Abstract
Glucose is the primary fuel to life on earth. Cellular uptake of glucose is a fundamental process for metabolism, growth, and homeostasis. Three families of secondary glucose transporters have been identified in human, including the major facilitator superfamily glucose facilitators GLUTs, the sodium‐driven glucose symporters SGLTs, and the recently identified SWEETs. Structures of representative members or their prokaryotic homologs of all three families were obtained. This review focuses on the recent advances in the structural elucidation of the glucose transporters and the mechanistic insights derived from these structures, including the molecular basis for substrate recognition, alternating access, and stoichiometric coupling of co‐transport.
Keywords: glucose transporter, GLUT, SGLT, SWEET, alternating access, structural biology
Introduction
Glucose is the primary energy source for life on earth. It also serves as an important precursor for biomolecule synthesis, and plays an important role in cell signaling. The dietary carbohydrates contain large amount of glucose in the form of monosaccharide or oligo‐ and polysaccharides, such as maltose, starch, and glycogen. The human brain, which generally represents about 2% of total body mass of an adult, consumes approximately 25% of glucose supply. The cellular uptake of glucose precedes other events concerning glucose metabolism and signaling. Multiple glucose transporters have evolved to shepherd the transmembrane movement of the water‐soluble glucose molecules.
Three families of solute carriers have been identified for glucose transport, including the most rigorously studied major facilitator superfamily (MFS) glucose facilitators GLUTs (SLC2), the sodium‐driven glucose symporters SGLTs (SLC5), and the recently characterized SWEET (SLC50).1, 2, 3, 4 The three glucose transporter families have distinct physiological functions and working mechanisms, which are illustrated in a simplified model [Fig. 1(A)]. GLUTs are ubiquitously distributed and catalyze facilitative diffusion of glucose down its concentration gradient. SGLTs are primarily expressed in the intestine and kidney cortex and harness the energy released from downhill flow of Na+ to drive the translocation of glucose against its concentration gradient across membrane.5, 6 The physiological function of sugars will eventually be exported transporters (SWEETs), which are mainly responsible for efflux and intracellular trafficking of sugars in plants, remains to be characterized in mammals.7
Figure 1.
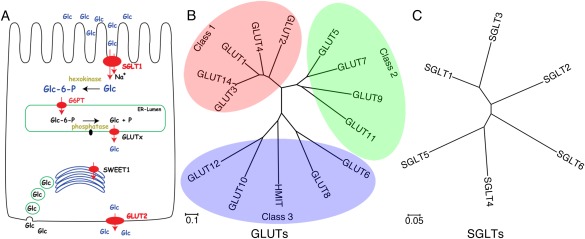
Glucose transporters in Homo sapiens. (A) The distribution of glucose transporters in human intestinal epithelial cell. The panel is adapted from the Supplementary Figure 22 by Chen et al.7 (B, C) The phylogenetic trees of GLUTs and SGLTs in Homo sapiens. Multiple sequence alignment was performed with ClustalW and the results were presented with MEGA6. Fourteen members of GLUTs and six SGLTs were shown here. The relevant NCBI sequence numbers of the 14 GLUTs and 6 SGLTs are listed below: GLUT1 (NP_006507.2), GLUT2 (NP_000331.1), GLUT3 (NP_008862.1), GLUT4 (NP_001033.1), GLUT5 (NP_003030.1), GLUT6 (NP_060055.2), GLUT7 (NP_997303.2), GLUT8 (NP_055395.2), GLUT9 (NP_064425.2), GLUT10 (NP_110404.1), GLUT11 (NP_001020109.1), GLUT12 (NP_660159.1), HMIT (NP_443117.3), GLUT14 (NP_703150.1), SGLT1 (NP_000334.1), SGLT2 (NP_003032.1), SGLT3 (NP_055042.1), SGLT4 (NP_001011547.2), SGLT5 (NP_689564.3), and SGLT6/SMIT2 (NP_443176.2).
To elucidate their mechanisms and to identify potentially novel targets for drug development, the structures of glucose transporters have been actively pursued. However, membrane proteins are notoriously known as the most challenging category for structural determination because of the insurmountable technical difficulties associated with expression, purification, and crystallization.8, 9, 10 The progress on the structural investigation of transporters is even slower due to their highly dynamic nature and low endogenous expression in general. The first structures of transporters were reported in 2002.11, 12, 13 Eukaryotic transporters represent qualitatively more challenging targets. For instance, among the over 1000 published structures of membrane proteins, only <20 are of eukaryotic transporters, among which merely three are of human origin.8, 14, 15, 16 Bacterial homologs have been exploited as surrogate for structural and mechanistic understanding of the corresponding eukaryotic targets.17
Among the three families of glucose transporters, structural investigation of GLUTs is most advanced with the structures resolved for human GLUT1and GLUT3,14, 15 two mammalian GLUT5 homologs,18 and the bacterial homologs XylE and GlcPSe.19, 20, 21, 22 For SGLTs and SWEETS, only structures of bacterial homologs, vSGLT and SemiSWEETs, were obtained.23, 24, 25, 26, 27
Here we briefly summarize the physiological function of the three glucose transporter families and review the recent progress on the structural investigations, upon which we discuss the updated understanding of the molecular basis for alternating access, substrate recognition, and coupled transport mechanism of these glucose transporters.
Physiological Functions of Glucose Transporters
GLUTs
GLUTs are encoded by the SLC2 genes. Fourteen members have been identified in human, which are classified into three classes [Fig. 1(B)].1 The most rigorously investigated GLUT1–4, together with GLUT14, constitute class 1.28 GLUT1 is one of the first membrane transporters to be cloned and has been comprehensively characterized in the past half century. It is a ubiquitous glucose transporter in all tissues, but predominantly functions in erythrocytes and blood–brain barrier.28, 29 GLUT1 was also suggested to be the receptor for human T‐lymphotropic virus (HTLV) and play an essential role in CD4 T cell activation.30, 31 The transport activity of GLUT1 is subjected to regulation by PKC and TXNIP.32, 33 Several dozens of SLC2A1 mutations have been identified in patients with the autosomal dominant genetic disease known as GLUT1 deficiency syndrome.34, 35, 36, 37, 38
GLUT2 is the major glucose transporter in hepatocytes and intestine and exhibits several distinctive characteristics compared to the other three GLUTs.28, 39 It is the only GLUT that catalyzes the bidirectional glucose flow depending on fed or fasting state. GLUT2 exhibits an extraordinary high K m value for glucose yet a high affinity for glucosamine.40 Correlated with its primary role in liver, defects of GLUT2 are associated with the Fanconi–Bickel syndrome, also known as glycogen storage disease type XI.41
GLUT3 is mainly expressed in neurons, thereby also referred to as the “neuronal glucose transporter.”42 It shows the lowest K m value and the highest turnover rate among GLUT1–4.43 These properties may ensure sufficient glucose uptake to neurons in the brain where the glucose concentration is considerably lower than in blood.44 Overexpression of GLUT1 and GLUT3 is observed in a variety of solid tumor cells for enhanced glucose uptake under aerobic (the Warburg effect).45, 46, 47, 48, 49, 50 Targeting the Warburg effect, isotope‐labeled glucose derivative 2‐deoxy‐2‐[18F]fluoroglucose (18F‐FDG) was used for the positron emission tomography (PET) for diagnosis of cancer.51, 52, 53, 54 Inhibitors that target GLUTs to reduce glucose uptake and inhibit glycolysis were shown to reduce cancer cell growth both in vitro and in vivo.55, 56
GLUT4 is highly expressed in adipose tissues and skeletal muscles.57 It is most notoriously known as the “insulin‐responsive glucose transporter” for its relocation from intracellular vesicles to the plasma membrane upon insulin signaling.57, 58 Disruption of the regulated GLUT4 trafficking is associated with obesity and the type II diabetes mellitus. Interestingly, GLUT4 can be inhibited by HIV protease inhibitors, which may cause acute insulin resistance and result in increased diabetes incidence among AIDS patients treated with these inhibitors.59, 60
GLUT5, 7, 9, 11 constitute class 2. Unlike GLUT1–4, the primary substrate for GLUT5 is fructose.61 GLUT5 is mainly responsible for fructose absorption into the epithelial cells of intestine, while it is also expressed in other organs including kidney and brain.28 GLUT7 and GLUT11, sharing approximately 50% sequence identities with GLUT5, transport both glucose and fructose. GLUT9 was recently identified to be a urate transporter that may play a critical role in the deposition of uric acid in joints.62, 63 GLUT6, 8, 10, 12, and HMIT (myoinositol:H+ symporter, also known as GLUT13) constitute the least understood class 3 [Fig. 1(B)].28 The substrates and physiological functions of most of them remain to be characterized.
SGLTs
There are six SGLT proteins identified in human, among which SGLT1 and SGLT2 have been extensively characterized in the past several years [Fig. 1(C)].5 Despite the prominent sequence homology between SGLT1 and SGLT2, they exhibit distinct physiological and biochemical properties.64 SGLT1 is primarily expressed in intestine, while SGLT2 is highly expressed in the kidney cortex and plays an important role in reabsorption of glucose in kidney.5 The major substrates of SGLT1 are both glucose and galactose, whereas SGLT2 prefers glucose to galactose. The stoichiometric ratios of sodium coupling of SGLT1 and SGLT2 were characterized to be 2:1 and 1:1, respectively.64
SGLT1 mutations are found in patients with glucose–galactose malabsorption.65 Mutations of SGLT2 are associated with the genetic disease Familial Renal Glucosuria.66, 67 SGLT2 inhibitors, including dapagliflozin, canagliflozin, ipragliflozin, tofogliflozin, luseogliflozin, and empagliflozin, have been tested at different stages of clinical trials for treatment of type II diabetes mellitus.68, 69, 70
The characterizations of other SGLTs are rather preliminary. Interestingly, SGLT3 has no glucose transport activity, but may serve as a glucose sensor in the enteric nervous system.71 SGLT4 exhibits higher affinity for mannose than glucose and may function as a mannose transporter.72 SGLT5, which is highly expressed in kidney cortex, also has a substrate preference for mannose.73 SGLT6, also known as SMIT2 (myoinositol:Na+ symporter), transports inositol instead of glucose.74
SWEETs
As a novel family of glucose transporters recently identified in plants, the physiological characterizations of SWEETs in animals are preliminary. In contrast to Arabidopsis thaliana in which up to two dozen SWEETs have been identified, animals usually have only one SWEET, with the exception of Caenorhabditis elegans where seven SWEET‐encoding genes are found.3, 7 Please refer to a recent review for the updated knowledge of SWEETs and their bacterial homologs SemiSWEETs.3
Structural Elucidation of Glucose Transporters
GLUTs and SGLTs belong to the major facilitator superfamily (MFS) and the amino acid‐polyamine:organocation (APC) superfamily, respectively, and display the typical MFS and APC folds [Fig. 2(A,B)].17 Eukaryotic SWEETs are predicted to contain 7 transmembrane segments (TMs), while the bacterial homologs have only 3 TMs, thereby named SemiSWEETs. SemiSWEETs are the smallest transporters known and function as parallel dimer. SWEETs have an additional TM4 that connects the two parallel triple‐helix bundles [Fig. 2(C)].
Figure 2.
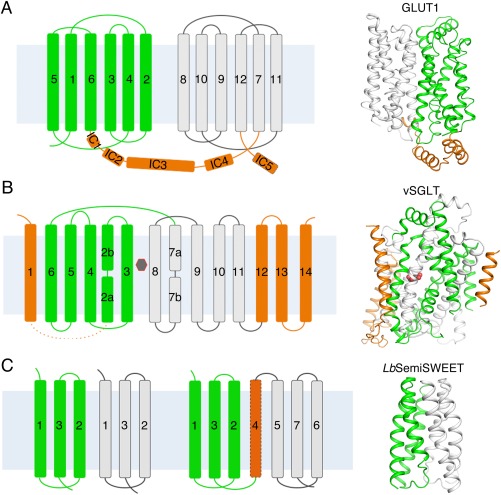
The structural folds and overall structures of glucose transporters. (A) Structure of GLUTs. The 12 transmembrane helices were divided into an N domain and a C domain, which are colored green and white, respectively. The intracellular helices domains (ICH) are colored orange. Shown on the right is the structure of the human GLUT1 (PDB accession code 4PYP). (B) Structure of SGLTs. The transmembrane helices TM2‐11 constitute the “5 + 5” inverted repeats of the LeuT‐fold of vSGLT. The two repeats are colored green and white. The additional transmembrane helices are colored orange. The substrate (galactose) is indicated by the black hexagon. Shown on the right is the structure of the inward‐occluded vSGLT (PDB accession code 3DH4). (C) Structure of SemiSWEETs. The topology of SemiSWEETs and predicted topology of SWEET1 in human are shown on the left. All SemiSWEETs form a functional dimer. Each protomer contains three transmembrane helices, which are arranged in a 1–3–2 pattern and colored as white or green. Shown on the right is the structure of LbSemiSWEET (PDB accession code 4QNC). In all the figures where overall structures are shown, the cytoplasmic side is at bottom if not otherwise indicated.
The structures of GLUTs have been pursued for nearly three decades ever since the cloning of GLUT1.29 Analysis of the primary sequence suggested a 12‐TM topology.54 Prior to the structure determination of GLUTs or its bacterial homologs, the structural information was derived from biochemical analysis,75 or modeling based on the structure of Escherichia coli lactose:H+ symporter LacY, a prototypical MFS protein that has little sequence similarity with GLUTs,76 and the glycerol:glycerol‐3‐phosphate antiporter GlpT.77 The first homology model of GLUT1 was obtained upon structural elucidation of XylE, a d‐xylose:H+ symporter in E. coli that shares approximately 30% identity and 50% similarity with GLUTs.19
The structures of XylE were first obtained in the outward‐occluded conformation in complex with three distinct ligands, the authentic substrate d‐xylose, the inhibitor glucose, and a glucose derivative.19 Then two more conformations of XylE were obtained, inward open and partly inward‐occluded.20 Conforming to the canonical MFS fold,6 12 TMs are organized into two discretely folded domains, the amino‐ (N) and carboxyl‐ (C) domains, which are related to each other around an axis that is perpendicular to the membrane plane. Each domain comprises a pair of 3‐TM bundles, which exhibit another fold of pseudosymmetry by approximate 180° rotation around an axis that is parallel to the membrane plane. Inverted repeats represent a common feature observed in a variety of membrane transporter families.17 In addition to the MFS fold in the transmembrane region, XylE, as later seen in GLUTs and GlcPSe, contains an intracellular helical (ICH) domain, which then proves to play an important role in the function of the transporters [Fig. 2(A) and Table 1]. GLUTs, XylE, and GlcPSe belong to the sugar porter (SP) subfamily within the MFS. The ICH domain may be a characteristic structural feature of the SP proteins because it hosts a number of the so‐called SP signature motifs.78, 79
Table 1.
Crystal Structures of Glucose Transporters and Related Proteins
| SLC family | Protein | Organism | Resolution (Å) | Ligands | Conformation | PBD accession code | Reference |
|---|---|---|---|---|---|---|---|
| SLC2 | GLUT1 | Homo sapiens | 3.2 | ‐ | Inward‐open | 4PYP | 14 |
| GLUT3 | Homo sapiens | 1.5 | d‐glucose | Outward‐occluded | 4ZW9 | 15 | |
| 2.4 | Maltose | Outward‐occluded | 4ZWB | 15 | |||
| 2.6 | Maltose | Outward‐open | 4ZWC | 15 | |||
| GLUT5 | Rattus norvegicus | 3.3 | Outward‐open | 4YBQ | 18 | ||
| Bos Taurus | 3.2 | Inward‐open | 4YB9 | 18 | |||
| XylE | Escherichia coli | 2.8 2.9 2.6 | d‐xylose d‐glucose 6‐Br‐glucose | Outward‐occluded | 4GBY 4GBZ 4GC0 | 19 | |
| 3.8 | ‐ | Partly inward occluded | 4JA3 | 20 | |||
| 4.2, 3.5 | ‐ | Inward‐open | 4J4A, 4QIQ | 20, 22 | |||
| GlcPse | Staphylococcus epidermidis | 3.2 | ‐ | Inward‐open | 4LDS | 21 | |
| SLC5 | vSGLT | Vebrio parahaemolyticus | 2.7 | d‐galactose | Inward‐occluded | 3DH4 | 23 |
| 2.7 | ‐ | Inward‐open | 2XQ2 | 24 | |||
| SLC50 | VsSemiSWEET | Vibrio sp. N418 | 1.7 | ‐ | Outward‐open | 4QND | 25 |
| LbSemiSWEET | Leptospira biflexa | 2.4 | ‐ | Occluded | 4QNC | 25 | |
| TySemiSWEET | Thermodesulfovibrio yellowstonii | 2.4 | ‐ | Occluded | 4RNG | 26 | |
| EcSemiSWEET | Escherichia coli | 3.0 | ‐ | Outward‐open | 4X5N | 27 | |
| 2.0, 3.0 | ‐ | Inward‐open | 4X5M, 4X5N | 27 |
The structure of human GLUT1 was eventually reported in 2014.14 The domain organization of GLUT1, which was captured in an inward‐open conformation, is similar to XylE [Fig. 2(A) and Table 1]. Recently, three structures of human GLUT3 were obtained, including that of the glucose‐bound GLUT3 in an outward‐occluded conformation at 1.5 Å resolution and those of GLUT3 in complex with the competitive inhibitor maltose in the outward‐open and ‐occluded states at 2.6 and 2.4 Å resolutions, respectively (Table 1).15 The structures of closely related GLUT1 and GLUT3 in three distinct conformations reveal the transport cycle of human glucose transporters and elucidate the molecular basis for substrate recognition. Most recently, the structures of rat and bovine GLUT5 were determined in the outward‐ and inward‐open states, respectively, and similar working model was proposed.18
The structural information of SGLTs came from the homolog vSGLT from Vibrio parahaemolyticus.23 The structures of vSGLT, which shares 32% sequence identity and 75% similarity with SGLT1, were obtained in two conformations, inward‐open and inward‐occluded, in the presence of a d‐galactose [Fig. 2(B) and Table 1].23, 24 SGLTs belong to the sodium solute symporter family (SSS), which is a member of the APC superfamily.80, 81 The core structure of APC members contains the so‐called “LeuT” fold, which consists of 10 TMs that are organized into a pair of “5 + 5” inverted repeats with both the N and C termini exposed to the cytoplasmic side of the membrane.82 Within each repeat, the first TM is usually a discontinuous helix, which is a common feature in secondary transporters and serves important functions in substrate recognition and transport.17, 83 In vSGLT, TM2‐6 and TM7‐11 constitute the LeuT fold, whereas four additional TMs are present on the periphery [Fig. 2(B)].
There was a structure boom for semiSWEETs since 2014 with five structures reported for four semiSWEETS homologs [Fig. 2(C) and Table 1], including VsSemiSWEET from Vibrio sp. N418 and LbSemiSWEET from Leptospira biflexa in the outward‐open and ‐occluded states, respectively,25 TySemiSWEET from Thermodesulfovibrio yellowstonii DSM 11347 in an occluded state,26 and EcSemiSWEET from E. coli in both inward‐ and outward‐open conformaitons.27 Despite the lack of significant sequence similarities, these five SemiSWEET homologs exhibit identical structural fold [Fig. 2(C)]. Biochemical studies support the structurally revealed dimeric organization of these SemiSWEETs. In each protomer, the three TMs, named the three‐helix bundle (THB), are arranged in a 1–3–2 pattern [Fig. 2(C)].25
Alternating Access of Glucose Transporters
In 1966, Jardetzky proposed an allosteric model for membrane transporters later known as the alternating access mechanism.84 Based on this generic model, the substrate binding site is alternately exposed to the two sides of the membrane, that is, substrate(s) bind to the transporter from one side of the membrane and are released from opposite side after conformational changes of the transporters. According to this model, a membrane transporter has to undergo cycles of conformational changes between outward‐open and inward‐open states. Multidisciplinary methods have been used to measure this process, including molecular dynamic simulation and single‐molecule FRET.85, 86, 87, 88 Nevertheless, capturing transporters in different conformations would provide the most straightforward evidence to test this model. The structural characterizations of glucose transporters, particularly those of GLUTs and semiSWEETS, have revealed the molecular basis for their respective alternating access mechanism (Fig. 3).
Figure 3.
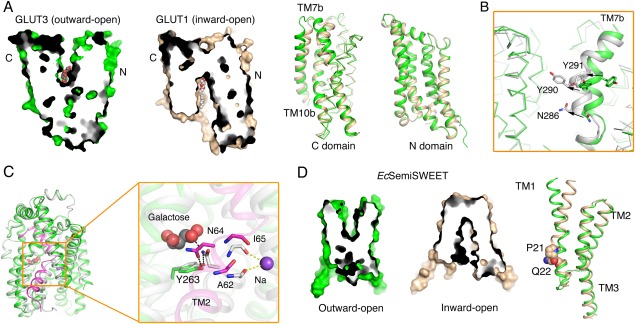
The alternating access mechanism of GLUTs, vSGLT, and SemiSWEET. (A) Structural comparison of the outward‐open GLUT3 and the inward‐open GLUT1 (PDB accession codes 4ZWC and 4PYP, respectively). Shown on the left are the cut‐open views of the surface representation of GLUT1 and GLUT3. Structural superposition of their C domains reveals local shifts in TM7b and TM10b, while the N domains remain nearly unchanged. (B) Local shifts of TM7b result in the conformational changes of GLUT3 from outward‐open to ‐occluded. The superposition of outward‐open GLUT3 (green) with the outward‐occluded (white) one reveals pronounced conformational changes of TM7b (PDB accession code for outward‐occluded GLUT3: 4ZWB). (C) Local rearrangement of TM2 results in the conformational switch from inward‐occluded to inward‐open of vSGLTs. The vSGLTs in the inward‐facing and inward‐open state are colored white and green, respectively. The TM2 of vSGLT in the inward‐open structure is highlighted by magenta. The rearrangement of the key residues (Ala62/Asn64/Ile65) in TM2 and the intracellular gate Try263 is shown on the right. (D) Structural comparison of EcSemiSWEET in the outward‐open and inward‐open states (PDB accession code 4X5N). The superposition of monomers of EcsemiSWEET in different conformations suggests that conserved PQ motif in each protomer serves as a hinge for the conformational bending of TM1.
As briefly mentioned above, XylE was captured in three different states, the ligand‐bound and outward‐occluded, inward‐open, and partly inward‐occluded. Nevertheless, the outward‐open conformation was missing.89 Fortunately, the structures of the closely related GLUT1 and GLUT3 were obtained in three important states—outward‐open, outward‐occluded, and inward‐open—which allow detailed examination of the conformational shifts involved in a transport cycle [Fig. 3(A) and Table 1]. Structural comparison elucidates relative rotations of the N and C domains as well as pronounced local structural shifts within C domain. From outward‐open to inward‐open, the two domains undergo a relative concentric rotation of approximately 16°. Meanwhile, TM7b and TM10b, two corresponding segments within the two inverted repeats in C domain, display marked local structural changes. From outward‐open to outward‐occluded, TM7b undergoes both bending and rotation of the short helix by about 60°, which results in the relocation of two key residues, Asn286 and Tyr290, into the central cavity [Fig. 3(B)]. From outward‐ to inward‐facing, TM10b undergoes an outward swing motion that results in the reorganization of the substrate‐binding site [Fig. 3(A)].15 In contrast, the N domain remains rigid during the alternating access cycle. Aside from the transmembrane region, the ICH domain undergoes prominent conformational changes, supporting the notion that it serves as a “door closer” on the intracellular side.15
The two structures of vSGLT provide two inward‐facing snapshots of SGLTs.23, 24 Whereas the overall structures remain similar, local rearrangement of TM1 and minor rigid body movement result in the switch from inward‐occluded to inward‐open. In particular, residues Asn64 on TM2 and Tyr263 on TM7, which are involved in substrate binding and intracellular gating in the inward‐occluded conformation, are dismissed in the inward‐open structure [Fig. 3(C)]. Notably, despite the completely different folds, the local conformational switches of GLUT3 and vSGLT both occur to discontinuous helices and involve a sugar‐coordinating Asn and a gating Tyr, suggesting a mechanistic conservation for sugar transporters in different families.
The crystal structures of SemiSWEETs were determined at distinct states of a transport cycle. In particular, the crystal structures of EcSemiSWEET were captured in both inward‐open and outward‐open conformations, allowing identification of the structural determinants for conformational switches [Fig. 3(D)].27 The alternating access of the dimeric SemiSWEET is achieved through a pronounced inward bending of TM1 in each protomer. A conserved PQ (Pro‐Gln) motif on TM1 serves as the hinge to promote the “binder clip‐like” motion [Fig. 3(D)].27 Another report suggested rigid‐body movements between the two protomers of SemiSWEET that result in the conformational switch.25 Additional biochemical and biophysical characterizations are required to elucidate the transport mechanism of SemiSWEETs.
Substrate Recognition of Glucose Transporters
Elucidating the molecular basis for substrate recognition is a key to the mechanistic understanding of transporters. The structures of XylE, GLUT1/3, and vSGLT were obtained in the presence of substrates or inhibitors, allowing detailed examination of the molecular basis for substrate selectivity. Despite the lack of a substrate‐bound SemiSWEET structure, the putative substrate binding site was suggested.
Structures of substrate‐bound XylE and GLUT3 as well as a substrate derivative‐bound GLUT1 reveal an interesting pattern in substrate recognition, whereby the substrate is primarily coordinated by the C domain and the N domain provides the conformational switch for alternating access [Figs. 3(A) and 4(A)].
Figure 4.
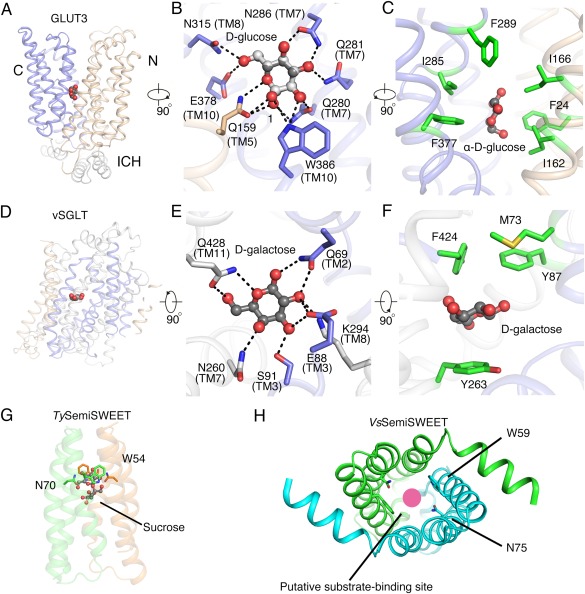
Substrate recognition by glucose transporters. (A) Glucose coordination by GLUT3. The d‐glucose molecule “stands” in the cavity between N, C domains and primarily coordinated by the C domain (PDB accession code 4ZW9). The hexose ring is approximately parallel to the membrane norm. (B) GLUT3 can recognize both anomers of d‐glucose. Each and every polar group of the bound d‐glucose is co‐ordinated through H‐bonds with surrounding polar residues, mainly from the C domain. The H‐bonds are represented by black dashed lines. (C) The carbon ring of d‐glucose is surrounded by hydrophobic residues. Shown here is the α‐anomer of d‐glucose only. (D) Substrate recognition by vSGLT. The bound galactose “lies” in the middle of the structures. Its sugar ring is approximately parallel to the membrane plane. (E) Polar contacts of galactose with vSGLT. (F) Galactose is sandwiched by hydrophobic residues, which serve as extracellular and intracellular gates. (G) Putative substrate binding site in TySemiSWEET. One sucrose molecule was tentatively assigned to the central pocket in TySemiSWEET (PDB accession code 4RNG). (E) Putative substrate binding pocket in the large cavity of VsSemiSWEET. The red circle indicates the putative substrate binding site (PDB accession code 4QND).
The structure of GLUT3 in complex with d‐glucose at 1.5 Å resolution provides unprecedented clarity for understanding substrate recognition.15 A serendipitous discovery bestowed by the high resolution is that both α‐ and β‐anomers of d‐glucose are unambiguously resolved in the structure [Fig. 4(B)]. The prevailing form of d‐glucose in aqueous solution is the β‐anomer; however, in the crystal structure of GLUT3, the α‐anomer exhibits a higher occupancy of 69%. The structure supports that GLUTs may recognize and transport both d‐glucose anomers. Both anomers are coordinated similarly except for the recognition of the C1–OH group. d‐glucose is primarily coordinated by polar residues from the C domain, whereas N domain only contributes one H‐bond by a polar residue on TM5 [Fig. 4(B)]. In addition to H‐bonds, the carbon backbone of the hexose ring is contoured by six hydrophobic residues with each domain contributing three [Fig. 4(C)].15
GLUT1 was purified and crystallized in the presence of the detergent n‐nonyl‐β‐d‐glucopyranoside (β‐NG), which is in fact a derivative of glucose. The presence of a β‐NG molecule in the central cavity of GLUT1 allows tentative examination of substrate binding by GLUTs in an inward‐open state.14 The glucoside is H‐bonded to C domain only, while N domain is out of reach. When the C domains of the outward‐occluded GLUT3 and inward‐open GLUT1 are superimposed, the sugar moieties can be overlaid despite slight rearrangement of the binding site. Due to the swing of TM10, Trp386 no longer participates in glucose binding. The dynamic rearrangement of the substrate binding site observed in the structures provides the molecular basis for the asymmetric binding affinities measured from the opposite sides of the membrane.90
Despite the differences of substrate selectivity and coupling mechanism, the coordination of d‐glucose by XylE is similar to that by GLUT3. However, d‐glucose is a competitive inhibitor, but not a substrate, to XylE despite it binds to XylE with a similar affinity as the authentic substrate d‐xyolse. It remains to be revealed why an additional 6‐hydroxylmethyl group would convert a substrate to an inhibitor for XylE.
In the structure of vSGLT, the galactose, which is located halfway of the membrane height, is coordinated by extensive polar and van der Waals interactions [Fig. 4(D)].23 It forms H‐bonds with Gln69, Glu88/Ser91, Asn260, Lys294, and Gln428 from TMs 2, 3, 7, 8, and 11, respectively [Fig. 4(E)]. All these residues, except for Ser91, are conserved in SGLTs. The sugar ring is further sandwiched by hydrophobic residues, which constitute the extracellular and intracellular gates in the inward‐occluded vSGLT [Fig. 4(F)].
It is noteworthy that whereas the d‐glucose “stands” along the surface of the C‐domain of GLUTs [Fig. 4(A)], the galactose lies in the central pocket of vSGLT [Fig. 5(A)]. That is, the sugar ring of glucose in GLUTs is nearly parallel, while that of galactose in vSGLT is perpendicular to the membrane norm. Nevertheless, the coordination of the monosaccharide in GLUT3 and vSGLT follows similar pattern with polar residues surrounding the hydroxyl groups and hydrophobic residues cast the contour of the carbon ring.
Figure 5.
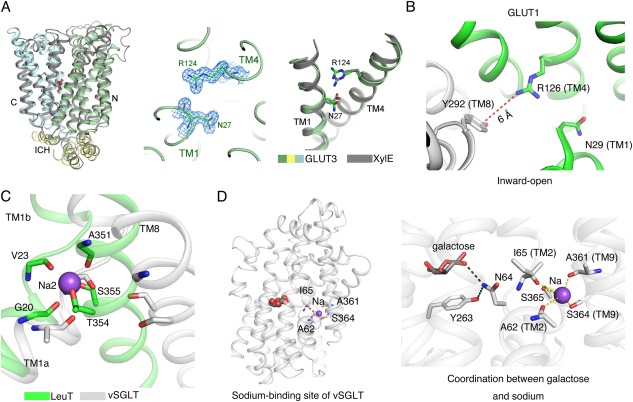
The coupling mechanism of sugar co‐transporters. (A) Proton coupling of the d‐xylose:H+ symporter XylE. Left panel: Structural superposition of the ligand‐bound, outward‐occluded GLUT3 and XylE (PDB accession codes 4ZW9 and 4GBY, respectively). Middle panel: The 2Fo‐Fc electron density of the side chains Asn27 and Arg124 in GLUT3 contoured at 1.5 σ. Right panel: Conformational differences of Asn27 and Arg124 in GLUT3 compared to the corresponding Asp27 and Arg133 in XylE. The N, C, and ICH domain of GLUT3 are pale green, pale cyan, and pale yellow, respectively, while XylE is colored dark grey. (B) In the inward‐open GLUT1, the freed Arg126 in N domain may form the cation–π interaction with Tyr292 in the C domain. (C) Structural comparison between vSGLT and LeuT suggests that the Na+ binding site in vSGLT is similar to the Na2 site in LeuT. LeuT and vSGLT are colored as green and white, respectively (PDB accession code 2A65). (D) Potential sodium coupling mechanism of vSGLT. The residues that may participate in sodium or galactose coordination are shown as sticks. The Na+ is shown as purple sphere.
The substrate‐bound structure of SemiSWEET is yet to be obtained. Nonetheless, the crystals of TySemiSWEET were obtained in the presence of 20 mM sucrose. The omit electron density in the central cavity may correspond to a small molecule, which was tentatively assigned as sucrose [Fig. 4(G)].26 Similarly, a large cavity in the outward‐open VsSemiSWEET was suggested to be the putative sugar‐binding site [Fig. 4(H)]. Two invariant residues, Trp and Asn (Trp54 and Asn70 in TySemiSWEET, and Trp59 and Asn75 in VsSemiSWEET), may be involved in substrate recognition in all SemiSWEET [Fig. 4(G,H)].27
Coupling Mechanism of Glucose Co‐Transporters
The human GLUTs are glucose uniporters, and (Semi)SWEETs may also be uniporters. In contrast, the bacterial homologs of GLUTs and SGLTs are symporters that employ the transmembrane gradient of H+ or Na+ to drive the uphill translocation of the substrate sugars against their concentration gradient across membrane. Structural and biochemical examinations have provided important clues to understanding the molecular mechanism that couples the two electrochemical gradients by the transporters.
GLUT1 and GLUT3 transport the substrate down its gradient, a process named facilitative diffusion. However, XylE is an obligatory proton symporter, that is, it cannot transport xylose without the translocation of proton, vice versa. Biochemical analysis suggested that a conserved residue Asp27 in XylE, which corresponds to Glu325 in LacY,91 Asp22 in GlcPse,21 and Asp32 in GalP,92 is critical for proton coupling, as neutralization of the residue with Asn or Ala abolished proton gradient‐dependent active transport, but retained counterflow activity.93
Structural comparison of XylE to GLUT1/3 provides important insight to the mechanistic interpretation of the biochemical observations. In the outward‐facing XylE structure, Asp27 on TM1 interacts with an invariant Arg133 on TM4 through extensive H‐bonds.19 In GLUT1 and GLUT3, the corresponding Asp residue is replaced by an Asn, which can be regarded as a mimetic of permanently protonated Asp. In the structure of outward‐facing GLUT3, both Asn27 and Arg124 (corresponding to Arg133 in XylE) undergo conformational changes of their side groups compared to those in XylE, resulting in the loss of interactions between the side chains of Asn27 with the invariant Arg124 [Fig. 5(A)]. It suggested that the guanidinium group of the invariant Arg would be released from the sequestration by Asp within the N domain upon protonation of the Asp residue [Fig. 5(A)]. In the structure of inward‐open GLUT1, the corresponding Arg126 appears to interact with a conserved aromatic residue Tyr292 in C domain through a cation–π interaction, which may stabilize the extracellular gate in the inward‐facing conformation [Fig. 5(B)]. Without protonation, the Arg is locked by the deprotonated Asp, unable to trigger the outward to inward switch without the closure of the extracellular gate. This observation in part addresses the critical role of protonation for conformational changes during alternating access cycle. However, the mechanism of deprotonation as well as the coupling between deprotonation and release of substrate by XylE awaits further investigations.
In the sodium symporter vSGLT, the Na+ and galactose are co‐transported at a 1:1 stoichiometric ratio. Despite that the resolution of vSGLT is insufficient to assign sodium ions,23 biochemical assay identified one Na+‐binding site in vSGLT. Comparison of the similar core domains of vSGLT and LeuT suggested that the Na+‐binding site in vSGLT may correspond to the Na2 site in LeuT [Fig. 5(C)].94 Accordingly, Ala62 and Ile65 in the unwound segment from TM2 and Ala361, Ser364, and Ser365 from TM9 may be involved in Na+ coordination [Fig. 5(D)]. Molecular dynamic simulations of LeuT suggested that the release of sodium from the Na2 site prompts the release of substrates.95 Similar results were obtained in the molecular dynamic simulations of vSGLT and the substrate would be passed to the intracellular gate.24 In vSGLT, Asn64 in the unwound segment of TM1, which stabilizes the intracellular gate residue Tyr263, bridges the sodium‐ and galactose‐binding sites together [Fig. 5(D), inset]. It may propagate the signal of sodium and sugar coupling, a speculation that awaits further experimental test.
In this review, we briefly summarize the recent breakthrough in the structural investigation of glucose transporters. Due to the wealth of data concerning the functional and mechanistic characterizations of sugar transporters, we confined our discussion to human glucose transporters GLUT1 and GLUT3, and the bacterial homologs of SGLTs and SWEETs. During the final revision of this review article, we noticed that the structure of a eukaryotic SWEET from Oryza sativa was reported.96 In the homotrimeric complex, each protomer exhibits the inward‐open conformation. The 7‐TM topology of SWEET is consistent with previous prediction [Fig. 2(C)].
Notwithstanding the exciting achievements in the structural elucidation of GLUTs, vSGLT, and SWEETs, the structures represent just a starting point toward a molecular deciphering of the working mechanism of these important transporters. The structures lay out the foundation for further dynamic and kinetic elucidation of the transport process.
Nieng Yan is the recipient of the Protein Society 2015 Protein Science Young Investigator Award.
References
- 1. Thorens B, Mueckler M (2010) Glucose transporters in the 21st Century. Am J Physiol Endocrinol Metab 298:E141–E145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wright EM (2013) Glucose transport families SLC5 and SLC50. Mol Aspects Med 34:183–196. [DOI] [PubMed] [Google Scholar]
- 3. Feng L, Frommer WB (2015) Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem Sci 40:480–486. [DOI] [PubMed] [Google Scholar]
- 4. Cesar‐Razquin A, Snijder B, Frappier‐Brinton T, Isserlin R, Gyimesi G, Bai X, Reithmeier RA, Hepworth D, Hediger MA, Edwards AM, Superti‐Furga G. (2015) A call for systematic research on solute carriers. Cell 162:478–487. [DOI] [PubMed] [Google Scholar]
- 5. Wright EM, Loo DD, Hirayama BA (2011) Biology of human sodium glucose transporters. Physiol Rev 91:733–794. [DOI] [PubMed] [Google Scholar]
- 6. Yan N (2015) Structural biology of the major facilitator superfamily transporters. Annu Rev Biophys 44:257–283. [DOI] [PubMed] [Google Scholar]
- 7. Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468:527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He Y, Wang K, Yan N (2014) The recombinant expression systems for structure determination of eukaryotic membrane proteins. Protein Cell 5:658–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi Y (2014) A glimpse of structural biology through X‐ray crystallography. Cell 159:995–1014. [DOI] [PubMed] [Google Scholar]
- 10. Bhattacharya A (2009) Protein structures: structures of desire. Nature 459:24–27. [DOI] [PubMed] [Google Scholar]
- 11. Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R (2002) X‐ray structure of a ClC chloride channel at 3.0 angstrom reveals the molecular basis of anion selectivity. Nature 415:287–294. [DOI] [PubMed] [Google Scholar]
- 12. Locher KP, Lee AT, Rees DC (2002) The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296:1091–1098. [DOI] [PubMed] [Google Scholar]
- 13. Murakami S, Nakashima R, Yamashita E, Yamaguchi A (2002) Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587–593. [DOI] [PubMed] [Google Scholar]
- 14. Deng D, Xu C, Sun P, Wu J, Yan C, Hu M, Yan N (2014) Crystal structure of the human glucose transporter GLUT1. Nature 510:121–125. [DOI] [PubMed] [Google Scholar]
- 15. Deng D, Sun P, Yan C, Ke M, Jiang X, Xiong L, Ren W, Hirata K, Yamamoto M, Fan S, et al. (2015) Molecular basis of ligand recognition and transport by glucose transporters. Nature 526:391–396. [DOI] [PubMed] [Google Scholar]
- 16. Shintre CA, Pike AC, Li Q, Kim JI, Barr AJ, Goubin S, Shrestha L, Yang J, Berridge G, Ross J, et al. (2014) Structures of ABCB10, a human ATP‐binding cassette transporter in apo‐ and nucleotide‐bound states. Proc Natl Acad Sci USA 110:9710–9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi Y (2013) Common folds and transport mechanisms of secondary active transporters. Annu Rev Biophys 42:51–72. [DOI] [PubMed] [Google Scholar]
- 18. Nomura N, Verdon G, Kang HJ, Shimamura T, Nomura Y, Sonoda Y, Hussien SA, Qureshi AA, Coincon M, Sato Y, et al. (2015) Structure and mechanism of the mammalian fructose transporter GLUT5. Nature 526:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun L, Zeng X, Yan C, Sun X, Gong X, Rao Y, Yan N (2012) Crystal structure of a bacterial homologue of glucose transporters GLUT1‐4. Nature 490:361–366. [DOI] [PubMed] [Google Scholar]
- 20. Quistgaard EM, Low C, Moberg P, Tresaugues L, Nordlund P (2013) Structural basis for substrate transport in the GLUT‐homology family of monosaccharide transporters. Nat Struct Mol Biol 20:766–768. [DOI] [PubMed] [Google Scholar]
- 21. Iancu CV, Zamoon J, Woo SB, Aleshin A, Choe JY (2013) Crystal structure of a glucose/H+ symporter and its mechanism of action. Proc Natl Acad Sci USA 110:17862–17867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wisedchaisri G, Park MS, Iadanza MG, Zheng H, Gonen T (2014) Proton‐coupled sugar transport in the prototypical major facilitator superfamily protein XylE. Nat Commun 5:4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Faham S, Watanabe A, Besserer GM, Cascio D, Specht A, Hirayama BA, Wright EM, Abramson J (2008) The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321:810–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Watanabe A, Choe S, Chaptal V, Rosenberg JM, Wright EM, Grabe M, Abramson J (2010) The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature 468:988–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu Y, Tao Y, Cheung LS, Fan C, Chen LQ, Xu S, Perry K, Frommer WB, Feng L (2014) Structures of bacterial homologues of SWEET transporters in two distinct conformations. Nature 515:448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang J, Yan C, Li Y, Hirata K, Yamamoto M, Yan N, Hu Q (2014) Crystal structure of a bacterial homologue of SWEET transporters. Cell Res 24:1486–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee Y, Nishizawa T, Yamashita K, Ishitani R, Nureki O (2015) Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter. Nat Commun 6:6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mueckler M, Thorens B (2013) The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med 34:121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mueckler M, Caruso C, Baldwin SA, Panico M, Blench I, Morris HR, Allard WJ, Lienhard GE, Lodish HF (1985) Sequence and structure of a human glucose transporter. Science 229:941–945. [DOI] [PubMed] [Google Scholar]
- 30. Manel N, Kim FJ, Kinet S, Taylor N, Sitbon M, Battini JL (2003) The ubiquitous glucose transporter GLUT‐1 is a receptor for HTLV. Cell 115:449–459. [DOI] [PubMed] [Google Scholar]
- 31. Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, Anderson SM, Abel ED, Chen BJ, Hale LP, et al. (2014) The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab 20:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Siska PJ, Rathmell JC (2015) PKCs sweeten cell metabolism by phosphorylation of Glut1. Mol Cell 58:711–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu N, Zheng B, Shaywitz A, Dagon Y, Tower C, Bellinger G, Shen CH, Wen J, Asara J, McGraw TE, et al. (2013) AMPK‐dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell 49:1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Giorgis V, Veggiotti P (2013) GLUT1 deficiency syndrome 2013: current state of the art. Seizure 22:803–811. [DOI] [PubMed] [Google Scholar]
- 35. Gras D, Roze E, Caillet S, Meneret A, Doummar D, Billette de Villemeur T, Vidailhet M, Mochel F (2014) GLUT1 deficiency syndrome: an update. Rev Neurol 170:91–99. [DOI] [PubMed] [Google Scholar]
- 36. Pearson TS, Akman C, Hinton VJ, Engelstad K, De Vivo DC (2013) Phenotypic spectrum of glucose transporter type 1 deficiency syndrome (Glut1 DS). Curr Neurol Neurosci Rep 13:342 [DOI] [PubMed] [Google Scholar]
- 37. Brockmann K (2009) The expanding phenotype of GLUT1‐deficiency syndrome. Brain Dev 31:545–552. [DOI] [PubMed] [Google Scholar]
- 38. Suls A, Mullen SA, Weber YG, Verhaert K, Ceulemans B, Guerrini R, Wuttke TV, Salvo‐Vargas A, Deprez L, Claes LR, et al. (2009) Early‐onset absence epilepsy caused by mutations in the glucose transporter GLUT1. Ann Neurol 66:415–419. [DOI] [PubMed] [Google Scholar]
- 39. Fukumoto H, Seino S, Imura H, Seino Y, Eddy RL, Fukushima Y, Byers MG, Shows TB, Bell GI (1988) Sequence, tissue distribution, and chromosomal localization of mRNA encoding a human glucose transporter‐like protein. Proc Natl Acad Sci USA 85:5434–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uldry M, Ibberson M, Hosokawa M, Thorens B (2002) GLUT2 is a high affinity glucosamine transporter. FEBS Lett 524:199–203. [DOI] [PubMed] [Google Scholar]
- 41. Santer R, Schneppenheim R, Dombrowski A, Gotze H, Steinmann B, Schaub J (1997) Mutations in GLUT2, the gene for the liver‐type glucose transporter, in patients with Fanconi‐Bickel syndrome. Nat Genet 17:324–326. [DOI] [PubMed] [Google Scholar]
- 42. Nagamatsu S, Kornhauser JM, Burant CF, Seino S, Mayo KE, Bell GI (1992) Glucose transporter expression in brain. cDNA sequence of mouse GLUT3, the brain facilitative glucose transporter isoform, and identification of sites of expression by in situ hybridization. J Biol Chem 267:467–472. [PubMed] [Google Scholar]
- 43. Simpson IA, Dwyer D, Malide D, Moley KH, Travis A, Vannucci SJ (2008) The facilitative glucose transporter GLUT3: 20 years of distinction. Am J Physiol Endocrinol Metab 295:E242–E253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Silver IA, Erecinska M (1994) Extracellular glucose concentration in mammalian brain: continuous monitoring of changes during increased neuronal activity and upon limitation in oxygen supply in normo‐, hypo‐, and hyperglycemic animals. J Neurosci 14:5068–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Amann T, Kirovski G, Bosserhoff AK, Hellerbrand C (2011) Analysis of a promoter polymorphism of the GLUT1 gene in patients with hepatocellular carcinoma. Mol Membr Biol 28:182–186. [DOI] [PubMed] [Google Scholar]
- 46. Amann T, Hellerbrand C (2009) GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin Ther Targets 13:1411–1427. [DOI] [PubMed] [Google Scholar]
- 47. Amann T, Maegdefrau U, Hartmann A, Agaimy A, Marienhagen J, Weiss TS, Stoeltzing O, Warnecke C, Scholmerich J, Oefner PJ, et al. (2009) GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol 174:1544–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shim BY, Jung JH, Lee KM, Kim HJ, Hong SH, Kim SH, Sun DS, Cho HM (2013) Glucose transporter 1 (GLUT1) of anaerobic glycolysis as predictive and prognostic values in neoadjuvant chemoradiotherapy and laparoscopic surgery for locally advanced rectal cancer. Int J Colorectal Dis 28:375–383. [DOI] [PubMed] [Google Scholar]
- 49. Ramani P, Headford A, May MT (2013) GLUT1 protein expression correlates with unfavourable histologic category and high risk in patients with neuroblastic tumours. Virchows Arch 462:203–209. [DOI] [PubMed] [Google Scholar]
- 50. Younes M, Lechago LV, Somoano JR, Mosharaf M, Lechago J (1996) Wide expression of the human erythrocyte glucose transporter Glut1 in human cancers. Cancer Res 56:1164–1167. [PubMed] [Google Scholar]
- 51. McGuire BB, Fitzpatrick JM (2009) Biomarkers in renal cell carcinoma. Curr Opin Urol 19:441–446. [DOI] [PubMed] [Google Scholar]
- 52. Kaira K, Serizawa M, Koh Y, Takahashi T, Yamaguchi A, Hanaoka H, Oriuchi N, Endo M, Ohde Y, Nakajima T, et al. (2014) Biological significance of F‐FDG uptake on PET in patients with non‐small‐cell lung cancer. Lung Cancer. 87:197–204. [DOI] [PubMed] [Google Scholar]
- 53. Gallamini A, Zwarthoed C, Borra A (2014) Positron Emission Tomography (PET) in oncology. Cancers 6:1821–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nelson DL, Cox MM (2008). Lehninger principles of biochemistry. New York: W. H. Freeman and Company, 1158 p. [Google Scholar]
- 55. Chan DA, Sutphin PD, Nguyen P, Turcotte S, Lai EW, Banh A, Reynolds GE, Chi JT, Wu J, Solow‐Cordero DE, et al. (2011) Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med 3:94ra70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Y, Cao Y, Zhang W, Bergmeier S, Qian Y, Akbar H, Colvin R, Ding J, Tong L, Wu S, et al. (2012) A small‐molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell‐cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol Cancer Ther 11:1672–1682. [DOI] [PubMed] [Google Scholar]
- 57. Huang S, Czech MP (2007) The GLUT4 glucose transporter. Cell Metab 5:237–252. [DOI] [PubMed] [Google Scholar]
- 58. Govers R (2014) Cellular regulation of glucose uptake by glucose transporter GLUT4. Adv Clin Chem 66:173–240. [DOI] [PubMed] [Google Scholar]
- 59. Murata H, Hruz PW, Mueckler M (2000) The mechanism of insulin resistance caused by HIV protease inhibitor therapy. J Biol Chem 275:20251–20254. [DOI] [PubMed] [Google Scholar]
- 60. Hresko RC, Hruz PW (2011) HIV protease inhibitors act as competitive inhibitors of the cytoplasmic glucose binding site of GLUTs with differing affinities for GLUT1 and GLUT4. PLoS One 6:e25237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Burant CF, Takeda J, Brot‐Laroche E, Bell GI, Davidson NO (1992) Fructose transporter in human spermatozoa and small intestine is GLUT5. J Biol Chem 267:14523–14526. [PubMed] [Google Scholar]
- 62. Anzai N, Ichida K, Jutabha P, Kimura T, Babu E, Jin CJ, Srivastava S, Kitamura K, Hisatome I, Endou H, et al. (2008) Plasma urate level is directly regulated by a voltage‐driven urate efflux transporter URATv1 (SLC2A9) in humans. J Biol Chem 283:26834–26838. [DOI] [PubMed] [Google Scholar]
- 63. Bibert S, Hess SK, Firsov D, Thorens B, Geering K, Horisberger JD, Bonny O (2009) Mouse GLUT9: evidences for a urate uniporter. Am J Physiol Renal Physiol 297:F612–F619. [DOI] [PubMed] [Google Scholar]
- 64. Hummel CS, Lu C, Loo DD, Hirayama BA, Voss AA, Wright EM (2011) Glucose transport by human renal Na+/D‐glucose cotransporters SGLT1 and SGLT2. Am J Physiol Cell Physiol 300:C14–C21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Turk E, Zabel B, Mundlos S, Dyer J, Wright EM (1991) Glucose/galactose malabsorption caused by a defect in the Na+/glucose cotransporter. Nature 350:354–356. [DOI] [PubMed] [Google Scholar]
- 66. Santer R, Kinner M, Lassen CL, Schneppenheim R, Eggert P, Bald M, Brodehl J, Daschner M, Ehrich JH, Kemper M, et al. (2003) Molecular analysis of the SGLT2 gene in patients with renal glucosuria. J Am Soc Nephrol 14:2873–2882. [DOI] [PubMed] [Google Scholar]
- 67. Santer R, Calado J (2010) Familial renal glucosuria and SGLT2: from a mendelian trait to a therapeutic target. Clin J Am Soc Nephrol 5:133–141. [DOI] [PubMed] [Google Scholar]
- 68. Chao EC, Henry RR (2010) SGLT2 inhibition–a novel strategy for diabetes treatment. Nat Rev Drug Discov 9:551–559. [DOI] [PubMed] [Google Scholar]
- 69. Kanwal A, Banerjee SK (2013) SGLT inhibitors: a novel target for diabetes. Pharm Pat Anal 2:77–91. [DOI] [PubMed] [Google Scholar]
- 70. Haas B, Eckstein N, Pfeifer V, Mayer P, Hass MD (2014) Efficacy, safety and regulatory status of SGLT2 inhibitors: focus on canagliflozin. Nutr Diabetes 4:e143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Diez‐Sampedro A, Hirayama BA, Osswald C, Gorboulev V, Baumgarten K, Volk C, Wright EM, Koepsell H (2003) A glucose sensor hiding in a family of transporters. Proc Natl Acad Sci USA 100:11753–11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tazawa S, Yamato T, Fujikura H, Hiratochi M, Itoh F, Tomae M, Takemura Y, Maruyama H, Sugiyama T, Wakamatsu A, et al. (2005) SLC5A9/SGLT4, a new Na+‐dependent glucose transporter, is an essential transporter for mannose, 1,5‐anhydro‐D‐glucitol, and fructose. Life Sci 76:1039–1050. [DOI] [PubMed] [Google Scholar]
- 73. Grempler R, Augustin R, Froehner S, Hildebrandt T, Simon E, Mark M, Eickelmann P (2012) Functional characterisation of human SGLT‐5 as a novel kidney‐specific sodium‐dependent sugar transporter. FEBS Lett 586:248–253. [DOI] [PubMed] [Google Scholar]
- 74. Lin X, Ma L, Fitzgerald RL, Ostlund RE Jr (2009) Human sodium/inositol cotransporter 2 (SMIT2) transports inositols but not glucose in L6 cells. Arch Biochem Biophys 481:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mueckler M, Makepeace C (2009) Model of the exofacial substrate‐binding site and helical folding of the human Glut1 glucose transporter based on scanning mutagenesis. Biochemistry 48:5934–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610–615. [DOI] [PubMed] [Google Scholar]
- 77. Huang Y, Lemieux MJ, Song J, Auer M, Wang DN (2003) Structure and mechanism of the glycerol‐3‐phosphate transporter from Escherichia coli. Science 301:616–620. [DOI] [PubMed] [Google Scholar]
- 78. Henderson PJ, Baldwin SA (2013) This is about the in and the out. Nat Struct Mol Biol 20:654–655. [DOI] [PubMed] [Google Scholar]
- 79. Henderson PJ, Maiden MC (1990) Homologous sugar transport proteins in Escherichia coli and their relatives in both prokaryotes and eukaryotes. Philos Trans R Soc Lond B Biol Sci 326:391–410. [DOI] [PubMed] [Google Scholar]
- 80. Turk E, Kerner CJ, Lostao MP, Wright EM (1996) Membrane topology of the human Na+/glucose cotransporter SGLT1. J Biol Chem 271:1925–1934. [DOI] [PubMed] [Google Scholar]
- 81. Wong FH, Chen JS, Reddy V, Day JL, Shlykov MA, Wakabayashi ST, Saier MH Jr (2012) The amino acid‐polyamine‐organocation superfamily. J Mol Microbiol Biotechnol 22:105–113. [DOI] [PubMed] [Google Scholar]
- 82. Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E (2005) Crystal structure of a bacterial homologue of Na+/Cl–dependent neurotransmitter transporters. Nature 437:215–223. [DOI] [PubMed] [Google Scholar]
- 83. Screpanti E, Hunte C (2007) Discontinuous membrane helices in transport proteins and their correlation with function. J Struct Biol 159:261–267. [DOI] [PubMed] [Google Scholar]
- 84. Jardetzky O (1966) Simple allosteric model for membrane pumps. Nature 211:969–970. [DOI] [PubMed] [Google Scholar]
- 85. Liu Y, Ke M, Gong H (2015) Protonation of Glu(135) facilitates the outward‐to‐inward structural transition of fucose transporter. Biophys J 109:542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Park MS (2015) Molecular dynamics simulations of the human glucose transporter GLUT1. PLoS One 10:e0125361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhao Y, Terry D, Shi L, Weinstein H, Blanchard SC, Javitch JA (2010) Single‐molecule dynamics of gating in a neurotransmitter transporter homologue. Nature 465:188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhao Y, Terry DS, Shi L, Quick M, Weinstein H, Blanchard SC, Javitch JA (2011) Substrate‐modulated gating dynamics in a Na+‐coupled neurotransmitter transporter homologue. Nature 474:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yan N (2013) Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem Sci 38:151–159. [DOI] [PubMed] [Google Scholar]
- 90. Barnett JE, Holman GD, Munday KA (1973) An explanation of the asymmetric binding of sugars to the human erythrocyte sugar‐transport systems. Biochem J 135:539–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gaiko O, Bazzone A, Fendler K, Kaback HR (2013) Electrophysiological characterization of uncoupled mutants of LacY. Biochemistry 52:8261–8266. [DOI] [PubMed] [Google Scholar]
- 92. Sanderson NM, Qi D, Steel A, Henderson PJ (1998) Effect of the D32N and N300F mutations on the activity of the bacterial sugar transport protein, GalP. Biochem Soc Trans 26:S306 [DOI] [PubMed] [Google Scholar]
- 93. Madej MG, Sun L, Yan N, Kaback HR (2014) Functional architecture of MFS D‐glucose transporters. Proc Natl Acad Sci USA 111:E719–E727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Krishnamurthy H, Piscitelli CL, Gouaux E (2009) Unlocking the molecular secrets of sodium‐coupled transporters. Nature 459:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Caplan DA, Subbotina JO, Noskov SY (2008) Molecular mechanism of ion‐ion and ion‐substrate coupling in the Na+‐dependent leucine transporter LeuT. Biophys J 95:4613–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tao Y, Cheung LS, Li S, Eom J‐S, Chen L‐Q, Xu Y, Perry K, Frommer WB, Feng L (2015) Structure of a eukaryotic SWEET transporter in a homotrimeric complex. Nature 527:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]


