Abstract
Background
Microbes are common inhabitants of floral nectar and are capable of influencing plant-pollinator interactions. All studies so far investigated microbial communities in floral nectar in plant populations that were located in natural environments, but nothing is known about these communities in nectar of plants inhabiting urban environments. However, at least some microbes are vectored into floral nectar by pollinators, and because urbanization can have a profound impact on pollinator communities and plant-pollinator interactions, it can be expected that it affects nectar microbes as well. To test this hypothesis, we related microbial diversity in floral nectar to the degree of urbanization in the late-flowering plant Linaria vulgaris. Floral nectar was collected from twenty populations along an urbanization gradient and culturable bacteria and yeasts were isolated and identified by partially sequencing the genes coding for small and large ribosome subunits, respectively.
Results
A total of seven yeast and 13 bacterial operational taxonomic units (OTUs) were found at 3 and 1 % sequence dissimilarity cut-offs, respectively. In agreement with previous studies, Metschnikowia reukaufii and M. gruessi were the main yeast constituents of nectar yeast communities, whereas Acinetobacter nectaris and Rosenbergiella epipactidis were the most frequently found bacterial species. Microbial incidence was high and did not change along the investigated urbanization gradient. However, microbial communities showed a nested subset structure, indicating that species-poor communities were a subset of species-rich communities.
Conclusions
The level of urbanization was putatively identified as an important driver of nestedness, suggesting that environmental changes related to urbanization may impact microbial communities in floral nectar of plants growing in urban environments.
Keywords: Nectar yeasts, Urbanization, Metschnikowia, Acinetobacter, Nestedness, Nectar microbial communities, Linaria vulgaris
Background
Numerous studies have shown that microbes are common inhabitants of floral nectar [1–8]. Nectar inhabiting microbes (NIMs) have been shown to modify important physicochemical properties of floral nectar, such as sugar and amino acid composition [9, 10], to alter nectar odor [11, 12], and even to increase the temperature of the flower itself [13]. Changes in physicochemical properties of floral nectar can, in turn, alter the attractiveness of a given flower to pollinators, resulting in increased visitation rates and plant reproductive success [14, 15].
Most research so far has shown that microbial diversity in floral nectar is low and often dominated by a limited number of culturable species [4, 7, 8, 16, 17]. However, the few studies that have investigated variation in nectar microbial communities among plant populations have shown that the distribution of NIMs is not uniform, but can vary substantially between populations. For example, bacterial communities in the floral nectar of the summer asphodel (Asphodelus aestivus) changed significantly along an aridity gradient [18]. Similarly, microbial communities in the floral nectar of several populations of the spring-flowering forest herb Pulmonaria officinalis showed large within-population variation and low among-population similarity, indicating that NIM community assembly may to some extent be context-dependent [5]. These results further suggest that both variation in the local species pool of microbes and in local environmental conditions can shape NIM communities.
All studies investigating the diversity and abundance of NIMs so far have focused on plant populations occurring in natural environments, and virtually nothing is known about the composition of nectar microbial communities in urban environments. Transformation of natural landscapes by urbanization can, however, be expected to have a profound impact on nectar microbial communities. Given that most NIMs are vectored from one flower to the next by insect pollinators, and that nectar yeasts in particular are thought to rely exclusively on them to colonize new flowers [19, 20], impoverishment of pollinator communities can negatively affect colonization rates and subsequent dispersal of NIMs and therefore affect NIM community composition. The typically increasing cover of impervious surfaces in urban environments has, for example, been shown to be correlated with decreased nesting density in Bombus vosnesenskii and with decreased abundance of other wild bees [21, 22]. It has also been shown that different bee species respond differently to urbanization, which results in varied pollinator guilds along an urban gradient [23]. Furthermore, plant populations may typically decrease in size and become more spatially isolated as a result of urbanization. The increased spatial isolation and reduced population size of co-flowering plant species in urban environments can impede the exchange of microorganisms between populations, which, in turn, results in a nested species distribution pattern, where only the most common species are present in the most isolated or the smallest habitat patches (see [24] for review of the nestedness concept).
To test the general hypothesis that microbial communities in nectar change along an urbanization gradient, we investigated microbial communities in the floral nectar of the late-flowering herb Linaria vulgaris (yellow toadflax), a species that occurs in both urban and rural habitats. After collecting nectar from twenty populations across an urbanization gradient and surveying its microbiota using culture dependent methods and Sanger sequencing, we specifically addressed the following questions:
How does urbanization affect microbial incidence in floral nectar of L. vulgaris?
Does urbanization lead to impoverished NIM communities, with the most heavily urbanized sites having the most impoverished communities?
Are impoverished NIM communities subsets of more species-rich communities?
Results and discussion
Yeast diversity and occurence
Following cultivation and isolation a total of 140 yeast isolates was obtained. When the sequences corresponding to these isolates were clustered according to a 3 % dissimilarity cutoff, seven fungal OTUs were identified (Table 1). Following BLAST analysis, these OTUs corresponded to five different validly named species from four families: Metschnikowiaceae, Dothioraceae, Sporidiobolales and Tremellales, indicating that Ascomycota were more frequently represented than Basidiomycota. Most isolates were related to Metschnikowia gruessi (101 isolates) and to a smaller degree to M. reukaufii (36 isolates), while other species were represented by single isolate (Table 1). Three out of the seven OTUs (OTU0.03Y2, OTU0.03Y3, and OTU0.03Y4) found were identified as M. reukaufii. The Chao2 estimator predicted a slightly higher number of OTUs, 12.7, while the ICE estimator predicted 18.61 OTUs (Fig. 1).
Table 1.
Yeast operational taxonomic units (OTUs) identified in the nectar of 20 L. vulgaris populations sampled along an urbanization gradient
| OTU 3 % | 118 | Phylum | Family | Closest match with GenBank entries | Accession number | Score | E value | Length | Identity (%) |
|---|---|---|---|---|---|---|---|---|---|
| OTU0.03Y1 | 22 | Ascomycota | Metschnikowiaceae | Metschnikowia gruessi | JX067745 | 665.914 | 0 | 360 | 100 |
| OTU0.03Y2 | 10 | Ascomycota | Metschnikowiaceae | Metschnikowia reukaufii | KM281795 | 682.534 | 0 | 369 | 100 |
| OTU0.03Y3 | 4 | Ascomycota | Metschnikowiaceae | Metschnikowia reukaufii | FJ455114 | 675.147 | 0 | 365 | 100 |
| OTU0.03Y4 | 1 | Ascomycota | Metschnikowiaceae | Metschnikowia reukaufii | JN642530 | 584.662 | 3.18e−163 | 332 | 98.5 |
| OTU0.03Y5 | 1 | Ascomycota | Dothioraceae | Aureobasidium pullulans | KP710217 | 774.886 | 0 | 419 | 100 |
| OTU0.03Y6 | 1 | Basidiomycota | Sporidiobolales | Sporobolomyces roseus | AM160644 | 250.48 | 1.32e−62 | 135 | 100 |
| OTU0.03Y7 | 1 | Basidiomycota | Tremellales | Cryptococcus aureus | JN004200 | 1059.25 | 0 | 583 | 99.5 |
The sequences were grouped into OTUs based on 97 % identity at the large ribosomal subunit gene. The BLAST search was conducted in June 2015, excluding uncultured/environmental samples. Only the closest matches are reported
Fig. 1.
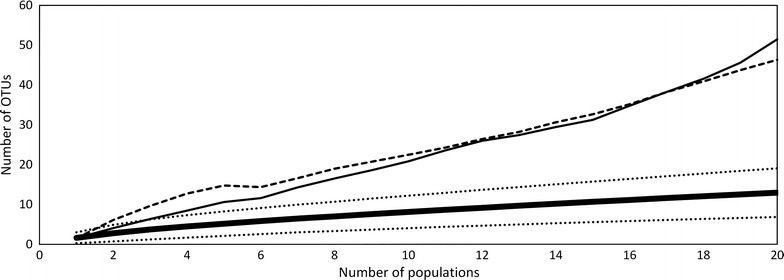
Rarefaction curve (bold, solid line) and the estimators Chao2 (thin solid line) and ICE (dashed line) of yeast operational taxonomic unit (OTU) richness found in the floral nectar of 20 Linaria vulgaris populations sampled across an urbanization gradient in the vicinity of Leuven, Belgium. The sequences were grouped into OTUs based on 97 % identity at the large ribosomal subunit gene
Yeasts were found in all studied L. vulgaris populations. On average, there were two yeast OTUs present per population (sd: 0.79), with M. gruessi present in 95 % of the populations, and M. reukaufii present in 70 % of the populations. There was no correlation between yeast incidence and ISI500 (Spearman-ρ = 0.06, p = 0.78) or plant population size (Spearman-ρ = 0.02, p = 0.98). The size of each population and impervious surface index pertaining to it were not inter-correlated (Spearman-ρ = −0.32, p = 0.15).
Thus, our results showed that yeast communities in urban environments were mainly dominated by the yeasts M. reukaufii and M. gruessi. However, unlike in many natural environments [17, 25–27] M. gruessi was more abundant than M. reukaufii. Assessment of the phenotypic landscape of both yeast species has recently shown that both species displayed a significantly different physiological profile, most likely facilitating co-occurrence of both species [28]. Moreover, comparison of utilization profiles in single vs mixed cultures indicated that M. reukaufii generally grows better in sucrose solutions, and that M. gruessi grows better in mixed cultures in glucose and fructose solutions [28]. On the other hand, out of the yeast strains isolated from 19 different nectars, the strains isolated from the nectar of Plantaginaceae, the family to which L. vulgaris belongs, exhibited the most dissimilar phenotypes for M. gruessi and M. reukaufii. This might mean that the nectar of Plantaginaceae exhibits strong differential constraints on yeast species growth, with which M. gruessi copes better in the specific case of L. vulgaris.
Bacterial diversity and occurence
In total, 52 bacterial isolates were obtained after cultivation. Thirteen bacterial OTUs were identified when clustered according to 1 % dissimilarity cutoff (Table 2). Based on BLAST results, these OTUs corresponded to 13 validly named species. The ICE and Chao2 estimators predicted a much higher number of 46.37 and 51.48 OTUs, respectively (Fig. 2). Similarly, the rarefaction curves showed that additional sampling would yield a higher number of OTUs, and this effect was more pronounced than in the case of yeast, probably due to higher number of bacterial singletons. The rarefaction curves for bacteria showed a similar level of saturation, however, to what has been found in earlier culture-dependent studies (i.e. 4, 5, 20). Acinetobacter nectaris comprised over 50 % (29 isolates) of bacterial isolates, with Rosenbergiellaepipactidis coming second (10 isolates). The remaining OTUs were represented by 1–3 isolates, coming mostly from the Enterobacteriaceae, but also from Pseudomonadaceae, Microbacteriaceae, Bacillaceae and Sphingomonadaceae (Table 2).
Table 2.
Bacterial operational taxonomic units (OTUs) identified in the nectar of 20 L. vulgaris populations sampled along an urbanization gradient
| OTU 1 % | Number of isolates | Phylum | Family | Closest match with GenBank entries | Accession number | Score | E value | Length | Identity (%) |
|---|---|---|---|---|---|---|---|---|---|
| OTU0.01B1 | 1 | Actinobacteria | Microbacteriaceae | Microbacterium testaceum | KP642087 | 1796.06 | 0 | 972 | 100 |
| OTU0.01B2 | 1 | Actinobacteria | Microbacteriaceae | Rathayibacter festucae | NR_042574 | 1768.36 | 0 | 957 | 100 |
| OTU0.01B3 | 1 | Firmicutes | Bacillaceae | Lysinibacillus odysseyi | NR_113881 | 1921.64 | 0 | 1043 | 100 |
| OTU0.01B4 | 1 | Proteobacteria | Sphingomonadaceae | Sphingomonas faeni | KM891564 | 1773.9 | 0 | 960 | 100 |
| OTU0.01B5 | 1 | Proteobacteria | Enterobacteriaceae | Pantoea ananatis | KC139412 | 1919.79 | 0 | 1048 | 99.7 |
| OTU0.01B6 | 1 | Proteobacteria | Enterobacteriaceae | Pantoea vagans | KP099965 | 1757.28 | 0 | 957 | 99.8 |
| OTU0.01B7 | 1 | Proteobacteria | Enterobacteriaceae | Pectobacterium carotovorum | GU129979 | 1628.02 | 0 | 885 | 99.9 |
| OTU0.01B8 | 1 | Proteobacteria | Enterobacteriaceae | Ewingella americana | KM891553 | 1842.23 | 0 | 997 | 100 |
| OTU0.01B9 | 1 | Proteobacteria | Pseudomonadaceae | Pseudomonas viridiflava | NR_117825 | 1899.48 | 0 | 1028 | 100 |
| OTU0.01B10 | 3 | Proteobacteria | Pseudomonadaceae | Pseudomonas moraviensis | KP165022 | 1892.09 | 0 | 1024 | 100 |
| OTU0.01B11 | 10 | Proteobacteria | Enterobacteriaceae | Rosenbergiella epipactidis | NR_126303 | 1816.38 | 0 | 989 | 99.8 |
| OTU0.01B12 | 1 | Proteobacteria | Moraxellaceae | Acinetobacter boissieri | NR_118409 | 1855.16 | 0 | 1004 | 100 |
| OTU0.01B13 | 29 | Proteobacteria | Moraxellaceae | Acinetobacter nectaris | JQ771134 | 1879.16 | 0 | 1023 | 99.8 |
The sequences were grouped into OTUs based on 99 % identity at the small ribosomal subunit gene. The BLAST search was conducted in June 2015, excluding uncultured/environmental samples. Only the closest matches are reported
Fig. 2.
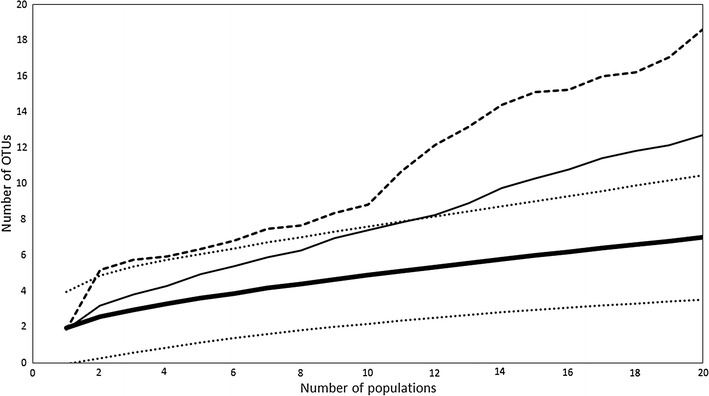
Rarefaction curve (bold, solid line) and the estimators Chao2 (thin solid line) and ICE (dashed line) of bacterial operational taxonomic unit (OTU) richness found in the floral nectar of 20 Linaria vulgaris populations sampled across an urbanization gradient in the vicinity of Leuven, Belgium. The sequences were grouped into OTUs based on 99 % identity at the small ribosomal subunit gene
Bacteria were present in all but four of the studied populations, with an average of 1.65 OTUs per population (sd: 1.26). A. nectaris was the most frequently found species and occurred in 60 % of the populations, while R.epipactidis was present in 35 % of the populations. Bacterial incidence was also not correlated to ISI500 (Spearman-ρ = −0.16, p = 0.49), but it was significantly correlated to plant population size (Spearman-ρ = 0.45, p = 0.04) (Table 3).
Table 3.
Properties of the 20 sampled Linaria vulgaris populations
| Population | Longitude | Latitude | ISI500 (%) | Population size | Nestedness rank | Yeast OTUs0.03 richness | Total yeast incidence (%) | Bacterial OTUs0.01 richness | Total bacterial incidence (%) | Total microbial richness |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 4.709365 | 50.864275 | 36.7 | 400 | 3 | 3 | 100 | 3 | 100 | 6 |
| B | 4.704492 | 50.863418 | 33.84 | 450 | 20 | 2 | 100 | 0 | 0 | 2 |
| C | 4.700154 | 50.86323 | 42.35 | 100 | 14 | 2 | 100 | 1 | 40 | 3 |
| D | 4.676421 | 50.885377 | 44.85 | 15 | 18 | 2 | 20 | 0 | 0 | 2 |
| E | 4.715524 | 50.867437 | 27.44 | 60 | 13 | 2 | 60 | 0 | 0 | 2 |
| F | 4.713976 | 50.85837 | 18.9 | 520 | 11 | 2 | 80 | 2 | 60 | 4 |
| G | 4.72284 | 50.850707 | 44.72 | 420 | 19 | 2 | 100 | 0 | 0 | 2 |
| H | 4.7281 | 50.843644 | 39.17 | 300 | 15 | 2 | 40 | 1 | 80 | 3 |
| I | 4.723781 | 50.852871 | 32.26 | 70 | 12 | 1 | 60 | 2 | 80 | 3 |
| J | 4.685015 | 50.888567 | 31.16 | 150 | 4 | 1 | 40 | 4 | 80 | 5 |
| K | 4.725668 | 50.858904 | 19.26 | 200 | 17 | 1 | 100 | 1 | 40 | 2 |
| L | 4.727276 | 50.861677 | 22.98 | 83 | 6 | 3 | 80 | 2 | 40 | 5 |
| M | 4.725735 | 50.860678 | 19.78 | 500 | 2 | 3 | 80 | 2 | 100 | 5 |
| N | 4.801452 | 50.822292 | 7.06 | 500 | 7 | 2 | 60 | 3 | 40 | 5 |
| O | 4.803312 | 50.823027 | 12.99 | 500 | 16 | 1 | 40 | 1 | 20 | 2 |
| P | 4.784239 | 50.826077 | 7.81 | 170 | 9 | 2 | 60 | 1 | 40 | 3 |
| Q | 4.79446 | 50.825404 | 5.98 | 400 | 10 | 1 | 40 | 2 | 60 | 3 |
| R | 4.790337 | 50.825352 | 6.65 | 1250 | 1 | 3 | 80 | 3 | 80 | 6 |
| S | 4.794894 | 50.823779 | 6.99 | 160 | 5 | 1 | 100 | 4 | 80 | 5 |
| T | 4.805983 | 50.852606 | 5.36 | 50 | 8 | 3 | 80 | 1 | 20 | 4 |
The approximate coordinates of each population are given in decimal degrees. The degree or urbanization of the surroundings of each population was assessed using ISI500, the impervious surface index within a radius of 500 m. For each of the sampled populations, the bacterial and yeast species richness (number of OTUs) and incidence (frequency of occurrence) are presented
Overall, the incidence of bacteria in L. vulgaris (which were present in 48 % of the nectar samples) was higher than the previously reported average of 19.9 % for Mediterranean species, the 6.5 % for Linaria viscosa in Spain [29], and closer to the 53.5 % reported from South African plant species [4]. Interestingly, we found a high incidence of the recently described bacterial species A. nectaris [29]. This bacterium was found originally in the nectar of Mediterranean plant communities [4]. However, our results indicate that it is not solely restricted to the Mediterranean area, but that it can also be found frequently in the floral nectar of plants growing in North-Western Europe. A. nectaris was previously isolated in Belgium in some orchid species, but by no means was it a nectar dominating bacterial species [6]. In another study on nectar microbial communities of P. officinalis in Belgium, it was not found either [5]. In Israel, Fridman et al. [30] found bacteria belonging to the Acinetobacter genus in at least half of the samples from each of the three Mediterranean plant species they studied. A. nectaris was later detected in nectar of Asphodelus aestivus in the same country [18]. This suggests that despite its broad distributional range, it might be plant species specific, either due to the nectar properties it requires, or due to its introduction route, which is unknown so far. It is noteworthy, however, that of the two recently described nectar specialist Acinetobacter taxa found in this study, A. nectaris was represented by 29 isolates, but A. boissieri, only by one isolate, which parallels the situation in the Metschnikowia genus, when one or the other of the two nectar specialist yeasts is usually dominant in any given plant species.
The second most abundant bacterium inhabiting the nectar of L. vulgaris was R. epipactidis, also a species previously isolated from nectar [31, 32]. Perhaps its lower abundance could be explained by its less effective dissemination method: it is speculated that thrips serve as its vectors [18]. Interestingly, although bacterial-yeast co-occurrence was common at the population level, it was less so at the plant level. This could be due to priority effects and that nectar colonized by yeasts could be less accessible to bacteria. However to confirm this, further co-occurrence studies on the nectary level are needed, as well as competition experiments [33].
Spatial distribution of yeasts and bacteria
Yeast OTU richness, measured as the number of OTUs per plant population, was not related to urbanization level (Spearman-ρ = 0, p = 1.00) nor to population size (Spearman-ρ = 0.005, p = 0.98) (Fig. 3). The number of bacterial OTUs in each L. vulgaris population was also not related to plant population size (Spearman-ρ = 0.30, p = 0.19), but was marginally negatively correlated to the ISI500 pertaining to this population (Spearman ρ = −0.43, p = 0.056) (Fig. 3). When bacterial and yeast OTU numbers were combined, the correlation with ISI500 was not significant (Spearman-ρ = −0.36, p = 0.11).
Fig. 3.
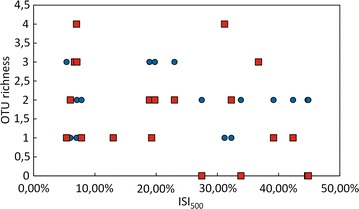
A scatter plot of urbanization versus microbial OTU richness found in the nectar of the 20 Linaria vulgaris populations located across an urbanization gradient in the vicinity of Leuven, Belgium. Urbanization was measured as an impervious surface percentage within a 500 m radius from each sampled population. Bacterial OTUs were defined as sequence clusters exhibiting at least 1 % dissimilarity from each other, and presented red squares while yeast OTUs were defined as sequence clusters exhibiting at least 3 % dissimilarity from each other, and presented as blue circles. There was no correlation between bacterial nor yeast OTU richness and urbanization
Co-occurrence of bacteria and yeasts was common at the population level, with all populations harboring yeasts, and only four lacking bacteria. A smaller degree of co-occurrence was observed at the plant level, where yeast and bacteria were found together in 33 % of the samples. A Mantel test showed no correlation between geographic distance and community dissimilarity for neither bacterial and yeast community composition separately (r = −0.08, p = 0.83 and r = 0.03, p = 0.34, respectively), nor when they were combined (r = −0.08, p = 0.82), NMDS also revealed no apparent clustering (Fig. 4). NMDS axis 1 was not correlated to ISI500 (Spearman-ρ = 0.2, p = 0.39), but NMDS axis 2 was (Spearman-ρ = 0.47, p = 0.03). Finally, the NIM communities showed significant nestedness (N = 0.83, p < 0.01; NODF = 42.19, p < 0.01), implying that species-poor communities were a subset of species-rich communities (Fig. 5). The nestedness rank was significantly correlated to ISI500 (Spearman-ρ = 0.49, p = 0.02), but not to plant population size (Spearman-ρ = −0.18, p = 0.42) (Fig. 6) . The nestedness rank correlation with ISI500 has to be treated with caution, since the rarefaction curves were not completely saturated, especially for bacteria (Figs. 1, 2). Additional OTUs would likely appear if the number of samples increased.
Fig. 4.
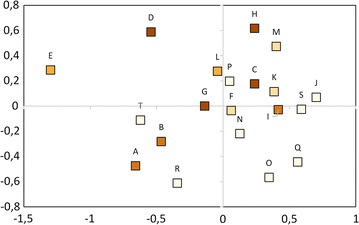
NMDS ordination of the total microbial community composition (bacteria and yeasts) in the floral nectar of 20 Linaria vulgaris populations located across an urbanization gradient in the vicinity of Leuven, Belgium. The populations have been color coded to represent the degree of impervious surfaces within 500 m radius of each population, as in Fig. 7
Fig. 5.
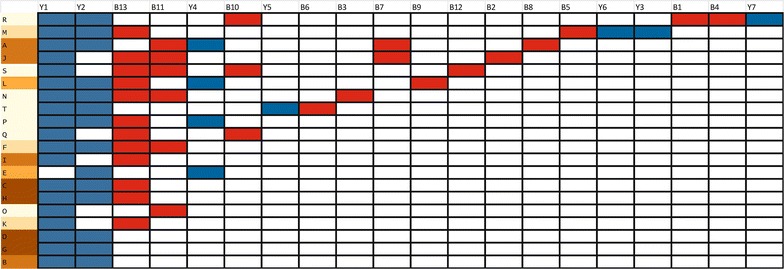
Matrix of incidence of nectar microbial communities in the 20 Linaria vulgaris populations located across an urbanization gradient in the vicinity of Leuven, Belgium. Rows are populations and columns are OTUs. The populations have been color coded to represent the degree of impervious surfaces within 500 m radius of each population, as in Fig. 7. The matrix was sorted according to ‘binmatnest’ algorithm as implemented in the R package ‘bipartite’. The presence of yeast communities is indicated in blue, while presence of bacterial communities is indicated in red. The communities showed significant nestedness (T = 16.81, p < 0.01, NODF = 42.19, p < 0.01), and their nestedness rank was correlated with impervious surface index (Spearman-ρ = 0.49, p = 0.02), which implies that nectar urban microbial communities are a subset of nectar rural microbial communities
Fig. 6.
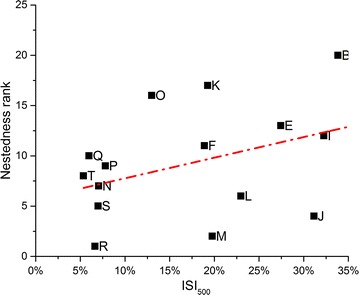
A scatter plot of urbanization versus nestedness rank of communities inhabiting each of the 20 Linaria vulgaris populations located across an urbanization gradient in the vicinity of Leuven, Belgium. Urbanization was measured as an impervious surface percentage within a 500 m radius from each sampled population. The nestedness rank was assigned according to ‘binmatnest’ algorithm as implemented in the R package ‘bipartite’. Urbanization and nestedness rank of microbial communities showed significant correlation (Spearman-ρ = 0.49, p = 0.02)
In contrast to our hypothesis, we did not find any clear changes in microbial incidence per se in the nectar of L. vulgaris across the studied urbanization gradient. OTU richness of urban and non-urban nectar communities was similar for yeasts, whereas urban bacterial nectar communities were marginally poorer than non-urban ones. Similarity in community composition did also not decrease with geographic distance, and no apparent clustering was detected, suggesting that the species pools from which NIMs colonize their nectary microhabitats are homogeneously throughout the area. These results further suggest that pollinators were still present in urban environments in sufficient numbers to effectively disperse bacteria and yeast. Moreover, the most urbanized plant populations we studied were still located in sites with only 45 % of impervious surface coverage within 500 m. It is therefore not impossible that the effect of such moderate urbanization on pollinator guilds is not drastic, and that microbial frequency would decrease sharply in more extremely urbanized areas.
On the other hand, the studied communities showed significant nestedness, indicating that OTU-poor communities were a subset of OTU-rich communities, and that generalist species occurred everywhere and specialist species were restricted to particular populations. The significant albeit weak correlation between urbanization and nestedness rank, and the lack of such correlation with plant population size may imply that the urban communities were a subset of the rural communities. Because many of the L. vulgaris populations located in sites with very low impervious surface coverage within 500 m were geographically relatively close to each other (Fig. 7), and because rarefaction analyses revealed potentially incomplete sampling, this correlation could be a result of some unknown spatial effect unrelated to urbanization. However, such correlation could also suggest that populations in urban environments may be less frequently visited by pollinators due the higher isolation of Linaria populations in urban environments or the lack of other co-flowering plant species that attract pollinators, which in turn limits the exchange of less prevalent species. Alternatively, these results may suggest that nectar conditions in urban environments are less suitable for microbial growth than in natural environments and that only generalist species are capable of growing in the floral nectar of plants growing in urban environments. This result is consistent with the hypothesis that nectar yeasts should be capable of exploiting a wide range of nectar microhabitats, as they rank as two first entries in the nestedness matrix (Pozo et al. [17]). Interestingly, most bacteria, including nectar specialists, rank further in the nestedness matrix, which suggests that they are less adapted to different nectar types than yeasts.
Fig. 7.
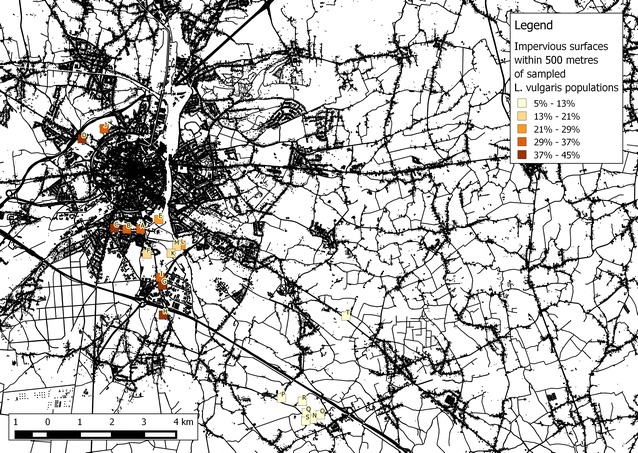
Map showing the locations of the 20 Linaria vulgaris populations studied in the vicinity of Leuven, Belgium. These populations have been color coded to represent the degree of impervious surfaces within 500 m radius of each population. From these populations, five shoots were chosen at random, and a single inflorescence was taken from each plant. Nectar was collected from four flowers belonging to each inflorescence
Conclusion
Overall, our results show that nectar yeasts are able to grow well in anthropogenically transformed environments. They grow equally well in autumn-flowering species L. vulgaris as in species that bloom earlier in the season on which previous studies were focused: Helleborus foetidus, Pulmonaria officinalis, Delphinium nuttalianum or Asphodelus aestivus. Moreover, our results indicate that not only nectar yeast communities, but also nectar bacterial communities are species poor and dominated by two specialist genera that seem to have a broad geographic range. Although urbanization did not seem to affect overall microbial richness nor incidence, microbial communities inhabiting the nectar of urban populations of L. vulgaris were nested in the communities inhabiting the nectar of its rural populations, although this result has to be treated with caution due to sampling limitations. Nevertheless, this might mean that urbanization may increase the isolation of plant populations harboring nectar microhabitats, resulting in reduced prevalence of rare species. Further research in this field will need to take into account two aspects that were outside of the scope of this study. Firstly, visitation rates, community composition and abundance of pollinators will have to be established more precisely in the chosen study system to give reliable estimations of their influence on nectar communities. Secondly, it is known that but a fraction of microbes can be successfully cultivated [34]. This is also reflected by the fact that rarefaction curves in studies relying on culture dependent methods are frequently not saturated, which decreases the level of confidence one can put into conclusions drawn from OTU richness and diversity. Perhaps more pronounced effects of urbanization could be detected with a more sensitive technique to measure their community composition, for instance through next-generation sequencing.
Methods
Study species and study area
Linaria vulgaris is a self-incompatible, perennial herb that is characterized by an extensive root system. It produces zygomorphic, bright yellow flowers with an orange nectar guide and a long spur between summer and late autumn [35]. The species is mainly pollinated by long-tongued bumblebees, such as B. hortorum and B. pascuorum [36, 37]. Short-tongued species, such as B. terrestris, behave like nectar robbers and access nectar illegitimately by biting a hole in the spur of a Linaria flower [37]. The main sugar found in L. vulgaris nectar is sucrose, but glucose, fructose and even trace amounts of raffinose have also been found [35].
Twenty L. vulgaris populations that grew in locations exhibiting different levels of urbanization were examined (Table 3). No sampling permissions were required to access the populations and sample them. The study area consisted of the city of Leuven (situated about 25 km east of Brussels, Belgium) and its immediate suburbs that gradually shift into areas with a more rural character. To assess the level of urbanization of the locations where each L. vulgaris population grew, for every of these locations we calculated an impervious surface index within a radius of 500 m (ISI500), as described previously [38] to serve as a proxy for urbanization (Fig. 7). In some cases, the 500 m radii overlapped. Some suburban areas also showed considerable amount of impervious surfaces within the 500 m radius, and no suitable populations were found in a 3 km stretch between populations P and H. Populations T, D and J were located outside of a linear transect encompassing all of the other populations. Given these deviations, our design was an approximation of sampling along an idealized urban–rural gradient. For each population, we also assessed its size by counting the number of inflorescences. Although the number of inflorescences does not necessarily reflect the total number of genets in the population, it does reflect the number of flowers that is perceived by pollinators.
Sampling and cultivation of microorganisms
From each population, five plants were randomly chosen, and a single inflorescence per plant was brought to the laboratory for further processing on the same day. Given that NIMs are often patchily distributed among flowers within an inflorescence [39], nectar from four flowers from one inflorescence was pooled. Within 8 h of collection, nectar was harvested. To this end, the spur was first cut with a sterile scalpel in order to avoid pollen contamination, and then 0.5 µl nectar was extracted using a micropipette. A total of 2 µl, pooled from four flowers, was diluted in 98 µl of water and kept at 4 °C. Within 24 h, 2 µl of the diluted nectar was plated on tryptone soy agar (TSA) supplemented with 0.01 % chloramphenicol or 0.01 % actidione in order to suppress the growth of bacteria and yeasts, respectively. The plates were incubated at 25 °C for 7 days. Subsequently, one colony of each morphotype per plate was isolated and purified on TSA. It has been shown in previous studies on nectar microbiota that identical morphotypes generally correspond to identical species [5, 6]. After purification, each isolate was again transferred to TSA after which it was stored at −80 °C in 96 well plates containing 40 % glycerol.
DNA extraction
Genomic DNA was extracted according to the procedure described by Pozo et al. [17]. Subsequently, partial ribosomal RNA genes were amplified using the primer pairs 27F/1492R and NL1/NL4 for bacteria and yeasts, respectively [4, 40]. When amplification failed using 27F/1492R, the forward primer 63F [41] was used instead of 27F. PCR amplification was performed using a Bio-Rad T100 thermal cycler in a reaction volume of 20 µl containing 0.5 µM of each primer, 0.25 mM of each dNTP, 1.25 units Takara Taq DNA polymerase, 1 × Takara Taq PCR buffer (Clontech Laboratories, Palo Alto, CA, USA), and 5 ng genomic DNA (as measured by a Nanodrop spectrophotometer). The reaction mixture was initially denatured at 94 °C for 2 min, followed by 35 cycles of 45 s at 94 °C, 45 s at 55 °C (yeasts) or 59 °C (bacteria), and 45 s at 72 °C, with a final extension at 72 °C for 10 min. Subsequently, obtained amplicons were sequenced by Macrogen Inc. using the same reverse primers as those used for amplification.
Data analysis
Obtained sequences were individually trimmed for quality, using a minimum Phred score of 20, and, in cases of ambiguous base calls, manually edited based on the obtained electropherograms. Subsequently, each sequence was assigned a taxonomic identity based on BLAST [42] results using the GenBank nucleotide (nt) database [43], excluding environmental/uncultured samples. Next, the bacterial and yeast sequences were separately aligned using the MUSCLE algorithm implemented in Geneious 7R. These alignments have been used to create genetic distance matrices, which served as a basis to assign the sequences to OTUs using Mothur v 1.32.1 at 1 % and 3 % dissimilarity cutoffs for bacteria and yeasts, respectively. For each OTU, a representative sequence as determined by Mothur has been deposited in GenBank (accession numbers: KT347518–KT347530 for bacteria and KU900119–KU900125 for yeast).
Using EstimateS v 9.1.0, rarefaction analyses were conducted to assess our sampling effort. EstimateS was also used to calculate the ICE and Chao2 estimators of species richness [44]. For each L. vulgaris population, OTU richness and incidence were calculated. Incidence in a given population was expressed as percentage of samples that contained microorganisms. OTU richness was defined as the total number of OTUs per given population. Both of these variables were related to the urbanization measure ISI500 and plant population size using a Spearman rank correlation.
To visualize differences in microbial community composition among populations, we applied the non-metric multidimensional scaling (NMDS) ordination technique using the vegan package [45] in R software. As distance measure, we used the Bray-Curtis coefficient. EstimateS software was used to create a dissimilarity in community composition matrix, using the Jaccard estimator. A Mantel test using the R software was then conducted to relate the dissimilarities in community composition with geographical distances between populations. The significance of the Mantel test was assessed by performing 9999 permutations.
Lastly, we tested the hypothesis that NIM communities were significantly nested, i.e. that species-poor communities were a subset of the more rich ones and that rare OTUs were only present in the most OTU rich communities [46–48]. Two different measures to estimate the degree of nestedness were applied. We first calculated N = (100−T)/100, where T is the matrix temperature, a measure of matrix disorder that varies between 0° (perfectly nested) and 100° (perfectly non-nested). Values of N close to one thus indicate a high degree of nestedness. However, because T may be dependent on the size and shape of the species presence matrix, we also calculated a second nestedness measure, based on overlap and decreasing fill (NODF), correcting for these flaws [48]. Conversely to T, high values of NODF signify a nested matrix structure.
Two different null models implemented in ANINHADO were used to test the significance of nestedness [47]. In the first null model, each cell in the interaction matrix has the same probability of being occupied. This null model is very general and does not take into account the fact that the number of connections per species may vary substantially. A more conservative null model would therefore be a model in which the probability of drawing an interaction is proportional to the degree of specialization [49]. In this null model, the probability of each cell being occupied is the average of the probabilities of occupancy of its row and column [48]. Finally, to identify the potential drivers behind the nestedness of our dataset, a nestedness rank was assigned to each sampling site according to the ‘binmatnest’ algorithm in the R package bipartite. Afterwards, plant population size and ISI500 was related to nestedness rank using the Spearman rank correlation.
Authors’ contributions
Conceived and designed the experiments: JB, BL, OH, HJ. Conducted the experiments: JB. Performed the analysis: JB. Wrote the paper: JB, BL, OH, HJ. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank Dr. Maria Pozo, the Editor and two Anonymous reviewers whose comments greatly improved this manuscript. The authors would also like to thank Belgian science policy office for providing funding necessary to carry out this investigation within the SPatial and Environmental determinants of Eco-Evolutionary dynamics (SPEEDY) project.
Availability of data and materials
Most data is available in the manuscript itself. The sequences of representative OTUs are available in GenBank. The dataset supporting the conclusions of this article is available in the Dryad repository doi:10.5061/dryad.2440v.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- BLAST
basic local alignment search tool
- ISI500
impervious surface index within 500 m radius
- OTU
operational taxonomic unit
- NIMs
nectar inhabiting microbes
- NMDS
non metric multidimensional scaling
- NODF
nestedness metric based on overlap and decreasing fill
- PCR
polymerase chain reaction
- TSA
tryptone soya agar
Contributor Information
Jacek Bartlewicz, Phone: +32 (0)16 32 15 20, Email: bartlewiczjacek@gmail.com.
Bart Lievens, Email: bart.lievens@kuleuven.be.
Olivier Honnay, Email: olivier.honnay@kuleuven.be.
Hans Jacquemyn, Email: hans.jacquemyn@kuleuven.be.
References
- 1.De Vega C, Herrera CM, Johnson SD. Yeasts in floral nectar of some South African plants: quantification and associations with pollinator type and sugar concentration. S Afr J Bot. 2009;75:798–806. doi: 10.1016/j.sajb.2009.07.016. [DOI] [Google Scholar]
- 2.Herrera CM, De Vega C, Canto A, Pozo M. Yeasts in floral nectar: a quantitative survey. Ann Botany. 2009;103:1415–1423. doi: 10.1093/aob/mcp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golonka AM, Vilgalys R. Nectar inhabiting yeasts in Virginian populations of Silene latifolia (Caryophyllaceae) and coflowering species. Am Mid Nat. 2013;169:235–258. doi: 10.1674/0003-0031-169.2.235. [DOI] [Google Scholar]
- 4.Alvarez-Pérez S, Herrera CM, De Vega C. Zooming-in on floral nectar: a first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol Ecol. 2012;80:591–602. doi: 10.1111/j.1574-6941.2012.01329.x. [DOI] [PubMed] [Google Scholar]
- 5.Jacquemyn H, Lenaerts M, Brys R, Willems K, Honnay O, Lievens B. Among-population variation in microbial community structure in the floral nectar of the bee-pollinated forest herb Pulmonaria officinalis L. PLoS ONE. 2013;8:e56917. doi: 10.1371/journal.pone.0056917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacquemyn H, Lenaerts M, Tyteca D, Lievens B. Microbial diversity in the floral nectar of seven Epipactis (Orchidaceae) species. Microbiol Open. 2013;2:644–658. doi: 10.1002/mbo3.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lievens B, Hallsworth JE, Pozo MI, Belgacem ZB, Stevenson A, Willems KA, Jacquemyn H. Microbiology of sugar-rich environments: diversity, ecology and system constraints. Environ Microbiol. 2015;17:278–298. doi: 10.1111/1462-2920.12570. [DOI] [PubMed] [Google Scholar]
- 8.Pozo MI, Lievens B, Jacquemyn H. Impact of microorganisms on nectar chemistry, pollinator attraction and plant fitness. In: Peck RL, editor. Nectar: production, chemical composition and benefits to animals and plants. 1. New York: Nova Publishers; 2015. pp. 1–45. [Google Scholar]
- 9.Peay KG, Belisle M, Fukami T. Phylogenetic relatedness predicts priority effects in nectar yeast communities. Proc Biol Sci. 2012;279:749–758. doi: 10.1098/rspb.2011.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrera CM, Garcia IM, Perez R. Invisible floral larcenies: microbial communities degrade floral nectar of bumble bee-pollinated plants. Ecology. 2008;89:2369–2376. doi: 10.1890/08-0241.1. [DOI] [PubMed] [Google Scholar]
- 11.Goodrich KR, Zjhra ML, Ley CA, Raguso RA. When flowers smell fermented: the chemistry and ontogeny of yeasty floral scent in pawpaw (Asimina triloba : Annonaceae) Int J Plant Sci. 2006;167:33–46. doi: 10.1086/498351. [DOI] [Google Scholar]
- 12.Pozo MI, De Vega C, Canto A, Herrera CM. Presence of yeasts in floral nectar is consistent with the hypothesis of microbial-mediated signaling in plant-pollinator interactions. Plant Signal Behav. 2009;4:1102–1104. doi: 10.4161/psb.4.11.9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera CM, Pozo MI. Nectar yeasts warm the flowers of a winter-blooming plant. Proc R Soc Lond. 2010;277:1827–1834. doi: 10.1098/rspb.2009.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaeffer RN, Irwin RE. Yeasts in nectar enhance male fitness in a montane perennial herb. Ecology. 2014;95:1792–1798. doi: 10.1890/13-1740.1. [DOI] [PubMed] [Google Scholar]
- 15.Vannette RL, Gauthier M-PL, Fukami T. Nectar bacteria, but not yeast, weaken a plant–pollinator mutualism. Proc Biol Sci. 2013;280:20122601. doi: 10.1098/rspb.2012.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrera CM, Canto A, Pozo M, Bazaga P. Inhospitable sweetnes: nectar filtering of pollinator borne inocula leads to impoverished, phylogenetically clustered yeast communities. Proc R Soc Lond. 2010;277:747–754. doi: 10.1098/rspb.2009.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pozo MI, Herrera CM, Bazaga P. Species richness of yeast communities in floral nectar of southern Spanish plants. Microb Ecol. 2011;61:82–91. doi: 10.1007/s00248-010-9682-x. [DOI] [PubMed] [Google Scholar]
- 18.Samuni-Blank M, Izhaki I, Laviad S, Bar-Massada A, Gerchman Y, Halpern M. The role of abiotic environmental conditions and herbivory in shaping bacterial community composition in floral nectar. PLoS ONE. 2014;9:e99107. doi: 10.1371/journal.pone.0099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lachance M-A, Starmer WT, Rosa CA, Bowles JM, Barke J, Stuart F, Janzen DH. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 2001;1:1–8. doi: 10.1111/j.1567-1364.2001.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 20.Aizenberg-Gershtein Y, Izhaki I, Halpern M. Do honeybees shape the bacterial community composition in floral nectar? PLoS ONE. 2013;8:e67556. doi: 10.1371/journal.pone.0067556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jha S, Kremen C. Resource diversity and landscape-level homogeneity drive native bee foraging. Proc Natl Acad Sci USA. 2013;110:555–558. doi: 10.1073/pnas.1208682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortel L, Henry M, Guilbaud L, Guirao AL, Kuhlmann M, Mouret H, Rollin O, Vaissiere BE. Decreasing abundance, increasing diversity and changing structure of the wild bee community (Hymenoptera: Anthophila) along an urbanization gradient. PLoS ONE. 2014;9:e104679. doi: 10.1371/journal.pone.0104679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banaszak-Cibicka W, Żmihorski M. Wild bees along an urban gradient: winners and losers. J Insect Conserv. 2012;16:331–343. doi: 10.1007/s10841-011-9419-2. [DOI] [Google Scholar]
- 24.Ulrich W, Almeida-Neto M, Gotelli NJ. A consumer’s guide to nestedness analysis. Oikos. 2009;118:3–17. doi: 10.1111/j.1600-0706.2008.17053.x. [DOI] [Google Scholar]
- 25.Giménez-Jurado G. Metschnikowia gruessi sp. nov., the teleomorph of Nectaromyces reukaufii but not a Candida reukaufii. Syst Appl Microbiol. 1992;15:432–438. doi: 10.1016/S0723-2020(11)80218-3. [DOI] [Google Scholar]
- 26.Brysch-Herzberg M. Ecology of yeasts in plant–bumblebee mutualism in Central Europe. FEMS Microbiol Ecol. 2004;50:87–100. doi: 10.1016/j.femsec.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Álvarez-Pérez S, Herrera C. Composition, richness and nonrandom assembly of culturable bacterial-microfungal communities in floral nectar of Mediterranean plants. FEMS Microbiol Ecol. 2013;83:685–699. doi: 10.1111/1574-6941.12027. [DOI] [PubMed] [Google Scholar]
- 28.Pozo MI, Herrera CM, Van den Einde W, Verstrepen K, Lievens B, Jacquemyn H. The impact of nectar chemical features on phenotypic variation in two related nectar yeasts. FEMS Microbiol Ecol. 2015;91:fiv055. doi: 10.1093/femsec/fiv055. [DOI] [PubMed] [Google Scholar]
- 29.Álvarez-Pérez S, Lievens B, Jacquemyn H, Herrera C. Acinetobacter nectaris sp. nov. and Acinetobacter boissieri sp. nov., two novel bacterial species isolated from floral nectar of wild Mediterranean insect-pollinated plants. Int J Syst Evol Microbiol. 2013;63:1532–1539. doi: 10.1099/ijs.0.043489-0. [DOI] [PubMed] [Google Scholar]
- 30.Fridman S, Izhaki I, Gercham Y, Halpern M. Bacterial communities in floral nectar. Environ Microbiol Rep. 2012;4:97–104. doi: 10.1111/j.1758-2229.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- 31.Halpern M, Fridman S, Atamna-Ismaeel N, Izhaki I. Rosenbergiella nectarea gen. nov., sp. nov., in the family Enterobacteriaceae, isolated from floral nectar. Int J Syst Evol Microbiol. 2013;63:4259–4265. doi: 10.1099/ijs.0.052217-0. [DOI] [PubMed] [Google Scholar]
- 32.Lenaerts M, Alvarez-Perez S, De Vega C, Van Assche A, Johnson SD, Willems KA, Herrera CM, Jacquemyn H, Lievens B. Rosenbergiella australoborealis sp. nov., Rosenbergiella collisarenosi sp. nov. and Rosenbergiella epipactidis sp. nov., three novel bacterial species isolated from floral nectar. Syst Appl Microbiol. 2014;37:402–411. doi: 10.1016/j.syapm.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Vannette RL, Fukami T. Historical contingency in species interactions: towards niche-based predictions. Ecol Lett. 2014;17:115–124. doi: 10.1111/ele.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart EJ. Growing unculturable bacteria. J Bacteriol. 2012;194:4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nepi M, Pacini E, Nencini C, Collavoli E, Franchi GG. Variability of nectar production and composition in Linaria vulgaris (L.) Mill. (Scrophulariaceae) Plant Syst Evol. 2003;238:109–118. doi: 10.1007/s00606-002-0275-0. [DOI] [Google Scholar]
- 36.Corbet SA, Bee J, Dasmahapatra K, Gale S, Gorringe E, La Ferla B, Moorhouse T, Trevail A, Van Bergen Y, Vorontsova M. Native or exotic? Double or single? Evaluating plants for pollinator-friendly gardens. Ann Botany. 2001;87:219–232. doi: 10.1006/anbo.2000.1322. [DOI] [PubMed] [Google Scholar]
- 37.Stout JC, Allen JA, Goulson D. Nectar robbing, forager efficiency and seed set: bumblebees foraging on the self incompatible plant Linaria vulgaris (Scrophulariaceae) Acta Oecolo. 2000;21:277–283. doi: 10.1016/S1146-609X(00)01085-7. [DOI] [Google Scholar]
- 38.Bartlewicz J, Vandepitte K, Jacquemyn H, Honnay O. Population genetic diversity of the clonal self-incompatible herbaceous plant species Linaria vulgaris along an urbanization gradient. Biol J Linnean Soc. 2015 [Google Scholar]
- 39.Pozo MI, Herrera CM, Alonso C. Spatial and temporal distribution patterns of nectar-inhabiting yeasts: how different floral microenvironments arise in winter-blooming Helleborus foetidus. Fungal Ecol. 2014;11:173–180. doi: 10.1016/j.funeco.2014.06.007. [DOI] [Google Scholar]
- 40.O’Donnell K. Fusarium and its near relatives. In: REYNOLDS, D. & TAYLOR, J. (eds.) The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics. Wallingford: CAB International: 1993. p. 000-000.
- 41.Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry C, Hiom SJ, Wade WG. Design and Evaluation of Useful Bacterium-Specific PCR Primers That Amplify Genes Coding for Bacterial 16S rRNA. Appl Environ Microb. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J. Sayers EW. Nucleic Acids Res D. 2013 doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colwell RK EstimateS: statistical estimation of species richness and shared species from samples. 2013; Version 9. – User’s Guide and application at < http://purl.oclc.org/estimates > .
- 45.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. Vegan: Community Ecology Package. 2013; R package version 2.0-10. http://CRAN.R-project.org/package=vegan.
- 46.Atmar W, Patterson BD. The measure of order and disorder in the distribution of species in fragmented habitat. Oecologia. 1993;96:373–382. doi: 10.1007/BF00317508. [DOI] [PubMed] [Google Scholar]
- 47.Guimaraes PR, Guimaraes P. Improving the analyses of nestedness for large sets of matrices. Environ Modell Softw. 2006;21:1512–1513. doi: 10.1016/j.envsoft.2006.04.002. [DOI] [Google Scholar]
- 48.Almeida-Neto M, Guimaraes P, Guimaraes PR, Loyola RD, Ulrich W. A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos. 2008;117:1227–1239. doi: 10.1111/j.0030-1299.2008.16644.x. [DOI] [Google Scholar]
- 49.Bascompte J, Jordano P, Melián CJ, Olesen JM. The nested assembly of plant-animal mutualistic networks. Proc of the Nat Aca of Sci USA. 2003;100:9383–9387. doi: 10.1073/pnas.1633576100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Most data is available in the manuscript itself. The sequences of representative OTUs are available in GenBank. The dataset supporting the conclusions of this article is available in the Dryad repository doi:10.5061/dryad.2440v.


