Individual resource specialisation is common in natural populations associated with competition and ecological opportunity (see Aroujo et al., 2011), and well known for the killer whale (where social groups specialise) and other delphinid cetaceans (see Hoelzel, 2002). Prey choice affects a predator's temporal and spatial pattern of habitat use. For the killer whale, social groups (pods) learn where prey resources are seasonally abundant, and the techniques required to exploit different resources efficiently. Some fish prey, especially anadromous species such as salmon, may provide predictable seasonally rich concentrations, whereas marine mammal prey may be more patchily distributed and show a different pattern of temporal abundance (and accessibility). However, these resources are found within the same waters, though the timing and technique for capture may differ. Foote and Morin (2015) suggest that the co-occurrence of populations in the same ocean doesn't necessarily imply that they differentiated in sympatry, which is clearly true. However, as Moura et al. (2015) and others (for example, Hoelzel et al., 1998, 2007) have discussed, it is the life history and behaviour of killer whales that suggest the potential for differentiation in sympatry. Although the proximity of resources brings killer whale pods into sympatry, the differential pattern of spatial and temporal habitat use, as well as fidelity to pods that forage by similar learned methods, could serve to isolate resource specialist communities reproductively.
It is conceivable that marine mammal and fish prey resources were more geographically isolated in the North Pacific during some relevant period in the past, but there are no data to indicate that this was the case. Foote and Morin (2015) propose that differentiation in sympatry is exceptional, restricted to ‘geographically isolated ‘island' populations, such as found in small crater lakes or on small remote oceanic islands'. They are not alone in this view, but alternative interpretations are well established (see review in Via (2001)), and indeed some of the most thoroughly investigated putative examples of sympatric speciation do not fit the scenarios proposed by Foote and Morin (2015), such as habitat-shift systems (for example, Filchak et al., 2000). Although Foote and Morin (2015) contend that killer whale ecotypes are too differentiated to represent even ‘sister taxa or a monophyletic endemic species flock' and therefore rule out speciation in sympatry, we point out that levels of differentiation are comparatively low, and propose that if speciation is relevant for killer whales it is at a very early stage (see Moura et al., 2014a, 2014b). However, Moura et al. (2014a) using restriction-associated DNA (RAD) sequencing to identify 3281 single-nucleotide polymorphisms (SNPs) compared differentiation at neutral loci and those putatively under directional selection based on outlier analysis and fixed differences, and found stronger differentiation at linked loci with relevant functions (associated with digestion, growth etc.). This is consistent with disruptive selection between ecotypes, thought to be an important mechanism in the process of sympatric speciation (see Via, 2001).
Foote and Morin (2015), following inference from Foote et al. (2011), propose that the demographic history indicated by mitochondrial DNA (mtDNA) phylogenies reflects the true history, and that it implies re-convergence of resource specialist populations that differentiated in allopatry. However, it is well-established (for example, see Shaw, 2002) that single gene trees can be misleading based on stochastic factors or introgression. Stochastic effects are especially likely for a tree that is as shallow for global diversity as that seen for the killer whale. Although some resolution by population was possible based on whole mitogenomes, the base of that tree represented only ~0.5% sequence divergence (Morin et al., 2010), and sampled individuals as geographically distant as either side of the North Pacific differed by just 1 out of 16 388 bp. Phylogenies based on less sequence data were mostly polytomous (for example, the control region phylogeny in Hoelzel et al. (2002)).
Foote et al. (2011), following inference from Morin et al. (2010), suggest that separation of the ‘transient' (North Pacific mammal predatory ecotype) and ‘resident' (North Pacific piscivorous ecotype) lineages (deepest node in the mitogenome tree) dates to ~750 Ka. However, this is based on a mutation rate estimate of 0.3% per million years, which is an order of magnitude slower than other estimates for cetacean mitogenomes (see Ho and Lanfear, 2010). At the higher mutation rate estimate (3% per million years), the timing of division becomes very recent (sometime during the last glaciation), consistent with other estimates for the timing of a bottleneck and subsequent expansion based on both nuclear and mtDNA (Hoelzel et al., 2002, 2007; Moura et al., 2014b). The timing then becomes very shallow for the proposition by Foote et al. (2011) that whales left the North Pacific, differentiated in isolation (to a species-level difference according to Morin et al. (2010)), and then returned, followed by further subdivision and then sufficient introgression to distort the nuclear DNA phylogeny (which under this scenario doesn't show up in the mtDNA lineages).
We tested the idea proposed by Foote and Morin (2015) that our nuclear DNA phylogeny presented in Moura et al. (2015) could be consistent with the mitogenome phylogeny if there was secondary contact between transients and ‘offshores' in the North Pacific. We had already shown that Marion Island (~2000 km south of Africa, representing Southern Ocean populations) had a higher likelihood value for being ancestral than transients based on allele frequency spectrum analysis of 2934 neutral SNPs (Moura et al., 2014a). That study also showed that the Marion Island population was most distinct from all other populations based on FCA analysis (except at loci under selection, for which the marine-mammal-eating population at Marion Island grouped with the transients). Here we test different historical scenarios using neutral SNP loci and ABC analyses (Figure 1; based on 1000 neutral SNPs from Moura et al. (2014a)), including one scenario where after a period of divergence the transient and offshore lineages are allowed to exchange gene flow.
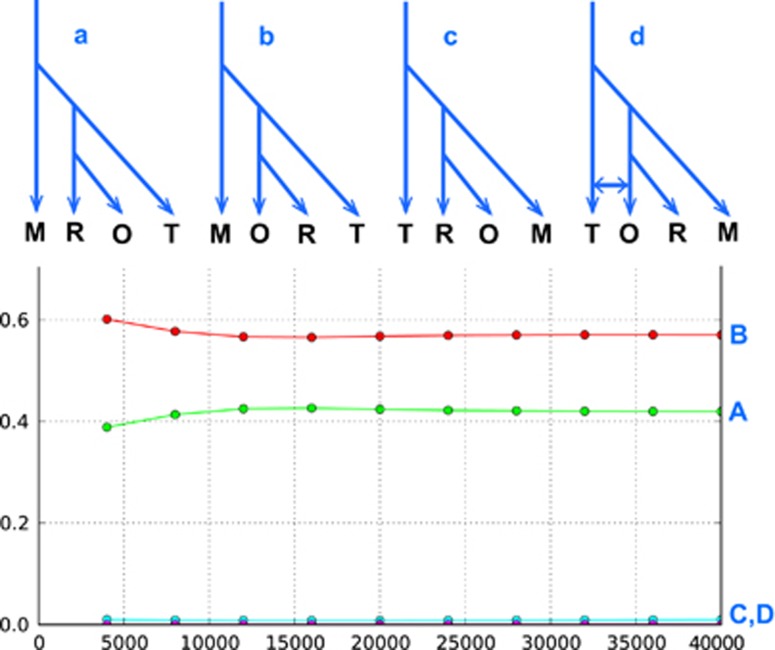
Figure 1.
Scenarios simulated using the DIY-ABC software, version 2.0.4 (Cornuet et al., 2014) and relative fit to the observed data. a–d illustrate the simulated scenarios along the top (M= Marion Island, R= residents, O= offshores, T= transients), while the relative proportions of each scenario found in the selected closest data sets are plotted below. The logistic regression is shown, but the direct estimate (not shown) gave similar results with a and b higher than c and d. At 40 000 data sets, scenario A is 0.4196 (95% confidence interval: 0.4053–0.4340), B is 0.5705 (0.5562–0.5848), C is 0.0097 (0.0000–0.0199) and D is 0.0002 (0.0000–0.0106). Branching order was coded by constraining the relative timings between split events, but giving wide priors (10–10 000) for the absolute timings. As estimating absolute splitting times was not the purpose of our simulations, this approach minimises bias resulting from uncertainty in splitting times. We coded different demographic scenarios for each split by including a change of Ne for the derived lineage (e.g., in scenario A, offshore separate from residents, while in scenario B, residents separate from offshores, even though the timing of the split is the same). Wide priors (10–10 000) were used for all Ne values again to minimise bias. Summary statistics associated with genic diversities, FST distances and Nei's genetic distance were included for all possible population combinations. 1 000 000 data sets were simulated for each scenario, and comparative assessment based on the 40 000 most similar simulations. Scenarios A and B reflect inferences obtained from the nuclear phylogenetic tree; scenario C represented inference from the mitogenome tree only; scenario D represents the mitogenomic tree, but allowing for gene flow (again with wide priors: 0.01–0.99) between offshores and transients after secondary contact, as proposed by Foote and Morin (2015). Simulations were generated based on a sample of 1000 SNPs from Moura et al. (2014a) and repeated twice for different sets of SNP's to check for consistency (no difference found and so only one version shown).
For simplicity (and to some extent by necessity), we compared models including only the populations directly relevant to this discussion. This is a limitation, and means that the results are preliminary. However, intermediate lineages (such as those in the North Atlantic) and further complexity in the North Pacific and Southern Oceans would have the same representation in each of the four models, and therefore their omission may not represent a bias. We also allow for free association (very wide priors for migration rate) between transients and offshores after secondary contact, giving full potential to the connectivity illustrated in Figure 1d of Foote and Morin (2015), and as may be suggested by overlapping clusters in the PCA plot based on 16 microsatellite DNA loci in Pilot et al. (2010); though not as clearly from the kinship analyses in that paper. However, similar overlap between transients and offshores is not seen for FCA plots based on 2934 neutral SNPs in Moura et al. (2014a), and Hoelzel et al. (2007) found roughly equivalent levels of gene flow when comparing all North Pacific ecotypes for both long-term and post-division migration estimates (always close to one migrant per generation).
As seen in Figure 1, the best supported scenarios reflect the inference from our nuclear DNA tree in Moura et al. (2015) where the Southern Ocean population is ancestral, with the North Pacific lineages separating more recently. The least well-supported scenarios approximate the mitogenome tree and the one proposed to explain the discrepancy between the nuclear and mtDNA trees by Foote and Morin (2015); where the transient population is ancestral and there is secondary contact in the North Pacific. To partly address some of the limitations of our approach, we compared two scenarios in a separate analysis. The first scenario was as illustrated in Foote and Morin (2015) Figure 1d (with Iceland representing the North Atlantic and Marion Island the Southern Oceans). The second scenario retained migration between offshores and transients (modelled as for scenario 1), but switched places for the transients and Marion Island (so that Marion Island became ancestral). The strongest support was for scenario 2 (using the logistic regression, 0.847; 95% confidence interval (CI): 0.840–0.854) compared with scenario 1 (0.153, 95% CI: 0.146–0.161; see Figure 1 caption for details of method), again consistent with our nuclear tree. Our approach for this two scenario comparison was to make everything the same except for the ancestry, so that this aspect in particular could be assessed. More complex analyses may provide different inference, but it will be difficult to control for all relevant factors in a complex model.
In Moura et al. (2015) we propose that a phylogeny based on 1.7 m nuclear bp representing many genes was likely to produce a more reliable topology than the shallow single gene tree represented by mtDNA. We furthermore believe that the evidence is strong for the genetic differentiation of resource specialists exhibiting differential spatial and temporal habitat use in sympatry, both in the North Pacific and elsewhere in the world (where no allopatric mechanisms similar to that suggested by Foote et al. (2011) for the North Pacific have been proposed). We further suggest that the relatively high diversity found in Southern Ocean populations (especially off South Africa; Moura et al., 2014b) is consistent with Southern Ocean ancestry, though it is true that neither the earlier mtDNA phylogenies nor our nuclear phylogeny are fully inclusive of killer whale lineages world-wide, and further inclusion could reveal further refinement. Given the strong signal for a population bottleneck in the northern hemisphere (Hoelzel et al., 2002; Moura et al., 2014b), the current pattern of mtDNA variation may reflect lineage sorting during post-bottleneck population expansion. Considering the alternative proposed by Foote and Morin (2015), we continue to believe that their ‘out of the Pacific and back again' scenario based on a single gene tree is the less parsimonious interpretation of the available data.
The authors declare no conflict of interest.
References
- Araujo MS, Bolnick DI, Layman CA. (2011). The ecological causes of individual specialisation. Ecol Lett 14: 948–958. [DOI] [PubMed] [Google Scholar]
- Cornuet JM, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R et al. (2014). DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30: 1187–1189. [DOI] [PubMed] [Google Scholar]
- Filchak KE, Roethele JB, Feder JL. (2000). Natural selection and sympatric divergence in the apple maggot Rhagoletis pomonella. Nature 407: 739–742. [DOI] [PubMed] [Google Scholar]
- Foote AD, Morin PA. (2015). Sympatric speciation in killer whales? Heredity 114: 357–338 doi:10.1038/hdy.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, Morin PA, Durban JW, Willerslev E, Orlando L, Gilbert MTP. (2011). Out of the Pacific and back again: insights into the matrilineal history of Pacific killer whale ecotypes. PLoS One 6: e24980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SY, Lanfear R. (2010). Improved characterisation of among-lineage rate variation in cetacean mitogenomes using codon-partitioned relaxed clocks. Mitochondrial DNA 21: 138–146. [DOI] [PubMed] [Google Scholar]
- Hoelzel AR. (2002) Resource specialization and the evolution of population genetic structure in delphinid species In: Cell and Molecular Biology of Marine MammalsPfeiffer CJ (ed). Krieger Publishing:: NY, USA. [Google Scholar]
- Hoelzel AR, Dahlheim M, Stern SJ. (1998). Low genetic variation among killer whales (Orcinus orca in the eastern North Pacific, and genetic differentiation between foraging specialists. J Hered 89: 121–128. [DOI] [PubMed] [Google Scholar]
- Hoelzel AR, Natoli A, Dahlheim M, Olavarria C, Baird RW, Black N. (2002). Low world-wide genetic diversity in the killer whale (Orcinus orca; implications for demographic history. Proc Royal Soc B 269: 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzel AR, Hey J, Dahlheim ME, Nicholson C, Burkanov V, Black N. (2007). Evolution of population structure in a highly social top predator, the Killer Whale. Mol Biol Evol 24: 1407–1415. [DOI] [PubMed] [Google Scholar]
- Morin PA, Archer FI, Foote AD, Vilstrup J, Allen EE, Wade P et al. (2010). Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca indicates multiple species. Genome Res 20: 908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura AE, Kenny JG, Chaudhuri R, Hughes MA, Welch A, Reisinger RR et al. (2014. a). Population genomics of the killer whale indicates ecotype evolution in sympatry involving both selection and drift. Mol Ecol 23: 5179–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura AE, Janse van Rensburg C, Pilot M, Tehrani A, Best PB, Thornton M et al. (2014. b). Killer whale nuclear genome and mtDNA reveal widespread population bottleneck during the last glacial maximum. Mol Biol Evol 31: 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura AE, Kenny JG, Chaudhuri R, Hughes MA, Reisinger RR, de Bruyn PJN et al. (2015). Phylogenomics of the killer whale indicates ecotype divergence in sympatry. Heredity 114: 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot M, Dahlheim ME, Hoelzel AR. (2010). Social cohesion among kin, gene flow without dispersal and the evolution of population genetic structure in the killer whale (Orcinus orca. J Evol Biol 23: 20–31. [DOI] [PubMed] [Google Scholar]
- Shaw KL. (2002). Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: What mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. PNAS 99: 16122–16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S. (2001). Sympatric speciation in animals: the ugly duckling grows up. TREE 16: 381–390. [DOI] [PubMed] [Google Scholar]