Abstract
Transitions in sexual system and reproductive mode may affect the course of sex chromosome evolution, for instance by altering the strength of sexually antagonistic selection. However, there have been few studies of sex chromosomes in systems where such transitions have been documented. The European tadpole shrimp, Triops cancriformis, has undergone a transition from dioecy to androdioecy (a sexual system where hermaphrodites and males coexist), offering an excellent opportunity to test the impact of this transition on the evolution of sex chromosomes. To identify sex-linked markers, to understand mechanisms of sex determination and to investigate differences between sexual systems, we carried out a genome-wide association study using restriction site-associated DNA sequencing (RAD-seq) of 47 males, females and hermaphrodites from one dioecious and one androdioecious population. We analysed 22.9 Gb of paired-end sequences and identified and scored >3000 high coverage novel genomic RAD markers. Presence–absence of markers, single-nucleotide polymorphism association and read depth identified 52 candidate sex-linked markers. We show that sex is genetically determined in T. cancriformis, with a ZW system conserved across dioecious and androdioecious populations and that hermaphrodites have likely evolved from females. We also show that the structure of the sex chromosomes differs strikingly, with a larger sex-linked region in the dioecious population compared with the androdioecious population.
Introduction
Sex chromosomes have evolved independently from autosomes numerous times in plants and animals and represent an extraordinary case of evolutionary convergence with shared features indicating the action of similar evolutionary forces (Fridolfsson et al., 1998; Lahn and Page, 1999; Skaletsky et al., 2003; Graves, 2008; Pease and Hahn, 2012). The established model of sex chromosome evolution states that genetic sex determination by a dominant allele coupled with sexually antagonistic selection can lead to the evolution of non-recombining, morphologically differentiated sex chromosomes (Charlesworth et al., 2005; Bergero and Charlesworth, 2009; Bachtrog et al., 2011). Sexually antagonistic selection, where alleles may benefit one sex and harm the other, is widespread in groups with genetically determined sexes (for example, Rice, 1992; Foerster et al., 2007; Cox and Calsbeek, 2009) and is a critical creative force in the evolution of morphologically differentiated sex chromosomes (Charlesworth and Charlesworth, 1980; Rice, 1987a; van Doorn and Kirkpatrick, 2010; Jordan and Charlesworth, 2012; Qiu et al., 2013; Charlesworth et al., 2014; Kirkpatrick and Guerrero, 2014; Otto, 2014). Transitions in sexual system and reproductive mode can alter the strength of sexually antagonistic selection, and so may potentially affect the course of sex chromosome evolution. Studying the sex chromosomes of species that have undergone such transitions in sexual system is therefore important, however, species with labile sexual systems have been little studied (Pires-daSilva, 2007; Bachtrog et al., 2014).
Tadpole shrimps (Crustacea: Notostraca), although renowned for their morphological stability (Fryer, 1988; Mathers et al., 2013a), show diverse sexual systems including dioecy (gonochorism), hermaphroditism and androdioecy (AD; self-fertile hermaphrodites and males) (Sassaman, 1991). AD is a sexual system where hermaphrodites and a small proportion of males coexist within populations (Weeks et al., 2006a). Phylogenetic character mapping of sexual systems has shown that AD is likely to have evolved from dioecy independently multiple times (Mathers et al., 2013b). The European tadpole shrimp, Triops cancriformis, has undergone one such transition. In this species, populations in Spain are dioecious and obligate outcrossers, whereas in the rest of Europe there are either obligatory selfing hermaphroditic populations or androdioecious populations with hermaphrodites and a reduced proportion of males, where both selfing and outcrossing occurs (Sassaman, 1991; Zierold et al., 2007, 2009). Importantly, phylogeographic analysis has revealed that hermaphroditism (hermaphrodite and androdioecious) has evolved from dioecy (Zierold et al., 2007; Mathers et al., 2013b).
This transition makes T. cancriformis potentially an illuminating model, but, currently there is no information on sex determination. A ZW genetic sex determination system (with heterogametic females) is the most likely model as in two other androdioecious branchiopod species, Eulimnadia texana and Triops newberryi, maleness is recessive to hermaphroditism (Sassaman, 1989, 1991; Sassaman and Weeks, 1993; Otto et al., 1993). However, karyological analysis identified 2n=12 chromosomes, with no morphologically distinct sex chromosomes in T. cancriformis (Ombretta et al., 2005). In addition, selfing experiments on E. texana and T. newberryi indicate two types of hermaphrodites in androdioecious populations: amphigenics (ZW), which produce males and hermaphrodites in a 1:3 ratio, and monogenics (WW), which produce only hermaphrodites (Sassaman, 1991; Sassaman and Weeks, 1993).
Investigating biologically interesting questions in non-model species with few genomic resources has been greatly aided by developments in next-generation sequencing. Restriction site-associated DNA sequencing (RAD-seq) is a genotyping-by-sequencing approach that has proved particularly useful (Davey and Blaxter, 2010; Reitzel et al., 2013). RAD-seq can generate thousands of single-nucleotide polymorphism (SNP) markers in tens to hundreds of individuals (Baird et al., 2008; Hohenlohe et al., 2010), and it has been successfully used to identify sex-linked markers (Anderson et al., 2012; Carmichael et al., 2013; Palaiokostas et al., 2013; Gamble and Zarkower, 2014).
Here, using a genome-wide association approach, we explicitly test a ZW model of genetic sex determination in T. cancriformis and investigate the effect of a transition in sexual system on patterns of sex chromosome evolution. We used RAD-seq to score thousands of genetic RAD markers (‘markers') in males and females from a dioecious population and males and hermaphrodites from an androdioecious population of Triops cancriformis to identity sex-linked markers. We show that sex is genetically determined in T. cancriformis, with a ZW system conserved across dioecious and androdioecious populations, and a smaller sex-specific region in the androdioecious population.
Materials and methods
Samples, rearing and individual sexing
Sediment samples containing T. cancriformis diapausing cysts were obtained from a dioecious population in Espolla (ESP), Spain, and an androdioecious population in pond 12 in Königswartha (KOE), Germany (Zierold et al., 2007, 2009). Cysts were hatched by mixing sediment with purified water in small tanks kept at 20 °C under 24-h illumination, with resulting hatchlings reared on Triops food (www.triops.es). Mature individuals were fixed in 100% ethanol and sexed before genetic analysis with individuals without ovisacs considered male and those with ovisacs females (ESP) or hermaphrodites (KOE) (Mathers et al., 2013b).
Sample preparation and DNA extraction
Pre-extraction, the digestive tract of each individual was removed from the abdominal section and the gut cavity flushed with 100% ethanol to minimise contamination with exogenous DNA. Genomic DNA was extracted from ~1-cm long abdominal sections using the DNAeasy Blood and Tissue Kit (Qiagen, Venlo, Netherlands) including an RNase A digestion step. DNA quality was assessed on 0.8% agarose gels and quantified by fluorimetry (Quant-iT PicoGreen Kit, Invitrogen, Paisley, UK). In total, 14 males and 12 females from the dioecious population (ESP) and 6 males and 15 hermaphrodites from the androdioecious population (KOE), each yielding at least 18 ng μl−1 of high molecular weight DNA, were used in the RAD libraries.
RAD library preparation and sequencing
Four paired-end RAD libraries were prepared by the NERC/NBAF facility at the University of Edinburgh (The GenePool) following Baird et al. (2008) with some modifications (Ogden et al., 2013). For each individual, 1 μg total genomic DNA was digested using the restriction enzyme SbfI followed by ligation of a barcoded P1 RAD adapter (for details, see Supplementary Table S1). Samples were then split into four multiplexed libraries before sonic shearing, size selection (300–700 bp) and ligation of P2 RAD adapters. Libraries were PCR amplified, quantified and sequenced in separate flow cells on an Illumina (San Diego, CA, USA) HiSeq 2000 platform with 100 bp, paired-end chemistry (v1 chemistry). Two sequencing runs were carried out and the raw reads concatenated in silico for each library.
Sequence quality control and trimming
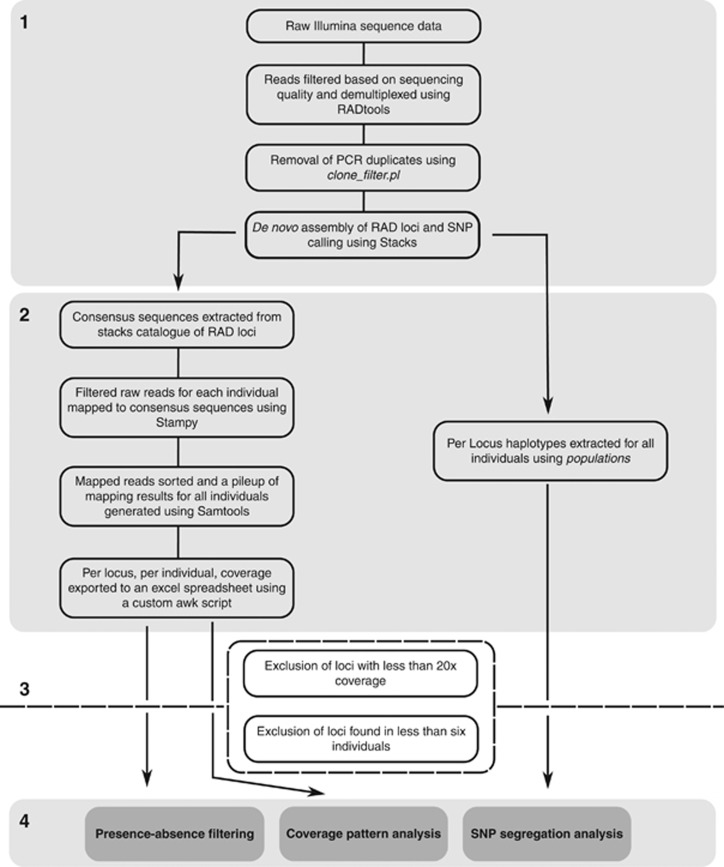
The pipeline used to process and analyse RAD-seq data is summarised in Figure 1. Sequencing quality of each RAD library was assessed with FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). We then used RADtools (Baxter et al., 2011) to trim reads to 70 bp and remove sequences lacking a correct barcode or SbfI restriction site, or with quality score <20 at any position. To account for sequencing errors, a fuzzy matching algorithm was used allowing 1- bp deviation from the expected restriction site and barcode sequences (Baxter et al., 2011). First- and second-end reads were then de-multiplexed based on their barcode sequence.
Figure 1.
Pipeline used to process the RAD-seq data.
De novo assembly of RAD markers and SNP calling
Genotyping-by-sequencing necessitates the grouping of sequences into homologous loci and allelic scoring at these loci. RAD-seq is a reduced representation method that generates sequences at either side of a restriction site (Baird et al., 2008), however, as there is no genomic reference for Triops cancriformis to map reads to, it was impossible to make this association. We therefore considered sequences with sufficient similarity (six mismatches or less) to be homologous, and we defined these groups of similar sequences as RAD markers. Variants within each RAD marker were defined as RAD alleles. We avoided the term ‘locus' as our RAD-seq approach provided a mixture of both presence–absence markers, likely caused by the presence–absence of a restriction site, and also allelic variation.
Assembly of RAD markers and SNP calling was performed using components of Stacks v. 0.99993 (Catchen et al., 2011). PCR duplicates were removed using clone_filter.pl. First-end reads for each individual were then clustered into candidate RAD markers using ustacks. At least 15 identical reads per individual were required to call an allele and up to six mismatches allowed between alleles of the same marker (we allowed six mismatches because our focus was the identification of sex-linked, and therefore potentially divergent, markers). SNPs were simultaneously called using the built in maximum likelihood diploid genotyping model (Hohenlohe et al., 2010). Candidate RAD markers with coverage two standard deviations above the average were removed as they likely were repetitive elements (Catchen et al., 2011, 2013). A catalogue of all RAD markers found across the sampled individuals was constructed using cstacks with markers considered to be homologous if they had six or fewer mismatches between individuals. Then, sstacks was used to identify genotypes for each RAD marker for every individual and the populations programme used to export genotypes for each marker of every individual and to calculate population summary statistics and population differentiation (FST).
To obtain coverage for each candidate RAD marker per individual, raw first-end reads from each individual were mapped back to consensus sequences from every RAD marker in the Stacks catalogue using Stampy (Lunter and Goodson, 2011) with default settings. SAMtools (Li et al., 2009) was then used to generate a pileup from which coverage information (per marker, per individual) was extracted.
Using this approach, we created a ‘filtered catalogue' containing high coverage RAD markers (>20 reads) that were found in six or more individuals (the number of males in KOE, the smallest gender sample) to be used in all downstream analyses. This filtered catalogue was used to test the expectations of marker presence–absence, RAD marker coverage and SNP segregation, predicted by a ZW sex determination model. As total per individual coverage varies in RAD libraries (Richards et al., 2013; Wagner et al., 2013), we normalised per marker coverage for each individual by dividing by the median coverage of the subset of RAD markers in the catalogue that were found in all 47 individuals. These were consider to be a large sample (800 markers, see Results section) of candidate autosomal markers.
Testing predictions for ZW chromosomal sex determination
Candidate completely sex-linked markers were first identified in females and males from ESP (dioecious) and hermaphrodites and males from KOE (androdioecious) based on patterns predicted by an explicit ZW model (Table 1). We used presence–absence of RAD markers, together with marker coverage, to identify markers specific to particular sex chromosomes (W- or Z-specific markers). We also used the distribution of alleles between the sexes to identify markers present on both sex chromosomes but with SNP alleles associated with a particular sex chromosome (sex-linked alleles).
Table 1. Predicted segregation patterns for completely sex-linked RAD markers given a ZW chromosomal sex determination system in dioecious (ESP) and androdioecious (KOE) populations.
| Population |
ESP |
KOE |
|||
|---|---|---|---|---|---|
| Sexual system |
Dioecious |
Androdioecious |
|||
| Sex | Female | Male |
Hermaphrodite |
Male | |
| Monogenic | Amphigenic | ||||
| Genetic model (sex-linked allele segregation pattern) | ZW | ZZ | WW | ZW | ZZ |
| W-specific markers | |||||
| Pattern | + | Ø | + | + | Ø |
| Coveragea | 0.5 | − | 1.0 | 0.5 | − |
| Polymorphism | Hem. | − | Hom. or het. | Hem. | − |
| Z-specific markers | |||||
| Pattern | + | + | Ø | + | + |
| Coveragea | 0.5 | 1.0 | − | 0.5 | 1.0 |
| Polymorphism | Hem. | Hom. or het. | − | Hem. | Hom. or het. |
Abbreviations: ESP, Espolla; hem., hemizygous; het., heterozygous; hom., homozygous; KOE, Königswartha; RAD, restriction site-associated DNA; SNP, single-nucleotide polymorphism.
For RAD markers with completely sex-linked alleles, SNP segregation patterns are shown. For RAD markers specific to either the Z or W, sex chromosome predicted patterns are shown for presence (+)/absence (Ø), marker coverage and zygosity (hem., hom. or het.).
Coverage relative to autosomal markers found in all individuals in both populations.
In addition, we compared the numbers of RAD markers in linkage disequilibrium (LD) with the completely sex-linked makers identified above between the dioecious and androdioecious populations. Genepop files containing genotypes for all markers from the filtered catalogue found in all individuals for each population were generated using populations (Catchen et al., 2011). Genepop On The Web version 4.2 (http://genepop.curtin.edu.au/) (Raymond and Rousset, 1995) was used to test for LD using default settings. We first investigated patterns of pairwise LD in both populations and then looked for pairs of markers in significant LD that involved at least one of our completely sex-linked markers.
Results
Sequencing
A total of 114 304 783 read pairs were obtained from the four RAD libraries comprising 22.9 Gb of sequence (Supplementary Table S2). After trimming, quality control and PCR duplicate removal we retained 28 986 269 70-bp read pairs (4.06 Gb). Individual coverage ranged from 246 930 (KOE_12_H2) to 1 390 021 reads (ESP_M11) with an average of 616 729 reads per sample (s=242 257) (Supplementary Table S3).
Assembly of RAD markers, SNP calling and population genetic diversity and differentiation
Stacks de novo assembled 20 902 candidate RAD markers. After filtering for RAD markers with >20 reads found in six or more individuals, the filtered catalogue contained 3822 markers, which accounted for 93% of the approximately 26 million retained reads that were mappable to the catalogue. Of these, 800 RAD markers were found in all individuals and had consistently high coverage (median=299x; see Supplementary Table S3). Based on 1559 markers found in all individuals of at least one of the two populations (which excludes sex chromosome-specific markers) FST between ESP and KOE was 0.71, in close agreement to microsatellite-based estimates between the same populations (Zierold et al., 2009) and confirming substantial population genetic differentiation. The level of within population polymorphism varied with 28.9% (351/1221) polymorphic markers (containing at least one SNP) in ESP and just 9.5% (122/1288) in KOE.
Dioecious and androdioecious populations of T. cancriformis have ZW chromosomal sex determination
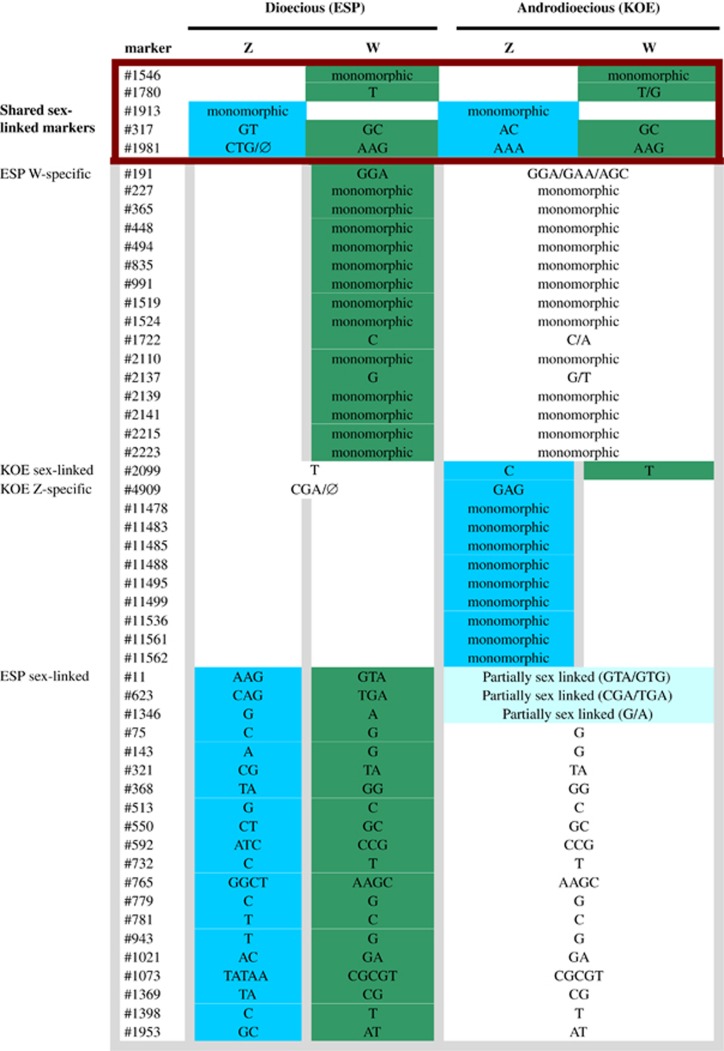
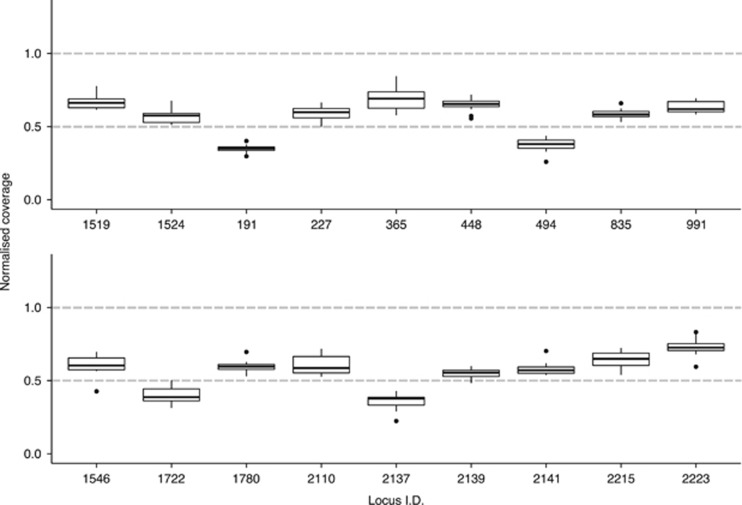
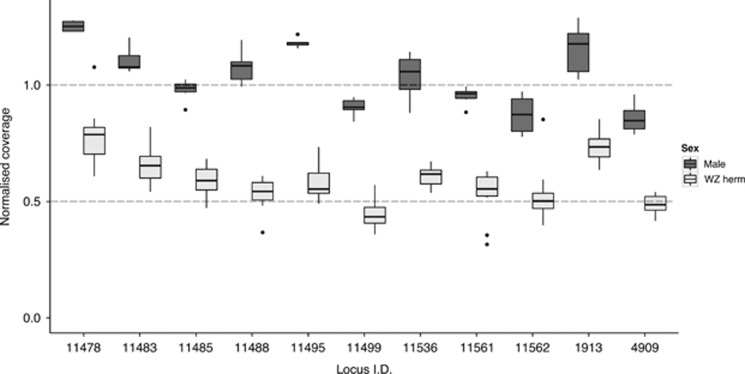
We searched the 3822 markers in the filtered catalogue according to our predictions for a ZW chromosomal sex determination system (Table 1) to identify completely sex-linked markers in the dioecious (ESP) and androdioecious (KOE) populations. In the dioecious population (ESP), 18 RAD markers met the criteria for putative W-specific markers (Figure 2): they were present in all ESP females (n=12, putatively ZW) and absent in all ESP males (n=14, putatively ZZ), had coverage consistent with being hemizygous (Figure 3) and all females had a single allele. Analysis of ESP genotype data recovered an additional 21 completely sex-linked RAD markers with alleles segregating in a manner consistent with a ZW chromosomal sex determination system, where females were always heterozygous and had female-specific (W-linked) alleles and the males always homozygous with Z-linked alleles (Figure 2). In contrast, our data were incompatible with a heterogametic XY male model because no markers were present in all males (Y specific) and absent in all females, nor were any markers found with sex-linked alleles where males were always heterozygous (Y linked).
Figure 2.
Summary of the candidate sex-linked RAD markers found in the dioecious (ESP) and androdioecious (KOE) populations. We note that this is an overview of the patterns of sex linkage found in our data set, not a genetic map. We show both markers specific for a particular sex chromosome (W and Z specific) and also markers with complete sex linkage (sex-linked markers). SNPs present in polymorphic RAD markers are indicated in the corresponding cells. Strong colours indicate complete sex linkage (blue for Z linked and green for W linked), whereas paler colours represents markers that are in LD to the sex-linked markers in the population-level LD analysis. Sex-linked markers shared between ESP and KOE are enclosed in a red box.
Figure 3.
Normalised coverage of 18 candidate W-specific markers in ESP females. Dotted lines show the expected coverage for autosomal markers (1.0), and hemizygous markers (0.5). All markers were identified based on patterns of presence–absence.
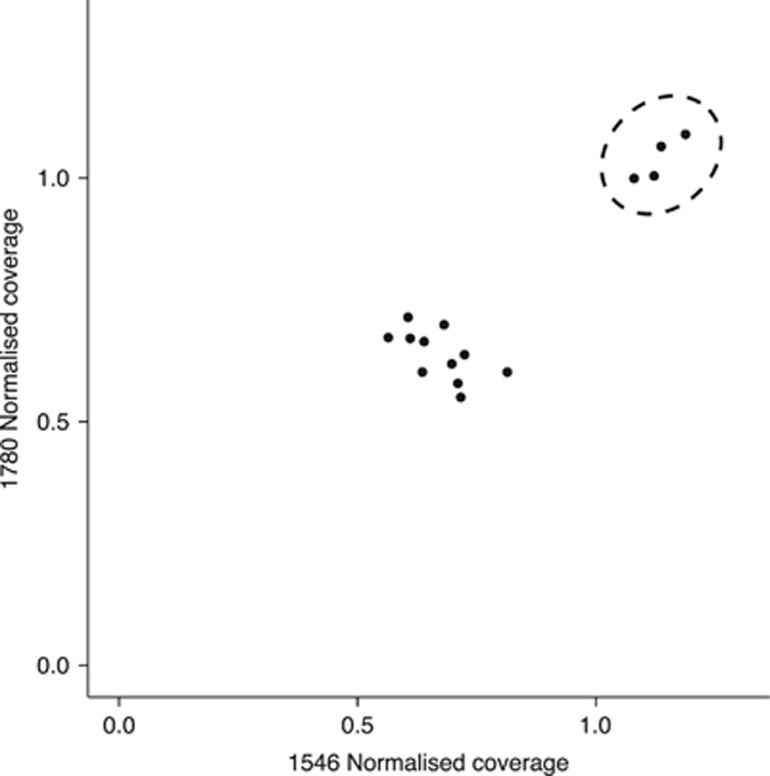
Just two markers, also previously identified by filtering in ESP, were putatively W specific (#1546 and #1780; see Figure 2) in the androdioecious population (KOE), being found in all hermaphrodites (n=15, putatively WW or ZW) but absent in all males (n=6, putative ZZ). Using these markers, we were able to test another expectation of a ZW chromosomal sex determinations system in an androdioecious population—the presence of monogenic (WW) and amphigenic (ZW) hermaphrodites. W-specific markers are expected to have approximately equivalent coverage to that of autosomal markers in monogenics (WW) and half that of autosomal markers in amphigenics (ZW) (Table 1). Plotting normalised coverage for the two W-specific markers against each other for all KOE hermaphrodites revealed a cluster of four putative monogenic (WW) hermaphrodites with coverage of around 1 for both markers and a second cluster of 11 putative amphigenic (WZ) hermaphrodites with coverage of around 0.5 for both markers (Figure 4).
Figure 4.
Identification of KOE monogenic (WW) and amphigenic (WZ) hermaphrodites. Normalised coverage of both W-specific markers (#1780 and #1546) found in KOE for 15 KOE hermaphrodites. The two clusters show amphigenic (ZW) hermaphrodites with coverage approximately half that of autosomal markers (as expected under hemizygosity) and monogenic (WW) hermaphrodites (dashed circle) with coverage equivalent to that of autosomal markers. Four monogenic (WW) hermaphrodites were identified: 1: KOE_12_H29, 2: KOE_12_H25, 3: KOE_12_H10, 4: KOE_12_H21.
Identification of Z-specific markers in KOE was facilitated by the identification of monogenic hermaphrodites (WW). Filtering the catalogue for markers present in all males (n=6, putatively ZZ) and all amphigenic hermaphrodites (n=11, putatively ZW) but absent in all monogenic hermaphrodites (n=4, putatively WW) recovered 11 candidate Z-specific markers. As expected, these markers had ~0.5 normalised coverage in amphigenic ZW hermaphrodites compared with normalised coverage of ~1 in ZZ males, further supporting the Z-specific status of these markers (Figure 5). Just 2 of the 11 candidate Z-specific markers in KOE were found in ESP. One of them, #1913, showed coverage expected for a Z-specific marker (0.5 normalised coverage in females, ZW, and 1 in males, ZZ), but patterns of presence–absence in the other marker, #4909, were incompatible with complete sex linkage (data not shown). In the dioecious population (ESP), patterns of presence–absence are uninformative to identify additional Z-specific markers as both males (ZZ) and females (ZW) are expected to carry at least one Z-linked allele.
Figure 5.
Male (ZZ, dark grey) vs amphigenic hermaphrodite (ZW, pale grey) normalised coverage for 11 candidate Z-specific markers in KOE. Dotted lines show the expected coverage for autosomal markers (1.0), and hemizygous markers (0.5). These markers were absent in monogenic (WW) KOE hermaphrodites.
Analysis of KOE genotype data revealed a further three completely sex-linked markers (#317, #1981 and #2099) with alleles segregating as expected for a ZW chromosomal sex determination system. These markers were heterozygous in all amphigenic hermaphrodites (n=11, putatively ZW) and homozygous in all monogenic hermaphrodites (n=4, putatively WW) and males (n=6, putatively ZZ), and all markers had hermaphrodite-specific (W-linked) alleles. All three sex-linked markers were also present in ESP, one was monomorphic (#2099), another was also sex-linked (#317) and the third one (#1981) had polymorphism and coverage patterns consistent with being completely sex linked, but with a null allele segregating in the Z chromosome, so it was not identified following our specific criteria in Table 1 (data not shown), so it was added to our list of sex-linked markers.
Overall, we identified 52 putative completely sex-linked markers across both populations (Figure 2) and confirmed ZW genetic sex determination in T. cancriformis, with two types of hermaphrodite in the androdioecious population (monogenics and amphigenics). Five markers are completely sex linked in both populations, suggesting conservation of a core sex-determining region. Of these, two markers are W specific (#1546 and #1780), one Z specific (#1913) and two have completely sex-linked alleles (#317 and #1981) with the phase preserved in both populations (Figure 2). Coverage across RAD markers identified as having completely sex-linked alleles was consistently high in all individuals with median coverage across markers ranging from 133 to 412 × .
The transition between dioecy and AD is associated with altered patterns of sex linkage
Although both dioecious (ESP) and androdioecious (KOE) populations show patterns of marker segregation consistent with ZW chromosomal sex determination, the size of the sex-linked region is smaller in the androdioecious population. All 41 completely sex-linked markers (Figure 2) identified in the dioecious population (ESP) are recovered in KOE but only five remain completely sex linked. Furthermore, only 12 additional completely sex-linked markers are found in KOE even including Z-specific markers, which cannot be reliably identified in ESP (Figure 2). The remaining 37 sex-linked markers in ESP were also found in KOE, but the sex-linked status could not be ascertained in 30 of them, which were monomorphic with the ESP W-specific allele and present in all individuals (Figure 2). The three polymorphic markers were in significant LD with sex-linked markers in the population analysis in KOE (markers #11, #1346 and #623; Figure 2, see below); two have the ESP W allele along with additional alleles not found in ESP; and the other has both the ESP W and Z allele (see Figure 2).
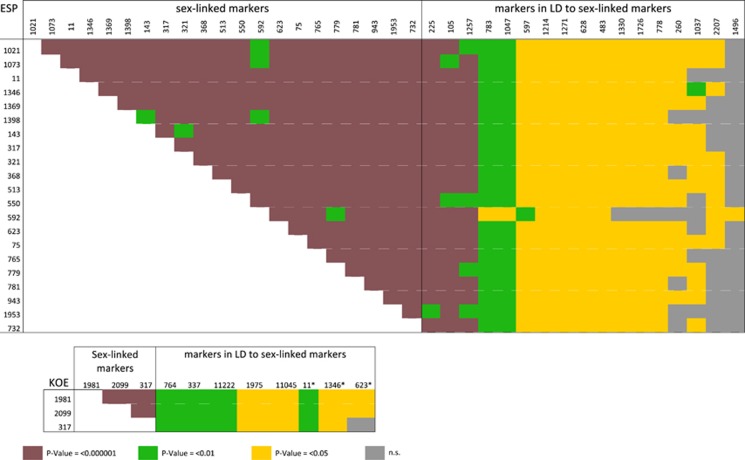
We investigated patterns of population sex linkage using tests of LD. In ESP, where, compared with KOE, there were a greater number of polymorphic markers (351 vs 122), just 5% of pairs of markers (2871 out of 58 311) were significantly in LD. In contrast, in KOE, 19% of pairs of markers (1204/6328) were significantly in LD, indicating higher levels of genome-wide population LD relative to ESP. We then looked for marker pairs in significant LD where one marker was known from our previous filtering to be sex linked. In ESP, 17% (497 out of 2871) involved at least one sex-linked marker, with 17 markers significantly linked to most of the 21 completely sex-linked markers, three of them highly significantly (#225, #105 and #1257). In KOE, in contrast, although a higher percentage of markers were in LD genome wide, just 2% of pairs of markers (24 out of 1204) included the three sex-linked markers. These involved a total of eight markers, three of them completely sex-linked in ESP (#11, #1346 and #623), which were significantly linked to the three completely sex-linked markers (Figure 6).
Figure 6.
Population linkage analysis. RAD markers in LD with identified sex-linked markers are shown for each population. Asterisks indicate markers that are sex linked in the ESP population.
We note that our criteria to discover completely sex-linked markers (Table 1) excluded sex-linked markers where polymorphism occurs within either sex chromosome (including null alleles, linked to restriction site polymorphism). However, this is likely to involve a small number of markers and will not affect our conclusions. Polymorphic markers within sex chromosomes, if they occur, should appear in LD to the completely sex-linked markers in the population. Therefore, we checked the haplotypes of all markers in LD with sex-linked markers in ESP and KOE in the population-level linkage analysis and did not find any that were polymorphic within either sex chromosome and completely sex linked. However, we identified one putative sex-linked marker (#1981; Figure 2), shared between populations, which had null alleles segregating in the Z chromosome.
Discussion
Our analysis confirmed chromosomal sex determination in T. cancriformis. Specifically, our prediction of a ZW sex determination system was upheld in both populations, with males from the dioecious and androdioecious populations homogametic (ZZ), females from the dioecious population heterogametic (ZW) and two types of hermaphrodites in the androdioecious populations (monogenic WW and amphigenic ZW). The percentage of monogenics (27%) is similar to that found in Eulimnadia (Weeks et al., 1999, 2014). Our data also showed that both T. cancriformis populations share a small set of completely sex-linked markers, suggesting conservation of a core sex-determining region, presumably containing the master sex determination gene. Finding ZW sex determination adds support to this sex determination mechanism being broadly conserved in Branchiopoda, as it is found not only in the androdioecious American species T. newberryi, where maleness is determined by a recessive locus (Sassaman 1991), but also in the more distantly related clam shrimp Eulimnadia texana (Sassaman and Weeks, 1993; Weeks et al., 2010) and the brine shrimp Artemia (Stefani, 1963; Legrand et al., 1987; Parraguez et al., 2009; De Vos et al., 2013).
Structure of Triops cancriformis sex chromosomes
Our results suggest striking differences between the structure of sex chromosomes in the androdioecious (KOE) and dioecious (ESP) populations, with fewer sex-linked markers in the androdioecious population, suggesting a smaller sex-specific region. We also found decreased differentiation between the putative W and Z chromosomes, as the Z chromosome in KOE contains a region almost identical to the W. In addition, we found a greater proportion of marker pairs in significant LD that contained a completely sex-linked marker (‘sex-linked pairs') in the dioecious (ESP) compared with the androdioecious (KOE) population. This was despite the greater proportion of marker pairs in LD over the whole genome in KOE compared with ESP. The higher LD seen over the whole genome in KOE is likely a direct effect of increased selfing by hermaphrodites (Nordborg, 2000) and a correlated reduction in effective population size. However, we also observed that a much higher proportion of sex-linked pairs were in LD compared with genome-wide pairs in ESP. Such a contrast suggests that the dioecious population (ESP) has a larger sex-specific, non-recombining region, despite lower levels of genome-wide LD.
The sizes of the sex-linked regions in both populations can be roughly estimated assuming that sex-linked markers are physically linked. Our RAD library was made using the restriction enzyme SbfI, which has an 8-bp recognition site. Given that Triops mean GC content is 0.47 (estimated from the RAD autosomal markers), this results in two RAD markers flanking the cut site approximately every 100 kb (see ‘RAD counter' on https://www.wiki.ed.ac.uk/display/RADSequencing/Home). As there are 41 completely sex-linked markers in the dioecious population (corresponding to ca 20 restriction sites), these sex-linked markers are expected to cover ca 2 Mb of W-linked sequence. In contrast, the W-specific region in the androdioecious population is much smaller, with just 3 W-linked RAD sites corresponding to around 300 kb. However, these estimated sizes should be interpreted with caution, given, that SbfI is expected to target GC-rich regions, and we do not know if there are any GC biases in the sex-linked regions.
Evolution of sex chromosomes
The strength of sexually antagonistic selection in the dioecious population is expected to be much higher than in the AD population. Given our ZW genetic model, the W is always found in females (ZW) in dioecious populations, so selection for female-benefitting mutations and recombination suppression between these markers and the sex-determining region is likely as is the case for male-benefitting mutations on the Y chromosome (Charlesworth and Charlesworth, 1980; Rice, 1987a; Bergero and Charlesworth, 2009; Kirkpatrick and Guerrero, 2014). The Z, meanwhile, would be subjected to mild male-biased sexually antagonistic selection as, assuming a 50:50 sex ratio, the Z is found 2/3 of the time in males and 1/3 in females. In contrast, in androdioecious populations, both the W and the Z are found mostly in hermaphrodites, which must balance both male and female function. The W is exclusively found in hermaphrodites (in both WW monogenics and ZW amphigenics), but the Z is largely found in hermaphrodites too because males (ZZ) are rare and the Z is also carried by amphigenic (ZW) hermaphrodites. Although some sexually antagonistic selection is expected to occur in hermaphroditic plants and animals (Bedhomme et al., 2009; Abbott, 2011), it seems highly likely that sexually antagonistic selection is a stronger force in dioecious compared with androdioecious populations. The smaller sex-linked region and the lower proportion of markers in LD to the sex-linked region in KOE compared with ESP (Figure 6) is broadly compatible with the hypothesis that recombination suppression is relaxed in KOE compared with ESP, a possibility that requires more research. The production of hermaphroditic individuals (effectively, putatively functional intersex individuals) in Eulimnadia has been attributed to a low level of recombination between sex chromosomes (Weeks et al., 2006b). However, and given that in many organisms, recombination depends on phenotypic and not genotypic sex (Kawamura and Nishioka, 1977; Matsuba et al., 2010), there is also the possibility that increased recombination could be brought about by the presence of Z and W chromosomes in the testis lobes of ZW hermaphrodites, which undergo spermatogenesis. In a ZW system with males and females, recombination is expected to be higher during spermatogenesis in ZZ males (the homogametic sex) than during oogenesis, so a transition to hermaphroditism may be directly followed by increased ZW recombination (Perrin, 2009; Guerrero et al., 2012). However, the patterns of variation between the Z and W in KOE, with the KOE Z having high similarity to the ESP W, suggest that differences in recombination patterns is not the complete story.
Surprisingly, most completely sex-linked markers found in ESP are also found in KOE, but in KOE they are fixed for the ESP W haplotype. This suggests there is a large region where the KOE Z and W chromosomes are very similar, with both strongly resembling the haplotype now found in ESP W. This is, in principle, surprising, as the W in ESP is non-recombining in the heterogametic sex, so under traditional models it would be expected to accumulate deleterious mutations that would be compensated by the Z in the heterogametic sex in ESP (Rice, 1987b, 1994; Bachtrog, 2006). The similarity of W and Z in KOE to the ESP W is therefore paradoxical as there would be no compensation.
A second surprising finding is that, in contrast to the pattern found in W chromosomes, the Z chromosomes from ESP and KOE show marked differentiation in the non-recombining region, with 10 Z-specific RAD markers found only in KOE. From the four shared markers with Z-specific alleles, only one of them (#1913) has the same haplotype in ESP and KOE. This is notwithstanding the large region now shared between KOE W and Z haplotypes. This suggests that while the W chromosome-determining hermaphroditism may have originated from a dioecious population, genetically similar to ESP, one or more regions of the Z chromosome may have originated from introgression from a population of a different geographic origin. This hypothesis is supported by the presence of two divergent mitochondrial lineages in T. cancriformis in Central Europe, including KOE, which are likely to originate in different glacial refugia (Zierold et al., 2007). Therefore, our findings are compatible with a stepwise evolution of AD, in which first, WW hermaphrodites evolved from a dioecious population related to ESP and expanded though Europe. The second step, the evolution of AD from hermaphroditism would follow. Indeed, the small geographical area where AD occurs in Europe (Zierold et al., 2007) is located in a contact zone between divergent mtDNA lineages, lending strength to the hypothesis that AD may have evolved from hermaphroditism because of male invasion.
Evolution of AD in Triops cancriformis
Given that the W sex chromosomes are highly similar between ESP females and KOE hermaphrodites and that the sex determination mode is conserved—with femaleness and hermaphroditism dominant over maleness—our data are consistent with hermaphrodites having evolved from females rather than males. It also agrees with the inference of a recent transition between dioecy and hermaphroditism/AD as proposed by Zierold et al. (2007). The evolution of hermaphrodites from females, rather than males in sexually dimorphic animals is expected to occur because of developmental constraints—it is simpler to evolve a self-fertilising hermaphrodite from a female rather than a male (Weeks et al., 2006c; Weeks, 2009, 2012). In females, only a mutation causing sperm production in the ovaries, to produce an ovotestis, is required for the evolution self-fertile hermaphrodites. In contrast, the evolution of hermaphrodites from males would be more complex as males lack important female traits, such as the ovisac and nest-building behaviour, and possess unimportant behaviours, such as mate searching.
Simple ovotestis, in which sperm-producing testis lobes are scattered in the ovary are found in androdioecious and hermaphroditic species of Triops, including T. cancriformis (Longhurst, 1955; Akita, 1971; Scanabissi et al., 2005; Garcia-Velazco et al., 2009; Murugan et al., 2009), suggesting this pathway to hermaphroditism could be conserved across the genus, possibly linked to a recombination event producing intersex individuals, as has been suggested for Eulimnadia (Weeks et al., 2006a). The low number of shared sex-linked RAD markers between the two populations, and the presence of only one additional W-linked marker in KOE suggests that these markers may be tightly linked to a possibly conserved master sex-determining locus, and likely also to the gene determining hermaphroditism in androdioecious populations. Further characterisation of these regions of the sex chromosomes, and investigation of these markers in related androdioecious taxa, could shed light on the genetic changes involved in the evolution of hermaphroditism in Triops and whether this mechanism is indeed conserved across the genus.
Conclusions
Evolutionary transitions from genetically determined separate sexes to AD offer a unique opportunity to investigate the evolution of sex chromosomes, including testing the role of sexually antagonistic selection in the evolution of sex-associated linkage. By de novo assembly of RAD markers and testing patterns of marker presence–absence, coverage, SNP association and linkage as predicted by a specific genetic model, we have uncovered the genetic mode of sex determination, identified putative sex-linked markers and established differences in the size of the sex-linked regions between populations, with lower differentiation between sex chromosomes in the androdioecious population. The presence of differentiated sex chromosomes in T. cancriformis alongside recently evolved diverse sexual systems (Zierold et al., 2007 2009) makes the species an excellent model for the study of sex chromosome evolution. Future work should extend the study of sex chromosomes across Notostraca, where multiple transitions in sexual system have occurred (Mathers et al., 2013b), to bring further insights into the genomic effects of labile sexual systems.
Data archiving
The sequence data produced in this study are available via the Sequence Read Archive through accession number PRJEB7851 (http://www.ebi.ac.uk/ena/data/view/PRJEB7851). Additional data are included in Dryad: http://doi.org/10.5061/dryad.4635t.
Acknowledgments
This work was part of TCM's PhD project ‘The genetics and evolutionary dynamics of reproductive mode in tadpole shrimps' funded by a NERC CASE Studentship (NE/G012318/1). We are grateful to Dani Boix, for sending us multiple sediment samples from Espolla Lake, and Thorid Zierold for her support and help when sampling German populations. RAD library preparation and Illumina sequencing was funded by a NERC-NBAF grant (NBAF400) and carried out at the NERC Biomolecular Analysis Facility at Edinburgh. We thank Karim Gharbi, Cathlene Eland, Urmi Trivedi and Tim Cezard at The GenePool for help with a pilot RAD library construction, bioinformatics analysis and sequencing. We thank Dave Lunt, Amir Szitenberg, Dan Jeffries and Steve Moss from the Evolution Group at Hull, for help with setting up bioinformatics analysis. AG was funded by an Advanced NERC fellowship (NE/B501298/1). Nicholas Perrin, Chiara Benvenuto, Lori Lawson-Handley, Steve Weeks and two anonymous reviewers gave valuable suggestions on an earlier version of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abbott JK. (2011). Intra-locus sexual conflict and sexually antagonistic genetic variation in hermaphroditic animals. Proc R Soc B Biol Sci 278: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akita M. (1971). On the reproduction of Triops longicaudatus (LECONTE). Doubutsugaku Zasshi 80: 242–250. [Google Scholar]
- Anderson JL, Marí AR, Braasch I, Amores A, Hohenlohe P, Batzel P et al. (2012). Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS One 7: e40701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. (2006). A dynamic view of sex chromosome evolution. Curr Opin Genet Dev 16: 578–585. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice W et al. (2011). Are all sex chromosomes created equal? Trends Genet 27: 350–357. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman T-L et al. (2014). Sex determination: why so many ways of doing It? PLoS Biol 12: e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA et al. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter SW, Davey JW, Johnston JS, Shelton AM, Heckel DG, Jiggins CD et al. (2011). Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS One 6: e19315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedhomme S, Bernasconi G, Koene JM, Lankinen Å, Arathi HS, Michiels NK et al. (2009). How does breeding system variation modulate sexual antagonism? Biol Lett 5: 717–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D. (2009). The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol 24: 94–102. [DOI] [PubMed] [Google Scholar]
- Carmichael SN, Bekaert M, Taggart JB, Christie HRL, Bassett DI, Bron JE et al. (2013). Identification of a sex-linked SNP marker in the salmon louse (Lepeophtheirus salmonis using RAD sequencing. PLoS One 8: e77832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. (2013). Stacks: an analysis tool set for population genomics. Mol Ecol 22: 3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. (2011). Stacks: building and genotyping loci de novo from short-read sequences. G3 Genes Genomes Genet 1: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Jordan CY, Charlesworth D. (2014). The evolutionary dynamics of sexually antagonistic mutations in pseudoautosomal regions of sex chromosomes. Evolution 68: 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. (1980). Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet Res 35: 205–214. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. (2005). Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. [DOI] [PubMed] [Google Scholar]
- Cox RM, Calsbeek R. (2009). Sexually antagonistic selection, sexual dimorphism, and the resolution of intralocus sexual conflict. Am Naturalist 173: 176–187. [DOI] [PubMed] [Google Scholar]
- Davey JW, Blaxter ML. (2010). RADSeq: next-generation population genetics. Brief Func Genomics 9: 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos S, Bossier P, Van Stappen G, Vercauteren I, Sorgeloos P, Vuylsteke M. (2013). A first AFLP-based genetic linkage map for brine shrimp Artemia franciscana and Its application in mapping the sex locus. PLoS One 8: e57585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster K, Coulson T, Sheldon BC, Pemberton JM, Clutton-Brock TH, Kruuk LE. (2007). Sexually antagonistic genetic variation for fitness in red deer. Nature 447: 1107–1110. [DOI] [PubMed] [Google Scholar]
- Fridolfsson A-K, Cheng H, Copeland NG, Jenkins NA, Liu H-C, Raudsepp T et al. (1998). Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc Natl Acad Sci USA 95: 8147–8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer G. (1988). Studies on the functional-morphology and biology of the Notostraca (Crustacea, Branchiopoda). Philos Trans R Soc Lond B Biol Sci 321: 27–142. [Google Scholar]
- Gamble T, Zarkower D. (2014). Identification of sex-specific molecular markers using restriction site-associated DNA sequencing. Mol Ecol Res 14: 902–913. [DOI] [PubMed] [Google Scholar]
- Garcia-Velazco H, Obregon-Barboza H, Rodriguez-Jaramillo C, Maeda-Martinez AM. (2009). Reproduction of the tadpole shrimp Triops (Notostraca) in Mexican waters. Curr Sci 96: 91–97. [Google Scholar]
- Graves JAM. (2008). Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu Rev Genet 42: 565–586. [DOI] [PubMed] [Google Scholar]
- Guerrero RF, Kirkpatrick M, Perrin N. (2012). Cryptic recombination in the ever-young sex chromosomes of Hylid frogs. J Evol Biol 25: 1947–1954. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. (2010). Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet 6: e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CY, Charlesworth D. (2012). The potential for sexually antagonistic polymorphism in differnt genome regions. Evolution 66: 505–516. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Nishioka M. (1977). Aspects of the reproductive biology of Japanese Anurans. In: Taylor D, Guttman S (eds) The Reproductive Biology of Amphibians. Springer: New York, NY, USA. pp 103–139. [Google Scholar]
- Kirkpatrick M, Guerrero RF. (2014). Signatures of sex-antagonistic selection on recombining sex chromosomes. Genetics 197: 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. (1999). Four evolutionary strata on the human X chromosome. Science 286: 964–967. [DOI] [PubMed] [Google Scholar]
- Legrand J, Legrand-Hamelin E, Juchault P. (1987). Sex determination in Crustacea. Biol Rev 62: 439–470. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhurst AR. (1955). The reproduction and cytology of the Notostraca (Crustacea, Phyllopoda). Proc Zool Soc Lond 125: 671–680. [Google Scholar]
- Lunter G, Goodson M. (2011). Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21: 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers TC, Hammond RL, Jenner RA, Hänfling B, Gómez A. (2013. a). Multiple global radiations in tadpole shrimps challenge the concept of ‘living fossils'. Peer J 1: e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers TC, Hammond RL, Jenner RA, Zierold T, Hänfling B, Gómez A. (2013. b). High lability of sexual system over 250 million years of evolution in morphologically conservative tadpole shrimps. BMC Evol Biol 13: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba C, Alho JS, Merilä J. (2010). Recombination rate between sex chromosomes depends on phenotypic sex in the common frog. Evolution 64: 3634–3637. [DOI] [PubMed] [Google Scholar]
- Murugan G, Obregon-Barboza H, Maeda-Martinez AM, Timms BV. (2009). Co-occurrence of two tadpole shrimp, Triops cf. australiensis (Branchiopoda: Notostraca), lineages in middle Paroo, north-western New South Wales, with the first record of Triops hermaphrodites for the Australian continent. Aust J Zool 57: 77–84. [Google Scholar]
- Nordborg M. (2000). Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 154: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden R, Gharbi K, Mugue N, Martinsohn J, Senn H, Davey JW et al. (2013). Sturgeon conservation genomics: SNP discovery and validation using RAD sequencing. Mol Ecol 28: 2732–2737. [DOI] [PubMed] [Google Scholar]
- Ombretta M, Cesari M, Eder E, Scanabissi F, Mantovani B. (2005). Chromosomes in sexual populations of Notostracan and Con-chostracan taxa (Crustacea, Branchiopoda). Caryologia 58: 164–170. [Google Scholar]
- Otto SP. (2014). Selective maintenance of recombination between the sex chromosomes. J Evol Biol 27: 1431–1442. [DOI] [PubMed] [Google Scholar]
- Otto SP, Sassaman C, Feldman MW. (1993). Evolution of sex determination in the conchostracan shrimp Eulimnadia texana. Am Naturalist 141: 329–337. [DOI] [PubMed] [Google Scholar]
- Palaiokostas C, Bekaert M, Davie A, Cowan ME, Oral M, Taggart JB et al. (2013). Mapping the sex determination locus in the Atlantic halibut (Hippoglossus hippoglossus) using RAD sequencing. BMC Genomics 14: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parraguez M, Gajardo G, Beardmore JA. (2009). The New World Artemia species A. franciscana and A. persimilis are highly differentiated for chromosome size and heterochromatin content. Hereditas 146: 93–103. [DOI] [PubMed] [Google Scholar]
- Pease JB, Hahn MW. (2012). Sex chromosomes evolved from independent ancestral linkage groups in winged insects. Mol Biol Evol 29: 1645–1653. [DOI] [PubMed] [Google Scholar]
- Perrin N. (2009). Sex reversal: a fountain of youth for sex chromsomes. Evolution 63: 3043–3049. [DOI] [PubMed] [Google Scholar]
- Pires-daSilva A. (2007). Evolution of the control of sexual identity in nematodes. Sem Cell Dev Biol 18: 362–370. [DOI] [PubMed] [Google Scholar]
- Qiu S, Bergero R, Charlesworth D. (2013). Testing for the footprint of sexually antagonistic polymorphisms in the pseudoautosomal region of a plant sex chromosome pair. Genetics 194: 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. (1995). GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249. [Google Scholar]
- Reitzel AM, Herrera S, Layden MJ, Martindale MQ, Shank TM. (2013). Going where traditional markers have not gone before: utility of and promise for RAD sequencing in marine invertebrate phylogeography and population genomics. Mol Ecol 22: 2953–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W. (1994). Degeneration of a nonrecombining chromosome. Science 263: 230–232. [DOI] [PubMed] [Google Scholar]
- Rice WR. (1987. a). The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914. [DOI] [PubMed] [Google Scholar]
- Rice WR. (1987. b). Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics 116: 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. (1992). Sexually antagonistic genes: experimental evidence. Science 256: 1436–1439. [DOI] [PubMed] [Google Scholar]
- Richards PM, Liu MM, Lowe N, Davey JW, Blaxter ML, Davison A. (2013). RAD-Seq derived markers flank the shell colour and banding loci of the Cepaea nemoralis supergene. Mol Ecol 22: 3077–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman C. (1989). Sex-ratio and sex determination in Western populations of Triops (Notostraca). Am Zoologist 29: A66–A66. [Google Scholar]
- Sassaman C. (1991). Sex-ratio variation in female-biased populations of notostracans. Hydrobiologia 212: 169–179. [Google Scholar]
- Sassaman C, Weeks SC. (1993). The genetic mechanism of sex determination in the Chonchostracan shrimp Eulimnadia texana. Am Naturalist 141: 314–328. [DOI] [PubMed] [Google Scholar]
- Scanabissi F, Eder E, Cesari M. (2005). Male occurrence in Austrian populations of Triops cancriformis (Branchiopoda, Notostraca) and ultrastructural observations of the male gonad. Invertebrate Biol 124: 57–65. [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG et al. (2003). The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825–U822. [DOI] [PubMed] [Google Scholar]
- Stefani R. (1963). La digametia femminile in Artemia salina Leach e la costituzione del corredo cromosomico nei biotipi diploide anfigonico e diploide partenogenetico. Caryologia 16: 625–636. [Google Scholar]
- van Doorn GS, Kirkpatrick M. (2010). Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 186: 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner CE, Keller I, Wittwer S, Selz OM, Mwaiko S, Greuter L et al. (2013). Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Mol Ecol 22: 787–798. [DOI] [PubMed] [Google Scholar]
- Weeks SC. (2009). Can males successfully invade hermaphroditic populations of clam shrimp (Eulimnadia texana? Curr Sci 96: 98–102. [Google Scholar]
- Weeks SC. (2012). The role of androdioecy and gynodioecy in mediating evolutionary transitions between dioecy and hermaphroditism in the Animalia. Evolution 66: 3670–3686. [DOI] [PubMed] [Google Scholar]
- Weeks SC, Benvenuto C, Reed SK. (2006. a). When males and hermaphrodites coexist: a review of androdioecy in animals. Integr Comp Biol 46: 449–464. [DOI] [PubMed] [Google Scholar]
- Weeks SC, Benvenuto C, Reed SK, Duff RJ, Duan ZH, David P. (2014). A field test of a model for the stability of androdioecy in the freshwater shrimp, Eulimnadia texana. J Evol Biol 27: 2080–2095. [DOI] [PubMed] [Google Scholar]
- Weeks SC, Benvenuto C, Sanderson TF, Duff RJ. (2010). Sex chromosome evolution in the clam shrimp, Eulimnadia texana. J Evol Biol 23: 1100–1106. [DOI] [PubMed] [Google Scholar]
- Weeks SC, Marcus V, Crosser BR. (1999). Inbreeding depression in a self-compatible, androdioecious crustacean, Eulimnadia texana. Evolution 53: 472–483. [DOI] [PubMed] [Google Scholar]
- Weeks SC, Reed SK, Cesari M, Scanabissi F. (2006. b). Production of intersexes and the evolution of androdioecy in the clam shrimp Eulimnadia texana (Crustacea, Branchiopoda, Spinicaudata). Invertebr Reprod Dev 49: 113–119. [Google Scholar]
- Weeks SC, Sanderson TF, Reed SK, Zofkova M, Knott B, Balaraman U et al. (2006. c). Ancient androdioecy in the freshwater crustacean Eulimnadia. Proc R Soc B Biol Sci 273: 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierold T, Hanfling B, Gomez A. (2007). Recent evolution of alternative reproductive modes in the 'living fossil' Triops cancriformis. BMC Evol Biol 7: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierold T, Montero-Pau J, Hanfling B, Gomez A. (2009). Sex ratio, reproductive mode and genetic diversity in Triops cancriformis. Freshwater Biol 54: 1392–1405. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.