Abstract
Understanding the genetic architecture of adaptive traits has been at the centre of modern evolutionary biology since Fisher; however, evaluating how the genetic architecture of ecologically important traits influences their diversification has been hampered by the scarcity of empirical data. Now, high-throughput genomics facilitates the detailed exploration of variation in the genome-to-phenotype map among closely related taxa. Here, we investigate the evolution of wing pattern diversity in Heliconius, a clade of neotropical butterflies that have undergone an adaptive radiation for wing-pattern mimicry and are influenced by distinct selection regimes. Using crosses between natural wing-pattern variants, we used genome-wide restriction site-associated DNA (RAD) genotyping, traditional linkage mapping and multivariate image analysis to study the evolution of the architecture of adaptive variation in two closely related species: Heliconius hecale and H. ismenius. We implemented a new morphometric procedure for the analysis of whole-wing pattern variation, which allows visualising spatial heatmaps of genotype-to-phenotype association for each quantitative trait locus separately. We used the H. melpomene reference genome to fine-map variation for each major wing-patterning region uncovered, evaluated the role of candidate genes and compared genetic architectures across the genus. Our results show that, although the loci responding to mimicry selection are highly conserved between species, their effect size and phenotypic action vary throughout the clade. Multilocus architecture is ancestral and maintained across species under directional selection, whereas the single-locus (supergene) inheritance controlling polymorphism in H. numata appears to have evolved only once. Nevertheless, the conservatism in the wing-patterning toolkit found throughout the genus does not appear to constrain phenotypic evolution towards local adaptive optima.
Introduction
Over the past decade, next-generation sequencing technologies have provided increased power to identify the genomic targets of selection: the loci, genes and genetic variants that control adaptive phenotypes (Stinchcombe and Hoekstra, 2007). These tools expand the frontiers beyond classical model species and, in particular, have provided powerful insights into convergent evolution (cf Arendt and Reznick, 2008), whereby the same phenotype evolves in two or more lineages independently, typically in response to similar environmental challenges. Natural systems displaying phenotypic convergence provide a robust framework to investigate whether this convergence derives from the recruitment of the same or different genes and genetic mechanisms, thus allowing a better understanding of the molecular basis of adaptive evolution (Stern, 2013).
The evolution of an adaptive trait is influenced by its genetic architecture. This term encapsulates the often complex genotype-to-phenotype relationship and includes the number and nature of genetic elements (genes and alleles), their absolute and relative genomic locations, their effect sizes and their interactions. These interactions can occur with the environment (for example, via epigenetic effects), between distinct genes (that is, epistasis, additivity), between variants at the same locus (that is, dominance) and in additional effects on other phenotypic traits (that is, pleiotropy). The genetic architecture of phenotypic variation can influence both convergence and diversification processes, and selective pressures may operate on any of its components, either singly or in combination (Hansen, 2006). A large number of theoretical studies described the evolution of these different features (Lande, 1980; Barton, 1995; Orr, 1998; Carter et al., 2005). Nevertheless, scant empirical data exist on the factors associated with the evolution of genetic architectures and on how heterogeneity in the genetic architecture of complex traits can influence their diversification (De Visser et al., 1997; Lair et al., 1997).
Butterflies in the genus Heliconius represent an excellent system to investigate the evolution of the genetic architecture of complex adaptive traits. This clade contains distinct lineages that display different wing colour patterns, show heterogeneity in the genetic architecture of these traits and permit a comparative approach across lineages owing to the occurrence of both convergent and divergent evolution. In addition, the ecological roles of wing patterns and the selection regimes shaping their variation have been relatively well studied in this genus (Brown, 1981; Kapan, 2001; Jiggins et al., 2001). Heliconius butterflies are unpalatable to predators and the spectacular wing colour patterns advertise their toxicity. Several species within and outside this genus converge in wing patterns, enjoying survival benefits in the face of predation by using similar signals of toxicity. This convergence is known as Müllerian mimicry. This adaptation to the local prey environment recognised by educated predators suggests that the genes controlling wing colour are subject to strong selective pressures.
Previous studies have defined a palette of genomic regions of large phenotypic effect shared by distinct Heliconius species and underlying the diversification of colour patterns (Joron et al., 2006; Kronforst et al., 2006a; Papa et al., 2008). This conserved ‘toolkit' of genes is mainly distributed across four of the 21 Heliconius chromosomes; however, several minor effect loci have also been detected (see summary Supplementary Table S1). Two of the causal genes that drive adaptive pattern variation have been identified. One is the WntA signalling ligand, a putative morphogen that determines the size and position of melanic patterns in the forewing median region (corresponding to the effects of loci Ac/Ac/Sd in H. melpomene, H. cydno and H. erato, respectively; Martin et al., 2012). Another gene is a transcription factor, homologous to the Drosophila gene optix, which prefigures the variety of red wing elements controlled by the cluster of loci B-D/D/G-Br in H. melpomene, H. erato and H. cydno (Reed et al., 2011; Martin et al., 2014). Causal genes at two other major loci are yet to be formally characterised at the gene level: K that controls the white/yellow switch in H. melpomene and H. cydno (Kronforst et al., 2006b), and a tight cluster of loci that controls most of the variations in yellow and white pattern elements. The latter is a complex of at least three linked loci (Yb, Sb and N) in H. melpomene, two of which have also been described in its sister species H. cydno (Yb and Sb). In the more distantly related H. erato, this region harbours the Cr locus that controls similar pattern variation (Jiggins and McMillan, 1997). Recombination occurs between loci Yb, Sb and N in H. melpomene, but Cr in H. erato segregates as a single genetic unit (Sheppard et al., 1985; Mallet, 1989; Ferguson et al., 2010).
This variation in the level of linkage reveals slight modifications in the genetic architecture, nested within an otherwise highly conserved multilocus architecture throughout the Heliconius genus for the control of pattern variation (Kronforst et al., 2006a; Papa et al., 2008). There are other subtle architectural differences. For instance, the red/yellow forewing band switch is caused by variation in a single locus, D, in H. erato, but by the interaction of two unlinked loci (B and N) in H. melpomene (Sheppard et al., 1985).
To date, almost all our knowledge about the architecture of colour pattern variation in Heliconius comes from studies of species displaying variable shapes of red, white and yellow elements within a mainly black wing (Jiggins and McMillan, 1997; Jiggins et al., 2005; Kronforst et al., 2006a; Reed et al., 2011; Nadeau et al., 2014). In contrast, the genetic basis of variation for ‘tiger' patterns, which are composed of a mosaic of black, orange and yellow/white elements, is less known. These patterns are widely used by species of the so-called ‘silvaniform' subclade of Heliconius, which contains 10 described species sharing mimicry relationships with other groups of butterflies, mainly in the Danainae subfamily. Within this clade, the genetic basis of colour pattern has only been characterised in species H. numata (Joron et al., 2006). Compared with what is known in other Heliconius species, H. numata shows a strikingly different genetic architecture of wing pattern variation. Indeed, a single locus (supergene P) virtually monopolises the control of wing pattern variation in this species. According to Thompson and Jiggins (2014), a supergene is ‘a genetic architecture involving multiple linked functional genetic elements that allows switching between discrete, complex phenotypes maintained in a stable local polymorphism'. The supergene P is positionally homologous to the Yb-Sb-N cluster of H. melpomene (Joron et al., 2006).
Mimetic selection regimes are largely determined by the distribution and abundance of distinct signals used by local prey communities. Most Heliconius species, including tiger-patterned species, display geographic races differentiated in wing patterning in response to directional selection imposed by positive frequency dependence favouring one single well-defended pattern in each locality (Brown, 1981). By contrast, H. numata displays a rich local polymorphism, and all populations harbour distinct forms mimicking multiple distinct tiger-patterned species (Brown and Benson, 1974). This polymorphism is believed to be driven by fine-scale variations in the abundance of alternative tiger-patterned mimicry rings, causing balancing selection at the regional level (Joron et al., 1999). The heterogeneity in selection regimes shaping Heliconius wing patterns, that is, local monomorphism under directional selection versus polymorphism under balancing selection, allows investigating the relationship between selection regimes and the evolution of distinct genetic architectures underlying complex adaptive traits.
Here, we focus on the silvaniform clade within Heliconius and ask whether the genetic architecture of colour pattern variation is associated with the phenotypic variation itself or with the selection regime shaping it. To this end, we carefully analyse wing pattern inheritance in two unexplored tiger-patterned species in this subclade, H. hecale and H. ismenius, which show geographic variation under local directional selection for mimicry. We combine traditional linkage mapping powered by next-generation sequencing, multivariate quantitative genetics and fine-mapping of candidate genes to identify the genomic regions controlling wing pattern variation in these two species and to explore the evolution of genetic architectures in a broader comparative framework.
Materials and methods
Crossing experiments
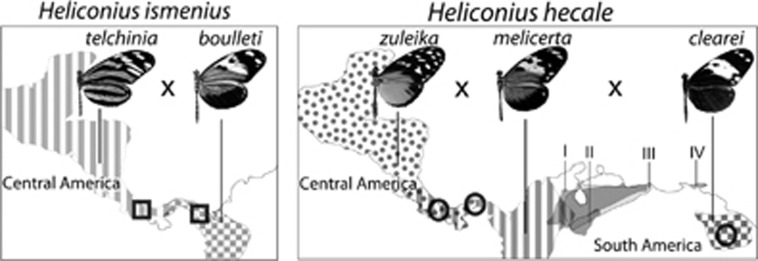
Intraspecific crosses were performed between geographic races of H. hecale and H. ismenius (Figure 1 and Supplementary Table S2). For H. hecale, we crossed subspecies melicerta (eastern Panama) with zuleika (western Panama), and melicerta with clearei (Venezuela) to obtain F1 males, which were backcrossed to melicerta females (Figures 1, 3aI and 3bI). For H. ismenius, we crossed boulleti (eastern Panama) with telchinia (western Panama), and then backcrossed F1 males to boulleti females (Figures 1 and 3cI). Breeding was performed at the Smithsonian Tropical Research Institute in Gamboa, Panama. Butterflies were kept in ~2 × 2 × 2 m cages and provided with ample sugared water and pollen. Passiflora vitifolia and P. edulis were used for H. hecale oviposition and as larval food plants, while P. quadrangularis was used for H. ismenius. The bodies of parents and progeny were preserved in NaCl-saturated dimethyl sulphoxide solution at −20 °C and wings were stored separately in glassine envelopes.
Figure 1.
Summary of crosses performed in H. hecale and H. ismenius. Geographic distribution of the subspecies used for the crosses are indicated by filling patterns, and sampling localities by circles and squares. The distribution of other H. hecale races found in Northern South America is also shown: H. h. annetta (I), H. h. rosalesi (II), H. h. anderida (III) and H. h. barcanti (IV).
Figure 3.
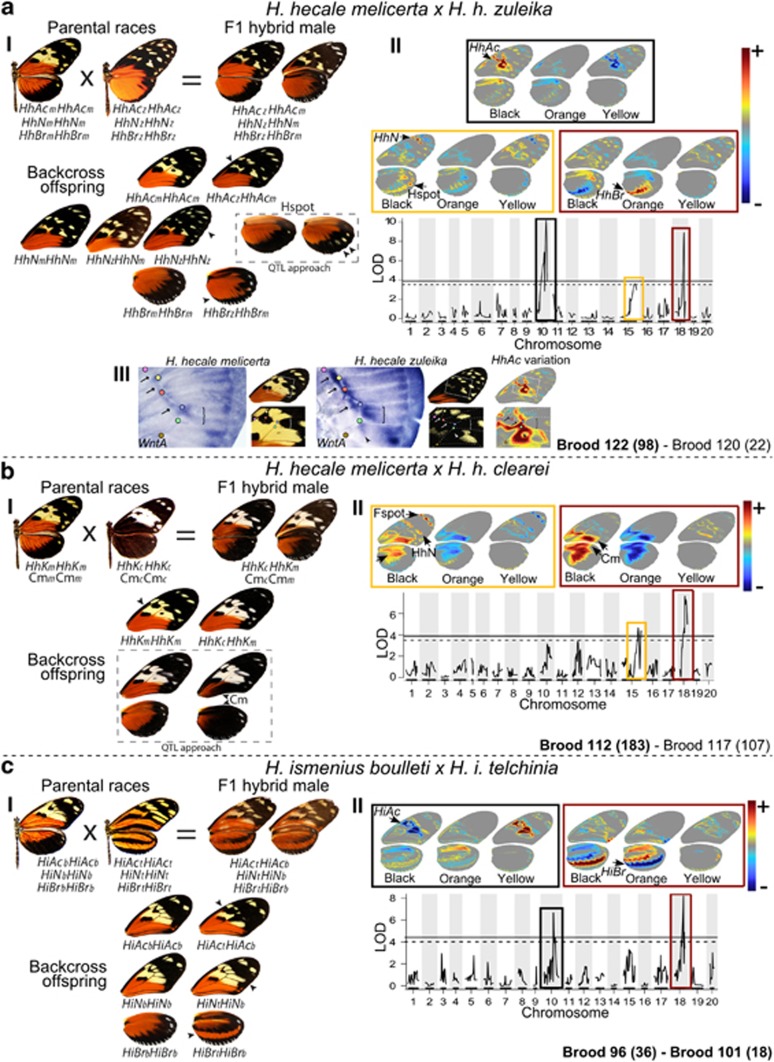
Phenotypic effect of Mendelian wing-patterning loci and major QTLs identified in H. hecale and H. ismenius crosses. For each type of cross (a–c), panel I (left) shows the crosses performed, the phenotypes associated with inferred genotypes at the major Mendelian loci (colour HhK; forewing melanisation HhAc/HiAc; forewing distal band layer/spot HhN/HiN; hindwing band HhBr/HiBr) and variation of the quantitative traits (dashed boxes: Hindwing spots Hspot, continuous melanisation Cm). Parental races (top left) are represented by their dorsal views, the F1 male siring the mapping families (top right) by its dorsal and ventral views and typical backcross specimens (bottom) have arrows pointing to the variable character. The name of the mapping families is written on the bottom of the panels of each cross type, with total number of offspring shown in brackets. Families labelled in bold were used to build the RAD libraries. Panel II (right) shows the genomic position and phenotypic effect of major QTLs. Coloured wing diagrams show the spatial distribution of individual QTL effects on pattern variation extracted from multivariate wing pattern analysis. Phenotypic variation is broken down into heatmaps for each of the three main colours (black, orange and yellow), representing, for every wing position, the strength of association between colour presence and allelic transition at the QTL (from blue to red). For analytical simplicity, both white and yellow elements in the H. hecale melicerta × H. h. clearei cross were considered as yellow elements. Genomic plots show genome-wide association (LOD) between wing pattern variation and markers along the 20 autosomes, with 5% (solid line) and 10% (dashed line) association thresholds. Panel aIII shows the detection of WntA transcripts by in situ hybridisation on wing imaginal discs of the last larval instar of H. h. melicerta and H. h. zuleika. WntA expression shows marked differences along the discal crossvein (arrows), in the M3-Cu2 intervein region (brackets) and in the Cu2-Cu1 intervein region (arrowheads). Colour dots indicate vein intersection landmarks. Phenotypic variation controlled by the HhAc locus is represented on the right.
Phenotypic analysis of the broods
Wing pattern variation was quantified in three distinct ways. First, variation segregating with largely discrete alternative phenotypic states (for example, presence/absence) was scored in all progeny. This included the number of marginal yellow spots in the dorsal and ventral views of the hindwing. However, much continuous variation was observed and hard to score by eye. Therefore, in order to get a comprehensive measure of colour pattern variation in the mapping families, we used morphometric quantification of pattern with the Colour Pattern Modelling tool (Le Poul et al., 2014). This method uses recursive alignment of wing outlines and image segmentation to identify conserved and homologous pattern elements. Briefly, Colour Pattern Modelling consists of a first colour-clustering step, where colours are treated as classes of pigments. Second, wing images are aligned on the basis of pattern and outline, on a modal wing ‘model' built recursively from the image stack. Finally, a principal component analysis of colour variation of homologous pixels across wings is performed to reduce the dimensionality of pattern variation. Hindwings and forewings were treated separately for the first two steps, but combined for the principal component analysis. For each cross, all offspring of the largest broods with intact wings were included.
Restriction site-associated DNA (RAD) library construction and sequencing
RNA-free genomic DNA was extracted from thoracic tissue using the Qiagen DNeasy Blood and Tissue kit following the manufacturer's instructions. Three RAD libraries were prepared from the backcross parents and 62 offspring of the largest broods of each type of cross, using the protocol adapted by Heliconius Genome Consortium (2012). Briefly, 300–350 ng of genomic DNA were digested with the 8-bp-cutter restriction enzyme, SbfI. We expected 1053 cutting sites on the basis of 5′-CCTGCAGG-3′ occurrences in the reference H. melpomene genome. For brood parents, reactions were scaled up to 1000–1400 ng inputs to increase representation. One of 64 Illumina P1 adapters, each with a unique 5-base barcode, was used to tag each specimen within a library. During the final PCR amplification step, 18 cycles of PCR were used, with eight independent amplifications pooled to minimise the contribution of PCR errors. Each library was paired-end sequenced in one lane of an Illumina HiSeq2000 with 100-base read length.
Bioinformatics analysis
The function process_radtags implemented in Stacks v0.9991 (Catchen et al., 2013) was used to demultiplex the separate libraries and apply basic quality filters. The processed reads of each individual were mapped to the reference genome of H. melpomene version 1.1 (Heliconius Genome Consortium, 2012) using Stampy v1.0.17 (Lunter and Goodson, 2011) with default parameters except for setting the substitution rate to 0.01. SAM/BAM file conversion, analysis and filtering were performed using SAMtools v0.1.18 (Li et al., 2009) and Picard Tools v1.67 (http://picard.sourceforge.net). To limit genotype miscalling due to PCR bias, PCR duplicates were removed using Picard Tools v1.67. At this stage, samples for each type of cross were combined in the same alignment file and processed together. Local realignment around indels was performed using the Genome Analysis Tool Kit (GATK) v1.6-2 (DePristo et al., 2011).
Single-nucleotide polymorphism genotypes were called using the GATK v1.6-2 UnifiedGenotyper with default parameters with the exception of setting expected heterozygosity to 0.015. We applied positional filters to exclude repetitive regions of the genome (Heliconius Genome Consortium, 2012). Filters for coverage (>10 × and <200 × /249 × for offspring/parents, respectively), genotype (GQ⩾30) and mapping quality (MQ⩾40) were applied using a custom Perl script (Kanchon Dasmahapatra, pers. comm.). After filtering, markers with genotype calls at fewer than 80% of individuals were excluded. Sites showing Mendelian inconsistencies were removed and missing genotype calls were imputed, scaffold-by-scaffold, using Beagle v.3.3.2 (Browning and Browning, 2009). Different subsets of individuals per brood were tested to define the data set that generated the best-quality downstream linkage maps. Final linkage maps were constructed from populations of 41, 42 and 29 offspring for the larger melicerta × zuleika, melicerta × clearei and boulleti × telchinia broods, respectively.
Linkage map construction
Crossing-over does not occur during oogenesis in Lepidoptera (Turner and Sheppard, 1975); therefore, an intact haplotype of each chromosome is passed from mother to offspring. Consequently, any female informative marker on a given autosome can inform on the segregation of linked maternal variation (‘chromosome print', cf. Jiggins et al., 2005). In contrast, male-informative single-nucleotide polymorphisms (heterozygous in father but homozygous in mother) and intercross sites (heterozygous in both parents) do recombine and inform on genetic distances within chromosomes. Genetic maps were computed independently for each cross using Joinmap v3.0 (Van Oijen and Voorrips, 2001). We filtered out single-nucleotide polymorphisms that deviated from the expected 1:1 segregation ratio (female and male-informative backcross markers) and 1:2:1 ratio (intercross markers) to generate a genotype matrix. Linkage groups corresponding to the 20 Heliconius autosomes were reconstructed using female informative markers and a logarithm of odds (LOD) threshold ⩾6 for all three data sets. The sex chromosome was not reconstructed. Male informative and intercross markers were collapsed to unique segregation patterns using a custom Perl script (John Davey, pers. comm.). Collapsed markers were combined with female-informative chromosome prints and clustered by linkage group (LOD⩾5). Individual linkage maps were built using the Kosambi mapping function and a LOD⩾1.
Mapping wing pattern loci
Phenotypes segregating with discrete alternative states were incorporated directly into map construction, alongside the collapsed marker sets, and were thus co-localised with the markers with which the phenotype was most strongly associated. A generalised linear model was used to test for an association between the number of spots on the margin of the hindwing and each of the 20 ‘chromosome prints', using R. This analysis was possible as the mother of the brood (and not the father) was heterozygous for this trait. The overall colour pattern variation quantified with Colour Pattern Modelling was mapped as quantitative trait locus (QTLs) by using a genome-wide Haley–Knott regression implemented in the R/qtl package (Broman et al., 2003). This analysis was extended by performing multivariate analysis on all principal components with an eigenvalue ⩾2% using the R/shapeQTL package (available on request; see Supplementary Methods for more details). Statistical thresholds for significant linkage were based on 1000 permutations. To further evaluate the identified QTLs, we ran a stepwise multiple QTL model search using the algorithm developed in Broman and Sen (2009) and implemented in the R/shapeQTL package for multivariate traits. The search was restricted to additive QTLs but epistatic interactions between discovered QTLs were evaluated in the final model. As colour pattern analysis is affected by sex (Jones et al., 2011), we used gender as an additive covariate. We employed only male informative markers on the 20 dense autosomal linkage maps for the different broods. The subsets of offspring in the final linkage maps having intact wings were used for this analysis. A conventional threshold of LOD⩾3 and other relaxed requirements were used to detect suggestive QTLs (Supplementary Methods).
Refining candidate intervals
To fine-map candidate intervals associated with discrete phenotypic variation, we genotyped additional markers within each region of interest in an extended panel of progeny. A combination of newly designed and previously published markers was used, generally targeting single-copy nuclear loci, but on occasion anchored in noncoding regions (Supplementary Table S3). Markers were first amplified in brood parents, and then in the progeny when allelic variation was found (see Supplementary Methods for more details about genotyping methods).
In situ hybridisation
Larval wing disc in situ hybridisations were performed following a previously described procedure (Martin et al., 2012). Wing imaginal discs of three H. hecale melicerta individuals and two H. hecale zuleika individuals originated from phenotypically pure stocks maintained in insectaries at Smithsonian Tropical Research Institute, in Gamboa, Panama. The WntA riboprobe was synthetised from a 885-bp cDNA amplicon previously cloned from the closely related species H. cydno (Martin et al., 2012).
Results
Mapping families show variable progeny
In each cross type, we obtained two families sired by the same F1 male crossed to unrelated mothers. In H. hecale, we reared a total of 120 (98+22) and 290 (183+107) butterflies for the melicerta × zuleika and melicerta × clearei crosses, respectively (Figure 3). H. ismenius was more difficult to rear, and we obtained 54 (36+18) offspring for the boulleti × telchinia crosses. The offspring of the broods showed segregation of discrete colour pattern characters affecting large portions of the wings (Figure 3), as well as some minor quantitative variations.
Four major loci segregated independently in Mendelian ratios (Supplementary Table S4) and were named according to the inferred homology with mapped loci of similar phenotypic effect in other Heliconius species. First, HhK governed the white (HhKc)/yellow (HhKm) switch for the forewing band in the H. h. melicerta × clearei families (Figure 3bI), white being dominant to yellow. Second, HiAc and HhAc shaped the size and position of black patterns close to the forewing discal cell in H. ismenius (Figure 3cI) and in the H. h. melicerta × zuleika families (Figure 3aI), respectively. The H. i. telchinia (HiAct) and H. h. zuleika (HhAcz) alleles were fully dominant over the H. i. boulleti (HiAcb) and H. h. melicerta (HhAcm) alleles, respectively. Dominant alleles break the continuity of the forewing yellow band by increasing the size of a black spot next to the discal cell. Third, HhN and HiN controlled presence veresus absence of small yellow dots in the largely black forewing submarginal area in H. hecale and H. ismenius, respectively. Although those loci are similar in the wing position and type of variation controlled, the two species showed differences in phenotypic action and dominance, which is detailed in Figures 3aI and cI. Because of its restricted phenotypic effect on the wing, HiN was considered a locus of minor effect. Fourth, in H. hecale and H. ismenius, respectively, HhBr and HiBr controlled the shape of the black marginal band of the hindwing, defined by the orange elements around this band. In our crosses, H. h. zuleika (HhBrz) and H. i. telchinia (HiBrt) alleles were strongly dominant over H. h. melicerta (HhBrm) and H. i. boulleti (HiBrb) alleles, respectively (Figures 3aI and cI). Dominant alleles produce a broken boundary of the black marginal band, whereas recessive homozygotes show a smooth, wide black band.
In addition, we recognised two presumably polygenic traits with continuous variation in our H. hecale families. Throughout this paper, trait names will be written in non-italics (in contrast to Mendelian loci) and will refer simultaneously to the quantitative trait and the QTL of major effect associated with it. First, melicerta × zuleika families showed segregation for the number of yellow spots (2–7) along the distal hindwing margin, a trait we called Hspot (Figure 3aI). The alleles were not fixed in specific parental races: the F1 father seemed to be homozygous and the mother of the backcross brood heterozygous for this trait. Second, melicerta × clearei crosses showed continuous variation in wing melanisation (Cm; Figure 3bI). Loci controlling Cm variation essentially determined the position of the boundary between black and orange areas on hindwings and in the proximal region of forewings.
Construction of RAD-sequence linkage maps
We obtained ~162, 247 and 343 million reads for each of the three libraries (Supplementary Table S5). The number of reads per individual ranged from 132 182 to 38 739 993, excluding four individuals in each library who were virtually absent because of presumed barcode failure. The intended over-representation of parental samples was observed. Supplementary Tables S5-S8 provides a detailed breakdown of RADseq library statistics. After applying basic quality filters, on average 82% of the raw read data set was retained (Supplementary Table S5), of which ~94% was mapped to the H. melpomene reference genome. PCR duplicates can potentially lead to biases towards a single allele and thus introduce genotyping errors, and so were excluded from our data set. Despite efforts to reduce library clonality during the preparation stages, we observed a drastic decrease in data quantity when excluding PCR duplicates from the mapped reads: only ~9% of the mapped reads were retained. To maximise genotype accuracy, the duplicate-removed data set was used for the map construction, with the corollary that some individuals were excluded from the analysis because of a high proportion of missing data. Subsets of individuals with the highest total number of high-quality calls were retained in the analysis: a subset of 41, 42 and 29 offspring plus the two backcross parents for the bigger melicerta × zuleika, melicerta × clearei and boulleti × telchinia broods (Supplementary Figure S1). At this stage, any remaining missing genotypes were imputed and markers with improbable segregation patterns, such as Mendelian inconsistencies, were excluded. On average, 2187 total polymorphic sites were informative for map construction: 963 female informative, 857 male informative and 367 intercross markers (Supplementary Table S9). These single-nucleotide polymorphisms were recovered from 460, 510 and 566 RAD tags among the 1053 expected cutting sites by SbfI enzyme, in the three broods, respectively. Thus, these sites are well distributed across the genome. The depth at each of these variable sites averaged 114 × for the parents and 50 × for the offspring (Supplementary Figure S2). The male informative and intercross markers together were collapsed to ~660 unique segregation patterns on average, 39.2% of which were supported by more than two markers (Supplementary Table S9). Around 450 markers on average were mapped to the 20 autosomes for each type of cross; however, they were heterogeneously distributed across the linkage groups (Supplementary Figures S3–S5).
Colour loci in H. hecale and H. ismenius map to previously identified regions
The loci segregating in our broods map to the same genomic regions where colour genes have previously been localised in other Heliconius species.
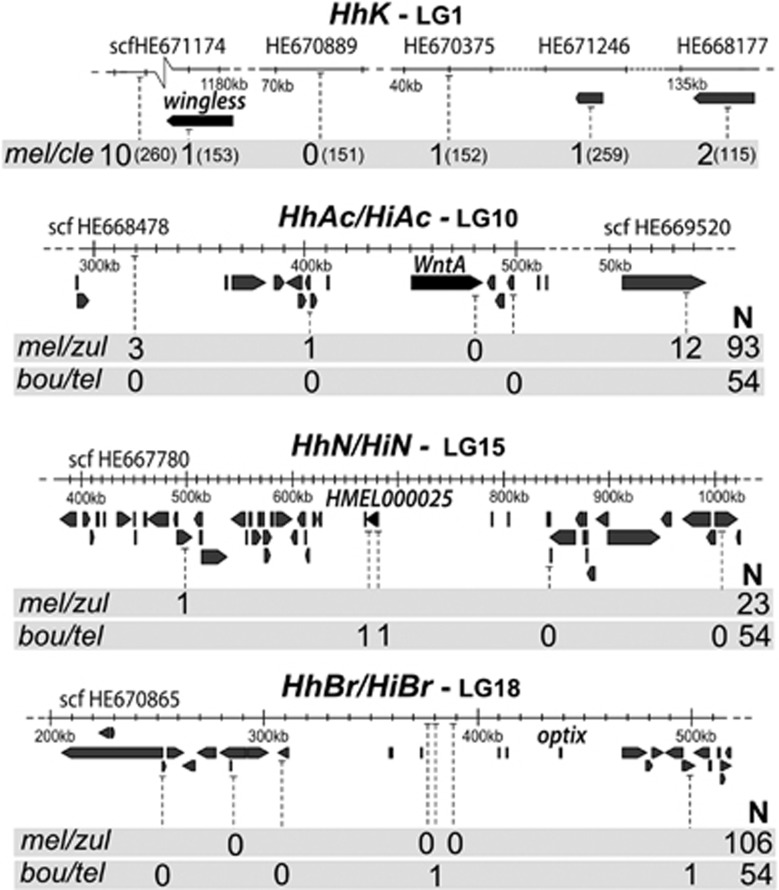
HhK maps to chromosome 1, and clusters with markers on the genomic scaffold that contains the developmental gene wingless (HE671174; Supplementary Figure S6). We found 1 recombinant in 153 genotyped offspring between HhK and wingless (Figure 2). The mapping interval is described on the basis of the order of the scaffolds defined for H. melpomene and may indicate the position of HhK between scaffolds HE670375 or HE671246 (relative position unresolved) and the scaffold containing wingless (Figure 2).
Figure 2.
Fine mapping of wing-patterning loci in H. hecale and H. ismenius. Grey-shaded boxes show recombinant individuals found in a total of N offspring in H. hecale melicerta × H. h. clearei (mel/cle), H. hecale melicerta × H. h. zuleika (mel/zul) and H. ismenius boulleti × H. i. telchinia (bou/tel) crosses. Annotated genes on each scaffold and candidate colour genes are represented by grey and black block arrows, respectively. Scaffolds on LG1 (top panel) are ordered according to the H. melpomene reference genome, but the order is unknown for the three scaffolds indicated on the right (HE670375, HE671246 and HE668177).
Both HhAc and HiAc map to chromosome 10. In both H. h. melicerta × zuleika and H. i. boulleti × telchinia crosses, these loci co-segregate with markers on scaffold HE668478 (Supplementary Figure S6), which contains the gene WntA. By genotyping additional markers around this locus for a larger number of offspring, we revealed a perfect association of HhAc/HiAc with WntA in our crosses (no recombinants in the vicinity of WntA; Figure 2).
The HhN and HiN loci map to chromosome 15 (Supplementary Figure S6). More specifically, they cluster with RAD markers placed close to the superscaffold containing Yb-Sb-N/Cr/P in other species (HE667780; Supplementary Figure S6). In H. ismenius we found one recombinant between HiN and gene HMEL000025 (Figure 2), one of the candidate genes for Yb in H. melpomene and putatively part of the P supergene in H. numata (Wu et al., 2010; Nadeau et al., 2012). In H. hecale, we found one recombinant between HhN and the only informative marker available for genotyping in 23 offspring (Figure 2). For this particular analysis, we only used individuals of genotype HhNzHhNz because of the difficulty of distinguishing HhNzHhNm and HhNmHhNm genotypes with certainty.
Finally, both HhBr and HiBr map to chromosome 18 (Supplementary Figure S6). HhBr/HiBr co-segregate with RAD markers on the scaffold containing optix (HE670865; Supplementary Figure S6). In the H. hecale families, we found no recombinants between HhBr and markers near optix in 106 offspring genotyped (Figure 2). In H. ismenius, a region that excludes the coding region of optix was delimited (Figure 2).
We found a significant association between the number of yellow spots on the margin of the hindwing (Hspot) and the maternal variation (chromosome prints) of linkage groups 15 (P=1.37 × 10−4), 6 (P=8.69 × 10−3) and 12 (P=1.21 × 10−2). The variation associated with this QTL is highly correlated with the discrete effect of HhN (τ=0,55; P=1,09 × 10−9), which suggests that the number of yellow spots is largely controlled by a locus located close to the Yb-Sb-N/Cr/P region on chromosome 15.
The genomic position and phenotypic effect of three of the mapped major Mendelian loci was confirmed through quantitative, multivariate analysis of whole-wing variation (Figures 3aII–cII and Supplementary Figure S7). We found peaks of significant association between the variation of specific wing areas and the genomic regions described above. Our morphometric analysis allowed visualising in the form of heatmaps the genotype-to-phenotype association, which enabled a fine description of the effects associated with each identified QTL (Figures 3aII–cII). The effect of these QTLs corresponded to that controlled by HhAc/HiAc (on LG10; Figures 3aII and cII), HhN (on LG15; Figure 3aII) and HhBr/HiBr (on LG18; Figures 3aII and cII). For all but one of those QTLs, confidence intervals included markers placed on the scaffolds containing known candidate colour genes (Supplementary Figure S7). These intervals are relatively precise, extending over 8.43±7.39 cM on each chromosome.
In both species, loci HhAc and HiAc essentially control variation in black patterns situated around the forewing discal crossvein and extending into the M3-Cu1 region (Figures 3aII and cII). In situ hybridisation assays showed that WntA mRNA is expressed in the median forewing region in H. hecale larval wing disks, overlapping with the presumptive position of the HhAc/HiAc-dependent pattern variations (Figure 3aIII).
In addition, our quantitative approach highlighted that the Cm trait is mainly explained by a major QTL mapped to a narrow region around optix on LG18 (Figure 3bII and Supplementary Figure S7B). This variation is also affected, albeit to a lower extent, by a second QTL mapping near HMEL000025 on LG15. The effect of this second QTL is mainly restricted to the medial region of the hindwing, similar to the region affected by Yb in H. melpomene and close allies. We did not detect significant epistasis between these two QTLs (F13,25=1.93, P=0.08). Furthermore, the QTL on LG15 in the melicerta × clearei family explains minor yellow/black variations in the distal area of the forewing band (HhN in Figure 3bII) in the same position as locus HhN. In addition, this QTL is associated with variation in yellow apical forewing spots (Fspot) in the same melicerta × clearei family (Figure 3bII). Finally, the Hspot trait was also highlighted in the melicerta × zuleika family in association with the QTL on LG15. We found a significant epistatic interaction between this QTL on LG15 and the QTL on LG10 in the melicerta × zuleika brood (F(7,23)=3.189, P=0.02). Our morphometric analysis allowed the detection of some cases where a genomic position is associated with multiple pattern elements on the wing (Figures 3aII and bII). However, this might be caused by different genetic elements, as suggested by the ‘apparent' pleiotropy of the HhN-Hspot cluster in Figure 3aII, which may be homologous to N-Sb of H. melpomene and therefore may represent the action of distinct, tightly linked loci (see Discussion below). Therefore, we do not claim pleiotropic effects of any mapped QTL, even though such effects have previously been reported in Heliconius (Supplementary Table S1).
Several suggestive QTLs (LOD⩾3) located across eight distinct chromosomes (including LG1, LG10 and LG15) were found to be modulating pattern element variations controlled by major QTLs (Supplementary Figure S8 and Supplementary Table S10). Remarkably, we find a suggestive QTL on LG15 in the boulleti × telchinia brood, which explains the variation controlled by the minor effect locus HiN in this brood (Supplementary Figure S8C).
Discussion
H. hecale and H. ismenius bear a multilocus architecture for the control of wing patterning
Using image analysis of wing patterns and linkage mapping on the basis of dense genome-wide genotyping, we have characterised the genetic architecture of mimicry variation in two species, H. hecale and H. ismenius, belonging to the underexplored ‘silvaniform' clade of Heliconius. These approaches revealed multiple, unlinked colour loci in those species, and showed that the combination of high-density genotyping, use of a reference genome, and multivariate phenotypic analysis can yield detailed information on the genetic underpinnings of the major components of adaptive traits, as well as a sensitive description of the effect of individual QTLs on the variation of such complex traits. Mapping was based both on Mendelian characters traditionally scored by eye and on a multivariate morphometric analysis of whole-wing pattern complexity. The latter does not rely on the subjective detection of variable elements, and proved powerful to extract major components of variation from the complexity of the entire wing pattern variation.
The power and precision of a QTL analysis relies on an accurate phenotypic description, a dense array of molecular markers and a sufficient number of offspring. The limiting factor here was the number of offspring genotyped (between 29 and 42 individuals), leading to an easier detection of QTLs of large effect. We retrieved each of the individual characters scored manually (mostly of major effect), which validates the relevance of our quantitative analysis and gives credit to the additional QTLs revealed. The credible intervals were relatively narrow around each mapped QTL (Supplementary Figure S7) and (with one exception) encompassed candidate genes known from other studies. This reflects the good resolution introduced by our phenotyping, despite the low number of offspring analysed. In addition, novel candidate minor effect genomic regions were identified as suggestive QTLs (loci detected with limiting statistical power; Supplementary Table S10). One of the strengths of the method used is that it permitted whole-wing visualisation of all phenotypic changes associated with each QTL separately.
Mapped loci include both Mendelian loci and QTLs affecting relatively large wing regions strongly differentiated between the variants used in our crosses. Minor effect loci (that is, suggestive QTLs) modify pattern elements also controlled by major loci, and modulate their phenotypic effects and the resemblance to local co-mimics. These findings are consistent with theoretical expectations concerning the distribution of gene effect sizes fixed during mimicry evolution (Baxter et al., 2008). Notably, our results confirm that a multilocus architecture of wing pattern variation spread out on multiple chromosomes is a feature shared by most species in the genus (Sheppard et al., 1985; Mallet, 1989; Naisbit et al., 2003; Jones et al., 2011; Papa et al., 2013).
The wing colour architecture in H. hecale and H. ismenius is largely homologous to the architecture found in other Heliconius species
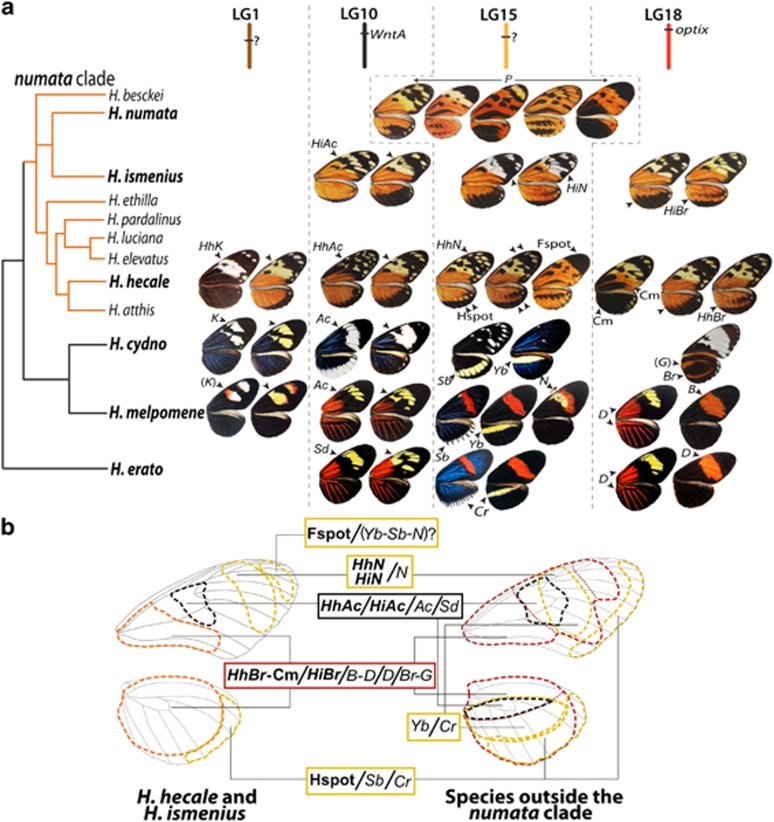
Major wing-patterning loci discovered here finely map in homologous positions to major colour loci previously identified in Heliconius, and the pattern elements and wing positions affected are generally conserved (Figure 4b). In some cases, their effects in H. hecale and H. ismenius are very similar to what is observed in other species. For instance, HhK causes a similar white/yellow switch in H. hecale as K in H. cydno and H. melpomene (Naisbit et al., 2003; Kronforst et al., 2006b; Figure 4a). Both loci map near to wingless on linkage group (LG) 1, and the combination of positional and phenotypic effect strongly argues for HhK to be the orthologue of K (Kronforst et al., 2006b; Figure 4a). K is not formally identified yet; however, our results for HhK exclude the coding region of wingless. Similarly, loci HhAc and HiAc on LG10 control melanisation of the discal region of the forewing, reminiscent of the variation controlled by Sd and Ac in other species (Figure 4, Supplementary Table S1) and identified to the WntA gene (Martin et al., 2012). Here, WntA is in perfect linkage with HhAc and HiAc (Figure 2), and its expression is markedly reduced in H. h. melicerta compared with H. h. zuleika around the discal crossvein and the adjacent M3-Cu1 and Cu2-Cu1 domains (Figure 3aIII). This strongly suggests that cis-regulatory variation of WntA expression causes the allelic effects of the HhAc and HiAc loci, revealing the molecular identity of HhAc/HiAc and confirming its homology at the gene level to one of the known ‘toolkit' colour loci in Heliconius.
Figure 4.
Conservatism and novelty in the genetic architecture underlying the diversity of Heliconius wing patterns. (a) Known genetic architectures underlying pattern diversity throughout the clade mapped onto an unscaled phylogeny. Orange tree branches represent nine of the ten species in the silvaniform clade. Major colour variation loci are located on four chromosomes (top) and control variation in similar wing regions (arrows) throughout the genus. Wing phenotypes are represented based on Holzinger and Holzinger (1994). Note that the effect of the Br locus in H. cydno is shown on the ventral side. Loci with names in brackets were described based exclusively on interspecific crosses. (b) Comparative diagram of the distribution of the gene effects across the wing for toolkit loci in the silvaniform clade (excepting H. numata; left) and in the H. melpomene and H. erato clades (right), showing the general conservatism of the regions affected by homologous elements of the multilocus architecture despite some flexibility.
In other cases, however, the phenotypic effects of toolkit loci were quite different in H. hecale and H. ismenius to their known effects in other species (see Figure 4a). The versatility in the effect of these loci across taxa is consistent with their developmental position as switch genes presumed to act relatively early in scale fate determination. Furthermore, this highlights the importance of the interaction of some components of genetic architecture in generating radically different phenotypes, despite an overall conserved multilocus architecture.
LG15 contains three linked loci related to distinct parts of the forewings and hindwings. HhN/HiN control the presence/absence of yellow elements in the forewing of the two species, and we hypothesise their homology to the H. melpomene N, also situated on LG15 and affecting a similar wing region. Other loci in other Heliconius species have been reported to affect the melanisation on the post-discal and subapical regions of the forewing (Fs and L in H. cydno, Ro in H. erato); however, they map to different chromosomes, or their location is unknown (Sheppard et al., 1985; Nijhout et al., 1990; Linares, 1996; Nadeau et al., 2014). Interestingly, locus Ro maps to LG13 in H. erato (Nadeau et al., 2014), which shows that similar wing-pattern elements can have a distinct underlying genetic basis in different species. In H. hecale, two QTLs (Hspot and Fspot) are also situated on linkage group 15. The phenotypic effect of Hspot (yellow or white spots along the hindwing margin) and its linkage to HhN suggest a homology to Sb, a locus tightly linked to N in H. melpomene. In H. numata silvana, the supergene P, situated on LG15 and presumed to contain the orthologue of Sb, also controls a very similar variation along the hindwing margin as in H. hecale. Regarding Fspot, no locus has been previously described to affect the forewing apical region, presumably because Heliconius species studied to date rarely show pattern variation in this wing region. Fspot may represent a new wing-patterning locus in Heliconius with a role for mimicry variation mainly in silvaniform species.
Despite the lack of functional analyses to pinpoint the causal gene(s) within the Yb-Sb-N/Cr/P cluster, molecular signals of selection were reported for the locus HMEL000025 (Wu et al., 2010; Nadeau et al., 2012). In H. ismenius, fine-mapping excludes the coding region of this gene from the interval for HiN (Figure 2), but does include its regulatory region, as well as other neighbouring genes. Taken together, fine-mapping and gene effects suggest that Fspot, Hspot and HhN form a cluster in H. hecale, partly homologous to the Yb-Sb-N cluster in H. melpomene, and possibly forming part of the elements participating in supergene P in H. numata (Joron et al., 2006, 2011).
Finally, LG18 contains HhBr/HiBr, mapping close to optix both in H. hecale and H. ismenius. Optix underlies the variation controlled by loci D, B-D and Br-G in H. erato, H. melpomene and H. cydno, respectively (Reed et al., 2011; Supple et al., 2013; Martin et al., 2014). Here, the conspicuous variation associated with HhBr and HiBr affect a similar wing position as Br in H. cydno (Gilbert, 2003; Figure 4a), albeit with slightly different phenotypic effects. In H. ismenius, mapping excludes the coding region of optix but includes a large intergenic region that has been proposed to contain 3′ enhancers of optix involved in its pattern-related cis-regulatory evolution (Pardo-Diaz et al., 2012; Supple et al., 2013). While a previous report failed to detect the expression of optix in the developing hindwings of H. hecale fornarina (Martin et al., 2014), this does not rule out a colour-patterning role for this gene in silvaniform species. Notably, species-specific delayed expression of optix could generate such a negative result in H. hecale. The region around optix also emerges as the one of largest effect in the melanisation of hindwings and forewings (Cm) in H. hecale, specifically at wing positions affected by the D locus in H. melpomene (Figure 4). Interestingly, hindwing melanisation is also associated with markers near HMEL000025 on LG15, especially at the position of the hindwing bar controlled by Yb in H. melpomene and H. cydno. The lack of significant epistasis between those markers may indicate that these genes largely act additively and relatively independently of each other, but could also be the result of a limited power to detect the epistatic interactions.
Epistasis and dominance are commonly used as criteria to infer gene homology between taxa (Naisbit et al., 2003). Here, we did not use these to infer homology as they show wide variations across the genus. For instance, no evident epistasis was detected between loci on LG15 and LG18, in contrast to other species (for example, N and B for the forewing submarginal band; Sheppard et al., 1985; Naisbit et al., 2003). Conversely, loci on LG10 and LG15 show epistasis in H. hecale. A similar interaction was also reported between loci Sd (LG10) and Cr (LG15) in H. erato (Mallet, 1989). These results suggest that gene interactions can differ between species and their detection depends on individual allele effects. Regarding dominance, the putative orthologues HhN (H. hecale) and HiN (H. ismenius) express codominance in different ways. Variation in dominance could be related to variation in mimicry selection pressures. Variable dominance relationships have been reported for some loci across Heliconius species (Joron et al., 2006) as well as within species (Le Poul et al., 2014).
The supergene P is restricted to H. numata and has evolved from a multilocus architecture
The multilocus architectures of mimicry found in the explored silvaniform species contrast with the single-locus architecture controlling mimicry polymorphism in H. numata. This indicates that the supergene evolved uniquely in the H. numata lineage from a multilocus architecture shared between its sister species H. ismenius and other more distant relatives.
Many Heliconius species show geographic variation in their mimicry associations and local populations are usually fixed for a given warning pattern, except in narrow hybrid zones where unlinked genes segregate freely. A few species such as H. cydno, H. hecale and H. ismenius maintain single-character polymorphisms in some parts of their range (for example, colour switch or presence/absence of a pattern element paralleling similar variation in the local mimicry community (Brown and Benson, 1974; Kapan, 2001). In contrast, H. numata shows rampant polymorphism across its entire range, involving highly differentiated wing patterns and the concerted co-variation of multiple colour elements across the wing. This polymorphism is controlled by a cluster of loci locked into a supergene, allowing the segregation of discrete mimicry types (Brown and Benson, 1974; Joron et al., 1999). Our data, therefore, argue for this peculiar architecture to be evolutionarily associated with the maintenance of polymorphism in H. numata and further confirm that balancing selection might be shaping the genetic architecture of wing patterns in this species.
Interestingly, single-locus architecture is not associated with a specific type of wing pattern, as both H. hecale and H. ismenius have ‘tiger-patterns' very similar to some of the H. numata forms (see Figure 4a). For instance, H. numata silvana, associated with the ancestral allelic class of the H. numata supergene (Joron et al., 2011), is phenotypically similar to H. ismenius boulleti and H. hecale melicerta. H. numata also participates in mimicry with H. hecale or closely related species in many parts of its range (Brown, 1981). Mimicry evolution can therefore involve distinct genetic architectures, even though some of the loci may be homologous (Jones et al., 2011). Recent research has revealed that similar phenotypes may not always show a parallel genetic basis at the nucleotide or gene level (Arendt and Reznick, 2008; Manceau et al., 2010; Elmer and Meyer, 2011). However, to our knowledge, no cases have been reported where different genetic architectures, in terms of linkage and gene effect size, underlie highly similar phenotypic variations.
Our mapping shows that the Heliconius ‘toolkit' of colour genes is used throughout the H. numata clade, where only the supergene architecture was previously known. Within H. numata, the large-effect toolkit loci not associated with the supergene play a minor role in pattern variation (Jones et al., 2011). The contrasting genetic architectures observed when comparing H. numata with other silvaniform species do not relate to differences in the identity of the colour loci themselves, but rather to large variations in the effect size of the loci participating in determining wing patterning. The increase in linkage between elements controlling different regions on the wings may explain the build-up of a large effect supergene in H. numata. As previously suggested, the H. melpomene loci Yb, N and Sb may be the orthologues of some elements composing the supergene in H. numata (Joron et al., 2006, 2011), and our data also suggest the existence of a gene cluster at this position in H. hecale. Those elements control variation in distinct regions of the wing (Figure 4b), and their co-variation in response to mimicry may participate to an initial build-up of co-adapted clusters in this region, later locked into a supergene in H. numata within the ~400-kb genomic region where recombination is suppressed by inversions (cf. the ‘sieve' hypothesis; Turner, 1977; Joron et al., 2011). This may be consistent with the observation that large gene effects may often result from the aggregation of independent small effect mutations (Martin and Orgogozo, 2013).
Conservation of genetic architectures does not constrain adaptation
Our results extend our knowledge of the homology of wing colour loci implicated in different adaptive radiations in the genus Heliconius, and show how shared genetic architectures are implicated both in mimicry convergence between species and in the diversification associated with local adaptation (Joron et al., 2006; Papa et al., 2008; Reed et al., 2011; Martin et al., 2012). The toolkit of Heliconius wing-patterning genes therefore stands as an ancestral architecture shared by species with radically different wing patterns and exposed to different mimicry selection pressures. If mimetic wing patterns are considered as an integrated complex trait, variation in the distribution of individual effect sizes and interactions among the contributing genes across the radiation demonstrates how profoundly malleable these traits are. Using a conserved set of switch genes, novel phenotypes appear to be explored via the effects of those genes on phenotypic variation, presumably through an evolution of the downstream wiring with effector genes, and the possible involvement of new modifiers, rather than by the recruitment of new switch genes. The conservatism in the wing-patterning toolkit does not appear to impose strict limits on the evolution of novel phenotypes, highlighting the power of selection regimes in bringing populations to local adaptive optima.
Here, we have mainly focused on primary components of genetic architecture such as number of loci, genome position and the distribution of gene effect sizes. However, the mapping populations restricted our capacity to investigate aspects related to interactions such as epistasis, dominance and pleiotropy, which may also respond to selection and contribute to the complexity of the adaptive responses (Le Poul et al., 2014). We encourage the use of detailed morphometric quantification of pattern variation on large mapping populations to examine the interaction of the components of genetic architecture and their role in adaptive evolution.
Data archiving
RAD-sequencing paired-end FASTQ files have been submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under accession number PRJEB8625. Input files used for QTL analysis are available in the Dryad repository (http://dx.doi.org/10.5061/dryad.n893m).
Acknowledgments
We thank Adriana Tapia, Andrés Orellana, Cristóbal Ríos, Catherine Brunton, Moisés Abanto, Elizabeth Evans, Nicola Nadeau and Patricio Salazar for help with butterfly collection and breeding; the Autoridad Nacional del Ambiente (Panamá) for collection permits; Lise Frézal, The Gene Pool (Edinburgh Genomics), the BoEM lab (MNHN) and the SSM (MNHN) for assistance with molecular work; Marcus Kronforst for providing primers; John Davey, Kanchon Dasmahapatra, Manmohan Sharma and Robert Jones for assistance with sequence analysis and bioinformatics; Violaine Llaurens and Chris Jiggins for help and comments on the manuscript. We are most grateful to the three anonymous referees whose in-depth reviews improved this work. This work was funded by a Smithsonian Tropical Research Institute short-term fellowship to BH, AM and BG and by a CNRS ATIP grant, an ERC grant (MimEvol) and an ANR grant (HybEvol) to MJ.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Arendt J, Reznick D. (2008). Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol Evol 23: 26–32. [DOI] [PubMed] [Google Scholar]
- Barton NH. (1995). A general model for the evolution of recombination. Genet Res 65: 123–145. [DOI] [PubMed] [Google Scholar]
- Baxter SW, Johnston SE, Jiggins CD. (2008). Butterfly speciation and the distribution of gene effect sizes fixed during adaptation. Heredity 102: 57–65. [DOI] [PubMed] [Google Scholar]
- Broman K, Sen S. (2009) A Guide to QTL Mapping with R/qtl. Springer: New York. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. (2003). R/qtl: QTL mapping in experimental crosses. Bioinforma Oxf Engl 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Brown KS. (1981). The biology of Heliconius and related genera. Annu Rev Entomol 26: 427–457. [Google Scholar]
- Brown KS Jr, Benson WW. (1974). Adaptive polymorphism associated with multiple Müllerian mimicry in Heliconius numata (Lepid. Nymph.). Biotropica 6: 205–228. [Google Scholar]
- Browning BL, Browning SR. (2009). A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84: 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AJR, Hermisson J, Hansen TF. (2005). The role of epistatic gene interactions in the response to selection and the evolution of evolvability. Theor Popul Biol 68: 179–196. [DOI] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. (2013). Stacks: an analysis tool set for population genomics. Mol Ecol 22: 3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Visser JA, Hoekstra RF, van den Ende H. (1997). An experimental test for synergistic epistasis and its application in Chlamydomonas. Genetics 145: 815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer KR, Meyer A. (2011). Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol Evol 26: 298–306. [DOI] [PubMed] [Google Scholar]
- Ferguson L, Lee SF, Chamberlain N, Nadeau N, Joron M, Baxter S et al. (2010). Characterization of a hotspot for mimicry: assembly of a butterfly wing transcriptome to genomic sequence at the HmYb/Sb locus. Mol Ecol 19: 240–254. [DOI] [PubMed] [Google Scholar]
- Gilbert LE. (2003). Adaptive novelty through introgression in Heliconius wing patterns: evidence for shared genetic ‘tool box' from synthetic hybrid zones and a theory of diversification. In: Boggs CL, Watt WB, Ehrlich PR (eds). Butterflies: Ecology and Evolution Taking Flight. University of Chicago Press: Chicago. [Google Scholar]
- Hansen TF. (2006). The evolution of genetic architecture. Annu Rev Ecol Evol Syst 37: 123–157. [Google Scholar]
- Heliconius Genome Consortium. (2012). Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger H, Holzinger R. (1994) Heliconius and Related Genera. Lepidoptera: Nymphalidae. The Genera Eueides, Neruda and Heliconius. Sciences Nat.: Vennette, France. 328 pages. [Google Scholar]
- Jiggins CD, Mavarez J, Beltrán M, McMillan WO, Johnston JS, Bermingham E. (2005). A genetic linkage map of the mimetic butterfly Heliconius melpomene. Genetics 171: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins CD, Naisbit RE, Coe RL, Mallet J. (2001). Reproductive isolation caused by colour pattern mimicry. Nature 411: 302–305. [DOI] [PubMed] [Google Scholar]
- Jiggins CD, McMillan WO. (1997). The genetic basis of an adaptive radiation: warning colour in two Heliconius species. Proc R Soc B Biol Sci 264: 1167–1175. [Google Scholar]
- Jones RT, Salazar PA, ffrench-Constant RH, Jiggins CD, Joron M. (2011). Evolution of a mimicry supergene from a multilocus architecture. Proc Biol Sci 279: 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Frezal L, Jones RT, Chamberlain NL, Lee SF, Haag CR et al. (2011). Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477: 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Papa R, Beltrán M, Chamberlain N, Mavárez J, Baxter S et al. (2006). A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. PLoS Biol 4: e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Wynne IR, Lamas G, Mallet J. (1999). Variable selection and the coexistence of multiple mimetic forms of the butterfly Heliconius numata. Evol Ecol 13: 721–754. [Google Scholar]
- Kapan DD. (2001). Three-butterfly system provides a field test of müllerian mimicry. Nature 409: 338–340. [DOI] [PubMed] [Google Scholar]
- Kronforst MR, Kapan DD, Gilbert LE. (2006. a). Parallel genetic architecture of parallel adaptive radiations in mimetic Heliconius butterflies. Genetics 174: 535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst MR, Young LG, Kapan DD, McNeely C, O'Neill RJ, Gilbert LE. (2006. b). Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proc Natl Acad Sci USA 103: 6575–6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lair KP, Bradshaw WE, Holzapfel CM. (1997). Evolutionary divergence of the genetic architecture underlying photoperiodism in the pitcher-plant mosquito, Wyeomyia Smithii. Genetics 147: 1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. (1980). The genetic covariance between characters maintained by pleiotropic mutations. Genetics 94: 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poul Y, Whibley A, Chouteau M, Prunier F, Llaurens V, Joron M. (2014). Evolution of dominance mechanisms at a butterfly mimicry supergene. Nat Commun 5: 5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares M. (1996). The genetics of the mimetic coloration in the butterfly Heliconius cydno weymeri. J Hered 87: 142–149. [Google Scholar]
- Lunter G, Goodson M. (2011). Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21: 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. (1989). The genetics of warning colour in peruvian hybrid zones of Heliconius erato and H. melpomene. Proc R Soc Lond B Biol Sci 236: 163–185. [Google Scholar]
- Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE. (2010). Convergence in pigmentation at multiple levels: mutations, genes and function. Philos Trans R Soc B Biol Sci 365: 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, McCulloch KJ, Patel NH, Briscoe AD, Gilbert LE, Reed RD. (2014). Multiple recent co-options of Optix associated with novel traits in adaptive butterfly wing radiations. EvoDevo 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Orgogozo V. (2013). The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67: 1235–1250. [DOI] [PubMed] [Google Scholar]
- Martin A, Papa R, Nadeau NJ, Hill RI, Counterman BA, Halder G et al. (2012). Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc Natl Acad Sci USA 109: 12632–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Ruiz M, Salazar P, Counterman B, Medina JA, Ortiz-Zuazaga H et al. (2014). Population genomics of parallel hybrid zones in the mimetic butterflies, H. melpomene and H. erato. Genome Res 24: 1316–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Whibley A, Jones RT, Davey JW, Dasmahapatra KK, Baxter SW et al. (2012). Genomic islands of divergence in hybridizing Heliconius butterflies identified by slarge-scale targeted sequencing. Philos Trans R Soc Lond B Biol Sci 367: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbit RE, Jiggins CD, Mallet J. (2003). Mimicry: developmental genes that contribute to speciation. Evol Dev 5: 269–280. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Wray GA, Gilbert LE. (1990). An analysis of the phenotypic effects of certain colour pattern genes in Heliconius (Lepidoptera: Nymphalidae). Biol J Linn Soc 40: 357–372. [Google Scholar]
- Orr HA. (1998). The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52: 935–949. [DOI] [PubMed] [Google Scholar]
- Papa R, Kapan DD, Counterman BA, Maldonado K, Lindstrom DP, Reed RD et al. (2013). Multi-allelic major effect genes interact with minor effect QTLs to control adaptive color pattern variation in Heliconius erato. PLoS ONE 8: e57033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa R, Martin A, Reed RD. (2008). Genomic hotspots of adaptation in butterfly wing pattern evolution. Curr Opin Genet Dev 18: 559–564. [DOI] [PubMed] [Google Scholar]
- Pardo-Diaz C, Salazar C, Baxter SW, Merot C, Figueiredo-Ready W, Joron M et al. (2012). Adaptive introgression across species boundaries in Heliconius Butterflies. PLoS Genet 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C et al. (2011). optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333: 1137–1141. [DOI] [PubMed] [Google Scholar]
- Sheppard PM, Turner JRG, Brown KS, Benson WW, Singer MC. (1985). Genetics and the evolution of muellerian mimicry in Heliconius Butterflies. Philos Trans R Soc Lond B Biol Sci 308: 433–610. [Google Scholar]
- Stern DL. (2013). The genetic causes of convergent evolution. Nat Rev Genet 14: 751–764. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. (2007). Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity 100: 158–170. [DOI] [PubMed] [Google Scholar]
- Supple MA, Hines HM, Dasmahapatra KK, Lewis JJ, Nielsen DM, Lavoie C et al. (2013). Genomic architecture of adaptive color pattern divergence and convergence in Heliconius butterflies. Genome Res 23: 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MJ, Jiggins CD. (2014). Supergenes and their role in evolution. Heredity 113: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR. (1977). Butterfly mimicry: the genetical evolution of an adaptation. Evol Biol 10: 163–206. [Google Scholar]
- Turner JRG, Sheppard PM. (1975). Absence of crossing-over in female butterflies (Heliconius. Heredity 34: 265–269. [DOI] [PubMed] [Google Scholar]
- Van Oijen JW, Voorrips RE. (2001). Joinmap Version 3.0, software for the calculation of genetic linkage maps. Plant Res Int. Wageningen, the Netherlands. [Google Scholar]
- Wu GC, Joron M, Jiggins CD. (2010). Signatures of selection in loci governing major colour patterns in Heliconius butterflies and related species. BMC Evol Biol 10: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.