Abstract
Background:
The GLIS family members are zinc fingers with transcriptional repression and activation function. GLIS3 is one of these family members, which aberrant expression of it revealed to be related to several different cancer types. Regarding to the role of GLIS3 in tumor genesis and its probable connection with β-catenin signaling pathway, one of the pathways that involves in both normal development and tumor genesis of breast tissue, the aim of this study is investigating the alteration of GLIS3 mRNA expression level in breast cancer.
Materials and Methods:
Real-time polymerase chain reaction performed with GLIS3 and GAPDH genes primer on the RNA which extracted from 15 fresh frozen breast tumor tissue samples and also 15 normal samples with slight distance from site of tumor.
Results:
The relative expression of GLIS3 in breast cancer tissues revealed a 4 times increase comparing normal breast tissues; with a significant difference between cancer and normal samples (P = 0.027) and in patients without lymph node involvement and tissues that had estrogen receptor (ER−) and progesterone receptor (PR−) statuses. We see no significant difference between cancer and normal tissues based on lobular or ductal origin of the tumor as well as the tumor grade.
Conclusions:
Our study suggested a probable relationship between GLIS3 overexpression and breast cancer. Furthermore, detection of a probable association between GLIS3 overexpression and triple-negative breast cancer (ER−/PR−/human epidermal growth factor receptor 2−) might be useful for prognostic and diagnostic uses or as a probable target for treatment of these patients.
Keywords: Breast cancer, gene expression, GLIS3
INTRODUCTION
Breast cancer is the most invasive form of cancer and the second leading cause of death in women in developed countries.[1,2] This cancer also causes the most cancer deaths in each year (besides lung, stomach, liver and colon cancer)[3] and it is expected that the number of affected people will have a global rise within the next two decades.[3,4] Cancer is a multifactorial disease that genetic, environmental, medical, and lifestyle factors interact in producing it.[5] Although, the precise pathogenesis of the disease remains poorly understood,[2] the genetic factors including: Loss or aberrant function of DNA repair genes, tumor suppressor genes[6] and signaling molecules shown to have an important role in breast cancer.[7,8] The Wnt/β-catenin signaling pathway is one of these pathways that involves in cell proliferation, migration and differentiation regulation[1] and also has shown to play key roles in both normal development and tumor genesis of breast tissue.[1,9,10] Aberrant activation of these pathways has shown in many of tumors including breast cancer.[10] β-catenin has a critical role in this signaling pathway.[11] Wnt proteins are extracellular signals that activate the Wnt/β-catenin signaling pathway by connecting with the cell surface receptors such as FZD and the LRP5/6. Subsequent activation of β-catenin in the cell cytoplasm and its nucleus entrance to induce the expression of downstream target genes with cell cycle and growth regulation actions are the next steps of Wnt/β-catenin signaling pathway.[1]
The GLIS family members are zinc fingers and can have transcriptional repression and activation functions.[11,12] Characteristic feature of GLIS family is the presence of two or more conserved C2H2 zinc finger domains. Members of this family involve in a broad range of cellular activities including proliferation, differentiation and development.[11] GLIS3 is one of these family members which aberrant expression of it, revealed to be related to some pathological statuses such as osteopenia and rib fracture,[11,13] hypothyroidism (in %85 of cases witch result of abnormal development of thyroid gland),[11,14] pancreatic insufficiency and diabetes,[11,15,16] degenerative liver disease, cystic renal dysplasia,[11,13] polycystic kidneys, facial dysmorphism and bilateral sensorineural deafness.[11] Increased GLIS3 expression also has been detected in several different cancer types such as ependymomas[11,17] and chromophobe renal cell carcinoma.[11,18] GLIS3 gene amplification has been observed in proneural glioblastomas.[11,19]
Recently was reported that GLIS3 interacts with the tumor suppressor and negative regulator of hedgehog (Hh) signaling pathway (SUFU).[11,20] SUFU also interacts with β-catenin in Wnt signaling pathway.[20] The interaction of GLIS3 with SUFU makes the possibility of connection between the GLIS3, Hh, and Wnt pathways.[11]
Regarding to the role of GLIS3 in tumor genesis and its probable connection with β-catenin; the aim of this study is investigating the alteration of GLIS3 mRNA expression level in breast cancer for the 1st time.
MATERIALS AND METHODS
Patients and tissue samples
Totally 15 tissue samples provided by national tumor bank of Iran had been obtained from women who are getting surgery because of breast cancer. Furthermore, 15 normal breast tissues were taken from the same patients that had partial or total mastectomy, with a slight distance from the site of tumor. Sampling performed based on convenient sampling method. Tissue samples were stored at − 80°C, immediately after collection. Histopathology examination performed on tissue samples to confirm their cancer diagnosis and grade of disease; the statuses of human epidermal growth factor receptor 2 (HER2) and estrogen receptor/progesterone receptor (ER−/PR−) also detected using immunohistochemistry method. For sample collection, consent was obtained from patients before surgery. None of these patients had treatment with radiation or chemotherapy before surgery.
Total RNA extraction
RNA extracted from 10 mg frozen tissue using high pure RNA tissue kit (Roche) following manufacturer's instructions. The quality of extracted RNAs checked by nano-spectrophotometer (German precision NANOLYTIK®) based on absorbance density in 260 nm/280 nm. Extracted RNAs were stored at −80°C before use.
cDNA synthesis
cDNA synthesis performed on 11 µg of extracted RNA, using F RevertAid™ First Strand cDNA Synthesis Kit (Fermentas) based on manufacturer's instructions. Oligo-dT primer was used to amplify all RNAs with the polyadenylate tail. A template free reaction used as negative control. Real-time polymerase chain reaction (PCR) performed on prepared cDNA using GAPDH primers for the analysis of quality and quantity of cDNA.
Real-time polymerase chain reaction
Real-time PCR performed with GLIS3 and GAPDH genes specific forward and reverse primers. The GLIS3 specific primers were: 5’-ACGTTTGAAGGTTGCGAGAAG-3’ (forward) and 5’-AGGTTTGGTGTCCAGATGCG-3’ (reverse) that detected both transcript variants of GLIS3 gene. The reaction efficiency was 100.22% for GAPDH primer and 99.5% for GLIS3 primer. The reaction performed on synthetized cDNA as template. SYBR Green master mix (thermo science; #RO581) used for quantitative investigation of reactions. Real-time PCR performed in a StepOnePlus™ v2.2 Real-time PCR system based on manufacturer's instructions and in the following conditions: 94°C for 2 min, 35 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec. The expression of GLIS3 studied in comparison with GAPDH as housekeeping the gene;[17,18] and also in tumor samples in comparison with normal samples.
Statistical analysis
Statistical analysis performed using the REST 2009 (Qiagen) software based on software instruction for efficiency and P value calculation[21]. In this study, P < 0.05 has been considered statistically significant.
RESULTS
Alteration of GLIS3 mRNA expression level in breast tumor
We analyzed GLIS3 mRNA expression level in 15 breast cancer tissues and 15 normal breast tissues. All of the patients were women between 33 and 74 years old with a mean of 48.73 years. Two of patients had family history of an unrelated cancer (one with gastric cancer in her father and sister, the other with renal cancer in her father) and none of them had history of breast cancer in her family. The patients hadn’t any treatment for their cancer disease before surgery. Cancer tissues classified in two subtypes based on their lobular or ductal origin. The tissues also had been analyzed by immunohistochemistry method for the detection of ER, PR and HER2 statuses. In this study, all of our tissue samples were negative for HER2 status.
The quantitative analysis of GLIS3 performed in comparison to GAPDH mRNA expression level as a control endogenous gene. All of the P values in this study calculated with comparison of normal and tumor samples in each of groups (e.g. group of ER/PR − samples). Besides, the differences between GLIS3 expression and GAPDH expression are separately calculated in normal and tumor sample for each patient. The P values then obtained from ΔΔCTs results.
Based on these calculations, the relative expression of GLIS3 in breast cancer tissues was 4 times higher than normal breast tissues [Figure 1] and there were a significant difference between cancer and normal samples (P = 0.027); although, no significant difference between cancer tissues obtained based on their lobular or ductal origin as well as their tumor grade (all of the ductal carcinomas was invasive ductal carcinoma type).
Figure 1.
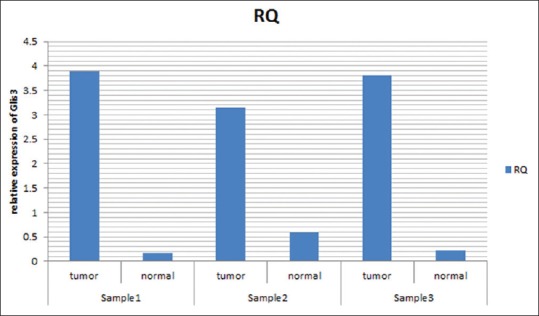
The relative expression of GLIS3 gene in tumor and normal breast tissues (ductal tissue samples). The figure shows the ΔCT in 3 of our samples (both of tumor and normal samples obtained from one patient indicated as paired charts). The RQ means relative quantity of GLIS3 gene expression in comparison to its control gene (GAPDH) in each sample that indicates the increased level of GLIS3 gene expression in tumor samples in comparison to normal samples
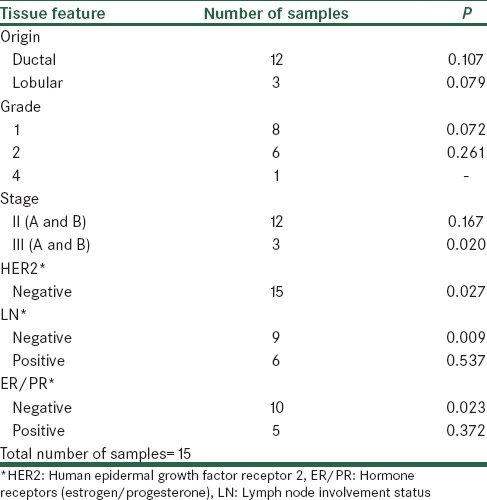
In turn, there was a significant difference between cancer and normal tissues in patients without lymph node involvement and tissues that were ER and PR negative. Table 1 summarized clinical data of patients based on the origin of the tumor, lymph node involvement, grade of tumor and ER/PR and HER2 statues of tumor.
Table 1.
Classification of samples based on their tumor property
DISCUSSION
Although the mechanisms of pathogenesis in breast cancer haven’t been understood clearly,[2] the key role of signaling pathways in tumor genesis has been recognized greatly.[7,22,23] One of these important signaling pathways, especially in breast cancer, is The Wnt/β-catenin signaling pathway.[1,3] Previous studies indicated that this essential pathway for breast cancer development has a probable relationship with GLIS3 a transcription regulator zinc finger protein.[11] The exact position of GLIS3 protein in signaling network of cells isn’t clear;[11] but, obvious role of aberrant activation of Wnt/β-catenin signaling pathway in breast cancer development[1,24] and its less known relationship with a zinc finger protein like GLIS3 with activator and repressor activity,[11] could be effective in a new recognition from both pathogenesis of cancer and GLIS3 function in it, and also from the different roles of Wnt/β-catenin signaling pathway in tumor genesis itself.
GLIS3 is a regulator of transcription, zinc finger protein. The best known downstream factors that related to GLIS3 are genes associated with different forms of diabetes[11] including (insulin) gene.[11,25] In addition, GLIS3 is related to other proteins with other functions, including: SUFU (suppressor of fused, a tumor suppressor gene and negative regulator of Hh signaling pathway) and (transcriptional coactivator, related to polycystic disease).[11] It seems that GLIS3 as a zinc finger protein can regulate transcription by interacting with GLIS-binding sites in the promoter of its target genes.[11]
Using the real-time PCR technique, we studied GLIS3 gene expression in 15 normal and 15 breast cancer tissue samples in a case-control study. This study indicated a statistically significant difference in GLIS3 gene expression between cancer and normal samples (P = 0.027) and we observed a 4 times higher expression of GLIS3 gene in cancer samples in comparison to normal ones. This significant difference has shown also in tumor samples with HER2, PR and ER-negative statuses and patients without lymph node involvement.
Breast cancers can be classified in up to 21 distinct subtype based on cell morphology, growth, and architecture patterns.[26] Triple negative breast cancer (ER, PR and HER2 with negative results) is one subtype of breast cancer with the characteristic, e.g., difficulty in treatment because of lack of a distinct target for treatment.[24] Studies have shown that Wnt/β-catenin signaling pathway activation is in association with these subtype of breast cancer and, as a result, is related to the poor clinical outcome observation.[1,24] Our observation of a statistically significant difference between case and control group in GLIS3 gene expression level in samples with negative statuses of ER, PR and HER2 might be related to triple negative subtype of breast cancer (TNBC) due to activation of Wnt/β-catenin signaling pathway. This result could be effective in finding a new target for facilitating treatment of TNBC. Although, it needs to more studies to be entirely validated.
Based on the TNM staging system, we also classified our samples to the 2 subgroup of stages as stage II or III. In this case, results of this study indicated a significant difference between GLIS3 expression in normal and tumor samples with higher stage of tumor (stage III-samples with T3 or T4 for primary tumor size, N0, N1 or N2 for regional lymph nodes and M0 for any metastases).[27] It can indicate a probable relationship between higher extension of tumor in advanced stages and GLIS3 gene overexpression. Having such an observation could be a result of GLIS3 gene role in the progression of breast cancer as a member of Wnt/β-catenin signaling pathway which should be confirmed by other researches.
In this study, we couldn’t show any significant difference in GLIS3 gene expression between samples with different origins (lobular or ductal) and grades. This might be indicated that GLIS3 gene isn’t related to differentiation of breast tissue, despite of Wnt/β-catenin signaling pathways role in this process.[24]
Previously, GLIS3 overexpression also had been shown to be related to some other types of cancer such as ependymomas[11,17] and chromophobe renal cell carcinoma.[11,18] These two types of cancer have ectodermal embryonic origin. Our study also suggested a probable relationship between GLIS3 overexpression and breast cancer; another cancer with ectodermal embryonic origin. Regarding these results, the next studies can show if the effect of GLIS3 expression enhancement is limited to cancers with this embryonic origin or not.
Results of previous studies and our study suggest a relationship between overexpression of GLIS3 and conversion of a normal tissue to a cancer tissue. This might be useful in declaring the molecular pathogenesis of breast cancer and improving the treatment of the patient affected by breast cancer. Furthermore, detection of a probable association between GLIS3 overexpression and TNBC might be useful for prognostic and diagnostic uses or as a target for treatment of these patients.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.King TD, Suto MJ, Li Y. The Wnt/ß-catenin signaling pathway: A potential therapeutic target in the treatment of triple negative breast cancer. J Cell Biochem. 2012;113:13–8. doi: 10.1002/jcb.23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu DY, Wang ZM, Li-Chen, Wang BL, Shen ZZ, Huang W, et al. Sox17, the canonical Wnt antagonist, is epigenetically inactivated by promoter methylation in human breast cancer. Breast Cancer Res Treat. 2010;119:601–12. doi: 10.1007/s10549-009-0339-8. [DOI] [PubMed] [Google Scholar]
- 3.Cancer: WHO Media Centre. [Last accessed on 2014 February 28]. Available from: http://www.who.int/mediacentre/factsheets/fs297/en/
- 4.Khazaee-pool M, Majlessi F, Foroushani AR, Montazeri A, Nedjat S, Shojaeizadeh D, et al. Perception of breast cancer screening among Iranian women without experience of mammography: A qualitative study. Asian Pac J Cancer Prev. 2014;15:3965–71. doi: 10.7314/apjcp.2014.15.9.3965. [DOI] [PubMed] [Google Scholar]
- 5.institue nc. Genetics of Breast and Ovarian Cancer (PDQR): National Institue of Health. [Last accessed on 2014 February 28]. Available from: http://www.cancer.gov/cancertopics/pdq/genetics/breast-and-ovarian/HealthProfessional .
- 6.Kobayashi H, Ohno S, Sasaki Y, Matsuura M. Hereditary breast and ovarian cancer susceptibility genes (review) Oncol Rep. 2013;30:1019–29. doi: 10.3892/or.2013.2541. [DOI] [PubMed] [Google Scholar]
- 7.Takebe N, Warren RQ, Ivy SP. Breast cancer growth and metastasis: Interplay between cancer stem cells, embryonic signaling pathways and epithelial-to-mesenchymal transition. Breast Cancer Res. 2011;13:211. doi: 10.1186/bcr2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al Saleh S, Sharaf LH, Luqmani YA. Signalling pathways involved in endocrine resistance in breast cancer and associations with epithelial to mesenchymal transition (Review) Int J Oncol. 2011;38:1197–217. doi: 10.3892/ijo.2011.942. [DOI] [PubMed] [Google Scholar]
- 9.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prosperi JR, Goss KH. A Wnt-ow of opportunity: Targeting the Wnt/beta-catenin pathway in breast cancer. Curr Drug Targets. 2010;11:1074–88. doi: 10.2174/138945010792006780. [DOI] [PubMed] [Google Scholar]
- 11.Lichti-Kaiser K, ZeRuth G, Kang HS, Vasanth S, Jetten AM. Gli-similar proteins: Their mechanisms of action, physiological functions, and roles in disease. Vitam Horm. 2012;88:141–71. doi: 10.1016/B978-0-12-394622-5.00007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song W, Chen YP, Huang R, Chen K, Pan PL, Li J, et al. GLIS1 rs797906: An increased risk factor for late-onset Parkinson's disease in the Han Chinese population. Eur Neurol. 2012;68:89–92. doi: 10.1159/000337955. [DOI] [PubMed] [Google Scholar]
- 13.Dimitri P, Warner JT, Minton JA, Patch AM, Ellard S, Hattersley AT, et al. Novel GLIS3 mutations demonstrate an extended multisystem phenotype. Eur J Endocrinol. 2011;164:437–43. doi: 10.1530/EJE-10-0893. [DOI] [PubMed] [Google Scholar]
- 14.Senée V, Chelala C, Duchatelet S, Feng D, Blanc H, Cossec JC, et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet. 2006;38:682–7. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Chang BH, Samson SL, Li MV, Chan L. The Krüppel-like zinc finger protein GLIS3 directly and indirectly activates insulin gene transcription. Nucleic Acids Res. 2009;37:2529–38. doi: 10.1093/nar/gkp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang HS, ZeRuth G, Lichti-Kaiser K, Vasanth S, Yin Z, Kim YS, et al. Gli-similar (Glis) Krüppel-like zinc finger proteins: Insights into their physiological functions and critical roles in neonatal diabetes and cystic renal disease. Histol Histopathol. 2010;25:1481–96. doi: 10.14670/hh-25.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukashova-v Zangen I, Kneitz S, Monoranu CM, Rutkowski S, Hinkes B, Vince GH, et al. Ependymoma gene expression profiles associated with histological subtype, proliferation, and patient survival. Acta Neuropathol. 2007;113:325–37. doi: 10.1007/s00401-006-0190-5. [DOI] [PubMed] [Google Scholar]
- 18.Yusenko MV, Kovacs G. Identifying CD82 (KAI1) as a marker for human chromophobe renal cell carcinoma. Histopathology. 2009;55:687–95. doi: 10.1111/j.1365-2559.2009.03449.x. [DOI] [PubMed] [Google Scholar]
- 19.Cooper LA, Gutman DA, Long Q, Johnson BA, Cholleti SR, Kurc T, et al. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS One. 2010;5:e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ZeRuth GT, Yang XP, Jetten AM. Modulation of the transactivation function and stability of Krüppel-like zinc finger protein Gli-similar 3 (GLIS3) by Suppressor of Fused. J Biol Chem. 2011;286:22077–89. doi: 10.1074/jbc.M111.224964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eroles P, Bosch A, Pérez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38:698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 23.García-Becerra R, Santos N, Díaz L, Camacho J. Mechanisms of Resistance to Endocrine Therapy in Breast Cancer: Focus on Signaling Pathways, miRNAs and Genetically Based Resistance. Int J Mol Sci. 2012;14:108–45. doi: 10.3390/ijms14010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King TD, Zhang W, Suto MJ, Li Y. Frizzled7 as an emerging target for cancer therapy. Cell Signal. 2012;24:846–51. doi: 10.1016/j.cellsig.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ZeRuth GT, Takeda Y, Jetten AM. The Krüppel-like protein Gli-similar 3 (GLIS3) functions as a key regulator of insulin transcription. Mol Endocrinol. 2013;27:1692–705. doi: 10.1210/me.2013-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dieci MV, Orvieto E, Dominici M, Conte P, Guarneri V. Rare breast cancer subtypes: Histological, molecular, and clinical peculiarities. Oncologist. 2014;19:805–13. doi: 10.1634/theoncologist.2014-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Compton CC, Byrd DR, Garcia-Aguilar J, Kurtzman SH, Olawaiye A, Washington MK. 2nd ed. New York, NY: Springer; 2012. Breast. AJCC Cancer Staging Atlas: A Companion to the Seventh Editions of the AJCC Cancer Staging Manual and Handbook; p. 419. [Google Scholar]


