Abstract
Farmed fish are typically genetically different from wild conspecifics. Escapees from fish farms may contribute one-way gene flow from farm to wild gene pools, which can depress population productivity, dilute local adaptations and disrupt coadapted gene complexes. Here, we reanalyse data from two experiments (McGinnity et al., 1997, 2003) where performance of Atlantic salmon (Salmo salar) progeny originating from experimental crosses between farm and wild parents (in three different cohorts) were measured in a natural stream under common garden conditions. Previous published analyses focussed on group-level differences but did not account for pedigree structure, as we do here using modern mixed-effect models. Offspring with one or two farm parents exhibited poorer survival in their first and second year of life compared with those with two wild parents and these group-level inferences were robust to excluding outlier families. Variation in performance among farm, hybrid and wild families was generally similar in magnitude. Farm offspring were generally larger at all life stages examined than wild offspring, but the differences were moderate (5–20%) and similar in magnitude in the wild versus hatchery environments. Quantitative genetic analyses conducted using a Bayesian framework revealed moderate heritability in juvenile fork length and mass and positive genetic correlations (>0.85) between these morphological traits. Our study confirms (using more rigorous statistical techniques) previous studies showing that offspring of wild fish invariably have higher fitness and contributes fresh insights into family-level variation in performance of farm, wild and hybrid Atlantic salmon families in the wild. It also adds to a small, but growing, number of studies that estimate key evolutionary parameters in wild salmonid populations. Such information is vital in modelling the impacts of introgression by escaped farm salmon.
Introduction
Intentional releases from hatcheries or unintentional escapes from aquaculture facilities can lead to genetic introgression between captive and wild fish populations where interbreeding occurs. Commercial farming of Atlantic salmon (Salmo salar) has increased markedly over the past few decades, raising concerns over the genetic and ecological impacts on native populations (Naylor et al., 2005). Escapes from open net-pen culture facilities regularly occur, either via chronic low-level ‘leakage' or acute events (for example, storms) that release thousands of fish at one time (Naylor et al., 2005). Many wild Atlantic salmon stocks are currently severely depleted (ICES, 2010) and in some regions farm escapees can account for a third or more of salmon caught at sea (Hansen et al., 1999) or on the spawning grounds (Fiske et al., 2006). A range of studies have demonstrated that escaped farm salmon can successfully spawn in the wild (Fleming et al., 1996) and hence may contribute one-way gene flow from farm to wild gene pools (Clifford et al., 1998; Skaala et al., 2006; Glover et al., 2012, 2013).
Farmed Atlantic salmon are often genetically different from wild conspecifics because of geographical origin, founder effects (Skaala et al., 2004), and especially domestication selection and genetic drift in captivity. For example, artificial selection for economically desirable traits such as faster growth and delayed maturity has been applied to many farm strains (Gjøen and Bentsen, 1997; Gjedrem, 2000). The domestication process can also lead to rapid genetic changes in farm populations as a result of unintentional selection on non-target traits, for example, increased aggression, higher risk-taking and altered feeding behaviours (Einum and Fleming, 1997; Fleming et al., 2002; Houde et al., 2010), or as a result of relaxed selection and genetic drift because of propagation with a limited number of broodstock (Lynch and O'Hely, 2001).
In the wild, salmon populations invariably exhibit hierarchical genetic structure, with substantial genetic differences apparent among regions, neighbouring catchments within regions and even tributaries within the same river (Dionne et al., 2008; Bourret et al., 2013). Some of this genetic divergence is thought to reflect adaptations to local environments (Garcia de Leaniz et al., 2007), although the magnitude of local adaptation varies with spatial scale (Fraser et al., 2011). If continued one-way gene flow occurs from farm to wild salmon populations at high rates, then genetic differences (both among wild populations and between wild and farm populations) could rapidly erode, although some populations may be less susceptible to ‘genetic invasion' than others (Glover et al., 2012, 2013).
Introgressive hybridisation between farm and wild salmon can also lead to a drop in mean individual fitness in the wild. Experimental studies involving artificial crosses between wild and farm fish have provided evidence that offspring with one or two farm parents display lower survival than those with two wild parents (McGinnity et al., 1997, 2003; Skaala et al., 2012). Larger, more aggressive farm and hybrid fish may also displace native fish or force them into suboptimal habitats, which increases average mortality (McGinnity et al., 1997, 2003; Fleming et al., 2000). These studies suggest that repeated introductions of farm fish may depress the productivity of wild populations through both ecological and genetic mechanisms, in addition to fostering genetic homogenisation (Skaala et al., 2006; Glover et al., 2012) and potential loss of local adaptations. Most studies of the effects of artificial immigration of non-native fish (whether from farms or hatcheries), however, tend to emphasise group-level performance differences and typically overlook family-level variation in performance (but see Skaala et al., 2012). Information on families can minimise analytical bias and yield important insights; for example, certain non-native or hybrid families may fortuitously perform much better than others in natural environments and therefore contribute disproportionately to introgression of non-native alleles/traits into wild populations (Garant et al., 2003).
A recent Norwegian study found substantial among-family differences in the freshwater growth and survival of Atlantic salmon in a natural stream setting, with progeny of farm parents exhibiting a broader range of survival rates (in addition to a lower mean survival) than hybrid or wild progeny (Skaala et al., 2012). As noted by these authors, patterns of variation in the performance of farm, wild and hybrid families are likely to vary across space and through time, given that rivers vary in habitat characteristics and performance depends on an interaction between genes and environment. The extent of genetic divergence between wild and farmed salmon (and hence the potential threat of outbreeding depression) is also expected to vary among locales depending on the farm strains used, patterns of differentiation in the local wild populations, and the extent of any prior gene flow from farm to wild populations. Additional data on family differences in survival and fitness-related traits (for example, size-at-age) of farm salmon and farm-wild hybrids (particularly F2 hybrids and backcrosses, which were not included in the Skaala et al., 2012 study and which provide extra information on the genetic basis of farm-wild differences and multi-generation consequences of interbreeding) from other geographic locations therefore would be highly valuable to estimate evolutionary consequences of introgression. On a more practical level, data on families can reveal if overall differences in mean performance between farmed, wild and hybrid groups are driven by one or two outlier families. Moreover, if phenotypic data are collected on related individuals (for example, half-siblings), the resulting pedigree can be exploited to estimate quantitative genetic parameters such as trait heritabilities and genetic correlations, for which there are still very few estimates from wild salmonid populations (Carlson and Seamons, 2008). Information on the extent to which variation in fitness-related traits (for example, size-at-age) is transmitted from parents to offspring is also crucial to predicting the genetic and demographic consequences of introgression.
Here, we reanalyse data from two experiments conducted in the west of Ireland (McGinnity et al., 1997, 2003) where survival and size-at-age of Atlantic salmon progeny originating from experimental crosses between farm and wild parents (in three different cohorts) were measured in a natural stream under common garden conditions. We have three primary objectives: (1) to reanalyse these data with modern mixed effects models that account for kin structure to test properly for group-level differences in mean survival and size-at-age, and to check whether patterns were driven by outlier families. (2) To test whether farm or hybrid families exhibited different patterns of variation in survival and size-at-age relative to wild families (that is, variance heterogeneity with respect to groups). For example, farm families may exhibit higher variance than wild families (Skaala et al., 2012), while outcrossing can lead to changes in additive genetic and residual (non-additive genetic and environmental) variance in hybrid groups (Lynch and Walsh, 1998; Debes and Hutchings, 2014). (3) To exploit the pedigree structure inherent in the experimental designs to estimate quantitative genetic parameters of interest in a wild setting. Effects of egg size on offspring performance, assumed to reflect environmental maternal effects, are also tested and controlled for statistically at different offspring ages.
Materials and methods
Study area and experimental design
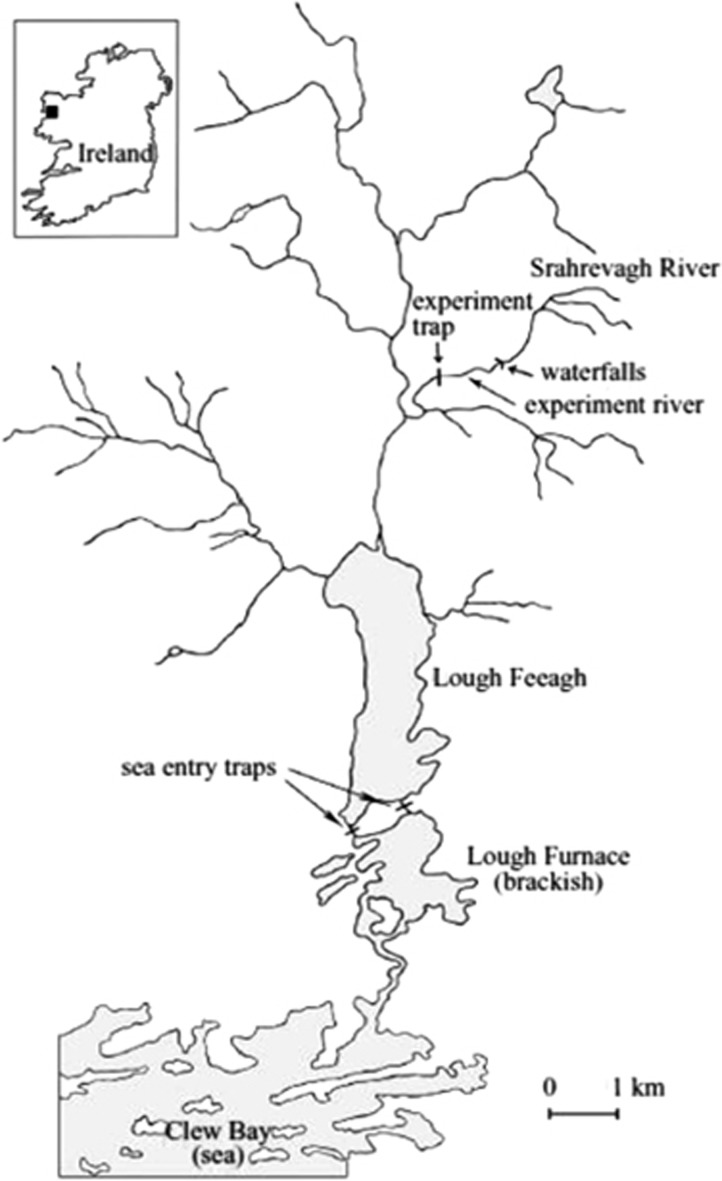
The experiments were undertaken in the Burrishoole system in the west of Ireland (Figure 1). A number of afferent rivers flow into Lough Feeagh (one of two major lakes in the catchment), one of which (the Srahrevagh River, hereafter ‘experiment river') was used for the freshwater stages of the experiment and was equipped with a trap (‘experiment trap') capable of capturing all downstream moving juveniles and upstream migrating adults. The first experiment involved artificial crosses between farm adults (a derivative of the Norwegian Mowi strain established in Ireland in 1983, which became known as the ‘Fanad' strain) and wild adults captured in the Burrishoole system in December 1992 and December 1993. By 1983, the Mowi strain had already experienced circa 15 years (3–5 generations) of domestication in Norway, and thereafter the selection trajectory of the Fanad strain, which has never received inputs from Irish wild strains, was likely different from that of the farm strains in Norway (Norris et al., 1999). Four cross-types (hereafter simply ‘groups') were made, involving pure farm, pure wild and both reciprocal hybrids (Table 1). The families established from the December 1992 broodstock, which hatched in spring 1993, are referred to as the 1993 cohort; similarly, the families established from the December 1993 broodstock, which hatched in spring 1994, are referred to as the 1994 cohort. To produce both the 1993 and 1994 cohorts, each farm dam was mated to one farm sire and one wild sire, and vice versa; thus all dams and sires were mated twice. For full details on the experimental design for the 1993 and 1994 cohorts, see (McGinnity et al., 1997) and Supplementary Table S1 (which includes a schematic on the mating design).
Figure 1.
Map of the Burrishoole river system showing location of experiment river, experiment trap and sea-entry traps.
Table 1. Experimental groups of Atlantic salmon in the 1993, 1994 and 1998 cohorts.
| Cohort | Group | Group code | No. dams | No. sires | No. families | No. eggs | Mean egg size (mm±s.d.) | % Farm genes |
|---|---|---|---|---|---|---|---|---|
| 1993 | Wild D × wild S | WW_93 | 6 | 6 | 6 | 5273 | 0.60 (0.04) | 0 |
| Wild D × farm S | WF_93 | 6 | 6 | 6 | 5886 | 0.60 (0.04) | 50 | |
| Farm D × wild S | FW_93 | 8 | 8 | 8 | 8659 | 0.61 (0.03) | 50 | |
| Farm D × farm S | FF_93 | 15 | 15 | 15 | 14 997 | 0.61 (0.04) | 100 | |
| 1994 | Wild D × wild S | WW_94 | 11 | 11 | 11 | 10 537 | 0.61 (0.04) | 0 |
| Wild D × farm S | WF_94 | 11 | 11 | 11 | 10 537 | 0.61 (0.04) | 50 | |
| Farm D × wild S | FW_94 | 11 | 11 | 11 | 10 537 | 0.64 (0.05) | 50 | |
| Farm D × farm S | FF_94 | 11 | 11 | 11 | 10 537 | 0.64 (0.05) | 100 | |
| 1998 | Wild D × wild S | WW_98 | 4 | 5 | 12 | 8787 | 0.61 (0.02) | 0 |
| F1 hybrid × wild | BC1W_98 | 14 | 5 | 41 | 9549 | 0.61 (0.02) | 25 | |
| F1 hybrid × F1 hybrid | F2Hy_98 | 14 | 2 | 26 | 8337 | 0.61 (0.02) | 50 | |
| F1 hybrid × farm | BC1F_98 | 14 | 5 | 42 | 9928 | 0.60 (0.03) | 75 | |
| Farm D × farm S | FF_98 | 7 | 5 | 33 | 9832 | 0.61 (0.02) | 100 |
Abbreviations: D, dam; S, sire.
Number of eggs=number of eyed–eggs planted out in the experiment river. Final column gives the expected percentage of farm genes per group.
In the autumn of 1997, returning F1 hybrid Atlantic salmon, which had been ranched (that is, released to the ocean as hatchery-reared smolts) from the 1994 cohort and had spent two winters at sea (2SW), were captured at the sea-entry traps (Figure 1). These were then used to produce F2 hybrids and BC1 backcrosses, whereas a new set of farm and wild adults were used as broodstock to produce pure and F1 hybrids (Table 1). Families thus established, which hatched in spring 1998, are referred to as the 1998 cohort. The mating design for the 1998 cohort was slightly different from the 1993 and 1994 cohorts (Supplementary Table S1). For full details on the experimental design for the 1998 cohort, see (McGinnity et al., 2003).
For each cohort, families were first mixed at the eyed-egg stage and then planted out in the experiment river in artificial redds (Donaghy and Verspoor, 2000). Juveniles were then sampled from the experiment river by electrofishing in August 1993, August 1994 and August 1998. The experiment trap was also inspected daily from 30 April 1993 to 20 April 1995, and from 24 April 1998 to 30 June 2011. A random subset of parr and smolts from the experiment river caught in the experiment trap during these periods were killed and preserved in 95% ethanol. Fish in their first calendar year of life were denoted as 0+ and in their second calendar year as 1+. For the 1993 and 1994 cohorts, subsamples of eggs from each family (250 eggs per family for 1993 cohort, 200 eggs per family for 1994 cohort, eggs measured at this point) were retained in the hatchery and reared to the smolt stage, denoted as ‘hatchery controls' (measured before being released to the ocean as smolts, and hence termed ‘pre-smolts'). A sample of 0+ parr from the 1993 cohort hatchery control group was sampled in August 1993, while further samples of mature male parr and pre-smolts were taken from the hatchery controls in November 1993 and March 1994, respectively. A sample of hatchery pre-smolts was also taken from the 1994 cohort in March 1995, just before their release to sea. In total, sampling of the 1993, 1994 and 1998 cohorts yielded 14 different data sets on size-related traits and survival. DNA profiling techniques based on microsatellite (1998 cohort) or minisatellite (1993 and 1994 cohorts) marker loci were used to assign sampled offspring back to their parents with close to 100% power, allowing individuals to be grouped into families (see McGinnity et al., 1997, 2003 for full details on the molecular methods and parentage assignment).
Statistical analyses
Representation
As the number of fish per family in some samples is determined by both emigration from the experiment stream and survival, counts are referred to simply as ‘representation', following McGinnity et al. (1997, 2003, 2004). A series of generalised linear mixed effects models (GLMMs) were constructed to examine variation in family-level representation at different life/sampling stages. Mixed effects models are a powerful statistical technique for making inferences about explanatory variables of interest (typically the fixed effects, that is, terms for which regression coefficients are estimated) while properly accounting for any sources of non-independence or hierarchical structure (random effects, that is, terms for which an estimate of the variance is obtained) in the data; GLMMs are used when the response variable is non-normal (Bolker et al., 2009). The GLMMs were fitted in R version 3.0.2 using the glmer function from the lme4 package (Bates et al., 2012). The binomial response variable considered in these models was a concatenated vector of the number of individuals represented per family and the number not represented (the initial number of eggs per family planted out minus the number of individuals represented) and a logit link function was used. ‘Dam' and ‘sire' (unique identifier codes for each mother and father) were included as random effects, which accounts for the kinship structure inherent in the data (full-sibs nested within half-sibs) and also provides estimates of the variance attributable to each parent.
For each model, fixed effects of group as a factor (that is, separate levels for each cross type) and eyed-egg diameter (mean-centred) were included. The latter was a single value per family (see McGinnity et al., 1997, 2003 for details on how this was measured) and was used as an index of maternal effects mediated via egg size (Einum and Fleming, 1999). Dam fork length (LF) and egg mass were also measured but both were strongly correlated with egg diameter (r>0.5 in all cohorts), so to avoid problems with collinearity of explanatory variables, only egg diameter was included in the models. Backwards model selection (Zuur et al., 2009) was performed on the fixed effects, by dropping each in turn and retaining only significant terms (as assessed using likelihood ratio tests) in the final model, whereas retaining the random effects of sire and dam regardless of their significance (which was necessary to properly account for kin structure in the data). Multiple contrasts with univariate P-values were then used to test whether each group differed significantly from the pure wild group (the reference group).
The existence of outlier families was checked by visually examining the family-level representation data. If a potential outlier was identified, its influence on the overall results was checked by re-running the analysis for that particular sample excluding that family and determining whether the results were changed qualitatively. To test for variance heterogeneity across groups in the raw representation data, the non-parametric Figner–Killeen test of homogeneity of variances was used. The null hypothesis was that all groups had equal variance; the alternative hypothesis was that the variance differed for at least two of them. Finally, to test whether representation was consistent from the 0+ to 1+ parr stages, representation of 1+ parr per family (sampled in June 1995) were plotted against representation of 0+parr per family (sampled in August 1994) and a standard regression performed.
Size-at-age
A series of linear mixed effects models (LMMs) were constructed to examine variation in the LF and mass of juveniles at different life stages (note that for some data sets, mass was not measured). Using LMMs is appropriate as ‘family' can be fitted as a random effect, which accounts for non-independence of measurements taken on individuals belonging to the same family (that is, accounts for ‘genetic pseudoreplication'). Failure to account for family structure can lead to inflated statistical significance of treatment (here group) effects, as the effective sample size per treatment level is lower than the number of observations per level (Zuur et al., 2009). The goals of these LMMS were to test for (1) group differences in mean LF and mass, (2) environmental maternal effects mediated via egg size (eyed-egg diameter) and (3) heterogeneity among groups in between-family variance and within-family variance. These goals were achieved by fitting a series of hierarchical models in two steps. In the first step, the most appropriate random effects structure was determined while including all candidate fixed effects, regardless of their statistical significance (Zuur et al., 2009). In the second step, backwards model selection was performed on the fixed effects (while retaining the best random effects structure identified in the first step) to determine which were significant. For each model, fixed effects of group and eyed-egg diameter were included. The response variables (LF, mass) were natural log transformed, which ensured that model residuals were normally distributed.
To determine the most appropriate random effects structure and test for variance heterogeneity across groups (for example, whether the variation in farm fish was less than that of wild fish), five different (increasingly complex) models were compared for each response variable. First, a common residual variance only was estimated using generalised least squares (the gls function in the R library nmle). Second, a random effect of family (common to all groups) was included (using the lme function). Third, the random effect of family was stratified by group, which allowed for different between-family variances for each group. Fourth, a common random effect of family (that is, not stratified by group) was fitted and the residual variance was stratified by group (which allowed for different within-family variances for each group). Fifth, both the random effect of family and the residual variance were stratified by group (which allowed for heterogeneity in both between-family and within-family variance). The model with the lowest Akaike information criterion was then chosen as the most appropriate model in terms of the random effects. To reduce the number of parameters to be estimated, all mixed ancestry groups were merged into a single ‘hybrids' group when stratifying the family or residual variance by group. That is, ‘group' was a three-level factor (pure, wild and hybrids) when included in the random effects part of the model, whereas hybrid groups were distinguished as separate levels when ‘group' was fitted as a fixed effect. Significance of the fixed effects were then tested via backwards selection, with P-values calculated by comparing models with and without the fixed effect of interest (fit by maximum likelihood) using likelihood ratio tests.
For the above LMMs, we focussed on size-at-age variation in the electrofishing and hatchery control samples only, where all individuals were measured on the same day. Variation in size-at-age was not examined for parr, pre-smolts and smolts caught in the experiment trap, as these fish were caught at different times of year and hence size differences could simply reflect age differences (age not being known accurately). The sample sizes were also insufficient to support more complex analyses of family variation in growth trajectories (for example, random regression) for the trap sample data. As for the representation analyses, the existence of outlier families was checked by visually examining the family-level size-at-age data. If a potential outlier was identified, its influence on the overall results was checked by re-running the analysis for that particular sample excluding that family and determining whether the results were changed qualitatively.
Quantitative genetic analyses
A Bayesian animal model approach was taken to estimate quantitative genetic parameters of interest, using the R package MCMCglmm (Hadfield, 2010). The animal model is a particular form of LMMs in which the breeding value, or ‘additive genetic merit', of each individual is treated as a random effect. An estimate of the additive genetic variance (VA), and in the case of multivariate models, also the additive genetic covariance (COVA) can be obtained by combining phenotypic data with a pedigree. In our case, sampled offspring were assigned back to their parents with almost complete certainty, as there were no unknown parents (see McGinnity et al., 1997, 2003). The resulting pedigree gives an expectation of how breeding values should co-vary among individuals of different genetic relatedness (in this case full-sibs and half-sibs; note that parental phenotypes were not measured at the same age and hence could not be included in the analysis), which then allows VA and COVA to be solved for algebraically (Kruuk, 2004; Hadfield, 2010).
Although it would have been possible to pool data from all groups to estimate quantitative genetic parameters, we chose not to, as outcrossing genetically divergent groups (that is, farm and wild fish) leads to changes in non-additive genetic components of variance (dominance and epistasis) in the hybrids (Lynch and Walsh, 1998). The data and pedigree structure were not sufficiently informative to separate out these non-additive components (which otherwise end up in the residual variance, VR) and hence obtaining clean estimates of heritability with the pooled data would be problematic, as both Va and VR are expected to vary among cross-types (groups). We therefore ran animal models separately for the pure wild and pure farm groups only and only for samples where at least 50 individuals were measured. Egg size was included as a continuous fixed effect in all cases to test for environmental maternal effects mediated via egg size.
Bivariate animal models were used to analyse variation in LF and mass simultaneously. Fixed effects of egg size were estimated for each trait in the same model (by including a trait × egg size interaction), and the phenotypic variance–covariance matrix was decomposed into an additive genetic matrix and a residual (environmental) matrix (Hadfield, 2010). The distribution of both traits was modelled as Gaussian and weakly informative inverse Wishart priors were used (posterior distributions were robust to alternative prior specifications). Samples were taken from the posterior distributions of the parameters every1000 iterations of the Markov chain, after an initial burn-in of 2.5 × 104 iterations, for a total of 1000 samples. In all cases, this was sufficient to achieve good convergence and acceptably low (<0.1) autocorrelation between adjacent Markov chain Monte Carlo samples. Posterior distributions of the narrow sense heritability h2 of each trait (for wild and farm groups separately) were calculated by dividing the posterior distribution of VA by the sum of the posterior distributions of VA and VR, and the mode and 95% credible intervals (CIs) of these posterior h2distributions are then presented. Posterior distributions of the genetic correlation between LF and mass were calculated as the posterior distribution of the genetic covariance divided by the square root of the product of the posterior distributions of the genetic variances. General maternal environmental effects not accounted for by egg size effects were also tested for in all models by including an additional random effect of ‘mother identity', but in all cases this variance component was estimated at close to zero (and the deviance information criterion did not drop by >2 units) and hence was not included in the final models.
Results
Representation
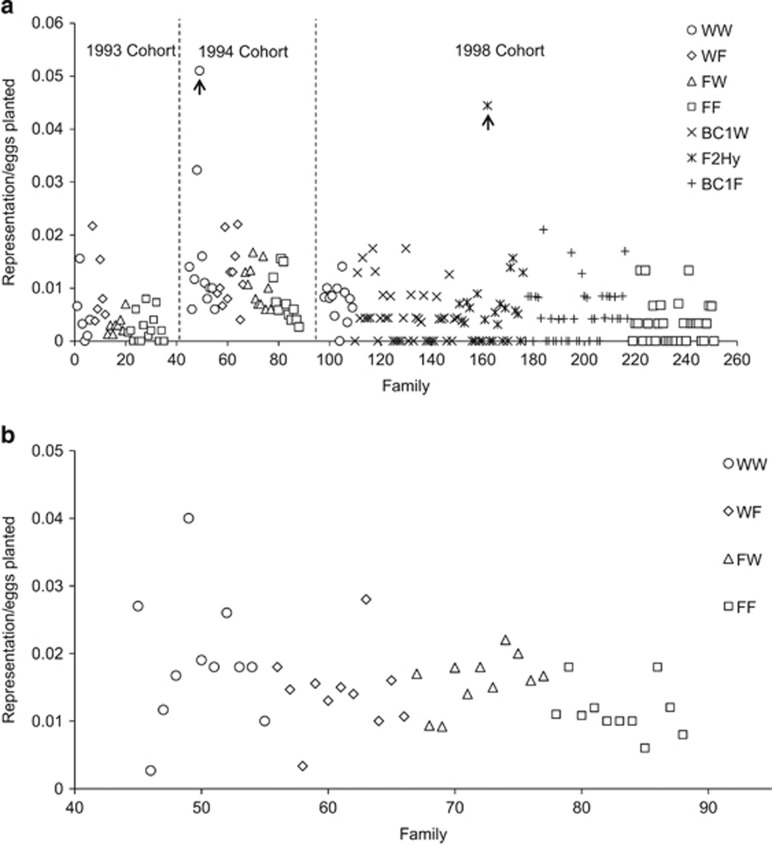
Overall group-level differences in representation were consistently found for 0+ parr in the electrofishing samples from each cohort, and for 1+ parr in the 1994 cohort (Table 2, full statistical results presented in Supplementary Table S2). In the 1993 cohort 0+ parr electrofishing sample, the WF (wild mother, farm father) group was significantly over-represented relative to the pure wild group (WW) reference group but the other groups were equally represented (Table 2, Figure 2). For the 1994 cohort, both 0+ and 1+ electrofished parr were significantly under-represented in the pure farm group (FF) group relative to the WW group, whereas 0+ parr were also under-represented in the FW (farm mother, wild father) group (Table 2, Figure 2). There was one obvious outlier in the WW group for the 1994 cohort 0+ parr electrofishing (family 49, Figure 2a); when this outlier was excluded, the results were qualitatively unchanged. Egg size had a significantly positive effect on representation of 0+ parr in the 1993 and 1994 cohort electrofishing samples and on the representation of 1+ parr in the June 1995 (1994 cohort) electrofishing sample (Supplementary Table S2, Table A2.1 and Supplementary Figure S1).
Table 2. Mean representation (number of fish in the sample divided by the initial number of eggs planted out/retained in hatchery) and mean LF for each group for the 1993, 1994 and 1998 cohorts.
| Cohort | Sample |
Representation |
Fork length (LF) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WW | WF | FW | FF | WW | WF | FW | FF | ||||
| 1993 | Electrofishing August 1993 0+parr | 0.005 (1.00) | 0.01 (1.97)a | 0.003 (0.61) | 0.002 (0.49) | 54.50±0.96 (1.00) | 57.43±0.58 (1.05)a | 54.78±1.00 (1.01) | 59.24±0.92 (1.09)b | ||
| 1994 | Electrofishing August 1994 0+parr | 0.016 (1.00) | 0.012 (0.76)c | 0.010 (0.65)d | 0.008 (0.48)d | 54.13±0.33 (1.00) | 55.67±0.43 (1.03)a | 56.89±0.43 (1.05)a | 57.41±0.52 (1.06)b | ||
| 1994 | Electrofishing June 1995 1+parr | 0.019 (1.00) | 0.014 (0.74) | 0.016 (0.84) | 0.011 (0.58)b | 94.02±0.71 (1.00) | 96.46±0.69 (1.03) | 98.09±0.66 (1.04)a | 98.69±0.78 (1.05)b | ||
| 1993 | Trapped parr May 1993–May 1994 | 0.017 (1.00) | 0.014 (0.82) | 0.008 (0.47)b | 0.005 (0.29)d | ||||||
| 1993 | Trapped pre-smolts+smolts September 1994–April 1995 | 0.007 (1.00) | 0.006 (0.86) | 0.003 (0.43)c | 0.003 (0.43)c | ||||||
| 1994 | Trapped parr May 1994–May 1995 | 0.023 (1.00) | 0.011 (0.48)b | 0.012 (0.52)b | 0.005 (0.22)d | ||||||
| 1994 | Trapped pre-smolts+smolts September 1995–April 1996 | 0.003 (1.00) | 0.002 (0.67) | 0.003 (1.00) | 0.002 (0.67) | ||||||
| 1993 | Hatchery controls 0+parr August 1993 | 0.013 (1.00) | 0.012 (0.92) | 0.009 (0.69) | 0.01 (0.77) | 106.14±2.72 (1.00) | 114.09±2.23 (1.07)a | 109.64±2.28 (1.03) | 111.41±1.54 (1.05)a | ||
| 1993 | Hatchery controls mature 0+parr November 1993 | 0.018 (1.00) | 0.006 (0.33)a | 0.011 (0.61) | 0.005 (0.28)a | 133.78±1.77 (1.00) | 135.89±5.07 (1.02) | 137.81±3.18 (1.03) | 133.47±2.48 (0.99) | ||
| 1993 | Hatchery controls smolts March 1994 | 0.023 (1.00) | 0.013 (0.57) | 0.019 (0.83) | 0.019 (0.83) | 170.35±1.72 (1.00) | 168.16±2.03 (0.99) | 173.21±1.73 (1.02) | 178.56±1.68 (1.05)a | ||
| 1994 | Hatchery controls smolts March 1995 | 0.015 (1.00) | 0.014 (0.93) | 0.013 (0.87) | 0.014 (0.93) | 136.34±5.29 (1.00) | 162.80±2.44 (1.19)d | 156.14±±3.11 (1.15)b | 170.20±3.61 (1.25)d | ||
| WW | BC1W | F2Hy | BC1F | FF | WW | BC1W | F2Hy | BC1F | FF | ||
| 1998 | Electrofishing August 1998 0+parr | 0.008 (1.00) | 0.005 (0.65)a | 0.006 (0.81)c | 0.005 (0.66)c | 0.003 (0.44)d | 64.60±0.61 (1.00) | 66.91±0.63 (1.04) | 69.99±0.91 (1.08)a | 70.49±0.73 (1.09)b | 73.77±0.74 (1.14) |
| 1998 | Trapped parr May 1998–May 1999 | 0.011 (1.00) | 0.007 (0.64)a | 0.002 (0.18)d | 0.001 (0.09)d | 0.001 (0.09)d | |||||
| 1998 | Trapped pre-smolts+smolts 1998 cohort | 0.003 (1.00) | 0.003 (1.00) | 0.005 (1.67) | 0.004 (1.33) | 0.003 (1.00) | |||||
Abbreviations: FF, pure farm group; GLMM, generalised linear mixed effects model; LF, fork length; LMM, linear mixed effects model; WW, pure wild group.
Also shown in parentheses are the means expressed relative to (divided by) the means for the WW reference group, and whether groups differ significantly (based on the GLMM for representation and the LMM for LF, both of which control for egg size variation) from the WW group: P-value bands.
P-value band: 0.05–0.01.
P-value band: 0.01–0.001.
P-value band: 0.1–0.05.
P-value band: <0.001.
Figure 2.
(a) Representation of 0+ parr in the August electrofishing samples for the 1993, 1994 and 1998 cohorts, scaled by the number of eyed-eggs planted per family. (b) Representation (scaled by eggs planted) of 1+ parr in the June 1995 electrofishing sample for the 1994 cohort. Families are labelled arbitrarily in each panel and family labels for 1994 cohort correspond between (a) and (b). Arrows indicate outlier families.
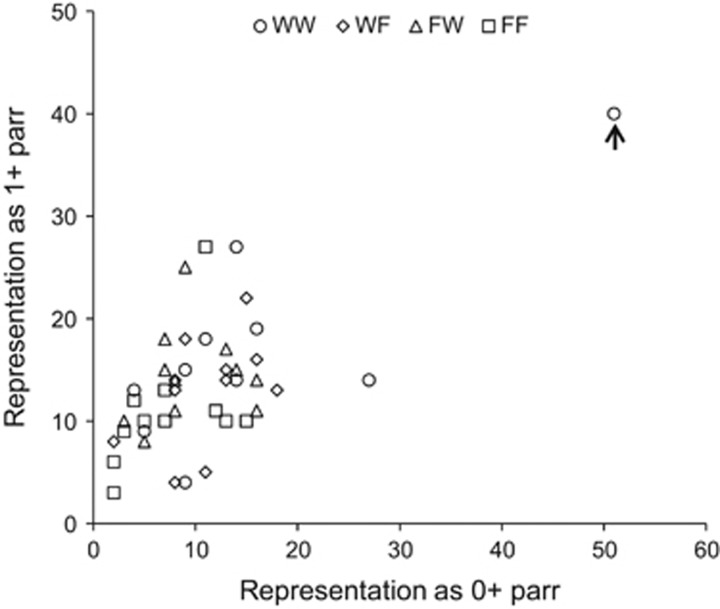
For the 1994 cohort, representation of 1+ parr per family in June 1995 was positively correlated with representation of 0+parr per family in August 1994 (Figure 3; r=0.674, P<0.001; no differences between groups in this relationship). A single outlier family (family 49, Figure 2) had a large influence on this relationship; however, the positive correlation remained significant when excluding this family (r=0.383, P=0.012).
Figure 3.
Representation of 1+ parr in the June 1995 electrofishing sample plotted against representation of 0+ parr in the August 1994 electrofishing sample (1994 cohort). Each data point is a family. The outlier family indicated with an arrow is family 49, which corresponds to the same outlier family identified in Figure 2a.
For the 1998 electrofishing sample, egg size did not have a significant effect on representation per family, but all groups were under-represented relative to the WW group, with the FF group having the lowest representation (Table 2, Figure 2). The other groups were approximately equally represented, but lower on average than the WW group (Table 2, Figure 2). There was one obvious outlier in the F2Hy group (family 162, Figure 2a), but excluding this family did not change the results qualitatively. The variances attributable to dam effects and sire effects for all representation models are given in Supplementary Table S3.
Parr belonging to the 1993 cohort were under-represented in the experiment trap in the FW and FF groups relative to the WW group (Table 2, Supplementary Figure S2A). Pre-smolts and smolts originating from this cohort were marginally under-represented in the FW and FF groups relative to the WW group (Table 2, Supplementary Figure S2B). The latter result was robust to excluding one outlier family (family 4, Supplementary Figure S2B). For the 1994 cohort, parr were under-represented in the experiment trap in the WF, FW and FF groups (in this order: WW>WF>FW>FF; Table 2, Supplementary Figure S2A). Results were qualitatively the same when a single outlier family belonging to the WW group (family 49, Supplementary Figure S2A) was excluded.
There were no significant differences among groups in representation of pre-smolts and smolts from the 1994 cohort in the experiment trap (Table 2). For the 1998 cohort, parr were under-represented in the experiment trap in the BC1W, F2Hy, BC1F and FF groups (in this order: WW>BC1W>F2Hy>BC1F>FF; Table 2, Supplementary Figure S2A). There were no significant differences among groups in representation of pre-smolts and smolts from the 1998 cohort in the experiment trap (Table 2), and this result was robust to excluding one outlier family (family 162, Supplementary Figure S2B). Egg size did not have a significant effect on representation in any of the experiment trap samples (Supplementary Table S2).
For the 1993 cohort, there were no significant representation differences between groups in the hatchery control 0+ parr August 1993 sample (Table 2, Supplementary Figure S3A). In the hatchery control mature male parr sample, the WF and FF groups were under-represented relative to the WW group (Table 2, Supplementary Figure S3B). There were no significant representation differences among groups in terms of smolts in the hatchery control groups for the 1993 and 1994 cohorts (Table 2, Supplementary Figures S3C and D). Egg size did not have a significant effect on representation in any of the hatchery control samples (Supplementary Table S2).
For most of the samples considered, no variance heterogeneity with respect to group was found (Supplementary Table S4), apart from a few exceptions. For the 1998 cohort electrofished 0+parr, the Fligner–Killeen test showed that at least two of the group variances were different (median χ2=11.65, df=4, P=0.020). The raw variance in representation (that is, not correcting for egg size variation) was highest for the F2Hy group (8.2 × 10−5), intermediate for the BC1W (2.6 × 10−5) and BC1F (2.7 × 10−5) groups and lowest for the FF (1.6 × 10−5) and WW (1.3 × 10−5) groups. Excluding the outlier in the F2Hy group (family 162, Figure 2a), the variance for this group dropped considerably (to 2.2 × 10−5), but the Fligner–Killeen test still showed that at least two of the groups were heterogeneous (median χ2=9.92, df=4, P=0.042). For the 1993 cohort trapped parr, the Fligner–Killeen test showed that at least two of the group variances were different (median χ2=11.93, df=3, P=0.008). The raw variance in representation was highest for the WW (7.6 × 10−5) and WF groups (8.2 × 10−5), and lower for the FW (2.7 × 10−5) and FF (8.1 × 10−6) groups. For the 1998 cohort trapped parr, the Fligner–Killeen test showed that at least two of the group variances were different (median χ2=56.6, df=4, P<0.001). The raw variance in representation was highest for the WW group (5.8 × 10−5), intermediate for the BC1W group (4.0 × 10−5) and lowest for the BC1F (6.5 × 10−6), F2Hy (6.3 × 10−6) and FF (2.8 × 10−6) groups.
Size-at-age variation
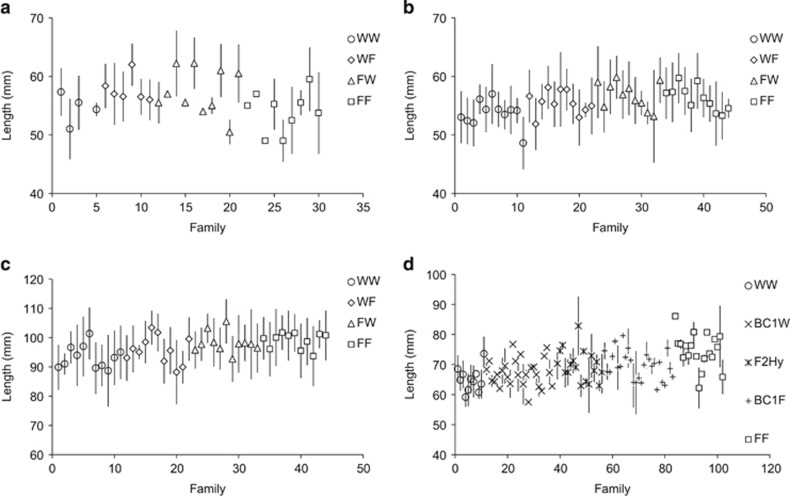
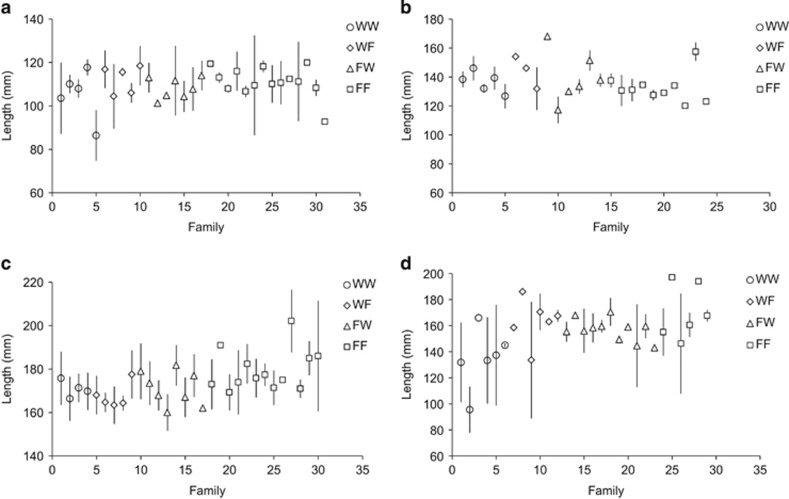
For the 1993 and 1994 cohorts, electrofished 0+ parr assigning to the FF group were significantly larger (LF) than those assigning to the WW group, whereas the hybrid groups (WF and FW) were intermediate (Table 2, Figures 4a and b). A similar pattern was found for electrofished 0+ parr from the 1998 cohort, with FF parr being larger than WW parr and the BC1W, F2Hy and BC1F groups being intermediate in size (Table 2, Figure 4d). The general pattern was an increase in LF of 0+ parr with an increase in the expected fraction of farm genes (that is, the order was WW<hybrids<FF). LF of 0+parr was also positively associated with egg size in all three cohorts (Supplementary Table S5 and Figure S4). Mass of 0+ parr showed similar patterns to LF, with farm fish being heavier than pure wild and hybrids being intermediate (Supplementary Figure S5). Egg size also had a positive effect on mass of 0+ parr in all three cohorts (Supplementary Table S5). LF and mass of 1+ parr in the 1994 cohort were also higher in the FF group compared with the WW group, with hybrids again being intermediate (Table 2, Figure 4c for LF and Supplementary Figure S5B for mass). Egg size did not have a significant effect on LF or mass of 1+ parr (Supplementary Table S5). There were no obvious outlier families in terms of LF and mass of electrofished parr (Figure 4).
Figure 4.
Fork length of (a) 0+ parr in August 1993 electrofishing sample (1993 cohort), (b) 0+ parr in August 1994 electrofishing sample (1994 cohort), (c) 1+ parr in June 1995 electrofishing sample (1994 cohort) and (d) 0+ parr in August 1998 electrofishing sample (1998 cohort). Error bars are s.d. around the mean per family.
Growth patterns were less consistent for 0+ parr measured in the hatchery controls (1993 cohort): FF fish were significantly larger than WW fish, as were WF fish, but FF fish were no larger than WF fish (standard errors largely overlapping, Table 2 and Supplementary Table S5). Parr from the FW group were not significantly larger than WW parr (Table 2, Figure 5a). Egg size did not have an effect on LF or mass of 0+parr in the hatchery controls (Supplementary Table S5). No significant differences in LF of mature male parr in the 1993 cohort hatchery controls were apparent (Table 2, Figure 5b), nor did egg size influence LF of mature male parr in the hatchery (Supplementary Table S5). For the 1993 cohort hatchery controls, FF pre-smolts were significantly larger and heavier than WW pre-smolts (Table 2, Figure 5c) but FW and WW pre-smolts were not significantly larger than WW pre-smolts (Table 2, Figure 5c). Egg size had no effect on the LF and mass of pre-smolts in the 1993 hatchery controls (Supplementary Table S5).
Figure 5.
Fork length of (a) 0+ parr in August 1993 hatchery control sample (1993 cohort), (b) mature male parr in November 1993 hatchery control sample (1993 cohort), (c) pre-smolts in March 1994 hatchery control sample (1993 cohort) and (d) pre-smolts in March 1995 hatchery control sample (1994 cohort). Error bars are s.d. around the mean per family.
For the 1994 cohort hatchery controls, WF, FW and FF pre-smolts were all significantly larger than WW pre-smolts, with FF being the largest and the two hybrid groups each intermediate between WW and FF (Table 2, Figure 5d). Egg size had only a marginally significant positive effect on the LF of pre-smolts in the 1994 cohort hatchery controls (Supplementary Table S5 and Figure S4). The patterns for mass in the hatchery controls were very similar to those for LF (Supplementary Table S5 and Figure S6). There were no obvious outlier families in terms of LF (Figure 5) and mass of hatchery control juveniles (Supplementary Figure S6).
For most of the samples considered, no variance heterogeneity in LF or mass with respect to group was found (Supplementary Table S6), apart from a few exceptions. In the 1994 cohort electrofished 1+ parr sample, there was heterogeneity among groups in the within-family variance in LF and mass; this variance was highest in the WW group (raw variance=10.39 g2) and lower in the other three groups (WF=6.66 g2; WF=7.58 g2; FW=7.34 g2). In the 1993 cohort hatchery controls (Supplementary Figure6), the variance in mass of pre-smolts was higher in the FF group (raw variance=242.93 g2) compared with the other groups (WW=102.06 g2; WF=85.59 g2; FW=112.29 g2). In the 1994 cohort hatchery controls (Figure 5d), the variance in LF of pre-smolts was higher in the WW group (raw variance=8.95 mm2) compared with the other groups (WF=1.51 mm2; FW=2.71 mm2; FF=4.03 mm2).
Quantitative genetic analyses
Moderate heritabilities were estimated for LF and mass, with a general trend for higher h2 estimates in the wild group than in the farmed group (Table 3). For LF, modal h2 estimates in the pure wild group ranged from 0.21 (Bayesian 95% CI: 0.07–0.75) in the June 1995 electrofished 1+ parr sample to 0.89 (CI: 0.23–0.96) in the August 1998 electrofished 0+ parr sample, whereas model h2 estimates in the pure farm group ranged from 0.10 (CI: 0.03–0.44; June 1995 electrofished 1+ parr sample) to 0.31 (CI: 0.04–0.86; August 1998 electrofished 0+ parr sample). For mass, modal h2estimates in the pure wild group ranged from 0.20 (CI: 0.09–0.77) in the June 1995 electrofished 1+ parr sample to 0.53 (CI: 0.15–0.94) in the August 1994 electrofished 0+ parr sample, whereas model h2 estimates in the pure farm group ranged from 0.08 (CI: 0.03–0.43; June 1995 electrofished 1+ parr sample) to 0.17 (CI: 0.05–0.86; August 1998 electrofished 0+ parr sample). The credible intervals for each h2 estimate were quite large, reflecting the relatively low samples sizes and simple pedigree structure. The genetic correlations between LF and mass of electrofished (0+ or 1+) parr were estimated to be very high (posterior modes of >0.85, with credible intervals not overlapping zero) in both the 1994 and 1998 cohorts, as were the environmental correlations (save for mass of August 1994 electrofished 0+ parr, where rE was low; Table 3).
Table 3. Quantitative genetic parameter estimates for size-at-age traits based on bivariate Bayesian animal models.
| Cohort | Variable and sample | Group | VP | VA | h2 | rP | rG | rE |
|---|---|---|---|---|---|---|---|---|
| 1994 | EF August 1994 0+parr LF | WW | 6.2 × 10−3 | 1.8 × 10−3 (0.3−8.1 × 10−3) | 0.29 (0.11−0.89) | 0.95 | 0.96 (0.73–0.98) | 0.94 (0.78–0.97) |
| 1994 | EF August 1994 0+parr mass | WW | 0.065 | 0.019 (0.005−0.085) | 0.53 (0.15−0.94) | — | — | — |
| 1994 | EF August 1994 0+parr LF | FF | 6.7 × 10-3 | 6.0 × 10−4 (0.2−5.7 × 10−3) | 0.15 (0.03−0.65) | 0.94 | 0.90 (0.27–0.97) | 0.06 (0.03–0.09) |
| 1994 | EF August 1994 0+parr mass | FF | 0.069 | 0.008 (0.002−0.060) | 0.11 (0.03−0.63) | — | — | — |
| 1994 | EF June 1995 1+parr LF | WW | 0.01 | 0.002 (0.001−0.01) | 0.21 (0.07−0.75) | 0.98 | 0.95 (0.82–0.99) | 0.97 (0.94–0.98) |
| 1994 | EF June 1995 1+parr mass | WW | 0.09 | 0.019 (0.005−0.091) | 0.20 (0.09−0.77) | — | — | — |
| 1994 | EF June 1995 1+parr LF | FF | 0.008 | 7.0 × 10-4 (0.2−4.2 × 10-3) | 0.10 (0.03−0.44) | 0.97 | 0.82 (0.49–0.98) | 0.97 (0.95–0.98) |
| 1994 | EF June 1995 1+parr mass | FF | 0.06 | 0.008 (0.002−0.033) | 0.08 (0.03−0.43) | — | — | — |
| 1998 | EF August 1998 0+parr LF | WW | 0.006 | 0.004 (0.001−0.009) | 0.89 (0.23−0.96) | 0.89 | 0.92 (0.47–0.97) | 0.56 (0.22–0.96) |
| 1998 | EF August 1998 0+parr mass | WW | 0.069 | 0.025 (0.006−0.105) | 0.39 (0.16−0.095) | — | — | — |
| 1998 | EF August 1998 0+parr LF | FF | 0.007 | 0.002 (0.001−0.009) | 0.31 (0.04−0.86) | 0.93 | 0.90 (0.38–0.98) | 0.93 (0.71–0.98) |
| 1998 | EF August 1998 0+parr mass | FF | 0.065 | 0.009 (0.003−0.084) | 0.17 (0.05−0.86) | — | — | — |
Abbreviations: EF, electrofished; FF, pure farm group; h2, narrow sense heritability; LF, fork length; rE, residual correlation between LF and mass; rP, raw phenotypic correlation between LF and mass; rG, additive genetic correlation between LF and mass; VP, raw phenotypic variance; VA, additive genetic variance; WW, pure wild group.
For VA, h2, rG and rE, estimates are posterior modes, with credible intervals in parentheses. LF and mass were natural log transformed in all models.
Discussion
Re-analysis of group-level performance differences accounting for family structure
The performances of individuals sharing one or two parents are not independent because of effects of shared genes and possible parental environmental effects. Earlier analyses of these experimental data (McGinnity et al., 1997, 2003) did not account for this family structure, but reassuringly the current results were largely congruent in terms of significant group-level differences (compare Table 2 here with Table 2 in McGinnity et al., 1997 and with Figure 2 in McGinnity et al., 2003) when hypothesis testing of parental genotypic effects was based on families rather than individuals (the former being tantamount to avoiding ‘genetic pseudoreplication'). Minor differences, however, were noted. For example, with the electrofishing August 1993 0+ parr sample, McGinnity et al. (1997) reported that the FF group was significantly under-represented relative to the WW group, whereas here that difference was not significant. However, the qualitative conclusions were largely unchanged when kin structure was accounted for, suggesting that either the kin structure was not strong enough for genetic pseudoreplication to be a major issue, and/or that covariation in the performance of individuals sharing one or two parents was relatively weak because of moderate trait heritabilities.
Focusing analyses in to the family level allowed us to uncover interesting biological patterns of variation and covariation in representation. Families highly represented at the 0+ parr stage in the experiment stream (caught by electrofishing) were also highly represented at the 1+ parr stage (Figure 3), implying consistent performance differences in the wild underpinned by genetic differences or persistent maternal effects. Outlier families were also obvious in some samples. For example, in the August 1994 0+ parr electrofishing sample, one pure wild (WW) family (family 49, see Figure 2a and Figure 3) was represented by 59 parr, which compares with an average representation of 11.4 parr per family excluding this family. Nevertheless, the overall group-level differences in this sample remained statistically significant after removing the outlier, instilling further confidence that the lower representation of offspring with one or two farm parents was a robust, biologically meaningful result, not driven simply by one or two highly performing wild families. Similarly, in the August 1998 0+ parr electrofishing sample, one F2 hybrid family (family 162, Figure 2a) was anomalously highly represented relative to all other families, but the inferences regarding group-level differences were robust to excluding this family. We can only speculate on the reasons as to why these particular families were so highly represented, but in the case of the F2 hybrid family, recombination between the divergent wild and farm parental genomes could have produced rare offspring genotypes that were fortuitously well adapted to the local conditions through heterosis.
Performance of farm and hybrid families: more or less variable than wild families?
Overall genetic diversity may be considerably lower in farm salmon compared with wild populations (Norris et al., 1999; Skaala et al., 2004), at least when considering highly polymorphic genetic markers, because of low effective population sizes in the farm and/or strong directional selection on target traits, which can deplete genetic variation (Lynch and Walsh, 1998). A priori, therefore, one may expect that offspring produced by farm parents should exhibit reduced phenotypic variation in the wild and therefore less variable survival rates compared with wild families. Skaala et al. (2012), however, reported the opposite: a larger range in survival rates (a ratio of 38:1 between the lowest and highest survival rates) in farm families compared with hybrid (7:1) or wild (8:1) families in a natural stream setting. In our case, however, no variance heterogeneity in representation with respect to group was found for most of the samples considered (Supplementary Table S2). In a few samples, we did find variance heterogeneity but the patterns were inconsistent; for example, in the 1998 cohort electrofished 0+parr, survival variation was greatest among F2Hy families (perhaps due to rare advantageous recombinants), while for the 1993 and 1998 cohort trapped parr samples, variance in representation was highest for pure wild families and lowest for pure farm families (as one would predict if farm families are genetically depauperate), with hybrid families being generally intermediate. Although Skaala et al. used Mowi strain salmon in one of their experimental cohorts, the Fanad Mowi strain used by us is likely to be divergent from theirs in its genetic make-up (because of lower broodstock numbers and a separate breeding programme); thus differences in genetic background and selection trajectories of the farm strains may explain the inconsistent results, in terms of variance in performance, between Skaala et al. (2012) and this study.
For offspring LF and mass, no heterogeneity in between-family variance was found (Supplementary Table S3), suggesting that each group had similar levels of additive genetic variance for these traits. In terms of within-family (residual) variance, which largely reflects environmental influences on the phenotype, no differences among groups were apparent in 9 out of 14 samples (Supplementary Table S3). In the other five samples, the residual variance was either highest in the WW group (for example, mass of 1994 cohort electrofished 1+ parr sample) or the FF group (for example, LF and mass of pre-smolts in 1993 cohort hatchery controls). Intriguingly, Debes and Hutchings (2014) found that within-family variation in body size of Atlantic salmon (measured in a hatchery setting) diminished with increasing generations of domestication (see also Solberg et al., 2013a). Under fully wild conditions, variance differences between wild, farmed and hybrid families may be largely unpredictable and context dependent, given that our findings did not match those of Skaala et al. (2012) despite very similar study designs (but different genetic backgrounds). One possibility is that farmed fish may lose their environmental sensitivity (that is, degree to which their phenotypes or performance is buffeted by prevailing conditions) in hatchery environments, but not wild environments, as they are only selected in the former.
Genetic basis of group and family differences in size-at-age
Directional selection in farm strains has resulted in higher intrinsic growth rates of farm salmon, which in a hatchery environment can grow up to three times faster than wild salmon (Glover et al., 2009; Solberg et al., 2013a, 2013b). However, these growth rate differences seem to be less pronounced in wild stream environments (Skaala et al., 2012) and in hatchery conditions simulating a semi-natural environment with restricted food (Solberg et al., 2013a). In our experiments, size-at-age differences between wild and farm offspring measured in the wild were statistically significant but moderate in magnitude (Table 2), with electrofished farm parr being in the order of 5–20% larger and heavier than wild parr, consistent with the findings of these previous studies. However, size differences between farm and wild juveniles were similar in the hatchery environment as in the wild (Table 2), which contrasts with the above-cited studies. This presumably reflects the fact that the Fanad farm strain used in our study had experienced a different selection trajectory in Ireland up until our experiments were carried out in the 1990 s than the Norwegian farm strains used in the more recent Norwegian studies had (Glover et al., 2009; Skaala et al., 2012; Solberg et al., 2013a, 2013b). The latter had also undergone more generations of targeted artificial (and/or inadvertent domestication) selection than our farm strain. These differences in historical selection regimes, as well as possible founder effect differences, may explain why we found only moderate size differences between our farm and wild groups in both the hatchery and wild environments, whereas the Norwegian studies observed much larger differences in hatchery environments (where their higher genetic growth potential is likely more easily realised) that were attenuated in the wild (where environmental influences on growth are likely larger and selection against extreme phenotypes also stronger). Interestingly, genetically based somatic growth differences between the farm and wild strains used in the Norwegian studies seem to be more important after the onset of exogenous feeding, with alevin lengths being similar once egg size differences between farm and wild strains are corrected for (Solberg et al., 2014).
VA is a crucial parameter influencing the rate of microevolution and thus the potential for genetic adaptation to a changing environment, while in the case of multivariate selection, COVA among characters determine the extent to which they can evolve along independent trajectories. Typically, VA is scaled relative to the phenotypic variance VP, which gives a measure of heritability h2, whereas COVA is scaled relative to the square root of the product of the VA in each trait to give a measure of the genetic correlation (rG). Estimates of both heritabilities and genetic correlations (including the sign of the latter) may depend, however, on the quality of the environment experienced by measured individuals, which can affect both VA and VR (that is, the residual, or environmental variance; Charmantier and Garant, 2005). In the case of salmonid fishes, quantitative genetic parameter estimates calculated under farm or hatchery conditions may have limited relevance for wild populations, given the environmental sensitivity of these parameters. Carlson and Seamons (2008) reported that only 2% of published h2 estimates in salmonids were from wild-reared populations, whereas no estimates of rG were available at the time for wild salmon reared in the wild. Since then, a few additional studies have been published that estimated quantitative genetic parameters in wild settings (Saura et al., 2010; Serbezov et al., 2010; Letcher et al., 2011) and this study adds to this small list. For the pure wild group, our estimates of h2of LF and mass (electrofished parr samples) were generally in the range of 0.20 to 0.50 (Table 3), which compares with a median h2 of 0.29 and 0.32 for length-at-age and mass-at-age, respectively, reported in Carlson and Seamons (2008). Saura et al. (2010) estimated the h2 of adult length (and also adult mass) of Atlantic salmon to be 0.32, while Serbezov et al. (2010) report h2 estimates between 0.16 and 0.31 for length-at-age for wild-living juvenile brown trout (Salmo trutta). Body size of salmon juveniles is positively related to their ability to acquire and defend feeding/nursing territories and has previously been shown to be under positive natural selection (Einum and Fleming, 2000). Thus, estimates of the h2 of size-at-age traits obtained under natural conditions are of evolutionary importance; moreover, these traits are known to vary between farm and wild populations and hence understanding how they are inherited can improve predictions of likely genetic consequences of introgression.
We also found that the modal h2 estimates for LF and mass were generally lower for the pure farm (FF) group, compared with the pure wild group (Table 3), although the uncertainty associated with each h2 estimate was relatively large and the posterior distributions for the wild and farm groups overlapped considerably. As these traits were first natural log transformed before running the animal models, the VA values reported in Table 3 (multiplied by 100) can also be interpreted as evolvabilities (that is, mean standardised additive genetic variances on the untransformed scale, see Hansen et al., 2011). Evolvability measures the expected proportional evolutionary change in a trait under a unit strength of selection and in many ways is a better measure of evolutionary potential than h2, particularly when comparing groups or populations that have very different VP. Thus, for example, when the mean standardised selection gradient is 1 (that is, very strong selection), the expected evolutionary response in LF for the wild 0+parr based on the August 1998 electrofishing sample would be 0.4% (that is, an evolvability of 0.4% for the WW group), whereas that for the farmed parr would be only 0.2% (Table 3). In general, we found that VA (and hence evolvability) was lower in the FF group compared with the WW group, which is in line with previous findings that genetic variation in farm salmon strains are often lower than in wild strains (Norris et al., 1999; Skaala et al., 2004). Interestingly, Solberg et al. (2013b) reported reduced heritability of juvenile mass in farm-provenance Atlantic salmon, compared with progeny of wild parents, when both were reared under standard hatchery conditions with unrestricted access to food. This pattern was reversed, however, when access to food was restricted, possibly reflecting selective mortality against the slowest-growing wild genotypes (Solberg et al., 2013b). We also found strong positive genetic correlations between LF and mass (>0.85 in all samples), which is higher than the median rG of +0.71 reported by Carlson and Seamons (2008) for pairs of morphological traits. Hence, positive selection on body size would be predicted to result in population-level increases in both mean LF and mean mass (Lynch and Walsh, 1998). We controlled for environmental maternal effects as far as possible by including egg size as a covariate (fixed effect) in the animal models. Larger females tend to produce larger eggs (as do farm females, see Table 1), and larger eggs can result in larger size of fry at emergence and higher early-life survival (Einum and Fleming, 1999; Heath et al., 1999). We found that egg size had a significant positive effect on the LF and mass of 0+ fry caught by electrofishing, whereas no egg size effect was found for electrofished 1+parr (Supplementary Table S3), consistent with previous findings in salmonids that egg size effects tend to attenuate with offspring age (Heath et al., 1999). However, positive effects of egg size on the representation (that is, survival) of both 0+ and 1+ electrofished parr were also found (Supplementary Table S2 and Figure S1). Future salmonid studies that disentangle maternal genetic and environmental effects from additive genetic effects in wild stream environments would be very revealing.
Data archiving
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.bk583.
Acknowledgments
This project was funded by the Marine Institute Ireland, the European Commission, the UK Natural Environment Research Council and the Beaufort Marine Research Award in Fish Population Genetics funded by the Irish Government under the Sea Change Programme. We thank Eric Verspoor for providing microsatellite primers and for assistance with experimental design, John Taggart for project design and parentage assignment, Natalie Baker (QUB) and Carol Stone (QUB) for DNA profiling and Ken Whelan, Declan Cooke, Niall O'Maoileidigh, Deirdre Cotter, Russell Poole, Brendan O'Hea, Tony Nixon (RIP), Ger Rogan and the field staff of MI Newport for technical and logistical support. Comments from Paul Debes, Tutku Aykanat, Kevin Glover and two anonymous referees greatly improved the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Bates D, Maechler M, Bolker B. (2012). lme4: linear mixed-effects models using S4 classes.
- Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH et al. (2009). Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol Evol 24: 127–135. [DOI] [PubMed] [Google Scholar]
- Bourret V, Kent MP, Primmer CR, Vasemägi A, Karlsson S, Hindar K et al. (2013). SNP-array reveals genome-wide patterns of geographical and potential adaptive divergence across the natural range of Atlantic salmon (Salmo salar). Mol Ecol 22: 532–551. [DOI] [PubMed] [Google Scholar]
- Carlson SM, Seamons TR. (2008). A review of quantitative genetic components of fitness in salmonids: implications for adaptation to future change. Evol Appl 1: 222–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier A, Garant D. (2005). Environmental quality and evolutionary potential: lessons from wild populations. Proc R Soc B Biol Sci 272: 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford SL, McGinnity P, Ferguson A. (1998). Genetic changes in an Atlantic salmon population resulting from escaped juvenile farm salmon. J Fish Biol 52: 118–127. [Google Scholar]
- Debes PV, Fraser DJ, Yates M, Hutchings JA. (2014). The between-population genetic architecture of growth, maturation, and plasticity in Atlantic Salmon. Genetics 196: 1277–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debes PV, Hutchings JA. (2014). Effects of domestication on parr maturity, growth, and vulnerability to predation in Atlantic Salmon. Can J Fish Aquat Sci 71: 1371–1384. [Google Scholar]
- Dionne Mel, Caron F, Dodson JJ, Bernatchez L. (2008). Landscape genetics and hierarchical genetic structure in Atlantic salmon: the interaction of gene flow and local adaptation. Mol Ecol 17: 2382–2396. [DOI] [PubMed] [Google Scholar]
- Donaghy MJ, Verspoor E. (2000). A new design of instream incubator for planting out and monitoring Atlantic salmon eggs. North Am J Fish Manag 20: 521–527. [Google Scholar]
- Einum S, Fleming IA. (1997). Genetic divergence and interactions in the wild among native, farmed and hybrid Atlantic salmon. J Fish Biol 50: 634–651. [Google Scholar]
- Einum S, Fleming IA. (1999). Maternal effects of egg size in brown trout (Salmo trutta: norms of reaction to environmental quality. Proc R Soc Lond B Biol Sci 266: 2095–2100. [Google Scholar]
- Einum S, Fleming IA. (2000). Selection against late emergence and small offspring in Atlantic salmon (Salmo salar. Evolution 54: 628–639. [DOI] [PubMed] [Google Scholar]
- Fiske P, Lund RA, Hansen LP. (2006). Relationships between the frequency of farmed Atlantic salmon, Salmo salar L., in wild salmon populations and fish farming activity in Norway, 1989–2004. ICES J Mar Sci J Cons 63: 1182–1189. [Google Scholar]
- Fleming IA, Agustsson T, Finstad B, Johnsson JI, Björnsson BT. (2002). Effects of domestication on growth physiology and endocrinology of Atlantic salmon (Salmo salar. Can J Fish Aquat Sci 59: 1323–1330. [Google Scholar]
- Fleming IA, Hindar K, MjÖlnerÖd IB, Jonsson B, Balstad T, Lamberg A. (2000). Lifetime success and interactions of farm salmon invading a native population. Proc R Soc Lond B Biol Sci 267: 1517–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming IA, Jonsson B, Gross MR, Lamberg A. (1996). An experimental study of the reproductive behaviour and success of farmed and wild Atlantic salmon (Salmo salar. J Appl Ecol 33: 893–905. [Google Scholar]
- Fraser DJ, Weir LK, Bernatchez L, Hansen MM, Taylor EB. (2011). Extent and scale of local adaptation in salmonid fishes: review and meta-analysis. Heredity 106: 404–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garant D, Fleming IA, Einum S, Bernatchez L. (2003). Alternative male life-history tactics as potential vehicles for speeding introgression of farm salmon traits into wild populations. Ecol Lett 6: 541–549. [Google Scholar]
- Garcia de Leaniz C, Fleming IA, Einum S, Verspoor E, Jordan WC, Consuegra S et al. (2007). A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biol Rev 82: 173–211. [DOI] [PubMed] [Google Scholar]
- Gjedrem T. (2000). Genetic improvement of cold-water fish species. Aquac res 31: 25–33. [Google Scholar]
- Gjøen HM, Bentsen HB. (1997). Past, present, and future of genetic improvement in salmon aquaculture. ICES J Mar Sci J Cons 54: 1009–1014. [Google Scholar]
- Glover KA, Otterå H, Olsen RE, Slinde E, Taranger G.L, Skaala Ø. (2009). A comparison of farmed, wild and hybrid Atlantic salmon (Salmo salar L.) reared under farming conditions. Aquaculture 286: 203–210. [Google Scholar]
- Glover KA, Quintela M, Wennevik V, Besnier F, Sørvik AG, Skaala Ø. (2012). Three decades of farmed escapees in the wild: a spatio-temporal analysis of Atlantic salmon population genetic structure throughout Norway. PLoS One 7: e43129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover KA, Pertoldi C, Besnier F, Wennevik V, Kent M, Skaala Ø. (2013). Atlantic salmon populations invaded by farmed escapees: quantifying genetic introgression with a Bayesian approach and SNPs. BMC Genet 14: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield JD. (2010). MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw 33: 1–22.20808728 [Google Scholar]
- Hansen LP, Jacobsen JA, Lund RA. (1999). The incidence of escaped farmed Atlantic salmon, Salmo salar L., in the Faroese fishery and estimates of catches of wild salmon. ICES J Mar Sci J Cons 56: 200–206. [Google Scholar]
- Hansen TF, Pélabon C, Houle D. (2011). Heritability is not evolvability. Evolutionary Biology 38: 258–277. [Google Scholar]
- Heath DD, Fox CW, Heath JW. (1999). Maternal effects on offspring size: variation through early development of chinook salmon. Evolution 53: 1605–1611. [DOI] [PubMed] [Google Scholar]
- Houde ALS, Fraser DJ, Hutchings JA. (2010). Reduced anti-predator responses in multi-generational hybrids of farmed and wild Atlantic salmon (Salmo salar L.). Conserv Genet 11: 785–794. [Google Scholar]
- ICES. (2010) Report of the ICES Advisory Committee, Book 10: North Atlantic Salmon Stocks, as reported to the North Atlantic Salmon Conservation Organization. ICES Headquarters: Copenhagen. [Google Scholar]
- Kruuk LE. (2004). Estimating genetic parameters in natural populations using the ‘animal model'. Philos Trans R Soc Lond B Biol Sci 359: 873–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letcher BH, Coombs JA, Nislow KH. (2011). Maintenance of phenotypic variation: repeatability, heritability and size-dependent processes in a wild brook trout population. Evol Appl 4: 602–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, O'Hely M. (2001). Captive breeding and the genetic fitness of natural populations. Conserv Genet 2: 363–378. [Google Scholar]
- Lynch M, Walsh B. (1998) Genetics and Analysis of Quantitative Traits. Sinauer Associates, Incorporated: Sunderland. [Google Scholar]
- McGinnity P, Prodöhl P, Ferguson A, Hynes R, ó Maoiléidigh N, Baker N et al. (2003). Fitness reduction and potential extinction of wild populations of Atlantic salmon, Salmo salar, as a result of interactions with escaped farm salmon. Proc R Soc Lond B Biol Sci 270: 2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnity P, Stone C, Taggart JB, Cooke D, Cotter D, Hynes R et al. (1997). Genetic impact of escaped farmed Atlantic salmon (Salmo salar L.) on native populations: use of DNA profiling to assess freshwater performance of wild, farmed, and hybrid progeny in a natural river environment. ICES J Mar Sci J Cons 54: 998–1008. [Google Scholar]
- Naylor R, Hindar K, Fleming IA, Goldburg R, Williams S, Volpe J et al. (2005). Fugitive salmon: assessing the risks of escaped fish from net-pen aquaculture. BioScience 55: 427–437. [Google Scholar]
- Norris AT, Bradley DG, Cunningham EP. (1999). Microsatellite genetic variation between and within farmed and wild Atlantic salmon (Salmo salar populations. Aquaculture 180: 247–264. [Google Scholar]
- Saura M, Moran P, Brotherstone S, Caballero A, Alvarez J, Villanueva B. (2010). Predictions of response to selection caused by angling in a wild population of Atlantic salmon (Salmo salar. Freshw Biol 55: 923–930. [Google Scholar]
- Serbezov D, Bernatchez L, Olsen EM, Vøllestad LA. (2010). Quantitative genetic parameters for wild stream-living brown trout: heritability and parental effects. J Evol Biol 23: 1631–1641. [DOI] [PubMed] [Google Scholar]
- Skaala Ø., Høyheim B., Glover K., Dahle G. (2004). Microsatellite analysis in domesticated and wild Atlantic salmon (Salmo salar L.): allelic diversity and identification of individuals. Aquaculture 240: 131–143. [Google Scholar]
- Skaala Ø, Wennevik V, Glover KA. (2006). Evidence of temporal genetic change in wild Atlantic salmon, Salmo salar L., populations affected by farm escapees. ICES J Mar Sci 63: 1224–1233. [Google Scholar]
- Skaala Ø, Glover KA, Barlaup BT, Svåsand T, Besnier F, Hansen MM et al. (2012). Performance of farmed, hybrid, and wild Atlantic salmon (Salmo salar families in a natural river environment. Can J Fish Aquat Sci 69: 1994–2006. [Google Scholar]
- Solberg MF, Skaala Ø, Nilsen F, Glover KA. (2013. a). Does domestication cause changes in growth reaction norms? A study of farmed, wild and hybrid Atlantic Salmon families exposed to environmental stress. PloS One 8: e54469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg MF, Zhang Z, Nilsen F, Glover KA. (2013. b). Growth reaction norms of domesticated, wild and hybrid Atlantic salmon families in response to differing social and physical environments. BMC Evol Biol 13: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg MF, Fjelldal PG, Nilsen F, Glover KA. (2014). Hatching time and alevin growth prior to the onset of exogenous feeding in farmed, wild and hybrid Norwegian Atlantic salmon. PloS One 9: e113697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuur A, Ieno EN, Walker N, Saveliev AA, Smith GM. (2009) Mixed Effects Models and Extensions in Ecology with R. Springer: New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.