Abstract
In many arthropods, maternally inherited endosymbiotic bacteria can increase infection frequency by manipulating host reproduction. Multiple infections of different bacteria in a single host population are common, yet few studies have documented concurrent endosymbiont phenotypes or explored their potential interactions. We hypothesized that spiders might be a particularly useful taxon for investigating endosymbiont interactions, because they are host to a plethora of endosymbiotic bacteria and frequently exhibit multiple infections. We established two matrilines from the same population of the linyphiid spider Mermessus fradeorum and then used antibiotic curing and controlled mating assays to demonstrate that each matriline was subject to a distinct endosymbiotic reproductive manipulation. One matriline was co-infected with Rickettsia and Wolbachia and produced offspring with a radical female bias. Antibiotic treatment eliminated both endosymbionts and restored an even sex ratio to subsequent generations. Chromosomal and fecundity observations suggest a feminization mechanism. In the other matriline, a separate factorial mating assay of cured and infected spiders demonstrated strong cytoplasmic incompatibility (CI) induced by a different strain of Wolbachia. However, males with this Wolbachia induced only mild CI when mated with the Rickettsia–Wolbachia females. In a subsequent survey of a field population of M. fradeorum, we detected these same three endosymbionts infecting 55% of the spiders in almost all possible combinations, with nearly half of the infected spiders exhibiting multiple infection. Our results suggest that a dynamic network of endosymbionts may interact both within multiply infected hosts and within a population subject to multiple strong reproductive manipulations.
Introduction
Maternally inherited endosymbiotic bacteria represent independent genetic elements within the cells and tissues of their arthropod hosts. Unlike virulent pathogens, the fate of these endosymbionts is wedded to their host's survival and reproduction (Engelstädter and Hurst, 2009), yet the interests of host and endosymbiont may not align. In particular, host and endosymbiont may come into conflict over offspring sex, as males are evolutionary dead ends for maternally inherited endosymbionts (O'Neill et al., 1997; Werren et al., 2008; Engelstädter and Hurst, 2009). Endosymbionts use a variety of mechanisms to manipulate host reproduction to promote the spread of infection within a host population, even if host fitness is reduced (Hoffmann and Turelli, 1997). The most common reproductive manipulation is cytoplasmic incompatibility (CI), which modifies the sperm of infected males such that it cannot produce a viable embryo unless the same or a closely related endosymbiont in the egg rescues sperm functionality (Werren et al., 2008). Alternatively, endosymbionts can directly skew the sex ratio of hosts by inducing parthenogenesis (Huigens and Stouthamer, 2003), killing males (Hurst and Jiggins, 2000) or feminizing genetic males (Negri et al., 2006).
Endosymbionts are very common; >30% of arthropods are infected with Wolbachia or other endosymbionts (for example, Rickettsia, Cardinium) known to cause reproductive anomalies (Duron et al., 2008a). The extent of endosymbiont infection is likely underestimated, as low frequency infections are easily missed in broad taxa screening studies (Hilgenboecker et al., 2008) and low titer infections often go undetected (Hughes et al., 2011). Rampant and assorted endosymbiont infections in arthropods suggest that endosymbionts employing different manipulative strategies will likely interact within the same host population or even the same host body. Although multiple endosymbiont infections have been documented across numerous host taxa (for example, Riegler and Stauffer, 2002; Duron et al., 2010), relatively few studies have empirically determined the phenotypic effects of potentially interacting endosymbionts on their hosts (Hiroki et al., 2004; Charlat et al., 2006; White et al., 2009; White et al., 2011; Zhu et al., 2012).
We suggest that spiders may be a particularly rich host taxon for investigating endosymbiont interactions. Though most endosymbiont investigation has focused on insects, recent surveys demonstrate that spiders harbor a wealth of endosymbionts (Goodacre et al., 2006; Baldo et al., 2008; Duron et al., 2008b; Perlman et al., 2010; Yun et al., 2011; Stefanini and Duron, 2012), with multiple endosymbionts co-occurring within the same population or co-infecting the same individual (Goodacre, 2011; Vanthournout et al., 2014). However, the functional role of these spider endosymbionts is not yet clearly defined. To date, only one reproductive manipulation has been reported in spiders: female bias thought to be male killing in Oedothorax gibbosus and O. retusus (Vanthournout et al., 2011, 2014). However, two novel behavioral alterations have also been associated with facultative bacterial symbionts in spiders (Goodacre et al., 2009; Gunnarsson et al., 2009), implying that endosymbionts may have a different or expanded suite of manipulative tools in spiders than their insect relatives (Goodacre, 2011).
To better understand what endosymbiont interactions may arise in a diverse endosymbiont community, we characterized the phenotypic function of different infection types in a population of the linyphiid spider, Mermessus fradeorum. We established two infection types of M. fradeorum in the laboratory, cured each infection type with antibiotics and determined endosymbiont effects on sex ratio and fecundity by comparing infected and uninfected spiders. Endosymbiont effects on genetic sex were determined via chromosomal observation. We then conducted mating assays to test for CI. Finally, we determined the natural frequency of endosymbiont infection in a field population of M. fradeorum to evaluate the potential for interaction between endosymbionts within the host body and among differentially infected hosts in the same population.
Materials and methods
Establishment of laboratory matrilines and determination of endosymbiont infection status
M. fradeorum is a small (average body length of 2.5 mm) sheet-weaving linyphiid spider common in Kentucky agroecosystems and generally found in agricultural fields and grasslands throughout North America (Welch, 2013). To investigate endosymbiont phenotypes in the laboratory, gravid M. fradeorum females were hand collected in February of 2012 from winter wheat at the Spindletop Research Farm in Lexington, KY, USA (38.1272°N, 84.5081°W). Each spider was placed in an individual plastic rearing cup (4.6 cm × 7.3 cm diameter) with a plaster base and imbedded plastic straws for web attachment, in a 24 h dark incubator with 98% relative humidity and a 16 h:8 h thermal cycle of 28 °C:18 °C. Egg masses were removed following deposition, and upon hatching, individual spiderlings were transferred to separate rearing cups. Spiders were fed twice a week: juveniles received Proisotoma minuta (Collembola), whereas adults received a combination of P. minuta, Sinella curviseta (Collembola) and Drosophila melanogaster (Diptera).
After egg mass deposition, mother spiders were screened for three endosymbionts common in spiders: Wolbachia, Rickettsia, and Cardinium. Prior to DNA extraction, spiders were surface sterilized with sequential 10 s rinses of ethanol, deionized water, 5% bleach solution and deionized water (adapted from Medina et al., 2011). DNA was extracted using a Chelex extraction protocol adapted from White et al. (2009): abdomens of adults were pulverized in 4 μl of Proteinase K, incubated in a 20% Chelex bead solution for 60 min at 37 °C and then heat shocked for 10 min at 95 °C to deactivate Proteinase K. Extraction quality was assessed by measuring DNA concentration using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and by PCR using universal eukaryotic primers NSF 18/19 or COI (Supplementary Table S1). To detect the endosymbionts, we used taxon-specific primers and protocols described in Supplementary Table S1 for segments of 16S rRNA of Rickettsia and Cardinium and the Wolbachia surface protein (wsp). PCR products were visualized under ultraviolet light on 1.5% agarose gels treated with Gel-Red (Biotium, Hayward, CA, USA). Endosymbiont identity for all positive PCR results was confirmed via Sanger Sequencing at the University of Kentucky's AGTC sequencing facility. Prey items were also screened for symbionts, none of which were detected in laboratory stocks. Morphological spider identification was confirmed by sequencing the COI barcoding region of each spider lineage (GenBank accession KJ561375).
Offspring of two females were used to establish two matrilines of differential endosymbiont infection types: ‘R1W1' bearing a double Rickettsia–Wolbachia infection, and ‘W2' singly infected with a different Wolbachia strain. Vertical transmission of the endosymbionts was confirmed in each generation with diagnostic PCR using new primers (Ricklong and mfwsp), which were designed using the Primer3 tool (Rozen and Skaletsky, 1998) in Geneious 5.6.4 (Drummond et al., 2012) to specifically amplify the Rickettsia and Wolbachia detected in M. fradeorum (Supplementary Table S1). We previously characterized the microbiomes of each matriline via 454-pyrosequencing of 16S bacterial diversity and did not find indication that other maternally inherited bacterial endosymbionts were present in the laboratory lines (Curry, 2013). Early rearing observation indicated that the R1W1 line produced only female offspring, whereas the W2 line produced equal numbers of male and female offspring. The R1W1 line was therefore propagated by matings with W2 line males. To prevent inbreeding depression, additional M. fradeorum infected with W2 were collected from a different Lexington, KY population in 2012 and reared in the laboratory for several generations. Sons from three different field-collected W2 mothers were introgressed equally into third generation R1W1 and W2 matrilines prior to antibiotic treatment.
Experimentally cured lines were generated for both R1W1 and W2 matrilines to investigate endosymbiont phenotype. To eliminate endosymbiotic bacteria in M. fradeorum, second and third instar spiderlings from each matriline were treated with antibiotics previously shown to be effective in spiders (Goodacre et al., 2009; Vanthournout et al., 2011). For 5 consecutive days, spiderlings (13 R1W1 and 25 W2) were sprayed with a fine mist of 0.1% ampicillin solution daily. Following 1 week of no treatment, the same spiderlings were sprayed with a 0.1% tetracycline solution daily for 5 days. Treated spiders were mated, and curing was confirmed in the subsequent generation by screening offspring for Rickettsia and Wolbachia. Antibiotic treatment successfully eliminated all symbiotic bacteria from R1W1 and W2 lines, establishing R1−W1− and W2− cured lines. Experiments were conducted with spiders at least two generations removed from antibiotic treatment to alleviate any direct negative fitness effects of the antibiotics.
Endosymbiont effects on sex ratio and fecundity
The effects of endosymbionts in the doubly infected R1W1 line and the singly infected W2 line were quantified by comparing life history parameters of naturally infected R1W1 and W2 lines with antibiotically cured R1−W1− or W2− lines. Females of the four lines were mated with cured males to eliminate the potential influence of paternal endosymbiont type (see Table 1 for sample sizes). Females were mated with males of either the R1−W1− or the W2− cured line. Mating was confirmed by visual observation, and males were removed 1 day after mating. Females were allowed to produce egg sacs for 10 days. We preserved all parents in 95% ethanol and stored them at −20 °C and then measured maternal size using patella–tibia length. All mothers and a subset of randomly selected fathers were subsequently screened for Rickettsia and Wolbachia to verify appropriate infection status.
Table 1. Mean±s.e. for all life history parameters observed in maternal endosymbiont effects and cytoplasmic incompatibility experiments with Mermessus fradeorum.
| n Mothers | Cross type: (female × male) | Hatched | Unhatched | Fecundity | Ratio female | Proportion undeveloped | Survivorship of hatched spiderlings |
|---|---|---|---|---|---|---|---|
| Maternal endosymbiont effects | |||||||
| 12 | R1W1 × uninfecteda | 10.7±1.1 | 1.0±0.8 | 20.7±2.4b | 1±0 | 0.16±0.11 | 0.97±0.01 |
| 10 | R1−W1− × uninfected | 11.7±1.4 | 0.1±0.1 | 21.8±2.2 | 0.62±0.06 | 0.02±0.02 | 0.97±0.02 |
| 7 | W2 × uninfected | 11.9±0.8 | 1.2±0.9 | 18.6±3.4 | 0.54±0.07 | 0.12±0.08 | 0.97±0.01 |
| 4 | W2− × uninfected | 12.0±1.8 | 0±0 | 16.4±3.6 | 0.42±0.17 | 0±0 | 1±0 |
| CI in W2 females | |||||||
| 12 | W2− × W2 | 7.3±2.4 | 14.0±3.3 | 21.3±2.5c | 0.53±0.08 | 0.63±0.063 | 1±0 |
| 15 | W2 × W2 | 24.9±1.9 | 1.5±1.2 | 26.4±1.6 | 0.54±0.03 | 0.05±0.052 | 0.87±0.04 |
| 11 | W2− × W2− | 28.1±1.9 | 1.0±0.7 | 29.1±1.5 | 0.40±0.03 | 0.04±0.063 | 0.94±0.03 |
| 11 | W2 × W2− | 27.6±2.3 | 0.7±0.6 | 28.3±1.8 | 0.54±0.07 | 0.05±0.063 | 0.91±0.03 |
| CI in R1W1 females | |||||||
| 4 | R1W1 × W2 | 25.5±5.4 | 8.0±4.3 | 33.5±1.3c | NA | 0.25±0.15 | NA |
| 4 | R1W1 × W2− | 39.5±3.8 | 0.5±0.3 | 40.0±3.7 | NA | 0.01±0.01 | NA |
Abbreviations: CI, cytoplasmic incompatibility; NA, not applicable.
Uninfected males are composed of R1−W1− and W2−.
Fecundity for maternal endosymbiont effects experiment includes embryo counts from egg sacs not allowed to hatch.
Fecundity is the sum of hatched and unhatched embryos; all egg sacs were allowed to hatch.
Females typically deposited two to three egg sacs in total. One egg sac per mother was randomly selected for rearing; the remaining egg sacs were removed and stored in 95% ethanol for embryo counting. Upon hatching, spiderlings were transferred to individual rearing cups, and each egg sac was checked for undeveloped embryos. Offspring were individually reared to adulthood to determine survivorship and sex. Fecundity was calculated as the total number of offspring, hatched and unhatched, in all egg sacs for each mother.
Statistically, we first considered whether paternal lineage (R1−W1− and W2−) influenced any of the response variables. We found no difference in the total production of offspring (one-way analysis of variance F=0.043, d.f.=1,42, P=0.836), sex ratio (logistic regression χ2=1.556, d.f.=1, P=0.212) or survivorship (logistic regression χ2=0.525, d.f.=1, P=0.217) between R1−W1− and W2− fathers, therefore cured males of both matrilines were pooled for subsequent analysis. We also did not find evidence that maternal size should be included in our models as a covariate, as there was no relationship between maternal size (patella–tibia length) and fecundity (regression R2=0.002, d.f.=42, P=0.7903). Therefore fecundity per mother was compared among matriline categories (R1W1, R1−W1−, W2, W2−) using a simple one-way analysis of variance in JMP 10 (SAS Institute Inc., 2012) followed by Tukey's honestly significant difference test to separate means. Endosymbiont effect on sex ratio was determined using an exact binomial two-tailed test for goodness of fit (McDonald and Delaware, 2009), testing each category against the expected 1:1 sex ratio. Differences in embryo development (hatched versus unhatched eggs) and survivorship of hatched spiderlings to adulthood among lines were analyzed with logistic regression using Arc 1.06 (Cook and Weisberg, 1999). When overall significance of the model was established, Wald statistics were used to make comparisons among categories.
Determination of genetic sex in R1W1 offspring
To determine endosymbiont effect on genetic sex, infected R1W1 eggs and uninfected eggs (both R1−W1− and W2−) at different stages of development were removed from their egg sacs using forceps and separately subjected to the staining protocol below. Eggs were pierced with forceps and soaked in hypotonic solution (0.075 M KCl) for 45 min at room temperature. The chorion was then removed, and egg contents were transferred to a freshly prepared solution of ethanol–acetic acid (3:1) and incubated for 45 min at room temperature. Egg contents were then transferred to a drop of 60% acetic acid on a clean glass slide and spread and dried on a slide drier at 45 °C. Dried slides were stained with either 5% giemsa in Sörensen buffer or aceto-orcein (2.5 g orcein per 100 ml 45% acetic acid) for 45 min at room temperature. Slides were then rinsed three times in distilled water, mounted in glycerol and sealed with nail polish. Slides were observed using a Zeiss Opton inverted microscope with phase contrast (Carl Zeiss Microscopy, Göttingen, Germany), and chromosomes from mitotically dividing cell spreads were counted. Although hundreds of spreads were prepared, spreads in which chromosomes were sufficiently dispersed so the total number could be reliably counted were rare. We counted chromosomes in spreads created from a total of 26 eggs: 12 eggs from 5 different egg sacs laid by uninfected females, and 14 eggs from 4 different egg sacs laid by R1W1 infected females. Eggs were categorized as male or female based on chromosome number, and an exact binomial two-tailed goodness-of-fit test was used to determine whether the sex ratio differed from the expected 1:1 in either infection category (McDonald and Delaware, 2009).
CI experiments
We tested whether W2 Wolbachia induces CI by conducting a two-way factorial cross, mating W2 and W2-females to W2 or W2-males (see Table 1 for sample sizes). Following mating, mothers and fathers were screened for Wolbachia. Matings, husbandry methods and fitness parameters for the CI cross were the same as those used in the previously described experiment, except that all egg sacs were allowed to hatch. One egg sac from each pair was randomly selected for full individual rearing of offspring to adulthood. Emerging spiderlings from the other egg sacs were immediately preserved in ethanol. One day following hatching, egg sacs were preserved in ethanol to prevent embryo desiccation and subsequently dissected to quantify undeveloped embryos. Hatching success was calculated as the number of spiderlings that successfully hatched relative to the total number of embryos produced. Hatching success, spiderling survivorship to adulthood and sex ratio were compared among the cross types using logistic regression in Arc 1.06 with Williams' correction (Williams, 1982) to address moderate overdispersion in the data when necessary.
We also investigated whether W2 in males induces CI when mated with R1W1 females; R1W1 females were mated with either W2 or W2− males (see Table 1 for sample sizes). For each mating pair, all conditions were identical to the previous CI cross, with the exception that no spiderlings were reared to adulthood. Hatching success was again analyzed using logistic regression in Arc v. 1.06.
Endosymbiont infection frequency in M. fradeorum
In August 2013, live M. fradeorum adults were collected from winter wheat at the same location as the original matriline collections. To eliminate false positives due to residual gut contents (Greenstone et al., 2013), spiders were reared on endosymbiont-free prey for at least 3 days and then starved for at least 2 additional days prior to DNA extraction. Upon morphological identification, specimens were dissected to remove epigynal and palpal vouchers. As described previously, adult spiders were surface sterilized and DNA extracted, and the 44 extractions that passed quality control were screened for Rickettsia, Wolbachia and Cardinium (Supplementary Table S1). Spider species (COI barcodes) and endosymbiont identity for all positive PCR results were confirmed via Sanger sequencing. Because two distinct Wolbachia strains were present in the laboratory M. fradeorum (W1 or W2), we also amplified and sequenced the fbpA gene from each Wolbachia-positive field specimen and the laboratory lines. Upon inspection, a number of specimens had double peaks in their Wolbachia chromatograms that perfectly corresponded to the single-nucleotide polymorphisms between Wolbachia strains W1 and W2 (Riegler and Stauffer, 2002). These specimens were categorized as doubly infected with W1 and W2; there was no evidence for additional Wolbachia strains. A few of the spiders had deposited eggs before being killed, and offspring of one Rickettsia-positive female were individually reared to adulthood to determine the sex ratio of offspring using the same rearing conditions described previously.
Results
Endosymbiont strain characterization in laboratory matrilines
Diagnostic PCR and Sanger sequencing confirmed that each of the laboratory-reared matrilines carried a different Wolbachia strain and that one matriline also carried Rickettsia. R1W1 and W2 matrilines were reared in the laboratory for nine generations with nearly perfect vertical endosymbiont transmission rates. Transmission of Rickettsia and Wolbachia in the R1W1 line from mothers to offspring exhibited 100% fidelity (19 mothers, 51 offspring). In the W2 line, we did not detect maternal Wolbachia infection in 2/86 offspring (from 17 mothers), indicating 98% transmission efficiency under laboratory conditions. Sequencing of a 590-bp region of 16S rRNA indicated that the Rickettsia present in the R1W1 line of M. fradeorum (KJ546642) is 99% similar to the Rickettsia of the leafhopper Nephotettix cincticeps (AB702995). The two Wolbachia strains in M. fradeorum diverge from one another by 8.8% for the wsp gene (W1: KJ561379, W2: KJ561380) and 2.8% for the fbpA gene (W1: KJ561362, W2: KJ561367). Sequences for both W1 and W2 wsp and fbpA genes were more similar to Supergroup A Wolbachia strains than strains from other supergroups.
Endosymbiont effects on sex ratio and fecundity
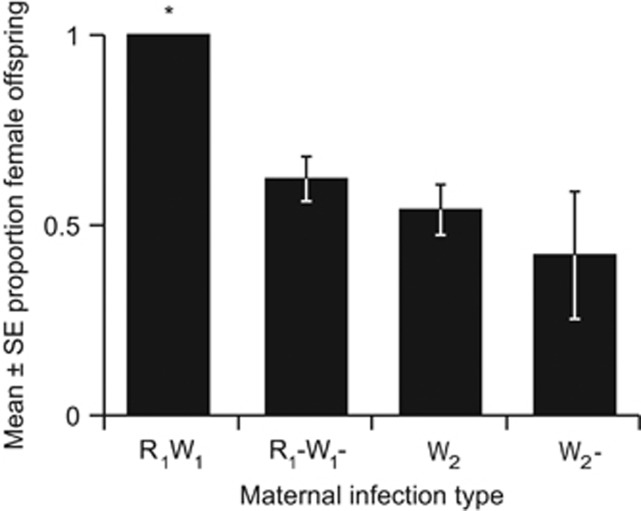
The R1W1 line offspring were entirely female and significantly differed from a 1:1 sex ratio (P<<0.001). In contrast, the antibiotically cured R1−W1− line produced male offspring and showed a restoration to the expected 1:1 sex ratio (P=0.064; Figure 1, Table 1). Neither the Wolbachia-infected W2 line (P=0.777) nor its cured W2− counterpart (P=0.618) had sex ratios that differed from 1:1, indicating that this strain of Wolbachia does not affect sex ratio (Figure 1, Table 1). The extreme female bias in the R1W1 line was observed not only in this experiment but also in nine generations of rearing. Only four males in total were produced out of the 328 non-experimental R1W1 line spiders individually reared in the laboratory. Of the four R1W1 males produced, all four were reproductively viable and successfully mated with females. Three of these males were screened for endosymbionts, and all three were positive for Rickettsia and Wolbachia.
Figure 1.
Mean±s.e. proportion female offspring from a controlled mating assay of each maternal infection type mated with uninfected males. Asterisk indicates a sex ratio that is significantly different from 1:1. R=Rickettsia, W=Wolbachia.
No significant difference in fecundity was detected among the four infection types (F=0.618, d.f.=3,39, P=0.608; Table 1) and few undeveloped embryos were found in hatched egg sacs, yielding no significant differences in proportion of undeveloped embryos among groups (χ2=2.98, d.f.=3, P=0.395; Table 1). Therefore, we found no evidence that endosymbiotic bacteria in the R1W1 and W2 mothers affect fecundity or embryo development. Furthermore, survivorship to adulthood for hatched offspring did not significantly differ among infection categories (χ2=2.98, d.f.=3, P=0.395; Table 1).
Determination of genetic sex in R1W1 offspring
Counting of chromosomes from testes spreads and observation of chromosomes in living cells suggest that the male number of chromosomes in M. fradeorum is 2n=24 and the female number is 2n=26, with a X1X1X2X2 (female)/X1X20 (male) sex determination system, similar to other spiders (Kořínková and Král, 2013). Examination of 12 uninfected eggs revealed 26 chromosomes in spreads from 7 eggs and 24 chromosomes in spreads from 5 eggs; indicating a sex ratio of 1.4:1 which does not differ significantly from 1:1 (P=0.387). Among the 14 eggs from R1W1 mothers, 8 eggs had 26 chromosomes and 6 had 24 chromosomes; this sex ratio of 1.3:1 also does not differ significantly from 1:1 (P=0.395). These data suggest that R1W1 females produce male eggs (24 chromosomes) but that these males are feminized into morphological females by bacterial endosymbionts in the all-female R1W1 line.
CI experiments
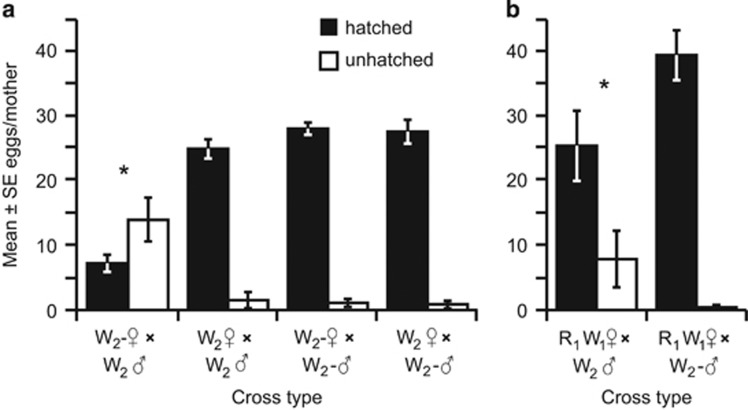
Wolbachia-infected W2 fathers induced CI when mated with cured W2− mothers, resulting in 73% fewer hatched spiderlings than any other mating combination (χ2=38.4, d.f.=3, P<0.001; Table 1,Figure 2a) with no significant differences among the other cross types. Cross type did not influence the sex ratio of offspring (χ2= 4.35, d.f.=3, P=0.22; Table 1). Survivorship of hatched offspring to adulthood differed significantly among the four different cross types (Wald χ2=10.12, d.f.=3, P=0.017; Table 1), with greater survival of offspring in the incompatible cross type than when W2-infected mothers mated with W2-infected fathers. Survivorship of hatched offspring in the other two crosses was intermediate and did not differ significantly from any other crosses. Anecdotally, visual observation of hatching spiderlings indicated that spiderlings from the incompatible cross often emerged later than other crosses, appeared larger and were observed consuming undeveloped embryos.
Figure 2.
Mean±s.e. hatched spiderlings (black) and unhatched embryos (white) from experimental matings of (a) W2 infected and W2− uninfected females and males and (b) R1W1 females with either W2 or W2− males. Asterisks indicate cross types that differed in proportion of hatched embryos from the other treatments in that experiment at α=0.05. R=Rickettsia, W=Wolbachia.
Though W2 induced strong CI when W2− females were crossed with W2 males, CI was relatively weak when R1W1 females were crossed with W2 males. Egg hatch in R1W1 × W2 matings was only 35% lower than R1W1 × W2− matings (χ2=5.06, d.f.=1, P=0.025; Figure 2b). This cross does not support the strong level of CI observed with the W2− mothers, suggesting that the endosymbionts or other elements present in R1W1 mothers may partially rescue the CI induced by W2.
Endosymbiont infection frequency in a field population of M. fradeorum
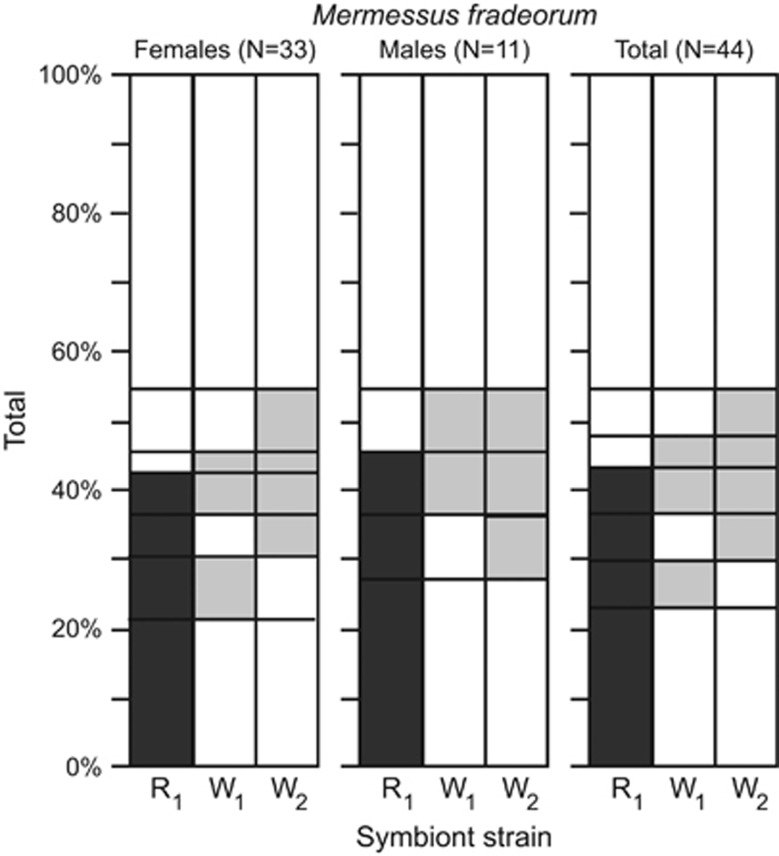
Diagnostic PCR screening indicated that 55% of all field-collected M. fradeorum individuals bore an endosymbiont infection (Figure 3). All three endosymbiont strains present in the laboratory populations (R1, W1, W2) were detected in the field-collected M. fradeorum in six of the seven possible infection combinations; W1 was never found as a single infection but was always found as a co-infection with R1 and/or W2 (Figure 3). Males were more difficult to collect and represented only 25% of our sample, precluding tests of statistical association between endosymbiont infection and sex ratio. However, males corresponding to all female infection types were found in the field, with the exception of R1W1 and W2. One R1 female reproduced in the laboratory prior to being screened and produced male and female offspring. A subsequent F1 mating of R1 siblings produced 23 male and 23 female offspring under rearing conditions analogous to all other experimental matings.
Figure 3.
Percentage of specimens positive for three endosymbiont strains detected in the Mermessus fradeorum spider survey. Female (N=33), male (N=11) and total (N=44) columns' order from left to right. Multiply infected individuals are indicated by more than one shaded region on the same horizontal plane. R=Rickettsia (dark grey), W=Wolbachia (light grey).
Discussion
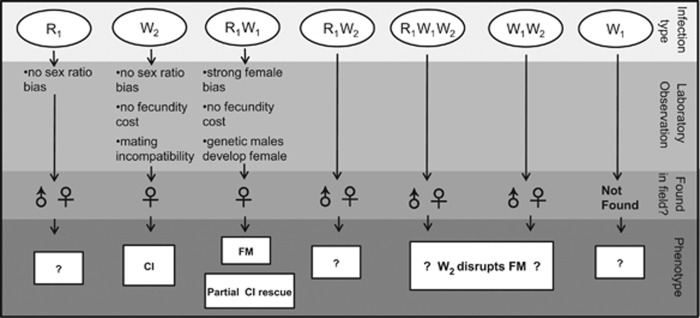
Multiple bacterially induced reproductive manipulations are simultaneously at work within a single population of the spider M. fradeorum (Figure 4). Some individuals, infected with Rickettsia and Wolbachia (R1W1), exhibit an almost complete female bias, whereas other individuals, infected with a different strain of Wolbachia (W2), experience CI. Both CI-inducing (Hoffmann and Turelli, 1997) and female-biasing (Taylor, 1990) endosymbionts can invade and come to dominate uninfected host populations. However, theoretical models suggest that competing endosymbionts may disrupt one another's spread (Engelstädter et al., 2004; Charlat et al., 2006). Indeed, the few previous empirical studies documenting multiple manipulations within a host population have primarily confirmed the dominance of one or the other manipulation (Hiroki et al., 2004; Charlat et al., 2006; Narita et al., 2006). In contrast, the presence of both manipulations at both time points for M. fradeorum (initial laboratory line collection and field survey 2 years later) suggests that both manipulations may persist in this system. Linyphiid spiders such as M. fradeorum are often subject to frequent disturbance in agroecosystems, necessitating high rates of dispersal (Weyman et al., 2002). It is possible that high migration rates may promote nonequilibrial coexistence of differentially infected hosts subject to conflicting reproductive manipulations (Engelstädter et al., 2004).
Figure 4.
Summary and implications of laboratory rearing and field collection for each endosymbiotic infection type for Mermessus fradeorum. FM=feminization, CI=cytoplasmic incompatibility, R=Rickettsia, W=Wolbachia.
In our field screen of M. fradeorum, we found these three endosymbiont strains in almost all possible combinations within hosts, indicating that endosymbiont infections in this host may be relatively dynamic, through imperfect vertical transmission and/or horizontal transmission of symbionts between hosts (Rowley et al., 2004; Goodacre et al., 2006). Vertical transmission rates in M. fradeorum were very high in the laboratory but may be lower in the field (Turelli and Hoffmann, 1995). High endosymbiont loss rates would be consistent with the relatively high proportion of uninfected individuals observed in the field (Figure 3). Horizontal transmission has previously been inferred among spiders, through hypothesized mechanisms such as shared vectors, cannibalism, paternal transmission (Baldo et al., 2008) or a predatory lifestyle (Rowley et al., 2004). Regardless of mechanism, the ‘mix and match' among endosymbionts in this system may lead to conflicting selection on bacterial strains, depending on the presence or absence of co-infecting strains.
In R1W1 spiders, the relative contribution of the two symbionts to the expressed phenotype is not entirely clear. Despite repeated attempts at low-dose antibiotics (for example, White et al. 2009), we were not able to differentially cure one or the other endosymbiont and therefore could not directly attribute feminization to either bacterial strain. However, 3 out of the 11 field males bore an R1 infection (Figure 3), and two generations of R1 rearing in the laboratory demonstrated an even sex ratio. These results indicate that R1 Rickettsia alone does not preclude production of males. It is possible that W1 alone is responsible for the female bias in R1W1, but no lone W1 infections were found in the field to allow evaluation of this hypothesis. It is also possible that both Rickettsia and Wolbachia are required to induce female bias in R1W1 spiders; previous studies investigating the respective roles of co-infecting symbionts have sometimes found interactive effects among the symbionts (Zhu et al., 2012) and sometimes have not (White et al., 2009).
This radical female bias induced by the R1W1 infection type is likely the result of feminization, rather than male killing or parthenogenesis. Fecundity and the number of undeveloped embryos in R1W1 versus R1−W1− spiders were similar (Table 1), whereas one would expect a substantial reduction in fecundity if male killing were at play in a population with a 1:1 ratio. Parthenogenetic reproduction is not supported, as we never observed virgin females to produce offspring during nine generations of laboratory rearing. Furthermore, the bimodal chromosome numbers in the R1W1 line of M. fradeorum is consistent with a mixture of genetic females (n=26 chromosomes) and feminized genetic males (n=24 chromosomes). Feminization is suspected to be very common in isopods (Cordaux et al., 2011) but has only rarely been reported in insects and mites (Engelstädter and Hurst, 2009). The present study is the first indication of feminization by endosymbionts in Araneae, but this mechanism of manipulation may be common in an order rife with female-biased populations (Eberhard, 2004).
On its own, W2 induces strong CI, reducing fecundity by 73% when W2 males were mated with uninfected W2− females. However, W2 CI strength may be undercut in two ways. First, the 27% of offspring that escaped CI in the incompatible cross enjoyed a higher chance of survival to adulthood, perhaps due to consumption of undeveloped siblings. Sibling cannibalism can provide a significant fitness benefit when resources are scarce, which is often the basis for the spread of male-killing endosymbionts in host populations (Freeland and McCabe, 1997). For a CI-inducing endosymbiont, however, a significant fitness boon for survivors of the incompatible cross is predicted to undermine its ability to spread or compete with other endosymbionts in the population (Freeland and McCabe, 1997). Second, W2 induced only weak CI in feminized R1W1 M. fradeorum. In this instance, we were unable to perform reciprocal crosses that would allow us to distinguish whether this partial rescue was attributable to the R1W1 endosymbionts; nevertheless, this lowered level of CI may help the R1W1 infection type persist in the same host population as the CI-inducing W2 infection.
As a further indication of potential endosymbiont interactions within hosts, our field collection data suggest that R1W1 feminization may be modulated by the additional presence of W2 within spiders. One of the three field-collected R1W1W2 spiders was male, as was one of the two field-collected W1W2 spiders, ratios that are unlikely if these spiders show a similar female bias as the R1W1 spiders in the laboratory (P<0.05, Fisher's exact test). Future studies to explore the phenotypes of other symbiont combinations within this system will provide important insights into the potential for conflict and collaboration among independent genetic elements within a shared host environment. Our results, in conjunction with a growing body of evidence for widespread multiple endosymbiont infection in spiders (Goodacre, 2011; Stefanini and Duron, 2012; Vanthournout et al., 2014), indicate that these infections are common and dynamic, suggesting that spiders are an ideal taxon for the investigation of interactions among endosymbionts.
Importantly, our results also affirm that the same mechanisms of reproductive manipulation found in insects are active in spiders. To the best of our knowledge, this is the first demonstration of CI and feminization in a spider, signifying that the larger body of endosymbiont knowledge based upon insects is likely applicable to spiders as well. In one population of M. fradeorum, we have found multiple endosymbionts, a high prevalence of multiply infected individuals and two distinct reproductive manipulations. Though the picture is far from complete, the potential importance of interactions among endosymbionts in population dynamics should not be ignored.
Data archiving
The data used in this study are available at the Dryad Digital Repository doi:10.5061/dryad.935hp.
Acknowledgments
We thank A Dehnel, Z Helton and E Squires of the University of Kentucky for help with spider rearing and diagnostic PCR. We also thank all reviewers for their constructive comments on previous drafts of this manuscript. This research was supported by the University of Kentucky College of Agriculture and Department of Entomology and USDA National Institute of Food and Agriculture, Hatch/Multistate project KY00856. The information reported in this paper (15-08-016) is part of a project of the Kentucky Agricultural Experiment Station and is published with the approval of the Director.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Baldo L, Ayoub NA, Hayashi CY, Russell JA, Stahlhut JK, Werren JH. (2008). Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol Ecol 17: 557–569. [DOI] [PubMed] [Google Scholar]
- Charlat S, Engelstädter J, Dyson EA, Hornett EA, Duplouy A, Tortosa P et al. (2006). Competing selfish genetic elements in the butterfly Hypolimnas bolina. Curr Biol 16: 2453–2458. [DOI] [PubMed] [Google Scholar]
- Cook RD, Weisberg S. (1999) Applied Regression Including Computing and Graphics. John Wiley & Sons: New York, NY, USA. [Google Scholar]
- Cordaux R, Bouchon D, Grève P. (2011). The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet 27: 332–341. [DOI] [PubMed] [Google Scholar]
- Curry MM. (2013). Endosymbiotic prevalence and reproductive manipulation of the spider Mermessus fradeorum. MSc thesis, University of Kentucky, Lexington, KY, USA.
- Duron O, Bouchon D, Boutin S, Bellamy L, Zhou LQ, Engelstädter J et al. (2008. a). The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol 6: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron O, Hurst GDD, Hornett EA, Josling JA, Engelstädter J. (2008. b). High incidence of the maternally inherited bacterium Cardinium in spiders. Mol Ecol 17: 1427–1437. [DOI] [PubMed] [Google Scholar]
- Duron O, Raymond M, Weill M. (2010). Many compatible Wolbachia strains coexist within natural populations of Culex pipiens mosquito. Heredity 106: 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C et al. (2012). Geneious v5.6. Available from http://www.geneious.com.
- Eberhard WG. (2004). Why study spider sex: special traits of spiders facilitate studies of sperm competition and cryptic female choice. J Arachnol 32: 545–556. [Google Scholar]
- Engelstädter J, Telschow A, Hammerstein P. (2004). Infection dynamics of different Wolbachia- types within one host population. J Theor Biol 231: 345–355. [DOI] [PubMed] [Google Scholar]
- Engelstädter J, Hurst GDD. (2009). The ecology and evolution of microbes that manipulate host reproduction. Annu Rev Ecol Evol Syst 40: 127–149. [Google Scholar]
- Freeland SJ, McCabe BK. (1997). Fitness compensation and the evolution of selfish cytoplasmic elements. Heredity 78: 391–402. [Google Scholar]
- Goodacre SL, Martin OY, Thomas CFG, Hewitt GM. (2006). Wolbachia and other endosymbiont infections in spiders. Mol Ecol 15: 517–527. [DOI] [PubMed] [Google Scholar]
- Goodacre SL, Martin OY, Bonte D, Hutchings L, Woolley C, Ibrahim K et al. (2009). Microbial modification of host long-distance dispersal capacity. BMC Biol 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodacre SL. (2011). Endosymbiont infections in spiders. Casas J (ed). Advances in Insect Physiology Vol 60, Academic Press: London, UK. pp 137–153. [Google Scholar]
- Greenstone MH, Payton ME, Weber DC, Simmons AM. (2013). The detectability half-life in arthropod predator-prey research: what it is, why we need it, how to measure it, and how to use it. Mol Ecol 23: 3799–3813. [DOI] [PubMed] [Google Scholar]
- Gunnarsson B, Goodacre SL, Hewitt GM. (2009). Sex ratio, mating behaviour and Wolbachia infections in a sheetweb spider. Biol J Linnean Soc 98: 181–186. [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. (2008). How many species are infected with Wolbachia?–a statistical analysis of current data. FEMS Microbiol Lett 281: 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroki M, Tagami Y, Miura K, Kato Y. (2004). Multiple infection with Wolbachia inducing different reproductive manipulations in the butterfly Eurema hecabe. Proc Soc Biol 271: 1751–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Turelli M. (1997). Cytoplasmic incompatibility in insects. O'Neill SL, Hoffmann AA, Werren JH (eds). Influential Passengers: Inherited Microorganisms and Arthropod Reproduction. Oxford University Press: Oxford, UK. pp 42–80. [Google Scholar]
- Hughes G, Allsopp P, Brumbley S, Woolfit M, McGraw E, O'Neill S. (2011). Variable infection frequency and high diversity of multiple strains of Wolbachia pipientis in Perkinsiella planthoppers. Appl Environ Microbiol 77: 2165–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huigens M, Stouthamer R. (2003). Parthenogenesis associated with Wolbachia. Bourtzis K, Miller TA (eds). Insect Symbiosis. CRC Press: Boca Raton, FL, USA. pp 247–266. [Google Scholar]
- Hurst GDD, Jiggins FM. (2000). Male-killing bacteria in insects: mechanisms, incidence, and implications. Emerg Infect Dis 6: 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kořínková T, Král J. (2013). Karyotypes, sex chromosomes, and meiotic division in spiders. Nentwig W (ed). Spider Ecophysiology. Springer: Heidelberg, NY, USA. pp 159–171. [Google Scholar]
- McDonald JH, Delaware UO. (2009) Handbook of Biological Statistics. Sparky House Publishing: Baltimore, MD, USA. [Google Scholar]
- Medina R, Nachappa P, Tamborindeguy C. (2011). Differences in bacterial diversity of host- associated populations of Phylloxera notabilis Pergande (Hemiptera: Phylloxeridae) in pecan and water hickory. J Evol Biol 24: 761–771. [DOI] [PubMed] [Google Scholar]
- Narita S, Nomura M, Kato Y, Fukatsu T. (2006). Genetic structure of sibling butterfly species affected by Wolbachia infection sweep: evolutionary and biogeographical implications. Mol Ecol 15: 1095–1108. [DOI] [PubMed] [Google Scholar]
- Negri I, Pellecchia M, Mazzoglio P, Patetta A, Alma A. (2006). Feminizing Wolbachia in Zyginidia pullula (Insecta, Hemiptera), a leafhopper with an XX/X0 sex-determination system. P Soc Biol 273: 2409–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill SL, Hoffmann AA, Werren JH. (1997) Influential Passengers: Inherited Microorganisms and Arthropod Reproduction. Oxford University Press: Oxford, UK, 214 pp. [Google Scholar]
- Perlman SJ, Magnus SA, Copley CR. (2010). Pervasive associations between Cybaeus spiders and the bacterial symbiont Cardinium. J Invertebr Pathol 103: 150–155. [DOI] [PubMed] [Google Scholar]
- Rowley SM, Raven RJ, McGraw EA. (2004). Wolbachia pipientis in Australian spiders. Curr Microbiol 49: 208–214. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. (1998) Primer3. Whitehead Institute: Cambridge, MA, USA. Available from http://www-genome.wi.mit.edu/genome_software/other/primer3.html. [Google Scholar]
- Riegler M, Stauffer C. (2002). Wolbachia infections and superinfections in cytoplasmically incompatible populations of the European cherry fruit fly Rhagoletis cerasi (Diptera, Tephritidae). Mol Ecol 11: 2425–2434. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc.. (2012) Using JMP 10. SAS Institute Inc.: Cary, NC, USA. [Google Scholar]
- Stefanini A, Duron O. (2012). Exploring the effect of the Cardinium endosymbiont on spiders. J Evol Biol 25: 1521–1530. [DOI] [PubMed] [Google Scholar]
- Taylor D. (1990). Evolutionary consequences of cytoplasmic sex ratio distorters. Evol Ecol 4: 235–248. [Google Scholar]
- Turelli M, Hoffmann AA. (1995). Cytoplasmic incompatibility in Drosophila simulans: dynamics and parameter estimates from natural populations. Genetics 140: 1319–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanthournout B, Swaegers J, Hendrickx F. (2011). Spiders do not escape reproductive manipulations by Wolbachia. BMC Evol Biol 11: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanthournout B, Vandomme V, Hendrickx F. (2014). Sex ratio bias caused by endosymbiont infection in the dwarf spider Oedothorax retusus. J Arachnol 42: 24–33. [Google Scholar]
- Welch KD. (2013). Selective utilization of microhabitats by web-building spiders. PhDSc dissertation, University of Kentucky, Lexington, KY, USA.
- Werren JH, Baldo L, Clark ME. (2008). Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6: 741–751. [DOI] [PubMed] [Google Scholar]
- Weyman G, Sunderland K, Jepson P. (2002). A review of the evolution and mechanisms of ballooning by spiders inhabiting arable farmland. Ethol Ecol Evol 14: 307–326. [Google Scholar]
- White JA, Kelly SE, Perlman SJ, Hunter MS. (2009). Cytoplasmic incompatibility in the parasitic wasp Encarsia inaron: disentangling the roles of Cardinium and Wolbachia symbionts. Heredity 102: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JA, Kelly SE, Cockburn SN, Perlman SJ, Hunter MS. (2011). Endosymbiont costs and benefits in a parasitoid infected with both Wolbachia and Cardinium. Heredity 106: 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DA. (1982). Extra-binomial variation in logistic linear models. Appl Stat 31: 144–148. [Google Scholar]
- Yun YL, Lei CL, Peng Y, Liu FX, Chen JA, Chen LB. (2011). Wolbachia strains typing in different geographic population spider, Hylyphantes graminicola (Linyphiidae). Curr Microbiol 62: 139–145. [DOI] [PubMed] [Google Scholar]
- Zhu L-Y, Zhang K-J, Zhang Y-K, Ge C, Gotoh T, Hong X-Y. (2012). Wolbachia strengthens Cardinium-induced cytoplasmic incompatibility in the spider mite Tetranychus piercei McGregor. Curr Microbiol 65: 516–523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.