Abstract
In vivo, as an advanced catalytic strategy, transient non‐covalently bound multi‐enzyme complexes can be formed to facilitate the relay of substrates, i. e. substrate channeling, between sequential enzymatic reactions and to enhance the throughput of multi‐step enzymatic pathways. The human thymidylate synthase and dihydrofolate reductase catalyze two consecutive reactions in the folate metabolism pathway, and experiments have shown that they are very likely to bind in the same multi‐enzyme complex in vivo. While reports on the protozoa thymidylate synthase‐dihydrofolate reductase bifunctional enzyme give substantial evidences of substrate channeling along a surface “electrostatic highway,” attention has not been paid to whether the human thymidylate synthase and dihydrofolate reductase, if they are in contact with each other in the multi‐enzyme complex, are capable of substrate channeling employing surface electrostatics. This work utilizes protein–protein docking, electrostatics calculations, and Brownian dynamics to explore the existence and mechanism of the substrate channeling between the human thymidylate synthase and dihydrofolate reductase. The results show that the bound human thymidylate synthase and dihydrofolate reductase are capable of substrate channeling and the formation of the surface “electrostatic highway.” The substrate channeling efficiency between the two can be reasonably high and comparable to that of the protozoa.
Keywords: metabolon, substrate channeling, electrostatic channeling, dihydrofolate reductase, thymidylate synthase, bifunctional DHFR‐TS, folate, one‐carbon metabolism
Introduction
Metabolons, transient non‐covalently bound multi‐enzyme complexes, are thought to be important for the organization of enzymes and the regulation of their reactions in vivo. Metabolons are usually composed of enzymes that catalyze the sequential reactions in an enzymatic pathway. One of the advantages of metabolon formation is substrate channeling.1 Substrate channeling is the direct transportation of a reaction intermediate from one enzyme active site to the next without prior release into the bulk solution. It can facilitate the enzymatic reactions in several ways. For example, it can reduce the lag time between two reactions and it can protect labile intermediates from the aqueous environment.2 While the existence of metabolons has been controversial,3 convincing evidence has been been established for substrate channeling.2, 4, 5 Among covalently bound enzymes, channeling has been observed in the protozoa bifunctional dihydrofolate reductase‐thymidylate synthase enzyme.6
Thymidylate synthase (TS) and dihydrofolate reductase (DHFR) are ubiquitous enzymes across organisms. TS is a homodimer; it catalyzes the reductive methylation of deoxyuridine monophosphate by H2C · H4folate to deoxythymidine monophosphate and H2folate.7 DHFR is a monomer and it catalyzes the reduction of H2folate to H4folate, replenishing the intra‐cellular H4folate pool.8 The production and regulation of DHFR and TS are closely coupled to the cell cycle because they participate in the sole de novo synthesis pathway of thymidylate, a building block of DNA.9, 10 Insufficient DHFR and TS activity leads to thymidylate deficiency, halted cell cycle and eventual cell death.11 Because of the crucial role that DHFR and TS play in living organisms, they have attracted much biological and biomedical interest. For example, they are drug targets in cancer treatments.12 Species‐specific inhibitors of DHFR and TS are also used to eliminate pathogens in the human body, such as the parasitic protozoa that cause severe epidemics like malaria.13, 14, 15, 16
While the protozoa DHFR and TS are known to be able to channel their intermediate substrate H2folate,6 such possibility for the human counterparts is unknown. The protozoa DHFR and TS are fused bifunctional enzymes translated from a single gene. Each monomer of the TS homodimer is covalently attached to one DHFR monomer (Supporting Information Fig. S1). The negatively charged H2folate produced by the protozoa TS is channeled directly to DHFR through a sequence of positively charged surface residues connecting the two active sites, a so‐called “electrostatic highway”.17, 18, 19 The human DHFR and TS are separate enzymes and each has its own gene.20, 21 However, it has been reported that the human DHFR and TS are distributed in the same cellular compartments22, 23 and may even participate in binding in the same metabolic multi‐enzyme complex, for example, the replitase complex.24, 25 Also, an experimental paper shows that, in vitro, one human TS dimer binds to up to six human DHFR monomers.26 Furthermore, there exists example of surface electrostatic substrate channeling between non‐covalently bound proteins in mammals, the porcine citrate synthase and malate dehydrogenase.27, 28, 29 Given the evidence above, it is possible that some form of substrate channeling exists between the bound human DHFR and TS in vivo. In this work, the simplest scenario of such possibility is explored computationally, where one human DHFR monomer binds to one human TS dimer.
Computational work on the protozoa DHFR‐TS bifunctional enzyme substrate channeling has previously been conducted30, 31 where the substrate channeling efficiency under different ionic strengths and charge mutations are calculated by Brownian dynamics methods.32, 33, 34 Here, a similar approach is taken, but since the human DHFR and TS are separate proteins, their possible bound‐states are first predicted by protein‐protein docking software.35 The substrate channeling behaviors of these bound‐state structures are then studied by electrostatics and Brownian dynamics calculations. It is found that there exist human DHFR‐TS bound‐states that are capable of channeling its substrate through surface electrostatics, and that their substrate channeling efficiencies can be as high as that of protozoa.
Results
Human DHFR‐TS binding poses
The bound‐states of the separated human DHFR and TS proteins are first predicted by the rigid‐body protein‐protein docking software ClusPro.36, 37 In the ClusPro run, the top 1000 lowest energy docking poses of DHFR‐TS are grouped into 30 clusters and the lowest energy poses of each cluster form the final 30 docking poses (Fig. 1). Twenty‐two of the 30 poses are on the top side of TS and 8 are on the bottom side. The “top side” is defined as the side through which the protozoa DHFR binds to the protozoa TS. Among the poses on the same side of TS, no two share similar orientations relative to TS.
Figure 1.
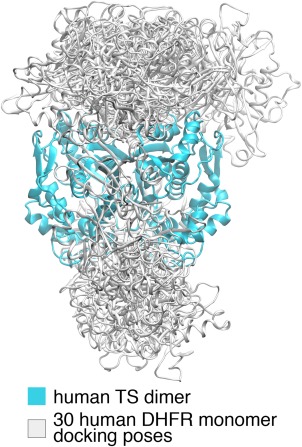
The 30 ClusPro docking poses. The cyan domain is the human TS dimer (PDB ID: 1HVY). The overlapping white domains are the ClusPro human DHFR (PDB ID: 1DHF) docking poses.
The “electrostatic highway”
The main substrate channeling mechanism of the protozoa DHFR‐TSs is the so called “electrostatic highway.”19 For the discussions in this work, the “electrostatic highway” is defined as a continuous region of positive electrostatic potential on the molecular surface that connects the DHFR and TS active sites.
In each human DHFR‐TS binding pose, the DHFR active site can take up substrates originated from either of the two TS active sites and so two different “electrostatic highways” may be formed. 30 ClusPro poses offer 60 DHFR‐TS active site pairs. The “electrostatic highway” is observed in 17 out of the 60 pairs. And in 1 of the 30 poses, a “highway” is formed between the DHFR active site and both of the TS active sites. An example of the human DHFR‐TS “electrostatic highway” is shown in Fig. 2. It is compared to the well‐studied Leishmania major “electrostatic highway.”19, 30, 31 Both “highways” not only connect the DHFR and TS active sites, but also cover the whole region between them completely. More illustrations of the “electrostatic highway” from the 17 DHFR‐TS active site pairs can be found in the Supporting Information Fig. S2. The electrostatic potentials in Fig. 2 are calculated by software APBS.38
Figure 2.
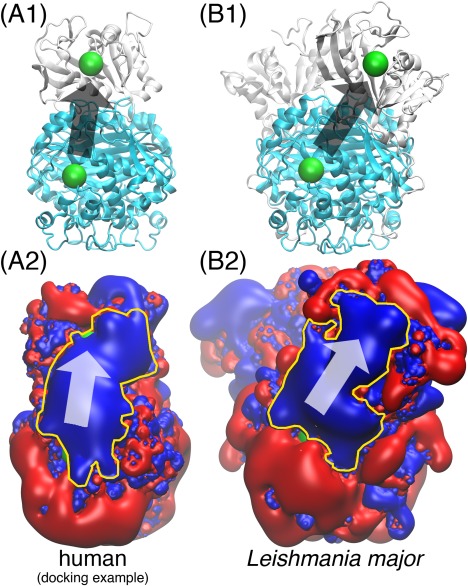
The human DHFR‐TS “electrostatic highway”. In (A1) and (B1), the cyan domain is the TS dimer; the white domain is DHFR (one monomer in (A1), two monomers in (B1)); the green spheres mark the location of the enzyme active sites. (A2) and (B2) display the electrostatics of the corresponding proteins in (A1) and (B1) with red and blue being the and electrostatic potential iso‐surfaces; The yellow contours highlight the “electrostatic highways.” The arrows in all four subfigures indicate the direction of substrate channeling. All of the proteins are displayed 90 degrees sideways compared to the front view in Fig. 1. The human DHFR and TS structures are the same as those used in Fig. 1; the Leishmania major DHFR‐TS structure is taken from Ref. 19. The docked human DHFR‐TS poses are capable of forming “electrostatic highways” for substrate channeling in a similar way to the Leishmania major DHFR‐TS, which is a well‐demonstrated case.
Channeling efficiency
After observing the “electrostatic highway” in a number of human DHFR‐TS docking poses, the “highways'” effects on substrate channeling is studied. Substrate channeling is quantified through the substrate channeling efficiencies calculated by Brownian dynamics software BrownDye.34 The substrate channeling efficiency is the percentage of substrates that started off at a TS active site and reach the DHFR active site by diffusion before escaping into the bulk solution. In the DHFR‐TS system, the diffusion of the charged substrate is heavily influenced by the electrostatic potential field exerted by the proteins. The “electrostatic highway,” which is a protein surface region with positive electrostatic potential, guides the diffusion of the substrate by opposite‐charge attractions and achieves the observed substrate channeling.
Fig. 3(A) shows that the channeling efficiency of the substrate is statistically significantly higher when there is an “electrostatic highway” connecting the DHFR and TS active sites. The channeling efficiency can be as large as 60%, comparable to the roughly 70% channeling efficiency of Leishmania major, which is known for its strong substrate channeling, calculated by BrownDye in previous work.31 Fig. 3(B) shows that if the substrate is neutral, the channeling efficiency distribution for the DHFR‐TS active site pairs connected by “electrostatic highways” (gray) overlaps with the distribution for the pairs that have no “highway” present (white). The “electrostatic highway” makes little contribution to the channeling of neutral substrates.
Figure 3.
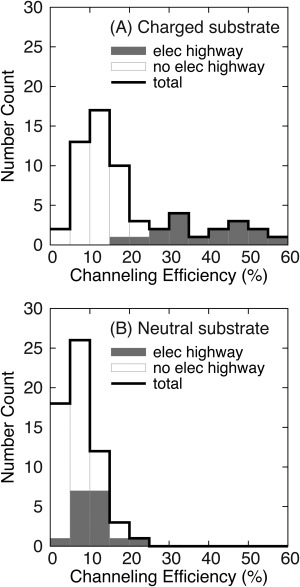
The influence of the “electrostatic highway” on the channeling of charged substrates. Among the 60 DHFR‐TS active site pairs from the 30 ClusPro poses, “elec highway” (gray) represents the 17 pairs with observed “highways”, and “no elec highway” (white) represents the other 43 pairs. This plot is a histogram with bin size of 5% and the first bin is (0%, 5%]. (A) For charged substrates, channeling is significantly enhanced by an “electrostatic highway.” B) When the substrate is neutral, the existence of an “electrostatic highway” does not have a noticeable impact on the substrate channeling efficiency.
Discussion
The human DHFR‐TS substrate channeling through “electrostatic highways” is studied with a simple model of one DHFR bound to one TS. The one‐to‐one binding between DHFR and TS is likely not the exact case in vivo. First, multiple DHFRs may bind to one TS.26 Second, other proteins may bind to DHFR and TS, for example those participating in the DNA synthesis.24, 25 Third, the human DHFR and TS are not guaranteed to bind to each other (although significant channeling may still occur since the two proteins are colocalized in the same cellular compartment39). This study does not aim to convince the readers that the electrostatic channeling observed here is the catalytic strategy that the human DHFR and TS use, but rather that the human DHFR‐TS has the capability of electrostatic substrate channeling if their in vivo binding conformations allow.
Rigid‐body protein–protein docking is employed here with the goal of searching for the possible formation of an “electrostatic highway” in the human DHFR‐TS bound‐states. It is only a minor goal of the docking calculations to predict the correct and stable binding poses between DHFR and TS. Without higher level calculations, such as molecular dynamics free energy calculations,40 the ranking of the stability of a binding pose cannot be conclusive. Note that the binding pose for the native protozoa DHFR‐TS (Supporting Information Fig. S1(B)) is different from what has been predicted by docking (Fig. 1). However, if the human DHFR is aligned to the Leishmania major binding pose by VMD MultiSeq,41, 42 an “electrostatic highway” is observed (Supporting Information Fig. S3). A likely reason why ClusPro does not consider the protozoa binding pose for the human proteins is a lack of steric complementarity. Leishmania major DHFR and TS each has about 35 residues on the binding interface while if the human proteins are docked similarly, there are only about 10 interface residues from each protein. This shows the limitation of rigid‐body protein–protein docking because induced conformational changes could happen upon DHFR‐TS binding.
The phrase “electrostatic highway” needs some clarification because, instead of literally speeding up the diffusion of the substrate, the “highway” serves as a “trap” to the oppositely charged substrate, limiting its escape into the bulk. In the Brownian dynamics calculations by BrownDye, the bulk extends to infinity and the substrate concentration in the bulk is zero, the chance of the substrate ever returning to DHFR quickly decreases to effectively zero as it diffuses away from the protein. Assuming the substrate reacts with the DHFR active site on contact, an “electrostatic highway” permanently “speeds up” the reaction by decreasing the number of escaped substrates. On the other hand, in small closed compartments, the “electrostatic highway” will only decrease the time it takes for the DHFR reaction to reach its full speed before the equilibrium of the bulk substrate concentration, and will not have a permanent speed‐up of the DHFR reaction. Also, the “electrostatic highway” in the DHFR‐TS system is itself not directional.30 A directional electrostatic channeling can theoretically be achieved by a monotonically increasing surface density of the charged amino acids along the path between the two active sites, but this is not observed for DHFR‐TS.
The enzyme specificity constants for the substrates for both of the human DHFR and TS are between 1 and 10 .43, 44 However, the relative reaction rates of the two enzymes do not affect the results of the BrownDye simulations, because the BrownDye simulations here observe the channeling efficiency at the initial time point of the DHFR‐TS reaction system way before it reaches equilibrium. To be more specific, at the initial moment, the substrates for TS are just added and the first few TS products are just produced, the concentrations of the products of TS in bulk are effectively zero. On the other hand, if one wants to simulate the concentration profiles of the DHFR and TS substrates and products over time, different reaction rates of the two enzymes will lead to different behaviors of the system.
The BrownDye Brownian dynamics calculations in this work are set up specifically to study the effect of electrostatics on substrate channeling; the calculations consider mainly the electrostatic and the two‐body hydrodynamic forces between the protein and the substrate. The protein and the substrate are also modeled as rigid bodies. Although electrostatics has been widely considered to be the cause of substrate channeling of the protozoa DHFR‐TS,18, 19, 30, 31 the BrownDye results do not rule out the possibilities of other channeling mechanisms. In fact, in the protozoa system, experiments have brought up the possibilities for more complicated electrostatic channeling mechanisms, on top of the simple “electrostatic highway trapping,” or additional non‐electrostatic channeling mechanisms.45, 46 Comprehensive mutation experiments on the charged residues along the electrostatic highway of the human DHFR‐TS are needed and it is hoped that the type of computational study in this work can be used to compare to those experiments to test whether and how mutations can alter the observed electrostatic restriction of substrate channeling and diffusion. Tests with molecular dynamics simulations that are capable of modeling the protein–ligand interactions more accurately and modeling the flexibility of proteins are also desirable.
In conclusion, with protein–protein docking, it is found that there exist bound‐state conformations of the human DHFR and TS proteins where a continuous positive surface potential region, an “electrostatic highway,” connecting the TS and DHFR active sites is formed. This “electrostatic highway” is formed in a similar way to what has been observed in the protozoa DHFR‐TS, which are known for substrate channeling through surface electrostatics. Brownian dynamics simulations have further shown that with the “electrostatic highway” a significantly greater number of negatively charged substrates are passed from the TS to the DHFR binding site without escaping into the bulk. The human DHFR‐TS have the capability of electrostatic substrate channeling if their in vivo binding conformations allow. However, higher level computations are needed for more thorough analysis and more convincing evidence on the DHFR‐TS electrostatic substrate channeling hypothesis.
Materials and Methods
Protein structures
The human DHFR and TS structures are taken from the Protein Data Bank (PDB)47 (PDB IDs 1DHF20 and 1HVY21). Because 1DHF is missing ligand NADPH, the NADPH binding pose in PDB ID 2W3M48 is aligned into the 1DHF structure by the VMD41 tool MultiSeq.42
The protein PDB structures submitted to the online docking server are prepared by Maestro Protein Preparation Wizard,49 and the protonation state of histidine residues is predicted by program PROPKA50 embedded in the Protein Preparation Wizard. Because the docking pose PDB files output by the protein‐protein docking software omit the hydrogens, program Reduce,51 due to its batch processing capability, is used to add the hydrogens back into the docking poses before the following electrostatics and Brownian dynamics calculations.
Protein–protein docking
Online rigid protein–protein docking server ClusPro36, 37 is used to dock one human DHFR monomer onto one human TS dimer. ClusPro is chosen because it is the best‐performing protein‐protein docking software in CAPRI52 rounds 22 to 27.53, 54 All of the docking jobs are submitted using the web server's default settings. The 30 ClusPro poses are scored using their “Balanced” scoring function.
The validity of ClusPro is tested by manually separating the known protozoa Leishmania major DHFR‐TS structure (obtained from the authors of Ref. 19) and docking the DHFR back onto the TS. ClusPro predicts the Leishmania major DHFR and TS native binding poses correctly. This result is included in the Supporting Information Fig. S4.
Brownian dynamics
Software BrownDye34 is used to quantify the substrate channeling efficiency of the human DHFR‐TS binding poses. To calculate the channeling efficiency between a TS active site and a DHFR active site, BrownDye is set up to run 10,000 Brownian dynamics32 trajectories of substrate H2folate starting at the TS active site diffusing under the influence of stochastic forces and the hydrodynamic and electrostatic forces between the substrate and the protein. The substrate channeling efficiency equals to the number of trajectories where the substrate reacted with the DHFR active site before escaping into the bulk over the total number of trajectories, 10,000. A Brownian dynamics trajectory is terminated on the first occasion of substrate reaction or escape. Substrate reaction is defined by the substrate diffusing into a spherical region of radius 12.5Å centered around the DHFR active site (Supporting Information Fig. S5); Substrate escape is defined by the substrate leaving a large spherical boundary in the bulk solution centered around the protein (Supporting Information Fig. S6). This “escape sphere” is significantly larger than the size of the protein and its radius is analytically determined.55 More simulation details can be found in the Supporting Information Fig. S5 and S6.
Electrostatic calculations
To calculate the electrostatic forces in BrownDye and to visualize the “electrostatic highway” along the DHFR‐TS surface, the electrostatic potential field of the DHFR‐TS system is needed. The Adaptive Poisson–Boltzmann Solver (APBS) software is used.38, 56 APBS takes in the charges and atomic radii of a protein and outputs the electrostatic potential field generated by the protein. The protein charge and atomic radii used in APBS are taken from the CHARMM27 force field57 and the protein is solvated in 0.15M NaCl solution. An example of the APBS input file is attached in the Supporting Information Section S7.
Supporting information
Supporting Information
Acknowledgments
Nuo Wang thanks Dr. Shenggao Zhou for the very helpful discussions on electrostatics calculations.
References
- 1. Srere PA (1987) Complexes of sequential metabolic enzymes. Annu Rev Biochem 56:89–124. [DOI] [PubMed] [Google Scholar]
- 2. Huang X, Holden HM, Raushel FM (2001) Channeling of substrates and intermediates in enzyme‐catalyzed reactions. Annu Rev Biochem 70:149–180. [DOI] [PubMed] [Google Scholar]
- 3. Williamson M. How proteins work. Chapter 9.4: the metabolon concept. Garland Science, New York and London, pp. 338–344. [Google Scholar]
- 4. Anderson KS (1999) Fundamental mechanisms of substrate channeling. Methods Enzymol 308:111–145. [DOI] [PubMed] [Google Scholar]
- 5. Miles EW, Rhee S, Davies DR (1999) The molecular basis of substrate channeling. J Biol Chem 274:12193–12196. [DOI] [PubMed] [Google Scholar]
- 6. Ivanetich KM, Santi DV (1990) Bifunctional thymidylate synthase‐dihydrofolate reductase in protozoa. FASEB J 4:1591–1597. [DOI] [PubMed] [Google Scholar]
- 7. Carreras CW, Santi DV (1995) The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem 64:721–762. [DOI] [PubMed] [Google Scholar]
- 8. Schnell JR, Dyson HJ, Wright PE (2004) Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct 33:119–140. [DOI] [PubMed] [Google Scholar]
- 9. Johnson LF (2012) Recombinant DNA and cell proliferation. Chapter 2: Expression of dihydrofolate reductase and thymidylate synthase genes in mammalian cells. Academic Press, Orlando, Florida, pp 25–48. [Google Scholar]
- 10. Zick A, Onn I, Bezalel R, Margalit H, Shlomai J (2005) Assigning functions to genes: identification of S‐phase expressed genes in Leishmania major based on post‐transcriptional control elements. Nucleic Acids Res 33:4235–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sienkiewicz N, Jarostawski S, Wyllie S, Fairlamb AH (2008) Chemical and genetic validation of dihydrofolate reductase‐thymidylate synthase as a drug target in African trypanosomes. Mol Microbiol 69:520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walling J (2006) From methotrexate to pemetrexed and beyond: a review of the pharmacodynamic and clinical properties of antifolates. Investig New Drugs 24:37–77. [DOI] [PubMed] [Google Scholar]
- 13. Gilbert IH (2002) Inhibitors of dihydrofolate reductase in leishmania and trypanosomes. Biochim Biophys Acta Mol Basis Dis 1587:249–257. [DOI] [PubMed] [Google Scholar]
- 14. Rathod PK, Phillips MA (2003) Prized malaria drug target nailed. Nat Struct Biol 10:316–318. [DOI] [PubMed] [Google Scholar]
- 15. Anderson AC (2005) Targeting DHFR in parasitic protozoa. Drug Discov Today 10:121–128. [DOI] [PubMed] [Google Scholar]
- 16. Yuthavonga Y, Tarnchompooa B, Vilaivanb T, Chitnumsuba P, Kamchonwongpaisana S, Charmanc SA, McLennanc DN, Whitec KL, Vivasd L, Bongardd E, Thongphanchanga C, Taweechaia S, Vanichtanankula J, Rattanajaka R, Arwona U, Fantauzzie P, Yuvaniyamaf J, Charmanc WN, Matthewse D (2012) Malarial dihydrofolate reductase as a paradigm for drug development against a resistance‐compromised target. Proc Nalt Acad Sci USA 109:16823–16828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meek TD, Garvey EP, Santi DV (1985) Purification and characterization of the bifunctional thymidylate synthetase‐dihydrofolate reductase from methotrexate‐resistant Leishmania tropica. Biochemistry 24:678–686. [DOI] [PubMed] [Google Scholar]
- 18. Liang P‐H, Anderson KS (1998) Substrate channeling and domain–domain interactions in bifunctional thymidylate synthase‐dihydrofolate reductase. Biochemistry 37:12195–12205. [DOI] [PubMed] [Google Scholar]
- 19. Knighton DR, Kan C‐C, Howland E, Janson CA, Hostomska Z, Welsh KM, Matthews DA (1994) Structure of and kinetic channelling in bifunctional dihydrofolate reductase‐thymidylate synthase. Nat Struct Biol 1:186–194. [DOI] [PubMed] [Google Scholar]
- 20. Davies JF, Delcamp TJ, Prendergast NJ, Ashford VA, Freisheim JH, Kraut J (1990) Crystal structures of recombinant human dihydrofolate reductase complexed with folate and 5‐deazafolate. Biochemistry 29:9467–9479. [DOI] [PubMed] [Google Scholar]
- 21. Phan J, Koli S, Minor W, Dunlap RB, Berger SH, Lebioda L (2001) Human thymidylate synthase is in the closed conformation when complexed with dUMP and raltitrexed, an antifolate drug. Biochemistry 40:1897–1902. [DOI] [PubMed] [Google Scholar]
- 22. Tibbetts AS, Appling DR (2010) Compartmentalization of mammalian folate‐mediated one‐carbon metabolism. Annu Rev Nutr 30:57–81. [DOI] [PubMed] [Google Scholar]
- 23. Stover PJ, Field MS (2011) Trafficking of intracellular folates. Adv Nutr 2:325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Prem veer Reddy G, Pardee AB (1980) Multienzyme complex for metabolic channeling in mammalian DNA replication. Proc Natl Acad Sci USA 77:3312–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murthy S, Prem veer Reddy G (2006) Replitase: complete machinery for DNA synthesis. J Cell Physiol 209:711–717. [DOI] [PubMed] [Google Scholar]
- 26. Antosiewicz A, Senkara E, Cieśla J (2015) Quartz crystal microbalance with dissipation and microscale thermophoresis as tools for investigation of protein complex formation between thymidylate synthesis cycle enzymes. Biosens Bioelectron 15:36–42. [DOI] [PubMed] [Google Scholar]
- 27. Elcock AH, McCammon JA (1996) Evidence for electrostatic channeling in a fusion protein of malate dehydrogenase and citrate synthase. Biochemistry 35:12652–12658. [DOI] [PubMed] [Google Scholar]
- 28. Shatalin K, Lebreton S, Rault‐Leonardon M, Vélot C, Srere PA (1999) Electrostatic channeling of oxaloacetate in a fusion protein of porcine citrate synthase and porcine mitochondrial malate dehydrogenase. Biochemistry 38:881–889. [DOI] [PubMed] [Google Scholar]
- 29. Wu F, Minteer S (2014) Krebs cycle metabolon: structural evidence of substrate channeling revealed by cross‐linking and mass spectrometry. Angew Chem Int Ed 54:1851–1854. [DOI] [PubMed] [Google Scholar]
- 30. Elcock AH, Potter MJ, Matthews DA, Knighton DR, McCammon JA (1996) Electrostatic channeling in the bifunctional enzyme dihydrofolate reductase‐thymidylate synthase. J Mol Biol 262:370–374. [DOI] [PubMed] [Google Scholar]
- 31. Metzger VT, Eun C, Kekenes‐Huskey PM, Huber GA, McCammon JA (2014) Electrostatic channeling in P. falciparum DHFR‐TS: Brownian dynamics and Smoluchowski modeling. Biophys J 107:2394–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ermak DL, McCammon JA (1978) Brownian dynamics with hydrodynamic interactions. J Chem Phys 69:1352–1360. [Google Scholar]
- 33. Madura JD, Briggs JM, Wade RC, Davis ME, Lutye BA, Ilin A, Antosiewicz J, Gilson MK, Bagheri B, Scott LR, McCammon JA (1995) Electrostatics and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics program. Comp Phys Commun 91:57–95. [Google Scholar]
- 34. Huber GA, McCammon JA (2010) Browndye: a software package for Brownian dynamics. Comput Phys Commun 181:1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vakser IA (2014) Protein–protein docking: from interaction to interactome. Biophys J 107:1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kozakov D, Brenke R, Comeau SR, Vajda S (2006) PIPER: an FFT‐based protein docking program with pairwise potentials. Proteins Struct Funct Bioinform 65:392–406. [DOI] [PubMed] [Google Scholar]
- 37. Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, Vajda S (2013) How good is automated protein docking? Proteins Struct Funct Bioinform 81:2159–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98:10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roberts CC, Chang CA (2015) Modeling of enhanced catalysis in multienzyme nanostructures: effect of molecular scaffolds, spatial organization, and concentration J Chem Theory Comput 11:286–292. [DOI] [PubMed] [Google Scholar]
- 40. Karplus M, McCammon JA (2002) Molecular dynamics simulations of biomolecules. Nat Struct Biol 9:646–652. [DOI] [PubMed] [Google Scholar]
- 41. Humphrey W, Dalke A, Schulten K (1996). VMD—visual molecular dynamics. J Mol Graph 14:33–38. [DOI] [PubMed] [Google Scholar]
- 42. Roberts E, Eargle J, Wright D, Luthey‐Schulten Z (2006). MultiSeq: unifying sequence and structure data for evolutionary analysis BMC Bioinform 7:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schweitzer BI, Srimatkandada S, Gritsman H, Sheridan R, Venkataraghavan R, Bertino JR (1989) Probing the role of two hydrophobic active site residues in the human dihydrofolate reductase by site‐directed mutagenesis J Biol Chem 264:20786–20795. [PubMed] [Google Scholar]
- 44. Salo‐Ahen OMH, Tochowicz A, Pozzi C, Cardinale D, Ferrari S, Boum Y, Mangani S, Stroud RM, Saxena P, Myllykallio H, Costi MP, Ponterini G, Wade RC (2015) Hotspots in an obligate homodimeric anticancer target. structural and functional effects of interfacial mutations in human thymidylate synthase. J Med Chem 58:3572–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Atreya CE, Johnson EF, Williamson J, Chang S‐Y, Liang P‐H, Anderson KS (2003) Probing electrostatic channeling in protozoal bifunctional thymidylate synthase‐dihydrofolate reductase using site‐directed mutagenesis. J Biol Chem 278:28901–28911. [DOI] [PubMed] [Google Scholar]
- 46. Sharma H, Landau MJ, Vargo MA, Spasov KA, Anderson KS (2013) First three‐dimensional structure of Toxoplasma gondii thymidylate synthase‐dihydrofolate reductase: insights for catalysis, interdomain interactions, and substrate channeling. Biochemistry 52:7305–7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leung AKW, Ross LJ, Zywno‐Van Ginkel S, Reynolds RC, Seitz LE, Pathak V, Barrow WW, White EL, Suling WJ, Piper JR, Borhani DW (2009) PDB entry 2W3M: Structural basis for selective inhibition of Mycobacterium avium dihydrofolate reductase by a lipophilic antifolate. DOI:10.2210/pdb2w3m/pdb
- 49. Schrödinger, LLC , New York, NY , Maestro, version 10.1, 2015. Schrödinger Release; 2015–1. http://www.schrodinger.com/Maestro/ [Google Scholar]
- 50. Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH (2011) PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput 7:525–537. [DOI] [PubMed] [Google Scholar]
- 51. Word JM, Lovell SC, Richardson JS, Richardson DC (1999) Asparagine and glutamine: using hydrogen atom contacts in the choice of sidechain amide orientation. J Mol Biol 285:1735–1747. [DOI] [PubMed] [Google Scholar]
- 52. Janin J, Henrick K, Moult J, Eyck LT, Sternberg MJE, Vajda S, Vakser I, Wodak SJ (2003) CAPRI: a critical assessment of predicted interactions. Proteins Struct Funct Bioinform 52:2–9. [DOI] [PubMed] [Google Scholar]
- 53. Lensink MF, Wodak SJ (2013) Docking, scoring, and affinity prediction in CAPRI. Proteins Struct Funct Bioinform 81:2082–2095. [DOI] [PubMed] [Google Scholar]
- 54. Rodrigues JPGLM, Bonvin AMJJ (2014) Integrative computational modeling of protein interactions. FEBS J 281:1988–2003. [DOI] [PubMed] [Google Scholar]
- 55. Luty BA, McCammon JA, Zhou HX (1992) Diffusive reaction rates from Brownian dynamics simulations: replacing the outer cutoff surface by an analytical treatment. J Chem Phys 97:5682. [Google Scholar]
- 56. Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res 35:W522–W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Foloppe N, MacKerell AD, Jr. (2000) All‐atom empirical force field for nucleic acids: I. parameter optimization based on small molecule and condensed phase macromolecular target data. J Comp Chem 21:86–104. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


