Abstract
The AbgT family of transporters was thought to contribute to bacterial folate biosynthesis by importing the catabolite p‐aminobenzoyl‐glutamate for producing this essential vitamin. Approximately 13,000 putative transporters of the family have been identified. However, before our work, no structural information was available and even functional data were minimal for this family of membrane proteins. To elucidate the structure and function of the AbgT family of transporters, we recently determined the X‐ray structures of the full‐length Alcanivorax borkumensis YdaH and Neisseria gonorrhoeae MtrF membrane proteins. The structures reveal that these two transporters assemble as dimers with architectures distinct from all other families of transporters. Both YdaH and MtrF are bowl‐shaped dimers with a solvent‐filled basin extending from the cytoplasm halfway across the membrane bilayer. The protomers of YdaH and MtrF contain nine transmembrane helices and two hairpins. These structures directly suggest a plausible pathway for substrate transport. A combination of the crystal structure, genetic analysis and substrate accumulation assay indicates that both YdaH and MtrF behave as exporters, capable of removing the folate metabolite p‐aminobenzoic acid from bacterial cells. Further experimental data based on drug susceptibility and radioactive transport assay suggest that both YdaH and MtrF participate as antibiotic efflux pumps, importantly mediating bacterial resistance to sulfonamide antimetabolite drugs. It is possible that many of these AbgT‐family transporters act as exporters, thereby conferring bacterial resistance to sulfonamides. The AbgT‐family transporters may be important targets for the rational design of novel antibiotics to combat bacterial infections.
Keywords: AbgT‐family transporters, antimetabolite transporters, membrane proteins, X‐ray crystallography, Neisseria gonorrhoeae MtrF, Alcanivorax borkumensis YdaH
Introduction
Across all organisms, folic acid is necessary for the methylation, repair, and synthesis of DNA, RNA, and certain amino acids. Folic acid is of particular importance in aiding cell growth and division. Although cellular requirement for folic acid is universal, the approaches that prokaryotes and eukaryotes use to obtain it are very different. Mammals cannot make folic acid themselves. Instead, they have to rely on active transport systems via membrane‐associated proteins to import this essential vitamin.1 In plants and most microorganisms, folic acid is synthesized de novo through the folate biosynthesis pathway.2 As this pathway is commonly found in bacterial pathogens, but absent from humans, it has been an attractive target for the design of novel antimicrobial drugs for treating infectious diseases.
De novo biosynthesis of the essential folic acid in bacteria can be simply interpreted as chemically linking pterin, p‐aminobenzoic acid (PABA), and glutamic acid together. In Escherichia coli, mutations or deletions of enzymes in the folate pathway are known to generate nonviable phenotypes.3 The feasibility of targeting the folate biosynthesis system for antimicrobial therapeutics was proposed several decades ago, based on the experimental results that sulfonamides were able to interrupt bacterial use of the metabolite PABA for folate synthesis.4 Indeed, as early as the 1930s, sulfonamides were widely used as potent antibiotics for treating of infectious diseases caused by a variety of pathogenic bacteria, including Neisseria meningitidis and Pneumococcal pneumonia.5, 6
As a consequence of the rapid adaption of bacterial pathogens to antimicrobials, drug resistance to currently administered antibiotics is a serious problem and is rising at an alarming rate. For example, it only took two years for N. meningitidis to develop resistant strains in the wake of the introduction of sulfonamides.7 The outlook is even more grim in the case of treating the parasitic malaria disease. A strong correlation has been found between mutations in Plasmodium falciparum and resistance to sulfonamides.8, 9, 10
The early studies of folate biosynthesis were largely carried out in the 1960 to 1970s, leading to the detailed description of this enzymatic pathway.11, 12, 13, 14, 15, 16 However, recent work demonstrated that E. coli AbgT17, 18 is capable of catalyzing the uptake of the catabolite p‐aminobenzoyl‐glutamate for de novo folic acid synthesis.18 Because of this finding, it was hypothesized that AbgT‐family transporters19 contribute to the bacterial folate biosynthesis pathway by importing p‐aminobenzoyl‐glutamate for producing this essential vitamin. On the other hand, it has been observed that Neisseria gonorrhoeae MtrF,20, 21 also belonging to the AbgT family, functions as an antimicrobial resistance protein, needed for the high‐level resistance of gonococci to hydrophobic antimicrobials.20, 21
N. gonorrhoeae is a Gram‐negative diplococcus, which is found only in humans and causes the sexually transmitted disease gonorrhea. Since it is a strictly human pathogen and can colonize both male and female genital mucosal surfaces and other sites, it has developed mechanisms to overcome antimicrobial systems of the host's innate defense. One major mechanism that N. gonorrhoeae uses to resist antimicrobial agents is the expression of multidrug efflux pumps that recognize and actively export a variety of structurally unrelated toxic compounds from the bacterial cell, including antibacterial peptides, long‐chain fatty acids and several clinically important antibiotics.22, 23, 24, 25, 26
The best characterized efflux system in N. gonorrhoeae is the MtrC‐MtrD‐MtrE multidrug efflux system.20, 27, 28, 29, 30, 31 This system is similar to other efflux pumps of the resistance‐nodulation‐cell division (RND)32 superfamily possessed by many Gram‐negative bacteria. MtrD29, 30, 33, 34 is the inner membrane transporter component of the tripartite RND pump. The complex is formed by interactions between MtrD, the periplasmic membrane fusion protein MtrC,30, 33, 35, 36 and the outer membrane channel MtrE.27, 36, 37, 38 This powerful efflux complex is presumed to assemble in the form of (MtrD)3(MtrC)6(MtrE)3 and mediates the export of several structurally diverse hydrophobic antimicrobial agents, such as antibiotics, nonionic detergents, antibacterial peptides, bile salts, and gonadal steroidal hormones.27, 36, 37, 38 Recently, our lab has determined the X‐ray structures of the N. gonorrhoeae inner membrane efflux pump MtrD34 and outer membrane channel MtrE.38 The structures of these two membrane proteins have provided us important clues about how this tripartite efflux system functions.
The architecture of the mtr locus consists of two back‐to‐back operons, mtrCDE and the divergently transcribed mtrR, which encodes the TetR family transcriptional regulator MtrR. Interestingly, downstream of mtrR lies another gene called mtrF, which is also regulated by the MtrR regulatory protein. The mtrF gene encodes an inner membrane protein MtrF,20, 21 which belongs to the AbgT family of transporters.19 This membrane protein was found to be required for high‐level resistance of N. gonorrhoeae to certain hydrophobic antimicrobials, including erythromycin and TX‐100. It has been proposed that MtrF cooperates with the MtrC‐MtrD‐MtrE complex to export certain antimicrobials by a yet unknown mechanism.20
In Alcanivorax borkumensis, the gene ydaH 39 encodes a membrane protein YdaH of previously unknown structure and function. Alignment of protein sequences suggests that this protein also belongs to the AbgT family of transporters19 (Fig. 1). To date, approximately 13,000 putative transporters of the AbgT family have been identified. AbgT‐type proteins are found in organisms spanning Gram‐negative bacteria (such as E. coli, N. gonorrhoeae, A. borkumensis, and Salmonella enterica), Gram‐positive bacteria (such as Staphylococcus aureus and Streptomyces coelicolor) as well as eukaryotes (such as the yeast Saccharomyces arboricola). Surprisingly, before our studies, only E. coli AbgT17, 18 and N. gonorrhoeae MtrF20, 21 have been partially characterized among proteins in this diverse family. There was no structural information available for this family of membrane proteins, obscuring the details of their function and mechanism.
Figure 1.
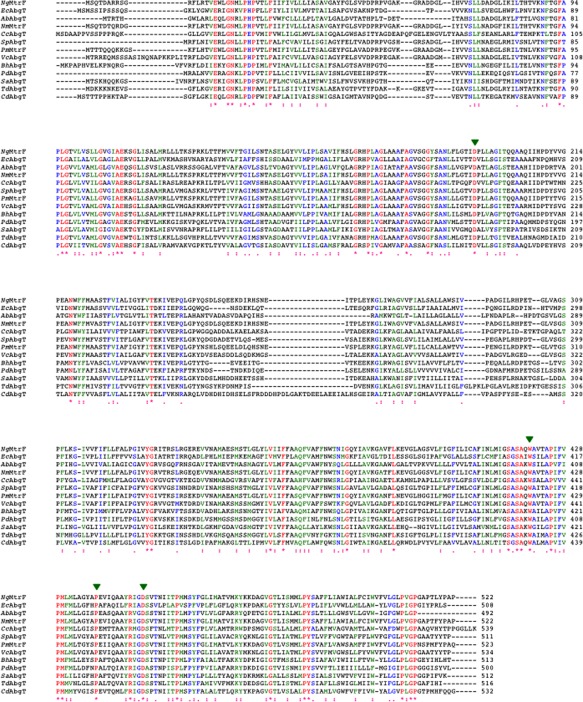
Alignment of amino acid sequences of the AbgT family of transporters. The alignments were done using CLUSTAL W. *identical residues: >60% homologous residues. The four conserved residues involved in lining the channel of the inner core of the protein are indicated with green arrows (Ng, Neisseria gonorrhoeae; Ec, Escherichia coli; Ab, Alcanivorax borkumensis; Nm, Neisseria meningitidis; Cc, Caulobacter crescentus; Sp, Shewanella putrefaciens; Pm, Pasteurella multocida; Vc, Vibrio cholerae; Bh, Bacillus halodurans; Pd, Peptoclostridium difficile; Sa, Staphylococcus aureus; Td, Treponema denticola; Cd, Corynebactrium diphtheria).
To understand how members of the AbgT family work, we have recently determined the crystal structures of the A. borkumensis YdaH40 and N. gonorrhoeae MtrF41 transporters. In this review, we summarize the structural and functional aspects of these two AbgT‐family proteins. Our studies indicate that both A. borkumensis YdaH and N. gonorrhoeae MtrF are exporters, capable of removing the folate metabolite p‐aminobenzoic acid from bacterial cells. Our experimental data strongly suggest that both YdaH and MtrF are antibiotic efflux pumps, which mediate bacterial resistance to sulfonamide antimetabolite drugs. The AbgT‐family transporters may be important targets for the rational design of novel antibiotics to combat bacterial infections.
Crystal structure of A. borkumensis YdaH
To begin understanding the transport functions of members of the AbgT family, we have determined the crystal structure of A. borkumensis YdaH, 492 amino acids, to a resolution of 2.96 Å (PDB ID: 4R0C)40 (Fig. 2). The asymmetric unit contained four YdaH molecules, which form two independent dimers. Superimposition of each dimer of YdaH gives a root mean squared deviation (RMSD) of 1.0 Å over 942 Cα atoms, indicating that their conformations are nearly identical. The crystal structure of YdaH is unique among the previously known membrane transporter proteins, revealing a bowl shaped dimer with a concave aqueous basin. The overall structure is approximately 60 Å tall, 80 Å wide, and 60 Å thick. The basin has a diameter as wide as 50 Å and deeply penetrates the inner leaflet of the cytoplasmic membrane by approximately 20 Å. This basin probably allows the aqueous solution to reach the midpoint of the membrane bilayer.
Figure 2.
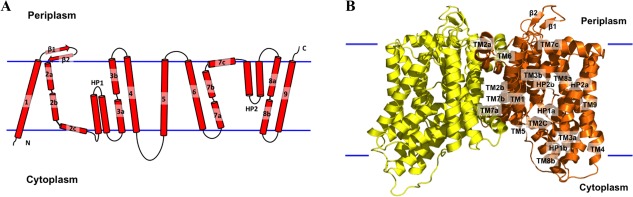
Structure of the A. borkumensis YdaH transporter. (A) Transmembrane topology of A. borkumensis YdaH. The transporter contains nine transmembrane helices (TMs) and two hairpins (HPs). (B) Ribbon diagram of a dimer of YdaH viewed in the membrane plane. The right subunit of the dimer is colored orange, whereas the left subunit is colored yellow. The YdaH dimer forms a bowl‐shaped structure with a concave aqueous basin facing the intracellular solution.
Each protomer of YdaH in the dimer contains nine transmembrane α‐helices and two helical hairpins, designated TM1 (21–42), TM2 (a (63–79), b (83–99) and c (101–112)), HP1 (a (115–131) and b (135–151)), TM3 (a (155–168) and b (178–194)), TM4 (204–228), TM5 (250–272), TM6 (290–314), TM7 (a (320–334), b (336–354) and c (356–371)), HP2 (a (375–392) and b (396–414)), TM8 (a (418–432) and b (441–452)) and TM9 (458–486). The two hairpins HP1 and HP2 span only half of the membrane. In addition, four of the nine transmembrane α‐helices (TM2, TM3, TM7, and TM8) are broken into segments within the membrane. It appears that the intramembrane loops of these segmented TMs and HPs allow the protein to form an internal cavity within the membrane.
Each YdaH protomer possesses only small periplasmic and cytoplasmic domains. The periplasmic domain is formed by two loops between TM1 and TM2 and between TM5 and TM6. Interestingly, the loop between TM1 and TM2 is structured, forming two anti‐parallel β‐strands (β1 and β2). In the cytoplasmic region, a long loop of 22 residues connects TM4 and TM5.
Based on our crystal structure of YdaH, each dimer can be roughly divided into inner and outer core regions (Fig. 3). The inner core comprises TM1, TM2, TM5, TM6, and TM7. Within the inner core, TM2a, TM2b, TM7a, TM7b, and the N‐terminal residues of TM1 and TM6 form a distinct dimerization domain [Fig. 3(A)]. Each YdaH protomer shares a substantial intersubunit interface, with an area of ∼2000 Å2 per protomer. The residues lining the dimer interface are predominantly hydrophobic in nature. The dimerization is secured by TM2a and TM2a′, which cross into each neighboring subunit, respectively.
Figure 3.
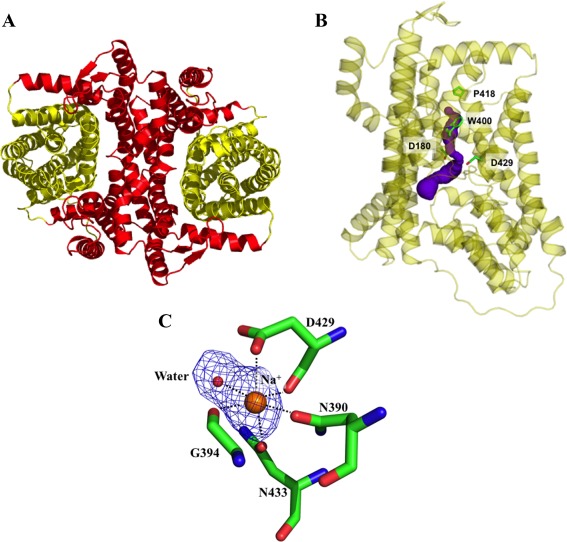
Inner and outer cores of YdaH. (A) The inner core of YdaH, comprising TMs 1, 2, 5, 6 and 7 (colored red), contributes to dimerization as well as formation of a frame‐like structure housing the outer core of the protomer. The outer core of YdaH is composed of TMs 3, 4, 8, 9 as well as HPs 1 and 2 (colored yellow). (B) The outer core of YdaH forms a channel (colored purple) spanning approximately from the middle of the inner membrane up to the periplasmic space. This channel was calculated using the program CAVER (http://loschmidt.chemi.muni.cz/caver). The secondary structural elements of the YdaH protomer are in yellow. Residues D180, N390, W400, P418 and D429 are in green sticks. (C) The bound Na+ (orange sphere) is found to coordinate with N390, G394, D429, N433 and a water molecule (red sphere). The F o–F c map, showing the bound Na+ and H2O, is contoured at 3.0 σ (blue mesh).
Comprising TM3, TM4, TM8, TM9, HP1, and HP2, the outer core likely forms the substrate‐binding and transport pathway domain. A channel was identified in each protomer connecting the middle of the inner membrane to the periplasmic domain of the YdaH dimer [Fig. 3(B)]. The entrance of this channel is formed by the internal cavity located at the basin of the bowl‐shaped structure. This cavity is formed by the loop regions of HP1, HP2, TM3, and TM8 and is accessible to the cytoplasm. It appears that the internal cavity may also form a substrate‐binding site of the transporter. Lining the inner walls of the channel are conserved residues D180, W400, P418, and D429 [Fig. 3(B)]. These residues are expected to play an important functional role of the transporter.
In each protomer of YdaH, the crystal structure revealed a strong spherical electron density contained by the flexible loop between HP2 and TM8 [Fig. 3(C)]. This extra electron density is likely a Na+ ion, which was present at all stages of purification and crystallization. Indeed, the results of valence calculations42 indicate that this site is most likely Na+ specific. The side chains of N390, D429, N433 and the backbone carbonyl oxygens of G394 and D429 form the binding site. A water molecule also participates in the interaction, forming another coordinate bond with the Na+ ion.
Crystal structure of N. gonorrhoeae MtrF
In addition to A. borkumensis YdaH, we have determined the crystal structure of N. gonorrhoeae MtrF, 522 amino acids, to a resolution of 3.95 Å (PDB ID: 4R1I)41 (Fig. 4). The asymmetric unit was found to contain two molecules of MtrF, which assemble as a dimer. Superimposition of these two MtrF molecules gives an RMSD of 0.5 Å over 506 Cα atoms, indicating that their conformations are nearly identical. Similar to YdaH, the crystal structure of the MtrF dimer reveals a bowl‐shaped concave aqueous basin, approximately 75 Å tall, 80 Å wide and 50 Å thick. The rim of the basin is as large as 45 Å in diameter and penetrates into the inner leaflet of the cytoplasmic membrane by approximately 25 Å. This deep basin probably allows aqueous solution to reach until the midpoint of the membrane bilayer.
Figure 4.
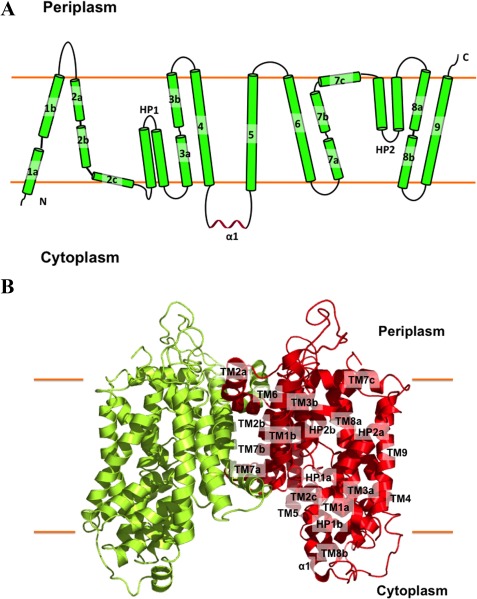
Structure of the N. gonorrhoeae MtrF transporter. (A) Transmembrane topology of N. gonorrhoeae MtrF. The transporter contains nine transmembrane helices (TMs) and two hairpins (HPs). (B) Ribbon diagram of a dimer of MtrF viewed in the membrane plane. The right subunit of the dimer is colored red, whereas the left subunit is colored green. The MtrF dimer forms a bowl‐shaped structure with a concave aqueous basin facing the intracellular solution.
Overall, the secondary structure of the MtrF dimer is very similar to that of A. borkumensis YdaH. A pairwise superimposition of the MtrF dimer onto YdaH results in overall RMSD of approximately 4 Å. Like YdaH, each molecule of MtrF comprises nine transmembrane α‐helices and two helical hairpins: TM1 (a (12–22) and b (26–47)), TM2 (a (78–92), b (94–112) and c (114–125)), HP1 (a (128–145) and b (147–164)), TM3 (a (168–182) and b (191–205)), TM4 (218–240), TM5 (269–292), TM6 (310–334), TM7 (a (341–353), b (356–374) and c (376–391)), HP2 (a (396–413) and b (417–434)), TM8 (a (438–451) and b (462–471)) and TM9 (480–506). In addition to HP1 and HP2, which are only long enough to span half of the membrane, the transmembrane helices TM1, TM2, TM3, TM7, and TM8 are broken into multiple segments within the membrane. As in YdaH, these segmented loops allow the transporter to form an internal cavity within the membrane.
Each protomer of MtrF contains a relatively small periplasmic domain. This domain is made up of two long loops formed between TMs 1 and 2, and TMs 5 and 6, respectively. Below the inner leaflet of the membrane, a small cytoplasmic domain links TMs 4 and 5 together. This domain is comprised by a relatively long random loop and helix (α1).
The MtrF dimer can also be divided into inner and outer core regions [Fig. 5(A)]. The outer core comprises TM3, TM4, TM8, TM9, HP1, and HP2. Like YdaH, the helices and hairpins of the outer core of MtrF form a tunnel spanning approximately from the middle of the inner membrane up to the periplasm [Fig. 5(B)]. Interestingly, this tunnel is also connected to the cytoplasm via an opening in the basin formed by the loop regions of HP1, HP2, TM3, and TM8. Importantly, several conserved residues, including D193, W420, P438, and D449, line the wall of the tunnel [Fig. 5(B)]. Similar to the case of YdaH, it is expected that these residues may play an important role for the function of this transporter.
Figure 5.
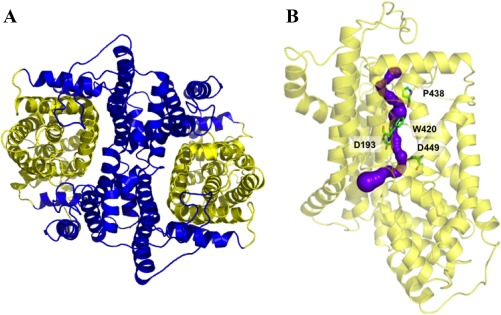
Inner and outer cores of MtrF. (A) The inner core of MtrF, comprising TMs 1, 2, 5, 6, and 7 (colored blue), contributes to dimerization as well as formation of a frame‐like structure housing the outer core of the protomer. The outer core of MtrF is composed of TMs 3, 4, 8, 9 as well as HPs 1 and 2 (colored yellow). (B) The outer core of MtrF forms a channel (colored purple) spanning approximately from the middle of the inner membrane up to the periplasmic space. This channel was calculated using the program CAVER (http://loschmidt.chemi.muni.cz/caver). The secondary structural elements of the MtrF protomer are in yellow. Residues D193, W420, P438, and D449 are in green sticks.
The inner core, comprising TM1, TM2, TM5, TM6, and TM7, creates a frame‐like housing for the inner core and contributes to the dimerization domain [Fig. 5(A)]. Involved in this dimerization interface are TM1b, TM2a, TM2b, TM6, TM7a, and TM7b, as well as the corresponding segments from the next subunit.
A. borkumensis YdaH and N. gonorrhoeae MtrF are capable of exporting p‐aminobenzoic acid from cells
As E. coli AbgT has been shown to enable uptake of the folate catabolite p‐aminobenzoyl‐glutamate, we investigated if YdaH or MtrF, expressed in E. coli cells, could function similarly. We first prepared an E. coli knockout strain BL21(DE3)ΔabgTΔpabA that lacked both the genes abgT 17, 18 and pabA,43 which impairs bacterial synthesis of the metabolite PABA. We then transformed this double knockout strain with either pET15bΩydaH (expressing A. borkumensis YdaH), pET15bΩmtrF (expressing N. gonorrhoeae MtrF) or the empty vector pET15b. Surprisingly, E. coli BL21(DE3)ΔabgTΔpabA cells transformed with either pET15bΩydaH, pET15bΩmtrF, or pET15b could not grow in minimal media (containing 90.4 mM Na2HPO4, 22.0 mM KH2PO4, 8.5 mM NaCl, 0.1 mM CaCl2, 1.0 mM MgSO4, 20.0 mM NH4Cl, and 22.2 mM glucose) supplemented with up to 1 mM p‐aminobenzoyl‐glutamate. However, each of these cells were capable of growing in liquid minimal medium when supplemented with as little as 30 nM of the folate metabolite PABA.
As PABA is an important precursor for folic acid synthesis, we thought that A. borkumensis YdaH and N. gonorrhoeae MtrF might enable uptake of this metabolite. It should be noted that PABA is capable of diffusing into bacterial cells through the membrane, participating as an intermediate in de novo biosynthesis of the essential vitamin folic acid. However, we decided to compare the radioactive PABA content over time in cells transformed with either pET15bΩydaH, pET15bΩmtrF or the empty vector pET15b. The data were normalized using a cell optical density of 1.0 at λ = 600 nm (OD). Surprisingly, E. coli BL21(DE3)ΔabgTΔpabA cells producing A. borkumensis YdaH or N. gonorrhoeae MtrF showed a significant decrease in the level of [3H]‐PABA, compared with cells transformed with the empty pET15b vector (Fig. 6). Instead of importing PABA, the data strongly suggested to us that both YdaH and MtrF may act as exporters, capable of expelling the intracellular PABA metabolite from the cell.
Figure 6.
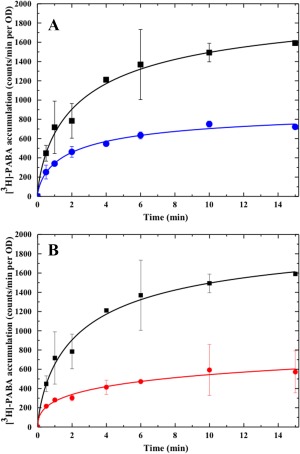
Accumulation of radioactive p‐aminobenzoic acid over time in cells expressing the YdaH or MtrF transporter. (A) Time course of [3H]‐PABA accumulation by E. coli BL21(DE3)ΔabgTΔpabA double knockout cells transformed with pET15bΩydaH or pET15b. Cells expressing ydaH (blue curve) show a significant decrease in [3H]‐PABA accumulation when compared with cells carrying the empty vector (black curve). Error bars represent standard deviation (n = 3). (B) Time course of [3H]‐PABA accumulation by E. coli BL21(DE3)ΔabgTΔpabA double knockout cells transformed with pET15bΩmtrF or pET15b. Cells expressing mtrF (red curve) show a significant decrease in [3H]‐PABA accumulation when compared with cells carrying the empty vector (black curve). Error bars represent standard deviation (n = 3). The data showed in (A) and (B) are the cumulative average of three successive recordings.
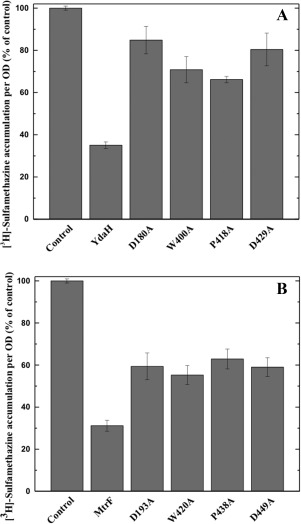
To determine whether the conserved YdaH residues, D180, W400, P418, and D429 (lining the inner wall of the tunnel formed by each protomer), are important for the transport function of the pump, we mutated each residue to alanine, individually. We then expressed these mutant transporters, D180A, W400A, P418A, and D429A, in BL21(DE3)ΔabgTΔpabA cells. Western analysis indicated that the expression levels of these mutant transporters were comparable with that of wild‐type YdaH. The accumulation of [3H]‐PABA per OD in cells expressing the mutant transporters D180A, W400A, P418A, and D429A showed an increase in the levels of [3H]‐PABA accumulation per OD, compared with cells expressing wild‐type YdaH [Fig. 7(A)]. The results indicate that YdaH is able to export PABA from the bacterial cell, and that the conserved residues D180, W400, P418, and D429 are important for this function.
Figure 7.
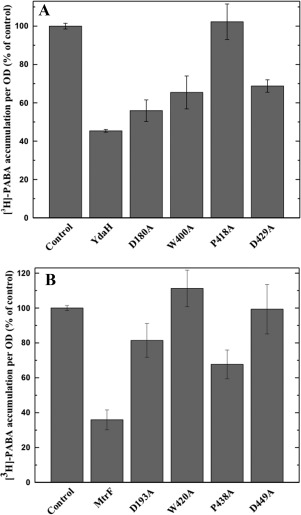
Accumulation of radioactive p‐aminobenzoic acid in cells expressing the mutant YdaH or MtrF transporter. (A) Mutants of the YdaH transporter. Cells possessing the mutant transporter D180A, W400A, P418A, or D429A show an increase in the level of [3H]‐PABA accumulations compared with cells expressing wild‐type YdaH. Error bars represent standard deviation (n = 3). (B) Mutants of the MtrF transporter. Cells possessing the mutant transporter D193A, W420A, P438A, or D449A show an increase in the level of [3H]‐PABA accumulations compared with cells expressing wild‐type MtrF. Error bars represent standard deviation (n = 3). The data showed in (A) and (B) are the cumulative average of three successive recordings.
Based on the crystal structure of Neisseris gonorrhoeae MtrF, we found that the conserved residues D193, W420, P438 and D449 similarly line the inner wall of the tunnel formed by each monomer of MtrF. Therefore, these four residues were also replaced by alanines, respectively. We expressed the mutant MtrF transporters, D193A, W420A, P438A, and D449A, in BL21(DE3)ΔabgTΔpabA cells. Western analysis also indicated that their expression levels were comparable with that of wild‐type MtrF. We then measured the accumulation of [3H]‐PABA in these cells. The results indicated an increase in the concentrations of [3H]‐PABA per OD in cells producing the single point mutants D193A, W420A, P438A, and D449A, compared with those expressing wild‐type MtrF [Fig. 7(B)].
The effect of alanine substitution on these amino acids is quite pronounced for intracellular PABA accumulation. For example, cells expressing the YdaH mutant (D180, W400, P418, or D429) accumulated between 25% and 125% more [3H]‐PABA per OD in comparison with cells expressing wild type YdaH. Similarly, E. coli cells producing the MtrF mutant (D193A, W420A, P438A, or D449A) have significantly higher accumulation of [3H]‐PABA per OD, between 85% and 205%, when compared with those possessing the wild‐type MtrF transporter. A recent study using computational structural analysis suggested that a substitution of residue 288 from glycine to aspartate in the AcrB transporter could heavily alter the specificity of the drug‐binding site.44 The mutation was then recreated in Salmonella typhimurium. It was found that S. typhimurium cells harboring the G288D mutant accumulated 25% less ciprofloxacin, as well as 70% and 80% more doxorubicin and minocycline compared with cells expressing the wild‐type AcrB tansporter.44 Thus, our data strongly suggest that both YdaH and MtrF are efflux pumps and these amino acids are important for their functions.
Expression of YdaH or MtrF decreases intracellular folic acid concentration
Our hypothesis is that both YdaH and MtrF are capable of catalyzing the efflux of PABA and related compounds. If this is correct, then cells expressing YdaH or MtrF should contain lower levels of folic acid. Therefore, we decided to measure the intracellular folic acid concentration of these cells microbiologically using Lactobacillus casei.45 E. coli BL21(DE3)ΔabgTΔpabA cells transformed with pET15bΩydaH, pET15bΩmtrF or pET15b were grown in liquid minimal medium supplemented with 30 nM PABA and harvested when the OD reached 0.5. Cells were then assayed to obtain their intracellular folic acid concentrations. Consistent with the results from the radioactive PABA accumulation assays, the folic acid concentration in E. coli BL21(DE3)ΔabgTΔpabA cells expressing either YdaH or MtrF were reduced in comparison with cells harboring the empty vector (Fig. 8). The data strongly support the idea that both YdaH and MtrF act as efflux pumps and participate in exporting PABA from the cell.
Figure 8.
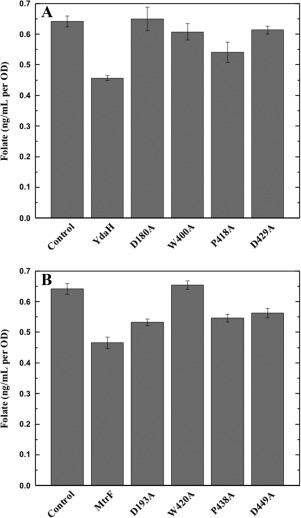
Intracellular folic acid concentration. (A) Folic acid concentration in E. coli BL21(DE3)ΔabgTΔpabA double knockout cells expressing YdaH were reduced in comparison with cells transformed with the empty vector. When transformed with plasmid expressing the mutant transporter, D180A, W400A, P418A, or D429A, folic acid production was increased in these cells. Each bar represents the mean of three separate cultures. (B) Folic acid concentration in E. coli BL21(DE3)ΔabgTΔpabA double knockout cells expressing MtrF were reduced in comparison with cells transformed with the empty vector. When transformed with plasmid expressing the mutant transporter, D193A, W420A, P438A, or D449A, folic acid production was increased in these cells. Each bar represents the mean of three separate cultures.
We also investigated how the YdaH single point mutants, D180A, W400A, P418A, or D429A, affect the intracellular folic acid concentration. When transformed with plasmid expressing the YdaH mutant transporter D180A, W400A, P418A, or D429A, folic acid production per OD was increased in these cells [Fig. 8(A)].
Likewise, the level of intracellular folic acid concentration in cells expressing the MtrF mutant D193A, W420A, P438A, or D449A was higher than that of the double knockout strain carrying wild‐type MtrF [Fig. 8(B)]. Again, the data indicate that these residues are important for the function of the YdaH and MtrF efflux pumps.
Both YdaH and MtrF behave as antibiotic efflux pumps
Both YdaH and MtrF appeared to lower the intracellular PABA concentration. However, as PABA is an important precursor for the production of the essential folic acid in bacteria, we did not understand why the function of YdaH and MtrF would export the PABA metabolite. Clearly, there was a significant reduction in PABA content, which led to the decrease in intracellular folic acid concentration in cells expressing the YdaH or MtrF efflux pump. We hypothesized that these efflux pumps may be able to protect bacterial cells by extruding antimetabolites that are structurally similar to PABA, such as sulfonamides. Sulfonamide antibiotics were popular in the late 1930s and early 1940s as a treatment for gonorrhea, but the rapid emergence of strains resistant to this class of drug resulted in its removal once penicillin became available.26 These resistant strains were later found to contain mutations in the folP gene,26 encoding the enzyme that catalyzes the last step of the folate synthesis pathway and the target for sulfonamide drugs. We therefore suspect that A. borkumensis YdaH and N. gonorrhoeae MtrF may act as drug efflux pumps, capable of extruding sulfonamide antimetabolites. Indeed, we found that the minimum inhibitory concentration (MIC) of sulfanilamide for the N. gonorrhoeae strain WV16 (an mtrF knockout strain) differed from that of the N. gonorrhoeae parental strain FA140 by twofold (MIC of 250 versus 500 μg/mL, respectively).41
To determine if A. borkumensis YdaH and N. gonorrhoeae MtrF behave as sulfonamide efflux pumps, we tested the effect of YdaH and MtrF expression on E. coli susceptibility to sulfonamides. Accordingly, we transformed BL21(DE3)ΔabgTΔpabA with pET15bΩydaH, pET15bΩmtrF or pET15b and tested the susceptibility of these transformants to four different sulfonamide drugs, sulfamethazine, sulfadiazine, sulfathiazole, and sulfanilamide (Tables 1 and 2). Specifically, sulfamethazine was chosen because of the availability of its radioactive analog. In many instances, expression of drug efflux pumps, including members of the RND32 and multidrug and toxic compound extrusion (MATE)46 families, can have only modest changes (twofold) in bacterial susceptibility to certain antimicrobials while more significant changes in susceptibility to other agents can be observed in the same system. We found that the BL21(DE3)ΔabgTΔpabA cells expressing A. borkumensis YdaH were 32‐fold less sensitive to sulfamethazine and eightfold more resistant to sulfanilamide when compared with BL21(DE3)ΔabgTΔpabA cells containing the empty pET15b vector. In addition, the susceptibilities of BL21(DE3)ΔabgTΔpabA/pET15bΩydaH cells to sulfadiazine and sulfathiazole were reduced by more than eight times in comparison with those of BL21(DE3)ΔabgTΔpabA cells carrying pET15b (Table 1). These data indeed show that YdaH functions as a drug efflux pump, which confers resistance to a variety of sulfonamides. Similar MIC values of these four sulfonamide drugs were also seen with the MtrF transporter (Table 2), supporting the idea that MtrF also functions as a sulfonamide efflux pump.
Table 1.
MICs of Sulfamethazine, Sulfadiazine, Sulfathiazole, and Sulfanilamide for Different YdaH Variants Expressed in E. coli BL21(DE3)ΔabgTΔpabA
| Gene in BL21(DE3)ΔabgTΔpabA | Sulfamethazine (μg/mL) | Sulfadiazine (μg/mL) | Sulfathiazole (μg/mL) | Sulfanilamide (μg/mL) |
|---|---|---|---|---|
| Empty vector | 62.5 | 31.25 | 62.5 | 500 |
| ydaH (wild‐type) | 2000 | >250 | >500 | 4000 |
| ydaH (D180A) | 1000 | 31.25 | 62.5 | 2000 |
| ydaH (W400A) | 62.5 | 31.25 | 62.5 | 2000 |
| ydaH (P418A) | 250 | 31.25 | 62.5 | 2000 |
| ydaH (D429A) | 62.5 | 31.25 | 62.5 | 2000 |
Table 2.
MICs of Sulfamethazine, Sulfadiazine, Sulfathiazole, and Sulfanilamide for Different MtrF Variants Expressed in E. coli BL21(DE3)ΔabgTΔpabA
| Gene in BL21(DE3)ΔabgTΔpabA | Sulfamethazine (μg/mL) | Sulfadiazine (μg/mL) | Sulfathiazole (μg/mL) | Sulfanilamide (μg/mL) |
|---|---|---|---|---|
| Empty vector | 62.5 | 31.25 | 62.5 | 500 |
| mtrF (wild‐type) | 2000 | >250 | >500 | 4000 |
| mtrF (D193A) | 1000 | 31.25 | 62.5 | 2000 |
| mtrF (W420A) | 125 | 31.25 | 62.5 | 2000 |
| mtrF (P438A) | 62.5 | 31.25 | 62.5 | 1000 |
| mtrF (D449A) | 62.5 | 31.25 | 62.5 | 1000 |
To determine the strength of transporter‐sulfonamide interaction, we used isothermal titration calorimetry (ITC) to quantify the binding affinity of sulfamethazine to YdaH. The data indicate an equilibrium dissociation constant, K D, of 0.41 ± 0.03 μM. The titration is characterized by a negative enthalpic contribution (ΔH = −1114.0 ± 16.2 kcal/mol), which yields a hyperbolic binding curve. The entropic contribution (ΔS) of this binding reaction was found to be 25.5 cal mol−1 deg−1 (Fig. 9). Similarly, ITC data suggest that the MtrF transporter specifically binds sulfanilamide with a K D of 1.14 ± 0.01 μM (Fig. 10).
Figure 9.
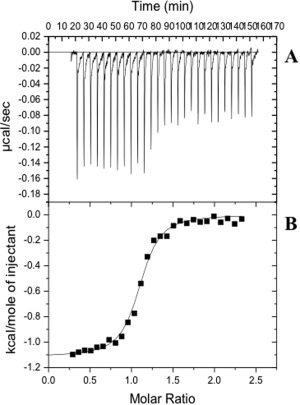
Representative isothermal titration calorimetry for the binding of sulfamethazine to YdaH. (A) Each peak corresponds to the injection of 10 μL of 20 μM monomeric YdaH in buffer containing 20 mM Tris‐HCl pH 7.5, 50 mM NaCl, and 0.03% DDM into the reaction containing 0.4 mM sulfamethazine in the same buffer. (B) Cumulative heat of reaction is displayed as a function of the injection number. The solid line is the least‐square fit to the experimental data, giving a K D of 0.41 ± 0.03 μM.
Figure 10.
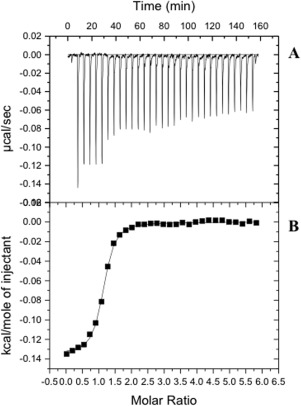
Representative isothermal titration calorimetry for the binding of sulfanilamide to MtrF. (A) Each peak corresponds to the injection of 10 μL of 40 μM monomeric MtrF in buffer containing 20 mM Tris‐HCl pH 7.5 and 0.03% DDM into the reaction containing 1.0 mM sulfanilamide in the same buffer. (B) Cumulative heat of reaction is displayed as a function of the injection number. The solid line is the least‐square fit to the experimental data, giving a K D of 1.14 ± 0.01 μM.
The binding affinities of sulfadiazine, sulfathiazole and sulfanilamide for the YdaH and MtrF transporters were also determined using ITC, suggesting that these drug‐transporter interactions are within the micromolar range (Tables 3 and 4). These ligand‐binding experiments indeed confirm that both YdaH and MtrF are capable of recognizing these antimetabolite sulfonamides.
Table 3.
Binding of Sulfamethazine, Sulfadiazine, Sulfathiazole, and Sulfanilamide by YdaH
| K D (μM) | ΔH (kcal mol−1) | ΔS (cal mol−1 deg−1) | |
|---|---|---|---|
| Sulfamethazine | 0.41 ± 0.03 | −1114.0 ± 16.2 | 25.5 |
| Sulfadiazine | 7.04 ± 0.54 | −403.1 ± 17.9 | 22.2 |
| Sulfathiazole | 0.60 ± 0.02 | −2540.0 ± 91.9 | 19.9 |
| Sulfanilamide | 4.97 ± 0.26 | −467.0 ± 22.6 | 22.7 |
Table 4.
Binding of Sulfamethazine, Sulfadiazine, Sulfathiazole, and Sulfanilamide by MtrF
| K D (μM) | ΔH (kcal mol−1) | ΔS (cal mol−1 deg−1) | |
|---|---|---|---|
| Sulfamethazine | 0.33 ± 0.02 | −580.2 ± 5.9 | 27.7 |
| Sulfadiazine | 12.74 ± 0.62 | −1900.0 ± 131.8 | 16.0 |
| Sulfathiazole | 1.52 ± 0.07 | −267.3 ± 8.0 | 25.7 |
| Sulfanilamide | 1.14 ± 0.01 | −135.2 ± 1.1 | 26.7 |
As a single point mutation on these transporters can have an impact on intracellular PABA and folic acid concentrations, we decided to investigate if these mutant transporters are able to alter the binding of antimetabolite drugs. Particularly, we determined the binding affinities of sulfamethazine, sulfadiazine, sulfathiazole, and sulfanilamide for the MtrF W420A mutant transporter using ITC. The data depict that the binding affinities of the W420A mutant for these sulfonamides are much weaker than those of the wild‐type MtrF transporter (Table 5), suggesting that residue W420 is critical for recognizing antimetabolite drugs.
Table 5.
Binding of Sulfamethazine, Sulfadiazine, Sulfathiazole, and Sulfanilamide by the MtrF W420A Mutant
| K D (μM) | ΔH (kcal mol−1) | ΔS (cal mol−1 deg−1) | |
|---|---|---|---|
| Sulfamethazine | 10.78 ± 1.17 | −233.4 ± 10.7 | 21.9 |
| Sulfadiazine | 105.82 ± 25.6 | −41103 ± 2065.0 | 4.4 |
| Sulfathiazole | 50.76 ± 8.9 | −713.5 ± 28.4 | 17.3 |
| Sulfanilamide | 6.80 ± 1.5 | −63.1 ± 3.0 | 23.4 |
In order to further test the drug efflux capability of YdaH, we expressed the mutant transporters D180A, W400A, P418A, and D429A in BL21(DE3)ΔabgTΔpabA and determined their ability to confer sulfamethazine, sulfadiazine, sulfathiazole, and sulfanilamide resistance. The mutation of each of these residues was found to produce hypersensitivity to sulfonamides in the cells that expressed them, compared with those expressing wild‐type YdaH (Table 1).
When the mutant MtrF transporters D193A, W420A, P438A, and D449A were expressed in BL21(DE3)ΔabgTΔpabA, we also found that the cells were more susceptible to all sulfonamides tested, compared with those expressing wild‐type MtrF (Table 2). Thus, these residues should be essential to the function of MtrF. Taken together, these data indicate that YdaH and MtrF function as a drug efflux pumps and reduce bacterial susceptibility to sulfonamides.
Both YdaH and MtrF are capable of reducing intracellular accumulation of sulfonamide
To support the drug susceptibility testing results, we measured the accumulation of radioactive sulfamethazine in BL21(DE3)ΔabgTΔpabA cells that carried pET15bΩydaH, pET15bΩmtrF or pET15b. For these experiments, we compared the accumulation of [3H]‐sulfamethazine per OD in these cells. As shown in Figure 11, the results indicated a lower level of [3H]‐sulfamethazine accumulation in BL21(DE3)ΔabgTΔpabA cells producing A. borkumensis YdaH or N. gonorrhoeae MtrF compared with controlled cells harboring the empty pET15b vector, suggesting that both YdaH and MtrF are capable of exporting sulfamethazine from cells.
Figure 11.
Accumulation of radioactive sulfamethazine. (A) E. coli BL21(DE3)ΔabgTΔpabA cells expressing YdaH show a significant decrease in [3H]‐sulfamethazine accumulation when compared with cells carrying the empty vector. When transformed with plasmids expressing the mutant transporters, D180A, W400A, P418A, and D429A, the levels of intracellular [3H]‐sulfamethazine accumulation were much higher than that of cells expressing wild‐type YdaH. Each bar represents the mean of three different cultures. (B) E. coli BL21(DE3)ΔabgTΔpabA cells expressing MtrF show a significant decrease in [3H]‐sulfamethazine accumulation when compared with cells carrying the empty vector. When transformed with plasmids expressing the mutant transporters, D193A, W420A, P438A, and D449A, the levels of intracellular [3H]‐sulfamethazine accumulation were much higher than that of cells expressing wild‐type MtrF. Each bar represents the mean of three different cultures.
When transformed with plasmid expressing the YdaH mutant transporter D180A, W400A, P418A or D429A, the level of intracellular sulfamethazine accumulation per OD was much higher than that of cells expressing wild‐type YdaH [Fig. 11(A)]. Similarly, cells transformed with plasmid expressing the MtrF mutant D193A, W420A, P438A, or D449A showed a much higher level of intracellular sulfamethazine accumulation per OD in comparison with cells expressing wild‐type MtrF [Fig. 11(B)]. Again, these data indicate that these conserved residues are critical for the function of the A. borkumensis YdaH and N. gonorrhoeae MtrF efflux pumps.
YdaH behaves as a drug efflux pump that is PMF and Na+ dependent
It has been suggested that members of the AbgT family use the proton‐motive‐force (PMF) to transport substrates across the membrane.19 Thus, loss of the PMF may inactivate pump activity, which results in enhanced accumulation of substrates. Accordingly, we measured the level of intracellular sulfamethazine accumulation in the presence of carbonyl cyanide m‐chlorophenylhydrazone (CCCP), an uncoupler of the membrane proton gradient. After the addition of CCCP into the assay solution, the accumulation of [3H]‐sulfamethazine increased drastically in the YdaH‐expressing cells [Fig. 12(A)], suggesting that the uncoupler CCCP was able to reduce the function of this pump.
Figure 12.
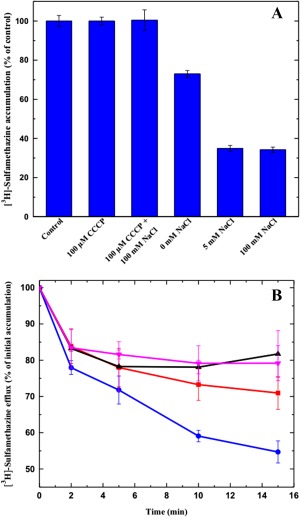
Sodium ion enhances sulfamethazine efflux via YdaH. (A) Accumulation of radioactive sulfamethazine in BL21(DE3)ΔabgTΔpabA/pET15bΩydaH cells with difference sodium ion concentrations. Cells showed a significant decrease in [3H]‐sulfamethazine accumulation in the presence of Na+. (B) Efflux of radioactive sulfamethazine in BL21(DE3)ΔabgTΔpabA/pET15bΩydaH cells in the presence of sodium or potassium ions. The presence of Na+ significantly enhances [3H]‐sulfamethazine efflux in BL21(DE3)ΔabgTΔpabA/pET15bΩydaH cells (black, control cells with the empty vector; red, 0 mM NaCl; magenta, 5 mM KCl; blue, 5 mM NaCl). The data showed in (A) and (B) are the cumulative average of three successive recordings. Error bars denote standard deviation (n = 3).
That CCCP appears to inhibit the pump activity indicates that YdaH is PMF‐dependent. However, it has been observed that proton mediated secondary transporters can also be regulated by Na+ ion. Indeed, our lab has shown that the multidrug and toxic compound extrusion (MATE)‐family transporter N. gonorrhoeae NorM is hindered by CCCP, but also sodium ion dependent.47 As a bound Na+ ion was found in the crystal structure of each subunit of the YdaH dimer, we investigated if the presence of Na+ affects intracellular drug accumulation. Specifically, we decided to determine the accumulation level of radioactive sulfamethazine in strain BL21(DE3)ΔabgTΔpabA/pET15bΩydaH in the presence of 5 or 100 mM NaCl. In both cases, the levels of accumulation of [3H]‐sulfamethazine were significantly attenuated when compared with that in the YdaH‐producing strain without the addition of Na+ ions [Fig. 12(A)].
To confirm that the presence of Na+ enhances the transport function of YdaH, we measured the efflux of radioactive sulfamethazine, both in the absence and presence of Na+ or K+, in the strain BL21(DE3)ΔabgTΔpabA/pET15bΩydaH, Cells were first loaded with [3H]‐sulfamethazine in the presence of CCCP, which inhibited the pump. Then, CCCP was removed, glucose was added to re‐energize these cells and cellular [3H]‐sulfamethazine levels were measured over time, both in the absence and presence of 5 mM NaCl or KCl. As shown in Figure 12(B), the addition of Na+ has a strong effect on sulfamethazine efflux in BL21(DE3)ΔabgTΔpabA/pET15bΩydaH. The data indicated that A. borkumensis YdaH functions more efficiently in the presence of Na+ ions. On the other hand, our results showed that the addition of K+ ions had no effect on sulfamethazine efflux.
MtrF behaves as a PMF, but not Na+, dependent drug efflux pump
To elucidate if N. gonorrhoeae MtrF is also a PMF‐dependent transporter, we used the same approach to measure the level of intracellular sulfamethazine accumulation. In the presence of CCCP, the accumulation of [3H]‐sulfamethazine increased drastically in the BL21(DE3)ΔabgTΔpabA cells expressing MtrF [Fig. 13(A)]. Thus, it is likely that MtrF is also a PMF‐dependent efflux pump.
Figure 13.
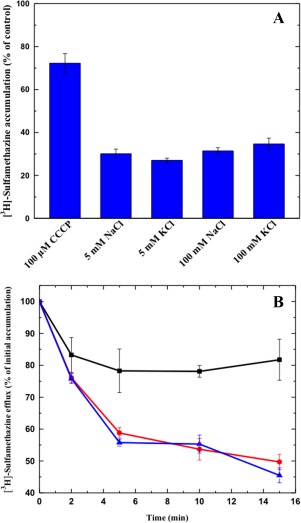
Transport of sulfamethazine via MtrF. (A) Accumulation of radioactive sulfamethazine in BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF cells with different sodium or potassium ion concentrations. The data indicate that the transport function of MtrF is independent of sodium or potassium ions. (B) Efflux of radioactive sulfamethazine in BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF cells in the absence and presence of sodium ions. The presence of Na+ does not affect [3H]‐sulfamethazine efflux in BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF cells (black, controlled cells transformed with empty vector; red, 0 mM NaCl; blue, 5 mM NaCl). The data showed in (A) and (B) are the cumulative average of three successive recordings. Error bars denote standard deviation (n = 3).
As the YdaH transporter is Na+ dependent, we decided to investigate the accumulation level of radioactive sulfamethazine in strain BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF in the presence of NaCl or KCl. The concentrations of these metal ions were either 5 or 100 mM. Surprisingly, in all cases, the levels of accumulation of [3H]‐sulfamethazine were similar to that in the MtrF‐producing strain without the addition of any metal ions [Fig. 13(A)]. The results indicated that the function of MtrF is independent of Na+ or K+. It appears that the mechanisms for energy coupling in the YdaH and MtrF pumps are quite different.
To further test the possibility that MtrF is a PMF‐dependent efflux pump, we next measured the efflux of [3H]‐sulfamethazine that had accumulated in strain BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF over time, both in the absence and presence of Na+. Like the case of YdaH, Cells expressing MtrF were first loaded with [3H]‐sulfamethazine and CCCP was added to inhibit the pump. Cells were then re‐energized by removing CCCP and adding glucose; thereafter, radioactive measurements were performed both in the absence and presence of 5 mM NaCl. As shown in Figure 13(B), the addition of Na+ essentially has no effect on sulfamethazine efflux in BL21(DE3)ΔabgTΔpabA/pET15bΩmtrF. These data suggest that MtrF is a PMF, but not Na+, dependent efflux pump.
Conclusion
The dimeric structures of A. borkumensis YdaH and N. gonorrhoeae MtrF represent the first two crystal structures of the AbgT family of transporters. They reveal overall unique folding that is distinct from other membrane proteins. We found that both YdaH and MtrF are drug efflux pumps, capable of removing sulfonamide antimetabolites from the bacterial cell and mediating resistance to this class of drugs in bacterial cells. Although both YdaH and MtrF belong to the same family, the energy coupling schemes are quite different from each other. On the basis of our experimental data, it is possible that the AbgT family of transporters is a novel family of antimetabolite efflux pumps, which is able to protect bacterial cells against these toxic compounds. The AbgT‐family transporters may be important targets for the design of novel antibiotics to fight against bacterial infections.
References
- 1. Henderson GB, Huennekens FM (1986) Membrane‐associated folate transport proteins. Methods Enzymol 122:260–269. [DOI] [PubMed] [Google Scholar]
- 2. Hyde JE, Dittrich S, Wang P, Sims PFG, de Crécy‐Lagard V, Hanson AD (2008) Plasmodium falciparum: a paradigm for alternative folate biosynthesis in diverse microorganisms? Trends Parasit 24:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pyne C, Bognar AL (1992) Replacement of the folC gene, encoding folylpolyglutamate synthetase‐dihydrofolate synthetase in Escherichia coli, with genes mutagenized in vitro. J Bacteriol 174:1750–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woods DD (1940) The relationship of p‐aminobenzoic acid to the mechanism of the action of sulphanilamide. Br J Exp Pathol 21:74–90. [Google Scholar]
- 5. Bermingham A, Derrick JP (2002) The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioassays 24:637–648. [DOI] [PubMed] [Google Scholar]
- 6. Singer M, Nambiar S, Valappil T, Higgins K, Gitterman S (2008) Historical and regulatory perspectives on the treatment effect of antibacterial drugs for community‐acquired pneumonia. Clin Infect Dis 47:S216–S224. [DOI] [PubMed] [Google Scholar]
- 7. Cartwright KAV, editor (1995). Meningococcal disease. Chichester: Wiley. [Google Scholar]
- 8. Wang P, Read M, Sims PFG, Hyde JE (1997) Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol Microbiol 23:979–986. [DOI] [PubMed] [Google Scholar]
- 9. Triglia T, Cowman AF (1994) Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum . Proc Natl Acad Sci USA 91:7149–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Triglia T, Menting JG, Wilson C, Cowman AF (1997) Mutations in dihydropteroate synthase are responsible for sulfon and sulfonamide resistance in Plasmodium falciparum . Proc Natl Acad Sci USA 94:13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown GM, Weisman RA, Molnar DA (1961) The biosynthesis of folic acid: I. Substrate and cofactor requirements for enzymatic synthesis by cell‐free extracts of Escherichia coli . J Biol Chem 236:2534–2543. [Google Scholar]
- 12. Brown GM (1962) The biosynthesis of folic acid: II. Inhibition by sulfonamides. J Biol Chem 237:536–540. [PubMed] [Google Scholar]
- 13. Reynolds JJ, Brown GM (1964) The biosynthesis of folic acid. IV. Enzymatic synthesis of dihydrofolic acid from guanine and ribose compounds. J Biol Chem 239:317–325. [PubMed] [Google Scholar]
- 14. Burg AW, Brown GM (1966) The biosynthesis of folic acid VI. Enzymatic conversion of carbon atom 8 of guanosine triphosphate to formic acid. Biochim Biophys Acta 117:275–278. [DOI] [PubMed] [Google Scholar]
- 15. Richey DP, Brown GM (1969) The biosynthesis of folic acid. IX. Purification and properties of the enzymes required for the formation of dihydropteroic acid. J Biol Chem 244:1582–1592. [PubMed] [Google Scholar]
- 16. Mathis JB, Brown GM (1970) The biosynthesis of folic acid. XI. Purification and properties of dihydroneopterin aldolase. J Biol Chem 245:3015–3025. [PubMed] [Google Scholar]
- 17. Hussein MJ, Green JM, Nichols BP (1998) Characterization of mutations that allow p‐aminobenzoyl‐glutamate utilization by Escherichia coli . J Bacteriol 180:6260–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carter EL, Jager L, Gardner L, Hall CC, Willis S, Green JM (2007) Escherichia coli abg genes enable uptake and cleavage of the folate catabolite p‐aminobenzoyl‐glutamate. J Bacteriol 189:3329–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prakash S, Cooper G, Singhi S, Saier MHJ (2003) The ion transporter superfamily. Biochim Biophys Acta 1618:79–92. [DOI] [PubMed] [Google Scholar]
- 20. Veal WL, Shafer WM (2003) Identification of a cell envelope protein (MtrF) involved in hydrophobic antimicrobial resistance in Neisseria gonorrhoeae . J Antimicrob Chemother 51:27–37. [DOI] [PubMed] [Google Scholar]
- 21. Folster JP, Shafer WM (2005) Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high‐level antimicrobial resistance. J Bacteriol 187:3713–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee EH, Shafer WM (1999) The farAB‐encoded efflux pump mediates resistance of gonococci to long‐chained antibacterial fatty acids. Mol Microbiol 33:7753–7758. [DOI] [PubMed] [Google Scholar]
- 23. Rouquette‐Loughlin C, Dunham SA, Kuhn M, Balthazar JT, Shafer WM (2003) The NorM efflux pump of by Neisseria gonorrhoeae and Neisseria meningitides recognizes antimicrobial cationic compounds. J Bacteriol 185:1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shafer WM, Qu X, Waring AJ, Lehrer RI (1998) Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci USA 95:1829–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shafer WM, Veal WL, Lee EH, Zarentonelli L, Balthazar JT, Rouquette C (2001) Genetic organization and regulation of antimicrobial efflux systems possessed by Neisseria gonorrhoeae and Neisseria meningitidis . J Mol Microbiol Biotechnol 3:219–225. [PubMed] [Google Scholar]
- 26. Unemo M, Shafer WM (2014) Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 27:587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delahay RM, Robertson BD, Balthazar JT, Ison CA (1997) Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology 143:2127–2133. [DOI] [PubMed] [Google Scholar]
- 28. Lucas CE, Hagman KE, Levin JC, Stein DC, Shafer WM (1995) Importance of lipooligosaccharide structure in determining gonococcal resistance to hydrophobic antimicrobial agents resulting from the mtr efflux system. Mol Microbiol 16:1001–1009. [DOI] [PubMed] [Google Scholar]
- 29. Hagman KE, Shafer WM (1995) Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae . J Bacteriol 177:4162–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hagman KE, Lucas CE, Balthazar JT, Snyder LA, Nilles M, Judd RC, Shafer WM (1997) The MtrD protein of Neisseria gonorrhoeae is a member of resistance/nodulation/division protein family constituting part of an efflux system. Microbiology 143:2117–2125. [DOI] [PubMed] [Google Scholar]
- 31. Warner DM, Shafer WM, Jerse AE (2008) Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC‐MtrD‐MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol Microbiol 70:462–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MHJ (1999) The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development protein. J Mol Microbiol Biotechnol 1:107–125. [PubMed] [Google Scholar]
- 33. Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM (1995) Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611–622. [DOI] [PubMed] [Google Scholar]
- 34. Bolla JR, Su C, Do SV, Radhakrishnan A, Kumar N, F. Long F, Chou T, Delmar JA, Lei H, Rajashankar KR, Shafer WM, Yu EW (2014) Crystal structure of the Neisseria gonorrhoeae MtrD inner membrane multidrug efflux pump. PLoS One 9:e97903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Veal WL, Nicholas RA, Shafer WM (2002) Overexpression of the MtrC‐MtrD‐MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae . J Bacteriol 184:5619–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Janganan TK, Bavro VN, Zhang L, Borges‐Walmsley MI, Walmsley AR (2013) Tripartite efflux pumps: energy is required for dissociation, but not assembly or opening of the outer membrane channel of the pump. Mol Microbiol 88:590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Janganan TK, Zhang L, Bavro VN, Matak‐Vinkovic D, Barrera NP, Burton MF, Steel PG, Robinson CV, Borges‐Walmsley MI, Walmsley AR (2011) Opening of the outer membrane protein channel in tripartite efflux pumps is induced by interaction with the membrane fusion partner. J Biol Chem 286:5484–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lei H, Chou T, Su C, Bolla JR, Kumar N, Radhakrishnan A, Long F, Delmar JA, Do SV, Rajashankar KR, Shafer WM, Yu EW (2014) Crystal structure of the open state of the Neisseria gonorrhoeae MtrE outer membrane channel. PLoS One 9:e97475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schneiker S, dos Santos VAPM, Bartels D, Bekel T, Brecht M, Buhrmester J, Chernikova TN, Denaro R, Ferrer M, Gertler C, Goesmann A, Golyshina OV, Kaminski F, Khachane AN, Lang S, Linke B, McHardy AC, Meyer F, Nechitaylo T, Pühler A, Regenhardt D, Rupp O, Sabirova JS, Selbitschka W, Yakimov MM, Timmis KN, Vorhölter F‐J, Weidner S, Kaiser O, Golyshin PN (2015) Genome sequence of the ubiquitous hydrocarbon‐degrading marine bacterium Alcanivorax borkumensis . Nat Biotech 24:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bolla JR, Su C, Delmar JA, Radhakrishnan A, Kumar N, Chou T, Long F, Rajashankar KR, Yu EW (2015) Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology. Nat Commun 6:6874. doi:10.1038/ncomms7874). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Su C, Bolla JR, Kumar N, Radhakrishnan A, Long F, Delmar JA, Chou T, Rajashankar KR, Shafer WM, Yu EW (2015) Structure and function of Neisseria gonorrhoeae MtrF illuminates a class of antimetabolite efflux pumps. Cell Rep 11:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nayal M, Di Cera E (1996) Valence screening of water in protein crystals reveals potential Na+ binding sites. J Mol Biol 256:228–234. [DOI] [PubMed] [Google Scholar]
- 43. Kaplan JB, Nichols BP (1986) Nucleotide sequence of Escherichia coli pabA and its evolutionary relationship to trp(G)D. J Mol Biol 168:451–468. [DOI] [PubMed] [Google Scholar]
- 44. Blair JMA, Bavro VN, Ricci V, Modi N, Cacciotto P, Klinekathöfer U, Ruggerone P, Vargiu AV, Baylay AJ, Smith HE, Brandon Y, Galloway D, Piddock LJV (2015) AcrB drug‐binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc Natl Acad Sci 112:3511–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilson SD, Horne WD (1982) Use of glycerol‐crypotected Lactobacillus casei for microbiological assay of folic acid. Clin Chem 28:1198–1200. [PubMed] [Google Scholar]
- 46. Brown MH, Paulsen IT, Skurray RA (1999) The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol 31:394–395. [DOI] [PubMed] [Google Scholar]
- 47. Long F, Rouquette‐Loughlin C, Shafer WM, Yu EW (2008) Functional cloning and characterization of the multidrug efflux pumps NorM from Neisseria gonorrhoeae and YdhE from Escherichia coli . Antimicrob Agents Chemother 52:3052–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]


