Abstract
Phenotypic plasticity is ubiquitous and generally regarded as a key mechanism for enabling organisms to survive in the face of environmental change. Because no organism is infinitely or ideally plastic, theory suggests that there must be limits (for example, the lack of ability to produce an optimal trait) to the evolution of phenotypic plasticity, or that plasticity may have inherent significant costs. Yet numerous experimental studies have not detected widespread costs. Explicitly differentiating plasticity costs from phenotype costs, we re-evaluate fundamental questions of the limits to the evolution of plasticity and of generalists vs specialists. We advocate for the view that relaxed selection and variable selection intensities are likely more important constraints to the evolution of plasticity than the costs of plasticity. Some forms of plasticity, such as learning, may be inherently costly. In addition, we examine opportunities to offset costs of phenotypes through ontogeny, amelioration of phenotypic costs across environments, and the condition-dependent hypothesis. We propose avenues of further inquiry in the limits of plasticity using new and classic methods of ecological parameterization, phylogenetics and omics in the context of answering questions on the constraints of plasticity. Given plasticity's key role in coping with environmental change, approaches spanning the spectrum from applied to basic will greatly enrich our understanding of the evolution of plasticity and resolve our understanding of limits.
Introduction
How do organisms meet the challenges inherent across spatially and temporally variable environments? Phenotypic plasticity has long been recognized as a key strategy enabling organisms to respond to varying environments both adaptively and non-adaptively (Bradshaw, 1965). Although the optimal response to a heterogeneous set of environmental challenges is perfect plasticity—in which an organism possesses perfect information on its current environment, and mechanisms to produce an appropriate phenotypic response at all points in development—the rarity or absence of this capacity in natural populations suggests that there are constraints on the evolution of plasticity. DeWitt et al. (1998) contrasted two types of restrictions: costs, which lead to reduced fitness when a trait is produced via plasticity rather than constitutively, and limits, an inability to produce the optimal trait value.
Investigations into the constraints on the evolution of adaptive plasticity (including costs and limits) have been conducted through two approaches: modeling and empirically. Models investigating the evolution of niche breadth or generalists vs specialists generally assume that advantages of being a specialist in one environment are accompanied by disadvantages in another (for example, van Tienderen, 1991; Lynch and Lande, 1993) and formally consider phenotypic plasticity as one component of being a generalist. Conversely, plastic generalists are considered to be ‘jacks-of-all-trades' but ‘masters-of-none' (for example, Richards et al., 2006). Experimental work to identify proposed trade-offs between generalists and specialists has led to inquiries into potential mechanisms in response to habitat heterogeneity, which may lead to plastic generalists or specialists (Callahan et al., 2008).
Both empirical and modeling approaches have employed the concept of a cost of plasticity as a cornerstone of arguments about why organisms are not infinitely and ideally plastic: the underlying assumption behind these arguments has been that organisms with an enhanced capacity to exhibit plasticity must pay a price in fitness (van Tienderen, 1991; Moran, 1992). This led to the expectation that costs or limits should be readily detectable. Dozens of investigations have explicitly looked for costs of plasticity, and many more have invoked them. Detection of costs of plasticity, however, has been infrequent (see meta-analyses by van Kleunen and Fischer, 2005; van Buskirk and Steiner, 2009). In the findings of van Buskirk and Steiner (2009), of 536 tests they report 262 positive coefficients and 262 negative coefficients, with 12 zeroes. This balanced distribution is fully consistent with the results of the regression of a random variable against fitness. Relative to the expectation from a truly random pattern, the van Buskirk and Steiner results do show a surplus of significant results (~20% in each direction). However, this might derive from a combination of (a) under-reporting of non-significant findings, (b) type I error (suggested by their evidence for a higher likelihood of significance with small sample sizes) and (c) accumulation of significant results due to replicated analyses on genetically correlated traits. Furthermore, investigation of the typical two-environment tests for costs has revealed flaws in the analytical framework (Auld et al., 2010; Roff, 2011). The lack of conformation of results to predictions has prompted us to take a fresh look at both the conceptual framework and the empirical approaches to evaluating constraints on the evolution of plasticity and highlight promising areas of investigation.
In this paper, we begin by addressing the distinction between costs of plasticity and costs of phenotype production, showing that these two types of costs are often conflated. We argue that costs of phenotype are more prevalent in many investigations of plasticity, and together with the environmental context may be a critical limit to the evolution of plasticity. We then refocus on examining constraints to the evolution of plasticity. In this discussion of constraints, we evaluate how costs of phenotypes might be offset, the potential influence of limits of plasticity, and offer discussion of promising research directions not previously evaluated in this context. In contrast to previous reviews distinguishing between costs of phenotype vs costs of plasticity, here we emphasize issues critical to evaluating potential costs of plasticity. We also draw attention to the importance of limits of plasticity in our overall discussion of constraints. We follow Auld et al. (2010) in describing the limits to plasticity as trait based, where a plastic genotype produces a phenotype further away from the optimum for a particular environment than a non-plastic genotype. In addition, we offer a synthesis of novel and integrative approaches that move beyond classic quantitative genetics models and set new goals for inquiry in plasticity.
Costs of phenotype and costs of plasticity
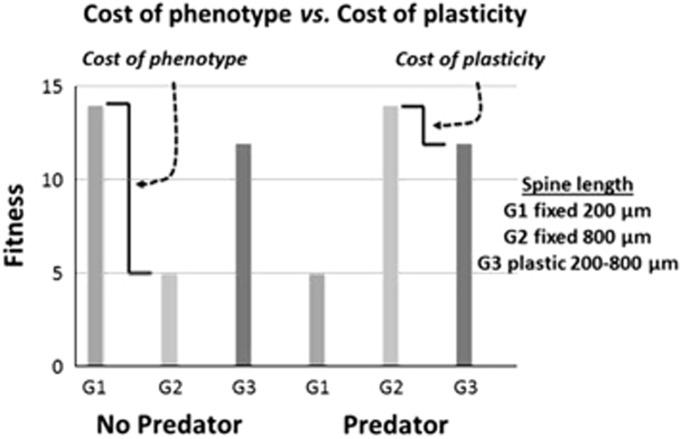
At the outset, it is critical to distinguish between costs of phenotypes and costs of plasticity (Callahan et al., 2008), which are often confounded in discussion of costs in the evolutionary literature. A cost of plasticity refers to the fitness decrement a highly plastic genotype pays relative to a less plastic genotype (DeWitt et al., 1998), thus the evolution of plasticity may be constrained by the cost function (Pigiucci, 2001). A cost of phenotype refers in part to the fitness trade-offs inherent in allocating resources to one trait vs another as well as the costs of obtaining information on the environment (Callahan et al., 2008). Consider the facultative production of a head spine in Daphnia in response to chemicals emitted by predators (kairomones). Suppose the Daphnia population consists of three genotypes: genotype 1 constitutively produces short spines (G1 fixed 200 μm spines legend Figure 1), genotype 2 constitutively produces long spines (G2 fixed 800 μm spines legend Figure 1) and genotype 3 produces a short spine (200 μm spines) in the absence of predator cues, but a long spine (800 μm spines) in their presence (thus, G3 has plastic spine length changing from 200 to 800 μm between environments, legend Figure 1). In an environment with no predators, genotype 1 has higher fitness than genotype 2; genotype 2 has incurred a phenotype cost (Figure 1), a situation reversed if predators were present. Thus, phenotype costs are genotype specific and environment dependent, that is, they are local costs. In each environment, we can evaluate the costs and fitness benefits of a particular phenotype in comparison to those of alternative phenotypes. A plasticity cost ensues in this example when two genotypes produce the same phenotype (for example, long spines 800 μm; Figure 1), but the more plastic genotype 3 has reduced fitness compared with the non-plastic long-spined genotype 2. If plasticity per se is costly this should be detectable even when the plasticity is not elicited (that is, in the no-predator treatment). Thus, costs of plasticity are genotype specific and global, that is, they exist in all environments. Note that plastic genotypes may also incur phenotype costs in situations where phenotypes are sub-optimal. Our example employs perfect symmetry in the fitness responses for simplicity of presentation. Many biological examples may be more complicated as in the case when fitnesses between the plastic and constitutive genotypes differ. In this scenario, the cost of plasticity (as described) and the cost of phenotype (the average of the fitness differences of G1 and G2 if the fitnesses are not equivalent in either environment) may both be playing a constraining role in the evolution of plasticity. Local costs of plasticity must be contextualized across the multiple environments in which selection on plasticity operates: thus, the net effects of costs and benefits will depend on the specific environments that the organism encounters. We distinguish these costs from limits of plasticity. Together, we consider costs of phenotype, cost of plasticity and limits to plasticity to be constraints on the evolution of plasticity.
Figure 1.
Classic framework for evaluating cost of plasticity, and contrast with a cost of phenotype. In each environment (no predator vs predator), genotypes 1 (G1) and 2 (G2) differ in the phenotypes (200 vs 800 μm) they produce, leading to differences in fitness—these represent costs to producing one phenotype rather than the other. In the predator environment, genotypes 2 (G2) and 3 (G3) both produce the same 800 μm phenotype yet differ in fitness. As genotype 3 is plastic, this represents a cost of plasticity.
Conditions that favor plasticity
Adaptive phenotypic plasticity is presumed to evolve in response to contrasting selection pressures that arise when organisms confront environmental heterogeneity (Bradshaw, 1965). Since Bradshaw's discussion, numerous models of the evolution of phenotypic plasticity indicate that plasticity is favored in heterogeneous environments where different phenotypic optima are present across environments experienced either within the lifetime of an organism or across generations (for example, Berrigan and Scheiner, 2004). Empirical evidence continues to accumulate demonstrating that heterogeneity in natural landscapes favors plasticity (for example, van Buskirk, 2002) but there is also the potential for variation in local environments to destabilize homeostasis of development and consequently disrupt the match between an organism's phenotype and environment (Whitman and Agrawal, 2009). The reliability of environmental cues is of critical importance for plasticity to be favored within and across generations (Scheiner and Holt, 2012). With reliable cues and machinery to sense the environment, phenotypic plasticity can reduce mismatches, and thereby enhance fitness (Getty, 1996).
Although the importance of environmental heterogeneity is known, relative environmental frequencies are rarely estimated (although large-scale studies are emerging, for example, Fournier-Level et al., 2011). The relative frequency of each environment and the overall diversity of environments experienced (in space and time) will shape the speed and likelihood of the evolution of adaptive reaction norms (for example, Sultan and Spencer, 2002; Scheiner, 2013). Our understanding of the importance of environmental novelty or rarity dates back to initial models of the evolution of plasticity that suggest that rarity strongly influences evolution of plasticity (for example, Levins, 1968; Via and Lande, 1985). For populations that experience rare or novel environments, plastic responses can have two important effects: they may move populations closer to the new phenotypic optimum and they may uncover phenotypic variation (cryptic genetic variation), influencing both intensity and response to selection (Chevin et al., 2010; 2008). Each effect is fundamentally interesting and of particular contemporary importance in terms of predicting organismal responses to global change (Quintero and Wiens, 2013). Such conditions that favor plasticity are widely discussed and form a foundation for our discussion of constraints.
Constraints on plasticity
Limited genetic variation
Under conditions in which plasticity is favored, an evolutionary response via phenotypic plasticity can be hindered by a lack of genetic variation (for example, through small population size or genetic drift), extensive gene flow and genetic correlation between genes for one trait and genes for plasticity of another trait. A lack of genetic variation or continued mixing can limit phenotypic evolution and evolution of plasticity (Schlichting and Pigliucci, 1998). What is novel in contemporary studies is that our knowledge of fine scale information on raw genetic variation and population genomic information is growing exponentially.
Many quantitative genetic studies suggest limited genetic variation for phenotypic plasticity compared with that for other phenotypic traits. However, in a recent meta-analysis designed to evaluate components of the reaction norm that differ between closely related pairs of species or populations demonstrated significantly greater evolution of plasticity (slope and curvature) than for mean trait values (Murren et al., 2014). This suggests that aspects of plasticity frequently evolve. Comparisons between environmental sensory networks of closely related populations (Tsuji et al., 2011; Long et al., 2013) offer evidence that novel genetic variation may be present in many systems—suggesting organisms frequently overcome such limits to plasticity.
Relaxed and variable selection
In the absence of continued environmental variation driving selection for plasticity, mutation accumulation and selection may erode plasticity (Maughan et al., 2007; Kvitek and Sherlock, 2013; Leiby and Marx, 2014). Selection that is not absent but relatively weak purifying or positive selection (relaxed selection) is also thought to be an important constraint in the evolution of plasticity (Snell-Rood et al., 2010). Relative to a specialist, a plastic generalist will experience less effective selection on developmental pathways specific to the range of environments they experience. When gene expression is specific to different environments (for example, Aubin-Horth and Renn, 2009), a specialist in one environment will purge deleterious mutations and fix beneficial mutations faster than a generalist, which experiences multiple environments (for example, Kawecki, 1994). The constraints imposed by relaxed selection are well established in the theoretical literature where specialists will often out-compete plastic generalists, but there are few data sets directly testing this idea. Genes specific to environmentally induced morphs in social insects and beetles show greater evolutionary divergence, consistent with weakened selection under conditions where the alternative morph is favored (Snell-Rood et al., 2011). Furthermore, some of these morph-specific genes show greater genetic variation, consistent with mutation accumulation due to less effective selection, although in some cases, strong sexual selection may offset this pattern (Kijimoto et al., 2014). Although there is strong theoretical evidence for relaxed selection on alternative developmental pathways, it is possible that the environment-specific gene expression assumed in these models is rare (for example, Snell-Rood et al., 2010) because of weakened selection on alternate developmental pathways. Emerging genomic data will clarify the mechanisms underlying plasticity and give insights to the degree of environment-specific expression and the likelihood of this weakened selection as a constraint on the evolution of plasticity.
Variation in the strength of selection across organisms, traits and generations can also influence the evolution of plasticity. In experimental evolution studies, the nature of variation in the selective environment influences the evolution of plasticity (that is, generalists vs specialists). Recent empirical studies have contrasted environments that differ in the scale of variation: coarse-grained environments, which are stable over the lifetime of an organism vs fine-grained environments, which are variable within an organism's lifetime (for example, Levins, 1968). Such comparisons of coarse- and fine-grained responses may be particularly informative to understanding the limits of the evolution of plasticity in organisms known to evolve over short timescales. Model systems to study plasticity over short timescales include invasive species (Ghalambor et al., 2007) and species exposed to anthropogenic contaminants (Hua et al., 2013).
Costs of plasticity
One potential character of universal relevance to both costs of plasticity or phenotype may be the costs associated with carrying around additional genetic machinery (Knight et al., 2005). One way of assessing this is through evaluating genome size. Indeed, evidence suggests that reduced genome size per se may be a target of selection for shorter generation times (for example, Price et al., 1980), and the repeated evolution of aneuploidization and variation in transposable element load suggests that removal of particular non-coding genetic material may have fitness benefits (for example, Hu et al., 2011). However, the costs of carrying unexpressed genes or additional regulatory elements is likely negligible in most cases—Latta et al. (2012) reported results of comparisons of expression plasticity of over 900 genes in response to salinity differences for both specialist (narrow tolerance) and generalist (plastic, broad tolerance) Daphnia genotypes. They found no differences between genotypes for three measures of cost: amount of transcription, protein length or ATP production.
We anticipate that costs of plasticity are generally negligible for the majority of organisms (that is, the fitness cost of the regulatory genetic machinery will typically be small), although for organisms with larger brains or sophisticated immune responses, costs of maintenance may become considerable (see Snell-Rood (2012) and discussion below on developmental selection in animals). Important avenues of research in the costs of plasticity remain. Employing theoretical approaches, we urge consideration of alternatives to quantitative genetics models (see Auld et al. (2010) for cautionary use of this model; further examinations of the limits of the model are developed in a companion manuscript, Schlichting et al. in preparation). Empirical approaches should take advantage of modern genomics and other ‘omics tools, particularly those which hold promise to disentangle the costs of plasticity from the costs of phenotype, and those which evaluate molecular machinery and regulation of plasticity and phenotypes. Together, these remain important avenues of research on the constraints of evolution of plasticity.
Experimental evolution can be used to directly assess costs of plasticity by looking for evidence of selection against plasticity: in a stable environment, selection should operate to reduce any plasticity that is inherently costly (Hall and Colegrave, 2008). Maughan et al. (2007) used this approach to examine costs of plasticity in the bacterium Bacillus subtilis. They documented the evolutionary loss of sporulation ability—a plastic response by some bacterial species to low-resource status—in an experiment with no selection for sporulation spanning 6000 generations. If costs of plasticity are significant, selection should drive the loss of sporulation ability; alternatively, sporulation ability could be lost via mutational degradation. Masel et al. (2007) showed that, in general, mutational degradation is a more important limit for rarely used plasticity whenever the functional mutation rate is greater than the selective coefficient. By directly measuring the functional mutation rate, Maughan et al. (2007) were able to predict how long until plasticity should be lost in the absence of costs, and compare this prediction to the observed rate of loss. All five populations lost plasticity, but four out of five showed no detectable cost, leaving only a single instance of selective loss that could also be explained by hitchhiking. Thus, costs of plasticity do not appear to limit the maintenance of plasticity in B. subtilis in a constant low-resource environment. Similar results were found for carbon usage in E. coli, where loss of plasticity was tightly correlated with mutation rate (Leiby and Marx, 2014). However, Kvitek and Sherlock (2013) found that many mutations to three growth control signaling networks in S. cerevisiae were adaptive in a constant environment. Although this last result is consistent with a cost of plasticity via regulatory sensing, it is also consistent with explanations that invoke a cost of a phenotype.
It may be informative to consider efforts to use experimental evolution approaches to test costs of plasticity together with growing theory on relaxed selection. Unfortunately, such empirical approaches require organisms that can be maintained under selection for hundreds or thousands of generations; such organisms may be the least likely to possess significant costs of plasticity. Possible tests might be made comparing laboratory strains of Drosophila (or other long-used laboratory animals or livestock) grown for generations in benign environments with their counterparts in nature, to evaluate evidence for reduction of costs of plasticity under relaxed selection. However, even in experimentally tractable systems, disentangling global costs of plasticity from local costs of phenotype remains a challenge.
Are the costs of phenotype and costs of plasticity for some complex characters inherently conflated?
For some forms of plasticity, costs of phenotype and costs of plasticity may be difficult to differentiate. In some cases, the costs of phenotype may be inherent in the costs of the ability to be plastic such as learning and acquired immunity in animals. Selective processes in development are costly particularly because a range of phenotypes are sampled during the developmental learning period and their performance evaluated through interactions with the environment (Snell-Rood, 2012). Phenotype sampling of such an animal generalist takes more time and energy relative to a specialist (for example, Laverty and Plowright, 1988), resulting in temporary phenotype–environment mismatches (for example, the cost of naïveté Dukas, 1998). In addition, information processing also requires additional investment in certain traits, for example, in larger costly brains (Laughlin et al., 1998). These costs of plasticity related to developmental selection can result in various trade-offs with allocation to reproduction or other tissues (for example, Isler and van Schaik, 2006). Regardless, learning-like mechanisms of plasticity may be truly special in that they come with clear costs and trade-offs.
Although important insights into costs have been uncovered with respect to learning, similar developmental costs should apply to any selective processes in development, such as clonal selection in the development of antibodies (vertebrate acquired immunity) or variability in gene expression and epigenetic stabilization of particular expression patterns (see Snell-Rood, 2012). In other types of organisms or traits, the constraints to plasticity can be more clearly evaluated as two distinct phenotypes: an environmental sensory machinery phenotype and a morphological or other focal phenotypic trait of interest. Such a perspective recasts the cost of plasticity as a localized cost of phenotype.
Opportunities to offset costs
Lifespan and ontogeny
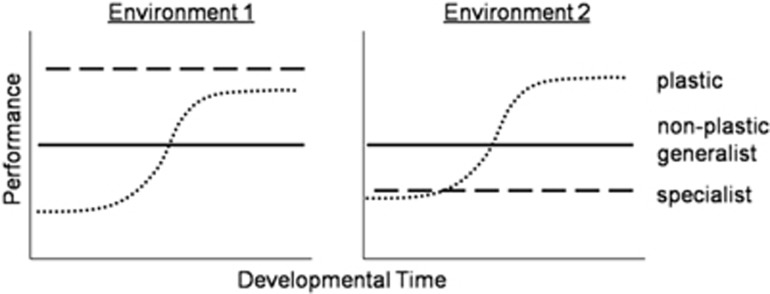
The developmental perspectives of Schlichting and Pigliucci (1998) and West-Eberhard (2003) have emphasized the importance of variation in responses to the environment throughout time, as selection on traits may vary throughout ontogeny (Figure 2) or as environments vary (for example, via physiological traits). The evolution of plasticity may be limited by opportunities to offset early costs (both phenotypic and plastic) later in life. These limits may be through altering compensation, redundancy or lag time (Sultan and Spencer 2002) and opportunities may vary with lifespan, life history and whether an organism has a final adult form. Modular organisms (such as plants and marine invertebrates) may compensate through the flexible addition of new modules, in conditions where a benefit would be realized through such investment. Organisms with determinate growth may be more likely to rely on other types of traits, such as behavior or have additional upfront costs of investing in plasticity. Further examination of how developmental variation limits the evolution of plasticity through experiments that contrast fitness components at different stages of development will help in understanding the mechanisms of compensation.
Figure 2.
Developmental timepoint and impacts on evaluation of costs. For learning-like mechanisms of plasticity, performance varies over development as several phenotypes are expressed and the optimal phenotype is gradually adopted. Because performance varies over time, the developmental timepoint at which phenotype costs are measured can have a major impact on their interpretation. A specialist (dashed line) has a fixed performance that is higher in environmental one. The non-plastic generalist (solid line) has the same performance in both environments. The plastic genotype (dotted line) varies performance throughout developmental time. Thus, relative performance of the plastic and specialist genotypes may vary through developmental time or across environments.
Amelioration of costs of phenotypes
Although models describing the evolution of plasticity assume a cost of plasticity must exist, we hypothesize that the cost of phenotype production may ultimately be more important in constraining the evolution of adaptive plasticity. However, over time strong selection may cause phenotypic costs to be reduced. This may take several forms: co-option of existing genetic machinery, compensatory mutations or evolution of novel genetic networks. One example of amelioration via compensatory mutation from single-celled organisms considers how yeast responds plastically to the availability of galactose (an environmentally variable resource) via upregulation of the GAL genes (with both metabolic and environmental sensory functions) for metabolizing galactose. The ancestral GAL1/3 gene product has two functions (Hittinger and Carroll, 2007): metabolize galactose and regulate transcription factors that lead to increased expression of GAL genes. GAL1/3 expression is constitutively low even when no galactose is present—that is, this ancestral gene does not respond to environmental variation in galactose. The cost of expression via GAL1/3 is a phenotypic cost that has been reduced in Saccharomyces evolution via gene duplication and subfunctionalization, which created two specialized genes. GAL3 is optimized for regulatory function with low levels of expression weakly tuned to galactose levels. GAL1 is optimized for metabolic function with much higher plastic levels of expression that depend strongly on regulatory signals. Thus, the evolutionary change that resulted in two genes with separate regulatory/environmental sensory and metabolic functions is one piece of evidence of reduced costs. The phenotype cost of low but constitutive expression of the optimized GAL3 is likely to be lower than the cost via constitutive expression of the ancestral gene GAL1/3 (Hittinger and Carroll, 2007). Such a division of labor among gene copies, separating responses to different cues, was proposed by Smith (1990) as a fundamental way of diversifying plastic responses, and may be a general solution to reducing the phenotypic costs associated with a lack of specificity of response (contrasting a gene always on GAL1/3, to one that is environmentally regulated GAL3). As data on gene expression patterns for closely related species are rapidly becoming available, the evolutionary history of changes in gene interactions and identification of co-option of function of existing regulatory sequences may lead to further insights into the evolution of novel plastic responses and these types of comparisons in multicellular organisms are encouraged.
van Noordwijk and de Jong (1986) demonstrated that life history trade-offs may be masked by condition-dependence of resource acquisition and allocation. We refer to this hypothesis as the condition-dependence hypothesis (see also Buchanan et al., 2013). It is also similar to Zahavi's (1975) handicap hypothesis for the evolution of elaborate male sexual traits. Such resource-based limits may be indications of costs of phenotype rather than costs of plasticity—as the costs are local and variation uncovered only in certain cases, and ameliorated in others. For example, Cothran et al. (2012) found that under low-resource environments where there was also an absence of predators, only some males were able to produce large sexual traits, but in resource-rich environments all males produce these traits. Further empirical investigations are warranted to determine the relative importance or sets of conditions where a condition-dependence hypothesis or relaxed selection will operate. For instance, will costs of plasticity be more apparent if we account for variation across genotypes in aspects of costs of phenotype such as ability to acquire and assimilate resources?
Moving forward
In reality, dissecting the genetic and epigenetic differences even among the three heuristic genotypes in Figure 1 may be a complex endeavor. However, new tools combined with classic methods in genetics and ecology will provide novel insights into the mechanisms behind differential performance of genotypes across environments. Below, we highlight potentially fruitful lines of inquiry. These recommendations include placing plasticity studies more securely within ecological and phylogenetic contexts, as well as employing simulation and ‘omics techniques to test hypotheses of reaction norm evolution.
Ecological context
A comprehensive approach to the study of plasticity includes the understanding of the adaptive nature of plasticity, whether there is heritable and ontogenetic variation of plastic responses within populations, and detailed knowledge of environments and their spatial and temporal frequencies (for example, Scheiner, 2013). Coordinated assessment of all these has been achieved in few systems. The documentation of environmental variation was historically a central area of research, and evaluating how well experimental environments mimic or simulate natural environments, particularly the relative frequency of each component of the environment, is still relevant today in assessing variation in evolutionary responses among populations (for example, Jacobs and Latimer, 2012).
The first obstacle to simulating the various abiotic dimensions of natural environmental variation under controlled conditions is the difficulty of quantifying fine- vs coarse-grained environmental variation in nature. Such efforts typically require long-term monitoring of conditions at the landscape scale. However, some abiotic measurements are becoming a more straightforward endeavor using monitoring equipment, and will proliferate as technology and access to weather and climate data continue to improve (for example, through NOAA: http://www.climate.gov/#dataServices; National Ecological Observation Network: http://www.neoninc.org). Similar efforts are also needed to obtain information on biotic environmental variation (that is, species interactions; Agrawal et al., 2007), which in many cases may be more challenging data to collect than abiotic environmental variation. Such data have been collected in some exemplary studies (for example, Werner et al., 2007), and advances in technology facilitate gathering biotic environmental variation for specific types of organisms (for example, remote sensing for vegetation).
Data on environmental variation and frequencies are also essential to experimental efforts to examine relaxed selection (Snell-Rood et al., 2010), to understand apparent patterns of selection on functional traits (Stinchcombe et al., 2012) or to forecast the likelihood of populations' evolutionary potential to match with rates of climate change (Quintero and Wiens, 2013). Quantifying environmental frequency and novelty (outside the typical range) will allow experiments to assess hidden reaction norms and contribute to a predictive theory of plastic responses (Snell-Rood, 2012; Hua et al., 2013). For example, are certain types of traits unlikely to respond appropriately to novel environments, revealing particular limits to plasticity? Experiments in arrays of environments may be able to generate data to successfully evaluate this important environmentally based limit of plasticity, and will simultaneously be of interest to applied ecologists managing populations.
Phylogenetic context and comparisons of closely related species
The ability to reconstruct evolutionary histories of traits and their plasticities is essential for understanding apparent limits to plasticity. Analyses of phenotypic responses of closely related species within a genus (Wund et al. 2008), or by employing a phylogenetically informed context, provide key information on the importance of both the evolution of phenotypic plasticity as a trait as well as the importance of phenotypic plasticity as a mechanism of diversification. Exemplary studies include comparison of physiological responses to light environments of 16 species in the plant genus Psychotria (Valladares et al., 2000), and of polyphenisms in response to food availability for example spadefoot toads (Ledon-Rettig et al., 2008).
Comparative (phylogenetic) approaches to the study of phenotype costs and genetic basis of plasticity may be more feasible from both genomic and phenotypic perspectives, thanks to advances in next-generation sequencing. Building data sets that combine genomic, phylogenetic and plasticity perspectives will be instrumental in developing a more complete picture of the evolution of plasticity. Such combined data sets will allow evaluation of aspects of evolutionary history, environment or genetic/genomic combinations that have contributed to situations where evolution of plasticity may be promoted or inhibited (Gompert et al., 2014).
Simulation
Modeling offers significant advantages for allowing us to understand how various factors might restrain evolution under a wide range of potential scenarios that might be difficult or impossible to experimentally manipulate. For example, in early investigations of this component of theory on phenotypic plasticity, van Tienderen (1991) compared non-plastic specialists and plastic generalists, exposing populations to selection while varying the strength of limits to plasticity. He delimited a parameter space where plastic generalists have an unequivocal advantage over non-plastic generalists, and other regions with more complex and contingent evolutionary dynamics. From van Tienderen's evolutionary modeling work, additional statistical procedures employing regression models were developed and adapted (for example, DeWitt et al., 1998). From this statistical framework, Auld et al. (2010) ran simulations to gain insight into how traditional regression analyses for costs of plasticity for multiple genotypes can lead to biased conclusions whenever there are strong correlations between the magnitudes of a trait and its plasticity. Other simulations have highlighted how maternal effects plasticity can alter performance in later generations (van den Heuvel et al., 2013), how plasticity allows individuals to improve fitness at the edges of the species' distribution while maximizing their fitness in the core (Pichancourt and van Klinken, 2012), and how unreliable environmental signals can favor the evolution of a population containing individuals that do and do not respond to the signal in a probabilistic manner (Arnoldini et al., 2012). Both models of plasticity evolution and statistical procedures for evaluating costs and environmental variation merit additional attention.
‘omics' approaches
As sequencing of whole genomes is increasingly employed for model and non-model organisms, the ability to evaluate reaction norms via transcriptomes, proteomes or metabolomes has arrived (Renn and Schumer, 2013; Grishkevich and Yanai, 2013). Our understanding of plasticity can be shaped by new tools and approaches including: (a) the ‘omic basis of phenotypes and plastic responses, (b) comparative genomics of the evolution of plastic responses, (c) epigenetic components of inheritance that may influence plastic responses (Glastad et al., 2011; Zhang et al., 2013), including the role of methylation and metabolic syndromes in predictive adaptive responses (Richards et al., 2010).
As ‘omics data accumulate, the matching data on phenotypes are typically lacking (White et al., 2012). Acquisition of extensive phenotypic data (phenomics) is now becoming a priority (Kuhl and Burghardt, 2013). Combining genomic and phenomic data, new bioinformatic and statistical tools, and clever experimental design will permit evaluation of the relative influences of genetic, environmental and epigenetic variation and further insights into the potential constraints to plasticity.
Understanding the genetic basis and allelic variation of traits and plasticity within and among populations for a broad set of biological systems can aid in uncovering critical constraints. For example, is a lack of genetic variation a fundamental limit to plasticity? Are there critical associations of particular aspects of genetic architecture and plastic traits? Do plasticity genes belong to particular gene ontology classes? In microbes, evolve-and-resequence approaches can be powerful (Kvitek and Sherlock, 2013). In sexual populations, genome-wide association studies (GWAS) allow the investigation of associations between single-nucleotide polymorphism and traits of interest. For example, in Arabidopsis thaliana, the adaptive plastic response of shade avoidance (increased hypocotyl length) was examined through GWAS uncovering known, de novo candidates and regions associated with genotype by environment interactions (Filiault and Maloof, 2012). Expanding from GWAS to phenotypic screens of mutant libraries (such as T-DNA mutants; O'Malley and Ecker, 2010; Murren, 2012) across environments will provide large phenotypic data sets to investigate questions of how particular genes influence plastic responses. As analytical tools in genomics and statistical informatics proliferate, there is promise of critical advances in our understanding of the limits of phenotypic plasticity (Table 1).
Table 1. Potential ‘omics approaches to investigate open questions on the evolution of plasticity: linking genomic, phenotypic and environmental data.
|
Unresolved questions in the evolution of and limits of phenotypic plasticity | |
|---|---|
| Topic and possible ‘omic × environment approaches | Example references of experimental approaches to date |
| To assess the role of habitat heterogeneity in limits to plasticity | |
| GWAS across multiple (3+) simulated environments | Uncovering de novo candidates and regions associated with genotype by environment interactions (see text for description of Filiault and Maloof, 2012) |
| Mutant library screens (including, but not limited to deletion collections, T-DNA; CRISPR) across multiple environments | Multiple environmental screens exposed complex mutant phenotypes, an approach which can be expanded to study plasticity, link between function and limits (Hillenmeyer et al., 2008) |
| To assess the roles of novel environments and relaxed selection on limits to plasticity | |
| Apply above GWAS or T-DNA screens in native simulated environments and novel environments | Bacterial isolates from multiple host sources, which vary in diet with a seven-gene region with a host-specific signal (Sheppard et al., 2013) |
| Sequencing of adaptive variants in defined experimental evolution conditions | Detecting, via overrepresented gene ontology groups, potentially adaptive loss of plasticity in a constant environment (Kvitek and Sherlock, 2013) |
| To assess the functional roles of candidate loci and assess whether certain functional roles contribute more/less as potential limits of plasticity | |
| Comparative genomics within and across species and expansion of model and non-model data sets, QTL and GWAS approaches | Root structure of natural Arabidopsis accessions and functional mutation experiments in a lab-line background indicate genetic component of GχE interactions (Rosas et al., 2013) |
| To assess the extent of epigenetic control on the expression of plasticity and the role of relaxed selection as a limit to plasticity or generalists | |
| epiRILs | Nearly isogenic lines that vary in DNA methylation pattern in response to drought and nutrient treatments varied in ecologically important plant traits (Zhang et al., 2013) |
| Gene expression and sequence evolution | Gene-specific expression to alternate environments or morphs evolve more quickly (Snell-Rood et al., 2011; Leichty et al., 2012); harbor more genetic variation (Moczek in revision) |
| DNA methylation signatures | Signatures of phenotype–environment mismatch (Gilbert and Epel, 2009) |
Abbreviation: epiRIL, epigenetic recombinant inbred line; GWAS, genome-wide association study; QTL, quantitative trait loci.
In tandem with basic research, applied evolutionary studies in agriculture (Vigueira et al., 2013), fisheries (Bradbury et al., 2013), conservation biology (Richards et al., 2006) and human health (Low et al., 2012) will provide new opportunities to examine the limits of plasticity from ecological genomic or epigenomic perspectives. Systems biology perspectives that scale from DNA to RNA to protein inform how intermediate traits (for example, RNA expression) may influence downstream traits and may further our conceptualization of plasticity and opportunities to offset costs and limits of plasticity. Investigations of a diversity of species that vary in mating system, geographic extent, habitat heterogeneity, and diversity of subspecies or landraces will be critically informative to examining evolution of the limits of plasticity over short timescales.
These new and growing knowledge bases together with carefully planned theoretical and empirical approaches such as Maughan et al. (2007) efforts to evaluate costs of plasticity or Snell-Rood et al. (2010) efforts to evaluate relaxed selection, offer the opportunity to shed new light on the constraints on plasticity evolution.
Conclusions
Our ability to develop robust theories of the evolution of plasticity requires further research on both the constraints and benefits of plasticity across systems. We advocate evaluating systems to examine the relative contributions of relaxed selection, variable selection and costs of plasticity on constraints on the evolution of phenotypic plasticity by employing both classical and new tools. We hypothesize that costs of phenotype and limits to plasticity outweigh costs of plasticity in many systems and warrant specific formalization in that context, yet animals that employ learning and single-celled organisms in constant environments may be special cases where costs may be detected. Even in these systems, costs of plasticity and costs of phenotype must be clearly and separately evaluated, and may often be difficult to differentiate. Costs and limits may be offset over the lifetime of an organism or in certain ecologies, thus developmental and ecological perspectives will shed new light on the response of organisms to heterogeneous landscapes.
Efforts to understand limits to ability of organisms to deploy appropriate plasticity have become more urgent in the face of pervasive and rapid environmental change such as habitat fragmentation, influx of invasive species and global climate change, in addition to human urban systems and understanding transmission of disease, and developing of agricultural methods and genotypes with reduced ecological footprints. Recent decades of research have amassed substantial data on the nature of plasticity and demonstrated that it is of fundamental importance in biological systems (for example, Pigiucci, 2001; West-Eberhard, 2003). The broad question of how generalists and specialists differ across environments, through development, and over evolutionary history remains central to understanding evolution in changing environments.
Acknowledgments
This work was supported by the National Evolutionary Synthesis Center (NESCent), NSF #EF-0423641 for a working group (project leaders: CJM and CDS) and a short-term fellowship to CJM. Support from the National Science Foundation and other sources is gratefully acknowledged (CJM and HSC: NSF IOS-1052262, CJM: IOS-1146977; CDS: NSF DEB-1242294, DEB-1046328, OISE-0623341; JM: John Templeton Foundation and NIH RO1 GM076041 and GM104040; CKG: NSF DEB-0846175; RAR: NSF DEB 07-16149, DEB 11-19430; ESR NSF IOS-1354737). We thank scholars whose work in this area past and present has contributed to the vibrancy of this area of inquiry, and note the journal's limits on the numbers of scholarly works that we were able to cite. Additionally we thank Allen Rodrigo (NESCent Director) and D Wilson and S Risbon at NESCent for their support for the working group. We further are grateful to the comments of Sam Scheiner, three anonymous reviewers and comments from the editors of this journal and special issue, which improved the manuscript.
The authors declare no conflict of interest.
References
- Agrawal AA, Ackerly DD, Adler F, Arnold AE, Caceres C, Doak DF et al. (2007). Filling key gaps in population and community ecology. Front Ecol Environ 5: 145–152. [Google Scholar]
- Arnoldini M, Mostowy R, Bonhoeffer S, Ackermann M. (2012). Evolution of stress response in the face of unreliable environmental signals. PLoS Comput Biol 8: e1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Horth N, Renn SCP. (2009). Genomic reaction norms: using integrating biology to understand molecular mechanisms of phenotypic plasticity. Mol Ecol 18: 3763–3780. [DOI] [PubMed] [Google Scholar]
- Auld JR, Agrawal AA, Relyea RA. (2010). Re-evaluating the costs and limits of adaptive phenotypic plasticity. Proc R Soc Ser B 277: 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrigan D, Scheiner SM: Modeling the evolution of phenotypic plasticity. DeWitt TJ, Scheiner SM. 2004) Phenotypic Plasticity: Function and Conceptual Approaches. New York, NY: Oxford University Press, 82–97. [Google Scholar]
- Bradbury IR, Hubert S, Higgins B, Bowman S, Borza T, Paterson IG et al. (2013). Genomic islands of divergence and their consequences for the resolution of spatial structure in an exploited marine fish. Evol Appl 6: 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw AD. (1965). Evolutionary significance of phenotypic plasticity in plants. Adv Genet 13: 115–155. [Google Scholar]
- Buchanan KL, Grindstaff JL, Pravosudov VL. (2013). Condition dependence, developmental plasticity, and cognition: implications for ecology and evolution. Trends Ecol Evol 28: 290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan HS, Maughan H, Steiner UK. (2008). Phenotypic plasticity, costs of phenotypes, and costs of plasticity. Ann NY Acad Sci 1133: 44–66. [DOI] [PubMed] [Google Scholar]
- Chevin LM, Lande R, Mace GM. (2010). Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol 8: e1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cothran RD, Stiff AR, Jeyasingh PD, Relyea RA. (2012). Eutrophication and predation risk affect sexual trait expression and mating success. Evolution 66: 708–719. [DOI] [PubMed] [Google Scholar]
- DeWitt TJ, Sih A, Wilson DS. (1998). Costs and limits of phenotypic plasticity. Trends Ecol Evol 13: 77–81. [DOI] [PubMed] [Google Scholar]
- Dukas R. (1998). Evolutionary ecology of learning. Ppin Dukas R (ed). Cognitive Ecology: the Evolutionary Ecology of Information Processing and Decision Making. University of Chicago Press: Chicago, 129–174. [Google Scholar]
- Filiault DL, Maloof JN. (2012). A genome-wide association study identifies variation underlying the Arabidopsis thaliana shade avoidance response. PLOS Genet 8: e1002589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier-Level A, Korte A, Coope MD, Nordborg M, Schmitt J, Wilczek AM et al. (2011). A map of local adaptation in Arabidopsis thaliana. Science 334: 86–89. [DOI] [PubMed] [Google Scholar]
- Getty T. (1996). The maintenance of phenotypic plasticity as a signal detection problem. Am Nat 148: 378–385. [Google Scholar]
- Ghalambor CK, McKay JM, Carroll SP, Reznick DN. (2007). Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct Ecol 21: 394–407. [Google Scholar]
- Gilbert SF, Epel D. (2009) Ecological Developmental Biology: Integrating Epigenetics, Medicine and Evolution. Sinauer Assoc: Sunderland MA. [Google Scholar]
- Glastad KM, Hunt BG, Yi SV, Goodisman MAD. (2011). DNA methylation in insects: on the brink of the epigenomic era. Insect Mol Biol 20: 553–565. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Comeault AA, Farkas TE, Feder JL, Prachman TL, Buerkle CA et al. (2014). Experimental evidence for ecological selection on genome variation in the wild. Ecol Lett 17: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishkevich V., Yanai I. (2013). The genomic determinants of genotype x environment interactions in gene expression. Trends Genet 29: 479–487. [DOI] [PubMed] [Google Scholar]
- Hall AR, Colegrave N. (2008). Decay of unused characters by selection and drift. J Evol Biol 21: 610–617. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W et al. (2008). The Chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. (2007). Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449: 677–681. [DOI] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng J-F, Clark F et al. (2011). The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Morehouse N, Relyea RA. (2013). Pesticide resistance in amphibians: Induced resistance in susceptible populations, constitutive tolerance in tolerant populations. Evol Appl 6: 1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isler K, van Schaik C. (2006). Costs of encephalization: the energy trade-off hypothesis tested on birds. J Human Evol 51: 228–243. [DOI] [PubMed] [Google Scholar]
- Jacobs BS, Latimer AM. (2012). Analyzing reaction norm variation in the field vs greenhouse: what the differences can tell us. Persp Plant Ecol Evol Sys 14: 325–334. [Google Scholar]
- Kawecki TJ. (1994). Accumulation of deleterious mutations and the evolutionary cost of being a generalist. Am Nat 144: 833–838. [Google Scholar]
- Kijimoto T, Snell-Rood EC, Pespeni MH, Rocha G, Kafadar K, Moczek AP et al. (2014). The nutritionally responsive transcriptome of the polyphonic beetle Onthophagus taurus and the importance of sexual dimorphism and body region. Proc Roy Soc B 281: 20142084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CA, Molinari NA, Petrov NA. (2005). The large genome constraint hypothesis: evolution, ecology and phenotype. Ann Bot 95: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl HS, Burghardt T. (2013). Animal biometrics: quantifying and detecting phenotypic appearance. Trends Ecol Evol 28: 432–441. [DOI] [PubMed] [Google Scholar]
- Kvitek DJ, Sherlock G. (2013). Whole genome, whole population sequencing reveals that loss of signaling networks is the major adaptive strategy in a constant environment. PLOS Genet 9: E10038972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta LC, Weider LJ, Colbourne JK, Pfrender ME. (2012). The evolution of salinity tolerance in Daphnia: a functional genomics approach. Ecol Lett 15: 794–802. [DOI] [PubMed] [Google Scholar]
- Laughlin SB, van Steveninck RRD, Anderson JC. (1998). The metabolic cost of neural information. Nat Neurosci 1: 36–41. [DOI] [PubMed] [Google Scholar]
- Laverty TM, Plowright RC. (1988). Flower handling by bumblebees: a comparison of specialists and generalists. Anim Behav 36: 733–740. [Google Scholar]
- Ledon-Rettig CC, Pfennig DW, Nascone-Yoder N. (2008). Ancestral variation and the potential for genetic accommodation in larval amphibians: implications for the evolution of novel feeding strategies. Evol Dev 10: 316–325. [DOI] [PubMed] [Google Scholar]
- Leiby N, Marx CJ. (2014). Metabolic erosion primarily through mutation accumulation not specific tradeoffs, drives limited evolution of substrate specificity in Escherichia coli. PLoS Biol 18: E1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichty AR, Pfennig DW, Jones CD, Pfennig KS. (2012). Relaxed genetic constraint is ancestral to the evolution of phenotypic plasticity. Integr Compar Biol 52: 16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levins R. (1968). Evolution in changing environments: Some theoretical expectations. Monogr Popul Biol. eds. Levin SA, Horn HS Princeton University Press: Princeton, New Jersey, USA. [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A et al. (2013). Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet 45: 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low FM, Gluckman PD, Hanson MA. (2012). Developmental plasticity, epigenetics and human health. Evol Biol 39: 650–665. [DOI] [PubMed] [Google Scholar]
- Lynch M, Lande R. (1993). Evolution and extinction in response to environmental change. in Karieva PM, Kingsolver JG, Huey RB eds. Biotic Interactions and Global Change. Sinauer Associates: Sunderland, MA, 234–250. [Google Scholar]
- Masel J, King OD, Maughan H. (2007). The loss of adaptive plasticity during long periods of environmental stasis. Am Nat 169: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan H, Masel J, Birky CW, Nicholson WL. (2007). The roles of mutation accumulation and selection in a loss of sporulation in experimental populations of Bacillus subtilis. Genetics 177: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. (1992). The evolutionary maintenance of alternative phenotypes. Am Nat 139: 971–989. [Google Scholar]
- Murren CJ. (2012). The integrated phenotype. Integr Compar Biol 52: 64–76. [DOI] [PubMed] [Google Scholar]
- Murren CJ, Maclean HJ, Diamond SE, Steiner UK, Heskel MA, Handelsman CA et al. (2014). Evolutionary change in continuous reaction norms. Am Nat 183: 453–467. [DOI] [PubMed] [Google Scholar]
- O'Malley RC, Ecker JR. (2010). Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61: 928–940. [DOI] [PubMed] [Google Scholar]
- Pichancourt J-B, van Klinken RD. (2012). Phenotypic plasticity influences the size, shape and dynamics of the geographic distribution of an invasive plant. PLoS ONE 7: e32323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigiucci M. (2001) Phenotypic Plasticity, Beyond Nature vs Nurture. Baltimore: John Hopkins Univ. Press. [Google Scholar]
- Price HJ, Bachmann K, Chambers KL, Riggs J. (1980). Detection of intraspecific variation in nuclear DNA content in Microseris douglasii. Bot Gaz 141: 195–198. [Google Scholar]
- Quintero I, Wiens JJ. (2013). Rates of projected climate change dramatically exceed past rates of climatic niche evolution among vertebrate species. Ecol Lett 16: 1095–1103. [DOI] [PubMed] [Google Scholar]
- Renn SCP, Schumer ME. (2013). Genetic accommodation and behavioural evolution: insights from genomic studies. Anim Behav 85: 1012–1022. [Google Scholar]
- Richards CL, Bossdorf O, Muth NZ, Gurevitch J, Pigliucci M. (2006). Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol Lett 9: 981–993. [DOI] [PubMed] [Google Scholar]
- Richards CL, Bossdorf O, Pigliucci M. (2010). What role does heritable epigenetic variation play in phenotypic evolution? BioScience 60: 232–237. [Google Scholar]
- Roff DA. (2011). Measuring the cost of plasticity: a problem of statistical non-independence. Proc R Soc B Biol Sci 278: 2724–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas U, Cibrian-Jaramillo A, Ristova D, Banta JA, Gifford ML, Fan AH et al. (2013). Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root system architecture. Proc Nat Acad Sci USA 110: 15133–15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiner SM. (2013). The genetics of phenotypic plasticity. XII.Temporal and spatial heterogeneity. Ecol Evol 3: 4596–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiner SM, Holt RD. (2012). The genetics of phenotypic plasticity. X. Variation versus uncertainty. Ecol Evol 2: 751–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting CD, Pigliucci M. (1998) Phenotypic Evolution: A Reaction Norm Perspective. Sunderland MA: Sinauer. [Google Scholar]
- Sheppard SK, Didelot X, Meric G, Torralbo A, Jolley KA, Kelly DJ et al. (2013). Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Nat Acad Sci USA 110: 11923–11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. (1990). Signal perception, differential expression within mutigene families and the molecular basis of phenotypic plasticity. Plant Cell Enviro 13: 585–594. [Google Scholar]
- Snell-Rood EC, Van Dyken JD, Cruickshank T, Wade MJ, Moczek AP. (2010). Toward a population genetic framework of developmental evolution: the costs, limits and consequences of phenotypic plasticity. BioEssays 32: 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell-Rood EC, Cash A, Han MV, Kijimoto T, Andrews J, Moczek AP et al. (2011). Developmental decoupling of alternative phenotypes insights from the transcriptomes of horn-polyphenic beetles. Evolution 65: 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell-Rood EC. (2012). Selective processes in development: implications for the costs and benefits of phenotypic plasticity. Integr Compar Biol 52: 31–42. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR Function-valued traits working groupKirkpatrick M. (2012). Genetics and evolution of function valued traits: understanding environmentally responsive phenotypes. Trends Ecol Evol 27: 637–647. [DOI] [PubMed] [Google Scholar]
- Sultan SE, Spencer HG. (2002). Metapopulation structure favors plasticity over local adaptation. Am Nat 160: 271s–283s. [DOI] [PubMed] [Google Scholar]
- Tsuji H, Taoka KL, Shimamoto K. (2011). Regulation of flowering in rice: two florigen genes, a complex gene network and natural variation. Curr Opin Biol 14: 45–52. [DOI] [PubMed] [Google Scholar]
- Valladares F, Wright SJ, Lasso E, Kitajima K, Percey RW. (2000). Plastic Phenotypic Response to Light of 16 Congeneric Shrubs from a Panamanian Rainforest. Ecology 81: 1925–1936. [Google Scholar]
- van Buskirk J, Steiner UK. (2009). The fitness costs of developmental canalization and plasticity. J Evol Biol 22: 852–860. [DOI] [PubMed] [Google Scholar]
- van Buskirk J. (2002). A comparative test of the adaptive plasticity hypothesis: relationships between habitat and phenotype in anuran larvae. Am Nat 160: 87–102. [DOI] [PubMed] [Google Scholar]
- van den Heuvel J, Saastamoinen M, Brakefield PM, Kirkwood TBL, Zwaan BJ, Shanley DP et al. (2013). The predictive adaptive response: modeling the life-history evolution of the butterfly Bicyclus anynana in seasonal environments. Am Nat 181: E28–E42. [DOI] [PubMed] [Google Scholar]
- van Kleunen M, Fischer M. (2005). Constraints on the evolution of adaptive phenotypic plasticity in plants. New Phytol 166: 49–60. [DOI] [PubMed] [Google Scholar]
- van Noordwijk AJ, de Jong G. (1986). Acquisition and allocation of resources: their influence on variation in life history tactics. Am Nat 128: 137–142. [Google Scholar]
- van Tienderen PH. (1991). Evolution of generalists and specialists in spatially heterogeneous environments. Evolution 45: 1317–1331. [DOI] [PubMed] [Google Scholar]
- Via S, Lande R. (1985). Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39: 505–522. [DOI] [PubMed] [Google Scholar]
- Vigueira CC, Olsen KM, Caicedo AL. (2013). The red queen in the corn: agricultural weeds as models of rapid adaptive evolution. Heredity 110: 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner EE, Yurewicz KL, Skelly DK, Relyea RA. (2007). Turnover in an amphibian metacommunity: the role of local and regional factors. Oikos 116: 1713–1725. [Google Scholar]
- West-Eberhard MJ. (2003) Developmental Plasticity and Evolution. Oxford University Press: New York. [Google Scholar]
- Whitman DA, Agrawal AA. (2009). What is Phenotypic Plasticity and why is it Important? Pages 1-63. in: Whitman DW, Ananthakrishna TN (eds) Phenotypic Plasticity of Insects: Mechanisms and Consequences. Science Publishers, Inc: Enfield, N.H. [Google Scholar]
- White JW, Andrade-Sanchez P, Gore MA, Bronson KF, Coffelt TA, Conley MM et al. (2012). Field-based phenomics for plant genetics research. Field Crops Research 133: 101–112. [Google Scholar]
- Wund MA, Baker JA, Clancy B, Golub JL, Foster SA. (2008). A test of the "Flexible stem" model of evolution: Ancestral plasticity, genetic accommodation, and morphological divergence in the threespine stickleback radiation. Am Nat 172: 449–462. [DOI] [PubMed] [Google Scholar]
- Zhang Y-Y, Fisher M, Colot V, Bossdorf O. (2013). Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol 197: 314–332. [DOI] [PubMed] [Google Scholar]
- Zahavi A. (1975). Mate Selection—a selection handicap. J Theor Biol 53: 205–214. [DOI] [PubMed] [Google Scholar]