Abstract
Magnetotactic bacteria (MTB) are a group of Gram‐negative microorganisms that are able to sense and change their orientation in accordance with the geomagnetic field. This unique capability is due to the presence of a special suborganelle called the magnetosome, composed of either a magnetite or gregite crystal surrounded by a lipid membrane. MTB were first detected in 1975 and since then numerous efforts have been made to clarify the special mechanism of magnetosome formation at the molecular level. Magnetosome formation can be divided into several steps, beginning with vesicle invagination from the cell membrane, through protein sorting, followed by the combined steps of iron transportation, biomineralization, and the alignment of magnetosomes into a chain. The magnetosome‐chain enables the sensing of the magnetic field, and thus, allows the MTB to navigate. It is known that magnetosome formation is tightly controlled by a distinctive set of magnetosome‐associated proteins that are encoded mainly in a genomically conserved region within MTB called the magnetosome island (MAI). Most of these proteins were shown to have an impact on the magnetism of MTB. Here, we describe the process in which the magnetosome is formed with an emphasis on the different proteins that participate in each stage of the magnetosome formation scheme.
Keywords: magnetotactic bacteria, magnetosome, biomineralization, magnetic nanoparticles, protein function
Abbreviations
- AMB‐1
Magnetospirillum magneticum strain AMB‐1
- BAR
Bin/Amphiphysin/Rvs
- CDF
cation diffusion facilitator
- CM
cytoplasmic membrane
- CTD
C‐terminal domain
- MAD
magnetosome‐associated Deltaproteobacteria
- MAI
magnetosome island
- Mam
magnetosome‐associated membrane
- MCP
methyl‐accepting chemotaxis proteins
- MM
magnetosome membrane
- Mms
magnetic particle membrane‐specific
- MSR‐1
Magnetospirillum gryphiswaldense MSR‐1
- MTB
magnetotactic bacteria
- NTD
N‐terminal domain
- TM
transmembrane
- TMD
transmembrane domain
- WT
wild‐type
Introduction
Magnetotactic bacteria (MTB) are a group of Gram‐negative microorganisms that can align along external magnetic fields.1 MTB were first described in Italian by Salvatore Bellini in 19632, 3 but remained untranslated into English; in 1975, they were independently discovered by Richard Blakemore in marine sediments4 and the worldwide MTB study was initiated.5 The ability of MTB to orient themselves along magnetic fields is achieved by a chain‐like organization of subcellular organelles, called magnetosomes, that are composed of a magnetic particle surrounded by a bilayer lipid membrane6, 7 (Fig. 1). Magnetosomes are able to biomineralize single crystals of magnetite or gregite8, 9 in strain‐dependent sizes and morphologies, wherein each strain these properties are conserved.10, 11, 12 The mineral crystal size is ∼30–120 nm, which fits the size of a single‐magnetic domain.10 The common theory in the MTB community suggests that magnetosome membranes (MMs) invaginate from the cytoplasmic membrane (CM) to form vesicles.5, 13 creating the optimal conditions for crystal nucleation and growth.10, 13, 14 Magnetosomes’ alignment into a fixed linear chain or multiple chains requires cytoskeletal actin‐like filamentous structures (Fig. 1B), and generates a permanent magnetic dipole moment. This enables the rotation of the entire cell to be aligned with the geomagnetic field lines, allowing the bacterium to move along these lines using their flagella. This behavior—magnetotaxis—increases their efficiency in finding suitable environmental conditions, usually the oxic‐anoxic zone in aquatic enviroments.4, 5, 7, 15, 16 The early model of magnetotaxis was based on the assumption that all MTB have a permanent polar preference to their swimming direction. In this model, north‐seeking bacteria swimming northward in the Northern Hemisphere and south‐seeking bacteria swimming southward in the Southern Hemisphere would migrate downward toward the sediments along the inclined geomagnetic field lines.4, 5, 16, 17 Later on, this model was shown not to be valid but only under specific conditions and cannot explain the taxis‐behaviors of some strains.18 A new model suggested that magnetotaxis together with aerotaxis enable the MTB to reach the appropriate environment, a behavior that was called “magneto‐aerotaxis.”18, 19 Two different mechanisms were proposed: (1) a polar magneto‐aerotaxis mechanism, in which the bacterium moves persistently in a specific direction (parallel or antiparallel to the magnetic field), depends on the oxic conditions, which results in an efficient aerotactic response in the vertical oxygen gradients, and (2) an axial magneto‐aerotaxis, in which the bacterium does not have a preference for the swimming direction and swims with frequent, spontaneous reversals of swimming directions (with no distinction between north‐seeking and south‐seeking bacteria).5, 13, 16, 18 Recently, six different magneto‐aerotactic behaviors were observed in different strains.20 Despite the above, the navigation mechanism of MTB does not depend only on oxygen concentration but is thought to be more complicated and to involve other mechanisms such as phototaxis21, 22 and chemotaxis.5, 13, 23, 24
Figure 1.
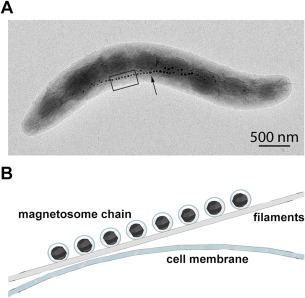
Magnetotactic bacterium. (A) Transmission electron microscope (TEM) image of Magnetospirillum gryphiswaldense MSR‐1, contributed by Dr. René Uebe and Dr. Dirk Schüler. The black arrow points toward the magnetosome chain. (B) Magnified illustration of the black box in A: magnetosomes are made of magnetic particles surrounded by a lipid membrane—which invaginate from the cell membrane—and organized as a chain on filaments.
From their rediscovery in 1975, MTB were greatly studied in many research groups around the world. Genetic studies showed that MTB are highly divergent: they are affiliated mainly with the Alpha‐, Gamma‐, and Deltaproteobacteria from the Proteobacteria phylum, as well as with the Nitrospirae 25, 26 and Omnitrophica 27, 28 phyla. The most characterized strains today include the cultivated Alphaproteobacteria species Magnetospirillum magneticum strain AMB‐1 (AMB‐1), Magnetospirillum gryphiswaldense MSR‐1 (MSR‐1), Magnetospirillum magnetotacticum strain MS‐1 and Magnetococcus marinus strain MC‐1.29 There are also cultivated, studied strains affiliated to Deltaproteobacteria and Gammaproteobacteria classes, such as Desulfovibrio magneticus strain RS‐130 and BW‐2,31 respectively.
The study of different strains showed that the formation of a functional magnetosome is a highly controlled process.32, 33, 34, 35 Most of the genes responsible for this process are located in the magnetosome island (MAI), a genomic segment (of 130 kb in MSR‐1) that is conserved among different species.32, 33, 35, 36, 37 This region contains a few operons; the most conserved and essential operon is mamAB, which can be found in all identified MTB, while other operons, such as mamGFDC, mamXY, and mms6 in MSR‐1, are specific to Alphaproteobacteria.25, 35, 38, 39, 40, 41 To simplify the information discussed in this review, we will focus on a set of identified genes within the MSR‐1 strain that are classified as MTB‐related and ‐specific genes, meaning that they share only slight or no similarities to the genes of other nonmagnetic organisms, respectively. These genes encode for most of the magnetosome‐associated membrane (Mam) proteins and magnetic particle membrane‐specific (Mms) proteins.5, 32 Examples of magnetosome‐related genes from other strains are the magnetosome‐associated Deltaproteobacteria (mad) group of genes that can be found in Deltaproteobacteria strains and in Nitrospirae and Omnitrophica phyla strains.25, 28, 40 Mam and Mms proteins can be classified approximately into several groups according to their roles during magnetosome formation processes, such as membrane invagination, protein sorting, magnetosome alignment into chains, biomineralization, and the control of mineral crystal shape and size.34, 42, 43, 44, 45 Here, by describing the known data regarding Mam and Mms proteins and focusing on their role (Table 1),14, 32, 34, 35, 42, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82 we present the process of magnetosome formation.
Table 1.
Key features of all discussed Mam and Mms proteins
| Protein | # Amino acidsa | Encoding operona | Suggested role | Main related articles |
|---|---|---|---|---|
| MamA | 217 | mamAB | Protein sorting | 14, 46, 47 |
| MamB | 297 | mamAB | Membrane invagination, iron transport | 48 |
| MamC | 125 | mamGFDC | Crystal size and shape control | 44, 49–52 |
| MamD | 314 | mamGFDC | Crystal size and shape control | 44, 50, 51 |
| MamE | 772 | mamAB | Protein sorting, redox control | 34, 53, 54 |
| MamF | 111 | mamGFDC | Crystal size control | 42, 44, 49, 55 |
| MamG | 84 | mamGFDC | Crystal size and shape control | 44, 49–51 |
| MamH | 427 | mamAB | Iron transport | 56 |
| MamI | 76 | mamAB | Membrane invagination | 34 |
| MamJ | 466 | mamAB | Magnetosome alignment | 57–61 |
| MamK | 359 | mamAB | Magnetosome alignment | 62–66 |
| MamL | 122 | mamAB | Membrane invagination | 34 |
| MamM | 318 | mamAB | Iron transport | 48, 67 |
| MamN | 437 | mamAB | pH control | – |
| MamO | 632 | mamAB | Crystal nucleation | 53, 54, 68 |
| MamP | 269 | mamAB | Redox control | 69–72 |
| MamQ | 271 | mamAB | Membrane invagination | – |
| MamR | 72 | mamAB | Crystal size and number control | – |
| MamS | 180 | mamAB | Crystal size and shape control | – |
| MamT | 174 | mamAB | Redox control | 70, 72 |
| MamX | 268 | mamXY | Redox control | 45, 56 |
| MamY | 370 | mamXY | Membrane invagination | 73, 74 |
| MamZ | 660 | mamXY | Iron transport, redox control | 56 |
| FtsZm | 323 | mamXY | Crystal size and shape control, denitrification | 75, 76 |
| Mms6 | 136 | mms6 | Crystal size and shape control | 77–79 |
| MmsF | 124 | mms6 | Crystal size and shape control | 55, 80 |
Membrane invagination
Magnetosome invagination is the first step in magnetosome formation,5, 13, 34 in which a few proteins are suspected to take part (Fig. 2A). Based on genetic dissection studies, mamB, I, L and Q deletion resulted in the lack of magnetosome vesicles in AMB‐134 and apart from mamI, also in no magnetic response in MSR‐1.42 This suggests that mamB, I, L, and Q are important for magnetosome invagination, although they are not sufficient by themselves in the ΔmamAB operon to restore the biogenesis of magnetosomes.34 MamB is suspected to have a dual role in MTB, in magnetosome invagination as well as in iron transportation,48 hence, it will be discussed in detail later in the biomineralization section. MamY was also suggested to have a role in membrane invagination,73 indicating that a set of at least five proteins control this process.
Figure 2.
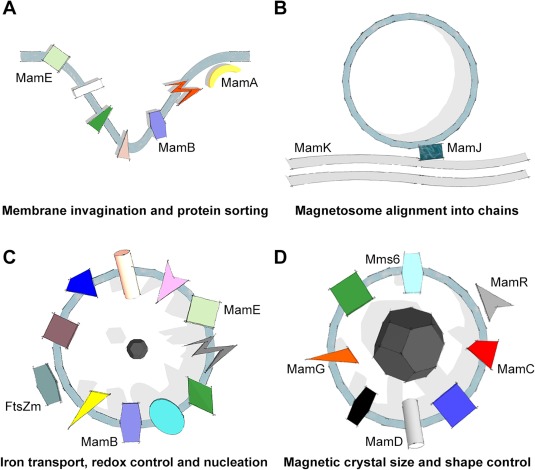
Schematic model of magnetosome formation. Proteins (each is presented in a different shape or color) can be roughly divided into different stages of magnetosome formation (TM proteins cross the membrane; proteins’ sizes, shapes, colors and locations are meaningless, unless specified): (A) MamB, I, L, Q, and Y were suggested to take part in magnetosome invagination, and MamA and E in protein sorting. (B) MamK and J participate in magnetosome alignment into chains. (C) MamB, E, H, M, N, O, P, T, X, Z, and FtsZm are involved in processes such as iron transport, nucleation and chemical environment control. (D) MamC, D, G, F, R, S, Mms6 and MmsF all influence the magnetic particle size and morphology. MamC, D, G and Mms6 locations correspond to the presumed locations in the magnetosome.
MamI and MamL are small, integral membrane proteins conserved within different strains and are unique to MTB.13, 32, 43 MamI was shown to be associated with the MM in AMB‐1, whereas MamL was shown to be associated mainly with the CM and less with the MM, suggesting a transient MM association of MamL.34 Deletion of mamI in MSR‐1 resulted in only a few, much smaller, nonmagnetic magnetosomes, in contrast to AMB‐1, which lacked magnetosome vesicles, suggesting it has a role also in early magnetite nucleation, perhaps by controlling the conditions for proper nucleation and growth.42 Both MamI and L are predicted to contain two transmembrane (TM) helices. The MamI connecting loop between the integral membrane helices is not predicted to bind magnetite, which further supports the proposed role of MamI in participating in MM bending.83 MamL has a basic C‐terminal tail that can bind the phospholipid heads on the inner MM, which can help in the membrane‐bending process.84
MamQ is a magnetosome‐integrated membrane protein conserved within different MTB phyla.28, 32, 34, 83 MamQ secondary structure prediction displays an integral helix followed by a C‐terminal domain (CTD), presumably located in the magnetosome lumen.83 MamQ is homologous to the LemA protein family that have no known function34, 84 and shares weak similarity to BAR (Bin/Amphiphysin/Rvs) proteins.42, 84 The BAR domain is a coiled‐coil, membrane‐bound domain that takes part in membrane deformation.42, 73, 83, 84, 85, 86 Since MamQ presumably contains a coiled‐coil domain similar to BAR proteins, it may have a role in membrane bending during magnetosome invagination.42, 84
MamY is an MTB‐specific protein.32 In AMB‐1, MamY was only present in magnetosomes containing small immature crystals (SM),73 whereas it was not identified in regular‐sized magnetite magnetosomes or in the CM.73, 74 When mamY was deleted, magnetosome vesicles were bigger but had a larger population of SM compared to wild type (WT).73 MamY is able to bind liposomes, to form tubules and to deform the membrane of both liposomes’ and magnetosomes’ lipid extract.73 MamY is predicted to contain two integral membrane helices followed by a large cytosolic CTD.83 MamY is similar to MCPs (methyl‐accepting chemotaxis proteins), BAR73, 83 and to talin, a cytoskeletal protein that links actin to the membrane.83 All of these together suggest that MamY participates in: (a) constriction of the cell membrane that leads to the invagination of the magnetosome membrane, (b) deformation and size control of the magnetosome membrane, and (c) magnetite nucleation or growth.73, 83
Protein sorting
Many of the proteins discussed in this review are uniquely found or enriched in the magnetosome membrane (containing at least one transmembrane domain [TMD]) or on the MM cytoplasmic side.6, 13, 35, 49, 87, 88 Some of these proteins, such as MamA and MamC, show dynamic localization.14, 49 Hence, an exclusive mechanism is needed to ensure that these proteins will be located in the MM or at the MM surface at the proper stage. This mechanism is not yet determined, but few proteins are thus far suspected to have a role in the protein sorting process (Fig. 2A).
MamA is one of the most conserved and abundant magnetosome‐associated proteins.13, 46, 87 In AMB‐1, MamA deletion does not abolish magnetosome formation, but not all magnetosomes are populated with magnetite.14 MamA has a dynamic localization during cell‐growth, which is independent of magnetite formation.14 MamA was shown to form in vitro globular homo‐oligomers with a central pore cavity.46 Moreover, MamA forms high‐molecular weight complexes that surround and cover the entire cytosolic side of the magnetosome membrane.47 Determined MamA structures from several species showed great structural similarity and interphyla conservation, despite the variances in their sequences.46, 89, 90 It contains five identified, and a total of putatively six, tetra‐tricopeptide repeat (TPR) motifs.89, 91, 92 Proteins with repetitive TPR motifs are known to take part in protein–protein interactions.93, 94 MamA has three proposed protein–protein interaction sites,46 two of which are assumed to participate in the oligomerization of MamA, whereas the third can bind other magnetosome‐associated proteins and by this may enable the association of MamA complexes to the magnetosome surface.46 In vivo and in vitro studies showed that MamA can bind different proteins,47, 57 which supports the assumption that the MamA homo‐oligomer surface forms a multiprotein interaction site.46
MamE is predicted to be an integral membrane protein with one TM region. MamE shares high identity with different Magnetospirillum strains53, 83 and it acts at two functionally distinct magnetosome formation steps, the protein sorting and the crystal biomineralization initiation (which will be discussed in the biomineralization section).53, 54 MamE contains a putative trypsin‐like serine protease domain, two putative PDZ domains belonging to the HtrA/DegP serine protease protein family and two putative CXXCH heme‐binding motifs.53, 54 The putative heme‐binding motifs are expected to take part in the iron redox chain, during magnetic particle biomineralization. Deletion of mamE in AMB‐1 lead to mislocalization of several proteins,34 indicating that the localization of magnetosome proteins can be accomplished through physical interaction of MamE with one or more magnetosome‐associated proteins at the MM. Furthermore, it was shown that magnetosome protein localization does not require MamE's protease acticity.54, 95
Magnetosome alignment into chains
Each magnetosome contains a crystal that is within the size of a single‐magnetic domain, which is not sufficient to sense magnetic fields. The passive alignment to a geomagnetic field is achieved by the alignment of magnetosomes into a linear chain, which creates a larger magnetic dipole moment than a single crystal.15 The current literature indicates that chain assembly formation is mediated mainly via two proteins, MamK and MamJ (Fig. 2B).
MamK is an MTB‐related protein that is homologous to the bacterial filaments‐forming actin‐like protein MreB.32, 62, 63 MamK forms long, linear filaments from one cell pole to the other, along the cell's inner curvature and aligned with the magnetosomes.62, 96, 97 In dividing cells, MamK was shown to have a role in positioning the magnetic dipole in each of the daughter cells.96 MamK polymerization into the filamentous chains is a dynamic process and is kinetically asymmetrical.58, 96 Thus, MamK is found not only next to the magnetosome chains but it is also dispersed throughout the cell.98 MamK contains an ATP‐binding site with ATPase activity, which is assumed to be related to the dissociation of filament aggregates and not directly to polymerization.58, 63, 64, 65, 96, 97 MamK's surface is largely hydrophobic or neutral, suggesting that the salts in the cell cytoplasm limit and control filament assembly.63 MamK monomers in solution assemble into two‐stranded helical filament structures, from unstaggered, parallel strands.64
In AMB‐1 ΔmamK cells, a MamK‐like protein creates filaments and magnetosomes are aligned into a chain, despite the lack of MamK65 (though not supported by another study62). MamK‐like is a protein encoded outside of the MAI region in AMB‐1 that is very similar to MamK,65 but MamK was found to have a more dominant function in magnetosome chain formation.99 In MSR‐1 ΔmamK cells, magnetosome chains were detected, but were smaller, misplaced, less organized and with less ability to self‐orient.66 MamK was suggested to serve as a track that magnetosomes can move on96, 98 and to have a role in the assembly and localization of mature magnetosomes into a chain in the midcell for some strains,58, 62, 66, 96 which was further supported by in silico analysis of magnetosome formation, assembly, and localization.100
AMB‐1 MamK was shown to interact with a few MCPs,57, 101 suggesting that magnetotaxis via MamK in AMB‐1 may relate in some way to the mechanism of chemotaxis.57 Also, flagella‐motor‐associated proteins were shown to interact with MamK, suggesting that the magnetosome torque produced by the magnetosome chain can impact on flagella rotation via the interaction with flagella motor proteins.57
MamJ is an MTB‐specific protein cotranscribed with mamK.32, 35, 59 MamJ is located in a linear structure that stretches between two ends of the cell, which extends beyond the magnetosome chain and is situated in the midcell.59 Deletion of mamJ in MSR‐1 and of mamJ together with its homologue limJ in AMB‐1 impaired magnetosome chain organization, suggesting MamJ has a role in this process.58, 60 MamJ's location is affected by the presence of other magnetosome proteins.59 MamJ was shown to physically interact with MamK in both MSR‐1 and AMB‐1,57, 60, 61 and it is assumed to be involved in the assembly of the magnetically attracted magnetosomes into chains by connecting them to the cytoskeletal structure formed by MamK.59 Supporting this, MamK was found to be associated with the MM of AMB‐1,74 possibly via the interactions with MamJ.60 Moreover, in MSR‐1, mamJ deletion does not affect the localization of MamK66 but MamK is needed for the proper localization of MamJ.59, 66 While MamK filaments are stable and not deformed under strong magnetic fields, strong magnetic fields can disturb MamJ function and/or the MamJ–MamK interaction, resulting in magnetosome alignment in the direction of the strong magnetic field.102 Additionally, MamJ and MamE were shown to interact, suggesting that this interaction anchors MamJ–MamK to the MM.57 Despite all of the above, MamJ is not conserved within all MTB, therefore, it is most likely that some other mechanisms are also involved in magnetosome alignment.98
Biomineralization
The MM, which is present before magnetic‐crystal formation, serves as a secluded compartment in the cell that provides a suitable environment for efficient crystal formation and growth into a proper mature crystal.5, 13, 14 This requires iron transfer into the magnetosome, adjustment of the chemical environment for crystal nucleation and maturation control.13, 34 Here, we describe the proteins participating in the biomineralization of magnetic nanocrystals (Fig. 2C). The proteins that are responsible for species‐specific size and shape control will be discussed in the next section.
MamM is an integral membrane protein that shares 47% sequence similarity to MamB.35 MamM was shown to be localized to the MM and to be involved in magnetite nucleation and crystal growth.48 It was recently proposed that MamM acts as an iron transporter, since it is homologous to the cation diffusion facilitator (CDF) protein family and its deletion or single point mutations have been shown to abolish magnetite biomineralization or cause alterations in magnetite crystals’ sizes and morphologies.34, 48, 67, 103 Deletion of mamM was shown to affect localization of other MM proteins, such as MamC‐GFP,104 in an elusive mechanism.34, 48 Recent structural and functional studies of MamM confirmed that its overall topology fits to the CDF protein family.67 MamM is suspected to share the TMD conserved fold, the putative metal‐binding sites and shares the CTD fold with other CDFs.67, 105 CDF proteins transport divalent transition‐metal‐cations by exploiting the proton motive force.106 Biophysical analyses of MamM demonstrated that a conformational change takes place upon binding of divalent cations to the CTD, which triggers the CTD conformation toward a compact fold that is believed to allow the activation of a two‐step ion transport mechanism through the TMD.67 This transport is essential not only for iron accumulation in the magnetosome lumen, but also for maintaining the high pH needed for magnetite/gregite biomineralization.
MamB is an integral membrane protein that is classified as a CDF protein.83, 107 Accordingly, MamB is expected to assemble as a dimer and to contain metal‐ion‐binding sites at its TMD and CTD. Additionally, deletion of mamB led to the lack of the intracellular MM,48 suggesting that MamB participates in two different magnetosome formation stages: first in membrane invagination and second in magnetosome iron accumulation.48 Of note, MamM is required for the stability of MamB in MSR‐1,48 consequently suggesting that MSR‐1 MamM and MamB interact with each other and might form heterodimers. MamB and MamM cannot functionally compensate for each other,48 supporting the hypothesis that these two proteins have different roles in the MTB.
MamO is a large integral membrane protein with eight predicted TM α‐helices that share high‐sequence identity with different Magnetospirillum strains.53, 83 MamO contains two domains: a domain of unknown function (DUF81)54 and a trypsin‐like serine protease domain.83 The MamO DUF81 domain may function as an anion transporter or as a magnetosome localization determinant via protein–protein interaction.53, 108, 109, 110 Deletion of mamO leads to empty magnetosomes with low magnetism and intracellular iron content.68 Similarly, insertion and deletion mutations within mamO were shown to be sufficient to abolish crystal biomineralization,53 whereas several point‐mutations at the trypsin‐like domain did not affect crystal formation.53, 54 Altogether, these results suggest that MamO may take part in crystal nucleation.
Iron oxidation‐reduction proteins have a key role in the magnetosome biomineralization process. Since magnetite and gregite nanocrystals’ formations require the oxidation of Fe2+ to Fe3+, it was suggested that some proteins take part in iron redox control.54, 56 The CXXCH motif, a typical c‐type cytochrome motif that is known to bind haem,111 is found in MamE, P, T, and X,54, 56 and can act in the reduction/oxidation of iron.69 The CXXCH‐containing domain in these proteins, the “magnetochrome,” seems to be specific to MTB, suggesting that it is a new, functional, unique class of cytochromes.69 Since magnetochrome‐containing proteins are the only redox proteins associated with the magnetosome, the possibility arises in which the presence of magnetochrome‐containing proteins could be related to the evolution of MTB and may be associated with magnetite/gregite crystal shape.112
Further to MamE's previously discussed role in protein sorting,53, 54 MamE plays a role in the biomineralization process. mamE deletion leads to empty magnetosome vesicles and to the loss of magnetite synthesis,34 whereas a mutation within MamE's heme‐binding site did not abolish biomineralization of mature magnetite‐crystals. These results indicate that MamE may act as a molecular switch to initiate crystal biomineralization. Biomineralization can be mediated via interaction with MamO or other proteins,53, 110 to create a network of redox activity in the center of the process.54, 56 Supporting this hypothesis, AMB‐1 MamE's magnetochrome domain was identified as a cytochrome c‐like domain.70 Since some serine proteases were shown to be capable of precipitating metal oxides, and since MamO and MamE contain this domain, a possibility arose in which MamO and MamE could play a direct role in the formation of iron‐oxide crystals.113
Additionally, MamE may have a role in the maturation of small 20 nm crystals. Once a magnetite crystal reaches the 20‐nm transition point, degradation of biomineralization inhibitors or proteolytical activation by MamE is essential for further crystal growth.54
MamP is an integral membrane protein with one predicted TM helix, two c‐type cytochrome motifs and a single PDZ domain.69, 70, 83 The structure for the soluble portion of MamP from magnetotactic ovoidal bacterium MO‐1 was determined, enabling the description of the magnetochrome domain structure.69 This structure confirms that the magnetochrome domain defines a single heme‐binding domain belonging to a new family of c‐type cytochrome; it folds as one of the smallest heme‐binding units known thus far.69 In vitro studies found that MamP can oxidize Fe2SO4 at alkaline pH efficiently, similarly to Fe2+ oxidation by the multihaem cytochrome c MtoA protein.69, 114 The optimal pH of MamP iron oxidase activity coincides with that found for the redox potential70 and for in vitro ferrihydrite and magnetite synthesis.69, 115 The combination of these data clearly demonstrates the identity of the magnetochrome domain as a cytochrome c‐like domain.70 Deletion of mamP resulted in smaller and irregular‐shaped crystals (suspected to be haematite) than the WT,34, 42 suggesting that MamP could play a role in controlling crystal size and number.34, 71 MamP was also proposed to optimize the stoichiometry of Fe2+ and Fe3+ so that the magnetite nanoparticle can be grown without defects,72 therefore, MamP was suggested to mediate the production of ferrihydrite or magnetite.69, 116 Beyond these roles, MamP may take part in the MTB cell‐cycle, since it was shown that its cell content is temporally regulated throughout the growth cycle and increased during the exponential growth phase.71
MamT is an integral membrane protein with a predicted N‐terminal TM helix.83 Furthermore, secondary structure prediction shows a double‐magnetochrome motif assumed to be located in the magnetosome lumen.70, 83 mamT deletion resulted in smaller particles compared to the WT34, 42. These phenotypes and the presence of magnetochrome motifs suggest that MamT has a role in magnetite crystal growth34, 72 and the electron redox chain.70, 72
MamX structure prediction is indicative for a TM helix at the protein's N‐terminus, a DNA‐binding domain at its CTD and two magnetochrome domains.45, 56, 117, 118 Deletion of either full‐length mamX or the substitution of its paired CXXCH motifs impaired magnetite biomineralization.56 Additionally, irregular small particles with no magnetic response and low‐iron content were observed in ΔmamX cells.45 MamX, thus, is likely involved in crystal maturation and shape control45 and in redox control needed for the synthesis of the mixed‐valence iron oxide Fe3O4 under oxidant‐limiting conditions.
MamH and MamZ (MamH‐like32) share high identity to the major facilitator superfamily (MFS) domain,43, 56 suggesting that these two proteins are involved in magnetosomal iron transport.48, 56 The MamH structural model supports this hypothesis—the model displays a negative cavity that can bind positive ions and transfer them through the magnetosome membrane.56, 83 Deletion of mamH leads to a decrease in magnetic response, suggesting it is also involved in magnetite biomineralization.34
MamZ contains a conserved ferric reductase TM component of the YedZ‐type that is assumed to bind heme B, therefore, MamZ might be involved in electron shuttling and redox reactions.119 In addition to the putative ferric reductase domain, MamZ contains an MFS transporter domain and represents the only known example in which this domain is fused to a ferric reductase domain.56, 120, 121 Based on this unique domain combination, MamZ was hypothesized to be an iron transporter120, 121 or to mediate ferric iron transport.56 Deletion of only the ferric reductase domain abolished the protein function.56
Double deletion of mamZ and mamH had larger effects than each individual deletion. Therefore, it was assumed that both proteins have partially redundant functions, and that the presence of at least one protein is necessary for the synthesis of regular magnetite crystals.56 However, it can be assumed that their functions are distinct from the functions of MamM and MamB.48, 56
MamN is a TM protein that is expected to share homology with Na+/H+ antiporters and to form a dimer, suggesting it has a role in increasing the pH within the magnetosome, a required condition for in vitro magnetite synthesis.13, 83 Deletion of mamN in AMB‐1 resulted in empty magnetosomes, which is also reflected in the absence of a magnetic response,34 whereas in MSR‐1 it resulted in half‐sized crystals.42 These findings suggest that MamN may be involved in pH regulation, by exporting the protons that are released during the magnetite precipitation.42
FtsZ are structural, bacterial tubulin‐like cell‐division proteins that assembles into ring‐like structures in a GTP‐dependent manner in the dividing cell septum.122, 123 In at least three Magnetospirillum species, there is a second copy of a conserved ftsZ‐like gene located in the mamXY operon32, 75 that encodes FtsZm, a truncated C‐terminal FtsZ protein.75, 76, 83 FtsZm's structural model consists of a GTP binding site in the N‐terminal domain (NTD) and a CTD, that is essential for FtsZ polymerization and interactions with other proteins.76, 83 In agreement with the model, FtsZm was shown to have both ATPase and GTPase activity.75, 76 Due to the gene's location in the MAI, the FtsZ‐like protein is suspected to be involved in magnetosome chain‐assembly with other actin‐like proteins or to have a role in the asymmetrical cell division in Magnetospirillum species.41, 76 In MSR‐1, FtsZm and FtsZ do not create filaments but instead a large number of spots at midcell.76 In the absence of FtsZm, magnetosome chains contained mature crystals in the chain center and flake crystals at the ends of the chain.76 Deletion of ftsZm had no significant impact on cell growth, which suggests that it does not have the cell‐division function of other bacterial FtsZ proteins.75, 76 Conversely, overexpression of FtsZm interfered in cell division similarly to other FtsZ proteins.76 Studies showed that ftsZm deletion caused changes in magnetite size and morphology only when cells grew in the presence of ammonia instead of nitrate.75, 76 This suggests FtsZm has a role in denitrification, redox control, and iron uptake.76
MamS is an MTB‐specific protein32, 35 that shares similarity to the putative serine‐protease domain of MamE and MamX in MSR‐1.42 In AMB‐1, and likewise in MSR‐1, the crystals of ΔmamS cells were small, mainly amorphous and with weak magnetic response, creating small clusters within the chain with irregular spacing.34, 42 This suggests that MamS has a function in the regulation of magnetosome size and morphology.34
MamR is an MTB‐specific protein that is predicted to be localized to the cytoplasmic side of the MM.32, 35 Deletion of mamR, together with its duplicated gene in AMB‐1, resulted in shorter magnetosome chains, smaller particles, and a weak magnetic response, which suggest that MamR plays a role in the control of the numbers and sizes of magnetosomes.34 MSR‐1 ΔmamR cells presented similar magnetic responses and magnetosome numbers to WT cells. In contrast, other phenotypes, such as magnetosome size and chain formation modifications, were similar to those of AMB‐1 ΔmamR cells, suggesting the same function for MamR as in AMB‐1.42
Crystals’ size and morphology
The last part that will be discussed here is the formation of functional magnetosomes. There is a large set of proteins that have a role in controlling the magnetite size and morphology, but are not essential for biomineralization (Fig. 2D). For example, in Alphaproteobacteria strains, there are at least six MTB‐specific proteins encoded by the mms6 and mamGFDC operons that have this role.32, 34, 35, 42, 43, 44, 50, 80 Deletion of both operons separately42, 43, 44 resulted in smaller magnetite crystals than WT, while their simultaneous deletion resulted in stronger phenotypes compared to each deletion alone.34, 43 In MSR‐1 ΔmamGFDC cells, magnetite crystals were 75% the size of WT crystals. Only complementation of three out of four of the proteins in any combination produced crystals of the size of the WT, suggesting these four proteins have a cumulative action in the regulation of crystal size.44 In AMB‐1, MamC, D and G (named Mms13, 7, and 5, respectively, in this strain) and Mms6 were shown to be tightly bound to magnetite.51 These genes are absent in all strains studied so far that synthesize bullet‐shaped magnetite, therefore, it is suggested that MamC, D, G, and Mms6 might have a role in the size control of octahedral‐shaped crystals.50 Moreover, these proteins are assumed to colocalize in the MM and to interact with the magnetite surface.50, 77 Mms6, MamD, and G all contain a hydrophobic leucine–glycine repeat region that is also found in other biomineralization proteins.35, 51, 83, 124
MamC (Mms13) is the most abundant protein found uniquely in the MM.35, 49, 51, 87, 125, 126 Deletion of mamC in MSR‐1 and AMB‐1 had only a small impact on magnetite size.44, 50, 127 MamC is predicted to contain two integral TM helices that are connected by an acidic alpha‐helical loop in the magnetosome lumen52, 83 that is suspected to be the magnetite binding site.128 MamC is proposed to influence magnetite formation via a specific mechanism: the high acidic region in the loop between the TM helices binds iron, which increases the local iron concentration and creates a favorable environment for magnetite nucleation.52 In vitro coprecipitation of magnetite with MamC resulted in magnetite particles that possessed missing corners, suggesting that the protein is bound to specific magnetite faces, preventing the crystals from growing in these directions.52
MamD (Mms7) is an abundant protein in the MM.32, 33, 44, 87 AMB‐1 MamD is composed of a hydrophobic NTD and hydrophilic CTD.51, 87, 124 The NTD integrates the protein into the magnetosome membrane and the CTD—which is located in the magnetosome lumen and contains acidic amino acids—is suspect to interact with the magnetite surface.51 In AMB‐1 ΔmamD cells, crystals had decreased size in the minor axis, and the crystal face was different from those in WT cells, which further supports MamD's role in controlling crystal morphology.50
MamG (Mms5) is an abundant protein, homologous to MamD in MSR‐1 and Mms6 and MamD in AMB‐1.33, 35, 51 AMB‐1 mamG deletion leads to the formation of smaller, spherical crystals, suggesting that MamG has a role in crystal growth via interaction with a specific crystal face.50 MamG is exclusively located in the magnetosome membrane along the chain.49 Secondary structure prediction suggest that MamG contain two integral TM helices and a charged, unstructured C‐terminal that faces the cytosol,83 which suggests that MamG interacts with the magnetite crystal via a connecting loop between the two TM helices.
MamF is the second most abundant MM protein.32, 35 In MSR‐1, MamF forms stable oligomers even in the presence of sodium dodecyl phosphate (SDS).35 MamF is encoded by the mamGFDC operon and is similar to MmsF (61% identity).32, 42 In MSR‐1, mamF deletion caused no change in crystal size or number, but its codeletion with mmsF increased the effect of mmsF deletion phenotypes, that is, a decrease in magnetosome numbers and sizes. This suggests a role for MamF in magnetosome size and number control.42 MamF is located in the MM exclusively49 and is predicted to contain three TM helices.44, 55, 83 The loop between the first two helices is rich in charged residues and is suspected to be located in the magnetosome lumen, hence it may interact with the magnetite crystal.83
MmsF is a protein encoded by the mms6 operon, in which its encoding gene deletion in MSR‐1 and AMB‐1 caused similar phenotypes of smaller crystals and lower magnetic responses.42, 80 MmsF participates in controlling the size and shape of magnetite, and was suggested to have a role in the control of crystal maturation after the nucleation stage.80 MmsF in AMB‐1 is predicted to be composed of three TM helices with a cytoplasmic N‐terminal and a C‐terminal in the magnetosome lumen. The C‐terminal and the loop between helices one and two, which are rich in acidic amino acids, are assumed to bind the magnetite crystal.55, 80 In vitro purification of MmsF without detergents led to oligomers or aggregates in the cell lysate soluble fraction.55 MmsF in solution was found to create artificial, doughnut‐shaped assemblies, named “proteinosomes,” that are probably high‐ordered aggregates. These proteinosomes coprecipitate iron in vitro into magnetic particles similar to those of AMB‐1. MmsF has an aspartate residue in the loop between helices one and two that is suspected to be a magnetite binder and has the same motif as Mms6 that is known to bind magnetite.55
Mms6 is a protein suspected to undergo proteolytic cleavage from its pro‐protein to create a shorter, C‐terminal, functional protein.35, 51 Not only the full protein but also the Mms6 C‐terminal peptide was shown to be tightly bound to magnetite and affect its size and shape in vitro.78 Mms6 NTD is suspected to have a random coil structure with a TM helix, while the C‐terminal is acidic and is suggested to form an alpha helix with a negative surface.51, 79, 83 The predicted helical CTD is suspected to bind the magnetite particles.51, 79, 83 Full‐Mms6 and C‐terminal‐Mms6 were shown to bind iron ions51, 79, 129 and to create homogenous‐sized and regular‐shaped magnetite particles in vitro.51, 79 These all occurred only if the C‐terminal was in its native form that contained the acidic amino acids,78, 129 thus suggesting that the CTD is responsible for iron binding. A negative surface charge composed of the CTD acidic amino acids enables iron nucleation at the Mms6 surface, and in turn specific magnetite face interaction with Mms6 controls the magnetite size and shape.79, 129, 130, 131, 132 Deletion of mms6 from AMB‐1 and MSR‐1 resulted in smaller crystals and in AMB‐1 also in different morphology compared to the WTs,42, 50, 77, 80 supporting the proposed role of Mms6 in magnetite maturation and orientation during magnetite crystal growth.77
From the different morphology phenotypes that were obtained in MamC, D, G, and Mms6 deletion studies in AMB‐1, the proteins’ locations were suggested. In a linear chain of n magnetosomes, MamC and G are mainly found at the location inside magnetosome k proximal to magnetosome k+1. In contrast, MamD and Mms6 are still within the magnetosome but are presumed to be located within the vesicle such that they face away from the magnetosome chain.50
The proteins discussed in this section are not conserved within all MTB branches.25 The mad group of 30 genes, which can be found in Deltaproteobacteria strains,40, 133 Nitrospirae phylum strains134, 135 and in Omnitrophica phylum strain,28 can be divided into three groups: magnetite biomineralization‐related genes, gregite magnetosome formation‐related genes and a third, unclassified group that can be found in all the Deltaproteobacteria strains that were studied.40 Most of the Mad proteins share no homology to any other proteins, and some were suggested to have a putative role based on their homology to other proteins, such as in iron uptake and in magnetosomes alignment into chains. Since these genes are unique to gregite and bullet‐shaped magnetite crystal‐synthesizing strains, they might have a role in size and morphology control specifically for these crystal types, which is analogous to the mamGFDC and mms genes in Alphaproteobacteria that are not found in those species.40
Concluding remarks
Here, we presented briefly the current known data about the main proteins involved in magnetosome formation. Yet, more data is needed to clarify this unique process. The revealing of magnetosome formation mechanisms in general, together with specific studies of relevant proteins, has an impact on many scientific fields that were not discussed here, such as nanotechnology, medicine and ecology. This makes magnetosome studies a “hot topic” that interests many groups around the world who continuously study different aspects of magnetosome‐related processes.
Acknowledgments
The authors thank Dr. Dirk Schüler and Dr. René Uebe for their permission to use their TEM image, to architect Nir Shmuely for his help in figures design and to Mr. Samuel Cronin for his help with the manuscript.
References
- 1. Bazylinski DA, Frankel RB (2004) Magnetosome formation in prokaryotes. Nat Rev Microbiol 2:217–230. [DOI] [PubMed] [Google Scholar]
- 2. Bellini S (2009) On a unique behavior of freshwater bacteria. Chin J Oceanol Limnol 27:3–5. [Google Scholar]
- 3. Bellini S (2009) Further studies on “magnetosensitive bacteria.” Chin J Oceanol Limnol 27:6–12. [Google Scholar]
- 4. Blakemore R (1975) Magnetotactic bacteria. Science 190:377–379. [DOI] [PubMed] [Google Scholar]
- 5. Faivre D, Schüler D (2008) Magnetotactic bacteria and magnetosomes. Chem Rev 108:4875–4898. [DOI] [PubMed] [Google Scholar]
- 6. Gorby YA, Beveridge TJ, Blakemore RP (1988) Characterization of the bacterial magnetosome membrane. J Bacteriol 170:834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balkwill DL, Maratea D, Blakemore RP (1980) Ultrastructure of a magnetotactic spirillum. J Bacteriol 141:1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frankel RB, Blakemore RP, Wolfe RS (1979) Magnetite in freshwater magnetotactic bacteria. Science 203:1355–1356. [DOI] [PubMed] [Google Scholar]
- 9. Mann S, Sparks NHC, Frankel RB, Bazylinski DA, Jannasch HW (1990) Biomineralization of ferrimagnetic greigite (Fe3S4) and iron pyrite (FeS2) in a magnetotactic bacterium. Nature 343:258–261. [Google Scholar]
- 10. Bazylinski DA, Garratt‐Reed AJ, Frankel RB (1994) Electron microscopic studies of magnetosomes in magnetotactic bacteria. Microsc Res Tech 27:389–401. [DOI] [PubMed] [Google Scholar]
- 11. Sparks NHC, Courtaux L, Mann S, Board RG (1986) Magnetotactic bacteria are widely distributed in sediments in the U.K. FEMS Microbiol Lett 37:305–308. [Google Scholar]
- 12. Frankel RB, Dunin‐Borkowski RE, Pósfai M, Bazylinski DA, Magnetic Microstructure of Magnetotactic Bacteria In: Baeuerlein E, Ed. (2008) Handbook of Biomineralization: Biological Aspects and Structure Formation. Vol. 1 Wiley‐VCH Verlag GmbH & Co. KGaA, pp 126–144. [Google Scholar]
- 13. Komeili A (2012) Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol Rev 36:232–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Komeili A, Vali H, Beveridge TJ, Newman DK (2004) Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci USA 101:3839–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frankel RB, Bazylinski DA (2006) How magnetotactic bacteria make magnetosomes queue up. Trends Microbiol 14:329–331. [DOI] [PubMed] [Google Scholar]
- 16. Frankel RB, Williams TJ, Bazylinski DA, Magneto‐Aerotaxis In: Schüler D, Ed. (2007) Magnetoreception and Magnetosomes in Bacteria. Berlin: Springer‐Verlag, pp 1–24. [Google Scholar]
- 17. Blakemore RP, Frankel RB, Kalmijn AJ (1980) South‐seeking magnetotactic bacteria in the Southern Hemisphere. Nature 286:384–385. [Google Scholar]
- 18. Frankel RB, Bazylinski DA, Johnson MS, Taylor BL (1997) Magneto‐aerotaxis in marine coccoid bacteria. Biophys J 73:994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Popp F, Armitage JP, Schüler D (2014) Polarity of bacterial magnetotaxis is controlled by aerotaxis through a common sensory pathway. Nat Commun 5:5398 [DOI] [PubMed] [Google Scholar]
- 20. Lefèvre CT, Bennet M, Landau L, Vach P, Pignol D, Bazylinski DA, Frankel RB, Klumpp S, Faivre D (2014) Diversity of magneto‐aerotactic behaviors and oxygen sensing mechanisms in cultured magnetotactic bacteria. Biophys J 107:527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen C, Ma Q, Jiang W, Song T (2011) Phototaxis in the magnetotactic bacterium Magnetospirillum magneticum strain AMB‐1 is independent of magnetic fields. Appl Microbiol Biotechnol 90:269–275. [DOI] [PubMed] [Google Scholar]
- 22. Shapiro OH, Hatzenpichler R, Buckley DH, Zinder SH, Orphan VJ (2011) Multicellular photo‐magnetotactic bacteria. Environ Microbiol Rep 3:233–238. [DOI] [PubMed] [Google Scholar]
- 23. Matsunaga T, Okamura Y, Fukuda Y, Wahyudi AT, Murase Y, Takeyama H (2005) Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB‐1. DNA Res 12:157–166. [DOI] [PubMed] [Google Scholar]
- 24. Alexandre G, Greer‐Phillips S, Zhulin IB (2004) Ecological role of energy taxis in microorganisms. FEMS Microbiol Rev 28:113–126. [DOI] [PubMed] [Google Scholar]
- 25. Lefèvre CT, Bazylinski DA (2013) Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol Mol Biol Rev 77:497–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amann R, Peplies J, Schüler D, Diversity and taxonomy of magnetotactic bacteria In: Schüler D, Ed. (2007) Magnetoreception and Magnetosomes in Bacteria. Berlin: Springer‐Verlag, pp 25–36. [Google Scholar]
- 27. Kolinko S, Jogler C, Katzmann E, Wanner G, Peplies J, Schüler D (2012) Single‐cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ Microbiol 14:1709–1721. [DOI] [PubMed] [Google Scholar]
- 28. Kolinko S, Richter M, Glöckner FO, Brachmann A, Schüler D (2015) Single‐cell genomics of uncultivated deep‐branching magnetotactic bacteria reveals a conserved set of magnetosome genes. Environ Microbiol, in press. PMID: 26060021 [PubMed]. [DOI] [PubMed] [Google Scholar]
- 29. Morillo V, Abreu F, Araujo AC, De Almeida LGP, Enrich‐Prast A, Farina M, De Vasconcelos ATR, Bazylinski DA, Lins U (2014) Isolation, cultivation and genomic analysis of magnetosome biomineralization genes of a new genus of South‐seeking magnetotactic cocci within the Alphaproteobacteria. Front Microbiol 5:72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sakaguchi T, Arakaki A, Matsunaga T (2002) Desulfovibrio magneticus sp. nov., a novel sulfate‐reducing bacterium that produces intracellular single‐domain‐sized magnetite particles. Int J Syst Evol Microbiol 52:215–221. [DOI] [PubMed] [Google Scholar]
- 31. Lefèvre CT, Viloria N, Schmidt ML, Pósfai M, Frankel RB, Bazylinski DA (2012) Novel magnetite‐producing magnetotactic bacteria belonging to the Gammaproteobacteria. Isme J 6:440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richter M, Kube M, Bazylinski DA, Lombardot T, Glöckner FO, Reinhardt R, Schüler D (2007) Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group‐specific genes implicated in magnetosome biomineralization and function. J Bacteriol 189:4899–4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schübbe S, Kube M, Scheffel A, Wawer C, Heyen U, Meyerdierks A, Madkour MH, Mayer F, Reinhardt R, Schüler D (2003) Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J Bacteriol 185:5779–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murat D, Quinlan A, Vali H, Komeili A (2010) Comprehensive genetic dissection of the magnetosome gene island reveals the step‐wise assembly of a prokaryotic organelle. Proc Natl Acad Sci USA 107:5593–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grünberg K, Müller EC, Otto A, Reszka R, Linder D, Kube M, Reinhardt R, Schüler D (2004) Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 70:1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fukuda Y, Okamura Y, Takeyama H, Matsunaga T (2006) Dynamic analysis of a genomic island in Magnetospirillum sp. strain AMB‐1 reveals how magnetosome synthesis developed. FEBS Lett 580:801–812. [DOI] [PubMed] [Google Scholar]
- 37. Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D (2005) A hypervariable 130‐kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol 187:7176–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schübbe S, Würdemann C, Peplies J, Heyen U, Wawer C, Glöckner FO, Schüler D (2006) Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 72:5757–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jogler C, Lin W, Meyerdierks A, Kube M, Katzmann E, Flies C, Pan Y, Amann R, Reinhardt R, Schüler D (2009) Toward cloning of the magnetotactic metagenome: Identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from different aquatic sediments. Appl Environ Microbiol 75:3972–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lefèvre CT, Trubitsyn D, Abreu F, Kolinko S, Jogler C, de Almeida LGP, de Vasconcelos ATR, Kube M, Reinhardt R, Lins U, Pignol D, Schüler D, Bazylinski DA, Ginet N (2013) Comparative genomic analysis of magnetotactic bacteria from the Deltaproteobacteria provides new insights into magnetite and greigite magnetosome genes required for magnetotaxis. Environ Microbiol 15:2712–2735. [DOI] [PubMed] [Google Scholar]
- 41. Jogler C, Schuler D, Schüler D (2009) Genomics, genetics, and cell biology of magnetosome formation. Annu Rev Microbiol 63:501–521. [DOI] [PubMed] [Google Scholar]
- 42. Lohße A, Borg S, Raschdorf O, Kolinko I, Tompa É, Pósfai M, Faivre D, Baumgartner J, Schülera D (2014) Genetic dissection of the mamAB and mms6 operons reveals a gene set essential for magnetosome biogenesis in magnetospirillum gryphiswaldense. J Bacteriol 196:2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lohße A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schüler D (2011) Functional analysis of the magnetosome Island in magnetospirillum gryphiswaldense: The mamAB operon is sufficient for magnetite biomineralization. PLoS One 6:e25561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scheffel A, Gärdes A, Grünberg K, Wanner G, Schüler D (2008) The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J Bacteriol 190:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang J, Li S, Huang X, Li J, Li L, Pan Y, Li Y (2013) MamX encoded by the mamXY operon is involved in control of magnetosome maturation in Magnetospirillum gryphiswaldense MSR‐1. BMC Microbiol 13:203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zeytuni N, Ozyamak E, Ben‐Harush K, Davidov G, Levin M, Gat Y, Moyal T, Brik A, Komeili A, Zarivach R (2011) Self‐recognition mechanism of MamA, a magnetosome‐associated TPR‐containing protein, promotes complex assembly. Proc Natl Acad Sci USA 108:E480–E487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamamoto D, Taoka A, Uchihashi T, Sasaki H, Watanabe H, Ando T, Fukumori Y (2010) Visualization and structural analysis of the bacterial magnetic organelle magnetosome using atomic force microscopy. Proc Natl Acad Sci USA 107:9382–9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Uebe R, Junge K, Henn V, Poxleitner G, Katzmann E, Plitzko JM, Zarivach R, Kasama T, Wanner G, Pósfai M, Böttger L, Matzanke B, Schüler D (2011) The cation diffusion facilitator proteins MamB and MamM of Magnetospirillum gryphiswaldense have distinct and complex functions, and are involved in magnetite biomineralization and magnetosome membrane assembly. Mol Microbiol 82:818–835. [DOI] [PubMed] [Google Scholar]
- 49. Lang C, Schüler D (2008) Expression of green fluorescent protein fused to magnetosome proteins in microaerophilic magnetotactic bacteria. Appl Environ Microbiol 74:4944–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arakaki A, Yamagishi A, Fukuyo A, Tanaka M, Matsunaga T (2014) Co‐ordinated functions of Mms proteins define the surface structure of cubo‐octahedral magnetite crystals in magnetotactic bacteria. Mol Microbiol 93:554–567. [DOI] [PubMed] [Google Scholar]
- 51. Arakaki A, Webb J, Matsunaga T (2003) A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB‐1. J Biol Chem 278:8745–8750. [DOI] [PubMed] [Google Scholar]
- 52. Valverde‐Tercedor C, Montalbán‐López M, Perez‐Gonzalez T, Sanchez‐Quesada MS, Prozorov T, Pineda‐Molina E, Fernandez‐Vivas MA, Rodriguez‐Navarro AB, Trubitsyn D, Bazylinski DA, Jimenez‐Lopez C (2015) Size control of in vitro synthesized magnetite crystals by the MamC protein of Magnetococcus marinus strain MC‐1. Appl Microbiol Biotechnol 99:5109–5121. [DOI] [PubMed] [Google Scholar]
- 53. Yang W, Li R, Peng T, Zhang Y, Jiang W, Li Y, Li J (2010) MamO and mamE genes are essential for magnetosome crystal biomineralization in Magnetospirillum gryphiswaldense MSR‐1. Res Microbiol 161:701–705. [DOI] [PubMed] [Google Scholar]
- 54. Quinlan A, Murat D, Vali H, Komeili A (2011) The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol Microbiol 80:1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rawlings AE, Bramble JP, Walker R, Bain J, Galloway JM, Staniland SS (2014) Self‐assembled MmsF proteinosomes control magnetite nanoparticle formation in vitro. Proc Natl Acad Sci USA 111:16094–16099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Raschdorf O, Müller FD, Pósfai M, Plitzko JM, Schüler D (2013) The magnetosome proteins MamX, MamZ and MamH are involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. Mol Microbiol 89:872–886. [DOI] [PubMed] [Google Scholar]
- 57. Pan W, Xie C, Lv J (2012) Screening for the interacting partners of the proteins MamK & MamJ by two‐hybrid genomic DNA library of magnetospirillum magneticum AMB‐1. Curr Microbiol 64:515–523. [DOI] [PubMed] [Google Scholar]
- 58. Draper O, Byrne ME, Li Z, Keyhani S, Barrozo JC, Jensen G, Komeili A (2011) MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol Microbiol 82:342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schüler D (2006) An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110–114. [DOI] [PubMed] [Google Scholar]
- 60. Scheffel A, Schüler D (2007) The acidic repetitive domain of the magnetospirillum gryphiswaldense MamJ protein displays hypervariability but is not required for magnetosome chain assembly. J Bacteriol 189:6437–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carillo MA, Bennet M, Faivre D (2013) Interaction of proteins associated with the magnetosome assembly in magnetotactic bacteria as revealed by two‐hybrid two‐photon excitation fluorescence lifetime imaging microscopy Förster resonance energy transfer. J Phys Chem B 117:14642–14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Komeili A, Li Z, Newman DK, Jensen GJ (2006) Magnetosomes are cell membrane invaginations organized by the actin‐like protein MamK. Science 311:242–245. [DOI] [PubMed] [Google Scholar]
- 63. Sonkaria S, Fuentes G, Verma C, Narang R, Khare V, Fischer A, Faivre D (2012) Insight into the assembly properties and functional organisation of the magnetotactic bacterial actin‐like homolog, MamK. PLoS One 7:e34189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ozyamak E, Kollman J, Agard Da, Komeili A (2013) The bacterial actin MamK: In vitro assembly behavior and filament architecture. J Biol Chem 288:4265–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rioux JB, Philippe N, Pereira S, Pignol D, Wu LF, Ginet N (2010) A second actin‐like mamk protein in magnetospirillum magneticum AMB‐1 encoded outside the genomic magnetosome island. PLoS One 5:e9151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schüler D (2010) Loss of the actin‐like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol Microbiol 77:208–224. [DOI] [PubMed] [Google Scholar]
- 67. Zeytuni N, Uebe R, Maes M, Davidov G, Baram M, Raschdorf O, Nadav‐Tsubery M, Kolusheva S, Bitton R, Goobes G, Friedler A, Miller Y, Schüler D, Zarivach R (2014) Cation diffusion facilitators transport initiation and regulation is mediated by cation induced conformational changes of the cytoplasmic domain. PLoS One 9:e92141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guo FF, Yang W, Jiang W, Geng S, Peng T, Li JL (2012) Magnetosomes eliminate intracellular reactive oxygen species in Magnetospirillum gryphiswaldense MSR‐1. Environ Microbiol 14:1722–1729. [DOI] [PubMed] [Google Scholar]
- 69. Siponen MI, Legrand P, Widdrat M, Jones SR, Zhang W‐J, Chang MCY, Faivre D, Arnoux P, Pignol D (2013) Structural insight into magnetochrome‐mediated magnetite biomineralization. Nature 502:681–684. [DOI] [PubMed] [Google Scholar]
- 70. Siponen MI, Adryanczyk G, Ginet N, Arnoux P, Pignol D (2012) Magnetochrome: a c‐type cytochrome domain specific to magnetotatic bacteria. Biochem Soc Trans 40:1319–1323. [DOI] [PubMed] [Google Scholar]
- 71. Taoka A, Eguchi Y, Mise S, Oestreicher Z, Uno F, Fukumori Y (2014) A magnetosome‐associated cytochrome MamP is critical for magnetite crystal growth during the exponential growth phase. FEMS Microbiol Lett 358:21–29. [DOI] [PubMed] [Google Scholar]
- 72. Jones SR, Wilson TD, Brown ME, Rahn‐Lee L, Yu Y, Fredriksen LL, Ozyamak E, Komeili A, Chang MCY (2015) Genetic and biochemical investigations of the role of MamP in redox control of iron biomineralization in Magnetospirillum magneticum. Proc Natl Acad Sci USA 112:3904–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tanaka M, Arakaki A, Matsunaga T (2010) Identification and functional characterization of liposome tubulation protein from magnetotactic bacteria. Mol Microbiol 76:480–488. [DOI] [PubMed] [Google Scholar]
- 74. Tanaka M, Okamura Y, Arakaki A, Tanaka T, Takeyama H, Matsunaga T (2006) Origin of magnetosome membrane: Proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics 6:5234–5247. [DOI] [PubMed] [Google Scholar]
- 75. Ding Y, Li J, Liu J, Yang J, Jiang W, Tian J, Li Y, Pan Y, Li J (2010) Deletion of the ftsZ‐like gene results in the production of superparamagnetic magnetite magnetosomes in Magnetospirillum gryphiswaldense . J Bacteriol 192:1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Müller FD, Raschdorf O, Nudelman H, Messerer M, Katzmann E, Plitzko JM, Zarivach R, Schüler D (2014) The FtsZ‐like protein FtsZm of Magnetospirillum gryphiswaldense likely interacts with its generic homolog and is required for biomineralization under nitrate deprivation. J Bacteriol 196:650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tanaka M, Mazuyama E, Arakaki A, Matsunaga T (2011) MMS6 protein regulates crystal morphology during nano‐sized magnetite biomineralization in vivo. J Biol Chem 286:6386–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Arakaki A, Masuda F, Amemiya Y, Tanaka T, Matsunaga T (2010) Control of the morphology and size of magnetite particles with peptides mimicking the Mms6 protein from magnetotactic bacteria. J Colloid Interface Sci 343:65–70. [DOI] [PubMed] [Google Scholar]
- 79. Amemiya Y, Arakaki A, Staniland SS, Tanaka T, Matsunaga T (2007) Controlled formation of magnetite crystal by partial oxidation of ferrous hydroxide in the presence of recombinant magnetotactic bacterial protein Mms6. Biomaterials 28:5381–5389. [DOI] [PubMed] [Google Scholar]
- 80. Murat D, Falahati V, Bertinetti L, Csencsits R, Körnig A, Downing K, Faivre D, Komeili A (2012) The magnetosome membrane protein, MmsF, is a major regulator of magnetite biomineralization in Magnetospirillum magneticum AMB‐1. Mol Microbiol 85:684–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schleifer KH, Schuler D, Spring S, Weizenegger M, Amann R, Ludwig W, Kohler M (1991) The genus magnetospirillum Gen‐Nov—description of magnetospirillum‐gryphiswaldense Sp‐Nov and transfer of aquaspirillum‐magnetotacticum to magnetospirillum‐magnetotacticum Comb‐Nov. Syst Appl Microbiol 14:379–385. [Google Scholar]
- 82. Wang X, Wang Q, Zhang W, Wang Y, Li L, Wen T, Zhang T, Zhang Y, Xu J, Hu J, Li S, Liu L, Liu J, Jiang W, Tian J, Li Y, Schüler D, Wang LLJ (2014) Complete genome sequence of Magnetospirillum gryphiswaldense MSR‐1. Genome Announc 2:e00171–e00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nudelman H, Zarivach R (2014) Structure prediction of magnetosome‐associated proteins. Front Microbiol 5:9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Greene SE, Komeili A (2012) Biogenesis and subcellular organization of the magnetosome organelles of magnetotactic bacteria. Curr Opin Cell Biol 24:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Arkhipov A, Yin Y, Schulten K (2009) Membrane‐bending mechanism of amphiphysin N‐BAR domains. Biophys J 97:2727–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303:495–499. [DOI] [PubMed] [Google Scholar]
- 87. Grünberg K, Wawer C, Tebo BM, Schüler D (2001) A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl Environ Microbiol 67:4573–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Okamura Y, Takeyama H, Matsunaga T (2000) Two‐dimensional analysis of proteins specific to the bacterial magnetic particle membrane from Magnetospirillum sp. AMB‐1. Appl Biochem Biotechnol 8486:441–446. [DOI] [PubMed] [Google Scholar]
- 89. Zeytuni N, Baran D, Davidov G, Zarivach R (2012) Inter‐phylum structural conservation of the magnetosome‐associated TPR‐containing protein, MamA. J Struct Biol 180:479–487. [DOI] [PubMed] [Google Scholar]
- 90. Zeytuni N, Cronin S, Lefèvre CT, Arnoux P, Baran D, Shtein Z, Davidov G, Zarivach R (2015) MamA as a model protein for structure‐based insight into the evolutionary origins of magnetotactic bacteria. PLoS One 10:e0130394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Okuda Y, Fukumori Y (2001) Expression and characterization of a magnetosome‐associated protein, TPR‐containing MAM22, in Escherichia coli. FEBS Lett 491:169–173. [DOI] [PubMed] [Google Scholar]
- 92. Okuda Y, Denda K, Fukumori Y (1996) Cloning and sequencing of a gene encoding a new member of the tetratricopeptide protein family from magnetosomes of Magnetospirillum magnetotacticum. Gene 171:99–102. [DOI] [PubMed] [Google Scholar]
- 93. Blatch GL, Lässle M (1999) The tetratricopeptide repeat: A structural motif mediating protein‐protein interactions. BioEssays 21:932–939. [DOI] [PubMed] [Google Scholar]
- 94. Zeytuni N, Zarivach R (2012) Structural and functional discussion of the tetra‐trico‐peptide repeat, a protein interaction module. Structure 20:397–405. [DOI] [PubMed] [Google Scholar]
- 95. Lee H‐J, Zheng JJ (2010) PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal 8:8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Pradel N, Santini C‐L, Bernadac A, Fukumori Y, Wu L‐F (2006) Biogenesis of actin‐like bacterial cytoskeletal filaments destined for positioning prokaryotic magnetic organelles. Proc Natl Acad Sci USA 103:17485–17489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Taoka A, Asada R, Wu LF, Fukumori Y (2007) Polymerization of the actin‐like protein MamK, which is associated with magnetosomes. J Bacteriol 189:8737–8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bennet M, Bertinetti L, Neely RK, Schertel A, Körnig A, Flors C, Müller FD, Schüler D, Klumpp S, Faivre D (2015) Biologically controlled synthesis and assembly of magnetite nanoparticles. Faraday Discuss 181:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Abreu N, Mannoubi S, Ozyamak E, Pignol D, Ginet N, Komeili A (2014) Interplay between two bacterial actin homologs, MamK and MamK like, is required for the alignment of magnetosome organelles in Magnetospirillum magneticum AMB‐1. J Bacteriol 196:3111–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Klumpp S, Faivre D (2012) Interplay of magnetic interactions and active movements in the formation of magnetosome chains. PLoS One 7:e33562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Philippe N, Wu LF (2010) An MCP‐Like Protein Interacts with the MamK Cytoskeleton and Is Involved in Magnetotaxis in Magnetospirillum magneticum AMB‐1. J Mol Biol 400:309–322. [DOI] [PubMed] [Google Scholar]
- 102. Ko A, Dong J, Bennet M, Widdrat M, Andert J, Mu FD, Schu D, Klumpp S, Faivre D (2014) Probing the mechanical properties of magnetosome vhains in living magnetotactic bacteria. Nano Lett 14:4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zeytuni N, Uebe R, Maes M, Davidov G, Baram M, Raschdorf O, Friedler A, Miller Y, Schüler D, Zarivach R (2014) Bacterial magnetosome biomineralization‐a novel platform to study molecular mechanisms of human CDF‐related Type‐II diabetes. PLoS One 9:e97154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kolinko I, Lohße A, Borg S, Raschdorf O, Jogler C, Tu Q, Pósfai M, Tompa E, Plitzko JM, Brachmann A, Wanner G, Müller R, Zhang Y, Schüler D (2014) Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nat Nanotechnol 9:193–197. [DOI] [PubMed] [Google Scholar]
- 105. Zeytuni N, Offer T, Davidov G, Zarivach R (2012) Crystallization and preliminary crystallographic analysis of the C‐terminal domain of MamM, a magnetosome‐associated protein from Magnetospirillum gryphiswaldense MSR‐1. Acta Cryst F 68:927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M (2007) Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 8:107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Grass G, Otto M, Fricke B, Haney CJ, Rensing C, Nies DH, Munkelt D (2005) FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Arch Microbiol 183:9–18. [DOI] [PubMed] [Google Scholar]
- 108. Weinitschke S, Denger K, Cook AM, Smits THM (2007) The DUF81 protein TauE in Cupriavidus necator H16, a sulfite exporter in the metabolism of C2 sulfonates. Microbiology 153:3055–3060. [DOI] [PubMed] [Google Scholar]
- 109. Mampel J, Maier E, Tralau T, Ruff J, Benz R, Cook AM (2004) A novel outer‐membrane anion channel (porin) as part of a putatively two‐component transport system for 4‐toluenesulphonate in Comamonas testosteroni T‐2. Biochem J 383:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Krojer T, Pangerl K, Kurt J, Sawa J, Stingl C, Mechtler K, Huber R, Ehrmann M, Clausen T (2008) Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc Natl Acad Sci USA 105:7702–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bowman SEJ, Bren KL (2008) The chemistry and biochemistry of heme c: functional bases for covalent attachment. Nat Prod Rep 25:1118–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Arnoux P, Siponen MI, Lefèvre CT, Ginet N, Pignol D (2014) Structure and evolution of the magnetochrome domains: No longer alone. Front Microbiol 5:117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Smith GP, Baustian KJ, Ackerson CJ, Feldheim DL (2009) Metal oxide formation by serine and cysteine proteases. J Mater Chem 19:8299–8306. [Google Scholar]
- 114. Liu J, Wang Z, Belchik SM, Edwards MJ, Liu C, Kennedy DW, Merkley ED, Lipton MS, Butt JN, Richardson DJ, Zachara JM, Fredrickson JK, Rosso KM, Shi L (2012) Identification and characterization of M to A: A decaheme c‐type cytochrome of the neutrophilic Fe(ll)‐oxidizing bacterium Sideroxydans lithotrophicus ES‐1. Front Microbiol 3:37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Baumgartner J, Bertinetti L, Widdrat M, Hirt AM, Faivre D (2013) Formation of magnetite nanoparticles at low temperature: From duperparamagnetic to stable single domain particles. PLoS One 8:e57070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Baumgartner J, Dey A, Bomans PHH, Le Coadou C, Fratzl P, Sommerdijk NaJM, Faivre D (2013) Nucleation and growth of magnetite from solution. Nat Mater 12:310–314. [DOI] [PubMed] [Google Scholar]
- 117. Söding J (2005) Protein homology detection by HMM‐HMM comparison. Bioinformatics 21:951–960. [DOI] [PubMed] [Google Scholar]
- 118. Slabinski L, Jaroszewski L, Rychlewski L, Wilson IA, Lesley SA, Godzik A (2007) XtalPred: A web server for prediction of protein crystallizability. Bioinformatics 23:3403–3405. [DOI] [PubMed] [Google Scholar]
- 119. Brokx SJ, Rothery Ra, Zhang G, Ng DP, Weiner JH (2005) Characterization of an Escherichia coli sulfite oxidase homologue reveals the role of a conserved active site cysteine in assembly and function. Biochemistry 44:10339–10348. [DOI] [PubMed] [Google Scholar]
- 120. Reddy VS, Shlykov Ma, Castillo R, Sun EI, Saier MH (2012) The major facilitator superfamily (MFS) revisited. Febs J 279:2022–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Von Rozycki T, Yen MR, Lende EE, Saier MH (2005) The YedZ family: Possible heme binding proteins that can be fused to transporters and electron carriers. J Mol Microbiol Biotechnol 8:129–140. [DOI] [PubMed] [Google Scholar]
- 122. Scheffers D, Driessen AJ (2001) The polymerization mechanism of the bacterial cell division protein FtsZ. FEBS Lett 506:6–10. [DOI] [PubMed] [Google Scholar]
- 123. Bi EF, Lutkenhaus J (1991) FtsZ ring structure associated with division in Escherichia coli . Nature 354:161–164. [DOI] [PubMed] [Google Scholar]
- 124. Arakaki A, Nakazawa H, Nemoto M, Mori T, Matsunaga T (2008) Formation of magnetite by bacteria and its application. J R Soc Interface 5:977–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Valverde‐Tercedor C, Abadía‐Molina F, Martinez‐Bueno M, Pineda‐Molina E, Chen L, Oestreicher Z, Lower BH, Lower SK, Bazylinski DA, Jimenez‐Lopez C (2014) Subcellular localization of the magnetosome protein MamC in the marine magnetotactic bacterium Magnetococcus marinus strain MC‐1 using immunoelectron microscopy. Arch Microbiol 196:481–488. [DOI] [PubMed] [Google Scholar]
- 126. Taoka A, Asada R, Sasaki H, Anzawa K, Wu LF, Fukumori Y (2006) Spatial localizations of Mam22 and Mam12 in the magnetosomes of Magnetospirillum magnetotacticum . J Bacteriol 188:3805–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kanetsuki Y, Tanaka M, Tanaka T, Matsunaga T, Yoshino T (2012) Effective expression of human proteins on bacterial magnetic particles in an anchor gene deletion mutant of Magnetospirillum magneticum AMB‐1. Biochem Biophys Res Commun 426:7–11. [DOI] [PubMed] [Google Scholar]
- 128. Yoshino T, Matsunaga T (2006) Efficient and stable display of functional proteins on bacterial magnetic particles using Mms13 as a novel anchor molecule. Appl Environ Microbiol 72:465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wang L, Prozorov T, Palo PE, Liu X, Vaknin D, Prozorov R, Mallapragada S, Nilsen‐Hamilton M (2012) Self‐assembly and biphasic iron‐binding characteristics of Mms6, a bacterial protein that promotes the formation of superparamagnetic magnetite nanoparticles of uniform size and shape. Biomacromolecules 13:98–105. [DOI] [PubMed] [Google Scholar]
- 130. Oestreicher Z, Valverde‐Tercedor C, Chen L, Jimenez‐Lopez C, Bazylinski DA, Casillas‐Ituarte NN, Lower SK, Lower BH (2012) Magnetosomes and magnetite crystals produced by magnetotactic bacteria as resolved by atomic force microscopy and transmission electron microscopy. Micron 43:1331–1335. [DOI] [PubMed] [Google Scholar]
- 131. Feng S, Wang L, Palo P, Liu X, Mallapragada SK, Nilsen‐Hamilton M (2013) Integrated self‐assembly of the Mms6 magnetosome protein to form an iron‐responsive structure. Int J Mol Sci 14:14594–14606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhang H, Liu X, Feng S, Wang W, Schmidt‐Rohr K, Akinc M, Nilsen‐Hamilton M, Vaknin D, Mallapragada S (2015) Morphological transformations in the magnetite biomineralizing protein Mms6 in iron solutions: A small‐angle X‐ray scattering study. Langmuir 31:2818–2825. [DOI] [PubMed] [Google Scholar]
- 133. Kolinko S, Richter M, Glöckner F‐O, Brachmann A, Schüler D (2014) Single‐cell genomics reveals potential for magnetite and greigite biomineralization in an uncultivated multicellular magnetotactic prokaryote. Environ Microbiol Rep 6:524–531. [DOI] [PubMed] [Google Scholar]
- 134. Jogler C, Wanner G, Kolinko S, Niebler M, Amann R, Petersen N, Kube M, Reinhardt R, Schüler D (2011) Conservation of proteobacterial magnetosome genes and structures in an uncultivated member of the deep‐branching Nitrospira phylum. Proc Natl Acad Sci USA 108:1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lin W, Deng A, Wang Z, Li Y, Wen T, Wu L‐F, Wu M, Pan Y (2014) Genomic insights into the uncultured genus “Candidatus Magnetobacterium” in the phylum Nitrospirae. Isme J 8:2463–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]


