Abstract
Nitric oxide synthase (NOS) is a multidomain enzyme that catalyzes the production of nitric oxide (NO) by oxidizing l‐Arg to NO and L‐citrulline. NO production requires multiple interdomain electron transfer steps between the flavin mononucleotide (FMN) and heme domain. Specifically, NADPH‐derived electrons are transferred to the heme‐containing oxygenase domain via the flavin adenine dinucleotide (FAD) and FMN containing reductase domains. While crystal structures are available for both the reductase and oxygenase domains of NOS, to date there is no atomic level structural information on domain interactions required for the final FMN‐to‐heme electron transfer step. Here, we evaluate a model of this final electron transfer step for the heme–FMN–calmodulin NOS complex based on the recent biophysical studies using a 105‐ns molecular dynamics trajectory. The resulting equilibrated complex structure is very stable and provides a detailed prediction of interdomain contacts required for stabilizing the NOS output state. The resulting equilibrated complex model agrees well with previous experimental work and provides a detailed working model of the final NOS electron transfer step required for NO biosynthesis.
Keywords: nitric oxide synthase, molecular dynamics, electron transfer, protein complex, redox partners
Abbreviations
- CaM
calmodulin
- EPR
electron paramagnetic resonance
- FAD
flavin adenine dinucleotide
- FMN
flavin mononucleotide
- MD
molecular dynamics
- NO
nitric oxide
- NOS
nitric oxide synthase
- RMSD
root mean squared displacement.
Introduction
Nitric oxide (NO) is an important signaling molecule required for fundamental physiological processes in the neuronal, immune, and cardiovascular systems.1, 2, 3 Disruptions to NO signaling have been linked to a wide variety of cardiovascular2 and neurodegenerative disorders.4, 5, 6, 7 NO biosynthesis requires the heme‐containing enzyme nitric oxide synthase (NOS), substrate l‐Arg, cofactor 6R‐5,6,7,8‐tetrahydrobiopterin (H4B), molecular oxygen, and electrons derived from NADPH.3, 8 At the NOS heme active site, l‐Arg is oxidized to l‐citrulline and NO. Electrons from NADPH are transferred to the oxygenase domain active site via the flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN)‐containing reductase domain of the opposing monomer in a calmodulin (CaM)‐dependent mechanism (Fig. 1).3 CaM facilitates interdomain electron transfer by binding to an α helical linker between the heme and FMN subdomain.9 It is generally thought that the required NOS conformational changes induced by CaM binding re‐orients the FMN subdomain from an electron accepting conformation (input state) to an electron donating conformation (output state) as illustrated in Fig. 1.10, 11 For both endothelial NOS (eNOS) and neuronal NOS (nNOS), the CaM‐induced conformational change between input and output states is dependent on the concentration of Ca2+. In sharp contrast, inducible NOS (iNOS) binds CaM very tightly and retains the ability to transition between the input and output states at basal Ca2+ concentrations.12
Figure 1.
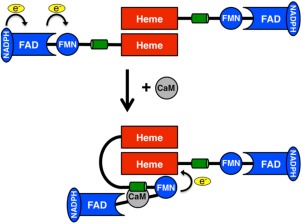
Nitric oxide synthase architecture. Each monomer of nitric oxide synthase consists of the oxygenase or heme domain (red) and the reductase domain (blue) which is composed of FMN and FAD containing subdomains. The oxygenase domain forms the heme active site and is the site of dimerization for holo‐NOS. In the input state (top), NADPH binds and reduces FAD, which in turn reduces FMN. Calmodulin (CaM, grey) binding on the α‐helical linker between the FMN and oxygenase domain (shown in green) is believed to induce a conformational switch towards the output state (bottom) to transfer electrons from FMN to the heme group.
The calmodulin‐dependent interdomain electron transfer between the NOS reductase and the oxygenase domains represents a key step in NO production. CaM is thought to function by first destabilizing the FMN–FAD interaction.13 Destabilization of the FMN–FAD subdomains interaction allows for the reductase domain to split and the FMN subdomain to approach the oxygenase domain in the output state. Electron transfer from the FMN subdomain to the oxygenase domain is gated by the required FMN conformational change from the input to the output state.10 Regulation of this step is critical because if NOS electron transfer is uncoupled, cell‐damaging peroxy species can form.9, 14, 15 Details on the mechanism of these CaM induced changes resulting in the output state remain unknown.
While crystal structures of individual NOS domains have been solved,16, 17, 18, 19, 20, 21, 22 a full‐length NOS crystal structure has remained elusive. Recent spectroscopic,23, 24 hydrogen–deuterium (HD) exchange,25 and single‐particle electron microscopy studies26, 27, 28 have for the first time begun to provide the structural and biophysical details of the interdomain electron transfer step in NOS and the required conformational changes of the NOS FMN domain. One key piece of evidence was the quantification of the FMN and heme group distance required for interdomain electron transfer at 18.8 Å using pulsed EPR.23
Molecular dynamics (MD) simulations that are consistent with the available data have proven to be a powerful approach for developing detailed atomic‐level complexes in the absence of experimental structures and probing important conformational changes.29, 30 In particular, such methods have been used to study similar reversibly binding electron transfer proteins.31, 32 The stability of such complexes, based on convergence of the model root mean squared displacement (RMSD), over a reasonably long trajectory can provide confidence that the model is reasonably close to a functionally relevant structure. These simulations also are useful to cross‐validate previous experimental data relative to the model being simulated. Therefore, we developed an output state model of the human iNOS oxy–FMN–CaM complex that is based on domain interactions resolved using HD exchange.25 The output state model remains stable over a 105‐ns MD simulation and agrees well with previous experimental data and provides new insights into the stability of this iNOS conformational state. Our model provides a structural framework for further experimental testing.
Results and Discussion
Molecular dynamics and architecture of iNOS oxy–FMN–CaM complex
Surface residues on the human iNOS oxygenase domain (defined in our model as residues 83–502) and CaM that were previously identified as complimentary binding sites by HD exchange25 were used to place CaM within our initial model. Similarly, the FMN subdomain (residues 536–699) was placed within the initial model based on previous HD exchange data.25 Linkers connecting the heme and FMN domains based on the iNOS sequence (residues 503–535) were added to the initial HD‐derived model and were modeled using φ,ψ peptide constraints to complete the oxy‐FMN model (residues 503–535). The constructed model (Fig. 2A) then was used as the starting point for the 105‐ns simulation.
Figure 2.
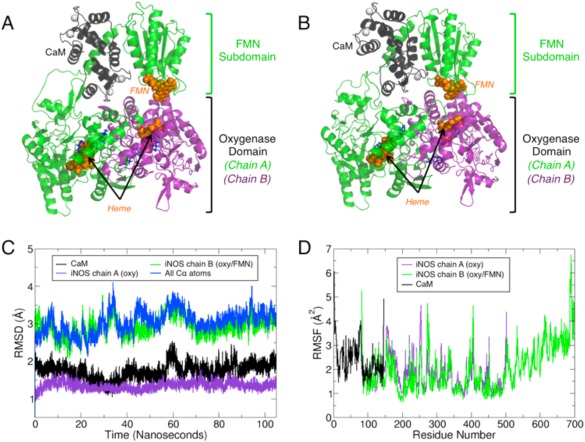
MD simulations of the oxy‐FMN‐CaM reveal complex stability. (A) The initial system for the oxy‐FMN‐CaM system, as described in the results and methods, is displayed and (B) a snapshot taken at 80 nanoseconds which represents an equilibrated complex structure. For both (A) and (B), chain A of the oxygenase domain is colored purple, chain B which includes a oxygenase domain, CaM‐binding helix and FMN subdomain is colored green, and CaM is shown in black. The heme and FMN cofactors are displayed as orange sticks, the substrate l‐Arg and H4B in blue sticks, and Ca2+ and Zn2+ as white spheres. The equilibrated CaM and FMN subdomain model is closer to chain A of the heme domain than in the initial starting model. (C) Root mean squared distance (RMSD) analysis over the full 105 nanosecond trajectory shows that the structure remains relatively unchanged over the course of the trajectory following initial minimization and initiation of the simulation for each major chain of the oxy‐FMN‐CaM. (D) Root mean square fluctuations (RMSF) measured for the sidechains for CaM (black), chain A heme domain (purple), and chain B heme/FMN domain (green).
The system underwent a quick conformational shift following the initial minimization and remained relatively unchanged for the remainder of the simulation as noted by RMSD analysis (Fig. 2C). For the initial 40 ns of the simulation, the FMN domain did not consistently interact with the heme domain. Visualization of the trajectory revealed that the FMN domain was constantly sampling different conformations at the FMN–heme domain interface as if it were undergoing a “bind and crawl” mechanism of association (Movie S1). Analysis of the FMN and heme cofactor positions indicates the distance between the heme iron and the N5 atom of FMN to be greater than the previously determined 18.8 Å distance during this initial phase of the trajectory23 (Fig. 3). However, after approximately 40 ns the FMN domain moved into a position where it formed close contacts with the heme domain that remained stable for the remainder of the simulation. This equilibrated output model coincides with the formation of several noncovalent contacts that likely function to facilitate formation of the output state (Table 1, Supporting Information Fig. S1). Moreover, the FMN to heme distance, as measured between the heme iron and the N5 atom of FMN, equilibrated to 18.0 ± 0.6 Å which agrees well with the previously EPR‐derived distance of 18.8 Å.23
Figure 3.
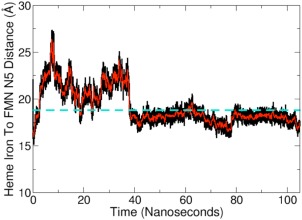
Distance measured between heme and FMN cofactors. The distance between the oxygenase heme iron and the N5 atom of the FMN group was calculated for the duration of the simulation. The experimentally determined heme to FMN distance of ∼18.8 Å23 is represented as a dashed turquoise line. A running average with a window of 100 ps is traced in red on the full dataset in black.
Table 1.
Major domain–domain interactions in the equilibrated oxy‐FMN‐CaM complex. Contacts are defined as a distance ≤3.5 Å
| Residue A | Residue B | Average contacts per picosecond | Selected atom–atom pair | MD distance and St. Dev. over final 50 ns b | Previous experimental work a |
|---|---|---|---|---|---|
| Chain A heme domain to chain B FMN domain | |||||
| HemeA‐Arg452 | FMN‐Glu546 | 4.331 | A‐NH2/B‐OE1 | 3.11 ± 0.51 Å | 35, 36 |
| HemeA‐Asn449 | FMN‐Glu546 | 1.891 | A‐ND2/B‐OE2 | 4.13 ± 1.10 Å | 35, 36 |
| HemeA‐Lys155 | FMN‐Glu603 | 1.626 | A‐NZ/B‐OE1 | 4.79 ± 2.14 Å | 37 |
| HemeA‐Glu156 | FMN‐Lys607 | 1.406 | A‐OE1/B‐NZ | 5.44 ± 2.74 Å | Reported here |
| HemeA‐Ser207 | FMN‐Asn595 | 1.219 | A‐O/B‐ND2 | 3.79 ± 0.94 Å | Reported here |
| HemeA‐Asn208 | FMN‐Gly594 | 1.063 | A‐ND2/B‐O | 3.58 ± 1.15 Å | 33 |
| Chain A of heme domain to calmodulin | |||||
| HemeA‐Arg86 | CaM‐Asp122 | 5.224 | A‐NH1/C‐OD2 | 2.78 ± 0.27 Å | 25 |
| HemeA‐Arg83 | CaM‐Glu127 | 4.357 | A‐NH2/C‐OE1 | 3.71 ± 1.88 Å | 38 |
| HemeA‐Gln97 | CaM‐Asp133 | 1.291 | A‐NE2/C‐O | 3.27 ± 0.63 Å | Reported here |
| Chain B FMN domain to calmodulin | |||||
| FMN‐Arg536 | CaM‐Glu47 | 5.076 | B‐NH2/C‐OE1 | 2.99 ± 0.34 Å | 22 |
| FMN‐Arg576 | CaM‐Glu83 | 4.568 | B‐NH1/C‐OE2 | 3.23 ± 0.66 Å | Reported here |
| FMN‐Glu551 | CaM‐Lys94 | 2.805 | B‐OE1/C‐NZ | 3.55 ± 1.22 Å | Reported here |
| FMN‐Lys568 | CaM‐Glu87 | 2.443 | B‐NZ/C‐OD2 | 3.32 ± 0.92 Å | 22 |
| FMN‐Lys574 | CaM‐Asp93 | 1.854 | B‐NZ/C‐OD2 | 2.67 ± 0.09 Å | Reported here |
Residue previously identified to be important for either enzyme activity or domain interactions as referenced in corresponding study in either iNOS or an analogous residue in either eNOS or nNOS.
For a more in depth view of the nature and dynamics of each contact, refer to Supporting Information Fig. S3.
Based on the previous NOS kinetic analyses, the two electron reduced hydroquinone, FMNH2, transfers electrons to the heme group of the opposing subunit via a conserved tryptophan.25, 33, 34 In our equilibrated MD model, the center of mass of this HemeAW372 residue (HemeAW366 in murine iNOS) sidechain is within 11.67 ± 0.7 Å of the N5 atom of the FMN group and 9.46 ± 0.2 Å of the heme iron. The distance between HemeAW372 and the redox partners is important, as recent work has implicated this conserved Trp residue to mediate electron transfer between the two redox cofactors and the observed distances between the cofactors and heme are similar to those predicted previously.25
In order to further test our model, we also docked our equilibrated model into a previously reported single‐particle NOS EM map (EMD 2748) at 74 Å resolution.26 Our model had a cross‐correlation value of 0.6824 using the “fit in map” function of Chimera with only 93 of 9,716 atoms falling outside the EM density at a map value of 1.287 (Supporting Information Fig. S2).
FMN and heme domain interface of the oxy–FMN–CaM complex
In the final equilibrated structure, several contacts were observed to facilitate binding of the FMN domain within close proximity to the heme domain (Fig. 4 and Supporting Information Fig. S3, Table 1). The observed domain contacts within our model are largely facilitated by side‐chain electrostatic interactions. Based on the average number of contacts per frame (where a contact is an atom‐to‐atom distance of ≤3.5 Å), the strongest of these interactions is an ion pair interaction between residues FMNE546 and HemeAR452. The FMNE546–HemeAR452 ion pair interaction was observed to form upon complex formation and remained stable for the remainder of the simulation. Previous mutagenesis analysis on the FMNE546N in iNOS shows that the FMN‐to‐heme electron transfer rate decreases by about 2.5‐fold owing primarily to a 3‐fold increase in the activation entropy.35, 36 In addition to FMNE546 contacting HemeAR452, FMNE546 contacts HemeAN449. Analysis of the FMNE546–HemeAN449 interaction indicates the HemeAN449 sidechain nitrogen to switch between contacting the FMNE546 carboxyl group and the carbonyl oxygen of FMNE456. Our model also indicates the heme–FMN interface of human iNOS to be further stabilized by electrostatic interactions between FMNK607–HemeAE156, FMNE603–HemeAK155, and FMNN595–HemeAS207. Human iNOS FMNE603 is analogous to rat nNOS FMNE819 that previously has been shown to facilitate charge‐pairing interactions within the FAD domain, which has been predicted to use the same FMN domain surface region to interact with the oxygenase domain.26, 37 Between our model and previous biochemical studies, FMNE603 (iNOS)/FMNE819 (rat nNOS) may also play a critical role in regulating electron transfer between the input and the output states.
Figure 4.
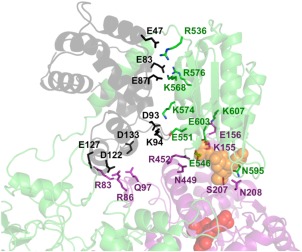
Interfacial contacts detected between calmodulin, oxygenase domain, and the FMN subdomain of iNOS. View of the NOS domain interface composed of the heme domain, calmodulin, and FMN subdomain (colored in the same fashion as Figure 2) in an equilibrated MD structure of the oxy‐FMN‐CaM complex, taking a snapshot from roughly 80 ns into the simulation. Residues listed in Table 1 are shown as sticks and labeled to illustrate the interactions that facilitate formation of the output state. The transient contacts observed throughout the simulation that averaged less than one contact per frame are not shown.
In our equilibrated model, we also observed a weak hydrogen‐bond interaction between the backbone oxygen of FMNG594 and the side chain of HemeAN208. These residues form a stable interaction between the two domains with an atom‐to‐atom distance of 3.6 ± 1.1 Å. To the best of our knowledge, these residues have not been experimentally validated by mutagenesis studies. However, both FMNG594 and HemeAN208 were broadly implicated to contribute toward domain–domain interactions based on HD exchange studies.25 A series of secondary or less frequent and longer‐range interactions at the interface of our output state model were also identified and displayed in heatmap form (Supporting Information Fig. S1). These secondary interactions may supplement the stronger contacts that facilitate binding of the FMN subdomain to the heme domain. When combined with the major contacts described above, the FMN–oxygenase interface averages 16.34 atom‐to‐atom contacts per frame over the final 50 ns of the simulation.
Possible role of CaM in the stabilization of the oxy–FMN–CaM complex
Our model is consistent with the proposed role of CaM in stabilizing the output state by directly interacting with the heme domain.25, 26 In our output state model, CaM binds to the FMN‐heme linker (Supporting Information Table S1) to facilitate formation of stable contacts between the oxygenase domain and FMN subdomain and is consistent with HD exchange data,25 as CaM was found to also interact directly with the heme domain (Figs. 4 and Supporting Information Fig. S3, Table 1). As a result, CaM works together with the FMN domain to form a complex with the heme domain. Analysis of the final 50 ns of the simulation demonstrates five residues of CaM to form noncovalent interactions with the FMN subdomain and three residues with the heme domain of iNOS. Of the CaM–FMN subdomain interactions observed in our model, ion pairs between charged residues dominate the noncovalent interactions, as detailed in Table 1. One ion pair in particular, CaMAsp122–HemeAArg86, is of interest as mutations at CaMAsp122 have previously been shown to lower NADPH oxidation rates, increase cytrochome‐c activity, and decrease NO production.25 These results provide additional support that CaMAsp122 plays an important role in facilitating interdomain electron transfer. Moreover, our data implicates HemeAArg86 of human iNOS as being required to stabilize CaM binding in the output state. We also observed several additional transient interactions that play a minor role in facilitating the CaM–FMN subdomain interaction, as demonstrated by the contact heatmaps (Supporting Information Fig. S1). When combined with the major interactions described in Table 1, the CaM–FMN subdomain interaction averages 21.23 atom‐to‐atom contacts per frame over the final 50 equilibrated nanoseconds of the trajectory, more than the contacts observed between the FMN and oxygenase domains. These data suggest that the CaM–FMN subdomain interactions are mediated by several noncovalent interactions that may play a more integral role in stabilizing the output state than previously predicted.
Following equilibration of the output state, NOS domain interactions were further stabilized by three noncovalent interactions between both CaM and chain A of the heme domain (Fig. 4, Table 1). Complimentary charged and polar residues dominate the contact interactions at this interface, similar to the aforementioned CaM–FMN interface. The entire CaM‐oxygenase domain interface interaction averages 13.79 atom‐to‐atom contacts per frame over the final 50 ns of the trajectory, making it the weakest of the domain–domain interactions observed in our MD simulation of the output state. One key difference between both our equilibrated model and the iNOS CaM–FMN crystal structure (PDB 3HR4) are the contacts made between the FMN subdomain and CaM that make up this final interface in the oxy–FMN–CaM complex. The resulting equilibrated complex structure conserves two contacts that include residues that were described by the previously described crystal structure,22 FMNR536–CaME47 and FMNK568–CaM87, and the addition of three novel contacts observed in our model are the result of a conformational change within the linker between the CaM‐binding helix of chain B and the FMN domain (Supporting Information Fig. S4). In short, our output state model requires more residue contacts between the FMN subdomain and CaM then previously observed by X‐ray crystallography. This is not too surprising since our model includes the heme domain together with information of recent HD and EM data,25, 26 while the crystal structure includes only CaM and the FMN domain.
In order to further probe the importance of CaM binding for the stability of the oxy–FMN domain complex, a second simulation was carried out with the same starting model where calmodulin was removed (Fig. 5A). It would be expected that if CaM does play a role in stability of the overall complex, than in the absence of CaM no stable complex would be formed between the FMN and the heme domain. Indeed, after 15 ns, the complex undergoes a large conformation change (Movie S2) as observed by RMSD analysis (Supporting Information Fig. S5A) corresponding to the FMN domain moving away from the heme domain dimer. After 20 ns, all significant contacts between the FMN and the heme domains were lost and the complex was broken for the remainder of the 100 ns simulation (Figs. 5B and Supporting Information Fig. S5A). The FMN‐to‐heme cofactor distance was recalculated as was done in Fig. 3 and shows that the distance remained much larger that the experimentally observed value of 18.8 Å (Supporting Information Fig. S6B). Taken together with the results of the initial simulation, this underscores the important role CaM plays in stabilizing the interactions between the FMN and the heme domains during the second electron transfer step.
Figure 5.
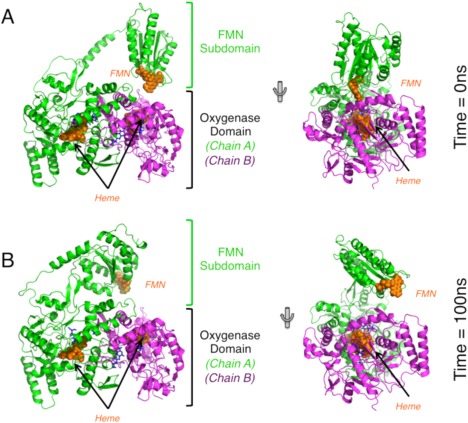
Overview of no‐CaM simulation of the iNOS oxy‐FMN complex. (A) The same initial starting model, shown here in two different orientations, was used in the no‐CaM simulation as was used in the full oxy‐FMN‐CaM simulation. (B) The resulting equilibrated no‐CaM complex following 100 ns of simulation. Over the course of 100 ns, the FMN domain moved away from the heme domain and no tight interdomain complex was formed. All domains are colored identically as displayed in Figure 2.
Conclusions
Through the implementation of modeling and MD, we have produced an atomic level model of the iNOS output state. Following equilibration, our output state model is in excellent agreement with previous biochemical, biophysical, and site‐directed mutagenesis studies. MD has also identified additional surface contacts that may play an important role in the final iNOS interdomain electron transfer step. One of the more important observations in this study is the role of CaM in activating iNOS catalysis. As first proposed by Smith et al.,25 CaM plays an active role by directly interacting with the heme domain. Thus, CaM may work synergistically with the FMN domain of iNOS to form an ET competent output state complex. However, the domain arrangement of the input state must be substantially different than our output state model. If we assume that the nNOS reductase domain structure, for which the FAD and FMN are within 3.4 Å,22 is a true representation of the input state then the CaM linker segment and heme domain must be positioned quite differently in the output state. Without such motion, there would be severe overlap between CaM and the FAD domain. The simplest dynamic model that emerges from these observations is of an equilibrium mix between the input state where the FMN and FAD are in close contact and the output state where CaM and the FMN domain interact with the heme domain. The function of CaM is thus not so much to destabilize the FMN–FAD interaction but rather to stabilize the ET competent output state long enough for heme reduction. Neither input nor output states must be very stable or long‐lived owing to the large domain rearrangements that are necessary for ET first from FAD to FMN and then FMN to heme. As a result, NOS is conformationally heterogeneous as evidenced by the recent cryoEM studies.26, 27, 28, 39 With this new information, it may now be possible to probe the NOS output state through additional biochemical and biophysical studies in order to better understand the dynamic equilibrium between the input and output states of NOS.
Methods
Construction of the initial oxy–FMN–CaM system
Atomic coordinates for iNOS oxy–FMN–CaM were constructed from PDB 3HR4 and 1NSI. The positioning of CaM bound to the CaM‐binding peptide (PDB 3HR4) relative to the iNOS oxygenase domain (PDB 1NSI) was based on the previously reported HD exchange data.17, 22, 25 Similarly, FMN subdomain (coordinates from PDB 3HR4) was then positioned relative to the oxygenase domain and CaM sequence. Domains of iNOS were positioned using Pymol. Peptide linkers determined from the human iNOS sequence were then modeled and built using COOT.40
Molecular dynamics of the oxy–FMN–CaM system
Hydrogens were added to the system using psfgen of VMD 1.9.1.41 The system was then hydrated with TIP3P water with a 15‐Å cushion for a total of 40,660 additional waters residues. Crystallographic waters from PDB 1NSI were also included within the initial start model. The final system had a unit cell box dimensions of (128 Å × 105 Å × 117 Å) and 141,497 atoms. Initial simulations were carried out on Greenplanet (gplogin1.ps.uci.edu) before productions runs were carried out on the Stampede supercomputer (stampede.tacc.utexas.edu) using NAMD 2.9.42 The CHARMM force fields used to model the protein, heme, FMN, and H4B cofactors were identical to those used in the previous studies.43, 44, 45 A smooth partial Ewald mesh method was used for the calculation of Columbic forces, a Langevin thermostat was used to maintain a constant temperature of 300 K, and a Nose–Hoover–Langevin piston employed for constant pressure control.46, 47
Positional restraints were placed on the central atoms of each heme group in order to reproduce the heme conformation that has been observed in crystal structures as well as an additional extra bond to preserve the hydrogen bonding of the H4B cofactor to the heme domain as has been done in a previous study.45 Similarly, extra bonds were added to the simulation to preserve the experimentally observed coordination of the heme domain Zn2+ and the four Ca2+ atoms in CaM. No restraints were placed on the protein backbone of the iNOS model nor were any interdomain restraints included in the simulation. Equilibration was carried out initially by minimizing the structure for 1000 fs before the simulation was allowed to propagate for 5 ns at 1.0 fs time step. After equilibration, the timestep was increased to 2.0 fs for the remainder of the simulation. Positional restraints were placed on central atoms of the heme to ensure that the heme coordination was in agreement with the previously solved crystal structures of the oxygenase domain. Frames were saved every 2 ps and saved for analysis which was carried out on local systems using locally developed tools and VMD41 and Chimera.48
Supporting information
Supporting Information
Supporting Information Movie 1.
Supporting Information Movie 2.
Acknowledgment
The authors acknowledge Dr. Douglas Tobias for many thoughtful discussions.
References
- 1. Moncada S, Higgs EA (1991) Endogenous nitric‐oxide—physiology, pathology and clinical relevance. Eur J Clin Invest 21:361–374. [DOI] [PubMed] [Google Scholar]
- 2. Forstermann U, Sessa WC (2012) Nitric oxide synthases: regulation and function. Eur Heart J 33:829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Poulos TL (2014) Heme enzyme structure and function. Chem Rev 114:3919–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dorheim MA, Tracey WR, Pollock JS, Grammas P (1994) Nitric‐oxide synthase activity is elevated in brain microvessels in Alzheimers‐disease. Biochem Biophys Res Commun 205:659–665. [DOI] [PubMed] [Google Scholar]
- 5. Norris PJ, Waldvogel HJ, Faull RLM, Love DR, Emson PC (1996) Decreased neuronal nitric oxide synthase messenger RNA and somatostatin messenger RNA in the striatum of Huntington's disease. Neuroscience 72:1037–1047. [DOI] [PubMed] [Google Scholar]
- 6. Sims NR, Anderson MF (2002) Mitochondrial contributions to tissue damage in stroke. Neurochem Int 40:511–526. [DOI] [PubMed] [Google Scholar]
- 7. Zhang L, Dawson VL, Dawson TM (2006) Role of nitric oxide in Parkinson's disease. Pharmacol Therapeut 109:33–41. [DOI] [PubMed] [Google Scholar]
- 8. Stuehr DJ, Griffith OW (1992) Mammalian nitric‐oxide synthases. Adv Enzymol Relat Areas Mol Biol 65:287–346. [DOI] [PubMed] [Google Scholar]
- 9. Alderton WK, Cooper CE, Knowles RG (2001) Nitric oxide synthases: structure, function and inhibition. Biochem J 357:593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li WB, Fan WH, Elmore BO, Feng CJ (2011) Effect of solution viscosity on intraprotein electron transfer between the FMN and heme domains in inducible nitric oxide synthase. FEBS Lett 585:2622–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Serafim RAM, Primi MC, Trossini GHG, Ferreira EI (2012) Nitric oxide: State of the art in drug design. Curr Med Chem 19:386–405. [DOI] [PubMed] [Google Scholar]
- 12. Balligand JL, Ungureanu‐Longrois D, Simmons WW, Pimental D, Malinski TA, Kapturczak M, Taha Z, Lowenstein CJ, Davidoff AJ, Kelly RA, et al. (1994) Cytokine‐inducible nitric oxide synthase (iNOS) expression in cardiac myocytes. Characterization and regulation of iNOS expression and detection of iNOS activity in single cardiac myocytes in vitro. J Biol Chem 269:27580–27588. [PubMed] [Google Scholar]
- 13. Ilagan RP, Tejero J, Aulak KS, Ray SS, Hemann C, Wang ZQ, Gangoda M, Zweier JL, Stuehr DJ (2009) Regulation of FMN subdomain interactions and function in neuronal nitric oxide synthase. Biochemistry 48:3864–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Calabrese V, Mancuso C, Calvani M, Rizzarelli E, Butterfield DA, Stella AM (2007) Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat Rev Neurosci 8:766–775. [DOI] [PubMed] [Google Scholar]
- 15. Piazza M, Guillemette JG, Dieckmann T (2015) Dynamics of nitric oxide synthase–calmodulin interactions at physiological calcium concentrations. Biochemistry 54:1989–2000. [DOI] [PubMed] [Google Scholar]
- 16. Fischmann TO, Hruza A, Niu XD, Fossetta JD, Lunn CA, Dolphin E, Prongay AJ, Reichert P, Lundell DJ, Narula SK, Weber PC (1999) Structural characterization of nitric oxide synthase isoforms reveals striking active‐site conservation. Nat Struct Biol 6:233–242. [DOI] [PubMed] [Google Scholar]
- 17. Li H, Raman CS, Glaser CB, Blasko E, Young TA, Parkinson JF, Whitlow M, Poulos TL (1999) Crystal structures of zinc‐free and ‐bound heme domain of human inducible nitric‐oxide synthase. Implications for dimer stability and comparison with endothelial nitric‐oxide synthase. J Biol Chem 274:21276–21284. [DOI] [PubMed] [Google Scholar]
- 18. Li H, Shimizu H, Flinspach M, Jamal J, Yang W, Xian M, Cai T, Wen EZ, Jia Q, Wang PG, Poulos TL (2002) The novel binding mode of N‐alkyl‐N'‐hydroxyguanidine to neuronal nitric oxide synthase provides mechanistic insights into NO biosynthesis. Biochemistry 41:13868–13875. [DOI] [PubMed] [Google Scholar]
- 19. Raman CS, Li H, Martasek P, Kral V, Masters BS, Poulos TL (1998) Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center. Cell 95:939–950. [DOI] [PubMed] [Google Scholar]
- 20. Crane BR, Arvai AS, Ghosh DK, Wu C, Getzoff ED, Stuehr DJ, Tainer JA (1998) Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science 279:2121–2126. [DOI] [PubMed] [Google Scholar]
- 21. Zhang J, Martasek P, Paschke R, Shea T, Siler Masters BS, Kim JJ (2001) Crystal structure of the FAD/NADPH‐binding domain of rat neuronal nitric‐oxide synthase. Comparisons with NADPH‐cytochrome P450 oxidoreductase. J Biol Chem 276:37506–37513. [DOI] [PubMed] [Google Scholar]
- 22. Xia C, Misra I, Iyanagi T, Kim JJ (2009) Regulation of interdomain interactions by calmodulin in inducible nitric‐oxide synthase. J Biol Chem 284:30708–30717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Astashkin AV, Elmore BO, Fan W, Guillemette JG, Feng C (2010) Pulsed EPR determination of the distance between heme iron and FMN centers in a human inducible nitric oxide synthase. J Am Chem Soc 132:12059–12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Astashkin AV, Chen L, Zhou X, Li H, Poulos TL, Liu KJ, Guillemette JG, Feng C (2014) Pulsed electron paramagnetic resonance study of domain docking in neuronal nitric oxide synthase: the calmodulin and output state perspective. J Phys Chem A 118:6864–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith BC, Underbakke ES, Kulp DW, Schief WR, Marletta MA (2013) Nitric oxide synthase domain interfaces regulate electron transfer and calmodulin activation. Proc Natl Acad Sci USA 110:E3577–E3586. 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Campbell MG, Smith BC, Potter CS, Carragher B, Marletta MA (2014) Molecular architecture of mammalian nitric oxide synthases. Proc Natl Acad Sci USA 111:E3614–E3623. 36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yokom AL, Morishima Y, Lau M, Su M, Glukhova A, Osawa Y, Southworth DR (2014) Architecture of the nitric‐oxide synthase holoenzyme reveals large conformational changes and a calmodulin‐driven release of the FMN domain. J Biol Chem 289:16855–16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Volkmann N, Martasek P, Roman LJ, Xu XP, Page C, Swift M, Hanein D, Masters BS (2014) Holoenzyme structures of endothelial nitric oxide synthase—an allosteric role for calmodulin in pivoting the FMN domain for electron transfer. J Struct Biol 188:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karplus M, McCammon JA (2002) Molecular dynamics simulations of biomolecules. Nat Struct Biol 9:646–652. [DOI] [PubMed] [Google Scholar]
- 30. Adcock SA, McCammon JA (2006) Molecular dynamics: survey of methods for simulating the activity of proteins. Chem Rev 106:1589–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salemme FR (1976) An hypothetical structure for an intermolecular electron transfer complex of cytochromes c and b5. J Mol Biol 102:563–568. [DOI] [PubMed] [Google Scholar]
- 32. Wendoloski JJ, Matthew JB, Weber PC, Salemme FR (1987) Molecular dynamics of a cytochrome c–cytochrome b5 electron transfer complex. Science 238:794–797. [DOI] [PubMed] [Google Scholar]
- 33. Li H, Das A, Sibhatu H, Jamal J, Sligar SG, Poulos TL (2008) Exploring the electron transfer properties of neuronal nitric‐oxide synthase by reversal of the FMN redox potential. J Biol Chem 283:34762–34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tejero J, Hannibal L, Mustovich A, Stuehr DJ (2010) Surface charges and regulation of FMN to heme electron transfer in nitric‐oxide synthase. J Biol Chem 285:27232–27240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sempombe J, Galinato MG, Elmore BO, Fan W, Guillemette JG, Lehnert N, Kirk ML, Feng C (2011) Mutation in the flavin mononucleotide domain modulates magnetic circular dichroism spectra of the iNOS ferric cyano complex in a substrate‐specific manner. Inorganic Chem 50:6859–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng C, Chen L, Li W, Elmore BO, Fan W, Sun X (2014) Dissecting regulation mechanism of the FMN to heme interdomain electron transfer in nitric oxide synthases. J Inorg Biochem 130:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haque MM, Bayachou M, Fadlalla MA, Durra D, Stuehr DJ (2013) Charge‐pairing interactions control the conformational setpoint and motions of the FMN domain in neuronal nitric oxide synthase. Biochem J 450:607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. George SE, Su Z, Fan D, Wang S, Johnson JD (1996) The fourth EF‐hand of calmodulin and its helix‐loop‐helix components: impact on calcium binding and enzyme activation. Biochemistry 35:8307–8313. [DOI] [PubMed] [Google Scholar]
- 39. Persechini A, Tran QK, Black DJ, Gogol EP (2013) Calmodulin‐induced structural changes in endothelial nitric oxide synthase. FEBS Lett 587:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Cryst 66:486–501. D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph Model 14:33–38. [DOI] [PubMed] [Google Scholar]
- 42. Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K (1999) NAMD2: greater scalability for parallel molecular dynamics. J Comput Phys 151:283–312. [Google Scholar]
- 43. Mackerell AD, Feig M, Brooks CL (2004) Extending the treatment of backbone energetics in protein force fields: limitations of gas‐phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 25:1400–1415. [DOI] [PubMed] [Google Scholar]
- 44. Madrona Y, Hollingsworth SA, Khan B, Poulos TL (2013) P450cin active site water: implications for substrate binding and solvent accessibility. Biochemistry 52:5039–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holden JK, Kang S, Hollingsworth SA, Li H, Lim N, Chen S, Huang H, Xue F, Tang W, Silverman RB, Poulos TL (2015) Structure‐based design of bacterial nitric oxide synthase inhibitors. J Med Chem 58:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Darden T, York D, Pedersen L (1993) Particle mesh Ewald—an N.Log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. [Google Scholar]
- 47. Martyna GJ, Tobias DJ, Klein ML (1994) Constant‐pressure molecular‐dynamics algorithms. J Chem Phys 101:4177–4189. [Google Scholar]
- 48. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information Movie 1.
Supporting Information Movie 2.


