Abstract
Aquaporins and aquaglyceroporins (AQPs) are membrane channel proteins responsible for transport of water and for transport of glycerol in addition to water across the cell membrane, respectively. They are expressed throughout the human body and also in other forms of life. Inhibitors of human AQPs have been sought for therapeutic treatment for various medical conditions including hypertension, refractory edema, neurotoxic brain edema, and so forth. Conducting all‐atom molecular dynamics simulations, we computed the binding affinity of acetazolamide to human AQP4 that agrees closely with in vitro experiments. Using this validated computational method, we found that 1,3‐propanediol (PDO) binds deep inside the AQP4 channel to inhibit that particular aquaporin efficaciously. Furthermore, we used the same method to compute the affinities of PDO binding to four other AQPs and one aquaglyceroporin whose atomic coordinates are available from the protein data bank (PDB). For bovine AQP1, human AQP2, AQP4, AQP5, and Plasmodium falciparum PfAQP whose structures were resolved with high resolution, we obtained definitive predictions on the PDO dissociation constant. For human AQP1 whose PDB coordinates are less accurate, we estimated the dissociation constant with a rather large error bar. Taking into account the fact that PDO is generally recognized as safe by the US FDA, we predict that PDO can be an effective diuretic which directly modulates water flow through the protein channels. It should be free from the serious side effects associated with other diuretics that change the hydro‐homeostasis indirectly by altering the osmotic gradients.
Keywords: ligand–protein interaction, aquaporin inhibitor, molecular dynamics
Abbreviations
- 1D
one‐dimensional
- 3D
three‐dimensional
- AQP
aquaporin
- ar/R
aromatic/arginine
- AZM
acetazolamide
- bw
body weight
- GRAS
generally recognized as safe
- hSMD
hybrid steered molecular dynamics
- IC50
half maximal inhibitory concentration
- MD
molecular dynamics
- NOAEL
no observed adverse effect level
- NPA
asparagine‐proline‐alanine
- PDB
protein data bank
- PDO
1,3‐propanediol
- PMF
potential of mean force
- SMD
steered molecular dynamics
- vdW
van der Waals.
Introduction
Aquaporins (AQPs)1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 constitute the cell's “plumbing system” in humans and in other forms of life. Among the 13 AQPs (AQP0–AQP12), AQPs 1, 2, 4, and 5 are known to be water channels that allow osmotic water current across the cell membrane. AQPs 3, 7, and 9 are glycerol channels that conduct glycerol and water as well. They are expressed in various cells from head to toe and govern a wide spectrum of physiological functions with broad clinical importance.15, 17, 18, 19, 20, 21, 22, 23, 24, 25 Therefore, great efforts have been invested to search for inhibitors of these membrane proteins. However, finding an efficacious and potent AQP inhibitor has proven to be challenging. So far, AQPs have remained elusive drug targets.15 This difficulty is related to the nature of AQPs whose channels are all very narrow and only allow single‐file lining up of waters or small solutes through the conducting pore. It is also related to the fact that the current computational methods are inadequate. For example, several research groups identified acetazolamide (AZM), an anti‐epilepsy drug, to bind strongly at the entry vestibule of AQP4 with a half maximal inhibitory concentration (IC50) at low µM.26 However, their computed results were in contradiction with the structural experiments and proteoliposome assays of AQP4 in the presence of mM amounts of this drug.8
Recently, we used a new computational method, the hybrid steered molecular dynamics (hSMD),27, 28 to compute the binding affinity of AZM to AQP4. On the basis of all‐atom CHARMM 36 force fields,29, 30 our computed value was in close agreement with the in vitro experiments.8 We also found that 1,3‐propanediol (PDO) binds deep inside the AQP4 channel. In this article, we present a computational investigation of PDO binding to six AQPs (bovine AQP1, human AQP 1, 2, 4, 5, and parasite PfAQP) whose atomic coordinates are available from the protein data bank (PDB). For bovine AQP1, human AQP2, AQP4, and AQP5, and parasite PfAQP whose structures were resolved to high‐resolution, our computed values are definitive with chemical accuracy. For human AQP1 whose structure resolution was lower, our computed value has a large uncertainty.
From these results, we are able to conclude that all water‐only channels are similar in their interactions with PDO. The dissociation constant is around 0.3 mM for all water‐only channels. The glycerol channel is different from the water‐only channels in two aspects: PDO can permeate through a glycerol channel but not water‐only channels; PDO binding to glycerol channels is weaker than to water‐only channels. These findings strongly indicate that PDO can be an efficacious aquaporin inhibitor for medical use because it is generally recognized as safe (GRAS) by the US FDA.
Results
In Table 1, we summarize our results on PDO binding to six AQPs. For convenience, we note a human AQP simply as AQP, bovine AQP1 as bAQP1, and Plasmodium falciparum AQP as PfAQP. Note that we take the z‐axis as perpendicular to the membrane surface pointing from the extracellular space to the cytoplasmic side. The aquaporin channels are approximately along the z‐axis albeit they are not exactly straight. Each channel curves to a different degree. For each of the six binding problems, we computed the one‐dimensional (1D) potential of mean force (PMF) of PDO inside the AQP channel from the region of the aromatic/arginine (ar/R) selectivity filter to the region of NPA motifs (NLA‐NPS in the case of PfAQP). From the 1D PMF, we obtain the partial partition . From the region of NPA motifs to the cytoplasmic bulk, we computed the three‐dimensional (3D) PMF curve from which we obtained the PMF difference . For each PDO‐AQP complex, we chose the interface between the 1D PMF and the 3D PMF at a z‐coordinate . On the xy‐plane of (x, y, z = ), we computed the PDO fluctuation and deviation in and . All these factors combine to give us the standard binding free energy or, equivalently, the dissociation constant . The numerical results are shown in Table 1. The curves are shown in Figures 1 and 2.
Table 1.
Computed Results of PDO‐AQP Complexes
| Protein | PDB code/resolution (Å) | (Å4) | (kcal/mol) | (Å) | (kcal/mol) | (kcal/mol) | (mM) |
|---|---|---|---|---|---|---|---|
| AQP1 | 1H6I/3.831 | 1.2 × 10−3 | 0.20 | 1.3 × 104 | −3.11 | −3.63 ± 3.0 | 2.19 |
| bAQP1 | 1J4N/2.232 | 1.6 × 10−4 | 0.08 | 1.3 × 102 | −7.87 | −4.9 ± 1.2 | 0.239 |
| AQP2 | 4NEF/2.7533 | 4.8 × 10−3 | 0.45 | 2.7 × 106 | −0.804 | −5.4 ± 1.2 | 0.172 |
| AQP4 | 3GD8/1.88 | 7.7 × 10−4 | 0.49 | 1.1 × 106 | −1.44 | −4.8 ± 1.2 | 0.328 |
| AQP5 | 3D9S/2.034 | 3.7 × 10−4 | 0.32 | 1.3 × 102 | −7.30 | −4.9 ± 1.2 | 0.245 |
| PfAQP | 3C02/2.0535 | 8.7 × 10−4 | 0.49 | 1.15 | −6.25 | −1.4 ± 1.2 | 89 |
Figure 1.
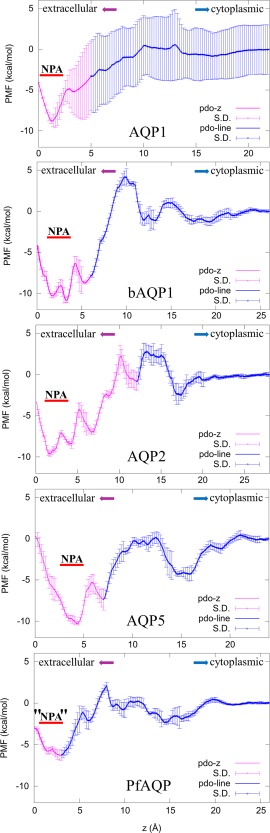
PMF curves of bAQP1, AQP 1, 2, 5, and PfAQP. 1D PMF is shown in purple and 3D PMF in blue.
Figure 2.
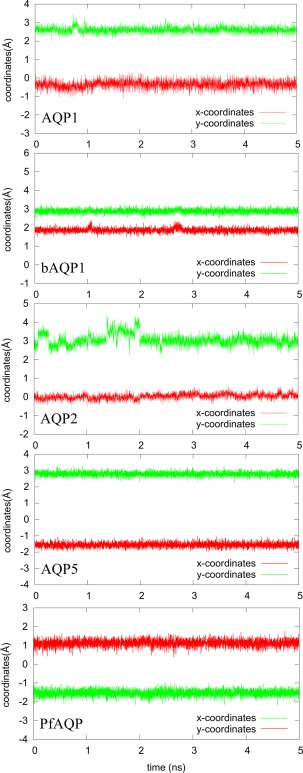
Fluctuations on the xy‐plane at the 1D–3D interface.
AQP1 is expressed in the epithelium of the renal proximal tubule and the thin descending limb of the loop of Henle, as well as in the endothelium in the descending vasa recta; mice that are deficient in AQP1 have a greatly impaired ability to concentrate urine.36, 37 Thus, inhibition of AQP1 was predicted to produce water diuresis by a mechanism that is different from conventional salt transport‐blocking diuretics. This suggests that AQP1 could have clinical potential in treating hypertension and also refractory edema associated with congestive heart failure and cirrhosis.
As the structure of human AQP1 (PDB: 1H6I) has not yet been resolved to atomistic accuracy and our computation ended up with very large uncertainty, we also investigated PDO binding to bovine AQP1 (PDB: 1J4N) whose crystal structure has been accurately determined. On the basis of bAQP1, our prediction is that PDO inhibits AQP1 with an IC50 of 0.239 mM.
AQP2 (PDB: 4NEF). The major expression site of AQP2 is the renal collecting duct.38, 39 An inhibitor of this water channel serves as a diuretic by simply reducing water reabsorption. We found that PDO inhibits AQP2 with an IC50 of 0.172 mM.
AQP4 (PDB: 3GD8). This water channel is expressed in multiple human organs, particularly in the central nervous system (CNS). It is an essential component in several physiological processes including water movement into and out of the brain, neuro‐excitation, as well as astrocyte migration toward injury sites.15 These key roles of AQP4 suggest that inhibitors of this particular water channel could potentially treat (cytotoxic) brain edema by reducing intracranial pressure and water content, epilepsy by increasing the seizure threshold, and CNS injuries (e.g., trauma or stroke) by accelerating neuronal regeneration via reducing the formation of glial scars.40, 41, 42, 43, 44, 45, 46, 47, 48 As such, AQP4 is an appealing drug target and the search for its inhibitors is being actively pursued by medical scientists.8, 15, 26, 49, 50, 51 PDO could be an answer to that search. It inhibits water conduction of AQP4 channel with an IC50 of 0.328 mM which would not cause any side effects.
AQP5 (PDB: 3D9S). This channel protein is richly expressed in salivary and airway submucosal glands. Mice lacking AQP5 were found to have defective secretion of saliva and airway mucus.52, 53 Inhibitors of this water channel can be used to reduce elevated salivation and airway mucus secretion caused by anesthesia. Our study shows that PDO inhibits AQP5 at an IC50 of 0.245 mM.
PfAQP (PDB: 3C02). We currently do not have atomistic structures of human glycerol channels AQP 3, 7, or 9. But we have a high‐resolution structure of the P. falciparum water–glycerol channel PfAQP. We study PfAQP as an analog of human AQPs 3, 7, and 9 and draw the following two conclusions: First, our computed PMF curve indicates that PDO can permeate through the glycerol channel as easily as glycerol. Second, PDO binds inside the glycerol channel with a moderately low affinity, k D = 89 mM. In contrast to this, erythritol binds to PfAQP very strongly.54 Both aspects are in agreement with the experimental measurement of glycerol transport across the membrane of erythrocytes.55
It should be noted that the study of PfAQP by itself is very important. Plasmodium falciparum, the malaria parasite, expresses only PfAQP on its plasma membrane.35, 56, 57 It does not have a dedicated water channel so it relies on this only aquaporin PfAQP for water transport, for glycerol transport, and for excreting metabolites, which are all essential for the parasite's survival. Inhibiting this multifunctional channel protein may hinder the growth of or even kill the deadly parasite that still causes over half a million deaths per year.
Discussion
Efficacy of PDO as an AQP inhibitor
Illustrated in Figure 3 are the geometric characteristics of the AQP water channel. The water pore has a diameter ranging from 1.5 to 4.2 Å and thus allows only single‐file lining of waters throughout the channel. No two waters can occupy the same z‐coordinate inside the channel [Fig. 3(A)]. However, near the NPA motifs, the pore is wide enough to host a PDO molecule [Fig. 3(B)]. Pulling PDO away from this site either way, to the cytoplasmic or the extracellular side, actually causes distortions to the pore‐lining sidechains of AQP because the channel is narrower on both sides of the binding site. When a PDO molecule binds there, it completely occludes the channel from water conduction. Additionally, its fluctuations inside the channel will not give space for water to squeeze by. Therefore, we conclude that PDO is an efficacious inhibitor of AQP water conduction.
Figure 3.
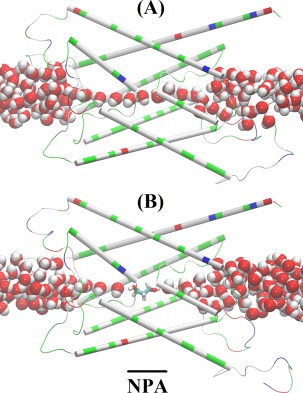
Illustration of binding means inhibition. Single file waters in the apo state versus water line interrupted by PDO in the holo state of the PDO‐AQP2 complex. Protein is represented as cartoons colored by residue types; PDO as licorices colored by atom names; and waters inside the channel and near the channel entry/exit as large spheres colored by atom names. Color schemes: hydrophilic, green; hydrophobic, white; negatively charged, red; positively charge, blue; hydrogen, white; oxygen, red; carbon, cyan (All molecular graphics in the article were rendered with VMD.59).
Potency of PDO as an AQP inhibitor
As a PDO bound to AQP totally occludes the water channel, the potency of PDO should be equal to the dissociation constant, IC50 = k D which is in the range of 0.2–0.3 mM for water channels. Considering that our PMF estimation has an error of ±1.2 kcal/mol for the systems with well‐resolved crystallographic structures, the IC50 of PDO should be less than 3.0 mM which indicates that PDO as an AQP inhibitor has sufficiently high potency. This is particularly true in light of the knowledge that PDO is nontoxic. Furthermore, the close agreement between the binding free energies across the studied water channels is a strong indicator of our method's reliability. This in turn strengthens our predictions regarding AQP1, as its poorly resolved crystallographic structure yields large error bars in the computed binding energy.
Interactions responsible for binding
In the bound state inside the AQP channel, PDO displaces two or three waters out of their places [Fig. 3(A) vs. (B) and Fig. 4(A)]. The three displaced waters, if not displaced, would form seven hydrogen bonds with the channel residues (three bonds) and with waters (four bonds). In their place, PDO forms two hydrogen bonds with the channel residues and two hydrogen bonds with two waters by its two hydroxyl groups [Fig. 4(B)]. Altogether, PDO in the bound state disrupts three or four hydrogen bonds on the average. In the dissociated state, when it is away from the protein, PDO forms four hydrogen bonds with waters. In terms of the hydrogen bonds PDO can form, there is no significant difference between its bound state and its dissociated state. However, in the dissociated state, PDO displaces four waters and disrupts 10 hydrogen bonds if we consider that each water forms 3.5 hydrogen bonds with other waters on the average. Therefore, the system of PDO‐AQP‐waters has six more hydrogen bonds with PDO in the bound state than in the dissociated state. Additionally, the van der Waals interaction between PDO and the protein is also favorable for PDO to reside at the binding site because it sterically fits in the channel near the NPA motifs. The van der Waals attraction between PDO and AQP and the hydrophobic effect (breaking more hydrogen bonds in the aqueous bulk than inside the protein) together are responsible for binding PDO inside the conducting pore of a water/glycerol channel. The overall free energy of binding PDO inside a water channel AQP is approximately = −4.8 to −5.4 kcal/mol in correspondence to 0.2–0.3 mM. Considering that the binding site of PDO is near the NPA motifs which are conserved across most AQPs (human or other forms of life), it is not surprising to observe that PDO binds to all water channel AQPs in similar ways with similar strength.
Figure 4.
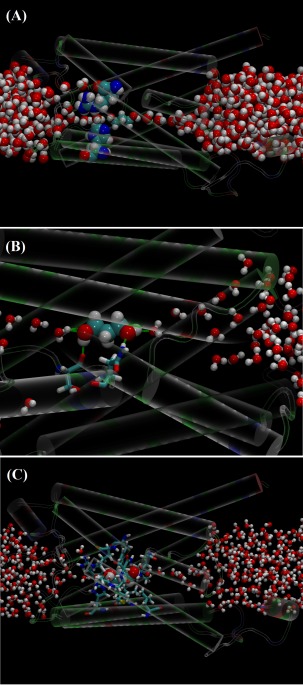
PDO at the binding site near the NPA motifs. (A) Protein in cartoons colored by residue types, ar/R selectivity filter Arg 187 and His 172 in large spheres colored by atom names, PDO and water in or near the channel in medium spheres colored by atom names. This panel shows PDO residing near the NPA motifs which is on the right‐hand side of the ar/R and where the two half membrane helices face each other. (B) Zoom‐in of PDO with near‐by residues (Ser 182 and Asn 184) and waters forming four hydrogen bonds (green dashed bars). (C) PDO (large spheres colored by atom names) in favorable (attractive) van der Waals contact with surrounding residues (licorices colored by atom names). Color schemes identical to Figure 3.
Furthermore, analyzing all the nonbonded terms in our simulation (both the electrostatic and the van der Waals interactions shown in Supporting Information Fig. S2), we note that the nonbonded interactions of PDO at the binding site are attractive in nature for all the water channels (AQPs 1, 2, 4, and 5) and the glycerol channel (PfAQP). These favorable interactions, the electrostatic parts, in particular, are stronger in the cases of water channels than the case of glycerol channel. Examining the conducting pore radii of all the six AQPs we studied, which are shown in Supporting Information Figure S3, we observe that all water channels are narrower at the NPA motifs than the glycerol channel at its “NPA” (NLA‐NPS) motifs. The fit of PDO inside a water channel at the NPA must be tighter/better than the fit inside a glycerol channel. Therefore, the electrostatic interaction (including hydrogen bonding) between PDO and a water channel is stronger than between PDO and a glycerol channel. Consequently, PDO can traverse a glycerol channel as easily as glycerol, which is in agreement with the in vitro finding of Ref. 55.
Possible medical use of 1,3‐propanediol
The reported no‐observed‐adverse‐effect‐levels (NOAELs) are: 1.8 mg/L in the 14‐day inhalation toxicity study; 1000 mg/kg bw/day in the 90‐day oral toxicity study; and 1000 mg/kg bw/day in the developmental toxicity study. Moreover, the US FDA approved DuPont Tate & Lyle's conclusion that PDO is GRAS under the intended conditions of use in food products at a level of 34 mg/kg bw/day. Considering all these data and our result that the IC50 of AQP inhibition by PDO is most probably around 0.2–0.3 mM and less than 3.0 mM, which is far less than the NOAELs, it should be safe to use 1,3‐propanediol as a drug for AQP‐correlated pathological conditions.
Conclusions
Using CHARMM 36 all‐atom force fields on the basis of the rigorous PMF formalism, we conducted hSMD simulations to compute the binding affinities of PDO to six AQPs. The reasonably strong affinity of the nontoxic PDO makes it a valid drug candidate. It is worth noting again that the location of the PDO bound state inside the single‐file channel of AQP renders PDO's efficacy so that the IC50 is simply equal to the dissociation constant k D. The computed dissociation constants of <1 mM for the water channel AQPs indicate its potency as an AQP inhibitor. Taking into account the error bars of the computation, we have 13 µM < k D < 3.0 mM (in accordance with the binding free energies between = −4.8 ± 1.2 and −5.4 ± 1.2 kcal/mol). Therefore, we conclude with high confidence that 1,3‐propanediol is an efficacious and potent AQP inhibitor, which could serve as a drug needed for various AQP‐correlated pathological conditions ranging from hypertension to neurotoxic brain edema.
Methods
Here, we briefly outline the essential information needed to reproduce the research presented in this article.
Procedures and parameters
We took the crystal structures of AQPs (PDB: 1J4N for bovine AQP1, 1H6I for human AQP1, 4NEF for AQP2, 3GD8 for AQP4, 3D9S for AQP5, and 3C02 for PfAQP), deleted glycerols (if present) from the structures, placed a PDO inside the conducting pore near the NPA motifs, and deleted waters from where they had steric collision with the PDO. In this manner, we built six individual PDO‐AQP complexes. Then we placed one PDO‐AQP monomer complex in a 150 mM NaCl saline box of 80 × 80 × 97 Å3 (of the fully equilibrated system). We also added an appropriate amount of counter ions (Na+ or Cl−) to each system to balance the charges carried by the protein. All together, we obtained six all‐atom systems for PDO‐AQP binding. We note that the system size used here is sufficiently large because PDO is not charged and the alpha carbons on the transmembrane helices are all fixed to their crystal structure coordinates. Possible errors are expected to come not from the smallness of the model systems but from the force field parameters, from the PDB coordinates, and from the statistics of our sampling procedures. It turned out that, in all our cases except human AQP1, our results have reached the expected chemical accuracy. The error bars are in the range of 2k BT for the binding free energies. (k B is the Boltzmann constant and T is the absolute temperature.)
We used the all‐atom force field CHARMM3629, 30 to represent all interactions. The van der Waals cut‐off distance was 10 Å with a switching distance of 9 Å. The electrostatic interactions were computed through the Particle‐Mesh Ewald method, and periodic boundary conditions were applied in every direction. The temperature was kept at 298 K and the pressure at 1.0 bar. The time‐step was 1.0 fs for the short range interactions and 2.0 fs for the long range interactions, and the Langevin damping coefficient was 5.0 ps−1. We used NAMD58 to perform the simulations, and VMD59 to set up simulation systems, analyze simulation results, and render molecular graphics.
Computing binding affinities with hSMD method
The formulas and validation of hSMD method can be found in Refs. 27 and 28. In this study, we only use the case of one pulling center, n = 1, and briefly outline the hSMD formulation below. Following the standard literature,60, 61 the binding affinity at one binding site is
| (1) |
where is the standard concentration. For clarity and for convenience of unit conversion, we use two different but equivalent forms, on the left‐hand side and on the right‐hand side of the equation. The 3D integrations are over the x‐, y‐, and z‐coordinates of the ligand's position that is chosen as the center of mass of PDO. The integral has the units of Å3 that renders the right‐hand side dimensionless as it should be. is the 3D PMF. The subscripts “site” and “bulk” indicate that is at the binding site (near the PMF minimum) and in the bulk region far away from the protein, respectively. Expressed in terms of the 3D PMF difference and the partial partition of the bound state , we have
| (2) |
Here, is the standard binding free energy. is the one bound state chosen from the bound state ensemble from which the ligand is pulled to the corresponding one state in the dissociated state ensemble. is the ligand displacement along the dissociation path from the binding site to the bulk region. The 3D PMF difference is computed from SMD runs following the multisectional scheme described in Ref. 62. The partial partition is computed as integration along the z‐axis ( is constant in the integral) and the integrand is simply related to the 1D PMF difference, , which is computed by conducting SMD runs in which only the z‐coordinate is steered while the x‐ and y‐coordinates are free to fluctuate inside the channel.62 The fluctuations on the xy‐plane are measured to compute the deviation of the initial state from the average state on the xy‐plane.
| (3) |
and the 2 × 2 fluctuation matrix
| (4) |
The brackets here represent statistical average on the xy‐plane.
Supporting information
Supporting Information
Acknowledgments
The authors acknowledge the computing resources provided by the Texas Advanced Computing Center at University of Texas at Austin. LY acknowledges the Jiangsu Overseas Research & Training Program for University Prominent Young & Middle‐aged Teachers and Presidents.
The authors declare no competing financial interest.
References
- 1. Agre P, Bonhivers M, Borgnia MJ (1998) The aquaporins, blueprints for cellular plumbing systems. J Biol Chem 273:14659–14662. [DOI] [PubMed] [Google Scholar]
- 2. Heymann JB, Engel A (1999) Aquaporins: phylogeny, structure, and physiology of water channels. Physiology 14:187–193. [DOI] [PubMed] [Google Scholar]
- 3. Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y (2000) Structural determinants of water permeation through aquaporin‐1. Nature 407:599–605. [DOI] [PubMed] [Google Scholar]
- 4. de Groot BL, Grubmüller H (2001) Water permeation across biological membranes: mechanism and dynamics of aquaporin‐1 and GlpF. Science 294:2353–2357. [DOI] [PubMed] [Google Scholar]
- 5. Engel A, Stahlberg H. Aquaglyceroporins: channel proteins with a conserved core, multiple functions, and variable surfaces In: Thomas Zeuthen WDS, Ed. (2002) International review of cytology. Academic Press, Cambridge, MA: pp 75–104. [DOI] [PubMed] [Google Scholar]
- 6. Gonen T, Walz T (2006) The structure of aquaporins. Q Rev Biophys 39:361–396. [DOI] [PubMed] [Google Scholar]
- 7. Carbrey JM, Agre P. Discovery of the aquaporins and development of the field aquaporins. In: Beitz E, Ed. (2009) Springer, Berlin, Germany: pp 3–28. [DOI] [PubMed] [Google Scholar]
- 8. Ho JD, Yeh R, Sandstrom A, Chorny I, Harries WEC, Robbins RA, Miercke LJW, Stroud RM (2009) Crystal structure of human aquaporin 4 at 1.8 Å and its mechanism of conductance. Proc Natl Acad Sci USA 106:7437–7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Branden M, Tabaei SR, Fischer G, Neutze R, Hook F (2010) Refractive‐index‐based screening of membrane‐protein‐mediated transfer across biological membranes. Biophys J 99:124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Benga G (2012) On the definition, nomenclature and classification of water channel proteins (aquaporins and relatives). Mol Aspects Med 33:514–517. [DOI] [PubMed] [Google Scholar]
- 11. Abramson J, Vartanian AS (2013) Watch water flow. Science 340:1294–1295. [DOI] [PubMed] [Google Scholar]
- 12. Gravelle S, Joly L, Detcheverry F, Ybert C, Cottin‐Bizonne C, Bocquet L (2013) Optimizing water permeability through the hourglass shape of aquaporins. Proc Natl Acad Sci USA 110:16367–16372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kosinska Eriksson U, Fischer G, Friemann R, Enkavi G, Tajkhorshid E, Neutze R (2013) Subangstrom resolution X‐ray structure details aquaporin‐water interactions. Science 340:1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oberg F, Hedfalk K (2013) Recombinant production of the human aquaporins in the yeast Pichia pastoris (Invited Review). Mol Membr Biol 30:15–31. PMID: 22908994 {Medline} [DOI] [PubMed] [Google Scholar]
- 15. Verkman AS, Anderson MO, Papadopoulos MC (2014) Aquaporins: important but elusive drug targets. Nat Rev Drug Discov 13:259–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horner A, Zocher F, Preiner J, Ollinger N, Siligan C, Akimov SA, Pohl P (2015) The mobility of single‐file water molecules is governed by the number of H‐bonds they may form with channel‐lining residues. Sci Adv 1:e1400083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. King LS, Kozono D, Agre P (2004) From structure to disease: the evolving tale of aquaporin biology. Nat Rev Mol Cell Biol 5:687–698. [DOI] [PubMed] [Google Scholar]
- 18. Krane C, Goldstein D (2007) Comparative functional analysis of aquaporins/glyceroporins in mammals and anurans. Mamm Genome 18:452–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verkman AS, Mitra AK (2000) Structure and function of aquaporin water channels. Am J Physiol Renal Physiol 278:F13–F28. PMID: 10644652 {Medline} [DOI] [PubMed] [Google Scholar]
- 20. Verkman AS (2002) Aquaporin water channels and endothelial cell function*. J Anat 200:617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Törnroth‐Horsefield S, Hedfalk K, Fischer G, Lindkvist‐Petersson K, Neutze R (2010) Structural insights into eukaryotic aquaporin regulation. FEBS Lett 584:2580–2588. [DOI] [PubMed] [Google Scholar]
- 22. Delporte C, Steinfeld S (2006) Distribution and roles of aquaporins in salivary glands. Biochim Biophys Acta 1758:1061–1070. PMID: 16537077 {Medline} [DOI] [PubMed] [Google Scholar]
- 23. Larsen HS, Ruus A‐K, Schreurs O, Galtung HK (2010) Aquaporin 11 in the developing mouse submandibular gland. Eur J Oral Sci 118:9–13. [DOI] [PubMed] [Google Scholar]
- 24. Noda Y, Sohara E, Ohta E, Sasaki S (2010) Aquaporins in kidney pathophysiology. Nat Rev Nephrol 6:168–178. [DOI] [PubMed] [Google Scholar]
- 25. Agre P, Preston GM, Smith BL, Jung JS, Raina S, Moon C, Guggino WB, Nielsen S (1993) Aquaporin CHIP: the archetypal molecular water channel. Am J Phys Renal Physiol 265:F463–F476. [DOI] [PubMed] [Google Scholar]
- 26. Huber VJ, Tsujita M, Kwee IL, Nakada T (2009) Inhibition of Aquaporin 4 by antiepileptic drugs. Biorg Med Chem 17:418–424. [DOI] [PubMed] [Google Scholar]
- 27. Chen LY (2015) Hybrid steered molecular dynamics approach to computing absolute binding free energy of ligand–protein complexes: a brute force approach that is fast and accurate. J Chem Theory Comput 11:1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodriguez RA, Yu L, Chen LY (2015) Computing protein–protein association affinity with hybrid steered molecular dynamics. J Chem Theory Comput 11:4427–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30:1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD (2010) CHARMM general force field: a force field for drug‐like molecules compatible with the CHARMM all‐atom additive biological force fields. J Comput Chem 31:671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Groot BL, Engel A, Grubmüller H (2001) A refined structure of human aquaporin‐1. FEBS Lett 504:206–211. [DOI] [PubMed] [Google Scholar]
- 32. Sui H, Han B‐G, Lee JK, Walian P Jap BK (2001) Structural basis of water‐specific transport through the AQP1 water channel. Nature 414:872–878. [DOI] [PubMed] [Google Scholar]
- 33. Frick A, Eriksson UK, de Mattia F, Öberg F, Hedfalk K, Neutze R, de Grip WJ, Deen PMT, Törnroth‐Horsefield S (2014) X‐ray structure of human aquaporin 2 and its implications for nephrogenic diabetes insipidus and trafficking. Proc Natl Acad Sci USA 111:6305–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Horsefield R, Nordén K, Fellert M, Backmark A, Törnroth‐Horsefield S, Terwisscha van Scheltinga AC, Kvassman J, Kjellbom P, Johanson U, Neutze R (2008) High‐resolution x‐ray structure of human aquaporin 5. Proc Natl Acad Sci USA 105:13327–13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Newby ZE, O'Connell J, 3rd , Robles‐Colmenares Y, Khademi S, Miercke LJ, Stroud RM (2008) Crystal structure of the aquaglyceroporin PfAQP from the malarial parasite Plasmodium falciparum . Nat Struct Mol Biol 15:619–625. PMID: 18500352 {Medline} [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma T, Yang B, Gillespie A, Carlson EJ, Epstein CJ, Verkman AS (1998) Severely impaired urinary concentrating ability in transgenic mice lacking aquaporin‐1 water channels. J Biol Chem 273:4296–4299. [DOI] [PubMed] [Google Scholar]
- 37. Schnermann J, Chou C‐L, Ma T, Traynor T, Knepper MA, Verkman AS (1998) Defective proximal tubular fluid reabsorption in transgenic aquaporin‐1 null mice. Proc Natl Acad Sci USA 95:9660–9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deen P, Verdijk M, Knoers N, Wieringa B, Monnens L, van Os C, van Oost B (1994) Requirement of human renal water channel aquaporin‐2 for vasopressin‐dependent concentration of urine. Science 264:92–95. [DOI] [PubMed] [Google Scholar]
- 39. Tamarappoo BK, Verkman AS (1998) Defective aquaporin‐2 trafficking in nephrogenic diabetes insipidus and correction by chemical chaperones. J Clin Investig 101:2257–2267. PMID: PMC508814 {Medline} [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Assentoft M, Larsen BR, Olesen ETB, Fenton RA, MacAulay N (2014) AQP4 plasma membrane trafficking or channel gating is not significantly modulated by phosphorylation at COOH‐terminal serine residues. Am J Physiol Cell Physiol 307:C957–C965. [DOI] [PubMed] [Google Scholar]
- 41. Hasegawa H, Ma T, Skach W, Matthay MA, Verkman AS (1994) Molecular cloning of a mercurial‐insensitive water channel expressed in selected water‐transporting tissues. J Biol Chem 269:5497–5500. [PubMed] [Google Scholar]
- 42. Jung JS, Bhat RV, Preston GM, Guggino WB, Baraban JM, Agre P (1994) Molecular characterization of an aquaporin cDNA from brain: candidate osmoreceptor and regulator of water balance. Proc Natl Acad Sci 91:13052–13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nagelhus EA, Ottersen OP (2013) Physiological roles of aquaporin‐4 in brain. Physiol Rev 93:1543–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Papadopoulos MC, Verkman AS (2005) Aquaporin‐4 gene disruption in mice reduces brain swelling and mortality in pneumococcal meningitis. J Biol Chem 280:13906–13912. [DOI] [PubMed] [Google Scholar]
- 45. Smith AJ, Jin B‐J, Ratelade J, Verkman AS (2014) Aggregation state determines the localization and function of M1– and M23–aquaporin‐4 in astrocytes. J Cell Biol 204:559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vella J, Zammit C, Di Giovanni G, Muscat R, Valentino M (2015) The central role of aquaporins in the pathophysiology of ischemic stroke. Front Cell Neurosci 9:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Badaut J, Fukuda AM, Jullienne A, Petry KG (2014) Aquaporin and brain diseases. Biochim Biophys Acta 1840:1554–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Papadopoulos MC, Verkman AS (2013) Aquaporin water channels in the nervous system. Nat Rev Neurosci 14:265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Papadopoulos MC, Verkman AS. Potential utility of aquaporin modulators for therapy of brain disorders In: Inga DN, Rainer L, Eds. (2008) Progress in brain research. Elsevier, New York, NY: pp 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huber VJ, Tsujita M, Yamazaki M, Sakimura K, Nakada T (2007) Identification of arylsulfonamides as aquaporin 4 inhibitors. Bioorg Med Chem Lett 17:1270–1273. [DOI] [PubMed] [Google Scholar]
- 51. Kato J, MKH, Aizu S, Yukutake Y Takeda J, Yasui M, (2013) A general anaesthetic propofol inhibits aquaporin‐4 in the presence of Zn2+. Biochem J 454:275–282. [DOI] [PubMed] [Google Scholar]
- 52. Ma T, Song Y, Gillespie A, Carlson EJ, Epstein CJ, Verkman AS (1999) Defective secretion of saliva in transgenic mice lacking aquaporin‐5 water channels. J Biol Chem 274:20071–20074. PMID: 10400615 {Medline} [DOI] [PubMed] [Google Scholar]
- 53. Song Y, Verkman AS (2001) Aquaporin‐5 dependent fluid secretion in airway submucosal glands. J Biol Chem 276:41288–41292. [DOI] [PubMed] [Google Scholar]
- 54. Chen LY (2015) Erythritol predicted to inhibit permeation of water and solutes through the conducting pore of P. falciparum aquaporin. Biophys Chem 198:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carlsen A, Wieth JO (1976) Glycerol transport in human red cells. Acta Physiol Scand 97:501–513. [DOI] [PubMed] [Google Scholar]
- 56. Hansen M, Kun JF, Schultz JE, Beitz E (2002) A single, bi‐functional aquaglyceroporin in blood‐stage Plasmodium falciparum malaria parasites. J Biol Chem 277:4874–4882. PMID: 11729204 {Medline} [DOI] [PubMed] [Google Scholar]
- 57. Hedfalk K, Pettersson N, Oberg F, Hohmann S, Gordon E (2008) Production, characterization and crystallization of the Plasmodium falciparum aquaporin. Protein Expr Purif 59:69–78. PMID: 18295508 {Medline} [DOI] [PubMed] [Google Scholar]
- 58. Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38. [DOI] [PubMed] [Google Scholar]
- 60. Zhou H‐X, Gilson MK (2009) Theory of free energy and entropy in noncovalent binding. Chem Rev 109:4092–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Woo H‐J, Roux B (2005) Calculation of absolute protein–ligand binding free energy from computer simulations. Proc Natl Acad Sci USA 102:6825–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen LY (2013) Glycerol modulates water permeation through Escherichia coli aquaglyceroporin GlpF. Biochim Biophys Acta 1828:1786–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


