown. This work identified ERF3 as an interacting partner of WOX11 (Figure 1). However, ERF3 transcripts were detectable in crown root initials and in emerging crown root meristem (Figures 2D to 2H), whereas WOX11 is expressed mostly in the active cell division region of the emerging crown root meristem (Zhao et al., 2009). This suggested that the expression pattern of ERF3 only partially overlapped with that of WOX11 during crown root development (Figure 2F; Zhao et al., 2009), and interaction between ERF3 and WOX11 most likely occurred after crown root emergence. Previous work showed that WOX11 represses RR2 in elongating crown roots (Zhao et al., 2009). The observations that RR2 mRNA levels was higher in crown root initials and gradually decreased during crown root growth (Zhao et al., 2009), suggesting that ERF3 and WOX11 might contribute to the spatial and temporal expression pattern of RR2 (Figure 9A). Because the WOX11/ERF3 interaction enhanced WOX11 binding to the RR2 promoter (Figure 6), and in amiERF3/wox11 roots, RR2 expression was even lower than in the wild type (Supplemental Figure 6), we speculated that in the elongating crown root meristem, the WOX11/ERF3 interaction might either enhance WOX11-mediated repression or inhibit ERF3-mediated activation of RR2.
Figure 9.
Proposed Models for ERF3, WOX11, and RR2 Expression Patterns and Their Functions in Controlling Crown Root Initiation and Elongation in Rice.
(A) Model of ERF3, WOX11, and RR2 expression patterns during crown root formation.
(B) Model of ERF3, WOX11, and RR2 functions controlling crown root initiation and elongation in rice.
During crown root initiation, ERF3 repressed cytokinin signaling by directly activating RR2 expression. During crown root elongation, WOX11 expression was turned on. WOX11-ERF3 interaction either enhances WOX11-mediated repression or inhibits ERF3-mediated activation of RR2, leading to cytokinin signaling activation and thereby promoting crown root growth.
Based on our data, we proposed a model of the regulatory pathway controlling crown root development in rice (Figure 9B). In crown root initials, ERF3 directly binds to RR2 and upregulates its expression, resulting in repression of cytokinin signaling and promotion of crown root initiation. In emerging crown roots, WOX11 expression was turned on and its binding to RR2 was enhanced by interaction with ERF3, leading to inhibition of ERF3 function or direct repression of RR2 and enhanced cytokinin signaling that promotes crown root elongation.
The CRL1/ARL1-CRL5 pathways affect crown root initiation by auxin (Inukai et al., 2005; Liu et al., 2005; Kitomi et al., 2011a). However, the expression of WOX11 is not regulated by CRL5 (Kitomi et al., 2011a). In this study, we found that CRL5 expression was unlikely to be regulated directly by ERF3 (Figure 4A). These results suggested that the crown root development pathway regulated by ERF3 and WOX11 might differ from the pathway regulated by CRL1/ARL1-CRL5. Further studies are needed to identify the interrelationship between these pathways in order to further understand the molecular regulatory mechanism of crown root development in rice.
Collectively, our data suggested a spatio-temporal sequence of events that may act in cytokinin signaling from crown root initiation to crown root elongation. This sequence involved a shift of RR2 activation toward RR2 repression, which linked to spatial expression of WOX11 and ERF3. Through the downstream effect on RR2 expression, this regulatory module determines the spatial frame for crown root initiation and consequently for crown root elongation. In conclusion, our discoveries revealed a regulatory mechanism underlying rice crown root development, which greatly advanced our understanding of adventitious root formation in crop plants, which appears to differ substantially from that in Arabidopsis. Thus, our work provided a foundation for further understanding crown root organogenesis and for crop improvement via targeting the ERF3/WOX11 pathway.
METHODS
Plant Materials and Growth Conditions
The rice variety Zhonghua11 (ZH11) (Oryza sativa subsp japonica) was used for transformation in this study. The wox11 mutant previously reported by Zhao et al. (2009) was introduced into the ZH11 background by backcrosses. All transgenic plants were produced in the ZH11 background, genotyped by PCR in the T2 segregating populations. Multiple independent segregating lines were used for molecular and phenotypic analysis. For in vitro cultures, rice seeds were surface-sterilized and germinated in media containing 0.8% agar supplemented with 3% (w/v) sucrose at 28°C (in light) and 24°C (in dark) with a 14-h-light/10-h-dark cycle or in fields.
Yeast Two-Hybrid Screening
The ProQuest system (Invitrogen) was used to screen for WOX11-interacting proteins following the manufacturer’s protocol. Briefly, a construct that contains the WOX11 full-length cDNA was cloned by PCR into the SalI and SpeI sites of Proquest’s pDBleu vector, and a rice cDNA library was built into the expression vector pEXP-AD502 of the ProQuest system using the restriction enzymes NotI and SalI with mRNA isolated from roots of ZH11 plants with two tillers. Total RNA was isolated using Trizol reagent (Invitrogen), and mRNA was then purified using the Absolutely mRNA purification kit (Stratagene) according to the protocol provided by the manufacturer. The primers WOX11-2YH F/R used in PCR amplification are listed in Supplemental Table 1.
In Vitro Pull-Down Assay
Pull-down was performed as described (Yang et al., 2008) with the following modifications: Equal volumes of GST or WOX3-His, and ERF3-GST or WOX11-His recombinant proteins were incubated for 6 h at 4°C with 400 μL of GST (GE Healthcare; 17-5132-01) or His (Promega; REF V8500) resin in a total volume of 1 mL of GST or His binding buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5% Lgepal CA-630, and protease inhibitor) for 2 to 3 h at 4°C, and the binding reaction was washed five times (10 min each time at 4°C) by the binding buffer. After extensive washing, the pulled down proteins were eluted by boiling, separated on 12% SDS-PAGE, and detected by immunoblots using an anti-GST antibody (abcam; ab19256) and anti-His antibody (abcam; ab9108), respectively.
Coimmunoprecipitation Assay
Coimmunoprecipitation assays were performed as previous described (Sun and Zhou, 2008). Total nuclear protein from roots of 10-d transformed plants and wild-type seedlings was extracted and ground in liquid nitrogen. Total nuclear proteins were extracted in nuclear extraction buffer 1 (2 mM EDTA, 2.5 mM DTT, 10 mM HEPES, pH 8.0, and 0.4 M sucrose) containing protease inhibitor cocktail (Roche). Cell debris was collected by centrifugation at 3000g for 10 min at 4°C and suspended with Nuclear Extraction Buffer 2 (10 mM MgCl2, 10 mM HEPES, pH 8.0, and 2.5 M sucrose containing protease inhibitor cocktail [Roche]), Nuclear Extraction Buffer 3, and coimmunoprecipitation buffer, respectively. The supernatant was incubated with anti-WOX11 (see below) or anti-FLAG (Sigma-Aldrich; F3165) specific antibody overnight at 4°C by gentle rotation. Then, 60 μL of protein G agarose beads (Millipore) was added. After 2 to 3 h of incubation at 4°C with gentle rotation, the beads were centrifuged and washed three times with 600 μL washing buffer (100 mM Tris-HCl, pH 7.4, 75 mM NaCl, 1 mM EDTA, 10% glycerol, 0.05% SDS, 0.1% Triton X-100, and protease inhibitor cocktail). FLAG proteins were eluted under acidic conditions and analyzed by immunoblots using anti-FLAG and anti-WOX11 antibodies.
The peptide-affinity polyclonal antibody against WOX11 (amino acids 177 to 191) was raised by Neweast Biotechnology. The specificity of this purified antibody was confirmed by immunoblot.
BiFC and Fluorescence Microscopy
BiFC assays were performed as described previously (Waadt et al., 2008). For generation of the BiFC vectors, the full-length cDNA of ERF3 was amplified with primer pairs ERF3BiFC-F/ R (Supplemental Table 1) and cloned at the SpeI-KpnI sites in pVYCE(R). The full-length cDNA of WOX11 was cloned into the SpeI-KpnI sites of pVYNE(R). Rice mesophyll protoplasts were prepared and the two fusion proteins were transiently cotransfected into rice protoplasts with Ghd7:CFP, as described (Zhou et al., 2009), with minor modifications. Fluorescence in the transformed protoplasts was imaged using a confocal laser scanning microscope (TCS SP2; Leica) after incubation at 23°C for 12 to18 h.
Vector Construction and Rice Transformation
For the construction of the fusion between the ERF3 promoter and the GUS coding sequence, the 2.561-kb ERF3 promoter was amplified from ZH11 genomic DNA by primer set ERF3pgus-F/R and inserted into pCAMBIA1381Xb (CAMBIA) at the BamHI and SalI sites.
To analyze the function of ERF3, the amiRNA strategy was used to knock down the ERF3 gene (Ossowski et al., 2008). For amiRNAs of the ERF3 construct, the 250-bp fragment containing 21-mers recognizing specifically the 39 untranslated region of ERF3 transcripts was amplified using universal primers G11491/G11494 and specific primers, including ERF3miR-s, ERF3miR-a, ERF3miR-*s, and ERF3miR-*a, which were designed in WMD2 (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) as described previously (Warthmann et al., 2008), and then cloned into KpnI-BamHI-digested pU1301 (Chu et al., 2006).
For overexpression and ERF3-FLAG fusion constructs, the full-length cDNA of ERF3 amplified with the primer set ERF3ox-F/R and ERF3flag-F/R was inserted into the KpnI and BamHI sites of pU1301 and pU2301, respectively (Dai et al., 2007; Sun and Zhou, 2008).
To construct the RR2 overexpression and RNAi vector, the full-length cDNA and a specific cDNA fragment of RR2 were amplified using the primer set OxRR2F/R and RiRR2-F/R and inserted into pU1301 and pDS1301 vectors, respectively (Chu et al., 2006; Dai et al., 2007).
All constructs were introduced into ZH11 plants by Agrobacterium tumefaciens (EHA105)-mediated transformation as previously described (Dai et al., 2007). All primers for genotyping and vector construction are listed in Supplemental Table 1.
In Situ Hybridization
The hybridization and immunological detection were performed as described by Zhao et al. (2009). The ERF3 probe was amplified using the gene-specific primers ERF3situ-F/R (Supplemental Table 1). The PCR fragment was inserted into the SpeI and BamHI sites of pGEM-T (Promega) and transcribed in vitro from either the T7 or SP6 promoter for sense or antisense strand synthesis using the Digoxigenin RNA labeling kit (Roche). The RR2 probe was previously described (Zhao et al., 2009).
Promoter Activity Detection and Histological Observation
Roots of ProOsERF3-GUS transgenic plants harvested 14 d after germination were incubated with X-gluc buffer overnight at 37°C (Jefferson et al., 1987) and directly photographed. Roots and the coleoptilar nodal region of different ERF3 transgenic lines and the wild type were fixed with 50% FAA (formalin/acetic acid/alcohol) at 4°C overnight. The staining and dehydration were performed according to the previously described method (Liu et al., 2005) and embedded with cold-curing resin (Heraeus Kulzer Dental) using the Technovit 7100 system. The microtome sections (4-μm thickness) were mounted on glass slides and stained with 0.25% toluidine blue. Sections were observed under a bright-field microscope (Zeiss AxioCam HRC) and photographed with a differential interference microscope (Nikon 80i). Root length was measured with Image J software (http://rsb.info.nih.gov/ij). Root meristem size was determined by measuring the length from the quiescent center to the first elongated epidermal cell. The average cell length in the root meristem was quantified with cells (as shown with red lines in the figures). For each quantification, at least 15 rice plants were analyzed.
EMSA
To produce the ERF3 protein, the full-length cDNA amplified with primers ERF3protein-F/R was inserted into the BamHI and SalI sites of the pGEX-4T-1 expression vector (GE Healthcare) and expressed in Escherichia coli DE3 (BL21) cells (GE Healthcare). The target protein was purified with GST four fast flow (GE Healthcare). The RR2 promoter DNA P1 (including the putative ERE binding site GCCGCC), 1st exon DNA P2 (including the putative ERE binding site GCCGCCGCC), P2 deletion (with nucleotide deletions in the ERE binding site), and 1st intron DNA P3 (including GCCGCC) were produced by annealing of oligonucleotides EMSAp1-F/R, EMSAp2-F/R, EMSAp2d-F/-R, and EMSAp3-F/R, respectively. The double-stranded oligonucleotides EMSAp1, EMSAp2, EMSAp2d, and EMSAp3 were labeled with [32P]dCTP using a Klenow fragment. DNA binding reactions were performed in the presence or absence of unlabeled P1, P2, P2d, and P3 fragments at different molar excess at room temperature for 20 min in 10 mM Tris, pH 7.5, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 1 mM MgCl2, 5% glycerol, and 50 mg/L poly(dI-dC) (Amersham Pharmacia Biotech). The reactions were resolved on 6% polyacrylamide gels in Tris-glycine (0.3% Tris and 1.88% glycine) buffer and visualized by autoradiography. The sequences of the primers used are listed in Supplemental Table 1.
In Vivo Binding Assay of ERF3 and WOX11 by ChIP
For ChIP assays, wild-type and ERF3-FLAG transgenic lines were used for chromatin extraction and immunoprecipitation as described by Huang et al. (2007). Briefly, roots were treated with formaldehyde and the nuclei were isolated and sonicated using an Ultrasonic Crasher Noise Isolating Chamber (Scientz). The soluble chromatin fragments were isolated and preabsorbed with sheared salmon sperm DNA/protein A-agarose (Sigma-Aldrich) to remove nonspecific binding. Immunoprecipitations with anti-FLAG (Sigma-Aldrich; F3165) or without any serum were performed as described. The precipitated DNA was analyzed by qPCR using specific primer sets ERF3ChIPs4F/R, ERF3ChIPs5F/R, and ERF3ChIPs6F/R (Supplemental Table 1). WOX11 ChIP qPCR primers are described in (Zhao et al., 2009). Typically, the precipitated and input DNA samples were analyzed by qPCR with gene-specific primers listed in Supplemental Table 1. Data normalized with input transcripts are means from three biological repeats ± sd. The values from nontreated samples were assessed as 1.
RT-qPCR
Total RNA was isolated using TRIzol reagent and reverse-transcribed according to the manufacturer’s instructions (Invitrogen). RT-qPCR was performed using gene-specific primers (Supplemental Table 1) and SYBR Premix Ex Taq on a real-time PCR 7500 system (Applied Biosystems). Data were collected using the ABI PRISM 7500 sequence detection system following the instruction manual. The rice ACTIN1 gene was used as the internal control. At least three biological replicates and three technical repeats were tested.
RNA Gel Blot and Immunoblot
For RNA gel-blotting analysis, 15 μg of total RNA samples extracted from tissues or organs harvested from field-grown rice plants was separated in 1.2% (w/v) formamide-denaturing agarose gels, before being transferred to nylon membranes. Gene-specific probes were labeled with [32P]dCTP using the Random Primer kit (Invitrogen) and hybridized to the RNA gel blots. The probe for ERF3 was digested from T/A plasmid with KpnI and BamHI, a fragment of 635 bp of the cDNA.
Rice root nuclear protein was extracted from wild-type and ERF3-FLAG transgenic plants and performed as described previously (Tariq et al., 2003). After being washed in acetone and dried, the proteins were resuspended in Laemmli sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 25% glycerol, 0.01% bromophenol blue, and 10% β-mercaptoethanol), then separated by 16% SDS-PAGE and transferred to an Immobilon-P polyvinylidene fluoride transfer membrane (Millipore). The membrane was blocked with 2% BSA in PBS (pH 7.5) and incubated overnight with primary antibodies, such as anti-FLAG (F3165; Sigma-Aldrich), in a 1:5000 dilution at 4°C. After three washes (30 min each), the secondary antibody (goat anti-mouse IgG [SouthernBiotech]) at 1:10,000 dilution was used. Visualization was performed using the Super Signal West Pico kit (Pierce) according to the manufacturer’s instructions.
Exogenous IAA, Naphthalene Acetic Acid, and 6-BA Treatment
Seeds were sown and germinated on agar medium. After 10 d, the seedlings were transferred to media with or without 10−6 M 2,4-D, or 10−5 M 6-BA. Total RNA was extracted after 0, 0.5, 1, 3, 4, 6, 9, and 12 h of treatment and analyzed by RT-qPCR with primers ERF3qPCR-F/R. For root growth tests, transgenic plants and wild-type seeds were germinated on agar medium containing 10−6 M 2,4-D, 10−6 M IAA, or 10−5 M 6-BA. Ten days after treatment, crown root phenotypes were recorded.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: ERF3, AK061380, LOC_Os01g58420; CRL1, AK064187 LOC_03g05510; CRL4, AK240747, LOC_Os03g46330; and CRL5, AK109848, LOC_Os07g03250. The accession numbers of other genes can be found in Zhao et al. (2009).
Supplemental Data
Supplemental Figure 1. Detection of ERF3 mRNA and protein in ERF3-FLAG transgenic and wild-type plants by RNA gel blot and immunoblot.
Supplemental Figure 2. Cell longitudinal length in root meristem zone and crown root primordium number in ERF3 transgenic plants and wild-type seedlings.
Supplemental Figure 3. Gel shift assay of ERF3 protein binding to P1, P2, and P3 regions of RR2 containing the ERF binding sites (underlined).
Supplemental Figure 4. Detection of expression level of ERF3 in wild type and wox11 and phenotype of crown root of amiERF3/wox11 transgenic plants at rooting stage.
Supplemental Figure 5. Auxin (2,4-D and IAA) and cytokinin (6-BA) rescued the root phenotype of ERF3 artificial microRNA (amiE3) transgenic plants.
Supplemental Figure 6. RT-qPCR and in situ hybridization detection of RR2 expression in wox11, ERF3:ERF3wox11, amiERF3wox11, and wild-type roots.
Supplemental Table 1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Qinglu Zhang and Xianghua Li for help in field experiments and management. This research was supported by grants from the National Natural Science Foundation of China (31371468), the Program for New Century Excellent Talents in University (NCET-12-0863), and the Fundamental Research Funds for the Central Universities (2013PY021).
AUTHOR CONTRIBUTIONS
Y.Z., D.-X.Z., and S.C. designed the research and analyzed the experimental data. S.C., Y.Z., Y.S., Y.H., and S.Z. performed the experiments. Y.Z. and D.-X.Z. wrote the article.
Glossary
- BiFC
bimolecular fluorescence complementation
- 6-BA
6-benzylaminopurine
- ERE
ethylene-responsive element
- EMSA
electrophoresis mobility shift assay
- ChIP
chromatin immunoprecipitation
- IAA
indole3-acetic acid
- amiRNA
artificial microRNA
Footnotes
Articles can be viewed online without a subscription.
References
- Benková E., Hejátko J. (2009). Hormone interactions at the root apical meristem. Plant Mol. Biol. 69: 383–396. [DOI] [PubMed] [Google Scholar]
- Bishopp A., Help H., Helariutta Y. (2009). Cytokinin signaling during root development. Int. Rev. Cell Mol. Biol. 276: 1–48. [DOI] [PubMed] [Google Scholar]
- Cheng X., Jiang H., Zhang J., Qian Y., Zhu S., Cheng B. (2010). Overexpression of type-A rice response regulators, OsRR3 and OsRR5, results in lower sensitivity to cytokinins. Genet. Mol. Res. 9: 348–359. [DOI] [PubMed] [Google Scholar]
- Chu Z., Yuan M., Yao J., Ge X., Yuan B., Xu C., Li X., Fu B., Li Z., Bennetzen J.L., Zhang Q., Wang S. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20: 1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudert Y., Périn C., Courtois B., Khong N.G., Gantet P. (2010). Genetic control of root development in rice, the model cereal. Trends Plant Sci. 15: 219–226. [DOI] [PubMed] [Google Scholar]
- Dai M., Hu Y., Zhao Y., Liu H., Zhou D.X. (2007). A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 144: 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Sabatini S. (2008). Emerging role of cytokinin as a regulator of cellular differentiation. Curr. Opin. Plant Biol. 11: 23–27. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682. [DOI] [PubMed] [Google Scholar]
- De Smet I., Jürgens G. (2007). Patterning the axis in plants--auxin in control. Curr. Opin. Genet. Dev. 17: 337–343. [DOI] [PubMed] [Google Scholar]
- Du L., Jiao F., Chu J., Jin G., Chen M., Wu P. (2007). The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics 89: 697–707. [DOI] [PubMed] [Google Scholar]
- Durbak A., Yao H., McSteen P. (2012). Hormone signaling in plant development. Curr. Opin. Plant Biol. 15: 92–96. [DOI] [PubMed] [Google Scholar]
- Gao S., Fang J., Xu F., Wang W., Sun X., Chu J., Cai B., Feng Y., Chu C. (2014). CYTOKININ OXIDASE/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol. 165: 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulini A., Wang J., Jackson D. (2004). Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430: 1031–1034. [DOI] [PubMed] [Google Scholar]
- Goh T., Joi S., Mimura T., Fukaki H. (2012). The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139: 883–893. [DOI] [PubMed] [Google Scholar]
- Hao D., Ohme-Takagi M., Sarai A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273: 26857–26861. [DOI] [PubMed] [Google Scholar]
- Hirose N., Makita N., Kojima M., Kamada-Nobusada T., Sakakibara H. (2007). Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 48: 523–539. [DOI] [PubMed] [Google Scholar]
- Huang L., Sun Q., Qin F., Li C., Zhao Y., Zhou D.X. (2007). Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 144: 1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Shibata Y., Gomi K., Umemura I., Hasegawa Y., Ashikari M., Kitano H., Matsuoka M. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17: 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46: 23–47. [DOI] [PubMed] [Google Scholar]
- Jain M., Kaur N., Garg R., Thakur J.K., Tyagi A.K., Khurana J.P. (2006). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 6: 47–59. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi Y., Ito H., Hobo T., Aya K., Kitano H., Inukai Y. (2011a). The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 67: 472–484. [DOI] [PubMed] [Google Scholar]
- Kitomi Y., Kitano H., Inukai Y. (2011b). Molecular mechanism of crown root initiation and the different mechanisms between crown root and radicle in rice. Plant Signal. Behav. 6: 1270–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi Y., Ogawa A., Kitano H., Inukai Y. (2008). CRL4 regulates crown root formation through auxin transport in rice. Plant Root 2: 19–28. [Google Scholar]
- Laplaze L., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19: 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.W., Kim N.Y., Lee D.J., Kim J. (2009). LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 151: 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Lee W.S., Kim S.H. (2013). Hormonal regulation of stem cell maintenance in roots. J. Exp. Bot. 64: 1153–1165. [DOI] [PubMed] [Google Scholar]
- Li P., Xue H. (2011). Structural characterization and expression pattern analysis of the rice PLT gene family. Acta Biochim. Biophys. Sin. (Shanghai) 43: 688–697. [DOI] [PubMed] [Google Scholar]
- Liu H., Wang S., Yu X., Yu J., He X., Zhang S., Shou H., Wu P. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43: 47–56. [DOI] [PubMed] [Google Scholar]
- Liu S., Wang J., Wang L., Wang X., Xue Y., Wu P., Shou H. (2009). Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 19: 1110–1119. [DOI] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon C., Paschold A., Hochholdinger F. (2013). Genetic control of root organogenesis in cereals. Methods Mol. Biol. 959: 69–81. [DOI] [PubMed] [Google Scholar]
- Moubayidin L., Perilli S., Dello Ioio R., Di Mambro R., Costantino P., Sabatini S. (2010). The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 20: 1138–1143. [DOI] [PubMed] [Google Scholar]
- Nakamura A., Umemura I., Gomi K., Hasegawa Y., Kitano H., Sazuka T., Matsuoka M. (2006). Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J. 46: 297–306. [DOI] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.J., Kim Y.S., Kwon C.W., Park H.K., Jeong J.S., Kim J.K. (2009). Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 150: 1368–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690. [DOI] [PubMed] [Google Scholar]
- Péret B., De Rybel B., Casimiro I., Benková E., Swarup R., Laplaze L., Beeckman T., Bennett M.J. (2009). Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14: 399–408. [DOI] [PubMed] [Google Scholar]
- Perilli S., Perez-Perez J.M., Di Mambro R., Peris C.L., Díaz-Triviño S., Del Bianco M., Pierdonati E., Moubayidin L., Cruz-Ramírez A., Costantino P., Scheres B., Sabatini S. (2013). RETINOBLASTOMA-RELATED protein stimulates cell differentiation in the Arabidopsis root meristem by interacting with cytokinin signaling. Plant Cell 25: 4469–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K., Simásková M., Duclercq J., Petrásek J., Zazímalová E., Simon S., Friml J., Van Montagu M.C.E., Benková E. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 106: 4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. (2002). Plant patterning: TRY to inhibit your neighbors. Curr. Biol. 12: R804–R806. [DOI] [PubMed] [Google Scholar]
- Schiefelbein J. (2003). Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6: 74–78. [DOI] [PubMed] [Google Scholar]
- Son O., et al. (2005). Induction of a homeodomain-leucine zipper gene by auxin is inhibited by cytokinin in Arabidopsis roots. Biochem. Biophys. Res. Commun. 326: 203–209. [DOI] [PubMed] [Google Scholar]
- Su Y.H., Liu Y.B., Zhang X.S. (2011). Auxin-cytokinin interaction regulates meristem development. Mol. Plant 4: 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Zhou D.X. (2008). Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. USA 105: 13679–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M., Saze H., Probst A.V., Lichota J., Habu Y., Paszkowski J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100: 8823–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten I., Beeckman T., Geelen D. (2013). Adventitious root induction in Arabidopsis thaliana as a model for in vitro root organogenesis. Methods Mol. Biol. 959: 159–175. [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Wang S., Xu Y., Li Z., Zhang S., Lim J.M., Lee K.O., Li C., Qian Q., de Jiang A., Qi Y. (2014). OsMOGS is required for N-glycan formation and auxin-mediated root development in rice (Oryza sativa L.). Plant J. 78: 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.F., He F.F., Ma X.X., Mao C.Z., Hodgman C., Lu C.G., Wu P. (2011). OsCAND1 is required for crown root emergence in rice. Mol. Plant 4: 289–299. [DOI] [PubMed] [Google Scholar]
- Warthmann N., Chen H., Ossowski S., Weigel D., Herve P. (2008). Highly specific gene silencing by artificial miRNAs in rice. PLoS One 3: e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Zhu L., Shou H., Wu P. (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46: 1674–1681. [DOI] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., Zhang Q. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yang L., Jiang Y., Wu S.F., Zhou M.Y., Wu Y.L., Chen G.Q. (2008). CCAAT/enhancer-binding protein alpha antagonizes transcriptional activity of hypoxia-inducible factor 1 alpha with direct protein-protein interaction. Carcinogenesis 29: 291–298. [DOI] [PubMed] [Google Scholar]
- Zhang W., To J.P., Cheng C.Y., Schaller G.E., Kieber J.J. (2011). Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. Plant J. 68: 1–10. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hu Y., Dai M., Huang L., Zhou D.X. (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21: 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., et al. (2009). BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J. 57: 446–462. [DOI] [PubMed] [Google Scholar]
- Zhu Z.X., Liu Y., Liu S.J., Mao C.Z., Wu Y.R., Wu P. (2012). A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Mol. Plant 5: 154–161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.