Abstract
For many organisms the ability to cold acclimate with the onset of seasonal cold has major implications for their fitness. In insects, where this ability is widespread, the physiological changes associated with increased cold tolerance have been well studied. Despite this, little work has been done to trace changes in gene expression during cold acclimation that lead to an increase in cold tolerance. We used an RNA-Seq approach to investigate this in two species of the Drosophila virilis group. We found that the majority of genes that are differentially expressed during cold acclimation differ between the two species. Despite this, the biological processes associated with the differentially expressed genes were broadly similar in the two species. These included: metabolism, cell membrane composition, and circadian rhythms, which are largely consistent with previous work on cold acclimation/cold tolerance. In addition, we also found evidence of the involvement of the rhodopsin pathway in cold acclimation, a pathway that has been recently linked to thermotaxis. Interestingly, we found no evidence of differential expression of stress genes implying that long-term cold acclimation and short-term stress response may have a different physiological basis.
Introduction
Insects have a range of tactics to deal with the onset of seasonal cold (Salt, 1961). One widespread tactic, cold acclimation, is an increase in an organism's cold tolerance that occurs over days or weeks when exposed to sub-lethal low temperatures, allowing insects to remain active for longer as the seasons change (Lee, 2010). The physiological changes leading to increased cold tolerance during cold acclimation have been well studied in insects and are known to primarily involve a shift in the metabolic profile of the insect, particularly sugars, polyols and amino acids (Lee, 1991; Lee, 2010). Accompanying this is a compositional change of the cell membrane primarily involving changes in phospholipids (Hazel, 1995; Koštál et al., 2003; Overgaard et al., 2008). These changes are cryoprotective, allowing cells to maintain their osmotic balance and thus to continue to function at low temperatures.
Cold acclimation is likely to be a homologous trait for many species of closely related insects as it has clear fitness benefits (Salt, 1961; Lee, 2010) and is thus unlikely to be lost and gained independently. Homology can be defined at several levels of biological organisation including genes, gene networks, and phenotypes, which may not necessarily coincide as the different levels may evolve at different rates. As a result it could be that although a trait is homologous and phenotypically similar across species, the genes that contribute to the production of the trait could be different as a result of evolutionary turnover, whereby genes involved in similar biological processes are co-opted in or out of influencing a trait as a result of orthologous selection or drift. This process of evolutionary turnover can thereby allow the genetic basis of a trait to change, whilst the trait itself remains conserved (see Canestro et al. (2007) and McCune and Schimenti (2012) for reviews).
Candidate gene approaches, whereby genes of interest identified in a model species are functionally examined in others, have often been successful and have indicated that gene function tends be conserved across species (Fitzpatrick et al., 2005; Reaume and Sokolowski, 2011; Martin and Orgogozo, 2013). In contrast, quantitative genetics studies have often suggested that traits important to adaptation are polygenic, and quantitative trait loci (QTL) may have low repeatability between species (Arbuthnott, 2009; Huang et al., 2012; Rockman, 2012). One reason for this disparity may be that candidate genes are typically those that have a large influence on a particular trait, and thus the evolutionary forces affecting them are likely to be stronger and more consistent than those for smaller effect loci. In particular, we expect that genes with a large influence on a particular trait are more likely to be conserved in a particular role (by purifying selection) than those with a smaller influence, which can be more easily co-opted in or out of influencing a trait.
As cold acclimation is an induced response, the gene expression changes involved with the trait can be studied using an RNA-seq approach. RNA-seq allows examination of the changes in gene expression independent of the strength of influence they have on cold acclimation, identifying both genes that have a major controlling influence on cold acclimation and genes involved in smaller secondary changes. Furthermore, it allows assessment of the proportion of genes that respond to cold acclimation that are the same between the two species, and what biological processes they are involved in.
Here we investigated gene expression changes in response to cold acclimation in Drosophila (D.) montana and D. virilis using an RNA-seq approach. These species belong to the D. virilis species group, and diverged from each other approximately 9–11 mya (Reis et al., 2008; Morales-Hojas et al., 2011). Both species show a high level of cold tolerance compared to other drosophilids, and D. montana is significantly more cold tolerant than D. virilis (Kellermann et al., 2012; Vesala et al., 2012b). These species have adapted to live in quite different environmental conditions (D. montana is found at high latitudes (30–70°N) and altitudes while D. virilis is a human commensal found at lower latitudes (south from 35°N) (Throckmorton, 1982)) but both exhibit a similar cold acclimation phenotype, whereby flies kept at sub-optimal temperatures show increased cold tolerance as measured by chill-coma recovery time (Vesala et al., 2012b). When insects are placed in cold (<0 °C) temperatures a chill-coma is induced whereby insects lose the ability to move temporarily, due to a loss of nerve and muscle excitability (MacMillan and Sinclair, 2011). The time it takes for an insect to regain its ability to move is referred to as chill coma recovery time, which can be used as an estimate of an insect's cold tolerance (see David et al. (1998)). The cold acclimation responses of D. montana and D. virilis are likely to represent homologous traits as this trait is present in both species and across Drosophila species in general (see Hoffmann et al. (2003)). Thus our aims were: (i) to identify the genes that are differentially expressed in response to cold acclimation in D. montana and D. virilis and (ii) to identify the molecular pathways and processes underlying the cold acclimation response in each species, allowing us to determine to what extent to which these are consistent between the species.
Materials and Methods
Samples
We used a single isofemale line established from the progenies of wild-caught fertilized females for each species: D. montana line 175OJ8 (originated from Oulanka, Finland, 2008) and D. virilis line TOY3F9 (originated from Toyama, Japan, 2003). The use of isofemale lines will reduce confounding effects of genetic variation within lines and cold acclimation responses (see discussion). Stock cultures have been maintained since their establishment on malt bottles in continuous light at 19±1 °C, 65% humidity. Flies for the experiment were collected within one day of eclosion using light CO2 anesthesia. Female flies were put into malt medium vials and transferred into a climate chamber (Sanyo MLR-351H) for 15 days at 16 °C for D. montana and 19 °C for D. virilis; see rationale for the temperature selection in discussion. Flies of each species were then split into two groups: a control group where flies were left in the same conditions for an additional 6 days, and a cold acclimation group where flies were maintained for 6 days at +5 °C. The light:dark (LD) cycle was 22:2 for all experimental groups and the samples for RNA extraction were flash frozen in liquid nitrogen 5 h after lights were turned on in the chambers (Zeitgeber=5).
RNA extraction and sequencing
Frozen flies were pooled into 6 samples for D. montana (3 acclimation group samples and 3 control group samples, with 10 whole flies in each sample) and 4 samples for D. virilis (2 acclimation group samples and 2 control group samples, with 20 whole flies in each sample). Different numbers of pooled individuals per sample is unlikely to influence our analysis as all flies are from isofemale lines, but this design could have more power to detect changes in D. montana than D. virilis (but, see later). RNA was extracted from each sample using Tri Reagent (Sigma-Aldrich) followed by RNeasy Mini kit purification with DNase treatment (Qiagen). Purity of the RNA was checked using NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) and integrity with 2100 Bioanalyzer (Agilent Technologies). Total RNA for each sample was approximately equal prior to sequencing.
Extracted RNA was sequenced using the SOLiD platform. For D. montana we used the SOLiD 5500 XL to produce 46 million 75+35 bp paired end reads and for D. virilis we used SOLiD V4 to produce 49 million 50 bp single end reads. Raw sequence reads were then trimmed using SOLiD TRIM (with run options: -p 3 -q 22 -y y -e 2 -d 10) to remove polyclonal errors from the data (Sasson and Michael, 2010). The reads that passed this filter were then error corrected using SOLiD Accuracy Enhancer Tools (SAET) to reduce the amount of color calling errors, or erroneous bases, in the sequence. Remaining low quality bases at the end of the reads were then trimmed using CLC Genomics Workbench 5.0.1 (CLC Bio http://www.clcbio.com/) (quality score: 0.02). Differences in the number of replicates and read type used between samples could influence our power to detect differential expression (DE), however we expect these issues to be small as we used a single model to detect DE and interactions. In addition we also repeated the analysis for D. montana using two replicates and with reads trimmed to be 50 bp and single end only (to make the samples comparable to those obtained for D. virilis). This produced very similar results as those with three replicates and paired reads (numbers of genes found to be DE 162 vs 177; correlation of the gene expression for DE genes from both analyses (rs)=0.94, P<2.2 × 10−16, see Supplementary Materials 1 for more details).
Mapping
Reads for each sample were mapped individually to the D. virilis genome (r1.2, available from http://flybase.org/) using CLC Bio. We also used this method to map reads to a de novo assembly of the D. montana transcriptome (see Supplementary Materials 2 for more details). HTSeq (Anders et al., 2014) was used to quantify the read counts mapping uniquely to the reference using the reference annotation available from Flybase (r1.2).
Expression analysis
Gene expression analysis was performed using the Bioconductor package EdgeR (Robinson et al., 2010) in R (R Core Team, 2013). Normalisation factors for each sample were computed using the TMM method. TMM normalisation allows variation in read depth (due to RNA quality, variation in sequencing reaction, etc.) to be accounted for, to prevent differences in read depth from influencing the detection of DE (Robinson and Oshlack, 2010). We then fitted a generalized linear model (GLM) with negative binomial distribution with the terms species, treatment and species * treatment (full model), and estimated dispersion using the Cox-Reid profile-adjusted likelihood (CR) method. We used a GLM likelihood ratio test to determine significance of a treatment effect for each gene by comparing appropriate model contrasts: the effect of treatment on D. montana, the effect of treatment on D. virilis and an interaction between species and treatment. The interaction term tests the extent to which gene expression changes differ between the species. The P-values from the GLM likelihood ratio tests were corrected for multiple testing using Benjamini and Hochberg's algorithm to control for false discovery rate (FDR) (Benjamini and Hochberg, 1995) with significance taken here to be <5% (FDR<0.05).
Functional classification
In order to functionally classify genes we used Gene Ontology (GO) annotation for orthologous genes in D. melanogaster (available from Flybase, version: FB2013_06). We used this approach instead of using GO terms from the D. virilis annotation due to the superior GO annotation available in D. melanogaster both in terms of the number of annotations and their specificity (Tweedie et al., 2009). Significant enrichment of single GO terms were determined using the ‘Gene Ontology Enrichment' function in FlyMine (www.flymine.org, v.37). The D. melanogaster orthologs of DE genes were also analysed using DAVID (Database for Annotation, Visualization and Integrated Discovery) v. 6.7 (Huang et al., 2009a; Huang et al., 2009b). DAVID clusters genes into functional groups using a ‘fuzzy' clustering algorithm, and then uses a Fisher's exact test to identify significantly enriched functional groups. A functional group was considered to be significantly enriched if its enrichment score (the geometric mean (in -log scale) of the P-values of the GO terms in the group) was >1 (P<0.1).
Results
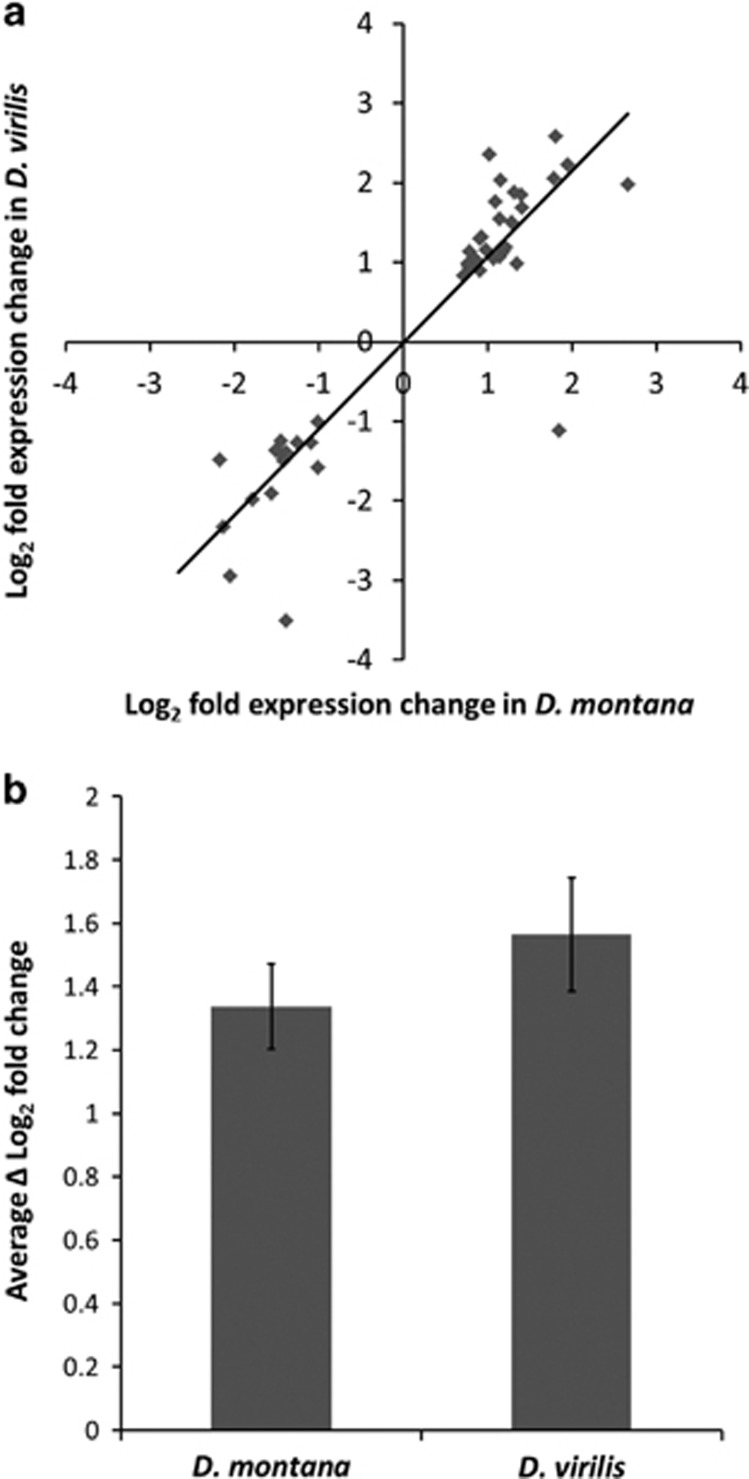
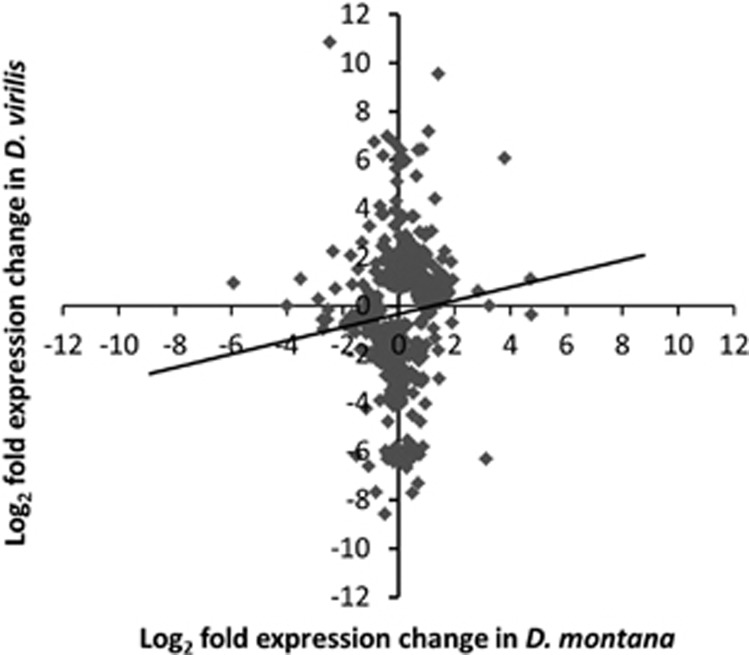
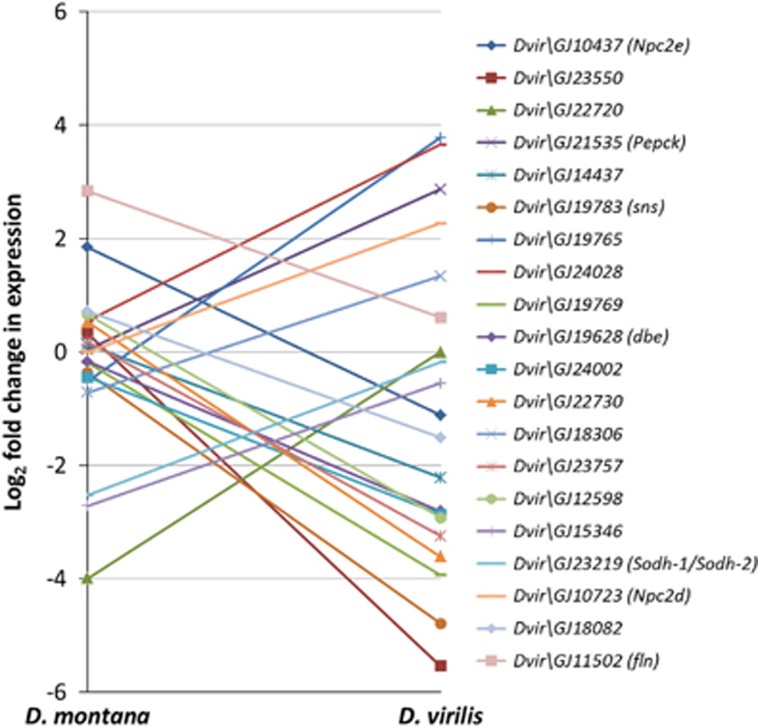
We obtained approximately 46 million reads for D. montana and 49 million reads for D. virilis, of which approximately 10% were found to map uniquely to a gene in the D. virilis reference genome. The number of reads mapping to each gene was highly correlated between the species (rs=0.90), suggesting the mapping efficiency per gene is approximately equal for the two species. The number of genes that were differentially expressed (DE) during cold acclimation was fewer in D. montana (177) than in D. virilis (458) at 0.05 FDR (representing 593 total unique genes DE across both species, see Supplementary Materials 3). Among these genes only 42 genes (7%) were DE in both species. The genes DE in both species had very similar expression changes in response to cold acclimation in terms of direction (rs=0.85, P<2.2 × 10−16) (Figure 1a) and fold change (Figure 1b) though changes were on average somewhat higher for D. virilis. One gene (Dvir\GJ10437) showed a significant interaction effect, being up-regulated in D. montana but down-regulated in D. virilis. The remaining genes, i.e. the genes that were DE in only one of the species, showed a much smaller correlation in expression levels (rs=0.20, P=1.18 × 10−6) (Figure 2) and 132 (24%) of them showed a significant species by treatment interaction, whereby genes which showed a strong response to acclimation in one species do not show a response in the other (Figure 3) (i.e. these genes showed species-specific responses).
Figure 1.
(a) Plot of log2 fold changes between control and cold acclimated flies for each of the genes that were DE in both D. montana and D. virilis (b) Average change in gene expression (Δ log2 fold change) between control and cold acclimated flies for genes that were DE in both species. Error bars represent approximate 95% confidence intervals.
Figure 2.
Plot of log2 fold changes between control and cold acclimated flies for each of the genes that are DE in either D. montana or D. virilis.
Figure 3.
Log2 fold change between control and cold acclimated flies for the top 20 genes which showed a significant species by treatment interaction in D. montana and D. virilis. D. virilis gene names are given in the legend with D. melanogaster orthologs in parentheses.
Gene function
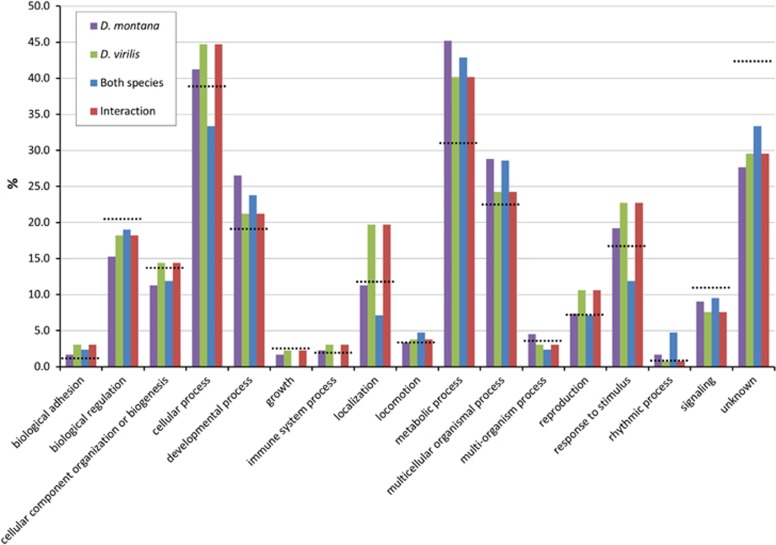
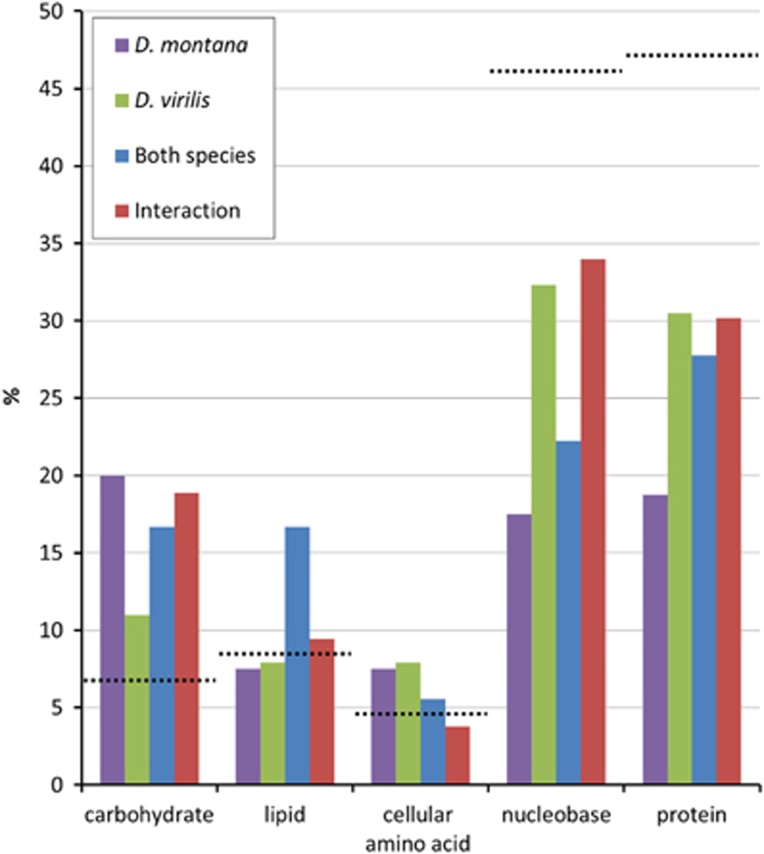
To examine the biological processes and pathways of the genes DE due to cold acclimation, we examined the genes in four subsets: those DE in D. montana, those DE in D. virilis, those DE in both species, and those that showed a significant treatment by species interaction. Broad level (2) GO term functional classification revealed that the majority of genes that showed significant differential expression due to cold acclimation in each of the subsets were involved in metabolic and cellular processes. We also found that the proportion of genes involved in a particular biological process was similar for each of the subsets (Figure 4). When the metabolic GO term was split into its constitute parts (Figure 5, Supplementary Materials 4–8) we found the proportion of genes annotated with each metabolic GO term was similar for each of the subsets.
Figure 4.
Proportion of genes annotated for each of the level 2 GO terms identified for each of the gene subsets (D. montana, D. virilis, both species and interaction). Black dashed lines indicate proportion of genes annotated for each of the level 2 GO terms in the D. virilis genome.
Figure 5.
Proportion of genes annotated for each of the primary metabolic processes for each of the gene subsets (D. montana, D. virilis, both species and interaction). Black dashed lines indicate proportion of genes annotated for each of the primary metabolic processes in the D. virilis genome.
GO term enrichment analysis
The single GO terms oxidation-reduction process, and single-organism metabolic process were significantly enriched in D. montana (P=0.002 and P=0.016 respectively). No single GO terms were significantly enriched in D. virilis. By using the functional clustering program DAVID we were able to identify significantly enriched functional clusters in three of the four subsets (D. montana, D. virilis, and interaction (Table 1)) but none were identified for the genes DE in both species. Significant clusters involved in metabolism or producing metabolites were found in the D. montana, D. virilis, and interaction subsets (Table 1, yellow) as well as in transmembrane transport/ion transport in D. montana and D. virilis (Table 1, green). 14 (8%) of the genes DE in D. montana and 39 (9%) of the genes DE in D. virilis were annotated with the GO terms for transmembrane transport/ion transport, with 2 genes DE in both species and 9 showing a significant interaction (Supplementary Materials 9).
Table 1. Functional clusters identified by DAVID for each of the gene groups for enrichment scores with P<0.1.
| Cluster | Enrichment Score | P | Term summary |
|---|---|---|---|
| D.montana | |||
| 1 | 2.45 | 0.004 | actin cytoskeleton, acetylation, myosin complex, muscle protein |
| 2 | 1.97 | 0.010 | sarcomere, actin cytoskeleton, myofibril, actin filament-based process, muscle protein |
| 3 | 1.74 | 0.018 | ion transmembrane transporter activity, mitochondrial, generation of precursor metabolites and energy, cytochrome-c oxidase activity, oxidoreductase activity, oxidative phosphorylation, phosphate metabolic process, phosphorylation |
| 4 | 1.55 | 0.028 | alkaline phosphatase, folate biosynthesis |
| 5 | 1.45 | 0.035 | mitochondrion, transit peptide |
| 6 | 1.20 | 0.063 | organic acid biosynthetic process, cellular amino acid biosynthetic process, amine biosynthetic process |
| 7 | 1.11 | 0.078 | calcium-binding EF-hand, calcium, calcium ion binding |
| 8 | 1.11 | 0.078 | oxidoreductase activity, nitrogen compound biosynthetic process, heterocycle biosynthetic process |
| 9 | 1.01 | 0.098 | ion transmembrane transporter activity, mitochondrial, generation of precursor metabolites and energy, oxidative phosphorylation, ATPase activity, hydrolase activity, phosphorylation |
| D. virilis | |||
| 1 | 1.97 | 0.011 | integral to membrane, intrinsic to membrane, transmembrane, membrane |
| 2 | 1.61 | 0.025 | CHK kinase-like, CHK |
| 3 | 1.45 | 0.035 | identical protein binding, protein dimerization activity, protein homodimerization activity |
| 4 | 1.29 | 0.051 | metabolism of xenobiotics by cytochrome P449, glutathione transferase activity, transferase activity, glutathione S-transferase/chloride channel, glutathione metabolism, posttranslational modification, protein turnover, chaperones |
| 5 | 1.2 | 0.063 | PAS fold, PAS |
| 6 | 1.19 | 0.065 | odorant binding protein, hormone binding, JHBP |
| 7 | 1.18 | 0.066 | glutamate receptor-related, NMDA receptor, ion transport, passive transmembrane transporter activity, metal ion transmembrane transporter activity, cell membrane |
| 8 | 1.12 | 0.076 | pteridine and derivative biosynthetic process, aromatic compound biosynthetic process, pteridine and derivative metabolic process, heterocycle biosynthetic process |
| Interaction | |||
| 1 | 1.27 | 0.054 | carbohydrate catabolic process, alcohol catabolic process |
| 2 | 1.27 | 0.054 | hexose metabolic process, monosaccharide metabolic process, glucose metabolic process |
Cluster colour code: orange=muscle-related, yellow=metabolic, green=transmembrane transport/ion transport, pink=protein signalling. A full color version of this table is available at the Heredity journal online.
In addition we identified a number of enriched gene clusters which were different between the species. In D. montana, we found two gene clusters which showed significant enrichment for muscle protein (Table 1, orange), while in D. virilis there were several clusters significantly enriched for protein signalling (Table 1, pink). Although the above mentioned functional clusters were only significantly enriched in one species, these processes are likely to be a component of cold acclimation in both of the species as we also found genes annotated with these functions DE in both species (Supplementary Materials 10).
Discussion
While our understanding of the genetic basis of many traits has advanced greatly in many model systems, whether these results translate across species remains an open question. Numerous candidate gene studies have demonstrated that genes appear to be conserved in their function between taxa (Fitzpatrick et al., 2005; Reaume and Sokolowski, 2011; Martin and Orgogozo, 2013). Candidate gene studies however generally focus upon a subset of genes which have a large influence on a particular trait. By using an RNA-Seq approach we were able to examine genes which altered expression in response to cold acclimation independent of the strength of influence they have on cold acclimation in two closely related Drosophila virilis group species where the trait is likely to be homologous. This approach captures both the genes that have a major influence on cold acclimation but also genes of smaller influence and those involved in secondary changes, which may be more susceptible to evolutionary turnover. Using this approach enabled us to identify genes that show differential expression in response to cold acclimation in both or only one of the species and to identify the molecular pathways and processes involved. Our study found evidence for both conservation and divergence in gene expression in response to cold acclimation between D. montana and D. virilis.
We identified 42 genes that were differentially expressed in both species. These genes (with one exception) showed very similar changes in terms of fold change and direction of expression changes, and thus may represent a ‘core set' of genes, which appear to be evolutionary conserved in response to cold acclimation. We also identified a large number of genes that were DE in one species but not the other with around a quarter of these showing a significant species by treatment interaction. Gene expression changes in these genes were only weakly correlated with each other, with only one of the 132 genes which showed a significant interaction DE in both species. This suggests that a large proportion of the genes that were DE in response to cold acclimation were different between D. montana and D. virilis. We also observed that many more genes were DE in D. virilis than D. montana. One potential reason for this may be that D. virilis is less cold tolerant than D. montana (Kellermann et al., 2012; Vesala et al., 2012b) and thus the cold acclimation treatment may be more stressful for D. virilis than D. montana, promoting a stronger cold acclimation response. In support of this we also note that the average fold change in genes DE in both species was also slightly larger in D. virilis than D. montana (Figure 1b).
Our focus in this study was to look at temperature-induced changes in gene expression in D. montana and D. virilis. Therefore we chose the conditions for the control treatments to represent non-cold-acclimating ‘summer' conditions for each species. A complication is that identical long day conditions for both species may have led to D. montana flies entering reproductive diapause which would have complicated the results markedly as D. virilis does not have such photoperiodic diapause (Throckmorton, 1982). According to temperature data obtained for the collection site of D. montana flies used in this experiment (years 2000–2009, Oulanka research station, University of Oulu, Finland), the average temperature of summer months was: June: 12.2 °C, July: 15.5 °C and August 12.6 °C. Based on this data we chose 16 °C to represent non-acclimating (control) temperature for D. montana strain and 19 °C for more southern and less cold tolerant D. virilis. As such, the temperature difference between the control (non-acclimating) and the acclimation temperatures was slightly larger for D. virilis than D. montana (14 and 11 °C respectively) which could partly influence the larger number of genes DE in D. virilis compared to D. montana. However, given the relatively small difference between the two control non-acclimating temperatures compared to the large difference between the control and the acclimation temperatures, we expect the potential influence of this to be small.
As we were primarily interested in species-specific responses to cold acclimation we used isofemale lines for both species to minimise confounding intra-population genetic variation within species. However, as we have only one line per species we cannot distinguish intra- and inter-specific sources of genetic variation. Yet, as the species separated 9–11 mya (Reis et al., 2008; Morales-Hojas et al., 2011) it seems most likely that interspecific differences will dominate the main differences described here. Future population-level work is required to explore the extent of differences within species.
Functional processes
As we found that the genes DE due to cold acclimation were often different between the species, we examined the functional processes involved in four subsets: genes DE in D. montana, in D. virilis, in both species, and genes which showed a significant treatment by species interaction. We discuss the main functional processes found in the DE gene subsets below (for full list of processes for all the genes found to be DE see Supplementary Materials 11).
Metabolic profile
Cold acclimation is known to involve a shift in the metabolic profile as well as the production of cyroprotectants which act to maintain osmotic balance and stabilise the membrane structures of a cell (Lee, 1991; Lee, 2010). Consistent with this previous work, we found that the majority of genes DE in both species were metabolic in function. Splitting the metabolic GO term into its constitute parts showed that there were a similar proportion of genes involved in each of the metabolic processes for the studied subsets (Figure 4). The single GO-terms ‘oxidation-reduction processes' and ‘single-organism metabolic processes' were significantly enriched in D. montana. We did not find any significantly enriched single GO terms for D. virilis, though D. virilis showed a similar (though slightly lower) proportion of metabolic genes involved in the cold acclimation response. Examination of the enriched functional clusters identified clusters of genes involved in metabolism/production of metabolites for three of the subsets (D. montana, D. virilis, and the species by treatment interaction, Table 1). We did not find any significantly enriched functional clusters for genes DE in both species, likely due to the small number of genes in this group. Although we identified a number of metabolic gene clusters for three of the subsets, the metabolic pathways implicated for each set were different. This suggests that although both species show significant enrichment for metabolic changes, the exact metabolic pathways involved differ.
Cell membrane composition
Changes to the composition of phospholipids in the cell membrane are thought to be particularly important for cold acclimation, as it allows cells to maintain their physiological function in sub-optimal temperatures (Hazel, 1995; Koštál et al., 2003). Our previous work has shown the major metabolite produced in overwintering D. montana flies to be myo-inositol (Vesala et al., 2012a), which functions in many processes including regulation of cell development and growth (Loewus and Loewus, 1983). It also is a precursor of inositol phospholipids leading to production of inositol phosphates that function as second messengers (Downes and Macphee, 1990). Myo-inositol has not been found to play a role in cold acclimation in other Drosophila species, but it has been shown to accumulate seasonally in few other insect species (Block and Sømme, 1983; Koštál et al., 1996). It has also been correlated with an increase in cold tolerance in some Coleoptera and Lepidoptera species (Watanabe and Tanaka, 1999; Watanabe, 2002; Liu et al., 2009) and we hypothesise that its accumulation in overwintering D. montana would act as a cryoprotectant. In the present study the ortholog of inos (Dvir\GJ20549) was upregulated in both D. montana and D. virilis. This is interesting as inos encodes the enzyme myo-inositol-1-phosphate synthase, which is part of the inositol biosynthetic pathway (GO:0006021), catalysing the conversion of D-glucose-6-phosphate into 1L-myo-inositol-1-phosphate, the first committed step into production of all inositol compounds (Majumder et al., 1997). It is not known what the major metabolite produced in overwintering D. virilis flies is but given the DE of inos ortholog in response to cold acclimation we suggest that the production of 1L-myo-inositol-1-phosphate is likely to be important for both species. Another gene annotated in the inositol metabolic pathway, Dvir\GJ15346 (D. melanogaster ortholog: CG6910), showed a significant species by treatment interaction during acclimation. This gene, which was significantly down-regulated only in D. montana, is annotated with the GO term inositol catabolic process and has been indicated to have the same function as myo-inositol oxygenase, an enzyme that is the first committed step in inositol catabolism in eukaryotes (Jones et al., 2012). Thus the downregulation of this gene may lead to higher concentrations of inositol compounds, enabling D. montana to accumulate proportionally more inositol compounds, than D. virilis. D. virilis, on the other hand, may increase its cold tolerance with the aid of sphingolipid and sterol compounds, which have previously been shown to act together to alter cell membrane fluidity (Guan et al., 2009), as genes annotated for the metabolism of these compounds were found to be DE in D. virilis but not in D. montana (Supplementary Materials 5).
An additional group of genes that may influence cell membrane composition are Niemann-Pick type C (npc) genes. In D. melanogaster npc genes have been shown to be involved in the homeostatic regulation of cholesterol, which influences the permeability and fluidity of cell membranes (Huang et al., 2007; Niwa and Niwa, 2011). We found that npc2e was DE in both species, though it was upregulated in D. montana and downregulated in D. virilis (interaction P-value<0.001). In addition, npc1b and npc2d were DE in D. virilis but not in D. montana (interaction P-values for both genes<0.001). This suggests that the changes in the regulation of cholesterol are important for cold acclimation in both species, though the genetic basis used to produce the change is different in each of the species.
Ion transport/transmembrane transport
Changes in temperature are known to affect the transport mechanisms involved in the maintenance of cellular ion balance (Heitler et al., 1977; Kivivuori et al., 1990). Failure to maintain the ionic balance of cells can lead to metabolic perturbations which can cause a wide range of negative consequences, in particular, the loss of nerve excitation (Hochachka, 1986; Kostal et al., 2004). We found enrichment of several functional gene clusters involved in ion transport/transmembrane transport in both species (Table 1) which suggests that changes in gene expression of these genes are acting to maintain cellular ion concentration at low temperatures. Although both species showed enrichment for ion transport/transmembrane transport the genes responsible were largely different, with only two genes shared between the species and nine showing a significant interaction (Supplementary materials 9).
Rhodopsin
The ortholog of a key gene in the rhodopsin bio-synthesis pathway santa-maria, Dvir\GJ17608, was up-regulated in both species in response to cold acclimation. Rhodopsin has long been known to be the primary pigment for phototransduction (see Katz and Minke (2009) for a review), however recently it has been shown that the rhodopsin signalling pathway also has a several additional light-independent roles including hearing (Senthilan et al., 2012) and thermosensory signalling (Shen et al., 2011). Shen et al. (2011) showed that by knocking out santa-maria in D. melanogaster, flies were unable to discriminate between differences in temperature. Our finding that Dvir\GJ17608 is upregulated in both species is intriguing as it suggests the rhodopsin pathway may act to detect changes in temperature and thus may cue the cold acclimation response. None of the other genes involved in the rhodopsin bio-synthesis pathway were found to be DE in either of the species suggesting that santa-maria may be the key gene involved in the detection of temperature change. We did, however, find that an ortholog of CdsA (Dvir\GJ13151), a gene involved in the regulation of lipid storage, was DE in D. virilis but not D. montana. CdsA also belongs to a rhodopsin mediated signalling pathway and thus could play a role in linking rhodopsin signalling and downstream metabolic changes.
Circadian clock
Consistent with previous candidate gene approaches in D. montana and D. virilis (Vesala et al., 2012b) we found that two of the core circadian clock genes, period and vrille, were DE in both species. These genes have a well-studied role in the regulation and maintenance of circadian rhythms in insects (Bell-Pedersen et al., 2005). In addition, we also found four other peripheral clock genes to be DE in either D. montana or D. virilis including one which showed a significant species by treatment interaction (Table 2). The circadian clock has been previously implicated in cold acclimation in several taxa (Fowler and Thomashow, 2002; Magnone et al., 2005; Vesala et al., 2012b). It is not clear however if the changes in these genes have a direct influence on the increase in cold tolerance or if changes of the expression levels of these genes affect changes to the circadian rhythms of the fly in anticipation of the change in seasonal photoperiod. Further functional genetic studies will allow disentanglement of these hypothesis but given extensive work showing how the clock genes have a large influence on the regulation of metabolic processes in Drosophila (Xu et al., 2008; Xu et al., 2011; Sahar and Sassone-Corsi, 2012) we suggest that clock genes are good candidates for orchestrating the shifts in metabolic profile seen during cold acclimation.
Table 2. Circadian genes differentially expressed in response to cold acclimation.
| Gene | D. melanogaster ortholog | Species | Interaction? |
|---|---|---|---|
| Dvir\per | per | Both species | |
| Dvir\GJ17539 | vri | Both species | |
| Dvir\GJ11819 | dysc | D. virilis | |
| Dvir\GJ16719 | CG2650 | D. virilis | |
| Dvir\GJ19065 | na | D. virilis | Yes |
| Dvir\GJ11425 | Pdp1 | D. montana |
Differences between the species
Despite the overall pattern of broadly similar processes occurring in both species we also identified differences between the species. In D. montana we found significant enrichment for actin filaments/muscle protein, while in D. virilis we found significant enrichment for several clusters of genes involved in protein signalling/modification (Table 1). Changes in actin filaments /muscle protein have previously been implicated in insects to prevent cold injury by preventing the depolymerisation of the cell membrane during diapause (Kim et al., 2006; Robich et al., 2007; Koštál, 2010). Thus, these changes may help to maintain cellular physiology at sub-optimal temperatures. The four clusters of genes involved in protein signalling/modification indicate that these processes may also be important in cold acclimation. These differences seen between the species are unlikely to represent the absence of one process in one species and presence in another, but rather a greater or lesser relative importance in the context of the other processes occurring during cold acclimation for that species. For each of these clusters we also found several genes in the other species involved in the same biological process (Supplementary Materials 10) suggesting that these processes are likely to be a component of cold acclimation in both of the studied species.
Cold acclimation and cold shock
Previous work on cold tolerance in insects (particularly in D. melanogaster) has mostly focused upon the response to short-term cold shock rather than longer periods of cold acclimation (Colinet and Hoffmann, 2012). These processes, although related, are distinct and likely to produce differing transcriptional responses. This is important to consider when comparing our findings to previous work. Broadly, work on short-term cold shock gene expression changes has shown that the predominant group of genes showing expression changes are those involved in the stress and immune response (e.g. heat shock proteins (Hsps), Turandot genes, etc) (Zhang et al., 2011; Sinclair et al., 2013; Storey and Storey, 2013). In addition, the neuropeptide CAPA has been shown to play an important role in both cold and desiccation resistance in several species of Drosophila, including D. montana, when assessed using cold shock (Terhzaz et al., submitted). We do not find these genes or others involved in heat-shock response to be DE during cold acclimation. This is consistent with the findings by Colinet et al. (2013) who did not detect changes in heat shock protein levels after a 5-day cold acclimation in D. melanogaster. One explanation for this may be that the Hsps are important in the early part of thermal stress but are not involved in longer-term cold acclimation per se (Colinet and Hoffmann, 2012; Teets and Denlinger, 2013; but see Vesala et al. 2012b).
Functional modules
We were able to identify several functional processes that contribute to the cold acclimation response, the most important of which are likely to include: cold detection, circadian genes, and metabolic and cellular membrane profile shifts. For each of these functional processes we found genes that are conserved between the species and others that have diverged. We suggest that the genes that were found to be DE in both species are likely to represent key genes with a large influence on the cold acclimation phenotype. Further functional genetic studies are needed to confirm this, although it should be noted that several of the genes DE in both species have previously been demonstrated to have a large influence on processes implicated in cold acclimation. For example, we found that orthologs of two of the major genes involved in the circadian clock (period and vrille) were DE in both species, whereas genes more peripheral to the clock were DE in one species but not in the other (Table 2). We also observed the ortholog of the gene inos, which is responsible for producing the main overwintering metabolite in D. montana, and thus presumably has a large influence on the cold acclimation, is DE in both species. Interestingly, we found santa-maria (Dvir\GJ17608) was DE in both species. As stated above, santa-maria has a large role in the rhodopsin bio-synthesis pathway (Shen et al., 2011). Although this pathway has been previously linked to the ability to discriminate between differences in temperature (Shen et al., 2011), it has not been implicated in the cold acclimation response. One possibility is that rhodopsin bio-synthesis pathway may act to detect the onset of cold and to subsequently cue the DE of genes which increase cold tolerance.
Conclusions
We examined the effect of cold acclimation on gene expression in two Drosophila virilis group species to determine what genes and pathways were involved and if these were the same for both species. We found that the transcriptional changes associated with cold acclimation were broadly different with only 42 genes DE in both species and 132 showing a significant species by treatment interaction. This suggests that when comparing homologous phenotypes the underlying genetic basis of the trait may differ. The genes that were DE in both species are likely to have been conserved in their role in cold acclimation since at least the time of the species split. These evolutionarily conserved genes are likely to have a large influence on cold acclimation, as the selective forces maintaining cold acclimation will be stronger on these genes, than on genes that have a smaller influence.
Although we found that many of the genes DE due to cold acclimation in each of the species were different, we also found that the biological processes they were involved in were broadly similar. This is consistent with the idea of evolutionary turnover co-opting functionally related genes of smaller influence in and out of the cold acclimation response. The result of such evolutionary turnover is that although a trait may be homologous and appear phenotypically similar between species, many of the genes underlying the trait can be different.
Data Archiving
All data has been deposited in NCBI's Gene Expression Omnibus (GEO). Accession ID: GSE58925 Databank URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE58925
Acknowledgments
We thank Mikko Merisalo for technical assistance, Tiina S. Salminen for help with sample preparation, and three anonymous reviewers for valuable comments. This work was financially support by a Marie Curie Initial Training Network grant, “Understanding the evolutionary origin of biological diversity” (ITN-2008–213780 SPECIATION), grants from the Academy of Finland to A.H. (project 132619) and M.K. (projects 268214 and 272927), a grant from NERC, UK to M.G.R. (grant NE/J020818/1), and NERC, UK PhD studentship to D.J.P. (NE/I528634/1).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Anders S, Pyl PT, Huber W. (2014). HTSeq–A Python framework to work with high-throughput sequencing data. bioRxiv 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuthnott D. (2009). The genetic architecture of insect courtship behavior and premating isolation. Heredity 103: 15–22. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL et al. (2005). Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet 6: 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate-a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57: 289–300. [Google Scholar]
- Block W, Sømme L. (1983). Low temperature adaptations in beetles from the sub-Antarctic island of South Georgia. Polar Biol 2: 109–114. [Google Scholar]
- Canestro C, Yokoi H, Postlethwait JH. (2007). Evolutionary developmental biology and genomics. Nat Rev Genet 8: 932–942. [DOI] [PubMed] [Google Scholar]
- Colinet H, Hoffmann AA. (2012). Comparing phenotypic effects and molecular correlates of developmental, gradual and rapid cold acclimation responses in Drosophila melanogaster. Funct Ecol 26: 84–93. [Google Scholar]
- Colinet H, Overgaard J, Com E, Sorensen JG. (2013). Proteomic profiling of thermal acclimation in Drosophila melanogaster. Insect Biochem Mol Biol 43: 352–365. [DOI] [PubMed] [Google Scholar]
- David R, Gibert P, Pla E, Petavy G, Karan D, Moreteau B et al. (1998). Cold stress tolerance in Drosophila: analysis of chill coma recovery in D. melanogaster. J Therm Biol 23: 291–299. [Google Scholar]
- Downes CP, Macphee CH. (1990). myo-Inositol metabolites as cellular signals. Eur J Biochem 193: 1–18. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick MJ, Ben-Shahar Y, Smid HM, Vet LEM, Robinson GE, Sokolowski MB et al. (2005). Candidate genes for behavioural ecology. Trends Ecol Evol 20: 96–104. [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XL, Souza CM, Pichler H, Dewhurst G, Schaad O, Kajiwara K et al. (2009). Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell 20: 2083–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel JR. (1995). Thermal adaptation in biological membranes-is homeoviscous adaptation the explanation? Annu Rev Physiol 57: 19–42. [DOI] [PubMed] [Google Scholar]
- Heitler WJ, Goodman CS, Fraserrowell CH. (1977). Effects of temperature on threshold of identified neurons in locust. J Comp Physiol 117: 163–182. [Google Scholar]
- Hochachka PW. (1986). Defense strategies against hypoxia and hypothermia. Science 231: 234–241. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Sorensen JG, Loeschcke V. (2003). Adaptation of Drosophila to temperature extremes: bringing together quantitative and molecular approaches. J Therm Biol 28: 175–216. [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. (2009. a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. (2009. b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang W, Richards S, Carbone MA, Zhu DH, Anholt RRH, Ayroles JF et al. (2012). Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci USA 109: 15553–15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Warren JT, Buchanan J, Gilbert LI, Scott MP. (2007). Drosophila Niemann-Pick Type C-2 genes control sterol homeostasis and steroid biosynthesis: a model of human neurodegenerative disease. Development 134: 3733–3742. [DOI] [PubMed] [Google Scholar]
- Jones MK, Eldon ED, Klig LS. (2012). myo-Inositol oxygenase identified in Drosophila melanogaster. Faseb J 26: 759. [Google Scholar]
- Katz B, Minke B. (2009). Drosophila photoreceptors and signaling mechanisms. Front Cell Neurosci 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann V, Loeschcke V, Hoffmann AA, Kristensen TN, Flojgaard C, David JR et al. (2012). Phylogenetic constraints in key functional traits behind species' climate niches: patterns of desiccation and cold resistance across 95 Drosophila species. Evolution 66: 3377–3389. [DOI] [PubMed] [Google Scholar]
- Kim M, Robich RM, Rinehart JP, Denlinger DL. (2006). Upregulation of two actin genes and redistribution of actin during diapause and cold stress in the northern house mosquito. Culex pipiens. J Insect Physiol 52: 1226–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivivuori L, Lehti S, Lagerspetz KYH. (1990). Effect of temperature-acclimation on thermal-dependence and hysteresis of the resting membrane-potential of the stretch-receptor neuron in crayfish Astacus astacus (L). J Therm Biol 15: 9–14. [Google Scholar]
- Koštál V. (2010). Cell structural modifications in insects at low temperatures. In: Denlinger DL, Lee RE (eds). Low Temperature Biology of Insects. Cambridge University Press: Cambridge, UK, pp 3–35. [Google Scholar]
- Koštál V, Nedved O, Simek P. (1996). Accumulation of high concentrations of myo-inositol in the overwintering ladybird beetle Ceratomegilla undecimnotata. Cryo Letters 17: 267–272. [Google Scholar]
- Kostal V, Vambera J, Bastl J. (2004). On the nature of pre-freeze mortality in insects: water balance, ion homeostasis and energy charge in the adults of Pyrrhocoris apterus. J Exp Biol 207: 1509–1521. [DOI] [PubMed] [Google Scholar]
- Koštál Vr, Berková P, Šimek P. (2003). Remodelling of membrane phospholipids during transition to diapause and cold-acclimation in the larvae of Chymomyza costata (Drosophilidae). Comp Biochem Physiol B-Biochem Mol Biol 135: 407–419. [DOI] [PubMed] [Google Scholar]
- Lee RE. (1991). Principles of insect low temperature tolerance. In: Lee RE, Denlinger DL (eds). Insects at Low Temperature. Chapman and Hall: London, UK, pp 17–46. [Google Scholar]
- Lee RE. (2010). A primer on insect cold-tolerance. In:Denlinger DL, Lee RE (eds). Low Temperature Biology of Insects. Cambridge University Press: Cambridge, UK, pp 3–35. [Google Scholar]
- Liu ZD, Gong PY, Heckel DG, Wei W, Sun JH, Li DM et al. (2009). Effects of larval host plants on over-wintering physiological dynamics and survival of the cotton bollworm, Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae). J Insect Physiol 55: 1–9. [DOI] [PubMed] [Google Scholar]
- Loewus FA, Loewus MW. (1983). myo-Inositol: its biosynthesis and metabolism. Annu Rev Plant Physiol 34: 137–162. [Google Scholar]
- MacMillan HA, Sinclair BJ. (2011). Mechanisms underlying insect chill-coma. J Insect Physiol 57: 12–20. [DOI] [PubMed] [Google Scholar]
- Magnone MC, Jacobmeier B, Bertolucci C, Foa A, Albrecht U. (2005). Circadian expression of the clock gene per2 is altered in the ruin lizard (Podarcis sicula when temperature changes. Mol Brain Res 133: 281–285. [DOI] [PubMed] [Google Scholar]
- Majumder AL, Johnson MD, Henry SA. (1997). 1L-myo-inositol-1-phosphate synthase. Biochim Biophys Acta-Lipids Lipid Metab 1348: 245–256. [DOI] [PubMed] [Google Scholar]
- Martin A, Orgogozo V. (2013). The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67: 1235–1250. [DOI] [PubMed] [Google Scholar]
- McCune AR, Schimenti JC. (2012). Using genetic networks and homology to understand the evolution of phenotypic traits. Curr Genomics 13: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Hojas R, Reis M, Vieira CP, Vieira J. (2011). Resolving the phylogenetic relationships and evolutionary history of the Drosophila virilis group using multilocus data. Mol Phylogenet Evol 60: 249–258. [DOI] [PubMed] [Google Scholar]
- Niwa R, Niwa YS. (2011). The fruit fly Drosophila melanogaster as a model system to study cholesterol metabolism and homeostasis. Cholesterol 2011: 176802–176802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard J, Tomčala A, Sørensen JG, Holmstrup M, Krogh PH, Šimek P et al. (2008). Effects of acclimation temperature on thermal tolerance and membrane phospholipid composition in the fruit fly Drosophila melanogaster. J Insect Physiol 54: 619–629. [DOI] [PubMed] [Google Scholar]
- R Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. URL http://www.r-project.org/. [Google Scholar]
- Reaume CJ, Sokolowski MB. (2011). Conservation of gene function in behaviour. Phil Trans R Soc B 366: 2100–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis M, Vieira CP, Morales-Hojas R, Vieira J. (2008). An old bilbo-like non-LTR retroelement insertion provides insight into the relationship of species of the virilis group. Gene 425: 48–55. [DOI] [PubMed] [Google Scholar]
- Robich RM, Rinehart JP, Kitchen LJ, Denlinger DL. (2007). Diapause-specific gene expression in the northern house mosquito, Culex pipiens L., identified by suppressive subtractive hybridization. J Insect Physiol 53: 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV. (2012). The QTN program and the alleles that matter for evolution: all that's gold does not glitter. Evolution 66: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. (2012). Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol Metab 23: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt RW. (1961). Principles of insect cold hardiness. Annu Rev Entomol 6: 55–74. [Google Scholar]
- Sasson A, Michael TP. (2010). Filtering error from SOLiD Output. Bioinformatics 26: 849–850. [DOI] [PubMed] [Google Scholar]
- Senthilan PR, Piepenbrock D, Ovezmyradov G, Nadrowski B, Bechstedt S, Pauls S et al. (2012). Drosophila auditory organ genes and genetic hearing defects. Cell 150: 1042–1054. [DOI] [PubMed] [Google Scholar]
- Shen WL, Kwon Y, Adegbola AA, Luo J, Chess A, Montell C et al. (2011). Function of rhodopsin in temperature discrimination in Drosophila. Science 331: 1333–1336. [DOI] [PubMed] [Google Scholar]
- Sinclair BJ, Ferguson LV, Salehipour-shirazi G, MacMillan HA. (2013). Cross-tolerance and cross-talk in the cold: relating low temperatures to desiccation and immune stress in insects. Integr Comp Biol 53: 545–556. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM. (2013). Molecular biology of freezing tolerance. Compr Physiol 3: 1283–1308. [DOI] [PubMed] [Google Scholar]
- Teets NM, Denlinger DL. (2013). Physiological mechanisms of seasonal and rapid cold-hardening in insects. Physiol Entomol 38: 105–116. [Google Scholar]
- Terhzaz S, Cabrero P, Teets NM, Henderson L, Ritchie MG, Nachman RJ et al, submitted. Neuroendocrine control of desiccation and cold stress responses.
- Throckmorton LH. (1982). The virilis species group. In: Ashburner M, Carson HL, Thompson JN (eds). Genetics and Biology of Drosophila. Vol. 3b. Academic Press: London, UK, pp 227–296. [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S et al. (2009). FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res 37: D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesala L, Salminen TS, Kostal V, Zahradnickova H, Hoikkala A. (2012. a). myo-inositol as a main metabolite in overwintering flies: seasonal metabolomic profiles and cold stress tolerance in a northern drosophilid fly. J Exp Biol 215: 2891–2897. [DOI] [PubMed] [Google Scholar]
- Vesala L, Salminen TS, Laiho A, Hoikkala A, Kankare M. (2012. b). Cold tolerance and cold-induced modulation of gene expression in two Drosophila virilis group species with different distributions. Insect Mol Biol 21: 107–118. [DOI] [PubMed] [Google Scholar]
- Watanabe M. (2002). Cold tolerance and myo-inositol accumulation in overwintering adults of a lady beetle Harmonia axyridis (Coleoptera: Coccinellidae). Eur J Entomol 99: 5–9. [Google Scholar]
- Watanabe M, Tanaka K. (1999). Seasonal change of the thermal response in relation to myo-inositol metabolism in adults of Aulacophora nigripennis (Coleoptera Chrysomelidae). J Insect Physiol 45: 167–172. [DOI] [PubMed] [Google Scholar]
- Xu K, Zheng X, Sehgal A. (2008). Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab 8: 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu KY, DiAngelo JR, Hughes ME, Hogenesch JB, Sehgal A. (2011). The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab 13: 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Marshall KE, Westwood JT, Clark MS, Sinclair BJ. (2011). Divergent transcriptomic responses to repeated and single cold exposures in Drosophila melanogaster. J Exp Biol 214: 4021–4029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.