Abstract
Pseudomonas aeruginosa is a Gram-negative bacterium that produces highly varied lipopolysaccharide (LPS) structures. The O antigen (O-Ag) in the LPS is synthesized through the Wzx/Wzy-dependent pathway where lipid-linked O-Ag repeats are polymerized by Wzy. Horizontal-gene transfer has been associated with O-Ag diversity. The O-Ag present on the surface of serotypes O5 and O16, differ in the intra-molecular bonds, α and β, respectively; the latter arose from the action of three genes in a serotype converting unit acquired from bacteriophage D3, including a β-polymerase (Wzyβ). To further our understanding of O-polymerases, the inner membrane (IM) topology of Wzyβ was determined using a dual phoA-lacZα reporter system wherein random 3′ gene truncations were localized to specific loci with respect to the IM by normalized reporter activities as determined through the ratio of alkaline phosphatase activity to β-galactosidase activity. The topology of Wzyβ developed through this approach was shown to contain two predominant periplasmic loops, PL3 (containing an RX10G motif) and PL4 (having an O-Ag ligase superfamily motif), associated with inverting glycosyltransferase reaction. Through site-directed mutagenesis and complementation assays, residues Arg254, Arg270, Arg272, and His300 were found to be essential for Wzyβ function. Additionally, like-charge substitutions, R254K and R270K, could not complement the wzyβ knockout, highlighting the essential guanidium side group of Arg residues. The O-Ag ligase domain is conserved among heterologous Wzy proteins that produce β-linked O-Ag repeat units. Taking advantage of the recently obtained whole-genome sequence of serotype O16 a candidate promoter was identified. Wzyβ under its native promoter was integrated in the PAO1 genome, which resulted in simultaneous production of α- and β-linked O-Ag. These observations established that members of Wzy-like family consistently exhibit a dual-periplasmic loops topology, and identifies motifs that are plausible to be involved in enzymatic activities. Based on these results, the phage-derived Wzyβ utilizes a different reaction mechanism in the P. aeruginosa host to avoid self-inhibition during serotype conversion.
Keywords: Pseudomonas aeruginosa, lipopolysaccharide, bacteriophage, serotype, polymerase, glycosyltransferase, O-antigen biosynthesis
Introduction
Pseudomonas aeruginosa is a highly successful opportunistic pathogen, due partly to its arsenal of virulence factors including, toxins, several secretion systems, and lipopolysaccharide (LPS). LPS is a complex glycolipid that comprises the majority of the outer-leaflet of the outer membrane (OM) and has three domains including lipid A, core oligosaccharide, and a distal O antigen (O-Ag). P. aeruginosa produces two forms of O-Ag, the homopolymeric common polysaccharide antigen (CPA), and the heteropolymeric O-specific antigen (OSA) made of a series of repeating sugar subunits (King et al., 2009). The diverse structures of OSA classify P. aeruginosa into 20 distinct serotypes in the International Antigen Typing Scheme (IATS) (Knirel et al., 1988; Stanislavsky and Lam, 1997).
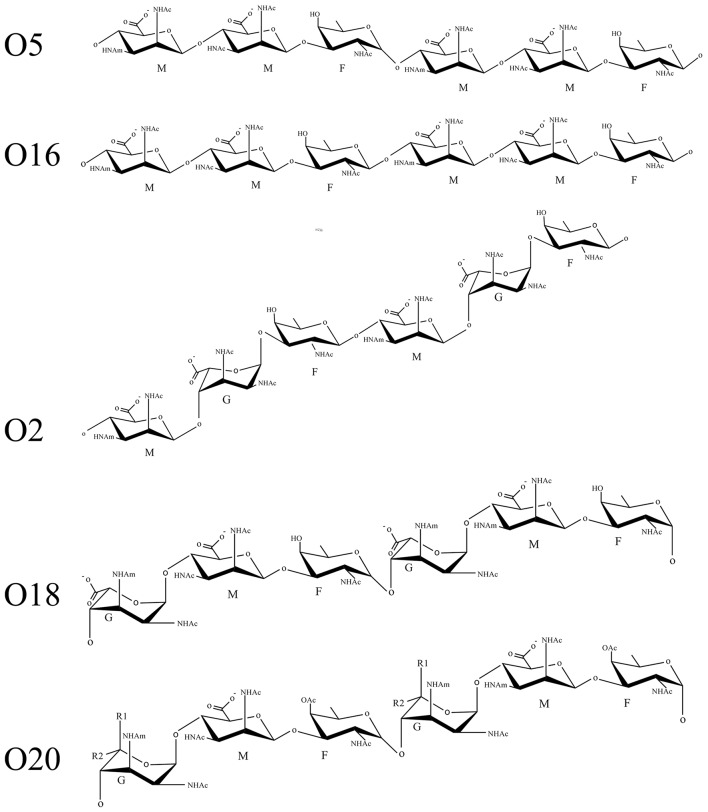
The OSA of P. aeruginosa is synthesized through the Wzx/Wzy-dependent pathway involving a series of inner membrane (IM) proteins that is highly conserved in Gram-negative and Gram-positive organisms that possess heteropolymeric glycans on the cell surface, which include O-antigen, spore coat, enterobacterial common antigen, and capsule (Islam and Lam, 2014). In this model, lipid-linked trisaccharide repeats are flipped from the cytoplasmic side to the periplasm side of the inner membrane by Wzx (O-flippase) (Islam et al., 2012), polymerized at the reducing-end by Wzy (O-polymerase) to a chain-length regulated by Wzz (polysaccharide co-polymerase, PCP) (Burrows et al., 1997). In P. aeruginosa, two distinct PCP proteins are encoded in the genome, named Wzz1 and Wzz2 (Daniels et al., 2002), regulating the synthesis of particular lengths of long and very-long repeats, respectively. The final step of LPS biosynthesis at the IM is ligation of the OSA to the lipid A-core by WaaL, by an inverting glycosyltransferase reaction, before the mature glycolipid is exported to the OM (Sadovskaya et al., 2000; Abeyrathne et al., 2005). The serotypes which comprise the O2 serogroup are: O2, O5, O16, O18, and O20. Although, serotype O5 and O16 of P. aeruginosa produce OSA with identical sugars, the structures of the two differ at the intra-molecular bond, wherein O5 is α-linked and O16 is β-linked (Figure 1).
Figure 1.
Schematic representation of serogroup O2 sugar repeats. The following abbreviations are used in the figure: G, guluronic acid; M, mannuronic acid; F, fucosamine; OAc, O-acetyl group; NHAc, acetamido group; NHAm, acetamidino group; R1, COOH or H; R2, H or COOH.
The biosynthesis clusters of these serotypes are identical; therefore, the genes responsible for this difference must be localized elsewhere within the genome. Previously it was determined that following infection of P. aeruginosa strain PAO1 (serotype O5) by the D3 bacteriophage, the lysogen, AK1380, undergoes serotype conversion to serotype O16 and produces β-linked OSA (Holloway and Cooper, 1962; Kuzio and Kropinski, 1983). The D3 bacteriophage is a temperate phage of P. aeruginosa isolated by Holloway in 1960 (Holloway et al., 1960), which has a polyhedral head containing linear double-stranded DNA (Miller et al., 1974). Infection by the D3 phage renders the bacteria resistant to superinfection by other phages (Holloway and Cooper, 1962). The genes responsible for this phenomenon were identified in the phage genome which encodes an inhibitor of α-polymerase (Iap) and a β-polymerase (Wzyβ) (Newton et al., 2001). The serotype converting genes are present in other P. aeruginosa phages: phi297 and pMG1 (Krylov et al., 2012, 2013). Further work by our group showed that in serotype O2 and O16 strains, the serotype converting unit is constitutively expressed resulting in active inhibition of the native Wzyα and only the production of β-linked OSA (Kaluzny et al., 2007). Although both polymerases recognize the same substrate they share 21% sequence identity to each other (Figure S1).
Prior investigations of Wzy by several laboratories relied on validating in silico topology maps through quantifying enzyme activity of fusions downstream of site-targeted truncations (Daniels et al., 1998; Mazur et al., 2003; Kim et al., 2010). A caveat of in silico predictions is the reliance on “low energy states,” which tends to bias the localization of charged residues to more soluble environments (Elofsson and von Heijne, 2007; Bañó-Polo et al., 2012). In addition, improper localization of the N− and C− terminal ends, or the number of TMS, detected for the protein of interest will affect the orientation of subsequent loop domains (Krogh et al., 2001). Our lab determined the topology of the cognate Wzyα from P. aeruginosa PAO1, which produces α-linked OSA, using an unbiased experimentally-derived approach based on a dual-enzyme reporter system pioneered by Alexeyev and Winkler (1999) in order to reveal previously unidentified domains. The experimental topology map revealed two novel periplasmic loops (PL) PL3 and PL5 in Wzyα. Intriguingly, the region of amino acid residues that make up PL3 had been predicted to localize within a transmembrane segment (TMS) in the in silico map. Further investigation of PL3 and PL5 determined that each of these loops contained a conserved RX10G tract of amino acids similar to the HX10G polymerase motif (Schild et al., 2005). Alanine-scanning mutagenesis of conserved Arg residues determined that their guanidinium side group, not just positive charge, is important for in vivo function of Wzyα. An earlier study has provided evidence that the guanidium side group binds carbohydrates (Dahms et al., 1993). Although the two loops possessed similar amino acid sequence, there was a drastic difference in their isoelectric point with PL3 at pI 8.59 and PL5 at pI 5.49; hence, under physiological pH, the two loops would have contrasting cationic and anionic charges, respectively. These observations and other data have led to the proposed “catch-and-release” mechanism of Wzyα wherein PL3 would act to recruit the newly-flipped OSA repeat with PL5 loosely interacting with the substrate as a retaining arm (Islam et al., 2011).
In a subsequent study, an extensive alanine-scanning screen of exposed charged and polar residues was undertaken which determined that only mutated residues localized within PL3 and PL5 would lead to abrogation of the function of Wzyα. These observations highlighted the importance of these domains. However, two amino acids within a cytoplasmic loop of Wzyα, N380 and R385, when modified by site-directed mutagenesis, conferred altered chain-length modality of the LPS produced by the mutant bacteria, suggesting that these residues are involved in interactions with the co-polymerase protein, Wzz1 (Islam et al., 2013a). Recently the Wang group (Zhao et al., 2014) produced chemo-enzymatically derived substrates to investigate the activity of Wzy to catalyze LPS assembly in vitro. They showed that Wzy adopts a distributive mechanism for polymerization of O-Ag in E. coli, which is consistent with the catch and release model that our group had proposed earlier for Wzy proteins (Islam et al., 2011).
In this study, we determined the topology of the bacteriophage-acquired Wzyβ from serotype O16 using the dual-reporter system as described above. Our data allowed us to build a topology map of Wzyβ with 10 TMS and cytoplasmic N- and C-terminal ends, plus the existence of two large periplasmic loops designated as PL3 and PL4, wherein PL3 possesses an RX10G motif and PL4 contains a Wzy_C motif, a conserved domain among members of the O-Ag ligase superfamily. Both motifs were deemed essential based on evidence from alanine-scanning mutagenesis in which critical Arg and His residues were identified in the two loops, respectively. In this investigation, we demonstrate that either class of Wzy proteins possesses a dual periplasmic loops topology and PL4 in Wzyβ contains an essential stretch of amino acids which likely perform inverting glycosyltransferase reactions and contributing to the O16 OSA to be assembled as a β-linked polymer. This finding substantiated the existence of a possible catalytic region in polymerase proteins localized to the C-terminal region which is involved in the formation of α- or β-linked OSA structures.
Materials and methods
In silico methods
TOPCONS and HMMTOP 2.0 were used to produce a de novo topology map of Wzyβ (Bernsel et al., 2009). The de novo map was generated using the web-based Protter software (Omasits et al., 2014). To generate the experimentally-derived topology map, once the truncations of wzyβ were localized, the designation was inputted using the Advanced setting of HMMTOP 2.0 (Tusnády and Simon, 1998).
Generating the truncation library upstream of the dual-reporter
The wzyβ sequence was amplified from a previously prepared plasmid construct (Kaluzny et al., 2007) and inserted into the pPLEO1 vector (Alexeyev and Winkler, 1999), a derivative of pBluescript II SK(+) containing the phoA-lacZα from pMA632. In order to generate the appropriate overhangs for exonuclease III nuclease digest, the pPLE01-wzyβ construct was digested with PstI and XbaI. Once exonuclease III was added, aliquots were removed at 30 s intervals and put in a stop solution containing 100 mM EDTA, which resulted in a collection of wzyβ truncations of varying lengths. To generate blunt-ends, Mung Bean Nuclease was added to remove the 5′ overhangs and finally the Klenow fragment combined with dNTPs formed blunt-ends suitable for ligation and subsequent transformation into E. coli DH10B. To ensure proper coverage of the entire wzyβ sequence, site-targeted truncations were selected at 10 amino acid intervals and generated by amplifying fragments using site-specific primers to the desired terminal residue. These constructs were then digested with SacI before inserting them into pPLE01. The random and site-targeted truncation library recoveries were plated onto dual-indicator plates containing 100 μg/ml ampicillin (Bio Basic), 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Bio Basic), 80 μg/ml 5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Fisher), and 100 μg/ml 6-chloro-3-indolyl-β-D-galactoside (Red-Gal) (Research Organics). After incubation, the colonies were color scored based on the breakdown of the specific substrates: Red-Gal produced a red pigment indicative of β-galactosidase activity, BCIP produced a blue pigment indicative of alkaline phosphatase activity and a purple combination of both. To avoid full-length constructs and repeated truncation lengths, colony PCR was performed on all colored colonies with Taq polymerase (Life Technologies). Selected clones were sent for sequencing to identify the terminal residues of each truncation.
Enzymatic assay
To assay the levels of enzyme activity, E. coli DH10B cells expressing the pPLEO1-wzyβ truncations were grown in triplicate overnight and subcultured in the presence of ampicillin. IPTG was added the morning after and the cells were grown to optical density at 600 nm (OD600) between 0.4 and 0.7. At this point, the culture was split into halves to assay for β-galactosidase using the Miller protocol and for alkaline-phosphatase following the Manoil protocol (Manoil et al., 1988). The normalized activity ratio (NAR) between these two enzymes was calculated as follows:
NAR = (alkaline phosphatase activity/highest alkaline phosphatase activity)/(β-galactosidase activity /highest β-galactosidase activity) (Lehane et al., 2005).
The resulting NAR determined the final localization and inputted into the HMMTOP 2.0 prediction algorithm (Islam et al., 2010).
Site-directed mutagenesis
Site-directed mutagenesis (SDM) was performed using the QuikChange™ protocol (Agilent). Briefly, wzyβ was cloned into pHERD26T, a pBAD-based l-arabinose-inducible vector, and primers designed with the specific mutation were used to amplify using KOD Hot Start Polymerase (Novagen). Non-methylated parental DNA was digested using DpnI (NEB) and the newly-formed nicked DNA was transformed into DH10B for sequencing (Table S1). In order to rapidly determine that mutant Wzyβ proteins do not demonstrate impaired membrane localization, the same mutations were generated within the pPLEO1-wzyβ-T363 backbone, a periplasmic truncation, and grown on dual-indicator plates to determine whether they retained their periplasmic (blue) localization.
Identifying a candidate wzyβ promoter
The wzyβ sequence was localized within the recently published P. aeruginosa O2 genome (Thrane et al., 2015); however, the contig identified to contain the target ORF for wzyβ was misassembled as it was organized within three duplicated genes. Therefore, a new de novo genome assembly of O16 was performed using SPAdes (Bankevich et al., 2012). The resulting assembly graph was visualized using Bandage (Wick et al., 2015) and used for identification of the duplicated genes and their flanking scaffolds for manual assembly of the region. The identified scaffolds were then aligned to the sequence from a multidrug-resistant P. aeruginosa strain NCGM2.S1 (Miyoshi-Akiyama et al., 2011), which contains the D3 serotype converting unit, for ordering of the correct flanking regions and generating a consensus sequence. This newly formed superscaffold contained a sizeable sequence upstream of wzyβ. Using this upstream nucleotide information as our reference sequence, a 1.65-kbp region containing the wzyβ and 500 bp upstream was amplified and then cloned into the mini-CTX2 integration plasmid for integration into P. aeruginosa cells (Hoang et al., 2000). The plasmid was transformed into P. aeruginosa using a standard electroporation protocol (Smith and Iglewski, 1989) and grown on LB-agar medium supplemented with 90 μg/ml tetracycline (Sigma). The unwanted backbone sequence was excised using the flippase encoded in the pFLP2 plasmid, which was then cured with growth on 5% sucrose (Hoang et al., 1998).
Lipopolysaccharide complementation analysis
To characterize the in vivo function of both wild-type wzyβ and mutant wzyβ, the plasmid containing each of these genes were transformed into a previously generated wzyβ chromosomal knockout (Kaluzny et al., 2007) and plated on LB agar medium supplemented with 90 μg/ml tetracycline. The additional serotypes analyzed in this study, including O1, O3, O5, O6, O8, O9, O10, O18, and O20 were grown overnight in the same LB medium. The transformation of pHERD26T-wzyβ was performed using the standard electroporation protocol and the transformants were plated onto LB agar supplemented with 90 μg/ml tetracycline. To observe varying OSA phenotypes, cultures were induced overnight with 0.1% l-arabinose, equilibrated the next morning to an OD600 of 0.45 and resuspended in Hitchcock and Brown lysis buffer for the preparation of LPS. The samples were heated at 100°C for 30 min and then treated with 5 μl of a 2 mg/mL proteinase K (Invitrogen) stock overnight at 55°C (Hitchcock and Brown, 1983). To observe OSA phenotypes, LPS was resolved by SDS-PAGE, and visualized based on silver staining and Western immunoblotting with monoclonal antibodies (mAb) specific to the serotypes. There are no monoclonal antibodies specific to serotype O18 and O20, therefore analysis of the OSA was done through silver staining and reactivity with the serotype O16 antibody.
Results
Wzyβ possesses an O-Antigen ligase superfamily motif
A characteristic of Wzyα proteins is the high sequence variability resulting in a lack of conserved domains. However, a BLAST search using the Wzyβ amino acid sequence identified a conserved Wzy_C superfamily motif (residues 189–320) within the C-terminal half of the protein. The Wzy_C domain is a conserved motif found within the distal region of the O-Ag ligase superfamily and in PilO. Topology studies has localized this region of WaaL to the periplasm and mutagenesis data showed that residues within the domain are essential for function (Abeyrathne and Lam, 2007; Qutyan et al., 2007; Pérez et al., 2008). Further work by our group (Abeyrathne and Lam, 2007) showed that the His residue within this Wzy_C motif is essential for the function of O-antigen ligase WaaL. It is worth noting that all WaaL protein reported today perform inverting glycosyltransferase reactions (Schild et al., 2005; Abeyrathne and Lam, 2007; Ruan et al., 2012).
Experimentally derived membrane topology Wzyβ
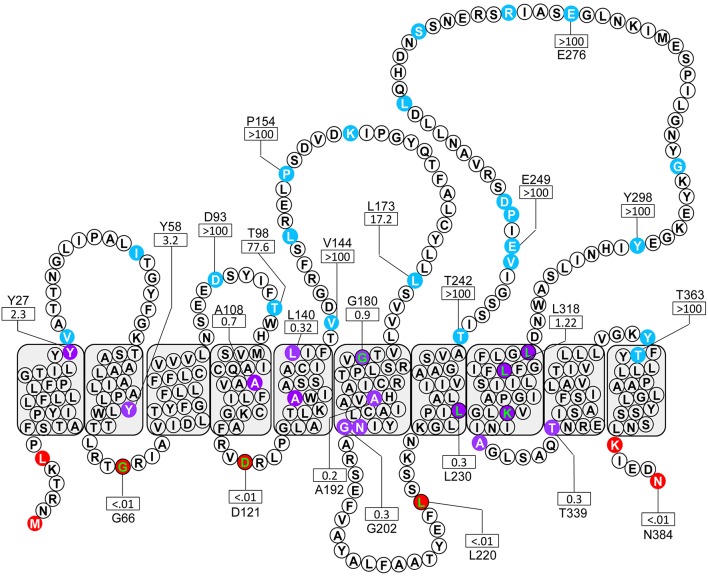
A total of 51 truncations were generated using ExoIII digestion (Alexeyev and Winkler, 1999) resulting in 36 unique localizations (the colony morphology showed 22 blue, 10 purple and 4 red). To ensure a sufficiently complete coverage of Wzyβ, seven site-targeted clones were generated resulting in three red and four purple colonies. Among the truncation constructs, 24 representative residues were selected for NAR evaluation: 17 from the random truncations and all seven of the site-targeted truncations (Table 1). Overall, the color scores of the truncations corresponded well with the anticipated enzyme values. The experimental-based topology map of Wzyβ hence developed is in stark contrast to the topology map derived from in silico analysis. The latter predicted that Wzyβ contained only a single large cytoplasmic loop between residues 246–302 (Figure S2). The experimental-based localizations were inputted into the HMMTOP 2.0 algorithm in order to produce the final map, which displayed the following structural features: (i) both the N- and C-terminal ends of Wzyβ are localized to the cytoplasm, (ii) 10 TMS span the length of the protein, (iii) two large periplasmic loops (PL) were observed, with PL3 spanning amino acids 142–178 and containing an RX10G motif and PL4 which spans amino acid positions 242–301 possesses an HX9G motif. The pI values are 4.68 for PL3 and 5.15 for PL4; and (iv) a cytoplasmic loop (CL) of considerable size was observed at amino acids 203–223 (Figure 2).
Table 1.
Normalized activities of AP and BG Wzyβ truncation fusions to PhoALacZα.
| Residuea | Avg APb | Avg BGc | %APd | %BGe | NAR (%AP/%BG)f | Colony Colorg | Localizationh |
|---|---|---|---|---|---|---|---|
| RANDOM | |||||||
| Y27 | 16.2 | 49 | 100 | 66.5 | 2.3 | Blue | TM |
| Y58 | 2.3 | 4.1 | 13.8 | 5.5 | 3.2 | Blue | TM |
| D93 | 8.7 | −0.38 | 53.3 | −0.5 | >100 | Blue | P |
| T98 | 12.9 | 0.7 | 79.9 | 1.03 | 77.6 | Purple | P |
| A108 | 3.6 | 22 | 22.4 | 29.9 | 0.7 | Purple | TM |
| L140 | 3.7 | 53 | 23.1 | 71.9 | 0.32 | Purple | TM |
| V144 | 1.7 | −4.1 | 10.3 | −5.6 | >100 | Blue | P |
| P154 | 8.1 | −8.7 | 49.6 | −11.7 | >100 | Blue | P |
| L173 | 3.2 | 0.9 | 19.8 | 1.15 | 17.2 | Purple | P |
| A192 | 3.2 | 73.7 | 19.8 | 100 | 0.2 | Purple | TM |
| G202 | 5.3 | 69.5 | 32.3 | 94.1 | 0.3 | Blue | TM |
| T242 | 8.2 | −2.3 | 50.4 | −3.1 | >100 | Blue | P |
| E249 | 10 | 0.3 | 61.5 | 0.4 | >100 | Blue | P |
| E276 | 4.8 | −5.9 | 29.7 | −8 | >100 | Blue | P |
| Y298 | 7.5 | −1.3 | 46.5 | −1.76 | >100 | Blue | P |
| T339 | 1.0 | 15.16 | 6.3 | 20.5 | 0.3 | Purple | TM |
| T364 | 2.48 | −0.9 | 15.3 | −1.1 | >100 | Blue | P |
| SITE TARGET | |||||||
| G66 | −2.7 | 27.7 | −30.1 | 25.9 | < 0.01 | Red | C |
| D121 | −0.7 | 76.5 | −6.4 | 67.5 | < 0.01 | Red | C |
| G180 | 5.7 | 40.1 | 63.7 | 35.5 | 0.95 | Purple | TM |
| L220 | −6.4 | 66 | −61.1 | 61.7 | < 0.01 | Red | C |
| L230 | 0.8 | 32.6 | 8.96 | 30.5 | 0.29 | Red | TM |
| L318 | 0.6 | 6.4 | 7.25 | 5.9 | 1.22 | Purple | TM |
| N384 | −0.8 | 107 | −8.67 | 100 | < 0.01 | Red | C |
Localization of wzyβ truncations based on the enzymatic ratio between alkaline phosphatase (AP) and β-galactosidase (BG).
Location of the terminal residue followed by the reporter.
AP and BG activity quantified in Miller units as described in the Materials and Methods.
Percentage of AP and BG activities of each fusion in relation to the maximum measured activity.
Normalized %AP / % BG activity ratio (NAR).
Color score of expressed clones grown on dual-indicator plates.
Final localization of terminal residues used to generate the topology map: periplasm (P), cytoplasm (C), transmembrane (TM).
Figure 2.
Topological map of Wzyβ based on the localization of 24 amino acids obtained from a phoA-lacZα fusion library. The colored residues designate the subcellular localization of the given truncation: blue, periplasm, purple, TM; red, cytoplasm. The green lettered residues were generated through site-targeted fusions. The NAR values for the enzyme ratios are shown for selected residues by a rectangle.
Positively-charged residues localized within PL3 are essential for Wzyβ function
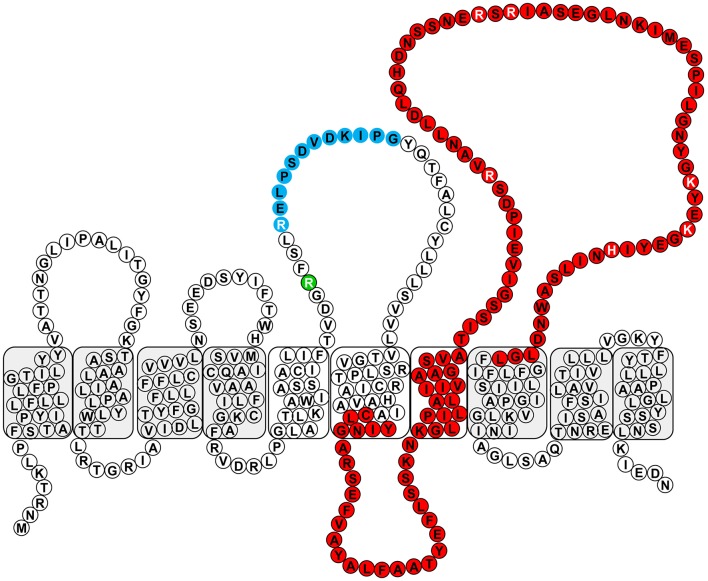
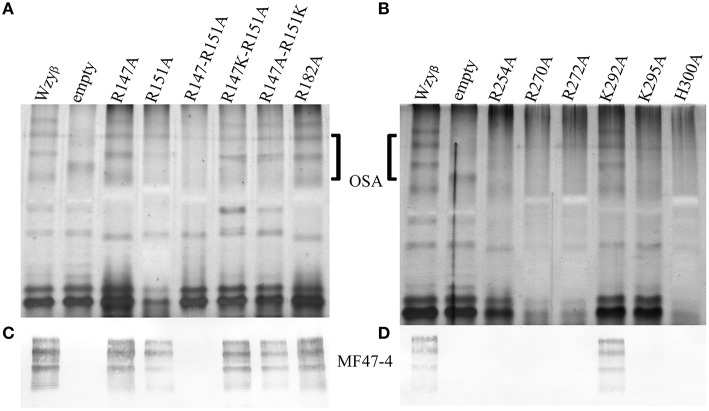
The experimentally-derived topology map revealed the presence of Arg residues within PL3 at positions 147 and 151, which make up the conserved RX10G motif, and an additional Arg at position 182 (Figure 3). Neither of the SDM mutants, R147A nor R151A, showed a disruption in OSA biosynthesis, though the latter mutation, R151A, caused a reduction in the level of OSA being produced. However, a double-mutation construct, R147A-R151A, exhibited a distinct lack of β-linked OSA (Figures 4A,C). Since, the guanidinium side group of Arg residues has been shown to be associated with carbohydrate binding (Dahms et al., 1993), we set out to screen whether positive-charge alone would suffice for function of the Wzyβ protein at these locations by producing R147K and R151K substitutions. In both cases, wild-type level of OSA was restored, demonstrating that a positive charge in PL3 is important for Wzyβ activity. (Figures 4A,C). Although the R147A-R151A is a double mutant, generating these mutations in the T363 truncation background did not affect periplasmic localization as evidenced by the blue coloration of the bacterial colonies on the dual-indicator plates. This observation indicated that the mutant version of Wzyβ was inserted into the IM (Figure S3).
Figure 3.
Conserved domains within Wzyβ. The RX10G motif is depicted in blue, and the Wzy_C domain is depicted in red. The residues selected for mutagenesis are written in white font and R147 is depicted in green.
Figure 4.
SDS-PAGE and Western immunoblotting analyses of LPS isolated from wzyβ::Gmr complemented with Wzyβ mutants generated through alanine-scanning mutagenesis. (A) Silver stain of residues localized in PL3. (B) Silver stain of residues localized in PL4. The OSA region is highlighted. (C,D) Western immunoblots probed with monoclonal antibodies to serotype O16 OSA (MF47-4) (Lam et al., 1987).
PL4 of Wzyβ contains residues essential for an inverting glycosyltransferase reaction
To establish that the Wzy_C domain (Figure 3) plays a significant role in Wzyβ activities and to identify amino acid residues that might be important for function, we decided to do alanine-scanning mutagenesis and substituted conserved residues throughout PL4 with Ala. These included Arg254, Arg270, Arg272, Lys292, Lys295, and His300. Note that H300 is an essential residue in the HX10G motif. When the SDM mutant constructs were expressed in the wzyβ knockout, all but K292A demonstrated impaired Wzyβ function (Figures 4B,D). Further investigation using like-charge substitutions revealed that the guanidinium side-group at positions Arg254 and Arg270 are important for Wzyβ activity, as evidenced by the inability of R254K and R270K to complement function of the wzyβ knockout mutant (Figure 5). Unlike the findings in the mutagenesis of WaaL of P. aeruginosa, a H300R substitution was unable to complement the wzyβ knockout and restore polymerase function. The mutants generated within the T363 truncation construct retained their periplasmic localization; hence the charged amino acids selected for mutagenesis within this region (250–363) of the protein are apparently not required for protein folding/membrane insertion (Figure S3).
Figure 5.
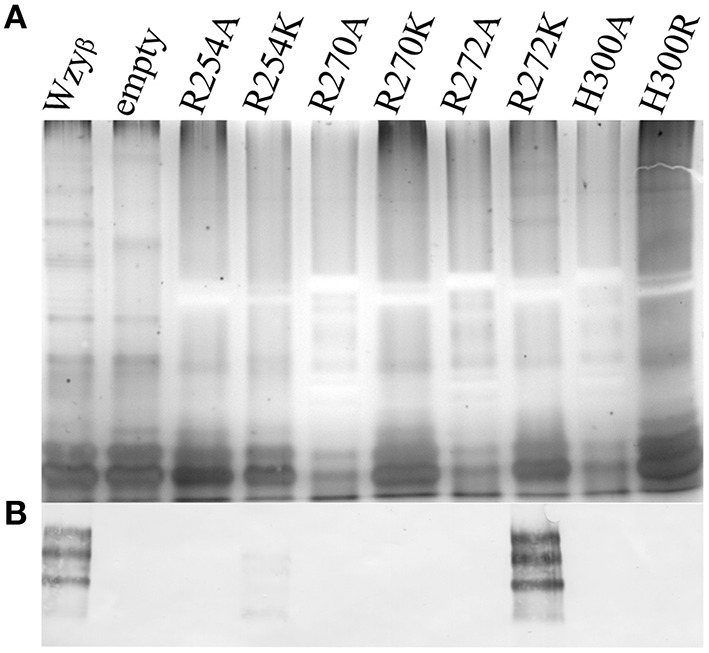
Analysis of LPS from wzyβ::Gmr complemented with like-charge residues by SDS-PAGE and Western immunoblot. (A) Silver stain with the OSA region highlighted and (B) Western immunoblot probed with monoclonal antibodies to serotype O16 OSA (MF47-4) (Lam et al., 1987).
Identification of the Wzy_C domain in heterologous Wzy proteins
We were able to take advantage of the increase in available information on O-Ag polysaccharide structures and O-Ag gene cluster sequences in public databases to investigate the prevalence of the Wzy_C domain within known Wzy amino acid sequences that we extracted from GenBank. This bioinformatic exercise revealed that a relationship between O-Ag structures and the occurrence of Wzy_C domain in Wzy homologs was conserved in 82% of the cases when the corresponding O-Ag structure is β-linked. Importantly, it was noted that the Wzy_C domain is absent in the amino acid sequences of Wzy proteins associated with α-linked O-Ag structures (Table 2).
Table 2.
Occurrence of the Wzy_C domain in organisms with β-linked O-Ag.
| Organism | Intramolecular linkage* | Wzy_C** | References | GenBank |
|---|---|---|---|---|
| P. aeruginosa O5 | (β1 → 3)-D-FucNAc-(α1 → | No | de Kievit et al., 1995 | AAM27801.1 |
| P. aeruginosa O16 | (β1 → 3)-D-FucNAc-(β1 → | Yes | Kaluzny et al., 2007 | ABM21470.1 |
| P. aeruginosa O9 | (α1 → 3)-D-QuiNAc(β1 → | No | Raymond et al., 2002 | AAM27879.1 |
| P. aeruginosa O13 | (α1 → 3)-D-QuiNAc-(β1 → | Yes | Raymond et al., 2002 | AAM27615.1 |
| P. aeruginosa O3 | (α1 → 3)-D-QuiNAc4NSHb-(β1 → | Yes | Raymond et al., 2002 | AAM27766.1 |
| P. aeruginosa O1 | (α1 → 3)-D-QuiNAc(α1 → | No | Raymond et al., 2002 | AAM27546.1 |
| Escherichia coli O86:B7 | (β1 → 3)-D-GalNAc(α1 → | No | Yi et al., 2006 | AY220982.1 |
| Shigella flexneri | (β1 → 3)-D-GlcNAc(α1 → | No | Morona et al., 1994; Knirel et al., 2013 | CAA50774.1 |
| Francisella tularensisi | (β1 → 2)-Qui4NFm(α1 → | No | Vinogradov et al., 1991; Kim et al., 2010 | ABU61108.1 |
| Streptococcus pneumoniae 9a | (β1 → 4)-Glc(β1 → | Yes | Bentley et al., 2006; Aanensen et al., 2007 | CAI32973.1 |
| Salmonella anatum | (1 → 4)-Rha-(α1 → | No | L'Vov et al., 1989 | AHW12776.1 |
| Epsilon 15 | (1 → 4)-Rha-(β1 → | Yes | Robbins et al., 1965; Kropinski et al., 2007 | AAO06084.1 |
| Yersinia pseudotuberculosis O:2a | (β1 → 3)-D-GlcNAc(α1 → | No | Kondakova et al., 2008; Kenyon and Reeves, 2013 | AAN23078.1 |
| Klebsiella pneumoniae K57 | (1 → 2)-α-Man-(β1 → | Yes | Kamerling et al., 1975; Pan et al., 2008 | BAF75760.1 |
| Vibrio vulnificus 27562 | (1 → 4)-β-L-Rha(β1 → | Yes | Gunawardena et al., 1998; Nakhamchik et al., 2007 | ADO64242.1 |
| Streptococcus pneumoniae 4a | (α1 → 3)GalNAc(α1 → | No | Bentley et al., 2006 | CAI32772.1 |
| Shigella boydii | (β1 → 3)-D-GlcNAc-(α1 → | No | Tao et al., 2004 | AAS98031.1 |
| Acetinobacter baumannii | (β1 → 3)-D-GalNAc(β1 → | No | Kenyon and Reeves, 2013 | AHM95427.1 |
| Staphylococcus aureus O5 | (β1 → 3)-d-FucpNAc-(β1 → | Yes | Moreau et al., 1990 | AAC46093.1 |
| Burkholderia cepacia K56-2 | (α1 → 3)-GalNAc(β1 → | Yes | Varga et al., 2013 | EPZ86155.1 |
The orientation of the intramolecular bond is indicated in bold.
Presence and absence of Wzy_C domain in the Wzy homolgos of various species and bacterial strains.
Wzyβ is specific to the O2 serogroup OSA structures
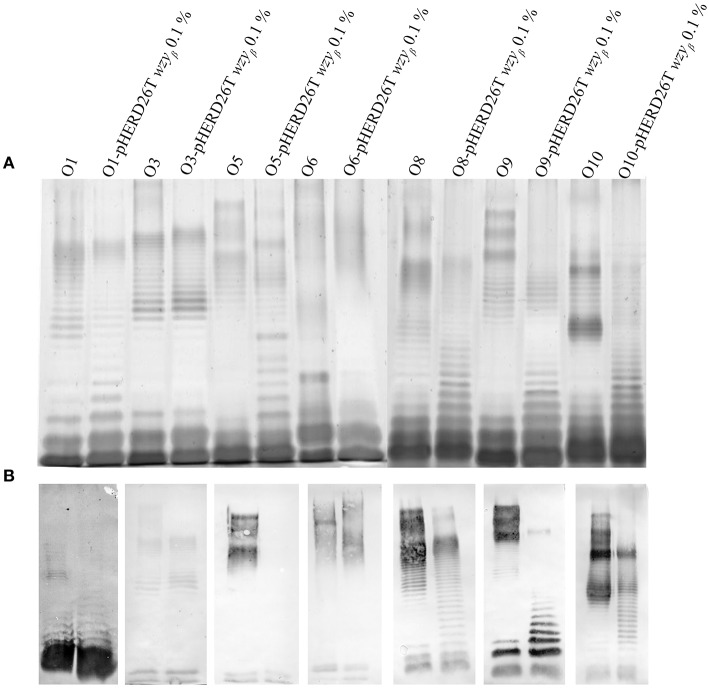
As Wzyβ was acquired from a yet unknown environmental source, it may have relaxed specificity to other O-antigen repeats. Wzyβ was therefore expressed in non-O5 serotypes, i.e., O1, O3, O6, O8, O9, and O10. The most obvious changes were observed in the serotypes which produce trisaccharide repeats: O8, O9, and O10 wherein the presence of Wzyβ affects the OSA biosynthesis as evidence by altered modal lengths in O8 and O10 and complete disruption of the native OSA in serotype O9 (Figure 6). The altered interaction between the native Wzy and Wzz demonstrates that Wzyβ localizes to their OSA biosynthesis machinery. This phenomenon is not observed when Wzyα from serotype O5 is expressed in serotype O9, which is consistent with Wzyα being highly specific to its native OSA structure (Figure S4). However, no evidence of altered intramolecular linkages was observed, demonstrating that although it can recognize the machinery, Wzyβ cannot utilize non-O2 serogroup OSA repeats. As a control for O2 serogroup specificity, the pHERD26T-wzyβ construct was expressed in O18 and O20 as both are members of the O2 serogroup. This heterologous expression resulted in the bacteria producing β-linked OSA (Figure 7).
Figure 6.
Wzyβ expression in various P. aeruginosa serotypes. LPS harvested from wild-type O1, O3, O5, O8, O9 and O10 and expressing the pHERD26T-wzyβ with 0.1% l-arabinose construct were electrophoresed by SDS PAGE and analyzed by (A) silver staining and (B) Western immunoblot of serotypes with antibodies specific to O1 (MF25-1), O3 (MF5-4), O5 (MF15-4), O8 (MF30-1), O9 (MF43-1), and O10 (MF54-1), and inner core 5c-7-4 (Lam et al., 1987).
Figure 7.
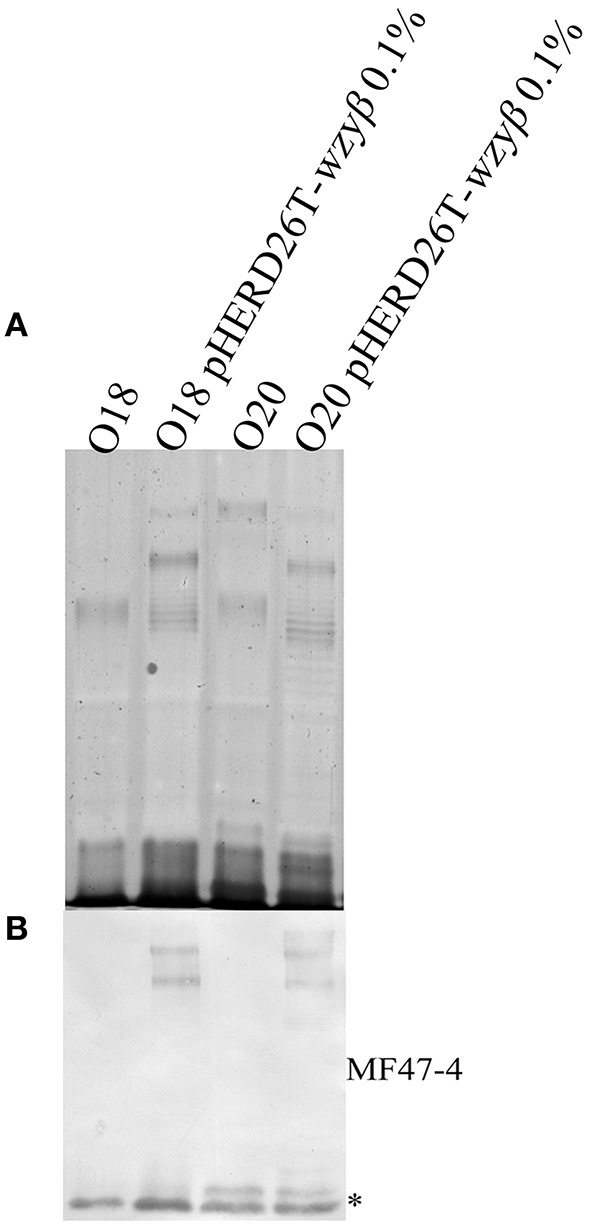
Expression of Wzyβ in serotype O18 and O20. (A) Silver staining and (B) Western immunoblot (MF47-4, O16-specific) of LPS harvested from wild-type O18 and O20 and expressing pHERD26T Wzyβ with 0.1% l-arabinose.
P. aeruginosa PAO1 can simultaneously produce OSA chains with distinct intramolecular linkages
Wzyβ and its native promoter were successfully amplified from the O16 chromosome based on the reconstruction of an O2 isolate genome utilizing the SPAdes system, confirming that these serotypes share an identical serotype-converting unit. The integration of wzyβ driven by its native promoter into the PAO1 chromosome using the miniCTX system resulted in the presence of two distinct OSA banding patterns as detected by silver staining (Figure 8A) and Western immunoblotting with serotype-specific mAbs, MF15-4 (O5 specific) (Figure 8B), and MF47-4 (O16-specific) (Figure 8C). The control lanes 1 and 2 demonstrate that PAO1 and serotype O16 produce α-linked and β-linked OSA repeats respectively. Lane 3 is the wzyβ knockout complemented with miniCTX2-wzyβ and the candidate promoter as indicated by the presence of β-linked OSA repeats. Therefore the included upstream region, identified during chromosomal reconstruction, must contain a promoter for wzyβ as the miniCTX system does not have a plasmid encoded promoter region. Lane 4 depicts LPS harvested from PAO1 that has the miniCTX2-wzyβ integrated within its chromosome. The presence of signal in both the O5-specific (B) and O16-specific (C) Western immunoblots is indicative of concomitant expression of the cognate Wzyα and the phage-derived Wzyβ. Hence two separate OSA polysaccharides α-linked and β-linked were produced in the same cell population (Figure 8).
Figure 8.
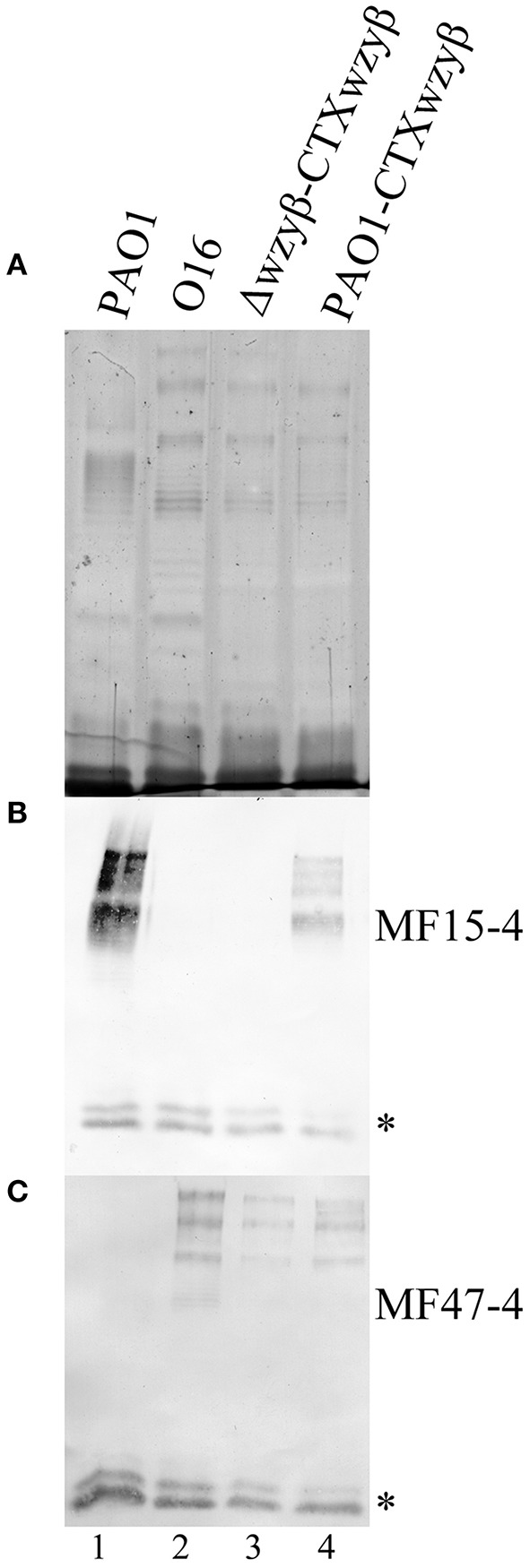
Analysis of the OSA structures after Wzyβ integration into the PAO1 chromosome. (A) Silver stain and Western immunoblot probed with mAbs [MF15-4, O5-specific (B), and MF47-4, O16-specific (C)]. mAb 5c-7-4 (specific to the inner core) was used as an internal standard for loading, and low molecular weight bands reacting to 5c-7-4 are inner core LPS bands and these are designated with *. The lanes are numbered 1 to 4.
Discussion
The mechanism used by the integral IM protein Wzy to polymerize long-chain OSA is currently unknown. The highly diverse P. aeruginosa OSA structures that make up the various serotypes provides an opportunity to study conserved features and differences among the membrane proteins associated with the assembly of LPS. A major hurdle to the investigation of the O-polymerase-mechanism is the lack of any published high-resolution structural data of Wzy proteins. Previous work in our lab established the genetic basis for the role of the phage-derived polymerase, Wzyβ, a component of the serotype-conversion locus in the D3 genome responsible for lysogenic conversion from serotype O5 to O16. (Newton et al., 2001; Kaluzny et al., 2007); however, no further investigations have been undertaken so far to decipher its function. In this study, we used the phoA-lacZα dual-reporter system and obtained experimental data to help elucidate the membrane topology of Wzyβ.
Unlike previous experimental investigations on Wzy (Daniels et al., 1998; Islam et al., 2010; Kim et al., 2010), a BLAST search has led to a novel observation that the Wzyβ sequence contains a Wzy_C domain, also called an O-Ag ligase domain due to its prevalence in WaaL proteins found in various Gram-negative species (Marchler-Bauer et al., 2013). In the case of Wzyβ, the Wzy_C domain spans amino acid residues 234–310. Importantly, this stretch of amino acids is conserved among members of the WaaL family of proteins, which are known to catalyze an inverting glycosyltransferase reaction to link O-Ag to the lipid A-core in a proposed metal-independent reaction (Schild et al., 2005; Abeyrathne and Lam, 2007; Ruan et al., 2012). It was observed that OSA trisaccharide units of P. aeruginosa PAO1 are linked via an α-bond to Und-P, their lipid-carrier, therefore Wzyβ must perform an inverting glycosyltransferase reaction to OSA repeats that confer β-linked intramolecular bonds in serotype O16 (Rocchetta et al., 1998; Sadovskaya et al., 2000; Bystrova et al., 2006; Woodward et al., 2010). To determine whether this domain is essential for Wzyβ, a topology map needs to be established such that appropriate experiments can be designed to probe the roles of specific amino acid residues and any potential functional motifs that might be involved in functional activities of this β-polymerase.
The existing topology maps of WaaL proteins all showed that the Wzy_C domain contains an essential His residue near the distal portion of the protein that forms a characteristic large periplasmic loop (Schild et al., 2005; Abeyrathne and Lam, 2007; Pérez et al., 2008; Islam et al., 2010). As such, the de novo topology map that was easily generated by using the in silico algorithms TOPCONS and HMMTOP 2.0 of Wzyβ was deemed unsatisfactory to utilize for downstream studies because two large periplasmic loops were absent; hence, this deficiency provides the rationale for developing an experimentally-based topology of Wzyβ as the polymerase reaction occurs in the periplasm (Figure S2) (Robbins et al., 1967; Whitfield, 1995). Our group has successfully used the phoA-lacZα dual-reporter system to determine the topology of Wzx, Wzyα, and WaaL from P. aeruginosa strain PAO1 (Islam et al., 2010). The knowledge of the topology of these membrane proteins is pivotal to our ability to initiate protein modeling (Islam et al., 2012) and biophysical experiments (Islam et al., 2013b) in order to examine the function of Wzx of P. aeruginosa. As a case in point, our group was able to develop a tertiary model of Wzx (Islam et al., 2012), generated based on comparison to the crystal structure of the closely-related protein called NorM of Vibrio cholerae (He et al., 2010). The topology of NorM showed strong similarity to the dual-reporter generated topology of Wzx, therefore gaining our confidence in the ability of this strategy to accurately localize particular domains in these membrane proteins (Islam et al., 2012). Although the presence of a prominent cytoplasmic loop was novel, no mutagenesis experiments have been pursued due to the previous report demonstrating that the Wzy protein of E. coli does not hydrolyze ATP (Woodward et al., 2010).
Based on the importance of the PL domains in Wzyα participating in the proposed “catch-and-release” mechanism of O-Ag polymerization, we undertook this project to obtain structure-function data of Wzyβ, a completely unrelated polymerase; our findings in this study provide further evidence for a conserved polymerase topology regardless of whether the resultant bond between O-units is α or β due to the activities of these two classes of Wzy proteins. Upon investigation of the net-charges across both Wzyβ PL regions, no particular amino acid stretch with notable positive charge was found within PL3 of Wzyβ, rather, a net-negative charge was seen. This does not discredit the catch-and-release mechanism as both the Shigella flexerni and Francisella tularensis Wzy proteins have a negatively-charged PL3 but in the case of S. flexneri the Arg residue in the noted RX15G motif was found to be essential for function (Kim et al., 2010; Nath and Morona, 2015). Previously, our lab used a position-based algorithm, Jackhmmer (Finn et al., 2011) and discovered distant homologs of Wzyα. Using this bioinformatic software alleviates the hurdle caused by the absence of strong sequence similarity among these proteins (Islam et al., 2013a). In the current study, the sequence alignment analysis using Jackhmmer was re-run, but this time, we included the peptide sequence of PL3 of Wzyβ. As expected, the RX10G motif in PL3 (R151) of Wzyβ aligned with the PL3 sequences from a number of other Wzy proteins that were not identified in the previous report (data not shown). Therefore, PL3 possesses essential positively-charged amino acids throughout which includes a conserved Arg residue that was identified in a large sequence alignment of heterologous Wzy-like proteins. With the observation of essential positive charge in PL3 of Wzyβ, it lends support to the role of a proximal periplasmic loop being utilized for substrate recruitment (Islam et al., 2011, 2013a; Nath and Morona, 2015).
A hallmark of the O-Ag ligase motif (Wzy_C) is the presence of a basic residue that is essential for inverting glycosyltransferase activities which serves to stabilize the leaving phosphate group once the transferase reaction occurs (Chiu et al., 2004; Lairson et al., 2008; Ruan et al., 2012). In the case of P. aeruginosa WaaL and Wzyβ, the residue in question is His of the HX10∕9G motif; a null mutation of this residue abrogates the function of these proteins (Abeyrathne and Lam, 2007; Lairson et al., 2008; Ruan et al., 2012). Although Wzyβ contains this motif, it is unable to be used to complement a waaL::Gmr mutant in a complementation assay (data not show). There could be a number of reasons why this complementation did not work, one of them being that WaaL and Wzy are vastly different proteins; although both share the HX10∕9G motif, cross complementation of the ligase reaction with a polymerase protein is not a natural phenomenon. However, the negative complementation outcome, could also suggest that the motif in Wzyβ is not involved in substrate recognition as in WaaL proteins, as the latter have high specificity in the recognition of lipid A-core as part of the substrate. A line of evidence that supports the interpretation that the presence of this motif attributes to inverting glycosyltransferase activities in Wzy polymerases was obtained from BLASTp analyses of a number of heterologous Wzy sequences from bacterial species with published O-Ag structures. With this exercise, the Wzy_C domain was identified in 9 of the 11 Wzy proteins from organisms that have corresponding β-linked O-Ag structures (82%).
Two Wzy sequences from organisms, which produce β-linked O-Ag but do not contain the Wzy_C domain include: Wzy from P. aeruginosa serotype O9 (WzyPaO9) and Acinetobacter baumannii (WzyAcb). These two proteins show higher levels of similarity to the “EspG” superfamily of membrane-bound glycosyltransferases than to other Wzy sequences. This is puzzling because EpsG family proteins have been reported to function as autophosphorylating tyrosine kinases, similar to Wzc, which are involved in chain-length regulation of bacterial capsules (Paiment et al., 2002; Ayabe-Chujo et al., 2012). Future work to investigate into the recently published O9 genome may reveal a previously uncharacterized Wzy (Thrane et al., 2015).
Of particular note is the presence of the Wzy_C domain in Wzyβ from the seroconverting unit of ϵ15, a bacteriophage that targets Salmonella species, which when infected with a phage would undergo lysogenic conversion through expression of an inhibitor of α-polymerase (Uetake et al., 1958; Losick, 1969). Hence, ϵ15 likely causes serotyping switch in Salmonella species via a similar mechanism as that observed in D3 bacteriophage, which causes serotyping switch of P. aeruginosa serotype O5 to O16 (Newton et al., 2001). Therefore it can be reasoned that the polymerase gene within the serotype converting units would perform a different mechanism (inverting) than the cognate Wzyα (retaining) to ensure resistance to the α-polymerase inhibitor. Unimpeded Wzyβ activity would lead to long-chain OSA formation with β-bonds between O-units and provide resistance to subsequent phage infection (Losick, 1969; Barksdale and Arden, 1974; Taylor et al., 2013). Through SDM experiments, our data showed that the distal arm of Wzyβ (PL4) contains essential residues, Arg270 and His300, which is conserved in the Wzy_C domain as the proposed stabilizing base. It is conceivable that these residues are involved in the transferase reaction. The consistent presence of this His residue in other β-polymerases suggests that this particular region of the protein could be the determinant of bond formation. The inability of H300R to restore glycosyltransferase function may be due to the essential nature of the imidazole ring as seen in GT-1 metal-independent glycosyltransferases where His acts as the base (Brazier-Hicks et al., 2007; Lairson et al., 2008).
The observation that Wzyβ is able to polymerize OSA repeats despite the presence of the cognate Wzyα helps to better understand the mechanism of serotype conversion. Unlike Wzyα from PAO1, the ability of Wzyβ to localize and interfere with the native Wzy/Wzz interaction in non O2 serogroup serotypes, demonstrates its relaxed specificity toward the OSA biosynthesis/assembly machinery. With the ability of Wzyβ to disrupt the cognate Wzy-Wzz interaction, we propose that both Wzyβ and Iap work in concert and not sequentially. If these two proteins worked sequentially, Wzyβ would not be able to polymerize OSA repeats while Wzyα was still functional. Once Wzyβ was expressed at native levels, it is apparent that the total amount of OSA is split between α- and β-linked polymers as indicated by the relative intensity of the OSA-LPS bands in lane 4 (Figure 8) compared to the WT O5 and O16. Our interpretation is that the Iap, which also affects the Wzz/Wzyα interaction (Taylor et al., 2013), must non-specifically localize to the OSA biosynthetic machinery, where it inhibits Wzyα but is unable to affect Wzyβ, which we propose is due to its differing glycosyltransferase mechanism. This is supported by the observation that the total amount of β-linked OSA produced never reaches the levels of WT α-linked OSA in serotype O5 even when Wzyβ is overexpressed on a plasmid. We propose that OSA repeats are still interacting with Wzyα and are therefore unable to enter in contact with the functional Wzyβ to be polymerized. The phenomenon of bacteria producing multiple OSA phenotypes on the cell surface is not uncommon within the context of O-Ag diversity as P. aeruginosa and, recently, Azosparillum and Salmonella have demonstrated concomintant expression of multiple O-Ag phenotypes (Knirel et al., 1982; Nghiêm et al., 1992; Sigida et al., 2015). Further, the ability of Wzyβ to alter the chain-length modality in serotype O5, O18, and O20 demonstrates that intra-molecular linkages affect OSA modal lengths.
In conclusion, the use of a dual-reporter approach to develop a topology map of Wzyβ revealed that it possesses two predominant PL loops, a structural feature we now confirm to be shared by other Wzy proteins. Another significant finding is the identification of essential amino acid residues within PL3 and in PL4. Through SDM and complementation experiments, we identified the amino acids, Arg270 and His300 that are associated with inverting glycosyltransferase activities of Wzyβ. Finally, we have obtained evidence to show that the two periplasmic loops are important structural characteristics of Wzy proteins, from both α- and β- families of these proteins. These new evidence is consistent with the proposed “catch-and-release” mechanism of substrate shuttling. The two periplasmic loops are purported to participate in substrate shuttling, despite catalyzing the formation of different stereochemical linkages. Future investigations into polymerase function could include obtaining three-dimensional structures of PL4 from Wzyβ and PL5 from Wzyα for an in-depth structure-function comparative study. This mechanistic difference between polymerase proteins further sheds light on the phenomenon of serotype conversion, a cause of O-Ag diversity in Gram-negative organisms.
Author contributions
VT designed the research and wrote the paper; VT, JH, ST and SH all contributed to executing experiments, VT, ST, and LJ performed data and bioinformatics analyses. JL and all other authors also contributed to writing, editing, and approval of the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
JL is supported by an operating grant from the Canadian Institutes of Health Research (CIHR) (MOP-14687). LJ is supported by a Villum Foundation (VKR023113). VT was the recipient of a Cystic Fibrosis Canada Doctoral Studentship, SH is the recipient of a CIHR Canadian Graduate Scholarship-M and an Ontario Graduate Scholarship. JL holds a Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00393
References
- Aanensen D. M., Mavroidi A., Bentley S. D., Reeves P. R., Spratt B. G. (2007). Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189, 7856–7876. 10.1128/JB.00837-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeyrathne P. D., Lam J. S. (2007). WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O antigen onto lipid A-core. Mol. Microbiol. 65, 1345–1359. 10.1111/j.1365-2958.2007.05875.x [DOI] [PubMed] [Google Scholar]
- Abeyrathne P. D., Daniels C., Poon K. K. H., Matewish M. J., Lam J. S. (2005). Functional characterization of WaaL, a ligase associated with linking O-Antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 187, 3002–3012. 10.1128/JB.187.9.3002-3012.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev M. F., Winkler H. H. (1999). Membrane topology of the Rickettsia prowazekii ATP/ADP translocase revealed by novel dual pho-lac reporters. J. Mol. Biol. 285, 1503–1513. 10.1006/jmbi.1998.2412 [DOI] [PubMed] [Google Scholar]
- Ayabe-Chujo Y., Usami Y., Yoshida T., Omori T., Nojiri H. (2012). Membrane topology and functional analysis of Methylobacillus sp. 12S genes epsF and epsG, encoding polysaccharide chain-length determining proteins. Biosci. Biotechnol. Biochem. 76, 608–612. 10.1271/bbb.110831 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañó-Polo M., Baeza-Delgado C., Orzáez M., Marti-Renom M. A., Abad C., Mingarro I. (2012). Polar/Ionizable residues in transmembrane segments: effects on helix-helix packing. PLoS ONE 7:e44263. 10.1371/journal.pone.0044263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barksdale L., Arden S. B. (1974). Persisting bacteriophage infections, lysogeny, and phage conversions. Annu. Rev. Microbiol. 28, 265–300. 10.1146/annurev.mi.28.100174.001405 [DOI] [PubMed] [Google Scholar]
- Bentley S. D., Aanensen D. M., Mavroidi A., Saunders D., Rabbinowitsch E., Collins M., et al. (2006). Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. 10.1371/journal.pgen.0020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsel A., Viklund H., Hennerdal A., Elofsson A. (2009). TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37, W465–W468. 10.1093/nar/gkp363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier-Hicks M., Offen W. A., Gershater M. C., Revett T. J., Lim E.-K., Bowles D. J., et al. (2007). Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 20238–20243. 10.1073/pnas.0706421104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows L. L., Chow D., Lam J. S. (1997). Pseudomonas aeruginosa B-band O-antigen chain length is modulated by Wzz (Ro1). J. Bacteriol. 179, 1482–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystrova O. V., Knirel Y. A., Lindner B., Kocharova N. A., Kondakova A. N., Zähringer U., et al. (2006). Structures of the core oligosaccharide and O-units in the R- and SR-type lipopolysaccharides of reference strains of Pseudomonas aeruginosa O-serogroups. FEMS Immunol. Med. Microbiol. 46, 85–99. 10.1111/j.1574-695X.2005.00004.x [DOI] [PubMed] [Google Scholar]
- Chiu C. P. C., Watts A. G., Lairson L. L., Gilbert M., Lim D., Wakarchuk W. W., et al. (2004). Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat. Struct. Mol. Biol. 11, 163–170. 10.1038/nsmb720 [DOI] [PubMed] [Google Scholar]
- Dahms N. M., Rose P. A., Molkentin J. D., Zhang Y., Brzycki M. A. (1993). The bovine mannose 6-phosphate/insulin-like growth factor II receptor. The role of arginine residues in mannose 6-phosphate binding. J. Biol. Chem. 268, 5457–5463. [PubMed] [Google Scholar]
- Daniels C., Griffiths C., Cowles B., Lam J. S. (2002). Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ. Microbiol. 4, 883–897. 10.1046/j.1462-2920.2002.00288.x [DOI] [PubMed] [Google Scholar]
- Daniels C., Vindurampulle C., Morona R. (1998). Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol. Microbiol. 28, 1211–1222. 10.1046/j.1365-2958.1998.00884.x [DOI] [PubMed] [Google Scholar]
- de Kievit T. R., Dasgupta T., Schweizer H., Lam J. S. (1995). Molecular cloning and characterization of the rfc gene of Pseudomonas aeruginosa (serotype O5). Mol. Microbiol. 16, 565–574. 10.1111/j.1365-2958.1995.tb02419.x [DOI] [PubMed] [Google Scholar]
- Elofsson A., von Heijne G. (2007). Membrane Protein Structure: prediction versus Reality. Annu. Rev. Biochem. 76, 125–140. 10.1146/annurev.biochem.76.052705.163539 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Clements J., Eddy S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena S., Reddy G. P., Wang Y., Kumar Kolli V. S., Orlando R., Morris J. G., et al. (1998). Structure of a muramic acid containing capsular polysaccharide from the pathogenic strain of Vibrio vulnificus ATCC 27562. Carbohydr. Res. 309, 65–76. 10.1016/S0008-6215(98)00115-3 [DOI] [PubMed] [Google Scholar]
- He X., Szewczyk P., Karyakin A., Evin M., Hong W.-X., Zhang Q., et al. (2010). Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467, 991–994. 10.1038/nature09408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. (1983). Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Hoang T. T., Kutchma A. J., Becher A., Schweizer H. P. (2000). Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72. 10.1006/plas.1999.1441 [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Cooper G. N. (1962). Lysogenic conversion in Pseudomonas aeruginosa. J. Bacteriol. 84, 1321–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B. W., Egan J. B., Monk M. (1960). Lysogeny in Pseudomonsa aeruginosa. Aust. J. Exp. Biol. Med. 38, 321–330. 10.1038/icb.1960.34 [DOI] [PubMed] [Google Scholar]
- Islam S. T., Lam J. S. (2014). Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can. J. Microbiol. 60, 697–716. 10.1139/cjm-2014-0595 [DOI] [PubMed] [Google Scholar]
- Islam S. T., Eckford P. D. W., Jones M. L., Nugent T., Bear C. E., Vogel C., et al. (2013b). Proton-dependent gating and proton uptake by Wzx support O-antigen-subunit antiport across the bacterial inner membrane. mBio 4:e00678–13. 10.1128/mbio.00678-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S. T., Fieldhouse R. J., Anderson E. M., Taylor V. L., Keates R. A. B., Ford R. C., et al. (2012). A cationic lumen in the Wzx flippase mediates anionic O-antigen subunit translocation in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 84, 1165–1176. 10.1111/j.1365-2958.2012.08084.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S. T., Gold A. C., Taylor V. L., Anderson E. M., Ford R. C., Lam J. S. (2011). Dual conserved periplasmic loops possess essential charge characteristics that support a catch-and-release mechanism of O-antigen polymerization by Wzy in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 286, 20600–20605. 10.1074/jbc.C110.204651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S. T., Huszczynski S. M., Nugent T., Gold A. C., Lam J. S. (2013a). Conserved-residue mutations in Wzy affect O-antigen polymerization and Wzz-mediated chain-length regulation in Pseudomonas aeruginosa PAO1. Sci. Rep. 3:3441. 10.1038/srep03441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S. T., Taylor V. L., Qi M., Lam J. S. (2010). Membrane topology mapping of the O-Antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures. mBio 1:e00189–10. 10.1128/mbio.00189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluzny K., Abeyrathne P. D., Lam J. S. (2007). Coexistence of two distinct versions of O-Antigen Polymerase, Wzy-α and Wzy-β, in Pseudomonas aeruginosa serogroup O2 and their contributions to cell surface diversity. J. Bacteriol. 189, 4141–4152. 10.1128/JB.00237-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerling J. P., Lindberg B., Lonngren J., Nimmich W. (1975). Structural studies of the Klebsiella type 57 capsular polysaccharide. Acta Chem. Scand. B Org. Chem. Biochem. 29, 593–598. 10.3891/acta.chem.scand.29b-0593 [DOI] [PubMed] [Google Scholar]
- Kenyon J. J., Reeves P. R. (2013). The Wzy O-antigen polymerase of Yersinia pseudotuberculosis O:2a has a dependence on the Wzz chain-length determinant for efficient polymerization. FEMS Microbiol. Lett. 349, 163–170. 10.1111/1574-6968.12311 [DOI] [PubMed] [Google Scholar]
- Kim T.-H., Sebastian S., Pinkham J. T., Ross R. A., Blalock L. T., Kasper D. L. (2010). Characterization of the O-antigen polymerase (Wzy) of Francisella tularensis. J. Biol. Chem. 285, 27839–27849. 10.1074/jbc.M110.143859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. D., Kocíncová D., Westman E. L., Lam J. S. (2009). Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15, 261–312. 10.1177/1753425909106436 [DOI] [PubMed] [Google Scholar]
- Knirel Y. A., Lan R., Senchenkova S. N., Wang J., Shashkov A. S., Wang Y., et al. (2013). O-antigen structure of Shigella flexneri serotype Yv and effect of the lpt-O gene variation on phosphoethanolamine modification of S. flexneri O-antigens. Glycobiology 23, 475–485. 10.1093/glycob/cws222 [DOI] [PubMed] [Google Scholar]
- Knirel Y. A., Vinogradov E. V., Kocharova N. A., Paramonov N. A., Kochetkov N. K., Dmitriev B. A., et al. (1988). The structures of O-specific polysaccharides and serological classification of Pseudomonas aeruginosa. Acta Microbiol. Hung. 35, 3–24. [PubMed] [Google Scholar]
- Knirel Y. A., Vinogradov E. V., Shashkov A. S., Dmitriev B. A., Kochetkov N. K., Stanislavsky E. S., et al. (1982). Somatic antigens of Pseudomonas aeruginosa: the structure of the O-specific polysaccharide chains of P. aeruginosa O:2 (Lanyi) lipopolysaccharides. Eur. J. Biochem. 125, 221–227. 10.1111/j.1432-1033.1982.tb06672.x [DOI] [PubMed] [Google Scholar]
- Kondakova A. N., Ho N., Bystrova O. V., Shashkov A. S., Lindner B., Creuzenet C., et al. (2008). Structural studies of the O-antigens of Yersinia pseudotuberculosis O:2a and mutants thereof with impaired 6-deoxy-d-manno-heptose biosynthesis pathway. Carbohydr. Res. 343, 1383–1389. 10.1016/j.carres.2008.04.003 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. L. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kropinski A. M., Kovalyova I. V., Billington S. J., Patrick A. N., Butts B. D., Guichard J. A., et al. (2007). The genome of ε15, a serotype-converting, group E1 Salmonella enterica-specific bacteriophage. Virology 369, 234–244. 10.1016/j.virol.2007.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov S. V., Kropinski A. M., Pleteneva E. A., Shaburova O. V., Burkal'tseva M. V., Mirosnnikov K. A., et al. (2012). Properties of the new D3-like Pseudomonas aeruginosa bacteriophage phiPMG1: genome structure and prospects for the use in phage therapy. Genetika 48, 902–911. 10.1134/s1022795412060087 [DOI] [PubMed] [Google Scholar]
- Krylov S. V., Kropinski A. M., Shaburova O. V., Miroshnikov K. A., Chesnokova E. N., Krylov V. N. (2013). New temperate Pseudomonas aeruginosa phage, phi297: specific features of genome structure. Genetika 49, 806–818. 10.1134/s1022795413080073 [DOI] [PubMed] [Google Scholar]
- Kuzio J., Kropinski A. M. (1983). O-antigen conversion in Pseudomonas aeruginosa PAO1 by bacteriophage D3. J. Bacteriol. 155, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008). Glycosyltransferases: Structures, Functions, and Mechanisms. Annu. Rev. Biochem. 77, 521–555. 10.1146/annurev.biochem.76.061005.092322 [DOI] [PubMed] [Google Scholar]
- Lam J. S., MacDonald L. A., Lam M. Y., Duchesne L. G., Southam G. G. (1987). Production and characterization of monoclonal antibodies against serotype strains of Pseudomonas aeruginosa. Infect. Immun. 55, 1051–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehane A. M., Korres H., Verma N. K.. (2005). Bacteriophage-encoded glucosyltransferase GtrII of Shigella flexneri: membrane topology and identification of critical residues. Biochem. J. 389, 137–143. 10.1042/BJ20050102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R. (1969). Isolation of a trypsin-sensitive inhibitor of O-antigen synthesis involved in lysogenic conversion by bacteriophage ε15. J. Mol. Biol. 42, 237–246. 10.1016/0022-2836(69)90040-0 [DOI] [PubMed] [Google Scholar]
- L'Vov V. L., Iakovlev A. P., Shashkov A. S. (1989). [Study of the structure of O-specific polysaccharide of Salmonella anatum using 1H- and 13C-NMR-spectroscopy]. Bioorg. Khim. 15, 1660–1663. [PubMed] [Google Scholar]
- Manoil C., Boyd D., Beckwith J. (1988). Molecular genetic analysis of membrane protein topology. Trends Genet. 4, 223–226. 10.1016/0168-9525(88)90154-0 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Zheng C., Chitsaz F., Derbyshire M. K., Geer L. Y., Geer R. C., et al. (2013). CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41, D348–D352. 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur A., Król J. E., Marczak M., Skorupska A. (2003). Membrane topology of PssT, the transmembrane protein component of the type I exopolysaccharide transport system in Rhizobium leguminosarum bv. trifolii strain TA1. J. Bacteriol. 185, 2503–2511. 10.1128/JB.185.8.2503-2511.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. V., Pemberton J. M., Richards K. E. (1974). F116, D3 and G101: temperate bacteriophages of Pseudomonas aeruginosa. Virology 59, 566–569. 10.1016/0042-6822(74)90466-8 [DOI] [PubMed] [Google Scholar]
- Miyoshi-Akiyama T., Kuwahara T., Tada T., Kitao T., Kirikae T. (2011). Complete genome sequence of highly multidrug-resistant Pseudomonas aeruginosa NCGM2.S1, a representative strain of a cluster endemic to Japan. J. Bacteriol. 193, 7010. 10.1128/JB.06312-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M., Richards J. C., Fournier J.-M., Byrd R. A., Karakawa W. W., Vann W. F. (1990). Structure of the type 5 capsular polysaccharide of Staphylococcus aureus. Carbohydr. Res. 201, 285–297. 10.1016/0008-6215(90)84244-O [DOI] [PubMed] [Google Scholar]
- Morona R., Mavris M., Fallarino A., Manning P. A. (1994). Characterization of the rfc region of Shigella flexneri. J. Bacteriol. 176, 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhamchik A., Wilde C., Rowe-Magnus D. A. (2007). Identification of a Wzy polymerase required for group IV capsular polysaccharide and lipopolysaccharide biosynthesis in Vibrio vulnificus. Infect. Immun. 75, 5550–5558. 10.1128/IAI.00932-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath P., Morona R. (2015). Mutational analysis of the major periplasmic loops of Shigella flexneri Wzy: identification of the residues affecting O antigen modal chain length control, and Wzz-dependent polymerization activity. Microbiology 161, 774–785. 10.1099/mic.0.000042 [DOI] [PubMed] [Google Scholar]
- Newton G. J., Daniels C., Burrows L. L., Kropinski A. M., Clarke A. J., Lam J. S. (2001). Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol. Microbiol. 39, 1237–1247. 10.1111/j.1365-2958.2001.02311.x [DOI] [PubMed] [Google Scholar]
- Nghiêm H. O., Himmelspach K., Mayer H. (1992). Immunochemical and structural analysis of the O polysaccharides of Salmonella zuerich. J. Bacteriol. 174, 1904–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U., Ahrens C. H., Müller S., Wollscheid B. (2014). Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886. 10.1093/bioinformatics/btt607 [DOI] [PubMed] [Google Scholar]
- Paiment A., Hocking J., Whitfield C. (2002). Impact of phosphorylation of specific residues in the tyrosine autokinase, Wzc, on its activity in assembly of Group 1 capsules in Escherichia coli. J. Bacteriol. 184, 6437–6447. 10.1128/JB.184.23.6437-6447.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.-J., Fang H.-C., Yang H.-C., Lin T.-L., Hsieh P.-F., Tsai F.-C., et al. (2008). Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J. Clin. Microbiol. 46, 2231–2240. 10.1128/JCM.01716-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez J. M., McGarry M. A., Marolda C. L., Valvano M. A. (2008). Functional analysis of the large periplasmic loop of the Escherichia coli K-12 WaaL O-antigen ligase. Mol. Microbiol. 70, 1424–1440. 10.1111/j.1365-2958.2008.06490.x [DOI] [PubMed] [Google Scholar]
- Qutyan M., Paliotti M., Castric P. (2007). PilO of Pseudomonas aeruginosa 1244: subcellular location and domain assignment. Mol. Microbiol. 66, 1444–1458. 10.1111/j.1365-2958.2007.06001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C. K., Sims E. H., Kas A., Spencer D. H., Kutyavin T. V., Ivey R. G., et al. (2002). Genetic variation at the O-Antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 184, 3614–3622. 10.1128/JB.184.13.3614-3622.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins P. W., Bray D., Dankert M., Wright A. (1967). Direction of chain growth in polysaccharide synthesis - work on a bacterial polysaccharide suggests that elongation can occur at reducing end of growing chains. Science 158, 1536–1542. 10.1126/science.158.3808.1536 [DOI] [PubMed] [Google Scholar]
- Robbins P. W., Keller J. M., Wright A., Bernstein R. L. (1965). Enzymatic and kinetic studies on the mechanism of O-Antigen conversion by bacteriophage ϵ15. J. Biol. Chem. 240, 384–390. [PubMed] [Google Scholar]
- Rocchetta H. L., Burrows L. L., Pacan J. C., Lam J. S. (1998). Three rhamnosyltransferases responsible for assembly of the A-band D-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol. Microbiol. 28, 1103–1119. 10.1046/j.1365-2958.1998.00871.x [DOI] [PubMed] [Google Scholar]
- Ruan X., Loyola D. E., Marolda C. L., Perez-Donoso J. M., Valvano M. A. (2012). The WaaL O-antigen lipopolysaccharide ligase has features in common with metal ion-independent inverting glycosyltransferases. Glycobiology 22, 288–299. 10.1093/glycob/cwr150 [DOI] [PubMed] [Google Scholar]
- Sadovskaya I., Brisson J.-R., Thibault P., Richards J. C., Lam J. S., Altman E. (2000). Structural characterization of the outer core and the O-chain linkage region of lipopolysaccharide from Pseudomonas aeruginosa serotype O5. Eur. J. Biochem. 267, 1640–1650. 10.1046/j.1432-1327.2000.01156.x [DOI] [PubMed] [Google Scholar]
- Schild S., Lamprecht A.-K., Reidl J. (2005). Molecular and functional characterization of O Antigen transfer in Vibrio cholerae. J. Biol. Chem. 280, 25936–25947. 10.1074/jbc.M501259200 [DOI] [PubMed] [Google Scholar]
- Sigida E. N., Fedonenko Y. P., Shashkov A. S., Zdorovenko E. L., Konnova S. A., Ignatov V. V., et al. (2015). Structure of the polysaccharides from the lipopolysaccharide of Azospirillum brasilense Jm125A2. Carbohydr. Res. 416, 37–40. 10.1016/j.carres.2015.08.011 [DOI] [PubMed] [Google Scholar]
- Smith A. W., Iglewski B. H. (1989). Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. 10.1093/nar/17.24.10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislavsky E. S., Lam J. S. (1997). Pseudomonas aeruginosa antigens as potential vaccines. FEMS Microbiol. Rev. 21, 243–277. 10.1111/j.1574-6976.1997.tb00353.x [DOI] [PubMed] [Google Scholar]
- Tao J., Feng L., Guo H., Li Y., Wang L. (2004). The O-antigen gene cluster of Shigella boydii O11 and functional identification of its wzy gene. FEMS Microbiol. Lett. 234, 125–132. 10.1111/j.1574-6968.2004.tb09523.x [DOI] [PubMed] [Google Scholar]
- Taylor V. L., Udaskin M. L., Islam S. T., Lam J. S. (2013). The D3 Bacteriophage α-Polymerase Inhibitor (Iap) Peptide disrupts O-Antigen biosynthesis through mimicry of the chain length regulator Wzz in Pseudomonas aeruginosa. J. Bacteriol. 195, 4735–4741. 10.1128/JB.00903-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrane S. W., Taylor V. L., Freschi L., Kukavica-Ibrulj I., Boyle B., Laroche J., et al. (2015). The widespread multidrug-resistant serotype O12 Pseudomonas aeruginosa clone emerged through concomitant horizontal transfer of serotype antigen and antibiotic resistance gene clusters. MBio 6:e01396–15. 10.1128/mBio.01396-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády G. E., Simon I. (1998). Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283, 489–506. 10.1006/jmbi.1998.2107 [DOI] [PubMed] [Google Scholar]
- Uetake H., Luria S. E., Burrous J. W. (1958). Conversion of somatic antigens in Salmonella by phage infection leading to lysis or lysogeny. Virology 5, 68–91. 10.1016/0042-6822(58)90006-0 [DOI] [PubMed] [Google Scholar]
- Varga J. J., Losada L., Zelazny A. M., Kim M., McCorrison J., Brinkac L., et al. (2013). Draft genome sequences of Burkholderia cenocepacia ET12 lineage strains K56-2 and BC7. Genome Announc. 1. 10.1128/genomea.00841-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov E. V., Shashkov A. S., Knirel Y. A., Kochetkov N. K., Tochtamysheva N. V., Averin S. F., et al. (1991). Structure of the O-antigen of Francisella tularensis strain 15. Carbohydr. Res. 214, 289–297. 10.1016/0008-6215(91)80036-M [DOI] [PubMed] [Google Scholar]
- Whitfield C. (1995). Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 3, 178–185. 10.1016/S0966-842X(00)88917-9 [DOI] [PubMed] [Google Scholar]
- Wick R. R., Schultz M. B., Zobel J., Holt K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. 10.1093/bioinformatics/btv383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward R., Yi W., Li L., Zhao G., Eguchi H., Sridhar P., et al. (2010). In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat. Chem. Biol. 6, 418–423. 10.1038/nchembio.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W., Bystricky P., Yao Q., Guo H., Zhu L., Li H., et al. (2006). Two different O-polysaccharides from Escherichia coli O86 are produced by different polymerization of the same O-repeating unit. Carbohydr. Res. 341, 100–108. 10.1016/j.carres.2005.11.001 [DOI] [PubMed] [Google Scholar]
- Zhao G., Wu B., Li L., Wang P. (2014). O-antigen polymerase adopts a distributive mechanism for lipopolysaccharide biosynthesis. Appl. Microbiol. Biotechnol. 98, 4075–4081. 10.1007/s00253-014-5552-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.