Abstract
The glycosylation of flavonoids increases their solubility and stability in plants. Flowers accumulate anthocyanidin and flavonol glycosides which are synthesized by UDP-sugar flavonoid glycosyltransferases (UFGTs). In our previous study, a cDNA clone (Fh3GT1) encoding UFGT was isolated from Freesia hybrida, which was preliminarily proved to be invovled in cyanidin 3-O-glucoside biosynthesis. Here, a variety of anthocyanin and flavonol glycosides were detected in flowers and other tissues of F. hybrida, implying the versatile roles of Fh3GT1 in flavonoids biosynthesis. To further unravel its multi-functional roles, integrative analysis between gene expression and metabolites was investigated. The results showed expression of Fh3GT1 was positively related to the accumulation of anthocyanins and flavonol glycosides, suggesting its potential roles in the biosynthesis of both flavonoid glycosides. Subsequently, biochemical analysis results revealed that a broad range of flavonoid substrates including flavonoid not naturally occurred in F. hybrida could be recognized by the recombinant Fh3GT1. Both UDP-glucose and UDP-galactose could be used as sugar donors by recombinant Fh3GT1, although UDP-galactose was transferred with relatively low activity. Furthermore, regiospecificity analysis demonstrated that Fh3GT1 was able to glycosylate delphinidin at the 3-, 4-′, and 7- positions in a sugar-dependent manner. And the introduction of Fh3GT1 into Arabidopsis UGT78D2 mutant successfully restored the anthocyanins and flavonols phenotypes caused by lost-of-function of the 3GT, indicating that Fh3GT1 functions as a flavonoid 3-O-glucosyltransferase in vivo. In summary, these results demonstrate that Fh3GT1 is a flavonoid 3-O-glycosyltransferase using UDP-glucose as the preferred sugar donor and may involve in flavonoid glycosylation in F. hybrida.
Keywords: Freesia hybrida, flavonoid, flavonoid 3-O-glycosyltransferase, substrate specificity, regiospecificity
Introduction
Flavonoids, the important polyphenolic secondary metabolites, are widely distributed in plant species and possess diverse biological functions including pollination, pigmentation, auxin transport inhibition, UV light protection and other defense mechanism (Brenda, 2001; Koes et al., 2005; Charles et al., 2010). In plants, some biological processes, such as cell-to-cell communication, signal transduction, and transcriptional regulation are also affected by flavonoids (Almudena et al., 2012). Flavonoids consist mainly of anthocyanins, phlobaphene pigments, and proanthocyanidins, as well as the flavonols, flavanones, and isoflavonoids (Grotewold, 2005; Taylor and Grotewold, 2005). Among them, anthocyanins are broadly existed in flowering plants to provide flowers and fruits with the red, purple, and blue pigmentation for attracting pollinators or seed dispersers. Flavonols, colorless co-pigments, affect the brightness and brilliance of colors and have vital roles in pollen germination (Ferreyra et al., 2012; Zhao and Tao, 2015). In addition, anthocyanins and flavonols also play an important role in human health and have potential medicinal uses, as the intake of them can protect against cardiovascular disease, cancer, and many other diseases (Petroni et al., 2014; Santos-Buelga et al., 2014).
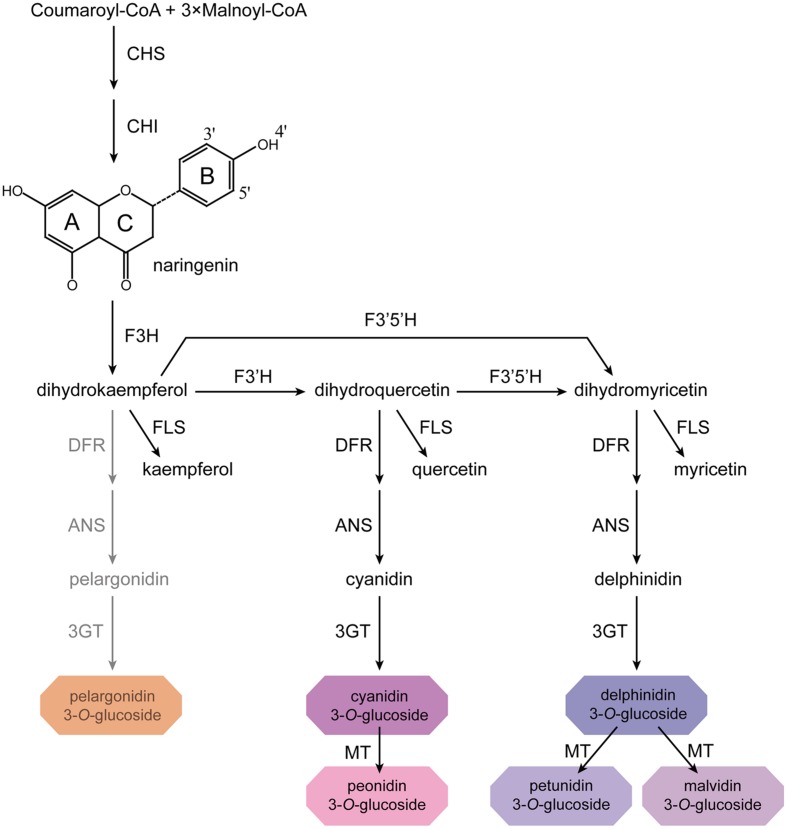
The biosynthesis of anthocyanins is derived from the flavonoid branch of the phenylpropanoid metabolic pathway and has been well studied in different plants including Zea mays, Petunia hybrida, Arabidopsis thaliana, and Antirrhinum majus (Gonzalez et al., 2008; Petroni and Tonelli, 2011; Wang et al., 2013). The anthocyanin biosynthetic pathway starts with the condensation of 4-coumaroyl-CoA with three molecules of malonyl-CoA, leading to the formation of naringenin chalcone by CHS (Figure 1). Next, the naringenin chalcone is isomerized to naringenin through the action of CHI. After hydroxylation at the three position of naringenin, dihydroflavonols are formed and will be further catalyzed to yield leucoanthocyanidins by DFR. Furthermore, FLS can also use dihydroflavonols as substrates to produce flavonols. Subsequently, the action of ANS generates the anthocyanidins which are then converted to glucosylated anthocyanins by UDP-glucose: 3GT (Tanaka et al., 2008). Recent studies have demonstrated that nearly all the enzymes involved in anthocyanidin biosynthesis have been isolated and functionally characterized (Quideau, 2006; Almudena et al., 2012), but the sequential modification of anthocyanidins metabolism, such as glycosylation, methylation and acylation, remains relatively unexplored.
FIGURE 1.
Proposed flavonoid pathway in Freesia hybrida flower. Hoary areas indicate the biosynthesis of pelargonidin-based pigments is blocked.
Many anthocyanins as well as other flavonoids are known to exist in glycosylated form in plants. During the biosynthesis of these glycosides, glycosylation is often the final step and serves a variety of roles in plant metabolism. For example, glycosylation can enhance the stability and solubility of the acceptor molecule and affect their subcellular localization (Thomas and Patrik, 2000; Wang, 2009). Meanwhile, it also regulates cellular homeostasis and plant growth, or may involve in the detoxification of exogenous toxins (Lim and Bowles, 2004; Brazier and Edwards, 2005). The enzymes that catalyze the formation of glycoside are known as uridine diphosphate (UDP): flavonoid glycosyltransferases (UFGTs), which transfer UDP-activated sugar moieties to low molecular-weight acceptor aglycone. UFGTs can be identified by the presence of PSPG (Plant Secondary Product Glycosyltransferase) box, a 44-amino acid conserved motif that is involved in binding substrates to the UDP moiety of the sugar donor (Joe et al., 2001; Gachon et al., 2005; Offen et al., 2006). Several different UDP-sugars have been reported to be donors for UFGTs including UDP-glucose, UDP-galactose, UDP-rhamnose, and UDP-xylose. Crystal structures of UFGTs and mutation analyses have demonstrated that sugar selectivity is relatively determined by the last residue in the PSPG motif (Wang, 2009). Moreover, these enzymes exhibit broad substrate specificity but exert strict regioselectivity in many cases (Thomas and Patrik, 2000). For example, 3GTs from petunia and Arabidopsis transfer sugar donors to only 3-position of both anthocyanidins and flavonols (Mami et al., 2002; Takayuki et al., 2005). To date, genes encoding UFGTs have been cloned and identified in a number of plant species. According to the specificity for substrates, they can be divided into two groups. The first group is responsible for attaching a sugar to flavonoids, including flavonoid 3-O-glycosyltransferase (Ford et al., 1998; Mami et al., 2002; Zhao et al., 2012) and flavonoid 5, 3-O-glucosyltransferase (Jun et al., 2005). The second group catalyzes the glycosyl transfer toward flavonoid glycoside leading to the formation of diglycoside, such as flavonoid 3-O-glucoside 2”-O-glucosyltransferase (Morita et al., 2005). Among these glycosyltransferases, flavonoid 3-O-glycosyltransferases are best characterized, because of their essential roles in flavonoids biosynthesis (Jun et al., 1998; Griesser et al., 2008; Hu et al., 2011).
Anthocyanin and flavonol accumulation has significant effects on flower quality, and many 3GT genes involved in the accumulation of anthocyanins and flavonols have been identified in dicotyledonous flower plant species, such as Antirrhinum majus (Martin et al., 1991), Gentiana triflora (Yoshikazu et al., 1996), Senecio × hybridus (Taylor and Grotewold, 2005), and petunia (Mami et al., 2002). However, only few studies have been reported on the characterization of 3GT genes in monocotyledonous ornamental plants. Until now, only two 3GT genes from monocotyledonous flower plants were confirmed to participate in the biosynthesis of anthocyanin (Yoshihara et al., 2005; Chen et al., 2011).
Freesia hybrida is one kind of monocotyledonous ornamental species that distributes widely in the world and belongs to the Iridaceae. Because of its diverse flower color, it could be chosen as an ideal material to study the biosynthesis of flavonoids. Moreover, functional characterization of 3GT gene from F. hybrida may also contribute to the study of evolution of the UFGT gene family considering that very few of them have been identified in monocotyledonous ornamental plant. In our previous study, Fh3GT1 was found to be responsible for cyanidin 3-O-glucoside biosynthesis (Sui et al., 2011). However, its roles in the biosynthesis of other anthocyanins and flavonols are still unknown. To further elucidate the versatile functionality of Fh3GT1 systematically, we firstly performed the correlation analysis between the flavonoid glycoside products accumulation and the expression profiles of Fh3GT1 in flowers at different pigmentation stages and in different plant tissues. Meanwhile, the biosynthetic pathway of anthocyanin in Fressia was also proposed on the basis of metabolites. In addition, substrate specificity and kinetic analyses were conducted to identify the activity of Fh3GT1 against various flavonoids. Furthermore, in order to confirm its function in vivo, Fh3GT1 was transformed into the Arabidopsis UGT78D2 mutant to restore the biosynthesis of anthocyanins and flavonols. In consequence, these results not only provide new insight to plant glycosyltransferase structure-function analysis but also useful for manipulating flavonoid biosynthesis in F. hybrida as well as in other monocotyledonous ornamental plants.
Materials and Methods
Plant Materials
Arabidopsis lines used in this study were in the Columbia ecotype background. Wild-type, T-DNA insertion line (UGT78D2), and transgenic seedlings of Arabidopsis (Arabidopsis thaliana) were grown on half-strength MS medium under a controlled condition (16 h of light/8 h of dark cycle at 22°C). For reverse transcription PCR (RT-PCR) and flavonoid analysis, whole seedlings were collected at 7 days after germination and stored at –80°C. Tissues and flowers at different developmental stages from F. hybrida were frozen in liquid nitrogen and kept at –80°C.
Chemicals
UDP-glucose, UDP-galactose, quercetin 3-O-glucoside, delphinidin 3-O-galactoside, cyanidin 3-O-galactoside, delphinidin, cyanidin and malvidin were purchased from Sigma–Aldrich (USA), petunidin and peonidin from Tokiwa Phytochemical (Japan), pelargonidin, delphinidin 3-O-glucoside, cyanidin 3-O-glucoside, malvidin 3-O-glucoside, pelargonidin 3-O-glucoside, petunidin 3-O-glucoside and peonidin 3-O-glucoside from Phytolab (Germany), quercetin, kaempferol from ChromaDex (USA), and 3-O-glucoside of kaempferol from Extrasynthese (France).
Subcellular Localization of Fh3GT1
GFP coding region without stop codon was amplified using primers (GFPF1 and GFPR1) that added XbaI sites to both the 5′end and 3′end of GFP. The fragments were then inserted into the XbaI-digested pBI121-Fh3GT1 (constructed as below). The full length coding region of GFP was also amplified using GFPF2 and GFPR2 (Supplementary Table S1) then subcloned into the pBI121 empty vector to serve as a control for localization experiments. Sequence integrity of the vectors was confirmed by sequencing. The two constructs were transformed to Agrobacterium tumefaciens strain GV3101 using a freeze-thaw method and then introduced into Arabidopsis plants using the floral dip method (Clough and Bent, 1998). Transformants were selected on 1/2 MS medium containing 50 mg L-1 kanamycin. For observation, a small piece of leaf tissue of T2 plants was cut out and mounted in deionized water followed by confocal microscopy.
HPLC Analysis of Anthocyanins and Flavonols Extracted from Freesia hybrida
For the analysis of anthocyanins and flavonols in flowers from F. hybrida, 0.3 g finely ground tissue was extracted with H2O:MeOH:HCl (75/24/1v/v/v) at 4°C for 12 h in the dark and subsequently centrifuged at 12,000 rpm for 10 min. After centrifugation, the supernatant was collected, filtered, and used for HPLC analysis. Chromatographic analysis was carried out on a Shimadzu HPLC system equipped with an autosampler with a 20 μl loop, a LC-6AD HPLC Pump and an ACCHROM XUnion C18 column (250 mm × 4.6 mm, 5 μm). The column was eluted with solvent systems A (5% formic acid in H2O) and B (methanol) under the following conditions: 0-10 min, 14–17% B; 10–35 min, 17–23% B; 35–60min, 23–47% B; 60–67 min, 47–14% B; 67–70 min, 14% B with a flow rate of 1 ml min-1. Detection was monitored at 520 and 360 nm for anthocyanins and flavonols, respectively, and the photodiode array spectra were recorded from 200 to 800 nm.
High performance liquid chromatography-electrospray ionization (ESI)-tandem mass spectrometry (MS) analysis used an API2000 mass spectrometer (AB Sciex) and a SPD-20AV UV/VIS Dectector (Shimadzu, Kyoto, Japan). This mass spectrometer was equipped with an ESI source. Ion Trap source parameters in positive mode were as follows: ESI source voltage, 4.5 kV; gas (N2) temperature, 450°C; declustering potential, +80 V; entrance potential, 10 V; and scan range, m/z 100–1000 units. Metabolites were identified by their retention times, mass spectra, and product ion spectra in comparison with the data of authentic standards. Quantitative analysis was done using the external standard curve calibration of delphinidin 3-O-glucoside and quercetin 3-O-glucoside standards (Fanali et al., 2011). The calibration curves used were linear (R2> 0.99) in the concentration ranges investigated. All the measurements were performed three times independently with three biological replicates.
Expression Analysis
For real-time qPCR, total RNA was isolated from petals, stamens, pistils, calyxes, toruses, scapes, leaves, roots and flowers at different stages of development using RNAiso Plus (TaKaRa). Then total RNA extracted from the above samples was treated with RNase-free DNase I (TaKaRa) and reverse transcribed using M-MLV reverse transcriptase (Promega). Gene-specific primers (PF1/PR1) were designed using Primer Premier 5 and summarized in the Supporting Information (Supplementary Table S1). To investigate the gene expression profiles, real-time qPCR was carried out with ABI StepOne Plus Real-Time PCR System (USA) and SYBR Master Mix (TOYOBO). Thermal cycling conditions were 95 °C for 60 s, then 40 cycles of 95°C for 5 s and 60°C for 60 s, followed by a melting temperature cycle, with constant fluorescence data acquisition from 60 to 95 °C. As an internal control, the 18s rRNA gene was used. To ensure a single amplicon had been generated, the real-time qPCR products were confirmed by agarose gel electrophoresis and DNA sequencing. Data of the gene expression was analyzed with the 2-ΔΔCTmethod (Livak and Schmittgen, 2001), and each data point represents the average of three independent experiments.
Heterologous Expression of Fh3GT1 in E. coli
To express Fh3GT1 in E. coli, the full length cDNA of Fh3GT1 was amplified by PCR using Premix TaqTM (TaKaRa) with the primers PF2 and PR2 (Supplementary Table S1). The PCR product was sub-cloned into the pET-28a (+) His-fusion protein expression vector, and clone authenticity was confirmed by sequencing. The constructed vector was transformed into E. coli BL-21 (DE3) cells for recombinant protein expression. Then, the transformants were pre-cultured at 37°C for 12–14 h in Luria Broth (LB) media containing 100 mg L-1 kanamycin. Two milliliter of the preculture was transferred to the fresh LB media (400 ml) containing the antibiotics and grown at 37°C until an A600 of 0.6 was reached. After the addition of 40 μl of 1 M isopropyl-β-d-thiogalactopyranoside (IPTG), the induced culture was further incubated at 16°C for 50 h. The cells were harvested by centrifugation, resuspended in 20 ml of phosphate-buffered saline (PBS, pH 7.4), and disrupted by sonication. After centrifugation at 12,000 rpm for 20 min, the supernatant was applied to a column containing 3 ml Ni Sepharose (GE Healthcare) that had been equilibrated with PBS. Bound protein was eluted from the column using 100 mM imidazole in PBS. The purified proteins were collected and analyzed on SDS-PAGE. Expressed proteins in the gels were stained with Coomassie brilliant blue R-250 for visualization.
Recombinant Enzyme Assays
The standard reaction mixture for Fh3GT1 enzyme assay consisted of 100 mM potassium phosphate buffer (pH 8.0), 100 μM flavonoid substrates (except for Km determination), 10 mM UDP-glucose and 30 μl enzyme extract (20–30 μg protein) in a total volume of 200 μl. The glucosyltransferase activity assays were performed at 30°C for 5 min and terminated by adding 50 μl HCl (5%). Reactions with protein extract obtained from BL-21 (DE3) cells that transformed with an empty pET-28a (+) vector were performed as controls. Subsequently, samples were centrifuged at 12,000 rpm for 5 min to collect the supernatant. After filtration through a 0.22 μm membrane filter, reaction products were analyzed by HPLC. Glucosylation products were determined by comparison of integrated peak areas of the glucoside to the corresponding standard curve which was verified to be linear over the range studied.
Enzyme Specificity
Substrate specificity of Fh3GT1 was also examined against eight potential substrates including cyanidin, delphinidin, pelargonidin, petunidin, peonidin, malvidin kaempferol, and quercetin using 10 mM UDP-glucose as a donor substrate. In addition, 10 mM UDP-galactose was also tested using cyanidin and delphinidin acceptors. Enzyme assays were performed as described above with 100 μM of acceptor substrate and stopped at 5 min.
Enzyme Kinetics
To determine apparent Km values, the concentration of peonidin, delphinidin, and quercetin was varied from 12.5 to 275 μM, and 10 mM UDP-glucose was used as donor substrate. Enzyme assays were conducted as described above with 8 μg of the purified enzyme but reactions were stopped at 2 min where the reaction velocity is liner. Km and Vmax values were obtained from Lineweaver-Burk plots of initial rate data.
Vector Construction and Arabidopsis Transformation
A pair of primer, PF3 and PR3 (Supplementary Table S1), was designed to amplify the full length coding regions of Fh3GT1 gene, using cDNA from red flowers of F. hybrida as templates. Fh3GT1 was cloned into XbaI/BamHI-digested pBI121 vector harboring the CaMV 35S constitutive promoter and confirmed by sequencing. Genetic transformation was performed as decribed above. T1 seeds were selected on 1/2 MS medium containing 50 mg L-1 kanamycin then transferred to soil to set T2 seeds. After 1 week of culture on anthocyanin gene induction media (Kovinich et al., 2010), three independent transgenic lines which accumulated visible levels of anthocyanins were subjected to further analysis. To confirm the expression of Fh3GT1 in the transformed mutants, total RNA was isolated from T2 transgenic seedlings and used for RT-PCR. And the Arabidopsis actin gene was used as a control (Penninckx et al., 1996).
Metabolite Analysis of T2 Transgenic Seedlings
Hundred microgram of 1-week-old Arabidopsis seedlings cultured on anthocyanin gene induction media (above) were ground in liquid nitrogen and submerged in 1 mL extraction solution at 4°C for 12 h. Extracts were centrifuged for 10 min at 12,000 rpm then filtered through a 0.22 μm filter, and 20 μl aliquots was analyzed by HPLC as described above. The content of anthocyanin and flavonol was determined by external standard curve calibration of cyanidin 3-O-glucoside and quercetin 3-O-glucoside standard, respectively (Fanali et al., 2011). Data were represented as mean values from three independent experiments with three replicates each. Statistical significances of the differences were determined using Student’s t-test. Differences between Arabidopsis lines were considered significant when P < 0.01.
Results
Analysis of Fh3GT1
Analysis of Fh3GT1 sequence for the N-terminal targeting signal or C-terminal membrane anchor signal using the SignalP and TargetP Web-based programs predicted a signal peptide localized between amino acids 20 and 21. Prediction of subcellular location with PSORT II1 yielded a chloroplast location. However, the TargetP version 1.1 program predicted the protein to be in the secretion pathway with the presence of a signal peptide (not a chloroplast transit peptide). In addition, due to the presence of a signal peptide at the N-terminal of Fh3GT1, it was also analyzed for the presence of glycosylated residues using the NetNGlyc 1.0 server2. The prediction revealed that two putative glycosylation sites were found in two Asn residues: Asn-244 (NPTL) and Asn-386 (NGTM). Finally, in order to examine the subcellular distribution of Fh3GT1, transgenic Arabidopsis plants expressing this enzyme fused to the C terminus of GFP were produced, and the results showed that this protein seemed to be localized in both the nuclei and the cytosol (Supplementary Figure S1).
Anthocyanin and Flavonol Analysis in Developing Flower
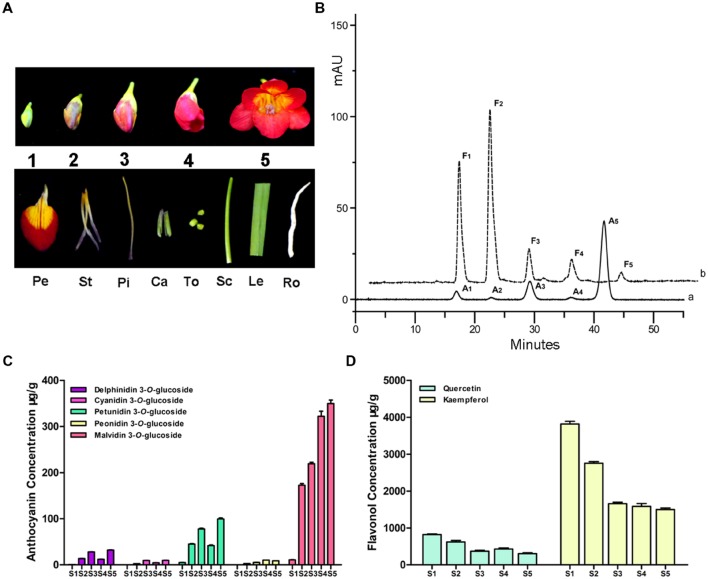
For determining the possible correlation between the presence of specific flavonoid glycoside derivatives and Fh3GT1 expression levels, anthocyanins and flavonols were identified and quantified at five flower developmental stages (S1–S5) (Figure 2A). For anthocyanin, a total of five peaks (A1–A5) were identified in flowers (Figure 2B). Based on the MS and comparison to authentic standards, these peaks were identified as delphinidin 3-O-glucoside, cyanidin 3-O-glucoside, petunidin 3-O-glucoside, peonidin 3-O-glucoside, and malvidin 3-O-glucoside, respectively (Supplementary Table S2). Among these anthocyanins, malvidin 3-O-glucoside was the most abundant anthocyanin detected at all times (accounting for 64.53–82.48% of the total content), followed by petunidin 3-O-glucoside, delphinidin 3-O-glucoside, cyanidin 3-O-glucoside and peonidin 3-O-glucoside (Figure 2C). But other kinds of basic anthocyanin derivatives, pelargonidin glycosides, were not detected. Furthermore, we also assayed flavonols (F1–F5), which were most abundant in S1 stage. Of these flavonols, kaempferol glycosides were predominant, with minor quantities of quercetin glycosides observed (Figure 2D). However, myricetin glycosides were undetectable throughout the flower development. In accordance with the flower deep coloration, the total anthocyanins levels peaked dramatically (0.54 mg g-1 fresh weight, Supplementary Figure S2A) in S5 stage. On the contrary, total flavonols concentrations continued to decline gradually from S1 to a minimum at S5 (1.8 mg g-1 fresh weight, Supplementary Figure S2B).
FIGURE 2.
Anthocyanin and flavonol component analysis in the flower of F. hybrida. (A) The phenotypes of different samples. S1–S5, represent the flowers of different developmental stages; Pe, petals; St, stamens; Pi, pistils; Ca, calyxes; To, toruses; Sc, scapes; Le, leaves; Ro, roots. (B) High performance liquid chromatography (HPLC) profiles of anthocyanins and flavonols in flowers. (a) monitored at 520 nm for anthocyan in (b) monitored at 360 nm for flavonol. (C) Quantitative analysis of individual anthocyanin during flower development. (D) Quantitative analysis of individual flavonol during flower development. Data represent means ± SD of three biological replicates.
Expression Profile of Fh3GT1 in Freesia hybrida
To examine whether the expression pattern of Fh3GT1 coincided spatially and temporally with anthocyanin and/or flavonol biosynthesis in F. hybrida, real-time PCR was performed to investigate its expression levels in flowers at five flower developmental stages and in different tissues. In flowers, Fh3GT1 was detected in all developmental stages and reached the highest level at stage 5 (Figure 3A). Transcript analysis in different tissues revealed that Fh3GT1 transcript was detected in all organs examined, and significantly higher levels of expression were observed in petals, pistils, and stamens. Comparatively, lower levels were seen in calyxes, toruses, scapes, leaves, and roots (Figure 3B) in which the anthocyanins were barely detected (data not shown). Taken together, the integrative expression pattern in combination with the accumulation of anthocyanins and flavonols suggest that Fh3GT1 may participate in both the anthocyanin and the flavonol glycoside biosynthesis in F. hybrida.
FIGURE 3.
Expression profiles of Fh3GT1 gene in Freesia hybrida. (A) Expression levels of Fh3GT1 in flowers at different developmental stages. (B) Expression profiles of Fh3GT1 in different tissues. Data represent means ± SD of three biological replicates.
Biochemical Characterization of Fh3GT1
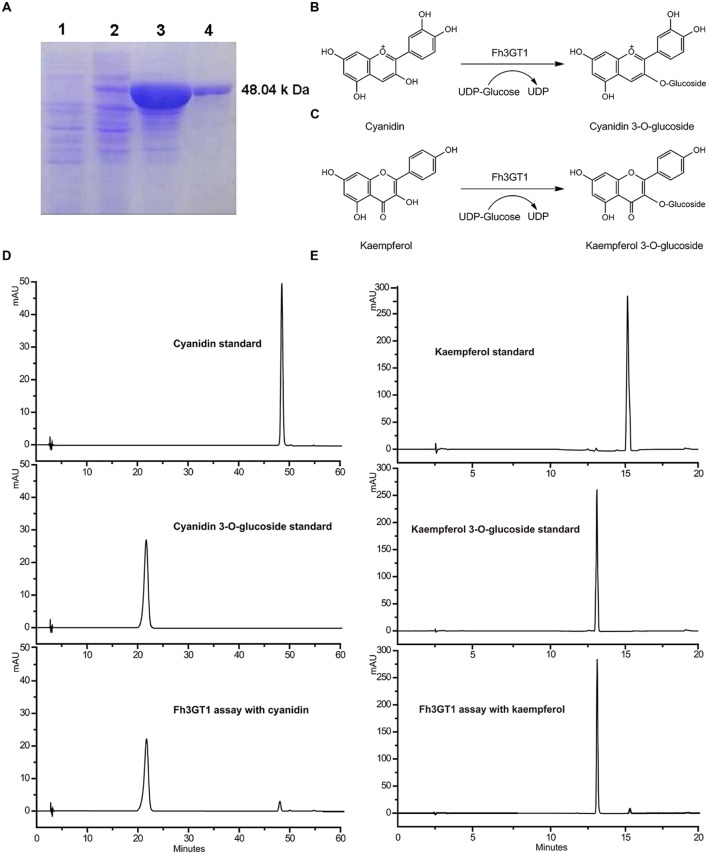
The full-length ORF of Fh3GT1 was cloned into the pET-28a (+) expression vectors with N-terminal His tags and introduced into BL-21 (DE3) Escherichia coli cells. A large amount of recombinant protein accumulated in inclusion bodies. However, only minor proportion of recombinant protein was detected in the soluble fraction by Western-blot analysis with an anti-His-tag antibody. Then the methods were optimized till sufficient soluble protein was obtained. Coomassie Blue -stained SDS-PAGE of the purified enzyme showed one major band of the expected size that matched well with the calculated molecular mass of Fh3GT1 (Figure 4A). Initial enzyme assays using UDP-glucose as the sugar donor demonstrated that the recombinant Fh3GT1 could catalyze the transfer of glucose to the 3-position of the anthocyanidin cyanidin (Figures 4B,D) and the flavonol kaempferol (Figures 4C,E). Correspondingly, recombinant protein expressing the empty vector did not glucosylate either of the substrates. The reaction products were monitored by HPLC-diode array detection (HPLC-DAD) and compared with the authentic standards. Subsequently, optimal reaction conditions for Fh3GT1 were determined using cyanidin and UDP-glucose. The recombinant enzyme showed maximum activity at 30°C and pH 8.0 which has been used in the analysis of the majority of plant UFGTs (Ford et al., 1998; Masayuki et al., 2000; Kim et al., 2006).
FIGURE 4.
Purification of recombinant Fh3GT1 and identification of reaction products. (A) Expression of Fh3GT1 in E. coli. (1) total soluble protein from E. coli expressing pET-28a (+) vector (2) total soluble protein from E. coli expressing Fh3GT1 prior to induction with IPTG (3) 50 h after induction (4) purified Fh3GT1. (B,C) Reaction scheme of the enzymatic synthesis of cyanidin/kaempferol 3-O-glucoside from cyanidin/kaempferol and UDP-glucose. (D,E) HPLC analysis of the activities of Fh3GT1 toward cyanidin and kaempferol.
Substrate specificity studies for Fh3GT1 with a range of substrates were tested, including several anthocyanidins and flavonols whose glycosylated products had been detected in F. hybrida. In the presence of UDP-glucose, the recombinant Fh3GT1 was able to glucosylate all tested anthocyanidin and flavonol aglycones at their 3-position. As shown in Table 1, quercetin and delphinidin exhibited the highest rates of flavonol and anthocyanidin glucosylation, respectively. Pelargonidin and petunidin served as substrate to a similar extent, whereas malvidin showed the lowest glucosylation rates. With regard to sugar donor preference, both UDP-glucose and UDP-galactose were accepted by Fh3GT1. But UDP-galactose was transferred to the 3-position of delphinidin and cyanidin with a lower level of galactosyl transfer activity, as determined by HPLC-DAD and compared to authentic standards (delphinidin 3-O-galactoside and cyanidin 3-O-galactoside). Moreover, the recombinant Fh3GT1 enzyme was unable to further glucosylate anthocyanidin 3-O-glucoside (data not shown), as the primary anthocyanins of F. hybrida in flowers are anthocyanidin 3-O-glucosides (Supplementary Table S2). These results revealed that Fh3GT1 worked well on anthocyanidins and flavonols in vitro preferentially as a glucosyltransferase with strict regiospecificity for the 3-position of anthocyanins and flavonols.
Table 1.
Relative activity of Fh3GT1 toward several substrates.
| Substrate | Sugar donor | Product | Relative activity (%) |
|---|---|---|---|
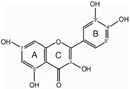 |
UDP-Glu | Quercetin 3-O-glucoside | 100a |
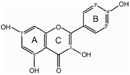 |
UDP-Glu | Kaempferol 3-O-glucoside | 42.30 |
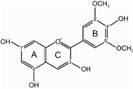 |
UDP-Glu | Malvidin 3-O-glucoside | 8.29 |
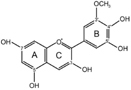 |
UDP-Glu | Petunidin 3-O-glucoside | 13.10 |
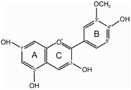 |
UDP-Glu | Peonidin 3-O-glucoside | 32.33 |
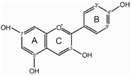 |
UDP-Glu | Pelargonidin 3-O-glucoside | 14.93 |
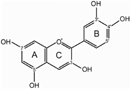 |
UDP-Glu | Cyanidin 3-O-glucoside | 28.94 |
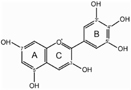 |
UDP-Glu | Delphinidin 3-O-glucoside | 33.25 |
 |
UDP-Gal | Cyanidin 3-O-galactoside | 19.23 |
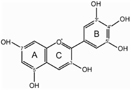 |
UDP-Gal | Delphinidin 3-O-galactoside | 29.64 |
a The relative activity was calculated by the activity toward quercetin as 100%.
100 μM substrate and 10 mM sugar were used and the reaction was incubated at 30°C for 5 min.
Activity values are the mean of three independent determinations.
The kinetic parameters of Fh3GT1 were examined using the best substrates of anthocyanidin and flavonol as determined by in vitro substrate specificity studies. By fixing the concentration of UDP-glucose and varying the concentration of substrates, hyperbolic saturation curves were obtained. Then the apparent Km values were calculated based on the Lineweaver-Burk plot and listed in Table 2. According to the results, recombinant Fh3GT1 enzyme used quercetin most efficiently, although a relatively high affinity toward both delphinidin and peonidin was observed.
Table 2.
Kinetic parameters for the recombinant Fh3GT1 protein.
| Substrate | Km (μM) | kcat (s-1) | kcat/Km (s-1M-1) |
|---|---|---|---|
| Quercetin | 98.47 ± 4.96 | 4.59 ± 0.035 | 48.51 × 103 |
| Delphinidin | 58.88 ± 1.68 | 0.53 ± 0.010 | 9.06 × 103 |
| Peonidin | 56.08 ± 2.54 | 0.50 ± 0.011 | 8.97 × 103 |
Data are the means ± SD of three independent experiments.
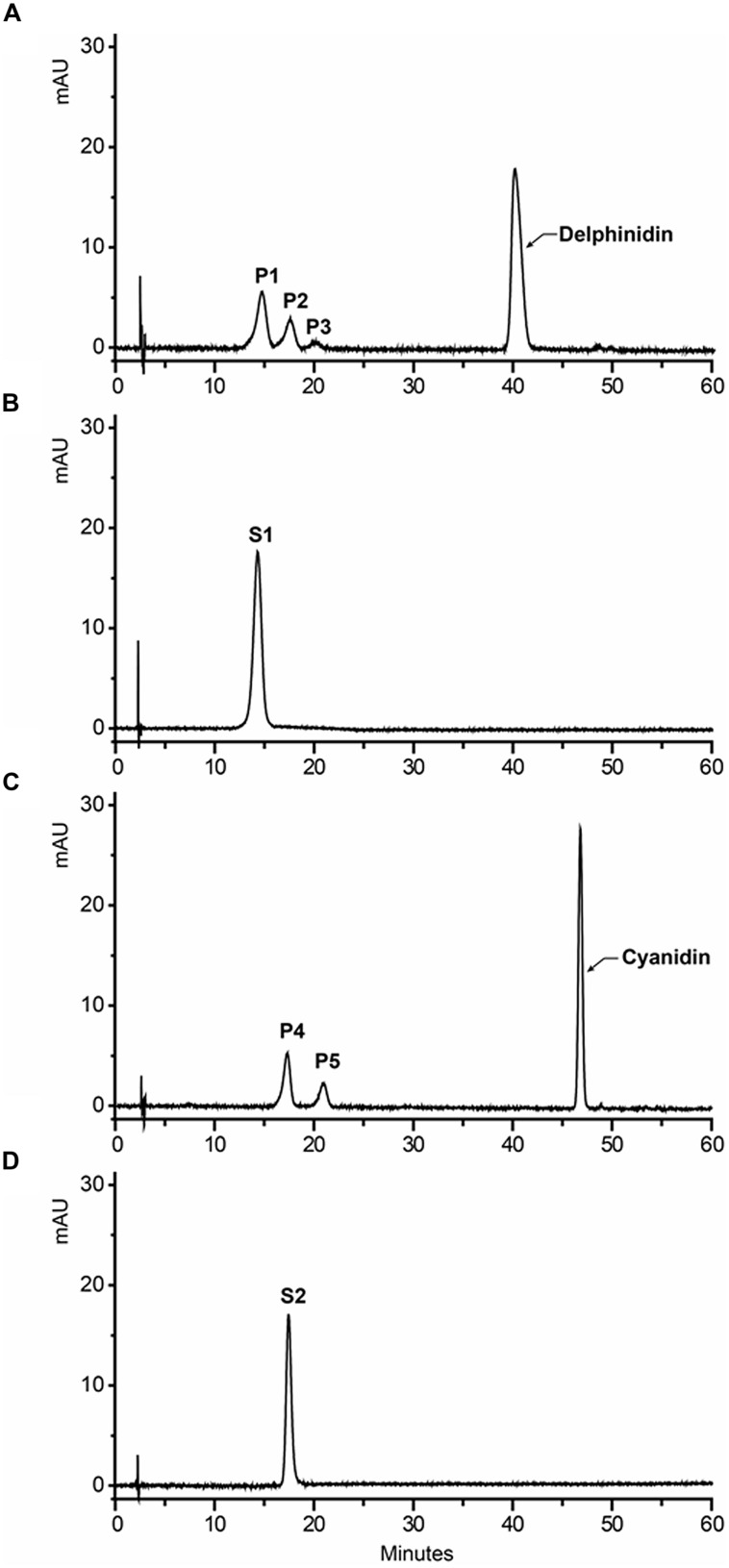
It has been suggested that concentration of the sugar donor could affect the regiospecificity of glycosyltransferase. Thus, 1 mM UDP-glucose and UDP-galactose were used as sugar donors with delphinidin and cyanidin as substrates to investigate the regiospecificity of Fh3GT1. In the case of UDP-glucose, one major reaction product was produced: the product was determined to be delphinidin and cyanidin 3-O-glucoside, while the amounts of other products were too low to be characterized (data not shown). However, HPLC analysis of the reaction products of UDP-galactose showed at least two peaks were different from substrates. HPLC-ESI-MS analyses of all reaction products were performed and the results indicated that molecular weight of each product increased 162 Da when compared to the weight of substrates. These results demonstrated that only one molecule glucose/galactose had been transferred to the substrates. In addition, the regiospecificity of Fh3GT1 could also be determined by observing the hypsochromic shift of UV spectra between Fh3GT1 reaction products and substrates. It has been reported that glycoslation at either C-3 or C-4′ hydroxyl group produce a hypsochromic shift, whereas glycoslation at the C-7 hydroxyl group showed no effect (Thomas et al., 1997; Kramer et al., 2003). The reaction products of delphinidin showed three peaks (P1-P3 in Figure 5A). The retention time of P1 was indistinguishable from that of the authentic delphinidin 3-O-galactoside (Figure 5B), revealing that P1 was 3-O-galactoside of delphinidin. P2 exhibited a hypsochromic shift, but P3 did not (Table 3). This suggested that P2 and P3 were likely to be delphinidin 4′-O-galactoside and delphinidin 7-O-galactoside, respectively. Fh3GT1 yielded two reaction products with cyanidin (P4–P5 in Figure 5C), P4 was determined to be cyanidin 3-O-galactoside by comparison with the authentic standard (Figure 5D) and P5 was likely cyanidin 4′-O-galactoside according to hypsochromic shift (Table 3).
FIGURE 5.
HPLC profiles of Fh3GT1 reaction products with delphinidin/cyanidin and UDP-galactose. (A) Delphinidin reaction products. (B) Authentic delphinidin 3-O-galactoside. (C) Cyanidin reaction products. (D) Authentic cyanidin 3-O-galactoside.
Table 3.
Structure assignment of Fh3GT1 reaction products.
| Compounds | Retention time (min) | UV (nm) | Structure assignment |
| Delphinidin | 40.12 | 254,531 | Delphinidin aglycon |
| S1 | 14.74 | 251,521 | Delphinidin 3-O-galactoside |
| P1a | 14.65 | 250,520 | Delphinidin 3-O-galactoside |
| P2b | 17.43 | 251,520 | Delphinidin 4′-O-galactoside |
| P3b | 19.92 | 251,533 | Delphinidin 7-O-galactoside |
| Cyanidin | 46.73 | 255,526 | Cyanidin aglycon |
| S2 | 17.44 | 254,515 | Cyanidin 3-O-galactoside |
| P4a | 17.17 | 255,515 | Cyanidin 3-O-galactoside |
| P5b | 20.84 | 255,516 | Cyanidin 4′-O-galactoside |
aThe reaction products were assigned according to comparison of HPLC retention time and UV spectra with authentic compounds.
bThe reaction products were assigned according to the hypsochromic shift.
Fh3GT1 Functions as A 3-O-glucosyltransferase in Anthocyanin and Flavonol Biosynthesis In Vivo
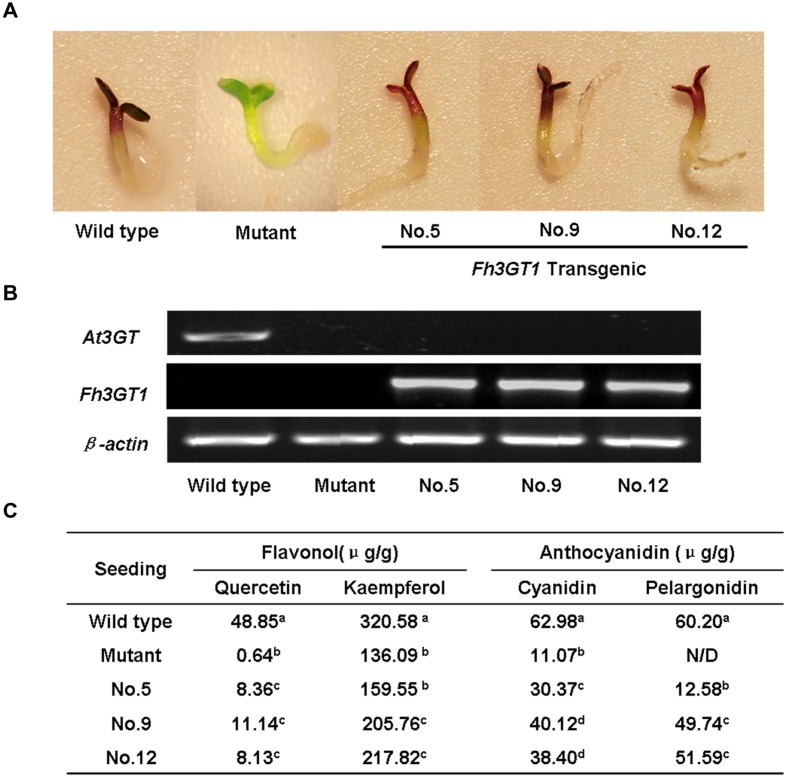
To further confirm that Fh3GT1 has 3GT activity in vivo, the cDNA under the control of 35S promoter was transferred into the Arabidopsis mutant (UGT78D2) with reduced anthocyanidin and flavonol 3-O-glucosyltransferase activity. After selecting on 1/2 MS medium with kanamycin, more than twelve independent transgenic Arabidopsis lines were produced, and three of them (NO.5, NO.9, NO.12) were chosen for further analysis. Seeds of the wild-type, Arabidopsis mutant, and T2 transgenic lines were germinated and grown on anthocyanin gene induction media. Seedlings of wild-type and transgenic plants expressing Fh3GT1 had anthocyanin pigmentation in the cotyledons and hypocotyls (Figure 6A), whereas the mutant transformed with the empty vector were green (not shown). Presence of the Fh3GT1 and Arabidopsis 3GT (At3GT) was verified by RT-PCR using primers PF1/PR1 and At3GTF/At3GTR. PCR results showed that all transgenic lines expressed Fh3GT1 and lacked the transcript of At3GT (Figure 6B).
FIGURE 6.
Complementation of the pigmentation of Arabidopsis mutant seedlings with Fh3GT1 gene. (A) Phenotypes of wild-type, mutant (UGT78D2) and transgenic Arabidopsis seedlings. (B) Expressional analysis of the At3GT and Fh3GT1 gene by reverse transcription polymerase chain reaction in wild-type, mutant and transgenic lines. (C) Contents of anthocyanins and flavonols in Arabidopsis seedlings. Data correspond to means of three biological replicates. Means with different letters within the same column are significantly different at the 0.01 level of probability. N/D, not detected.
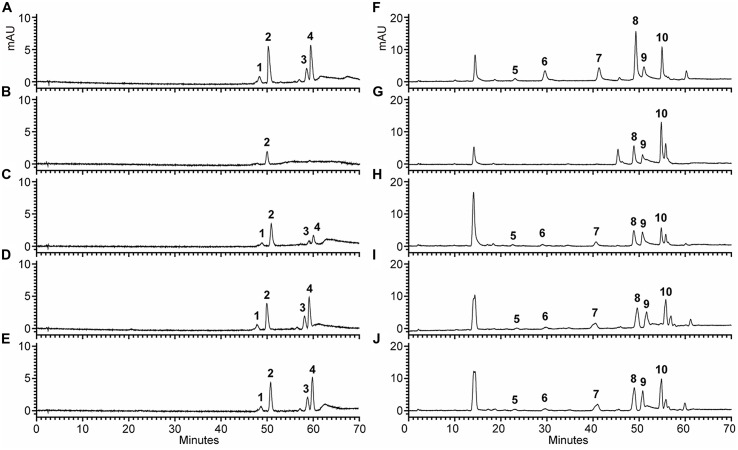
To examine the change of anthocyanin and flavonol more clearly in transgenic lines, T2 seedlings from each line cultured on anthocyanin induction media were extracted with extraction solvent mixture and analyzed by HPLC, and the MS data of these peaks were obtained and summarized in Supplementary Table S3. As shown in Figure 7, the Arabidopsis mutant had reduced peak area for some peaks of anthocyanin (monitored at 520) and flavonol (monitored at 360) relative to the wild type control. As expected, the pooled seedlings carrying Fh3GT1 cDNA could restore these peaks, though lower levels of anthocyanin and flavonol were detected in transgenic plants (Figure 6C). Overall, our results strongly suggested that Fh3GT1 had similar activity to At3GT, a 3GT gene participated in anthocyanin and flavonol biosynthesis in vivo.
FIGURE 7.
High performance liquid chromatography analyses of anthocyanins and flavonols in transgenic Arabidopsis seedlings. (A–J). HPLC chromatograms of the samples from seedlings of wild type, mutant and transgenic lines. (A,F) wild type, (B,G) Mutant, (C,H) NO.5, (D,I) NO.9, (E,J) NO.12. (A–E) Absorbance at 520 nm for analysis of anthocyanins. (F–J) Absorbance at 360 nm for analysis of flavonols.
Discussion
Sequence analysis revealed that Fh3GT1 was most closely related to anthocyanidin 3GT from Iris hollandica (IhAn3GT) and they were clustered into the same clade of the phylogenetic tree (Sui et al., 2011). However, these two enzymes differ considerably in their substrate specificities. For example, Fh3GT1 showed highest activity toward quercetin, whereas IhAn3GT used malvidin as its best substrate (Yoshihara et al., 2005). The lack of congruence between substrate specificities and phylogenetic position has been observed in other UFGTs like VLOGT2 from grape (Vitis labrusca) and two UFGTs from onion (Kramer et al., 2003; Hall et al., 2011). These results are in agreement with the past suggestions that function and specificity of UFGT can not predict based on the primary sequence alone (Dhaubhadel et al., 2008). Coupling the tissue-specific expression with metabolites analysis has been known as an efficient method to confirm the glycosyltransferase functions. In this study, metabolite analysis and molecular approaches were used to investigate Fh3GT1-metabolite correlations for determining the role of Fh3GT1 in Freesia. Furthermore, analysis of substrate specificity in vitro and the use of transgenic Arabidopsis lines expressing Fh3GT1 provided strong evidence demonstrating the biochemical function of Fh3GT1.
Based on the metabolites found in the flowers, the anthocyanin biosynthetic pathway in Freesia was proposed and presented in Figure 1, all of the six basic anthocyanin aglycons could be synthesized except pelargonindin derivatives. In previous studies, several plant species, such as Petunia hybrida and Cymbidium hybrida, have been found unable to produce pelargonidin derivatives because of the DFR substrate specificities. In these two plants, DFRs showed almost undetectable catalytic activities against dihydrokaempferol (DHK) (Joseph et al., 1998; Johnson et al., 1999), and we also found that the DFRs isolated from the Freesia flowers could not utilize DHK as substrates (unpublished date). On the other hand, DHK might be converted to dihydromyricetin (DHM) by F3′5′H before reduced by DFR or it fails to compete substrates with FLS.
Freesia hybrida accumulates both anthocyanins and flavonols, while anthocyanins accumulate to visible levels only in flower tissues. During the flower development, the level of anthocyanin gradually increased and peaked at stage 5, whereas flavonols reached its highest level at stage 1 (Supplementary Figure S2). Transcripts of Fh3GT1 were detected in all examined samples, not just the sites of visible anthocyanin accumulations. This expression pattern is similar to that of ANL1 in Arabidopsis (Kubo et al., 2007) and may suggest that Fh3GT1 has a role in biosynthesis of anthocyanin and flavonol glycoside in vivo. Moreover, Fh3GT1 was the most highly expressed at stage 5 (Figure 3A) and in petals (Figure 3B), respectively, and this expression profile showed the strongest correlation with the presence of anthocyanins rather than flavonols. It is therefore likely that Fh3GT1 prefers to glycosylate the endogenous anthocyanidin in F. hybrida. Similar to 3GT from black soybean (Kovinich et al., 2010), transfer of Fh3GT1 into the Arabidopsis mutant also restored the biosynthesis of both anthocyanin and flavonol glycoside, confirming the functionality of Fh3GT1 as a 3GT in vivo. As shown in Figure 6C, anthocyanin levels were restored to a more degree (20.89–85%) than flavonols (7.31–25.49%) in transgenic Arabidopsis compared to the wild type control. These results further demonstrate that Fh3GT1 prefers anthocyanidin as substrates in vivo.
To date, all recombinant 3GTs characterized in vitro display unique substrate specificities. Some prefer anthocyanidins (Ford et al., 1998; Jun et al., 2004; Yoshihara et al., 2005; Almeida et al., 2007), while others prefer flavonols or other flavonoids (Modolo et al., 2007; Owens and McIntosh, 2009). On the basis of our in vitro biochemical assays, Fh3GT1 recombinant enzyme recognizes both anthocyanidins and flavonols as substrates, and has a clear preference for flavonols (Table 1). However, these results are inconsistent with those presented above demonstrating Fh3GT1 prefers to catalyze the glucosylation of anthocyanidin in vivo. Similar arguments have been proposed in UGT78K1, a 3GT of Medicago truncatula showed relatively low activity to cyanidin in vitro when compared to other flavonoid substrates (Modolo et al., 2007), but was demonstrated to play a critical role in the biosynthesis of anthocyanin in vivo (Peel et al., 2009). These results indicate that the enzymes involved in natural product biosynthesis are promiscuous, and the relative concentrations of potential substrates might be one of the most critical factors to determine their in vivo activity. Additionally, the recombinant Fh3GT1 could catalyze the transfer of a glycosyl moiety from sugar donor to the 3-position of pelargonidin in vitro, but no pelargonidin 3-O-glucoside was detected in F. hybrida. This suggests that the substrate recognition of Fh3GT1 is broader in vitro, and the branch of pelargonindin biosynthesis is blocked before Fh3GT1.
Fh3GT1 utilizes both UDP-glucose and UDP-galactose as sugar donors. The preference of UDP-glucose as the sugar donor is consistent with recent demonstration that the last residue of the PSPG box is a glutamine (Kubo et al., 2004). Meanwhile, the utilization of UDP-galactose as sugar donor also suggests that sugar donor specificity between glucose and galactose is probably determined not only by the last residue but also by other residue(s) in the PSPG box (Montefiori et al., 2011). Apparent Km values for characterized 3GTs from various species vary greatly with values ranging from 1.5 to 400 μM (Saleh et al., 1976; Veljanovski and Constabel, 2013). The Km of Fh3GT1 is within this range, and the relatively low Km value for peonidin suggests it is likely to be the natural substrate for this enzyme in vivo. However, quantitative analyses of anthocyanidin glycosides in F. hybrida revealed that malvidin 3-O-glucoside was the most abundant anthocyanin (Figure 2C). This implies that F. hybrida has another type of 3GT recognizing malvidin as its natural substrate, because UFGTs in the flavonoid pathway are usually encoded by a multi-gene family in many plant species (Caputi et al., 2012).
Enzymatic studies of recombinant protein revealed some unique properties of Fh3GT1 toward its substrate. It has been reported that many glycosyltransferases involved in the flavonoid biosynthesis display high regiospecificity to a specific position of the hydroxy group but low substrate specificity to the flavonoid structure (Hansen et al., 2003). For instance, 3GTs of gentian, grape and petunia can glycosylate the 3-position of cyanidin, pelargonidin, and delphinidin (Yoshikazu et al., 1996; Ford et al., 1998; Mami et al., 2002). The gentian 5GT catalyzed the glycosylation of 5-hydroxy group of several anthocyanidin 3-glycosides (Nakatsuka et al., 2008). However, the regiospecificity of Fh3GT1 is determined by both the concentration and the kind of sugar donor. In contrast to the data of 1 mM sugar dornor, only anthocyanidin 3-O-glycoside was detected when 10 mM sugar dornor was used (Table 1). Experiments with recombinant Fh3GT1 showed galactosylation of the 3-OH, 4′-OH, and 7-OH with a preference for the 3-OH (Table 3). While in the case of glucose, Fh3GT1 glucosylated predominantly in the 3-position, and other products were produced only in trace amounts. This result is congruent with the fact that Freesia contains mainly anthocynidin 3-O-glucoside which is glucosylated in the 3-position. Further analyses suggest that 7-OH galactosylation is governed by the presence or absence of a hydroxyl group at C-5′. When this group is present (delphinidin), 7-OH galactosylation is observed, when absence (cyanidin) 7-OH galactosylation is not detected. Further investigation should be performed to confirm whether the presence of 5′-OH is a determinant for 7-OH galactosylation.
Conclusion
The enzyme reported in this study can be identified as flavonoid 3-O-glycosyltransferase with UDP-glucose as the preferred sugar donor. Biochemical analysis together with the in planta data reveal its involvement in biosynthesis of flavonoid glucosides in F. hybrida. Moreover, this enzyme exhibits broad range substrate specificity toward flavonoids, including substrate (pelargonidin) that does not naturally occur in Freesia. This suggests the substrate versatility of Fh3GT1 could be even broader than described here. Interestingly, Fh3GT1 was also found to preferentially transfer a galactose group to the 3-OH, 4′-OH, and 7-OH of flavonoids. Due to the wide range substrate specificity and low regiospecificity of Fh3GT1, it could be an attractive enzyme to engineer the flavonoid diversity.
Author Contributions
WS performed the research, wrote, and revised this manuscript; LL, XM, YL, FG, SW, and XL performed the experiments and helped analyze data. XG and LW designed the experiments, discussed results, and revised this manuscript. All authors have participated in this research and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31170276, 31300271), Jilin provincial Government of the People’s Republic of China (20130102061JC, 201201074, 20125035) and the Fundamental Research Fund for the Central Universities (2412015ZH006).
Abbreviations
- 3GT
Flavonoid 3-O-glucosyltransferase
- 5GT
Anthocyanin 5-O-glucosyltransferase
- ANS
Anthocyanidin synthase
- CHI
Chalcone isomerase
- CHS
Chalcone synthase
- DFR
Dihydroflavonol-4-reductase
- F3′5′H
Flavonoid 3′ 5′-hydroxylase
- FLS
Flavonol synthase
- HPLC
High performance liquid chromatography
- UFGT
UDP flavonoid glycosyltransferases
Funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00410
References
- Almeida J. R., D’Amico E., Preuss A., Carbone F., de Vos C. H., Deiml B., et al. (2007). Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria xananassa). Arch. Biochem. Biophys. 465 61–71. 10.1016/j.abb.2007.04.040 [DOI] [PubMed] [Google Scholar]
- Almudena T., Oussama A., Angela R. M., Maria L. J., Maria D. G., Lourdes G. G. (2012). Characterization of a glucosyltransferase enzyme involved in the formation of kaempferol and quercetin sophorosides in crocus sativus. Plant Physiol. 159 1335–1354. 10.1104/pp.112.198069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier H. M., Edwards R. (2005). Functional importance of the family 1 glucosyltransferase ugt72b1 in the metabolism of xenobiotics in Arabidopsis thaliana. Plant J. 42 556–566. 10.1111/j.1365-313X.2005.02398.x [DOI] [PubMed] [Google Scholar]
- Brenda W. S. (2001). Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126 485–493. 10.1104/pp.126.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi L., Malnoy M., Goremykin V., Nikiforova S., Martens S. (2012). A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69 1030–1042. 10.1111/j.1365-313X.2011.04853.x [DOI] [PubMed] [Google Scholar]
- Charles S. B., Nijat I., Michael A. D. (2010). flavonoids: new roles for old molecules. J. Integr. Plant Biol. 52 98–111. 10.1111/j.1744-7909.2010.00905.x [DOI] [PubMed] [Google Scholar]
- Chen W. H., Hsu C. Y., Cheng H. Y., Chang H., Chen H. H., Ger M. J. (2011). Downregulation of putative UDP-glucose: flavonoid 3-O-glucosyltransferase gene alters flower coloring in Phalaenopsis. Plant Cell Rep. 30 1007–1017. 10.1007/s00299-011-1006-1 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S., Farhangkhoee M., Chapman R. (2008). Identification and characterization of isoflavonoid specific glycosyltransferase and malonyltransferase from soybean seeds. J. Exp. Bot. 59 981–994. 10.1093/jxb/ern046 [DOI] [PubMed] [Google Scholar]
- Fanali C., Dugo L., D’Orazio G., Lirangi M., Dacha M., Dugo P., et al. (2011). Analysis of anthocyanins in commercial fruit juices by using nano-liquid chromatography-electrospray-mass spectrometry and high-performance liquid chromatography with UV-vis detector. J. Sep. Sci. 34 150–159. 10.1002/jssc.201000665 [DOI] [PubMed] [Google Scholar]
- Ferreyra M. L. F., Rius S. P., Casati P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3:222 10.3389/fpls.2012.00222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford C. M., Boss P. K., Hoj P. B. (1998). Cloning and characterization of vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J. Biol. Chem. 273 9224–9233. 10.1074/jbc.273.15.9224 [DOI] [PubMed] [Google Scholar]
- Gachon C. M., Langlois M. M., Saindrenan P. (2005). Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 10 542–549. 10.1016/j.tplants.2005.09.007 [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J. M., Lloyd A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53 814–827. 10.1111/j.1365-313X.2007.03373.x [DOI] [PubMed] [Google Scholar]
- Griesser M., Hoffmann T., Bellido M. L., Rosati C., Fink B., Kurtzer R., et al. (2008). Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol. 146 1528–1539. 10.1104/pp.107.114280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. (2005). Plant metabolic diversity: a regulatory perspective. Trends Plant Sci. 10 57–62. 10.1016/j.tplants.2004.12.009 [DOI] [PubMed] [Google Scholar]
- Hall D., Kim K. H., Luca V. D. (2011). Molecular cloning and biochemical characterization of three Concord grape (Vitis labrusca) flavonol 7-O-glucosyltransferases. Planta 234 1201–1214. 10.1007/s00425-011-1474-0 [DOI] [PubMed] [Google Scholar]
- Hansen K. S., Kristensen C., Tattersall D. B., Jones P. R., Olsen C. E., Bak S., et al. (2003). The in vitro substrate regiospecificity of recombinant UGT85B1, the cyanohydrin glucosyltransferase from Sorghum bicolor. Phytochemistry 64 143–151. 10.1016/S0031-9422(03)00261-9 [DOI] [PubMed] [Google Scholar]
- Hu C., Gong Y., Jin S., Zhu Q. (2011). Molecular analysis of a UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT) gene from purple potato (Solanum tuberosum). Mol. Biol. Rep. 38 561–567. 10.1007/s11033-010-0141-z [DOI] [PubMed] [Google Scholar]
- Joe R., Yi L., Eng-Kiat L., Dianna J. B. (2001). Higher plant glycosyltransferases. Genome Biol. 2 3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. T., Yi H., Shin B., Oh B. J., Cheong H., Choi G. (1999). Cymbidium hybrida dihydroflavonol 4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type anthocyanins. Plant J. 19 81–85. 10.1046/j.1365-313X.1999.00502.x [DOI] [PubMed] [Google Scholar]
- Joseph M., Erich G., Ronald K. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3 212–217. 10.1016/S1360-1385(98)01242-4 [DOI] [Google Scholar]
- Jun O., Susumu T., Kunijiro Y. (1998). Isolation and Characterization of UDP-glucose: cyanidin 3-O-glucosyltransferase from the Flower Buds of Senecio x hybridus. J. Plant Res. 111 213–216. 10.1007/BF02512172 [DOI] [Google Scholar]
- Jun O., Yoshiaki K., Yoshio I., Hidehito T., Masahiko S. (2005). Anthocyanin biosynthesis in roses. Nature 435 757–758. 10.1038/nature435756a [DOI] [PubMed] [Google Scholar]
- Jun O., Yoshio I., Ishida M., Yoshida H., Ozeki Y. (2004). Cloning and heterologous expression of cDNAs encoding flavonoid glucosyltransferases from Dianthus caryophyllus. Plant Biotechnol. 21 367–375. 10.5511/plantbiotechnology.21.367 [DOI] [Google Scholar]
- Kim J. H., Kim B. G., Park Y., Ko J. H., Lim C. E., Lim J., et al. (2006). Characterization of flavonoid 7-O-glucosyltransferase from Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 70 1471–1477. 10.1271/bbb.60006 [DOI] [PubMed] [Google Scholar]
- Koes R., Verweij W., Quattrocchio F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10 236–242. 10.1016/j.tplants.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Kovinich N., Saleem A., Arnason J. T., Miki B. (2010). Functional characterization of a UDP-glucose:flavonoid 3-O-glucosyltransferase from the seed coat of black soybean (Glycine max (L.) Merr.). Phytochemistry 71 1253–1263. 10.1016/j.phytochem.2010.05.009 [DOI] [PubMed] [Google Scholar]
- Kramer C. M., Prata R. T. N., Willits M. G., De Luca V., Steffens J. C., Graser G. (2003). Cloning and regiospecificity studies of two flavonoid glucosyltransferases from Allium cepa. Phytochemistry 64 1069–1076. 10.1016/S0031-9422(03)00507-7 [DOI] [PubMed] [Google Scholar]
- Kubo A., Arai Y., Nagashima S., Yoshikawa T. (2004). Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophys. 429 198–203. 10.1016/j.abb.2004.06.021 [DOI] [PubMed] [Google Scholar]
- Kubo H., Nawa N., Lupsea S. A. (2007). Anthocyaninless1 gene of Arabidopsis thaliana encodes a UDP-glucose:flavonoid-3-O-glucosyltransferase. J. Plant Res. 120 445–449. 10.1007/s10265-006-0067-7 [DOI] [PubMed] [Google Scholar]
- Lim E. K., Bowles D. J. (2004). A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 23 2915–2922. 10.1038/sj.emboj.7600295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mami Y., Emiko Y., Gong Z., Masako F. M., Yuko F., Yoshikazu T., et al. (2002). Two flavonoid glucosyltransferases from Petunia hybrida: molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol. Biol. 48 401–411. 10.1023/A:1014043214943 [DOI] [PubMed] [Google Scholar]
- Martin C., Prescott A., Mackay S., Bartlett J., Vrijlandt E. (1991). Control of anthocyanin biosynthesis in flowers of Antirrhinurn majus. Plant J. 1 37–49. 10.1111/j.1365-313X.1991.00037.x [DOI] [PubMed] [Google Scholar]
- Masayuki K., Yutaka H., Takaya M., Tomoko E. I., Ryoji M., Shin H., et al. (2000). Molecular cloning and characterization of a novel gene encoding limonoid UDP-glucosyltransferase in Citrus1. FEBS Lett. 469 173–178. 10.1016/S0014-5793(00)01275-8 [DOI] [PubMed] [Google Scholar]
- Modolo L. V., Blount J. W., Achnine L., Naoumkina M. A., Wang X., Dixon R. A. (2007). A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol. Biol. 64 499–518. 10.1007/s11103-007-9167-6 [DOI] [PubMed] [Google Scholar]
- Montefiori M., Espley R. V., Stevenson D., Cooney J., Datson P. M., Saiz A., et al. (2011). Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J. 65 106–118. 10.1111/j.1365-313X.2010.04409.x [DOI] [PubMed] [Google Scholar]
- Morita Y., Hoshino A., Kikuchi Y., Okuhara H., Ono E., Tanaka Y., et al. (2005). Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2”-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J. 42 353–363. [DOI] [PubMed] [Google Scholar]
- Nakatsuka T., Sato K., Takahashi H., Yamamura S., Nishihara M. (2008). Cloning and characterization of the UDP-glucose:anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian. J. Exp. Bot. 59 1241–1252. 10.1093/jxb/ern031 [DOI] [PubMed] [Google Scholar]
- Offen W., Fleites C. M., Yang M., Kiat-Lim E., Davis B. G., Tarling C. A., et al. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25 1396–1405. 10.1038/sj.emboj.7600970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens D. K., McIntosh C. A. (2009). Identification, recombinant expression, and biochemical characterization of a flavonol 3-O-glucosyltransferase clone from Citrus paradisi. Phytochemistry 70 1382–1391. 10.1016/j.phytochem.2009.07.027 [DOI] [PubMed] [Google Scholar]
- Peel G. J., Pang Y., Modolo L. V., Dixon R. A. (2009). The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J. 59 136–149. 10.1111/j.1365-313X.2009.03885.x [DOI] [PubMed] [Google Scholar]
- Penninckx I. A., Eggermont K., Terras F. R., Thomma B. P., De Samblanx G. W., Buchala A., et al. (1996). Pathogen-lnduced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-lndependent pathway. Plant Cell 8 2309–2323. 10.2307/3870470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K., Pilu R., Tonelli C. (2014). Anthocyanins in corn: a wealth of genes for human health. Planta 240 901–911. 10.1007/s00425-014-2131-1 [DOI] [PubMed] [Google Scholar]
- Petroni K., Tonelli C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181 219–229. 10.1016/j.plantsci.2011.05.009 [DOI] [PubMed] [Google Scholar]
- Quideau S. (2006). Flavonoids. Chemistry, Biochemistry and Applications. Edited by Øyvind M. Andersen and Kenneth R. Markham. Angew Chem. Int. Edit. 45 6786–6787. 10.1002/anie.200685399 [DOI] [Google Scholar]
- Saleh N. A. M., Fritsch H., Witkop P., Grisebach H. (1976). UDP-Glucose: Cyanidin 3-O-Glucosyltransferase from cell cultures of Haplopappus gracilis. Planta 133 41–45. 10.1007/BF00386004 [DOI] [PubMed] [Google Scholar]
- Santos-Buelga C., Mateus N., Freitas V. D. (2014). Anthocyanins. Plant pigments and beyond. J. Agric. Food Chem. 62 6879–6884. 10.1021/jf501950s [DOI] [PubMed] [Google Scholar]
- Sui X., Gao X., Ao M., Wang Q., Yang D., Wang M., et al. (2011). cDNA cloning and characterization of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in Freesia hybrida. Plant Cell Rep. 30 1209–1218. 10.1007/s00299-011-1029-7 [DOI] [PubMed] [Google Scholar]
- Takayuki T., Yasutaka N., Masami Y. H., Mitsuru Y., Jun-ichiro N., Motoko A., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an myb transcription factor. Plant J. 42 218–235. 10.1111/j.1365-313X.2005.02371.x [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Sasaki N., Ohmiya A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54 733–749. 10.1111/j.1365-313X.2008.03447.x [DOI] [PubMed] [Google Scholar]
- Taylor L. P., Grotewold E. (2005). Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 8 317–323. 10.1016/j.pbi.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Thomas V., Elke Z., Rudi G., Michael M., Dieter S. (1997). Are the characteristics of betanidin glucosyltransferases from cell-suspension cultures of Dorotheanthus bellidiformis indicative of their phylogenetic relationship with flavonoid glucosyltransferases. Planta 203 349–361. 10.1007/s004250050201 [DOI] [PubMed] [Google Scholar]
- Thomas V., Patrik J. (2000). Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 5 380–386. [DOI] [PubMed] [Google Scholar]
- Veljanovski V., Constabel C. P. (2013). Molecular cloning and biochemical characterization of two UDP-glycosyltransferases from poplar. Phytochemistry 91 148–157. 10.1016/j.phytochem.2012.12.012 [DOI] [PubMed] [Google Scholar]
- Wang H., Fan W., Li H., Yang J., Huang J., Zhang P. (2013). Functional characterization of Dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS ONE 8:e78484 10.1371/journal.pone.0078484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2009). Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 583 3303–3309. 10.1016/j.febslet.2009.09.042 [DOI] [PubMed] [Google Scholar]
- Yoshihara N., Imayama T., Fukuchi-Mizutani M., Okuhara H., Tanaka Y., Ino I., et al. (2005). cDNA cloning and characterization of UDP-glucose: Anthocyanidin 3-O-glucosyltransferase in Iris hollandica. Plant Sci. 169 496–501. 10.1016/j.plantsci.2005.04.007 [DOI] [Google Scholar]
- Yoshikazu T., Keiko Y., Masako F. M., Yuko F., Hiroyuki F., Toshihiko A., et al. (1996). Molecular and biochemical characterization of three anthocyanin synthetic enzymes from Gentiana triflora. Plant Cell Physiol. 37 711–716. 10.1093/oxfordjournals.pcp.a029004 [DOI] [PubMed] [Google Scholar]
- Zhao D., Tao J. (2015). Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 6:261 10.3389/fpls.2015.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. C., Hu G. B., Hu F. C., Wang H. C., Yang Z. Y., Lai B. (2012). The UDP glucose: flavonoid-3-O-glucosyltransferase (UFGT) gene regulates anthocyanin biosynthesis in litchi (Litchi chinesis Sonn.) during fruit coloration. Mol. Biol. Rep. 39 6409–6415. 10.1007/s11033-011-1303-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.