Abstract
With increasing structural information on proteins, the opportunity to understand physical forces governing protein folding is also expanding. One of the significant non‐covalent forces between the protein side chains is aromatic–aromatic interactions. Aromatic interactions have been widely exploited and thoroughly investigated in the context of folding, stability, molecular recognition, and self‐assembly processes. Through this review, we discuss the contribution of aromatic interactions to the activity and stability of thermophilic, mesophilic, and psychrophilic proteins. Being hydrophobic, aromatic amino acids tend to reside in the protein hydrophobic interior or transmembrane segments of proteins. In such positions, it can play a diverse role in soluble and membrane proteins, and in α‐helix and β‐sheet stabilization. We also highlight here some excellent investigations made using peptide models and several approaches involving aryl–aryl interactions, as an increasingly popular strategy in protein and peptide engineering. A recent survey described the existence of aromatic clusters (trimer, tetramer, pentamer, and higher order assemblies), revealing the self‐associating property of aryl groups, even in folded protein structures. The application of this self‐assembly of aromatics in the generation of modern bionanomaterials is also discussed.
Keywords: aromatic cluster, side chain interaction forces, membrane proteins, peptide model, protein engineering, extremophiles, bionanomaterials
Abbreviations
- CRABP1
cellular retinoic acid‐binding protein 1
- FtE
face‐to‐edge.
Introduction
The fundamental principle of how a linear polypeptide folds into a compact three‐dimensional structure is a central question that has become paramount with its increasing correlation to several diseases caused by protein misfolding.1, 2 With the development of advanced tools and techniques, such as high‐resolution NMR spectroscopy, molecular imaging, single molecule spectroscopy, improved crystallization methods for difficult biomolecules including membrane proteins, the understanding of protein dynamics and functions through structural information is rapidly expanding. The eventual goal is to understand the physical forces that govern protein folding and function, and which can then be utilized in modeling de novo biomolecular structures for medical applications. One such non‐covalent force is aromatic interactions.
Aryl interaction geometries are not only under active investigation but are also mostly exploited in biomolecule folding, stability, recognition, and molecular self‐assembly. Since the first report highlighting the potential significance of aromatic‐aromatic interactions in proteins was recognized in 1985,3 numerous investigations have drawn attention to the importance of π–π interactions involving phenylalanine, tyrosine, and tryptophan. Through this review, we recapitulate various facets of aromatic interactions, their contribution to the structural stability of proteins and peptides, and finally discuss the application of aromatics in synthetic biomolecules.
Noncovalent Interactions Involving Aromatic Amino Acids
Non‐covalent interactions involving aromatic amino acids are ubiquitous in nature, and facilitate most of the chemical and biological processes. For example, the interactions of aromatic amino acids with nucleobases, forming protein/DNA or protein/RNA complexes, are crucial in many processes, particularly transcription and translation.4 A statistical analysis by Baker and Grant revealed that the interaction involving phenylalanine in protein‐DNA complexes and tryptophan in protein‐RNA complexes are essential for initiation of transcription.5 It was purported that this difference in the choice of aromatic amino acid also allows the protein to differentiate the two nucleic acid types.
Carbohydrate‐protein interactions are another class of non‐covalent interactions through which one cell communicate with another cell. With considerable evidence for the occurrence of aromatic amino acids in the carbohydrate binding sites of proteins, the significance of C‐H···π interaction in sugar recognition is now widely acknowledged.6 This knowledge can be exploited in the design of novel carbohydrate receptors. Excellent articles and reviews have discussed carbohydrate‐pi interactions in detail.6, 7, 8
Pioneering work on cation‐pi interaction by the Dougherty group has led to its recognition as a significant non‐covalent force by which proteins mediate functional activities, such as ligand‐receptor interactions.9, 10, 11 For instance, nicotine binds to acetylcholine receptor through cation‐pi interactions. Multiple cation‐pi interactions also provide stability to protein structures.10 While the role of cation‐pi interaction is well established, with our increasing understanding of anion receptors and transporters, anion‐pi interactions are also being considered as key players. An excellent tutorial review on anion‐pi interaction has been written by Dunbar and co‐workers.12
Sulfur‐pi interactions are yet another class of non‐covalent interaction involving aromatic residues. They are prevalent in protein structures and contribute significantly to protein stability and recognition. A recent survey by Valley et al., using a 7 Å distance cut‐off, reported that 33% of protein structures had at least one Met sulfur‐aromatic motif.13 The stabilizing contribution of this interaction is comparable with that of a salt bridge. Most importantly, however, sulfur‐aromatic interactions can also occur at a distance much larger than the salt bridge. The geometric modes and role of sulfur‐aromatic interactions have been extensively discussed.14, 15
CH···π interactions involving aromatic residues are increasingly being identified in proteins. Although small and weak in magnitude, these interactions can act together to contribute in the folding and stability of proteins, and have a significant additive effect. Using NMR spectroscopy, Boisbouvier and co‐workers16 provided direct evidence of CH···π interaction between the side chains of aliphatic methyl group and aromatic residues in proteins. Using the general peptide sequence Ac‐TXPN‐NH2 and Ac‐PPX‐NH2 (where X = aromatic residue), Basu17 and Zondlo18, respectively, reported the extensive investigation of CH···π interactions between proline and aromatic residues. They provided direct evidence for the existence of CH···π interaction in these peptides, and their importance in stabilizing Pro‐cisPro peptide bonds. A recent study from our laboratory using NMR spectroscopy of de novo designed peptides also supported the structure stabilizing role of CH···π interactions.19 Figure 1 illustrates representative CH···π interactions seen in model systems.
Figure 1.

CH···π interactions as a class of non‐covalent interaction seen in proteins. CH···π interactions contribute to three‐dimensional structure, stability and in biomolecular functioning of the protein. Reprinted (adapted) with permission from Zondlo, Acc Chem Res, 2013, 46, 1039‐1049, © ACS Publications 18 for (a). Reprinted (adapted) with permission from Ganguly et al., J Am Chem Soc, 2012, 134, 4661‐4669, © ACS Publications 17 for (b). Reprinted (adapted) with permission from Makwana and Mahalakshmi, J Phys Chem B, 2015, 119, 5376–5385, © ACS Publications 19 for (c).
The striking feature that makes aromatic‐aromatic interactions unique candidates for investigation is the geometrical ways in which aromatic rings can interact with each other. Such stereospecific interactions can provide specificity to the protein in its folding, stability and recognition. An early survey of aromatic pairs by Burley and Petsko in 1985 reported that “phenyl ring centroids are separated by a preferential distance of between 4.5‐7.0 Å, and dihedral angles approaching 90° are most common”.3 Later in the same year, Singh and Thornton also found from the survey of protein structures that the geometry of Phe‐Phe interaction has a preference for the perpendicular arrangement.20 But they also concluded that “interaction with other side chains can interfere with and obviously overcome the preference for perpendicular interaction between the aromatic rings”.20
Hunter et al.,21, 22 found a good correlation when comparing the electrostatic interaction between benzene molecules as a function of orientation, with the experimental geometries of the Phe‐Phe interaction in proteins surveyed by Singh and Thornton (Fig. 2). Completely stacked arrangements resulting in repulsive interactions were found to be unfavorable, while a full range of edge‐to‐face and offset stacked orientation resulting in attractive interactions were seen as favorable.21, 22 Not surprisingly, Sun and Bernstein discussed from theoretical work on the benzene dimer that aromatic dimers are better viewed as dynamic systems, and should not be represented as well‐defined structures.23
Figure 2.

Benzene and Phe interaction geometries. (a) Variation of the electrostatic interaction calculated for a benzene dimer as a function of orientation and distance. The shaded region corresponds to favorable or attractive forces and the unshaded regions are unfavorable or repulsive. (b) Phe‐Phe geometries in proteins, as observed by Singh and Thornton20. Reproduced with permission from Hunter, J Mol Biol, 1991, 218, 837‐846, © Elsivier.21
From Psychrophilic to Thermophilic Proteins
Psychrophilic and thermophilic microorganisms predominantly inhabit cold and hot environments on earth, respectively. Their ability to survive in such extreme conditions is attributed to their repertoire of proteins.24 In general, when the structures of a protein homolog from thermophilic, mesophilic, and psychrophilic microorganisms are superposed, we find that a similarly folded conformation is retained across the three proteins. A closer examination of the structures, however, reveals interesting differences on structural features that thermophilic proteins have evolved to function at high temperatures. A greater number of interactions, such as salt bridges, disulfides, hydrophobic interactions etc., tight packing, greater structural content and shorter loops, etc., are seen in these proteins.25, 26 A study compiled by Siddiqui and Cavicchioli27 and Feller28 suggested the optimization to low‐temperature is evolutionary achieved by adopting structural features that result in the weakening of almost all types of interactions, particularly aromatic‐aromatic interactions, for improved structural flexibility in the cold‐adapted enzymes.29
The number and strengths of covalent and non‐covalent interactions vary between proteins of thermophilic, mesophilic, and psychrophilic origins, and all of these interactions are important in achieving the observed characteristics (structure, stability and dynamicity, and function) of these proteins. However, in keeping with the focus of this review, we only compare the role of aromatic interactions across these three classes of proteins. For example, in the case of cold‐adapted subtilisin, a non‐specific serine protease, the protein lacks 11 aromatic‐aromatic interactions when compared with its thermophilic homolog.30 Figure 3 highlights one such cluster comparing aromatic interactions across thermophilic, mesophilic and psychrophilic subtilisin. Feller and co‐workers substituted specific amino acids in psychrophilic α‐amylase with the corresponding residues found in its thermophilic homolog. They found that these substitutions resulted in increased thermodynamic stability, compactness, and resistance towards chemical denaturation, at the expense of kinetic (functional) optimization to low temperature activity.31 Another study on T4 lysozyme (of mesophilic origin) showed that the introduction of an aromatic amino acid increased protein stability at the expense of function,32 but this is context‐dependent, as demonstrated in another study where the incorporation of an aromatic amino acid was destabilizing.33
Figure 3.

Aromatic‐aromatic interactions observed across thermophilic, mesophilic, and psychrophilic subtilisin structures. A representative cluster comparing aromatic interactions is shown. PDB IDs are provided in the legend.
Many studies have been carried out with the aim of increasing the thermostability of enzymes, owing to their greater industrial applications. Kannan and Vishveshwara systematically investigated the role of aromatic interactions in thermostability of thermophilic proteins.34 They analyzed 24 protein families and compared the number of aromatic clusters in thermophiles to its mesophilic homologs. They found that 17 of the 24 thermophilic protein families showed an increase in aromatic residues. Ten protein families had greater aromatic clusters in thermophiles, and these additional clusters were found to be surface exposed. An additional seven of the thermophilic proteins had a larger number of aromatic residues in the cluster, which resulted in expanded aromatic clusters. Three had equal number of aromatic residues in protein families from thermophiles and mesophiles. Mesophilic proteins had more aromatic clusters in only four families. They attributed the high thermophilic nature of these proteins to the possession of at least one additional aromatic cluster, situated proximal to the active site of the thermophile.
These investigations, therefore, suggest the analogous contributions of aromatic–aromatic interactions: thermodynamically favorable for stability and kinetically adjusted for functionality. However, as mentioned earlier, other non‐covalent interactions and disulfide bonds, are also very important in stabilizing thermophilic proteins,25, 26, 35 and these interactions cannot be ignored. Overall, an optimal balance of aromatic‐aromatic interactions would, therefore, help in the rational design of peptide or protein structures with temperature‐sensitive properties.
Similarities and Differences in Aromatics of Soluble and Membrane Proteins
The process of sequestering apolar residues from the outer environment, termed as the hydrophobic effect, is the fundamental principle by which most of the proteins achieve the final compactly folded three‐dimensional structure.36, 37 The environment in which soluble protein exist (aqueous medium) is significantly different from where the membrane proteins reside (lipid system). Thus, while the overall hydrophobicities of the membrane and water‐soluble proteins may be quite similar, the difference arises from the surface‐exposed residues (Fig. 4).38 The membrane‐exposed residues in membrane proteins are more hydrophobic than the buried interior and the aqueous‐exposed residues in water soluble proteins are more hydrophilic than buried interiors. This observation had led to the belief that membrane proteins are “inside‐out”. However, studies on membrane protein structures that share similar scaffolds with soluble proteins indicate that optimal van der Waal's forces, and not hydrophobic exclusion, are better descriptors of these interactions.39, 40
Figure 4.
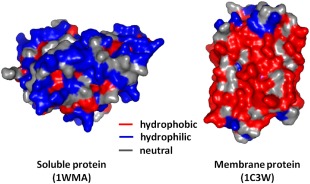
Differential arrangement of hydrophobic and hydrophilic residues in soluble versus membrane proteins. Shown, as an example, are human carbonyl reductase 1 as a representative of soluble proteins, and bacteriorhodopsin from Halobacterium salinarum as a representative of membrane proteins. Hydrophobic residues, shown in red, are situated towards the protein exterior in membrane proteins, and favor its accommodation in the lipid milieu. Hydrophilic residues are present on the exterior in soluble proteins or in the solvent‐exposed interior in membrane proteins, and are depicted here in blue. Redrawn with permission from Fiedler et al., Cell Mol Life Sci, 2010, 67:1779‐1798, © Springer Basel.40
In soluble proteins, phenylalanine, tyrosine, and tryptophan, owing to their hydrophobic nature, tend to pack tightly within hydrophobic protein interior, and play a significant role in driving rapid protein folding and scaffold stability.37 In an excellent investigation into the role of aromatic‐aromatic interactions in the hydrophobic core, for determining folding and stability, the McKnight group examined the autonomously folding small 35‐residue villin headpiece subdomain.41 The aromatic cluster contains three highly conserved phenylalanine residues F47, F51, and F58, which form the hydrophobic core of this protein (Fig. 5). When individually substituted with Leu, the native fold of the protein was retained. However, all the substitutions resulted in destabilization, wherein the mid‐point of thermal denaturation drops from ∼70°C to ∼35–50°C. The studies also revealed that F58 was important to attain the native fold. The authors concluded that residue rearrangement in the protein core can compensate for the substitution of an aromatic amino acid whereas aromatic clusters are essential for stability.
Figure 5.
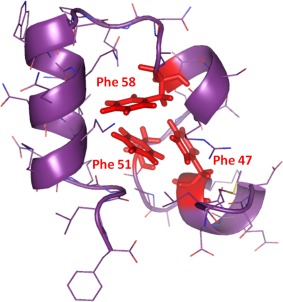
A highly conserved phenylalanine cluster forms a hydrophobic core in the autonomously folding villin headpiece subdomain. Figure generated from PDB ID: 1VII using PyMOL,42 and adapted with permission from Frank et al., Protein Sci, 2002, 11:680‐687, © Wiley‐Blackwell.41
Another interesting finding that emerged from the Gierasch group involves aromatic‐aromatic interactions in β‐sheets in the cellular retinoic acid‐binding protein 1 (CRABP1).43 The 10‐stranded β‐barrel contains three Phe residues forming an array of two aromatic pairs: F50‐F65 across strands 3 and 4, and F65‐F71 across the 4th and 5th strands. By substituting Phe to Met (an aliphatic with similar hydropathy and side chain size as Phe), they dissected the differential contribution of the two aromatic pairs. They observed that F50‐F65 involved in hydrophobic interactions, while aromatic interaction plays a significant role in the F65‐F71 pair. This finding also emphasizes the fact that proximal aromatic pairs do not necessarily engage only in aromatic interactions. Their kinetic analysis also revealed that aromatic interactions are formed late in the protein folding process, and thus they play a role in stabilizing the folded state.43
In the case of membrane proteins, aromatic residues show preferential occupancy toward the ends of transmembrane segments or at lipid‐facing surfaces.44, 45, 46 Aromatics determine the precise position of the transmembrane protein region in lipid membranes and contribute to lipid‐protein or protein‐protein interactions.47 Aromatic residues are also over‐represented near the ends of transmembrane helices,45 where they serve as anchors to stabilize the transmembrane regions, through interaction with other transmembrane segments or with the lipid head groups.47, 48 Tryptophan, in particular, also acts as stop‐transfer sequences during co‐translational folding of these proteins in the membrane.49 The position of aromatic residues in the helices is also a rate‐limiting step in membrane insertion: they are considered less favorable when placed centrally and become energetically favorable when moved apart.50, 51, 52, 53
Tamm, Kleinschmidt, and co‐workers were the first to investigate the role of aromatic interactions in the folding and stability of integral β‐barrel membrane proteins.54, 55, 56, 57 Using the bacterial outer membrane protein A as their model, the Tamm group observed that aromatic side chains can form a girdle and interact with each other even if they are 7 Å apart (Fig. 6). Tyrosine imparts the strongest contribution to the stability of OmpA. From the survey of three α‐helical and three β‐barrel membrane proteins they also found that the number of lipid‐facing aromatic clusters is higher in β‐barrel membrane proteins. On the contrary, α‐helical membrane proteins have more protein‐facing aromatic clusters that significantly involve in helix packing in these proteins.57 Studies from our group also support these observations that aromatics, particularly tryptophan, play a key role in the refolding and stability of transmembrane β‐barrels.58, 59, 60
Figure 6.
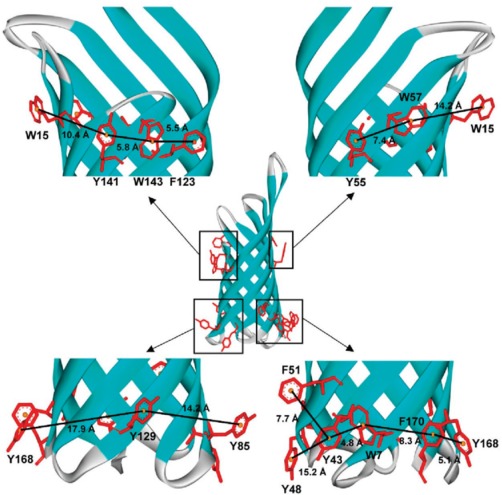
Aromatic girdle in the outer membrane protein OmpA, highlighting the arrangement of Phe, Tyr and Trp side chains around the β‐barrel. Most of the aromatic side chain face outside and are localized at the solvent‐membrane interface, so that it can form favorable contacts with the surrounding lipid chain, headgroup, and solvent. Reprinted (adapted) with permission from Hong et al., J Am Chem Soc, 2007, 129:8320‐8327, © ACS Publications.57
Overall, aromatic interactions in soluble proteins can assist in the early folding event, stabilize the protein core, and engage in various interactions including aromatic geometries and hydrophobic forces. They contribute significantly to the observed stability of proteins while they generally bear a lesser influence on the protein activity. In the case of membrane proteins, aromatic amino acids perform the dual function of defining the solvent‐membrane interface and act as stop‐transfer sequences during the coupled membrane protein synthesis and folding. Additionally, aromatic networks seen at the membrane interface add to scaffold stability in these proteins, similar to their role in the soluble counterparts.
Aromatic Interactions in Scaffold Stabilization: From β‐Sheets to α‐Helices
The chemical structure of the amino acid plays a critical role in determining the propensities towards a particular secondary structure. Being β‐branched, the aromatic amino acids Phe and Tyr (and Trp; discussed separately later in this section) have a higher intrinsic tendency to occur in β‐rich structures than α‐helices. Indeed, in a dataset of 593 proteins,61 Thomas et al. observed that “74% of the aromatic pairs are between residues separated by a difference of > 5 residues and of that one‐third aromatic pairs exist in β‐structure”. Similarly, the Gierasch group supported the interesting idea through experimental and bioinformatics analysis that nature employs aromatic interactions as a substitute for interstrand hydrogen bonds in β‐strand pairing.43
β‐Hairpins, the simplest unit of β‐sheets, consist of alternating hydrogen bonding and non‐hydrogen bonding sites across the two anti‐parallel strands. The side‐chain residues at the non‐hydrogen bonding position face opposite to each other and are spatially proximal. Aromatic pairs, when placed at the non‐hydrogen bonding position, usually result in stabilizing such scaffolds. The geometrical angle between the two rings approaching close to 90° (T‐shaped) is the most common and preferred mode of interaction. Experimental studies using isolated peptide systems from our work has yielded an asymmetric contribution (stabilizing/destabilizing) for a pair of interacting aromatics in β‐strands and sheets.19, 62 Such differential contributions arise from a distinct preference of one aromatic ring to interact with another via only its “face” or “edge”.19, 62, 63
α‐Helices are the other, more ubiquitous, class of structural motifs, in which aromatic interactions are shown to be prevalent for providing helical fold stability and in helical bundle assembly.64 The Chakrabarti group first reported, from extensive investigation using a dataset of 434 proteins, that the geometry of aromatic‐aromatic interaction in α‐helices is dependent on the sequence difference (Δ) between two residues. The preferred geometry was found to be edge‐to‐face when Δ = 1 and the geometry is tilted offset when Δ=4 having nearest contact.65
Chakrabarti group further examined the specific interaction and packing preferences of tryptophan with other aromatic amino acids.63 From the survey of protein structures, they found that a Trp interacting with Phe in a face‐to‐edge (FtE) geometry was more stable as compared to when the Trp edge points to the Phe face (EtF geometry). However, Trp‐Trp pairs were still found to adopt a T‐shaped geometry, with no difference in FtE or EtF interactions. The bulky indole side chain of this amino acid is capable of establishing multiple interactions, including π···π, C‐H···π, and N‐H···π, in which the indole nitrogen can also act as a hydrogen bond donor.66, 67, 68 In an investigation of indole side chain interactions, the Balaram group reported structures of several short peptide helices containing tryptophans.66, 67, 69 Their results revealed critical intermolecular interactions mediated by the indole side chain: aromatic‐amide and aromatic‐aromatic interactions, which stabilized and promoted self‐assembly of α‐helices.66
One of the significant roles of aromatic‐aromatic interactions is in the assembly and stability of the helical bundle proteins. DeGrado and co‐workers reported the solution NMR structure of a de novo designed 35‐residue peptide α2D.64 This peptide adopts a helix‐loop‐helix scaffold that dimerizes to form a 4‐helix bundle.64 In this case, the packing of helices takes place due to Phe‐Phe stacking interactions as illustrated in the solution NMR structure of this peptide in Figure 7, and the interaction between Phe‐Trp provides stability to the protein. A similar contribution of aromatic interactions in helical scaffolds is also seen for transmembrane helix assembly (discussed earlier).
Figure 7.
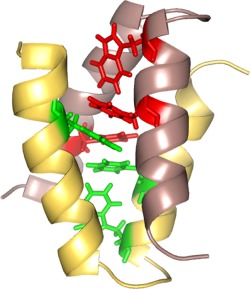
Solution NMR structure of a de novo designed native‐like protein α2D, highlighting the role of aromatic‐aromatic interactions in stability and in the packing of helices to form a four helix bundle. Figure rendered from PDB ID 1QP664 using PyMOL.42 Re‐drawn with inspiration from Hill and DeGrado.64
Experimental and structural analyses suggest a secondary structure‐dependent contribution of aromatic interactions to protein scaffolds. Local, intrastructure interactions are more predominant in β‐sheets whereas aromatic interactions in α‐helices facilitate supersecondary structure assemblies. Such understanding of the differential roles, modes and geometries of aromatic interactions in the two secondary structure elements have led to improved design of peptide mimetics and stable protein scaffolds.
Aromatic Interactions: Peptide Models to Protein Engineering
In the past few decades, numerous groups have investigated aromatic‐aromatic interactions using peptide model systems.8, 67, 68, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90 These investigations have led to a better insight in the context of fundamental forces driving protein folding, stability and in biomolecular recognition. On the basis of secondary structure, the interaction between the aromatic rings has been thoroughly investigated in both isolated α‐helix and β‐hairpin model peptide systems.
Waters and co‐workers investigated the incorporation of Phe residues at i and i+4 positions of designed helical peptides and provided experimental evidence for the stabilizing role of aromatic interactions.73 They also showed that this interaction is stronger when placed near the C‐terminus than in the center of a helix. Balaram and co‐workers also reported several short helical peptides containing Trp or Phe residues involved in intra‐ and interhelix aromatic interactions.66, 67, 76 They observed that peptides in which Phe side chains were on the same face of the helix showed both intrahelix and interhelix aromatic interactions [Fig. 8(a)].76 However, the peptides in which Phe side chains were placed on opposite faces of the helix resulted in only inter‐helix aromatic interactions. Their studies using peptide models showed that “the energy landscape for a pair of interacting phenyl rings consists of a broad, relatively flat minimum, which appears to be somewhat rugged, with several local minima separated by small energy barriers”.76 Supramolecular assembly is also possible in peptide structures. One such example from the DeGrado group of a peptide dimer of α2D64 has already been discussed earlier.
Figure 8.
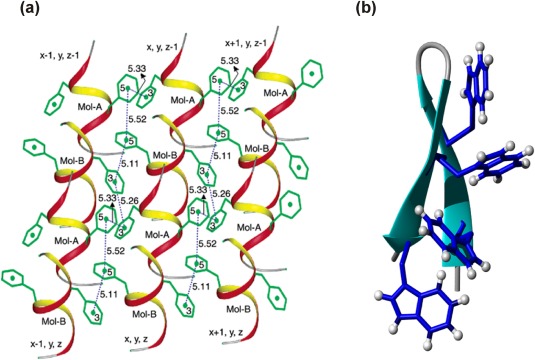
Aromatic interactions in peptide models. (a) A short helical synthetic peptide (Boc‐Aib‐Ala‐Phe‐Aib‐Phe‐Ala‐Val‐Aib‐OMe) displays strong inter‐helix aromatic interactions in the crystal. Reprinted (adapted) with permission from Aravinda et al., J Am Chem Soc, 2003, 125:5308‐5315, © ACS Publications.76 (b) Designed β‐hairpin Trpzip peptide with high thermostability, due to the incorporation of two pairs of Trp‐Trp interactions at the non‐hydrogen bonding position. Figure rendered from PDB ID 1LEO92 using MolMol.91
Since the first report of Trpzip β‐hairpin peptides, shown in Figure 8(b), by the Cochran group,92 Trp‐Trp pairs at the non‐hydrogen bonding position has proved to be excellent hairpin stabilizing elements.68 Numerous hairpin scaffolds incorporating Trp‐Trp, Trp‐non‐Trp and other aromatic pairs have since been designed by us,19, 62, 93, 94, 95 and groups of Andersen,80, 83 Jimenez,68, 82, 84, 96 Keiderling,85, 86, 87, 90, 97 Kelly,98, 99 Waters78, 89 and others. These extensive investigations using peptide models have led us to a thorough understanding of aromatic‐aromatic interactions. Excellent reviews have been published recently in this area.68, 75, 79
Aromatic interactions have also proved as useful structure stabilizing elements in protein engineering. The Kelly group has shown that the incorporation of a single cross‐strand Trp‐Trp pair at the non‐hydrogen bonding position in an autonomously folded protein hPin1 WW domain significantly increased its thermodynamic stability.99 However, the protein lost its function due to restricted backbone motion caused by highly stabilizing Trp‐Trp interaction, suggesting that proteins have evolved to balance stability against functional demands in various cases.
In a designed three‐stranded peptide β‐sheet nucleated by DPro‐Gly segment, Balaram and co‐workers incorporated β‐phenylalanine at positions facing each other.88 The structural fold of the β‐sheet promoted the N‐terminal and C‐terminal β‐phenylalanines to participate in long‐range aromatic‐aromatic interactions. Such strategies incorporating backbone modified β‐ and γ‐ aromatic amino acid residues has gathered substantial interest in the de novo design of proteolytically resistant bioactive peptides and proteins. Aromatic residues further stabilize such scaffolds through cross‐strand interactions.
Fluorination of aromatic amino acids is also now widely exploited in protein engineering; excellent work100 and review101 in this area has been reported by the Gao group. In general terms, fluorination increases the hydrophobicity of the molecule, thus favoring a “hydrophobic effect” in protein folding and stability. The Gellman group probed the effect of substituting Phe‐Phe interactions in a small protein villin headpiece subdomain with perfluoro phenylalanine.102 They found that the substitution of aryl side chains with fluoro‐aryl side chains could result in stabilizing the folded conformation of proteins; however the effect cannot be generalized. Figure 9 illustrates one such successful example using α2D.64 Tatko and Waters also reported that the halogen substituent in aromatic amino acid side chains can enhance edge‐to‐face aromatic interactions, resulting in increased strand stability.78
Figure 9.
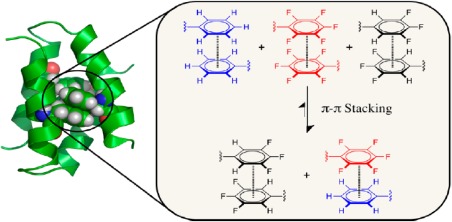
Schematic representation of α2D, a de novo designed model system that forms a four helix bundle. This system has been widely exploited to study aryl‐perflouroaryl interaction as an attractive strategy in protein engineering. Reprinted (adapted) with permission from Pace and Gao, Acc Chem Res, 2013, 46:907‐915, © ACS Publications.101
The incorporation of aromatic pairs is therefore increasingly attracting attention in protein engineering and the design of peptide‐based bio‐nanomaterials (see next sections).
Prevalence of Higher Order Aromatic Clusters
The early survey of aromatic pairs in proteins by Burley and Petsko revealed that “on an average 60% of aromatic side chain in proteins are involved in aromatic pairing and 80% of that forms a network of three or more interacting aromatic side chains”.3 A more recent survey by Turjanski and co‐workers reported that almost half of the proteins crystallized so far have higher order aromatic clusters (trimer, tetramer, and even larger).103 This survey also highlighted the preference of aromatic trimers to be in helical structures and usually reside in the protein interior, forming a part of the hydrophobic core.
Thus, apart from π‐π dimer pairing, aromatic residues also involve in forming extended π‐π networks. Such a network with larger aromatic clusters is proposed to arise from pre‐existing smaller clusters, and can exist in various geometrical conformations. As shown in Figure 10, an aromatic tetramer with chain‐like conformation can form three aromatic interactions while, in another highly synergic conformation, it can involve in a total of six aromatic interactions.103
Figure 10.
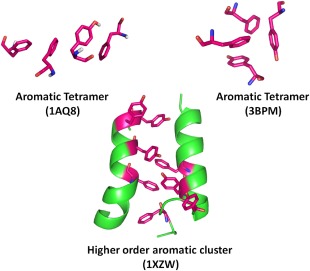
Higher order aromatic clusters in proteins. Aromatic tetramer in linear arrangement can involve in 3 interactions (top left). An aromatic tetramer found in circular symmetry can, however, establish a total of 6 interactions (top right). Larger aromatic clusters can show preferred occurrence in helical structures. Reprinted (adapted) with permission from Lanzarotti et al., J Chem Inf Model, 2011 51:1623‐1633, © ACS Publications.103
It is important to understand how aromatic amino acids in a cluster coordinate among themselves as well as with the structural fold to achieve a globally favorable conformation with preferred modes of aromatic interactions. If this coordination is erroneous, it is vital to deduce how it affects the structural fold and stability of the molecule. We have designed and studied several β‐hairpin peptides incorporating one or two aromatic pairs with the combination of Phe, Trp, and Tyr residues. Our investigation reveals the existence of an asymmetric contribution of aromatic interactions.19, 62 This asymmetry stems from spatial positioning of aromatic pairs in β‐strands and side chain positioning of individual aromatic residues in pairs plays a significant role in the overall structural stability of a β‐hairpin scaffolds. Such asymmetry can exist in nature. Less favorable aryl geometries can be supplemented with additional neighboring interactions to offset their destabilizing effect.
The observation of higher order aromatic clusters even in folded protein conformations reveals its natural self‐associating property, and suggests that the protein can adopt its structure to exploit the maximum benefit from aromatic residues. In a very recent survey of protein structures by Gray and Winkler, they revealed that a precise positioning of Tyr and Trp side chain can protect proteins from oxidative damage, through a 'hole hopping' mechanism.104 They also found that one‐third of all proteins in the protein database have extended Tyr/Trp chains even in proteins that do not perform redox reactions. Thus, Gray and Winkler propose that the property of redox‐active Tyr/Trp to protect from oxidative damage may be useful even for proteins that are not directly involved in redox reactions.104
Future Prospects
With our progress in understanding the cause of several brain diseases including Alzheimer's, it is becoming apparent that aromatic residues do play a role in biomolecular self‐association. The Gazit group was the first to report the self‐assembly of diphenylalanine, which can itself form well‐ordered structures.105 Gazit suggested that “restricted geometry and the attractive forces of aromatic moieties provide order and directionality”.105 As exceptions to this generalization, there are studies which indicate that aromatic interactions are dispensable for amyloidogenesis of some proteins, for example, the islet amyloid polypeptide.106, 107 However, the presence of aromatic amino acids in such sequences considerably augments aggregation of the amyloid fibrils.107
A recent computational analysis of all 8000 possible tripeptides by Uljin and co‐workers was used to identify aggregating prone sequences.108 They found out that all highly aggregating peptides sequences contained a pair of aromatic residues, and at least one phenylalanine. Furthermore, they found that the order of arrangement of the aromatic residues also had an impact on the outcome, by influencing the aggregation propensity and the rate of amyloid formation. These findings can have high biomedical significance, as it provides insight on amyloidogenesis, and may find its use in identifying aggregation‐prone regions in proteins. Further studies in this direction can help us design small molecule inhibitors for diseases caused by protein aggregation. Attempts in this direction have yielded promising results.109, 110, 111
A recent study by Pellach et al. reported the ingenious design of peptides that form bioinspired self‐assembled nanostructures.112 The design of such decapeptides was based on architectures of phospholipids with a hydrophilic head consisting of charged residues and phosphoserine, and the hydrophobic tail containing valine and phenylalanine residues. Aromatic residues were placed at both hydrogen and non‐hydrogen bonding positions of the β‐strands (hydrophobic tail) resulting in inter‐ and intramolecular aromatic interactions, respectively. The study showed that such a design provided the driving force for peptide self‐assembly and stability. Similarly, designed nanostructures can find diverse application in nanotechnology and biomedical fields.
In another functional aspect using aromatic interactions, Mahajan and Bhattacharjya reported a de novo design of membrane active β‐hairpin and α‐helical scaffolds stabilized by aromatic interactions.113, 114 Tryptophan and histidine residues were also utilized to anchor the scaffold into lipid micelles and bind heme. This study was the first report of designed membrane active scaffolds that showed porphyrin binding and performing peroxidase activity. Such strategies can be readily utilized to build novel membrane active enzymes, bearing potential application in electron transport and energy production across the membranes. They can also be extended to the design and development of peptide‐based enzyme mimetics, which can now be engineered with increased catalytic efficiency to produce novel drug libraries.
The self‐associating property of aromatic residues is now believed to find extensive use in modern nanostructures research and the generation of robust hydrogels, catalytic antibodies, etc. (Fig. 11). Through this review, we have discussed a few outstanding examples wherein our understanding of aromatic interactions has successfully been translated to biomaterial design and peptide engineering. Our increasing realization of the benefits of aromatic interactions can be exploited in bionanomaterials, for modern tissue engineering and plastic surgery.115 Examples of applications of aromatic interactions for diverse processes are growing rapidly, and we surmise that this can be used in the successful design of several small molecule next generation drugs and drug delivery cargoes.
Figure 11.
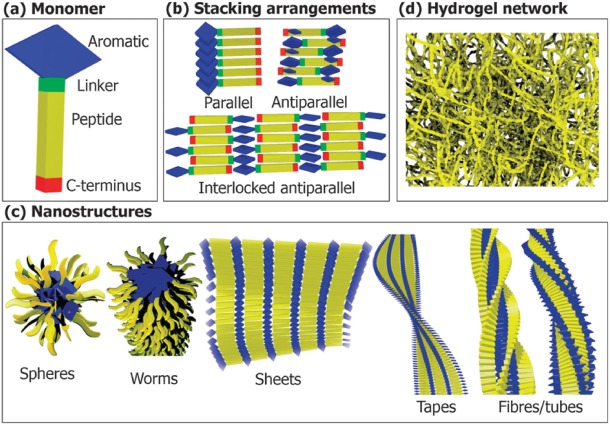
Applying the self‐assembly property of aromatics in the generation of several modern bionanomaterials. These amphiphilic self‐assemblies may have applications in biomedicine, including tissue engineering, plastic surgery and next generation drugs, and drug delivery cargoes. Figure reproduced with permission from Fleming and Ulijn, Chem Soc Rev, 2014, 43:8150‐8177, RSC Publishing.115
Acknowledgments
While our compilation is fairly comprehensive, there are many more exquisite examples from various groups, which might have escaped mention; however, we duly acknowledge their contribution as well. K.M.M. is a Senior Research Fellow of the University Grants Commission, Govt. of India. R.M. is a Wellcome Trust/DBT India Alliance Intermediate Fellow. The authors declare no conflict of interest.
References
- 1. Chiti F, Dobson CM (2006) Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75:333–366. [DOI] [PubMed] [Google Scholar]
- 2. Gregersen N, Bross P, Vang S, Christensen JH (2006) Protein misfolding and human disease. Annu Rev Genomics Hum Genet 7:103–124. [DOI] [PubMed] [Google Scholar]
- 3. Burley SK, Petsko GA (1985) Aromatic‐aromatic interaction: a mechanism of protein structure stabilization. Science 229:23–28. [DOI] [PubMed] [Google Scholar]
- 4. Riley KE, Hobza P (2013) On the importance and origin of aromatic interactions in chemistry and biodisciplines. Acc Chem Res 46:927–936. [DOI] [PubMed] [Google Scholar]
- 5. Baker CM, Grant GH (2007) Role of aromatic amino acids in protein‐nucleic acid recognition. Biopolymers 85:456–470. [DOI] [PubMed] [Google Scholar]
- 6. Asensio JL, Arda A, Canada FJ, Jimenez‐Barbero J (2013) Carbohydrate‐aromatic interactions. Acc Chem Res 46:946–954. [DOI] [PubMed] [Google Scholar]
- 7. del Carmen Fernandez‐Alonso M, Canada FJ, Jimenez‐Barbero J, Cuevas G (2005) Molecular recognition of saccharides by proteins. Insights on the origin of the carbohydrate‐aromatic interactions. J Am Chem Soc 127:7379–7386. [DOI] [PubMed] [Google Scholar]
- 8. Kiehna SE, Laughrey ZR, Waters ML (2007) Evaluation of a carbohydrate‐pi interaction in a peptide model system. Chem Commun 4026–4028. [DOI] [PubMed] [Google Scholar]
- 9. Dougherty DA (1996) Cation‐pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 271:163–168. [DOI] [PubMed] [Google Scholar]
- 10. Ma JC, Dougherty DA (1997) The cation‐p interaction. Chem Rev 97:1303–1324. [DOI] [PubMed] [Google Scholar]
- 11. Dougherty DA (2013) The cation‐pi interaction. Acc Chem Res 46:885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schottel BL, Chifotides HT, Dunbar KR (2008) Anion‐pi interactions. Chem Soc Rev 37:68–83. [DOI] [PubMed] [Google Scholar]
- 13. Valley CC, Cembran A, Perlmutter JD, Lewis AK, Labello NP, Gao J, Sachs JN (2012) The methionine‐aromatic motif plays a unique role in stabilizing protein structure. J Biol Chem 287:34979–34991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reid K, Lindley P, Thornton J (1985) Sulphur‐aromatic interactions in proteins. FEBS Lett 190:209–213. [Google Scholar]
- 15. Zauhar RJ, Colbert CL, Morgan RS, Welsh WJ (2000) Evidence for a strong sulfur‐aromatic interaction derived from crystallographic data. Biopolymers 53:233–248. [DOI] [PubMed] [Google Scholar]
- 16. Plevin MJ, Bryce DL, Boisbouvier J (2010) Direct detection of CH/pi interactions in proteins. Nat Chem 2:466–471. [DOI] [PubMed] [Google Scholar]
- 17. Ganguly HK, Majumder B, Chattopadhyay S, Chakrabarti P, Basu G (2012) Direct evidence for CH…pi interaction mediated stabilization of Pro‐cisPro bond in peptides with Pro‐Pro‐aromatic motifs. J Am Chem Soc 134:4661–4669. [DOI] [PubMed] [Google Scholar]
- 18. Zondlo NJ (2013) Aromatic‐proline interactions: Electronically tunable CH/pi interactions. Acc Chem Res 46:1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Makwana KM, Mahalakshmi R (2015) NMR analysis of tuning cross‐strand Phe/Tyr/Trp‐Trp interactions in designed beta‐hairpin peptides: Terminal switch from L to D amino acid as a strategy for beta‐hairpin capping. J Phys Chem B 119:5376–5385. [DOI] [PubMed] [Google Scholar]
- 20. Singh J, Thornton J (1985) The interaction between phenylalanine rings in proteins. FEBS Lett 191:1–6. [Google Scholar]
- 21. Hunter C, Singh J, Thornton J (1991) Pi‐pi interactions: the geometry and energetics of phenylalanine‐phenylalanine interactions in proteins. J Mol Biol 218:837–846. [DOI] [PubMed] [Google Scholar]
- 22. Hunter C, Lawson K, Perkins J, Urch C (2001) Aromatic interactions. J Chem Soc Perkin Trans 2:651–669. [Google Scholar]
- 23. Sun S, Bernstein E (1996) Aromatic van der Waals clusters: Structure and nonrigidity. J Phys Chem B 100:13348–13366. [Google Scholar]
- 24. Goldstein RA (2007) Amino‐acid interactions in psychrophiles, mesophiles, thermophiles, and hyperthermophiles: insights from the quasi‐chemical approximation. Protein Sci 16:1887–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taylor TJ, Vaisman II (2010) Discrimination of thermophilic and mesophilic proteins. BMC Struct Biol 10 (Suppl 1):S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meruelo AD, Han SK, Kim S, Bowie JU (2012) Structural differences between thermophilic and mesophilic membrane proteins. Protein Sci 21:1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siddiqui KS, Cavicchioli R (2006) Cold‐adapted enzymes. Annu Rev Biochem 75:403–433. [DOI] [PubMed] [Google Scholar]
- 28. Feller G (2013) Psychrophilic enzymes: from folding to function and biotechnology. Scientifica 2013:512840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feller G, Arpigny J, Narinx E, Gerday C (1997) Molecular adaptations of enzymes from psychrophilic organisms. Camp Biochem Physiol 118:495–499. [Google Scholar]
- 30. Davail S, Feller G, Narinx E, Gerday C (1994) Cold adaptation of proteins. Purification, characterization, and sequence of the heat‐labile subtilisin from the antarctic psychrophile Bacillus TA41. J Biol Chem 269:17448–17453. [PubMed] [Google Scholar]
- 31. Cipolla A, D'Amico S, Barumandzadeh R, Matagne A, Feller G (2011) Stepwise adaptations to low temperature as revealed by multiple mutants of psychrophilic alpha‐amylase from Antarctic bacterium. J Biol Chem 286:38348–38355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anderson DE, Hurley JH, Nicholson H, Baase WA, Matthews BW (1993) Hydrophobic core repacking and aromatic aromatic interaction in the thermostable mutant of T4 lysozyme Ser 117→Phe. Protein Sci 2:1285–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mooers BH, Baase WA, Wray JW, Matthews BW (2009) Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme. Protein Sci 18:871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kannan N, Vishveshwara S (2000) Aromatic clusters: a determinant of thermal stability of thermophilic proteins. Protein Eng 13:753–761. [DOI] [PubMed] [Google Scholar]
- 35. Szilagyi A, Zavodszky P (2000) Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure 8:493–504. [DOI] [PubMed] [Google Scholar]
- 36. Baldwin R (1989) How does protein folding get started? Trends Biochem Sci 14:291–294. [DOI] [PubMed] [Google Scholar]
- 37. Dill KA (1990) Dominant forces in protein folding. Biochemistry 29:7133–7155. [DOI] [PubMed] [Google Scholar]
- 38. Rees DC, DeAntonio L, Eisenberg D (1989) Hydrophobic organization of membrane proteins. Science 245:510–513. [DOI] [PubMed] [Google Scholar]
- 39. Stevens TJ, Arkin IT (1999) Are membrane proteins “inside‐out” proteins? Proteins 36:135–143. [DOI] [PubMed] [Google Scholar]
- 40. Fiedler S, Broecker J, Keller S (2010) Protein folding in membranes. Cell Mol Life Sci 67:1779–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Frank BS, Vardar D, Buckley DA, McKnight CJ (2002) The role of aromatic residues in the hydrophobic core of the villin headpiece subdomain. Protein Sci 11:680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schrödinger LLC (2010) The PyMOL Molecular Graphics System, Version 12r3pre.
- 43. Budyak IL, Zhuravleva A, Gierasch LM (2013) The role of aromatic‐aromatic interactions in strand‐strand stabilization of beta‐sheets. J Mol Biol 425:3522–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wimley WC, White SH (1996) Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol 3:842–848. [DOI] [PubMed] [Google Scholar]
- 45. White SH, Wimley WC (1999) Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct 28:319–365. [DOI] [PubMed] [Google Scholar]
- 46. Killian JA, von Heijne G (2000) How proteins adapt to a membrane‐water interface. Trends Biochem Sci 25:429–434. [DOI] [PubMed] [Google Scholar]
- 47. Schlebach JP, Sanders CR (2015) The safety dance: biophysics of membrane protein folding and misfolding in a cellular context. Q Rev Biophys 48:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cymer F, von Heijne G, White SH (2015) Mechanisms of integral membrane protein insertion and folding. J Mol Biol 427:999–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Braun P, von Heijne G (1999) The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry 38:9778–9782. [DOI] [PubMed] [Google Scholar]
- 50. Chang YC, Bowie JU (2014) Membrane protein folding stability and kinetics in bilayers. Protein Sci 23:233–233. [Google Scholar]
- 51. Fleming KG (2014) Energetics of membrane protein folding. Annu Rev Biophys 43:233–255. [DOI] [PubMed] [Google Scholar]
- 52. Hong H (2014) Toward understanding driving forces in membrane protein folding. Arch Biochem Biophys 564:297–313. [DOI] [PubMed] [Google Scholar]
- 53. Otzen D (2014) Membrane protein folding and stability. Arch Biochem Biophys 564:262–264. [DOI] [PubMed] [Google Scholar]
- 54. Kleinschmidt JH, Tamm LK (1996) Kinetic studies on the refolding and membrane insertion of outer membrane protein a (Omp A) of E‐coli. Biophys J 70:Mp139–Mp139. [Google Scholar]
- 55. Kleinschmidt JH, Tamm LK (1996) Folding intermediates of a beta‐barrel membrane protein. Kinetic evidence for a multi‐step membrane insertion mechanism. Biochemistry 35:12993–13000. [DOI] [PubMed] [Google Scholar]
- 56. Kleinschmidt JH, den Blaauwen T, Driessen AJM, Tamm LK (1999) Outer membrane protein A of Escherichia coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry 38:5006–5016. [DOI] [PubMed] [Google Scholar]
- 57. Hong H, Park S, Jimenez RH, Rinehart D, Tamm LK (2007) Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J Am Chem Soc 129:8320–8327. [DOI] [PubMed] [Google Scholar]
- 58. Chaturvedi D, Mahalakshmi R (2014) Juxtamembrane tryptophans have distinct roles in defining the OmpX barrel‐micelle boundary and facilitating protein‐micelle association. FEBS Lett 588:4464–4471. [DOI] [PubMed] [Google Scholar]
- 59. Gupta A, Zadafiya P, Mahalakshmi R (2014) Differential contribution of tryptophans to the folding and stability of the attachment invasion locus transmembrane beta‐barrel from Yersinia pestis. Sci Rep 4:6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mahalakshmi R (2015) Folding and stability of transmembrane beta‐barrels of bacterial and human origin: Probing underlying similarities and principal differences using in vitro systems. Proc Indian Natn Sci Acad 81:463–478. [Google Scholar]
- 61. Thomas A, Meurisse R, Charloteaux B, Brasseur R (2002) Aromatic side‐chain interactions in proteins. I. Main structural features. Proteins 48:628–634. [DOI] [PubMed] [Google Scholar]
- 62. Makwana KM, Mahalakshmi R (2014) Asymmetric contribution of aromatic interactions stems from spatial positioning of the interacting aryl pairs in beta‐hairpins. ChemBioChem 15:2357–2360. [DOI] [PubMed] [Google Scholar]
- 63. Samanta U, Pal D, Chakrabarti P (1999) Packing of aromatic rings against tryptophan residues in proteins. Acta Cryst D55:1421–1427. [DOI] [PubMed] [Google Scholar]
- 64. Hill R, DeGrado W (1998) Solution structure of α2D, a nativelike de novo designed protein. J Am Chem Soc 120:1138–1145. [Google Scholar]
- 65. Bhattacharyya R, Samanta U, Chakrabarti P (2002) Aromatic‐aromatic interactions in and around alpha‐helices. Protein Eng 15:91–100. [DOI] [PubMed] [Google Scholar]
- 66. Mahalakshmi R, Sengupta A, Raghothama S, Shamala N, Balaram P (2005) Tryptophan‐containing peptide helices: interactions involving the indole side chain. J Pept Res 66:277–296. [DOI] [PubMed] [Google Scholar]
- 67. Sengupta A, Mahalakshmi R, Shamala N, Balaram P (2005) Aromatic interactions in tryptophan‐containing peptides: crystal structures of model tryptophan peptides and phenylalanine analogs. J Pept Res 65:113–129. [DOI] [PubMed] [Google Scholar]
- 68. Santiveri CM, Jimenez MA (2010) Tryptophan residues: scarce in proteins but strong stabilizers of beta‐hairpin peptides. Biopolymers 94:779–790. [DOI] [PubMed] [Google Scholar]
- 69. Mahalakshmi R, Sengupta A, Raghothama S, Shamala N, Balaram P (2007) Tryptophan rich peptides: influence of indole rings on backbone conformation. Biopolymers 88:36–54. [DOI] [PubMed] [Google Scholar]
- 70. Zhao CX, Polavarapu PL, Das C, Balaram P (2000) Vibrational circular dichroism of beta‐hairpin peptides. J Am Chem Soc 122:8228–8231. [Google Scholar]
- 71. Tatko CD, Waters ML (2001) Cross strand aromatic interactions in beta‐turn peptides. Abstr Pap Am Chem Soc 222:U122–U122. [Google Scholar]
- 72. Andrew CD, Bhattacharjee S, Kokkoni N, Hirst JD, Jones GR, Doig AJ (2002) Stabilizing interactions between aromatic and basic side chains in alpha‐helical peptides and proteins. Tyrosine effects on helix circular dichroism. J Am Chem Soc 124:12706–12714. [DOI] [PubMed] [Google Scholar]
- 73. Butterfield S, Patel P, Waters ML (2002) Contribution of aromatic interactions to α‐helix stability. J Am Chem Soc 124:9751–9755. [DOI] [PubMed] [Google Scholar]
- 74. Tatko CD, Waters ML (2002) Selective aromatic interactions in beta‐hairpin peptides. J Am Chem Soc 124:9372–9373. [DOI] [PubMed] [Google Scholar]
- 75. Waters ML (2002) Aromatic interactions in model systems. Curr Opin Chem Biol 6:736–741. [DOI] [PubMed] [Google Scholar]
- 76. Aravinda S, Shamala N, Das C, Sriranjini A, Karle IL, Balaram P (2003) Aromatic‐aromatic interactions in crystal structures of helical peptide scaffolds containing projecting phenylalanine residues. J Am Chem Soc 125:5308–5315. [DOI] [PubMed] [Google Scholar]
- 77. Aravinda S, Harini VV, Shamala N, Das C, Balaram P (2004) Structure and assembly of designed beta‐hairpin peptides in crystals as models for beta‐sheet aggregation. Biochemistry 43:1832–1846. [DOI] [PubMed] [Google Scholar]
- 78. Tatko CD, Waters ML (2004) Effect of halogenation on edge‐face aromatic interactions in a beta‐hairpin peptide: enhanced affinity with iodo‐substituents. Org Lett 6:3969–3972. [DOI] [PubMed] [Google Scholar]
- 79. Waters ML (2004) Aromatic interactions in peptides: impact on structure and function. Biopolymers 76:435–445. [DOI] [PubMed] [Google Scholar]
- 80. Andersen NH, Olsen KA, Fesinmeyer RM, Tan X, Hudson FM, Eidenschink LA, Farazi SR (2006) Minimization and optimization of designed beta‐hairpin folds. J Am Chem Soc 128:6101–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Palermo NY, Csontos J, Owen MC, Murphy RF, Lovas S (2007) Aromatic‐backbone interactions in model alpha‐helical peptides. J Comput Chem 28:2510–2510. [DOI] [PubMed] [Google Scholar]
- 82. Santiveri CM, Leon E, Rico M, Jimenez MA (2008) Context‐dependence of the contiribution of disulfide bonds to beta‐hairpin stability. Chem Eur J 14:488–499. [DOI] [PubMed] [Google Scholar]
- 83. Eidenschink L, Kier BL, Huggins KNL, Andersen NH (2009) Very short peptides with stable folds: Building on the interrelationship of Trp/Trp, Trp/cation, and Trp/backbone‐amide interaction geometries. Proteins 75:308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mirassou Y, Santiveri CM, Perez de Vega MJ, Gonzalez‐Muniz R, Jimenez MA (2009) Disulfide bonds versus TrpTrp pairs in irregular beta‐hairpins: NMR structure of vammin loop 3‐derived peptides as a case study. ChemBioChem 10:902–910. [DOI] [PubMed] [Google Scholar]
- 85. Takekiyo T, Wu L, Yoshimura Y, Shimizu A, Keiderling TA (2009) Relationship between hydrophobic interactions and secondary structure stability for Trpzip beta‐hairpin peptides. Biochemistry 48:1543–1552. [DOI] [PubMed] [Google Scholar]
- 86. Wu L, McElheny D, Huang R, Keiderling TA (2009) Role of tryptophan‐tryptophan interactions in Trpzip beta‐hairpin formation, structure, and stability. Biochemistry 48:10362–10371. [DOI] [PubMed] [Google Scholar]
- 87. Wu L, McElheny D, Takekiyo T, Keiderling TA (2010) Geometry and efficacy of cross‐strand Trp/Trp, Trp/Tyr, and Tyr/Tyr aromatic interaction in a beta‐hairpin peptide. Biochemistry 49:4705–4714. [DOI] [PubMed] [Google Scholar]
- 88. Sonti R, Gopi HN, Muddegowda U, Ragothama S, Balaram P (2013) A designed three‐stranded beta‐sheet in an alpha/beta hybrid peptide. Chem Eur J 19:5955–5965. [DOI] [PubMed] [Google Scholar]
- 89. Matsumoto M, Lee SJ, Gagne MR, Waters ML (2014) Cross‐strand histidine‐aromatic interactions enhance acyl‐transfer rates in beta‐hairpin peptide catalysts. Org Biomol Chem 12:8711–8718. [DOI] [PubMed] [Google Scholar]
- 90. Popp A, Wu L, Keiderling TA, Hauser K (2014) Effect of hydrophobic interactions on the folding mechanism of beta‐hairpins. J Phys Chem B 118:14234–14242. [DOI] [PubMed] [Google Scholar]
- 91. Koradi R, Billeter M, Wuthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14:51–55. 29‐32. [DOI] [PubMed] [Google Scholar]
- 92. Cochran AG, Skelton NJ, Starovasnik MA (2001) Tryptophan zippers: stable, monomeric beta ‐hairpins. Proc Natl Acad Sci USA 98:5578–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Makwana KM, Raghothama S, Mahalakshmi R (2013) Stabilizing effect of electrostatic vs. aromatic interactions in diproline nucleated peptide beta‐hairpins. Phys Chem Chem Phys 15:15321–15324. [DOI] [PubMed] [Google Scholar]
- 94. Makwana KM, Mahalakshmi R (2014) Comparative analysis of cross strand aromatic‐Phe interactions in designed peptide beta‐hairpins. Org Biomol Chem 12:2053–2061. [DOI] [PubMed] [Google Scholar]
- 95. Makwana KM, Mahalakshmi R (2015) Nature of aryl‐tyrosine interactions contribute to beta‐hairpin scaffold stability: NMR evidence for alternate ring geometry. Phys Chem Chem Phys 17:4220–4230. [DOI] [PubMed] [Google Scholar]
- 96. Viguera AR, Jimenez MA, Rico M, Serrano L (1996) Conformational analysis of peptides corresponding to beta‐hairpins and a beta‐sheet that represent the entire sequence of the alpha‐spectrin SH3 domain. J Mol Biol 255:507–521. [DOI] [PubMed] [Google Scholar]
- 97. Setnicka V, Huang R, Thomas CL, Etienne MA, Kubelka J, Hammer RP, Keiderling TA (2005) IR study of cross‐strand coupling in a beta‐hairpin peptide using isotopic labels. J Am Chem Soc 127:4992–4993. [DOI] [PubMed] [Google Scholar]
- 98. Nesloney CL, Kelly JW (1996) A 2,3'‐substituted biphenyl‐based amino acid facilitates the formation of a monomeric beta‐hairpin‐like structure in aqueous solution at elevated temperature. J Am Chem Soc 118:5836–5845. [Google Scholar]
- 99. Jager M, Dendle M, Fuller AA, Kelly JW (2007) A cross‐strand Trp Trp pair stabilizes the hPin1 WW domain at the expense of function. Protein Sci 16:2306–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pace CJ, Kim D, Gao J (2012) Experimental evaluation of CH‐pi interactions in a protein core. Chem Eur J 18:5832–5836. [DOI] [PubMed] [Google Scholar]
- 101. Pace CJ, Gao J (2013) Exploring and exploiting polar‐pi interactions with fluorinated aromatic amino acids. Acc Chem Res 46:907–915. [DOI] [PubMed] [Google Scholar]
- 102. Woll MG, Hadley EB, Mecozzi S, Gellman SH (2006) Stabilizing and destabilizing effects of phenylalanine ‐> F5‐phenylalanine mutations on the folding of a small protein. J Am Chem Soc 128:15932–15933. [DOI] [PubMed] [Google Scholar]
- 103. Lanzarotti E, Biekofsky RR, Estrin DA, Marti MA, Turjanski AG (2011) Aromatic‐aromatic interactions in proteins: beyond the dimer. J Chem Inf Model 51:1623–1633. [DOI] [PubMed] [Google Scholar]
- 104. Gray HB, Winkler JR (2015) Hole hopping through tyrosine/tryptophan chains protects proteins from oxidative damage. Proc Natl Acad Sci USA 112:10920–10925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Reches M, Gazit E (2003) Casting metal nanowires within discrete self‐assembled peptide nanotubes. Science 300:625–627. [DOI] [PubMed] [Google Scholar]
- 106. Marek P, Abedini A, Song B, Kanungo M, Johnson ME, Gupta R, Zaman W, Wong SS, Raleigh DP (2007) Aromatic interactions are not required for amyloid fibril formation by islet amyloid polypeptide but do influence the rate of fibril formation and fibril morphology. Biochemistry 46:3255–3261. [DOI] [PubMed] [Google Scholar]
- 107. Tu LH, Raleigh DP (2013) Role of aromatic interactions in amyloid formation by islet amyloid polypeptide. Biochemistry 52:333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Frederix PW, Scott GG, Abul‐Haija YM, Kalafatovic D, Pappas CG, Javid N, Hunt NT, Ulijn RV, Tuttle T (2015) Exploring the sequence space for (tri‐)peptide self‐assembly to design and discover new hydrogels. Nature Chem 7:30–37. [DOI] [PubMed] [Google Scholar]
- 109. Sievers SA, Karanicolas J, Chang HW, Zhao A, Jiang L, Zirafi O, Stevens JT, Munch J, Baker D, Eisenberg D (2011) Structure‐based design of non‐natural amino‐acid inhibitors of amyloid fibril formation. Nature 475:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bram Y, Lampel A, Shaltiel‐Karyo R, Ezer A, Scherzer‐Attali R, Segal D, Gazit E (2015) Monitoring and targeting the initial dimerization stage of amyloid self‐assembly. Angew Chem Int Ed Engl 54:2062–2067. [DOI] [PubMed] [Google Scholar]
- 111. Tu LH, Young LM, Wong AG, Ashcroft AE, Radford SE, Raleigh DP (2015) Mutational analysis of the ability of resveratrol to inhibit amyloid formation by islet amyloid polypeptide: critical evaluation of the importance of aromatic‐inhibitor and histidine‐inhibitor interactions. Biochemistry 54:666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Pellach M, Atsmon‐Raz Y, Simonovsky E, Gottlieb H, Jacoby G, Beck R, Adler‐Abramovich L, Miller Y, Gazit E (2015) Spontaneous structural transition in phospholipid‐inspired aromatic phosphopeptide nanostructures. ACS Nano 9:4085–4095. [DOI] [PubMed] [Google Scholar]
- 113. Mahajan M, Bhattacharjya S (2013) Beta‐hairpin peptides: heme binding, catalysis, and structure in detergent micelles. Angew Chem Int Ed Engl 52:6430–6434. [DOI] [PubMed] [Google Scholar]
- 114. Mahajan M, Bhattacharjya S (2014) Designed di‐heme binding helical transmembrane protein. ChemBioChem 15:1257–1262. [DOI] [PubMed] [Google Scholar]
- 115. Fleming S, Ulijn RV (2014) Design of nanostructures based on aromatic peptide amphiphiles. Chem Soc Rev 43:8150–8177. [DOI] [PubMed] [Google Scholar]


