Abstract
RAD-tag sequencing is a promising method for conducting genome-wide evolutionary studies. However, to date, only a handful of studies empirically tested its applicability above the species level. In this communication, we use RAD tags to contribute to the delimitation of species within a diverse genus of deep-sea octocorals, Chrysogorgia, for which few classical genetic markers have proved informative. Previous studies have hypothesized that single mitochondrial haplotypes can be used to delimit Chrysogorgia species. On the basis of two lanes of Illumina sequencing, we inferred phylogenetic relationships among 12 putative species that were delimited using mitochondrial data, comparing two RAD analysis pipelines (Stacks and PyRAD). The number of homologous RAD loci decreased dramatically with increasing divergence, as >70% of loci are lost when comparing specimens separated by two mutations on the 700-nt long mitochondrial phylogeny. Species delimitation hypotheses based on the mitochondrial mtMutS gene are largely supported, as six out of nine putative species represented by more than one colony were recovered as discrete, well-supported clades. Significant genetic structure (correlating with geography) was detected within one putative species, suggesting that individuals characterized by the same mtMutS haplotype may belong to distinct species. Conversely, three mtMutS haplotypes formed one well-supported clade within which no population structure was detected, also suggesting that intraspecific variation exists at mtMutS in Chrysogorgia. Despite an impressive decrease in the number of homologous loci across clades, RAD data helped us to fine-tune our interpretations of classical mitochondrial markers used in octocoral species delimitation, and discover previously undetected diversity.
Introduction
The advent of next-generation sequencing tools has permitted significant advances in our understanding of evolutionary processes such as speciation (for example, Ekblom and Galindo, 2011), but some other practical applications of genomic data have been less explored, including phylogenomics and species delimitation. Among genomic approaches that are applicable to these fields, the usefulness of restriction site-associated DNA tag (RAD tag; Baird et al., 2008) sequencing has been investigated in few studies to date. This methodology typically provides short sequences (~100–150 bp) flanking the cut sites of a restriction enzyme (or several enzymes), generally yielding thousands of loci distributed throughout the genome. This approach does not require a reference genome and can therefore be applied to non-model organisms. However, some technical difficulties remain for groups where very little genomic knowledge is available (see Davey et al., 2011). For instance, the choice of restriction enzyme(s) and methodology (single-digest versus double-digest RAD) is key to estimating the number of expected cut sites and coverage, but relies on prior knowledge of genome size and GC content.
Despite these difficulties, RAD-tag sequencing constitutes one of the reduced genomic approaches that are suitable for investigating interspecific evolutionary questions. Published RAD-tag sequencing research beyond the species level includes in silico studies (Drosophila, mammals and yeasts in Rubin et al., 2012; Drosophila in Cariou et al., 2013) and empirical work (for example, Restionaceae flowering plants in Lexer et al., 2013; cetaceans in Viricel et al., 2014), which both suggest this approach is promising for taxa having diverged up to 60 million years (Myr) ago. For instance, RAD-tag sequencing has proven useful in species delimitation and phylogenies within recently and rapidly diverged groups (for example, Orobanchaceae flowering plants in Eaton and Ree, 2013; swordtails in Jones et al., 2013; Heliconius butterflies in Nadeau et al., 2013; cichlids in Wagner et al., 2013; geckos in Leaché et al., 2014). Comparatively, reconstructing the phylogeny of more distantly related taxa has been the topic of two study (Carabus beetles, Cruaud et al., 2014; oak trees, Hipp et al., 2014). Herein, we use this approach on a group of deep-sea octocorals for which little genomic data are available. Thus, our contribution constitutes one of the first studies investigating the use of RAD-tag sequencing for practical species delimitation within a taxonomic group composed of divergent species (up to 16 Myr ago).
Deep-sea octocorals are one of the groups for which RAD-tag sequencing can significantly advance our understanding of evolutionary patterns. As for shallow-water octocorals, deep-water octocorals present significant challenges for taxonomists, with few morphological characters being available for species delimitation (for example, McFadden et al., 2010). In addition, several studies have shown conflicting patterns of morphological and molecular data (France, 2007; Dueñas and Sánchez, 2009; Pante and France, 2010), suggesting that an integrative approach to taxon delimitation must be applied in this group (for example, Schlick-Steiner et al., 2010). Octocorals, as with other anthozoans (for example, scleractinians and sea anemones), are also plagued with remarkably low levels of mitochondrial genome evolution that renders the use of classical barcoding gene regions such as cox1 of limited use (McFadden et al., 2011). Comparatively, a few studies have successfully used nuclear markers within octocoral species (for example, Concepcion et al., 2008; Mokhtar-Jamaï et al., 2011), but these are either not widely useable across octocorals (for example, SRP54; France and Pante, unpublished observations), or not informative at multiple phylogenetic scales (for example, microsatellites). Multicopy markers have been employed (for example, Herrera et al., 2010); however, their use implies that lack of concerted evolution within and across genomes will not blur the phylogenetic signal (Vollmer and Palumbi, 2004; Calderón et al., 2006). In this group, RAD-tag genotyping may therefore offer a panel of markers to help describe patterns of population structure, delimit species and investigate phylogenetic relationships. This technique may however be difficult to implement in this group. Indeed, the composition of the deep-sea octocoral genome is unknown (size, GC content, prevalence of cut sites for restriction enzymes and so on); the size of known cnidarian genomes, for instance, varies between 224 Mb and 1.8 Tb (Animal Genome Size Database; Gregory, 2014). In addition, sampling of deep-sea animals can be associated with a loss of quality of genomic DNA samples, particularly when sampling in tropical waters using trawls or dredges.
The genus Chrysogorgia (Calcaxonia: Chrysogorgiidae) is a noteworthy model for testing the utility of RAD sequencing for delimiting octocoral species, as it is diverse (62 nominal species described, 93% of which were based solely on morphology), widely distributed and can be locally abundant (Watling et al., 2011). The large geographic, bathymetric and ecological distributions of some Chrysogorgia species (Pante et al., 2012b) question whether taxa are appropriately delimited, and whether cryptic diversity is important in the group. In the northwestern Atlantic, congruence exists between morphological and genetic data, suggesting that a relatively short fragment of the mitochondrial mtMutS gene can be used to formulate ‘Primary Species Hypotheses' (Pante and Watling, 2012). It is suspected that little to no intraspecific variation exists for this marker within the group (McFadden et al., 2011), but the null hypothesis that single mutations at mtMutS are diagnostic of species limits must be evaluated using genetic data from markers informative within and above the species level. RAD loci allow to test whether lineages that putatively belong to different species do not exchange genes.
In this communication, we test the utility of RAD-tag genotyping for delimiting species in Chrysogorgia using the genealogical criterion defined by Taylor et al. (2000). More specifically, we test whether single mutations on the mitochondrial mtMutS gene can be used as a criterion for grouping Chrysogorgia colonies into separate, putative species (or, more specifically, ‘Primary Species Delimitation hypotheses' as in Puillandre et al., 2012). We compare the results from two analysis pipelines, Stacks (Catchen et al., 2013) and PyRAD (Eaton, 2014), which significantly differ in the method employed for detecting homologous loci.
Materials and methods
Specimen collection and mtDNA typing
Chrysogorgia specimens were collected from the SE slope of New Caledonia (NC) and adjacent seamounts of the Norfolk Ridge (82 colonies; Terrasses cruise, 2008), from Papua New Guinea (PNG; 8 colonies; BioPapua cruise, 2010) and from the northwestern Atlantic (1 colony, Extreme Coral 2010 cruise; Table 1 and Supplementary Table S1). Pacific specimens were retrieved from dredges and trawls (details on cruises of the Tropical Deep Sea Benthos research program: Bouchet et al., 2008; details on the BioPapua cruise: Pante et al., 2012a); the Atlantic specimen was collected using the Jason II ROV (Woods Hole Oceanographic Institution). Specimens were fixed in 80% ethanol as soon as possible after collection. Genomic DNA was extracted using a CTAB protocol according to France et al. (1996). A 700-bp fragment of the mitochondrial mtMutS gene (identified as more informative than cox1 or 18S in chrysogorgiids, Pante et al., 2012b) was amplified using the ND4L2475F–MUT3458R primer pair and sequenced using an ABI PRISM (R) 3100 or 3130xl Genetic Analyzer (primer information, PCR and sequencing conditions: Pante et al., 2012b). Sequences were checked for quality and edited in Sequencher (TM) 4.7 (Gene Codes), aligned by eye (a single, 3-bp indel was present in the alignment) and haplotypes were submitted to GenBank (Supplementary Table S1). Divergence times among putative species were estimated using the molecular clock from Lepard (2003), which was calculated for the shallow-water octocoral genus Leptogorgia based on mtMutS genetic distances for clades located on either sides of the Isthmus of Panama (0.14–0.25% per Myr).
Table 1. Summary table of haplotype information (sample size, geographical spread, depth range, habitat (seamounts vs slopes) and mtMutS vs RAD delimitation.
| Haplotype | No. of colonies | Geography | Habitat | Depth range (m) | Delimitation |
|---|---|---|---|---|---|
| J | 1 | Atlantic | Slope | 627–627 | mtMutS/RAD congruence |
| 2 | 11 | NC | Slope | 390–500 | mtMutS/RAD incongruence |
| 4 | 20 | NC | Slope and seamount | 150–330 | mtMutS/RAD congruence |
| 6 | 2 | NC | Seamount | 270–310 | mtMutS/RAD congruence |
| 7 | 8 | NC-PNG | Slope and seamount | 300–880 | mtMutS/RAD incongruence |
| 8 | 20 | NC | Slope | 390–500 | mtMutS/RAD incongruence |
| 9 | 18 | NC | Slope | 390–450 | mtMutS/RAD congruence |
| 10 | 3 | NC | Slope and seamount | 458–880 | mtMutS/RAD congruence |
| 11 | 3 | NC | Seamount | 750–840 | mtMutS/RAD congruence |
| 13 | 1 | NC | Slope | 460–490 | mtMutS/RAD incongruence |
| 14 | 1 | NC | Slope | 400–420 | mtMutS/RAD congruence |
| 30 | 3 | PNG | Slope | 220–1020 | mtMutS/RAD congruence |
Abbreviations: NC, New Caledonia, PNG, Papua New Guinea.
Library construction, RAD sequencing and quality control
Genomic DNA quality was evaluated by 1% agarose gel electrophoresis and quantified using a Thermo Scientific (Waltham, MA, USA) Nanodrop ND-1000 spectrophotometer. DNA was sent to Eurofins Genomics (Ebersberg, Germany) for RAD-tag library preparation and sequencing. Libraries were constructed from 1–2 μg of DNA per colony using the SbfI restriction enzyme. This enzyme was chosen because it was successfully used in RADseq experiments with marine invertebrates (sea anemones, Reitzel et al., 2013; abalone, Gruenthal et al., 2014), and was expected to allow an acceptable compromise between prevalence of cut sites and depth of coverage, based on RADcounter (the University of Edinburgh, https://www.wiki.ed.ac.uk/display/RADSequencing/Home). As the genome size and GC content of Chrysogorgia (or other octocorals, to the best of our knowledge) are not known, we estimated the prevalence of SbfI cut sites based on a range of genome sizes and GC content, based on information from the Animal Genome Size Database (see Introduction) and with a GC content of 40% (for example, Soza-Ried et al., 2010). Barcodes 6–9 nt long and differing by at least 2 nt were used to differentiate multiplexed samples (Supplementary Table S1). Sequencing was performed on two lanes of the Illumina (R) HiSeq (TM) 2000 instrument (Illumina Inc., San Diego, CA, USA) using the single read, 100 nucleotide configuration. Raw HiSeq output was processed using the CASAVA v1.8.2 software pipeline (Illumina Inc.) and demultiplexed and quality filtered using the process_radtags.pl module (default quality settings) of the Stacks v.0.99994 pipeline (Catchen et al., 2013). A single sequencing error was tolerated in the barcode. Reads were truncated to 91 nt. Quality (as measured by phred scores and percentage of sequence overrepresentation) was checked before and after treatment by process_radtags using FastQC v.0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Exploration of the divergence parameter space
Two main pipelines specifically designed for analysis of RADseq data are currently available. The most used to date is the Stacks pipeline. It constructs a catalog of loci for a set of samples mainly based on three parameters: the minimum stack depth parameter m (that is, the minimum number of reads allowed per allele), the intraindividual divergence parameter M (that is, the maximum number of mutations that can be observed between stacks within a sample), and the interindividual divergence parameter n (that is, the maximum number of mutations that can be observed between loci across samples).
PyRAD (Eaton, 2014) is a more recently developed pipeline and differs from Stacks in several ways, the most important one being that it allows the presence of insertions and deletions (indels), as the clustering process of reads into loci uses alignment tools. This is anticipated to be an advantage compared with the first pipeline when considering more phylogenetically distant species. PyRAD relies on a large number of parameters used at different steps of the process. Most of them are related to reads quality control, detection of homology and filtering of paralogs. Two main parameters are of particular importance: the minimum depth coverage Mindepth (minimum depth necessary to make a statistical base call at each position of a cluster) and the similarity threshold Wclust (similarity value to be used for the alignment during both the within and across-sample clustering).
For both pipelines, these parameter settings are expected to influence greatly the number of markers available for intra- and interspecific comparisons and it is necessary to explore which parameter combinations maximize the number of orthologous loci (Viricel et al., 2014). To explore the effect of these parameters at different phylogenetic depths, we randomly selected pairs of specimens that (1) were separated by 0–16 mutations at mtMutS (representing different levels of phylogenetic divergence) and (2) were characterized by 1–1.5 million reads (to alleviate potential effects of depth of coverage on the number of assembled loci). For each level of divergence, we used three replicate pairs of specimens. We refer to specimens with mtMutS haplotypes differing by few mutations as pairs of closely related colonies, and those with haplotypes differing by many mutations as distantly related colonies.
In Stacks, m was kept to 3 (the default value); M was incremented from 1 to 10 in two cases (specimens separated by 0 and 12 mutations at mtMutS) and from 1 to 7 in all other cases. Similarly, n was incremented from 1 to 10 (0 and 12 mutations cases) and from 1 to 8 (all cases). All combinations of M and n were not tested: only similar values of M and n were used together (two settings were used: M=n and M+1=n), as to (1) keep maximum levels of intra- and interindividual divergence levels close and (2) keep the number of Stacks analyses to a reasonable number. A total of 408 Stacks catalog construction tests were therefore performed using the denovo_map.pl script available in Stacks. Catalogs were parsed with the populations.pl script, where each sample was considered as a separate population, no missing data were allowed and a minimum of 10 reads per single-nucleotide polymorphism (SNP) was set.
In PyRAD v. 2.0, combinations of two values for Mindepth (3 and 6) and 3 values for Wclust (0.89, 0.93 and 0.96) were tested, resulting in 156 analyses. For these analyses, the maximum number of sites per read with a quality <20 (NQual) was set to 4, the minimum number of samples in a final locus (MinCov) was set to 1 and the maximum proportion of shared polymorphic sites in a locus (MaxSH) was set to 10%. For this last parameter, which aims at detecting paralogs, preliminary tests showed that in our case, changing this value did not drastically affect the number of loci and SNPs detected. Finally, optional parameters were kept to default values.
Comparison of Stacks and PyRAD
To evaluate what proportion of loci was detected by both PyRAD and Stacks, a custom BLASTN search was performed (BLAST toolkit v. 2.2.25; Zhang et al., 2000). Local BLAST databases were constructed using PyRAD sequences (locus file containing consensus sequences for each individual; PyRAD parameters m=6 and Wclust=93 and 89%) for three groups of specimens with different numbers of reads (Table 2). Stacks loci for these specimens (based on the locus file produced by the populations script, for which a single allele was retained per locus; denovo_map parameters m=3, M=4, n=4, and m=3, M=10, n=12) were then compared with the PyRAD database using BLASTN (percent identity set to 93 and 89%, word size 80 and 84 nt, ungapped alignments). The XML output of BLASTN searches was then parsed in bash using grep.
Table 2. Results of the BLASTN alignments performed between Stacks and PyRAD sequences.
| Specimen | Haplotype | Read category | No. of reads (M) |
89% Divergence |
93% Divergence |
||||
|---|---|---|---|---|---|---|---|---|---|
| No. of loci (PyRAD) | No. of loci (Stacks) | Intersect (%) | No. of loci (PyRAD) | No. of loci (Stacks) | Intersect (%) | ||||
| TER2044 | 11 | High | 5.82 | 6580 | 866 | 7.84 | 6720 | 607 | 5.54 |
| JAC1018 | J | High | 5.49 | 3305 | 1851 | 24.57 | 2717 | 1202 | 21.46 |
| TER7092 | 7 | High | 4.04 | 6867 | 1363 | 13.03 | 6862 | 1246 | 11.40 |
| TER130424 | 9 | Median | 1.61 | 6151 | 1198 | 12.73 | 6323 | 850 | 8.86 |
| TER13064 | 8 | Median | 1.61 | 6876 | 4183 | 39.89 | 6584 | 4607 | 42.72 |
| TER13087 | 9 | Median | 1.60 | 5959 | 1131 | 13.81 | 6189 | 821 | 9.26 |
| TER11101 | 4 | Low | 0.09 | 1046 | 228 | 1.15 | 944 | 138 | 0.64 |
| TER13047 | 9 | Low | 0.08 | 1145 | 396 | 9.96 | 1107 | 297 | 8.67 |
| TER11108 | 4 | Low | 0.04 | 441 | 50 | 2.49 | 384 | 32 | 1.04 |
The number of loci detected within nine individuals (with high-, medium- and low-read numbers) is presented for the two analyses performed on the entire set of 91 specimens. The number of quality-filtered reads is given in million.
Phylogenetic reconstruction and species delimitation
RAxML v. 8.0.9 (Stamatakis, 2006; Stamatakis et al., 2008) was used on the CIPRES Portal (Miller et al., 2010) to infer phylogenetic relationships among Chrysogorgia colonies, based on mitochondrial and nuclear sequences, using the GTRCATI model and automating boot-stopping. The mitochondrial phylogeny was inferred from the first 700 nt of the mtMutS gene (see above); the nuclear phylogeny was inferred using concatenated RAD loci obtained based on two parameter sets in Stacks, and one parameter set in PyRAD. The first Stacks set (‘m3M4n4', denovo_map parameters m=3, M=4, n=4; populations script parameters m=6, P=2, r=0.5) corresponds to parameters that maximize the total number of loci detected while minimizing the divergence parameters (see ‘Exploration of the divergence parameter space' section above). For this analysis, each mtMutS haplotype was considered as a separate population. The Stacks populations script parameters that were used signify that 50% missing data were allowed within each population, a locus had to be present in at least two populations to be included in the output and a minimum of six reads per SNP was required. The second Stacks set (‘m3M10n12', Stacks script denovo_map parameters m=3, M=10, n=12; populations script parameters m=6, P=2, r=0.5) allowed more divergence between loci. The PyRAD data set (‘m6s93') was constructed with m=6 and Wclust=93% (details above). In all analyses, the Atlantic colony JAC1018 was used as the outgroup.
Once clades were delimited with RAxML, a Discriminant Analysis on Principal Components (DAPC, Jombart et al., 2010) was used to explore genetic structure within three clades represented by 18–31 colonies (see below). This method takes into account the multilocus genotype of each individual and forms clusters based on genetic similarity without considering a model of evolution. We also used TESS (Durand et al., 2009) to investigate population structure using the conditional auto-correlative admixture model with a spatially explicit, Bayesian framework. In TESS, the deviance information criterion was used to compare population structure in the presence of different numbers of clusters (the maximum number of cluster K was set to the total number of individual in the tested clade; for example, K was set from 2 to 18 for clade 1). Five replicate runs were used per K, with 1200 MCMC steps and a 200-step burn-in. The best K was determined by minimizing deviance information criterion and its variance; once the best K was determined, a longer analysis with 12 000 steps and a 2000-step burn-in was run to obtain reliable individual assignments. The populations script in Stacks was rerun to keep only one SNP per locus, to minimize the probability of coanalyzing linked markers. The Stacks m3M4n4 data set was chosen for these analyses for two reasons: (1) the DAPC and TESS analyses are run within clades at shallow phylogenetic depths and (2) as only one SNP/locus is retained, divergence level should be kept minimal to prevent the inclusion of non-homologous loci. The DAPC analysis was run using adegenet in R (Jombart, 2008; R Development Core Team, 2014).
Results
Mitochondrial typing and RAD-tag sequencing
A total of 12 mtMutS haplotypes were detected among the 91 colonies investigated, 10 of which were from NC, 3 from PNG, 1 from the northwestern Atlantic and 2 being shared between NC and PNG. The biogeography of these mitochondrial haplotypes at these locations is further discussed in Pante et al. (2012a, 2012b). A total of 236 million raw reads, corresponding to 35 463 Mbp were produced on two HiSeq2000 lanes. The number of quality-filtered reads (in millions) per colony varied between 0.04 (TER11108) and 5.82 (TER2044), with a median of 1.6. There was a significant correlation between the number of quality-filtered reads per colony and haplotypes (Kruskal–Wallis X2-test=25.11, df=13, P=0.02), haplotypes 6 and 10, for instance, yielded fewer reads than other haplotypes (haplotype-10 colonies were sampled from depths down to 880 m, and haplotype-6 colonies had remarkably small polyps that may have been particularly sensitive to prolonged times to preservation).
Loci, SNPs and indel cataloging using Stacks and PyRAD
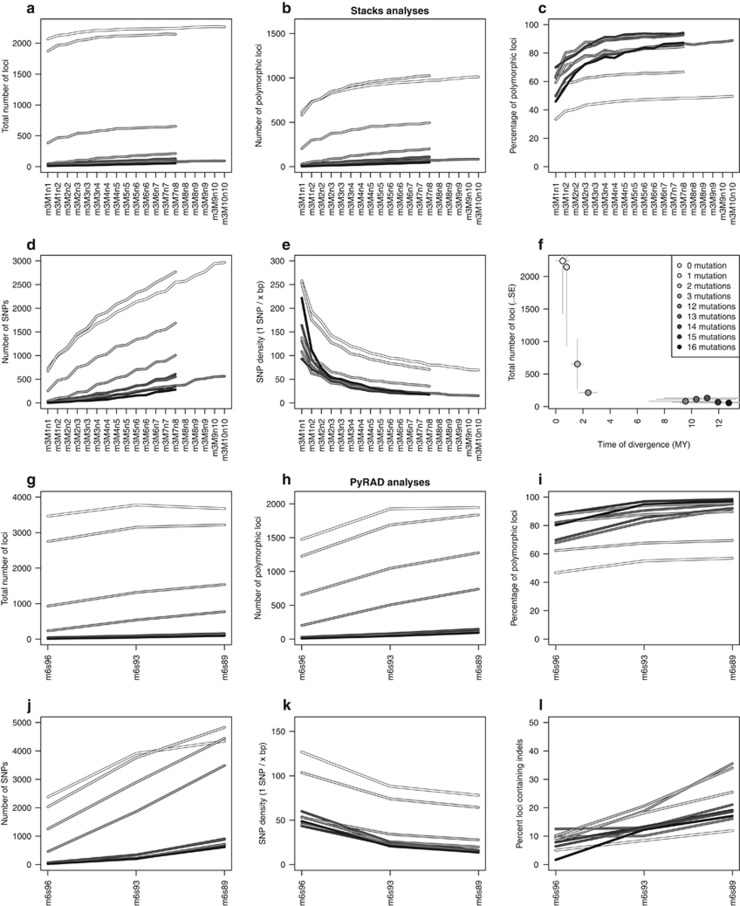
Results from both pipelines (Stacks and PyRAD) show variations in the number of loci and SNPs depending on the set of parameters used (Figures 1a–e and 1g–k), as well as the mitochondrial genetic distance between samples (Figure 1f). For Stacks, as the mitochondrial genetic distance among included samples decreases, both the total number of loci and the number of polymorphic loci increases (Figures 1a and b). The former ranges from a few loci to more than 2000, whereas the latter ranges from a few loci to ~1000, depending on the set of parameters used. When related to the time of divergence (in Myr, based on mtDNA), the total number of loci obtained decreases exponentially (Figure 1f). Inversely, the percentage of polymorphic loci is lower for more closely related colonies (~40%) than for distantly related colonies (~90% Figure 1c). These three measures (number of loci, number of polymorphic loci and percentage of polymorphic loci) show the same response to an increase in divergence parameters M and n, namely a rapid increase followed by a plateau. This plateau is reached for the m3M4n4 set of parameters. Conversely, the number of SNPs increases drastically without reaching a plateau, from a few SNPs for the most stringent set of parameters and the most distantly related colonies to around 3000 for the most closely related colonies and the most relaxed set of parameters (Figure 1d). Thus, the effect of increasing mitochondrial genetic distance among samples or decreasing stringency of parameters is to increase SNPs densities, from one SNP every 250 bp to one SNP every 20 bp (Figure 1e).
Figure 1.
Comparison of locus detection for Stacks (a–f) and PyRAD (g–l). The number of loci, SNPs and indels detected for specimens separated by 0–16 mutations at the mitochondrial mtMutS gene are shown for the different read coverage (m parameter) and divergence levels (M and n parameters, see text). In PyRAD analyses, ‘s' corresponds to the ‘Wclust' parameter. A full color version of this figure is available at the Heredity journal online.
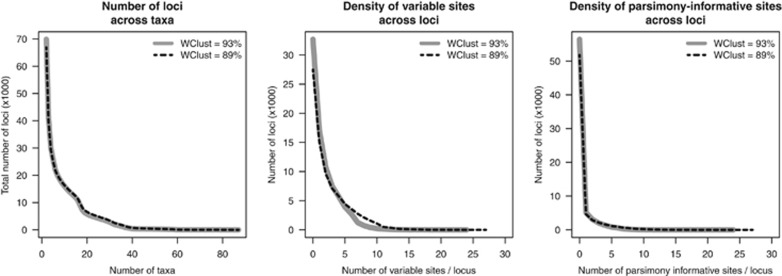
Results of the PyRAD analyses follow the general trends observed for the Stacks pipeline. These trends are an increase in total number of loci and polymorphic loci (Figures 1g and h) for more relaxed parameters sets, as well as for more closely related colonies. As for Stacks, more distantly related specimen pairs have fewer loci than for closely related ones, but a larger proportion of those is polymorphic (Figure 1i). Although the percentage of polymorphic loci shows similar ranges of values for Stacks and PyRAD, the total number of loci as well as the number of polymorphic loci are almost doubled (from 2000 to almost 4000 and from 1000 to almost 2000, respectively). The same pattern is observed for the number of SNPs and SNP densities (Figures 1j and k): PyRAD output differs from Stacks output by a factor of almost two, resulting in SNPs densities twice as high (from one SNP every 130 bp to one SNP every 20 bp). Finally, unlike Stacks, PyRAD allows for indels within loci. The percentage of loci containing indels increases with less stringent sets of parameters (Figure 1l). Depending on the pair of samples considered, this measure varies from a few percent to almost 40%. For PyRAD, the number of cataloged loci decreased rapidly with the number of specimens included in the analysis (with significant drops corresponding to the number of individuals in the haplotype clades revealed by the phylogenetic reconstruction, see below; Figure 2). Most loci bore less than three SNPs even when 10 polymorphisms were allowed on a single RAD locus (Figure 2).
Figure 2.
Information content of the locus catalog built by PyRAD for all 91 Chrysogorgia specimens. Wclust: percent divergence permitted between loci within and across specimens; in addition to the 93% Wclust level used to infer the Chrysogorgia phylogeny, the 89% Wclust level was tested here. A full color version of this figure is available at the Heredity journal online.
We measured the proportion of loci cataloged by Stacks that was also detected by PyRAD using custom BLASTN database searches. Overall, 0.6–42.7% of loci detected by Stacks were present in the PyRAD catalog. This pattern is partly explained by the proportion of PyRAD loci with indels (see above), but might also be influenced by the differential detection of repeated regions (that is, deleveraging algorithm in Stacks), or the number of reads per individual (the proportion of loci in common between Stacks and PyRAD was lower for individuals with fewer reads; Table 2).
Phylogenetic reconstruction and species delimitation
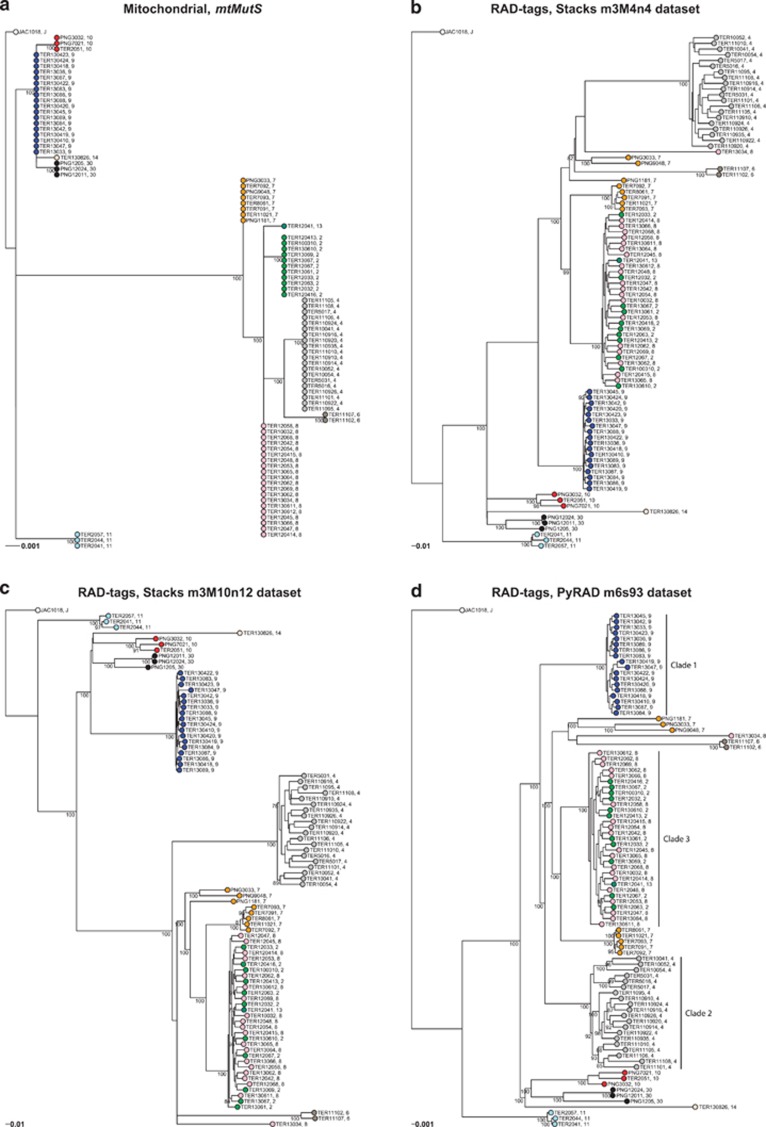
The automatic boot-stopping method implemented in RAxML yielded 1000 bootstrap replicates for the mitochondrial phylogeny (91 taxa × 700 nt), 500 replicates for the Stacks RAD phylogenies (91 taxa × 1 080 352 nt, 11 872 loci for the first data set and 1 146 054 nt, 12 594 loci for the second data set), and 200 replicates for the PyRAD phylogeny (91 taxa × 6 120 523 nt, 69 851 loci). The proportion of gaps and undetermined characters ranged between 83 and 84% for Stacks and was 92% for PyRAD. The three RAD phylogenies were similar but not identical, the second Stacks data set being better resolved than the first, and the PyRAD data set being better resolved than the Stacks sets (nodes with bootstrap >70%: 19% for m3M4n4, 29% for m3M10n12, 40% for m6s93; Figure 3). Divergence levels were much higher in the RAD phylogenies compared with the mitochondrial phylogeny. For instance, the groups composed of haplotypes 9 and 10 were separated by a distance of 0.001 substitution/site on the mtMutS tree, whereas these clades were separated by 0.27 and 0.25 substitutions/sites on the m3M4n4 and m3M10n12 RAD phylogenies, respectively (Figure 3).
Figure 3.
Maximum likelihood phylogenetic trees inferred using RAxML for the mitochondrial mtMutS data (a) and RAD loci (b–d). Bootstrap node support (1000 replicates for a, 500 replicates for b, c and 200 for d) is presented only for nodes with ⩾70% support. At the tips, colored dots, which represent mtMutS haplotype membership (each color represents a unique haplotype), are followed by specimen identifiers and haplotype numbers. Each tree was rooted to the Atlantic specimen (JAC1018, haplotype J). Genetic structure within clades 1, 2 and 3 were further investigated using a DAPC and TESS (see text and Supplementary Figure S1). Scale bars: substitution/site. A full color version of this figure is available at the Hypertension Research journal online.
Out of nine mitochondrial haplotypes represented by more than one individual, six formed well-supported monophyletic groups on the RAD phylogenies, for all data sets. One of these clades (corresponding to haplotype 10) contained specimens from both NC and PNG. The group formed by mitochondrial haplotype 7 was polyphyletic on the RAD phylogenies, with specimens grouping in two well-supported clades on the PyRAD phylogeny: one composed of five closely related NC specimens and one composed of three more divergent PNG colonies (this clade was split in two on the Stacks phylogenies). Specimens characterized by mtMutS haplotype 7 may therefore belong to at least three distinct species. On the other hand, specimens characterized by three distinct mitochondrial haplotypes (2, 8, 13) clustered into a single, well-supported clade (with the exception of one individual, TER13034, haplotype 8, which clusters well outside this clade). These three haplotypes, which form a paraphyletic group on the mitochondrial phylogeny and are one to two mutations different from each other, would therefore be considered as one evolutionary unit based on the RAD phylogenies (and population clustering analyses with DAPC and TESS failed to detect structure within this clade; see below). Finally, out of three singleton haplotypes (J, 13, 14), two (J, 14) sit on long branches and are clearly differentiated from other haplotypes using RAD-tag data.
We ran a DAPC on the three clades that contained the most colonies (clade 1: 18 colonies of haplotype 9; clade 2: 20 colonies of haplotype 4; clade 3: 31 colonies of haplotypes 2, 8, 13). Within these clades, 3685, 1470 and 8201 loci were retained (with 25, 42 and 55% missing data, respectively). In all three cases, DAPC failed to detect intraclade genetic structure, as the most likely number of group (based on BIC, discounting the scenario in which each sample belongs to its own group), in each case, was one (Supplementary Figure S1). The spatially explicit admixture model implemented in TESS also failed to detect genetic structure within clades 1 and 3, but suggested the presence of three clusters in clade 2, these clusters being composed of colonies sampled (1) on the slope of New Caledonia, (2) Munida Seamount (Norfolk Ridge) and (3) Jumeaux Ouest Seamount (Norfolk Ridge; Supplementary Figure S1). The population genetics of Chrysogorgia will be further discussed in a separate study.
Detection of environmental contaminants
As octocoral DNA was extracted from whole polyps rather than dissected, internal tissue, some loci may come from environmental contaminants such as bacteria. To evaluate the prevalence of such loci, we blasted all the loci that were cataloged for the m3M4n4 Stacks data set from individual JAC1018 (n=1202). The BLASTN algorithm (Altschul et al., 1997) was used to match RAD loci to the non-redundant NCBI nucleotide database, using 10−3 as a statistical significance threshold (e-value). Most sequences (92.6%) could not be assigned to a match in the nucleotide database and 4.5% of loci were similar to bacterial sequences (78–100% similarity between match and query). A single locus matched human mitochondrial DNA (84% similarity); other matches (n=34) included other invertebrates and plant sequences. Given (1) the small prevalence of potential contaminants, (2) our inability to determine whether these loci really belong to contaminant DNA or correspond to coral sequences which closest matches are non-cnidarian taxa and (3) the large number of Stacks analyses performed (>400), we decided to run our analyses without trying to filter loci from exogenous DNA sources.
Discussion
A critical decision in RAD analyses is the way the sequencing data are filtered to get to the final SNP data set. This process goes through several steps to ensure that the final loci will correspond to homologous sequences. The main filters involve several quality filters (sequencing quality, sequencing depth) as well as several similarity thresholds aimed at identifying the different allelic states of homologous loci. Finally, for each sample, an algorithm is used to tell apart sequencing errors from real mutations to conduct the final SNP calling. Even though the overall process is quite similar for Stacks and PyRAD analyses pipelines, a strict comparison of their results is not straightforward as they use sets of parameters that differ to some extent. A main difference between these two pipelines is in the assessment of similarity of loci: Stacks uses a strict similarity criterion (maximum number of mutations) to cluster reads into loci, whereas PyRAD uses an overall similarity criterion, after an alignment step, allowing for the presence of indels within clusters. This should be a critical difference when comparing genetically more-distant samples as indels are more likely to occur, and would thus result in sequences being assigned to different loci using Stacks (which will then be excluded from the final catalog as not present in all individuals), whereas PyRAD would theoretically allow these reads to be considered as homologous loci.
Our results show that more loci are recovered using the PyRAD pipeline. Despite these differences, general trends are similar using both pipelines. First, fewer loci and SNPs are recovered when comparing more genetically distant samples. This result is expected and has been anticipated through simulation (Cariou et al., 2013) and observed empirically (Cruaud et al., 2014). Our data show an exponential decay of the number of loci recovered as a function of divergence time of samples. Second, the stringency of the filtering process has a significant effect on the number of loci and SNPs identified. Indeed, higher minimum depth of sequencing thresholds and higher similarity threshold lead to fewer loci being identified. This trend is observed regardless of the level of genetic divergence between samples, but it seems to be accentuated when samples are more closely related.
Despite the similarities in general trends, quantitative and qualitative differences are observed in the outputs of each pipeline. Indeed, whatever the set of parameters used, almost twice as many loci are identified using PyRAD compared with Stacks. This difference cannot be solely attributed to the management of indels as our results show that the percentage of loci containing indels is usually around 5–20% and never reaches 40% whatever be the genetic distance between samples and the parameters set. Another interesting result is that PyRAD is not simply adding extra loci to the total loci identified by Stacks: only half of the loci identified using Stacks are also present in the PyRAD loci catalogs. It is thus necessary to invoke other filtering processes and differences in algorithm to explain these differences in output. More thorough analyses would be needed to identify precisely what are the main sources of divergence in the processing of raw data, in addition to the treatment of indels.
One major result is the remarkable loss of homologous loci with increasing divergence among specimens with different mitochondrial haplotypes. For instance, compared with specimens sharing the same haplotype, specimens two mutations apart at mtMutS (estimated divergence of 1–2 Myr) had on average 70% fewer homologous loci (Stacks analysis at m3M7n8). Within the genus, specimens from mitochondrial clades 16 mutations apart (that is, the highest divergence level included in our study, estimated between 9 and 16 Myr) share 97% fewer loci. This rate of loss of homologous RAD tags is far greater than what has been observed in cetaceans (Viricel et al., 2014), for which 66% of homologous loci were retained at the interfamilial level (short-beaked common dolphins, Delphinus delphis, vs harbor porpoise, Phocoena phocoena; estimated divergence of 14–19 Myr) compared with the intraspecific level (within Delphinus delphis). Comparisons within cetaceans were performed using the same custom pipeline as used in the present study, using Stacks parameters m3M3n3 (the results for corals were similar when comparing m3m3n3 to m3M7n8).
The differences observed between our study and that of Viricel et al. (2014) may be explained by various factors. For example, the choice of restriction enzyme was different (Sbf1 here, Not1 for Viricel et al.), and differences in genome composition (most importantly GC content and size) are unknown. Although both studies were conducted with two lanes of Illumina HiSeq2000 sequencing (conducted by Eurofins Genomics in both cases), throughput may have been influenced by the quality of genomic DNA (trawled deep-sea samples here, stranded animals for Viricel et al.). These various factors may have significantly influenced the number of cut sites. Our comparisons might also be significantly affected by the precision of the molecular clocks available. Divergence times between cetacean families were inferred based on fossil evidence (see references in Viricel et al., 2014), whereas no such fossil-calibrated molecular clock exists, to the best of our knowledge, for octocorals. The mtMutS divergence rates estimated by Lepard (2003) are based on a group of shallow-water octocorals that may evolve faster than the deep-sea Chrysogorgia (a long standing question in deep-sea biology is whether evolutionary process take longer in deeper water, compared with shallower waters; for example, Wilson and Hessler, 1987), and rely on a geological event (rising of the Isthmus of Panama), which can introduce further bias.
The exploration of divergence parameter space, as outlined above, was made using pairs of specimens, and not allowing any missing data. Stacks and PyRAD can build catalogs with loci shared by a set proportion of individuals within predefined groups. Hence, our phylogenetic matrix based on over 12K loci (Stacks parameters m3M10n12) resolved most deeper nodes of the tree despite 83–84% of missing data. Similarly, Cruaud et al. (2014) constructed a phylogeny of 18 species of the beetle genus Carabus, and found that the deepest node of the tree (17 Myr divergence between species) was characterized by 67% of missing data but strong statistical support. Jones et al. (2013) reconstructed phylogenetic relationships among congeneric species of swordtail and platyfish (Xiphophorus sp.) that diverged <3 Myr, and estimated up to 70% missing data (ingroup data). They noted, however, that missing data had little effect on tree topology and branch support. The rate of loss of homologous loci observed in swordtail and platyfish is more on par with what we observed for Chrysogorgia than what was reported for cetaceans and Carabus beetles, and further emphasizes that (1) the utility of RAD sequencing for phylogenetic reconstruction may be taxon dependent and (2) molecular clocks must be critically interpreted. It must be underlined, however, that notable differences in tree topologies were observed between the three inferred RAD phylogenies, such as deep but well-supported nodes (for example, relative positions of clade 3 and haplotypes 6, 7 and 8).
RAD-tag sequencing has also proven very useful in testing the criterion used for our primary species delimitation hypotheses, namely that single mitochondrial mtMutS haplotypes discriminate species that fit within the General Lineage Concept of species as defined by de Queiroz (1998). Indeed, a large numbers of variable loci could be cataloged within and among closely related colonies (sharing the same mtMutS haplotype, and therefore putatively belonging to the same species) and more distantly related colonies (separated by 1–16 mutations at mtMutS, putatively belonging to different species), allowing us (1) to plot our primary delimitation hypotheses onto well-supported phylogenies and (2) to explore the spatial structure of populations. Three patterns were evidenced from the data: (1) in the majority of cases, we noted a complete congruence between mtMutS haplotypes and RAD clades (6/9 non-singleton haplotypes and 2/3 singleton haplotypes); (2) in one case, incomplete congruence was noted (with PyRAD, haplotype 7 corresponding to two RAD clades (one NC, one PNG) that did not form a monophyletic group; (3) in one case, a single RAD clade included specimens with different (but closely-related) haplotypes. This result is significant for octocoral taxonomy and systematics, as mtMutS has been widely used to assist species delimitation across a large number of families (for example, review of McFadden et al., 2010). Although morphological, mitochondrial (Pante and Watling, 2012) and genomic data (this study) all point to the utility of mtMutS for delimiting Chrysogorgia species, its resolution should be interpreted in two ways. First, as we did not find 100% congruence between RAD clades and mtMutS haplotypes, and tested only a restricted set of putative species, mtMutS should still be considered as one of the first steps in an integrative taxonomic loop incorporating more variable markers (for example, Schlick-Steiner et al., 2010; Kekkonen and Hebert, 2014). Second, the evolutionary speed of mtMutS may well vary among octocorals, and its resolving power may therefore vary from one group to another (for example, Baco and Cairns, 2012). Nevertheless, combining mitochondrial markers such as mtMutS and RAD-tag data will without doubt be of tremendous value for testing the large number of outdated species hypotheses within the Octocorallia.
Data archiving
Mitochondrial haplotypes were deposited on GenBank (Supplementary Table S1). Phylogenetic data were deposited on Dryad: doi:10.5061/dryad.rp8d0.
Acknowledgments
We thank the participants of the National Research group ‘Génomique Environnementale' for stimulating discussions on the use of RAD tags for inferring phylogenies, in particular R Debruyne and A Cruaud. Samples used in this study were collected during the Terrasses and BioPapua cruises (PIs S Samadi and L Corbari) as part of the MNHN/IRD Tropical Deep Sea Benthos program, and during the Extreme Corals 2010 cruise in the northwestern Atlantic (PIs SW Ross and SD Brooke; funding from NOAA's Deep Sea Coral Research and Technology Program). We warmly thank the participants and crew members on these cruises for they indispensable help at sea. This study was funded by the ‘Institut Écologie et Environnement' of the CNRS (‘Appel à Projets en Génomique Environnementale,' organizers Dominique Joly and Denis Faure), the French Muséum national d'Histoire naturelle (Action Thématique du Muséum ‘Taxonomie moléculaire: DNA Barcode et gestion durable des collections') and by the Agence National de la Recherche (ANR 12-ISV7-0005-01 French-Taiwanese project TF-DeepEvo). Parts of the analysis were run on the YMIR super-computer (partly funded by the European Union, contract 31031-2008, European Regional Development Fund) of the University of La Rochelle (many thanks to Mikael Guichard, Marc-Henri Boisis-Delavaud and Frédéric Bret). We also thank Julio Pedraza (UMS 2700, MNHN) for his help with bioinformatics. Salary for EP was covered by a grant to the Poitou-Charentes region (Contrat de Projet État-Région 2007-2013). Finally, we thank the editor and two anonymous reviewers for their constructive comments.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baco AR, Cairns SD. (2012). Comparing molecular variation to morphological species designations in the deep-sea coral Narella reveals new insights into seamount coral ranges. PLoS ONE 7: e45555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA et al. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet P, Héros V, Lozouet P, Maestrati P. (2008). A quarter-century of deep-sea malacological exploration in the South and West Pacific: Where do we stand? How far to go? In: Héros V, Cowie RH, Bouchet P (eds). Tropical Deep-Sea Benthos 25, Vol 196, Muséum national d'Histoire naturelle: Paris. pp 9–40. [Google Scholar]
- Calderón I, Garrabou J, Aurelle D. (2006). Evaluation of the utility of COI and ITS markers as tools for population genetic studies of temperate gorgonians. J Exp Mar Biol Ecol 336: 184–197. [Google Scholar]
- Cariou M, Duret L, Charlat S. (2013). Is RAD-Seq suitable for phylogenetic inference? An in silico assessment and optimization. Ecol Evol 3: 846–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. (2013). Stacks: an analysis tool set for population genomics. Mol Ecol 22: 3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion GT, Crepeau M, Toonen RJ. (2008). An alternative to ITS, a hypervariable, single-copy nuclear intron in corals, and its use in detecting cryptic species within the octocoral genus Carijoa. Coral Reefs 27: 323–336. [Google Scholar]
- Cruaud A, Gautier M, Galan M, Foucaud J, Sauné L, Genson G et al. (2014). Empirical Assessment of RAD sequencing for interspecific phylogeny. Mol Biol Evol 31: 1272–1274. [DOI] [PubMed] [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML et al. (2011). Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12: 499–510. [DOI] [PubMed] [Google Scholar]
- de Queiroz K. (1998). The General Lineage Concept of Species, Species Criteria, and the Process of Speciation. In: Howard DJ, Berlocher SH (eds). Endless Forms: Species and Speciation. Oxford University Press: Oxford, UK. pp 57–75. [Google Scholar]
- Dueñas L, Sánchez J. (2009). Character lability in deep-sea bamboo corals (Octocorallia, Isididae, Keratoisidinae). Mar Ecol Prog Ser 397: 11–23. [Google Scholar]
- Durand E, Jay F, Gaggiotti OE, François O. (2009). Spatial inference of admixture proportions and secondary contact zones. Mol Biol Evol 26: 1963–1973. [DOI] [PubMed] [Google Scholar]
- Eaton D. (2014). PyRAD: assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics 30: 1844–1849. [DOI] [PubMed] [Google Scholar]
- Eaton DAR, Ree RH. (2013). Inferring phylogeny and introgression using RADseq data: an example from flowering plants (Pedicularis: Orobanchaceae). Syst Biol 62: 689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblom R, Galindo J. (2011). Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- France SC. (2007). Genetic analysis of bamboo corals (Cnidaria: Octocorallia: Isididae): does lack of colony branching distinguish Lepidisis from Keratoisis? Bull Mar Sci 81: 323–333. [Google Scholar]
- France SC, Rosel PE, Agenbroad JE, Mullineaux LS, Kocher TD. (1996). DNA sequence variation of mitochondrial large-subunit rRNA provides support for a two-subclass organization of the Anthozoa (Cnidaria). Mol Mar Biol Biotechnol 5: 15–28. [PubMed] [Google Scholar]
- Gregory TR. (2014). Animal genome size database. Available at http://www.genomesize.com.
- Gruenthal KM, Witting DA, Ford T, Neuman MJ, Williams JP, Pondella DJ et al. (2014). Development and application of genomic tools to the restoration of green abalone in southern California. Conserv Genet 15: 109–121. [Google Scholar]
- Herrera S, Baco A, Sánchez JA. (2010). Molecular systematics of the bubblegum coral genera (Paragorgiidae, Octocorallia) and description of a new deep-sea species. Mol Phylogenet Evol 55: 123–135. [DOI] [PubMed] [Google Scholar]
- Hipp AL, Eaton DAR, Cavender-Bares J, Fitzek E, Nipper R, Manos PS. (2014). A framework phylogeny of the American oak clade based on sequenced RAD data. PLoS ONE 9: e93975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24: 1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, Fan S, Franchini P, Schartl M, Meyer A. (2013). The evolutionary history of Xiphophorus fish and their sexually selected sword: a genome-wide approach using restriction site-associated DNA sequencing. Mol Ecol 22: 2986–3001. [DOI] [PubMed] [Google Scholar]
- Kekkonen M, Hebert PD. (2014). DNA barcode-based delineation of putative species: efficient start for taxonomic workflows. Mol Ecol Res 14: 706–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché AD, Fujita MK, Minin VN, Bouckaert RR. (2014). Species delimitation using genome-wide SNP data. Syst Biol 63: 534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepard A. (2003). Analysis of variation in the mitochondrial encoded msh1 in the genus Leptogorgia (Cnidaria: Octocorallia) and implications for population and systematics studies. Master's thesis, College of Charleston, Charleston, SC.
- Lexer C, Mangili S, Bossolini E, Forest F, Stölting KN, Pearman PB et al. (2013). ‘Next generation' biogeography: towards understanding the drivers of species diversification and persistence. J Biogeogr 40: 1013–1022. [Google Scholar]
- McFadden CS, Benayahu Y, Pante E, Thoma JN, Nevarez PA, France SC et al. (2011). Limitations of mitochondrial gene barcoding in Octocorallia. Mol Ecol Res 11: 19–31. [DOI] [PubMed] [Google Scholar]
- McFadden CS, Sánchez JA, France SC. (2010). Molecular phylogenetic insights into the evolution of Octocorallia: A review. Integr Comp Biol 50: 389–410. [DOI] [PubMed] [Google Scholar]
- Miller M, Pfeiffer W, Schwartz T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, United States. pp 1–8. [Google Scholar]
- Mokhtar-Jamaï K, Pascual M, Ledoux JB, Coma R, Féral JP, Garrabou J et al. (2011). From global to local genetic structuring in the red gorgonian Paramuricea clavata: the interplay between oceanographic conditions and limited larval dispersal. Mol Ecol 20: 3291–3305. [DOI] [PubMed] [Google Scholar]
- Nadeau NJ, Martin SH, Kozak KM, Salazar C, Dasmahapatra K, Davey JW et al. (2013). Genome-wide patterns of divergence and gene flow across a butterfly radiation. Mol Ecol 22: 814–826. [DOI] [PubMed] [Google Scholar]
- Pante E, Corbari L, Thubaut J, Chan TY, Mana R, Boisselier MC et al. (2012. a). Exploration of the deep-sea fauna of Papua New Guinea. Oceanography 25: 214–225. [Google Scholar]
- Pante E, France SC, Couloux A, Cruaud C, McFadden CS, Samadi S et al. (2012. b). Deep-sea origin and in-situ diversification of chrysogorgiid octocorals. PLoS ONE 7: e38357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante E, France SC. (2010). Pseudochrysogorgia bellona n. gen. n. sp.: a new genus and species of chrysogorgiid octocoral (Coelenterata: Anthozoa) from the Coral Sea. Zoosystema 32: 595–612. [Google Scholar]
- Pante E, Watling L. (2012). Chrysogorgia from the New England and Corner Seamounts: Atlantic – Pacific connections. J Mar Biol Assoc UK 92: 911–927. [Google Scholar]
- Puillandre N, Modica MV, Zhang Y, Sirovich L, Boisselier MC, Cruaud C et al. (2012). Large-scale species delimitation method for hyperdiverse groups. Mol Ecol 21: 2671–2691. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2014). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available at http://www.R-project.org. [Google Scholar]
- Reitzel AM, Herrera S, Layden MJ, Martindale MQ, Shank TM. (2013). Going where traditional markers have not gone before: utility of and promise for RAD sequencing in marine invertebrate phylogeography and population genomics. Mol Ecol 22: 2953–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BER, Ree RH, Moreau CS. (2012). Inferring phylogenies from RAD sequence data. PLoS ONE 7: e33394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH et al. (2010). Integrative taxonomy: a multisource approach to exploring biodiversity. Annu Rev Entomol 55: 421–438. [DOI] [PubMed] [Google Scholar]
- Soza-Ried J, Hotz-Wagenblatt A, Glatting K-H, del Val C, Fellenberg K, Bode HR et al. (2010). The transcriptome of the colonial marine hydroid Hydractinia echinata. FEBS J 277: 197–209. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Viricel A, Pante E, Dabin W, Simon-Bouhet B. (2014). Applicability of RAD-tag genotyping for interfamilial comparisons: empirical data from two cetaceans. Mol Ecol Res 14: 597–605. [DOI] [PubMed] [Google Scholar]
- Vollmer S, Palumbi S. (2004). Testing the utility of internally transcribed spacer sequences in coral phylogenetics. Mol Ecol 13: 2763–2772. [DOI] [PubMed] [Google Scholar]
- Wagner CE, Keller I, Wittwer S, Selz OM, Mwaiko S, Greuter L et al. (2013). Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Mol Ecol 22: 787–798. [DOI] [PubMed] [Google Scholar]
- Watling L, France SC, Pante E, Simpson A. (2011). Biology of deep-water octocorals. Adv Mar Biol 60: 41–123. [DOI] [PubMed] [Google Scholar]
- Wilson GDF, Hessler R. (1987). Speciation in the deep sea. Annu Rev Ecol Evol Syst 18: 185–207. [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. (2000). A greedy algorithm for aligning DNA sequences. J Comput Biol 7: 203–214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.