Abstract
Compared with diploid species, haplodiploids suffer less inbreeding depression because male haploidy imposes purifying selection on recessive deleterious alleles. However, alleles of genes only expressed in the diploid females are protected in heterozygous individuals. This leads to the prediction that haplodiploids suffer more from inbreeding effects on life-history traits controlled by genes with female-limited expression. To test this, we used a wild population of the haplodiploid mite Tetranychus urticae. First, negative effects of inbreeding were investigated by comparing maturation rate, juvenile survival, oviposition rate and longevity between lines created by three generations of either outbreeding or mother–son inbreeding. Second, purging through inbreeding was investigated by comparing the intensity of inbreeding depression between outbred families with known inbreeding/outbreeding mating histories. Negative effects of inbreeding and evidence for purging were found for the female trait oviposition rate, but not for juvenile survival and longevity. Both male and female maturation rate were negatively affected by inbreeding, most likely due to maternal effects because inbred offspring of outbred mothers was not affected. These results support the hypothesis that, in haplodiploids inbreeding effects and genetic variation due to deleterious recessive alleles may depend on gender.
Introduction
Mating between relatives often leads to reduced fitness of the offspring. Such inbreeding depression is on average more intense in diploid species than in haplodiploid species (Hedrick and Parker, 1997; Hente, 2003). This difference in fitness effects is related to the extent to which selection can act against recessive deleterious alleles, one of the sources of inbreeding depression (the other being heterozygote advantage; Lynch and Walsh, 1998; Roff, 2002; Charlesworth and Willis, 2009). In diploids, recessive deleterious alleles are protected against purifying selection in the heterozygotes. In this way, quantitative genetic variance attributable to recessive deleterious alleles is maintained in traits that are under directional selection (Lynch and Walsh, 1998, chapter 10), most notably life-history traits. In haplodiploids, the deleterious alleles are expressed in the hemizygous males and hence exposed to continuous selection, which greatly reduces their frequency in the population (Avery, 1984; Werren, 1993).
In dioecious plants and animals, cross-gender genetic correlations are usually large and positive, but typically smaller for life-history traits than behavioural or morphological traits (Poissant et al., 2009). Most life-history traits are present in both genders and their genetic control in the females is probably related to that in the males. For such traits, low effects of inbreeding may be expected due to purging of recessive deleterious alleles via the haploid gender, just like selection on the haploid gametes of plants reduces inbreeding depression (Charlesworth and Charlesworth, 1992). Some traits—such as oviposition—are female traits and thus potentially controlled by many genes with female-limited expression. Recessive alleles for such genes can remain protected in heterozygotes against selection and thus exist at higher frequencies than those also or exclusively expressed in males (Crozier, 1976; Werren, 1993). Hence, the effect of inbreeding in haplodiploid species may be related to gender due to the degree to which life-history traits are controlled by genes with female-limited expression. Alternatively, if a trait present in both genders is controlled by a different suite of genes in either gender, the effects of inbreeding may also be larger in the females. Furthermore, if a trait present in both genders is genetically controlled by maternal factors, an inbred mother may cause lower trait values in both her sons and daughters. Here, we determined the effects of inbreeding on a suite of life-history traits of a haplodiploid mite to systematically discern whether and how these effects are gender related.
Purging of deleterious alleles can also take place during an episode of inbreeding. Mating between relatives increases homozygosity, which exposes recessive alleles to selection. The intensity of purging depends on the impact of such alleles on fitness and on their degree of recessiveness (Hedrick, 1994; Wang et al., 1999). These two factors are related: strongly deleterious alleles are usually more recessive than weakly deleterious alleles (Simmons and Crow, 1977; Charlesworth and Charlesworth, 1987). Theory predicts that during bouts of strong inbreeding, alleles that are weakly deleterious and recessive will not be effectively eliminated from the population, but alleles that are strongly deleterious and recessive will be quickly purged (Hedrick, 1994; Wang et al., 1999). To assess whether the life-history traits of a haplodiploid may be affected by strongly deleterious and recessive alleles, we examined not only direct negative effects of inbreeding but also the occurrence of purging through inbreeding.
We used the haplodiploid spider mite Tetranychus urticae Koch to test for inbreeding depression and purging. Four pivotal life-history traits of this herbivore were examined: maturity rate, juvenile survival, oviposition rate and longevity. Maturity rate was measured for males and females separately, but also calculated for both genders together. Juvenile survival was measured for both genders together, because gender is very hard to determine phenotypically in the embryonic and early juvenile stages. Oviposition rate was of course only measured in females. It is also the only female trait among the life-history traits of T. urticae. Longevity was measured for females only, because males are restless and therefore hard to isolate in experiments for life-span measurements. Inbred lines were created by three rounds of mother–son mating, which led to an inbreeding coefficient for the females of F=0.875 (where F is the probability that an individual has two alleles that are identical by descent). The outbred lines were subjected to the same rearing conditions to minimise differences in laboratory conditions and thus in potential adaptation to these conditions. The life-history traits of the two groups of lines were directly compared to determine whether negative effects of inbreeding occurred. To determine whether purging had occurred during inbreeding, we compared the intensity of inbreeding depression (at F=0.5) in life-history traits of the T. urticae lines with a history of inbreeding with that of the lines without a history of inbreeding depression: purging is expected to lead to a lower intensity in the lines with a history of inbreeding (Willis, 1999).
Materials and Methods
T. urticae population
A sample of ~200 spider mites was collected along a transect of 5 m of spindle bushes (Euonymus europaeus) in the dunes bordering the Dutch coast near the town of Castricum. The sampled field population is known to have been stably present for many years with large numbers of mites (personal observation N Tien and M Egas). Analysis of five microsatellite markers has consistently shown much variation over the years, and no departure from the Hardy–Weinberg equilibrium (unpublished data M Egas).
The laboratory population was maintained at a minimal population size of 300 individuals under climate-controlled conditions in the laboratory (19 °C, 55% humidity, light:dark (L:D)=16:8 h). Bean leaves (Phaseolus vulgaris) served as a food source for the spider mites. The leaves were placed on wet cotton wool surrounded by water in an open plastic container. During the experiments the spider mites were kept on bean leaf discs (diameter=1.5 cm) placed on wet cotton wool under controlled conditions similar to those present during rearing, except for the temperature (26 °C, unless stated otherwise). The experiment was carried out in two blocks, where the second block (B) started during the first day of the second round of inbreeding (see below) in the first block (A). Block A consisted initially of 109 families, block B of 98 families.
Inbreeding depression
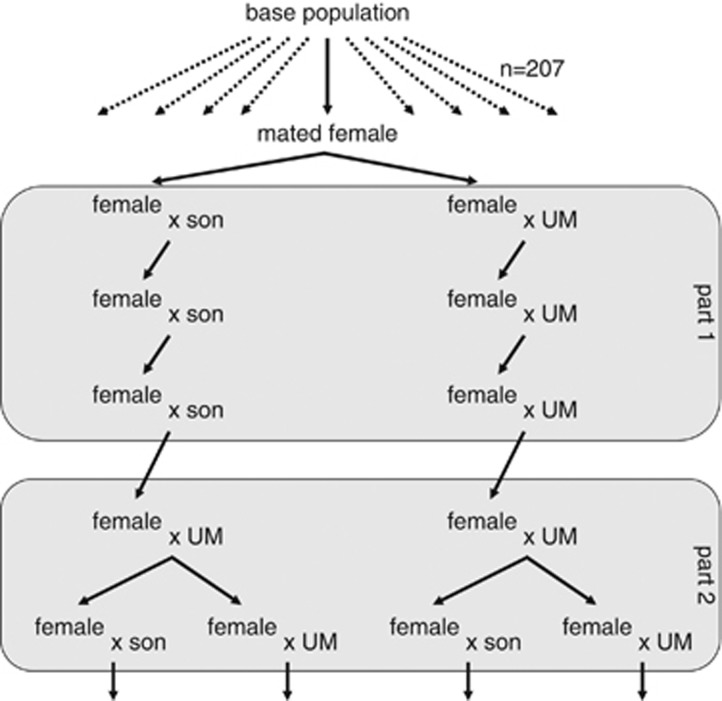
From the base population, (mated) adult females were collected. Each female formed the basis for two lines: one outbred line and one inbred line (Figure 1, part 1). The females were kept on individual leaf discs for 2 days to oviposit. Two of their daughters were collected in the pre-adult moulting stage and placed on individual leaf discs to oviposit. As they were virgins, their eggs were unfertilized and developed into sons. The females were then placed at 19 °C. At this low temperature their fertile life span is increased, which increases their chance for mating with a son. When the sons matured, the first round of inbreeding started: two sons (for the inbred line) or two unrelated males of another line (for the outbred line) were placed 1 day together with the female on a leaf disc to mate, after which the female was transferred to a fresh leaf disc to oviposit. Of their subsequent offspring, a daughter was collected in the pre-adult moulting stage and then the next round of inbreeding/outbreeding commenced. For the inbred lines, the three rounds of mating were between mother and son. For the outbred lines, females were mated to males of a different outbred line in every round. After the first round of mating, oviposition rate was determined in both treatments: females were collected in the last moulting stage and 5 days later they were placed on individual leaf discs to oviposit for 24 h (n=181).
Figure 1.
Breeding scheme: 207 females were taken from the base population and per female an inbred and an outbred line were set up. UM=unrelated male from the same treatment. At the end of part 1 and of part 2, the life-history traits of the offspring of the last breeding pairs were measured to investigate the presence of inbreeding depression (part 1) and purging (part 2).
After the last round of mating, the mated females were placed on a fresh leaf disc to oviposit for 24 h. The eggs were counted and of each individual offspring the maturity rate and juvenile survival were determined, and of each female offspring longevity and oviposition rate were determined. Indices for maturity rate were created for the two genders together and for each gender separately. (i) Overall maturity rate was measured as the fraction of adults that had reached adulthood on the 11th day, which can be separated into (ii) female maturity rate and (iii) male maturity rate (note that the sex ratio in this species is female-biased. The overall maturity rate is thus not the average of the male and female maturity rate). Juvenile survival was taken as the fraction of offspring that survived from egg to adulthood. To determine longevity and oviposition rate, each female offspring was isolated on a fresh leaf disc upon reaching the pre-adult moulting stage. After 4 days, the oviposition rate was determined by transferring the female to a fresh leaf disc and counting the number of eggs laid, 24 h later. Due to logistical problems, the oviposition rate was measured in block B only. Thereafter, the female was provided with fresh leaf discs every 2–3 days and longevity was measured as the number of days the adult female stayed alive. The effect of inbreeding was determined by comparing the trait values of the inbred lines with that of the outbred lines.
Purging
Two virgin females per line were collected and mated to males from another line of the same treatment (Figure 1, part 2). Of their (outbred) female offspring, two virgin sisters (in the pre-adult moulting stage) were transferred, each to a separate leaf disc to oviposit for 24 h, after which the females were placed on fresh leaf discs at 19 °C. To provide opportunities to mate, one of the two sisters was kept together with two of her sons and the other sister with two unrelated males of another line of the same treatment. These mated females were placed on fresh leaf discs and they were allowed to oviposit for 24 h, after which the eggs were counted and the life-history traits of their offspring were measured as above.
Statistics
All statistical analyses were performed in R (R Development Core Team, 2009). To determine whether inbreeding depression occurred with regard to oviposition rate at F=0.5 (after the first round of mating), a mixed-effect generalized linear model was constructed with treatment (inbred/outbred) as a fixed factor and block (A/B) as a random factor. A normal error distribution was assumed. The P-value for treatment was determined by removing treatment as a factor from the full model and comparing the two models using a likelihood ratio test (Crawley, 2007).
To determine whether inbreeding depression occurred at F=0.875 (after three rounds of mating), for each trait (except oviposition rate) a mixed-effect generalized linear model was constructed with treatment (inbred/outbred) as a fixed factor and block (A/B) as a random factor. For oviposition rate, a generalized linear model was constructed with treatment as only (fixed) factor, because data on oviposition rate were only available for block B. The P-value per treatment was determined by removing treatment as a factor from the full model and comparing the two models using a likelihood ratio test (Crawley, 2007). All data expressed as fractions (juvenile survival, female/male/overall maturity rate) were analyzed assuming a binomial error distribution. Longevity and oviposition rate were analyzed assuming a normal error distribution.
To determine whether the intensity of inbreeding depression differed between the lines with and without a history of inbreeding, indices for the intensity of inbreeding depression were used that allow calculations on a family basis. The classic index for the intensity of inbreeding depression is defined as δ=(wo−wi)/wo where (wi)=the phenotypic inbred value and (wo)=phenotypic outbred value. This index cannot be reliably used for per-family comparisons, because its asymmetric nature (from −∞ to 1) can cause a few families with higher inbred than outbred fitness to greatly diminish the average δ of the group as a whole (Johnston and Schoen, 1994). Therefore, a group-wise index for δ should be used, with a bootstrapped standard error. As an alternative, we preferred to use two indices calculated on a family basis. First, relative performance (RP) was calculated following (Ågren and Schemske, 1993): if wi⩽wo, then RP=1−wi/wo. If wi>wo, then RP=wo/wi−1. RP is scaled from −1 to 1, where RP=1 represents complete inbreeding depression and RP=−1 represents complete outbreeding depression. When both the inbred and outbred sisters have a trait value of zero, the RP cannot be calculated. This occurred a number of times within the data set of the female (nine cases), male (one case) and overall (two cases) maturity rates. These data were discarded from further analysis. The second index is given by the absolute difference (AD)=wo−wi (Willis, 1999). Positive values of AD represent inbreeding depression and negative values outbreeding depression. A mixed-effect generalized linear model was performed per trait (assuming a normal error distribution), with treatment (population with/without history of inbreeding) as a fixed factor and block (A/B) as a random factor, for both indices of inbreeding depression. The P-value for treatment effect was determined by removing treatment as a factor from the full model and comparing the two models in a χ2-test (Crawley, 2007).
To determine whether inbreeding depression occurred at F=0.50 within the lines with or without a history of inbreeding, a paired t-test was carried out per treatment (with or without a history of inbreeding), with the trait values of the offspring of the two sisters paired together. Female and overall maturity rate were arcsin (√x)-transformed and oviposition rate was log-transformed, to achieve a normal frequency distribution. Although a t-test is quite robust to violations of its assumptions (Quinn and Keough, 2006), we also tested the data with the non-parametric Wilcoxon signed-rank test, but this led to similar results as the t-test (data not shown).
Results
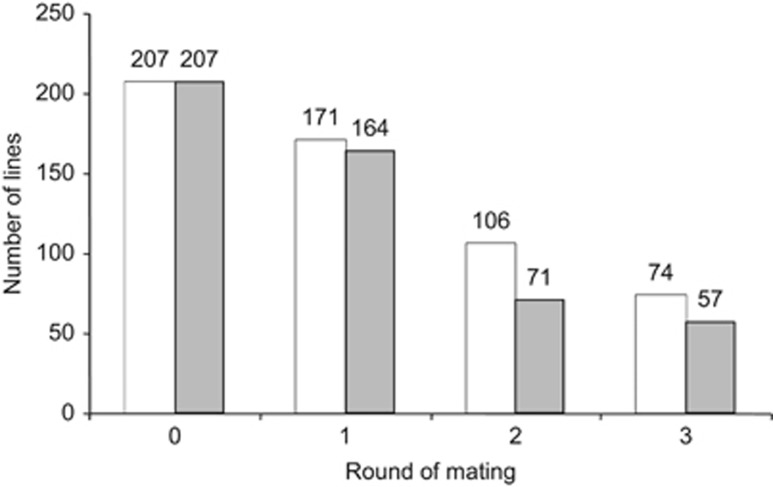
In total, 207 inbred and outbred lines were created. During the three rounds of mating, the number of lines decreased in both treatments (Figure 2). After three rounds of mating, 72% of the inbred lines and 64% of the outbred lines were lost. In most cases, loss of lines was due to females that did not survive until mating, whereas in some cases mating was unsuccessful or the offspring failed to reach adulthood. Significantly more inbred lines than outbred lines were lost in the second and third round (G-tests with William's corrected G-values for round 1, 2 and 3: G=1.6 and P=0.2, G=24.0 and P<0.001, G=6.3, P=0.01, respectively (Sokal and Rohlf, 1995, chapter 17)). After the three rounds of mating there were 23% less inbred lines than outbred lines (N=57 and 74, respectively). After the first round of mating, the inbred lines had a significantly lower mean oviposition rate than the outbred lines (χ21=9.3, P<0.01): inbred females (F=0.5) laid on average 10.24 eggs per 24 h (s.e.=0.17, n=82), the outbred females 10.98 eggs per 24 h (s.e.=0.17, n=99).
Figure 2.
Number of inbred (grey bars) and outbred (white bars) lines over the rounds of mating (mother–son mating for inbred lines and random mating for outbred lines). The number depicted on the x-axis represents the rounds of mating, prior to reaching the remaining number of lines (y-axis). Data labels above bars indicate the number of lines.
Inbreeding depression
The life-history traits of inbred and outbred lines (see Figure 1, part 1) were compared to determine whether negative effects of inbreeding occurred. The average juvenile survival and longevity of these offspring did not differ between the inbred and outbred lines (P=0.39 and P=0.43, respectively, Table 1). Negative effects of inbreeding did occur with respect to the oviposition rate: the inbred females (F=0.875) had a mean oviposition rate of 9.7 eggs/24 h (±s.e.=0.6), whereas the outbred females laid on average 11.3 eggs/24 h (±s.e.=0.5) (Table 1). Also the overall maturity rate was significantly lower in the inbred lines than in the outbred lines (0.67±0.06 and 0.82±0.04, respectively, see Table 1). This lower maturity rate in the inbred lines applied to both genders (for the females: 0.62±0.06 and 0.79±0.04, for the inbred and outbred lines, respectively, and for the males: 0.77±0.07 and 0.92±0.04, respectively).
Table 1. Statistics of the life-history traits of the inbred lines (F=0.875) and outbred lines (F=0).
| Trait |
Inbred |
Outbred |
X21 | P-value | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | s.e. | n | Mean | s.e. | n | |||
| Female maturity rate | 0.62 | 0.06 | 45 | 0.79 | 0.04 | 58 | 28.7 | <0.001 |
| Male maturity rate | 0.77 | 0.07 | 36 | 0.92 | 0.04 | 54 | 5.1 | <0.05 |
| Overall maturity rate | 0.67 | 0.06 | 46 | 0.82 | 0.04 | 58 | 33.8 | <0.001 |
| Juvenile survival | 0.86 | 0.03 | 46 | 0.85 | 0.03 | 60 | 0.7 | 0.39 |
| Longevity | 25.6 | 1.3 | 37 | 27.4 | 1.2 | 45 | 0.6 | 0.43 |
| Oviposition rate | 9.7 | 0.6 | 15 | 11.3 | 0.5 | 17 | 4.2a | <0.05 |
The sample size n refers to the number of independent lines.
F-value instead of X2-value.
Purging
To determine whether purging had occurred during the prior period of inbreeding (Figure 1 part 1), the intensity of inbreeding depression was compared between the populations of lines with and without a history of inbreeding (Figure 1 part 2). The two indices (AD and RP) for intensity of inbreeding depression showed results comparable to each other for all life-history traits (Table 2). The intensity of inbreeding depression with regard to oviposition rate was lower (c. 4/5 lower) in the population with a history of inbreeding than in the one without a history of inbreeding (P<0.05, see Table 2). No significant difference in intensity of inbreeding depression was found with regard to juvenile survival, longevity or maturity rate (P>0.1 in all cases, Table 2).
Table 2. Statistics for two indices (RP and AD) of the intensity of inbreeding depression in the life-history traits of the population of lines with and without a history of inbreeding.
| Trait | Index |
Inbreeding history |
No inbreeding history |
X21 | P-value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | s.e. | n | Mean | s.e. | n | ||||
| Female maturity rate | RP | −0.027 | 0.117 | 34 | 0.083 | 0.104 | 44 | 0.5 | 0.47 |
| AD | −0.053 | 0.067 | 30 | 0.023 | 0.046 | 39 | 1.0 | 0.31 | |
| Male maturity rate | RP | −0.113 | 0.080 | 26 | 0.084 | 0.091 | 35 | 2.4 | 0.12 |
| AD | −0.079 | 0.068 | 25 | 0.022 | 0.078 | 35 | 0.8 | 0.36 | |
| Overall maturity rate | RP | −0.163 | 0.099 | 34 | 0.058 | 0.091 | 44 | 2.7 | 0.10 |
| AD | −0.087 | 0.063 | 32 | 0.001 | 0.048 | 44 | 1.4 | 0.23 | |
| Juvenile survival | RP | 0.072 | 0.055 | 37 | 0.013 | 0.040 | 44 | 0.7 | 0.41 |
| AD | 0.075 | 0.052 | 37 | 0.014 | 0.039 | 44 | 0.8 | 0.36 | |
| Longevity | RP | −0.008 | 0.059 | 16 | −0.028 | 0.046 | 22 | 0.1 | 0.77 |
| AD | −0.250 | 2.044 | 16 | −1.091 | 1.662 | 22 | 0.1 | 0.74 | |
| Oviposition rate | RP | 0.027 | 0.022 | 29 | 0.114 | 0.027 | 33 | 6.0 | 0.01 |
| AD | 0.276 | 0.310 | 29 | 1.424 | 0.348 | 33 | 5.8 | 0.02 | |
Abbreviations: AD, absolute difference; RP, relative performance.
The sample size n refers to the number of independent lines; average values of the indices per line were used in the statistical tests.
In the population with a history of inbreeding, none of the examined traits differed between the inbred and outbred offspring of the pairs of sisters (P>0.1 in all cases, Table 3): one generation of inbreeding had no negative effect on oviposition rate, maturity rate, juvenile survival or longevity. However, in the population without a history of inbreeding, the mean oviposition rate of the inbred females (10.52±0.44) was significantly lower than that of their outbred cousins (11.94±0.44) (P<0.001, Table 4). This is a confirmation of the previous finding of reduced oviposition rate after one round of inbreeding in the base population, as in both cases the lines have no history of inbreeding. No negative inbreeding effects occurred in juvenile survival or in longevity (P>0.5, Table 4). Also, no negative effects were found in maturity rate (P⩾0.46, Table 4).
Table 3. Statistics of the life-history traits of the population of lines with a history of inbreeding, measured in inbred and outbred offspring of pairs of sisters.
| Trait |
Inbred |
Outbred |
n | t | P-value | ||
|---|---|---|---|---|---|---|---|
| Mean | s.e. | Mean | s.e. | ||||
| Female maturity rate | 0.47 | 0.06 | 0.41 | 0.06 | 34 | 0.85 | 0.40 |
| Male maturity rate | 0.86 | 0.05 | 0.78 | 0.08 | 26 | 1.15 | 0.26 |
| Overall maturity rate | 0.55 | 0.05 | 0.48 | 0.06 | 34 | 1.53 | 0.14 |
| Juvenile survival | 0.79 | 0.04 | 0.86 | 0.03 | 37 | −1.43 | 0.16 |
| Longevity | 29.44 | 1.47 | 29.19 | 1.31 | 16 | 0.12 | 0.90 |
| Oviposition rate | 11.48 | 0.49 | 11.76 | 0.46 | 29 | −1.21 | 0.24 |
The sample size n refers to the number of independent lines; average values per line were used in the statistical tests.
Table 4. Statistics of the life-history traits of the population of lines without a history of inbreeding, measured in inbred and outbred offspring of pairs of sisters.
| Trait |
Inbred |
Outbred |
n | t | P-value | ||
|---|---|---|---|---|---|---|---|
| Mean | s.e. | Mean | s.e. | ||||
| Female maturity rate | 0.39 | 0.05 | 0.41 | 0.05 | 44 | −0.44 | 0.66 |
| Male maturity rate | 0.78 | 0.07 | 0.80 | 0.06 | 35 | −0.28 | 0.78 |
| Overall maturity rate | 0.50 | 0.05 | 0.50 | 0.05 | 44 | −0.22 | 0.83 |
| Juvenile survival | 0.88 | 0.02 | 0.87 | 0.02 | 43 | −0.34 | 0.73 |
| Longevity | 30.68 | 1.39 | 29.59 | 1.30 | 22 | −0.66 | 0.52 |
| Oviposition rate | 10.52 | 0.44 | 11.94 | 0.44 | 33 | −4.14 | <0.001 |
The sample size n refers to the number of independent lines; average values per line were used in the statistical tests.
Discussion
Inbreeding depression
In populations of haplodiploids recessive deleterious alleles will differ in frequency depending on whether they are limited to expression in females or not. We investigated whether effects of inbreeding on life-history traits of T. urticae emerge from the extent to which relevant coding genes are uniquely expressed in females. Unlike longevity and juvenile survival, the oviposition rate, that is, the only trait unique to females, was negatively affected by strong inbreeding (F=0.875). This negative effect also arose after a single round of inbreeding (F=0.5) in females from the base population, as well as in females from lines without a history of inbreeding. Given that there are probably more genes with female-limited expression controlling oviposition than the two generic traits, the contrasting effects of inbreeding are likely due to differential genetic control.
For the maturity rate our results are less easy to interpret, as negative effects of inbreeding on this trait were present in both sexes. A negative effect on male maturity rate cannot emerge from increased homozygosity, as males are hemizygous. More likely the maturity rate in males and females is (partially) controlled by their mother, who was also inbred in our experimental setup. This explanation is supported by the experiment on purging, in which the offspring was inbred but the mother was outbred (in the population without a history of inbreeding, Table 4), because, here, there was no negative effect on male or female maturity rate. Admittedly, the degree of inbreeding was lower in this experiment (F=0.5 instead of F=0.875), but probably enough for negative effects to arise, as the oviposition rate was affected by inbreeding in each of the two setups. Taking into account that inbreeding to F=0.875 had slightly less impact on the oviposition rate than on the maturity rate (RP=0.14 and on average 0.19, respectively; Table 1), roughly the same potential for an effect would be expected at F=0.5 (assuming similar epistatic variation in both traits, Lynch and Walsh, 1998, chapter 10). However, at F=0.5 decreases were found in oviposition rate but not in maturity rate. Probably, the maturity rate is under (partial) maternal control thereby determining the impact of inbreeding on both sexes. As there is no evidence for maternal care in T. urticae, the underlying mechanism may well depend on how mothers allocate nutrients to eggs.
To our best knowledge, only two published studies on haplodiploids considered inbreeding depression with regard to gender (Saito et al., 2000; Henter, 2003). In the parasitoid Uscana semifumipennis inbreeding led to more or less equal reduction in reproductive performance (number of adult offspring out of eggs produced in 48 h) and female longevity (Henter, 2003). To explain this, Henter (2003) considered differential genetic control of longevity in the two genders (resulting in a female-specific longevity trait) or a weak relationship of longevity with fitness (resulting in weak purifying selection on longevity). A difficulty in interpreting her data is that performance includes both oviposition rate and juvenile survival, but their effects cannot be separated. In another study on the mite Stigmaeopsis miscanthi, inbreeding lines were compared with outcrossing lines (Saito et al., 2000; Mori et al., 2005). The oviposition rate decreased in some of the inbreeding lines and no effect was found on the survival rate. This is consistent with the results we found in T. urticae, but it is unclear to what degree the S. miscanthi results are caused by inbreeding depression in the inbred lines or by heterosis in the outcrossed lines, which may have led to artificially high levels of heterozygosity. Our data on gender-related inbreeding depression in haplodiploids are therefore more amenable to interpretation.
Purging during inbreeding
During periods of inbreeding, increased homozygosity will emerge in diploid organisms and this causes recessive deleterious alleles to be exposed to selection, thereby purging the population from these alleles. We subjected lines of T. urticae to a strong inbreeding regime and found indirect evidence for purging with respect to the oviposition rate (Table 2). It appears that recessive deleterious alleles for oviposition rate in the lines had been purged during the imposed inbreeding episode to such a degree that there were no more detectable negative effects of inbreeding at F=0.5. The absence of inbreeding depression after the inbreeding episode appears not to be a consequence of maternal effects. A comparison of data in Tables 3 and 4 for outbred offspring of mothers with and without a history of inbreeding, respectively, shows that there were no significant differences for any of the traits we measured: the mean value of one category differed always less than one s.e. from the mean value of the other category.
Whether purging occurred in the (female or male) maturity rate is unclear. As we hypothesize that negative inbreeding effects occur via maternal control of the trait, the setup we used was not suitable for investigating purging in maturity rate, because the mothers in both treatments were outbred. This may explain why there was no difference in intensity of inbreeding depression with regard to maturity rate between the populations with and without a history of inbreeding (Table 2).
In the literature, most proof for purging has been sought in the form of higher outbred fitness of inbred families than of outbred families or a rebound of fitness with increasing inbreeding (reviewed in Crnokrak and Barrett, 2002). However, this type of result leaves room for the alternative interpretation of adaptation to laboratory conditions in the inbred families (Willis, 1999), especially because the outbred families used are often not subjected to the same growing conditions as the inbred families, because the outbred families are taken from the base population (Crnokrak and Barrett, 2002). In our experiments we reduced the potential for a differential influence of laboratory adaptation by subjecting the outbreeding lines to the same laboratory conditions and the same procedure as the inbreeding lines. However, outbred lines may still adapt faster to laboratory conditions as they are expected to harbour more genetic variation (Willis, 1999). Both treatments led to a considerable loss of lines (Figure 2), which suggests that laboratory conditions imposed significant selection in this inbreeding experiment.
The influence of laboratory adaptation can be controlled for by comparing the intensity of inbreeding depression between populations with and without a history of inbreeding (Willis, 1999). To our best knowledge, we are the first to use this method and to demonstrate purging in a haplodiploid species. In diploid species, the few studies carried out show mixed results. No evidence for purging was found for any life-history trait in the common monkey-flower Mimulus guttatus (Willis, 1999) or for fecundity in the bulb mite Rhizoglyphus robini (Radwan, 2003). However, evidence for purging was found for egg production in the fruitfly Drosophila melanogaster in two cases (Swindell and Bouzat, 2006a, 2006b) and for some life-history traits of the bean weevil Stator limbatus (Fox et al., 2008). Theory on purging predicts that under a regime of brother–sister mating, purging of deleterious alleles for female fecundity will only be possible if they are strongly recessive with large negative effects (Hedrick, 1994; Byers and Waller, 1999; Wang et al., 1999). The lines we tested experienced an even stronger inbreeding regime (mother–son mating) and purging still occurred. This suggests that strongly recessive and deleterious alleles affecting the oviposition rate were present in the base population of T. urticae.
Conclusion
Thus, inbreeding affects life-history traits of both females and males, and these effects depend on the extent of unique female expression of genes affecting the traits (see also the concept of narrow expression breadth and its effect on purging; Szövenyi et al., 2013). Hence, our results support the hypothesis that in haplodiploids the degree of female genetic control determines the amount of genetic variation for a life-history trait due to deleterious recessive alleles. Ideally, this should be tested using various female and various generic life-history traits within a species. However, in T. urticae as well as in many other haplodiploids there is only one female life-history trait: egg production. Therefore, we advocate interspecific or interpopulation comparisons to further test this hypothesis.
Data archiving
Data available from the Dryad Digital Repository: doi:10.5061/dryad.cb58n.
Acknowledgments
This work was made possible by an ALW-grant from the Netherlands Organisation for Scientific Research (NWO).
The authors declare no conflict of interest.
References
- Ågren J, Schemske DW. (1993). Outcrossing rate and inbreeding depression in 2 annual monoecious herbs, Begonia hirsuta and B. semiovata. Evolution 47: 125–135. [DOI] [PubMed] [Google Scholar]
- Avery PJ. (1984). The population genetics of haplo-diploids and X-linked genes. Genet Res 44: 321–341. [Google Scholar]
- Byers DL, Waller DM. (1999). Do plant populations purge their genetic load? Effects of population size and mating history on inbreeding depression. Annu Rev Ecol Syst 30: 479–513. [Google Scholar]
- Charlesworth D, Charlesworth B. (1987). Inbreeding depression and its evolutionary consequences. Annu Rev Ecol Syst 18: 237–268. [Google Scholar]
- Charlesworth D, Charlesworth B. (1992). The effects of selection in the gametophyte stage on mutational load. Evolution 46: 703–720. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Willis JH. (2009). The genetics of inbreeding depression. Nat Rev Genet 10: 783–796. [DOI] [PubMed] [Google Scholar]
- Crawley MJ. (2007). The R Book. John Wiley & Sons Ltd: Chichester, UK. [Google Scholar]
- Crnokrak P, Barrett SCH. (2002). Perspective: purging the genetic load: a review of the experimental evidence. Evolution 56: 2347–2358. [DOI] [PubMed] [Google Scholar]
- Crozier RH. (1976). Why male-haploid and sex-linked genetic systems seem to have unusually sex-limited mutational genetic loads. Evolution 30: 623–624. [DOI] [PubMed] [Google Scholar]
- Fox CW, Scheibly KL, Reed DH. (2008). Experimental evolution of the genetic load and its implications for the genetic basis of inbreeding depression. Evolution 62: 2236–2249. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. (1994). Purging inbreeding depression and the probability of extinction: full-sib mating. Heredity 73: 363–372. [DOI] [PubMed] [Google Scholar]
- Hedrick PW, Parker JD. (1997). Evolutionary genetics and genetic variation of haplodiploids and X-linked genes. Annu Rev Ecol Syst 28: 55–83. [Google Scholar]
- Henter HJ. (2003). Inbreeding depression and haplodiploidy: experimental measures in a parasitoid and comparisons across diploid and haplodiploid insect taxa. Evolution 57: 1793–1803. [DOI] [PubMed] [Google Scholar]
- Johnston MO, Schoen DJ. (1994). On the measurement of inbreeding depression. Evolution 48: 1735–1741. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. (1998). Genetics and Analysis of Quantitative Traits. Sinauer Associates Inc: Sunderland, MA, USA. [Google Scholar]
- Mori K, Saito Y, Sakagami T, Sahara K. (2005). Inbreeding depression of female fecundity by genetic factors retained in natural populations of a male-haploid social mite (Acari: Tetranychidae). Exp Appl Acarol 36: 15–23. [DOI] [PubMed] [Google Scholar]
- Poissant J, Wilson AJ, Coltman DW. (2009). Sex-specific genetic variance and the evolution of sexual dimorphism: a systematic review of cross-sex genetic correlations. Evolution 64: 97–107. [DOI] [PubMed] [Google Scholar]
- Quinn GP, Keough MJ. (2006). Experimental Design and Data Analysis for Biologists. Cambridge University Press: Cambridge, UK. [Google Scholar]
- R Development Core Team. (2009). R: A language and environment for statistical computing. R-Foundation-for-Statistical-Computing, Vienna, Austria, http://www.R-project.org/.
- Radwan J. (2003). Inbreeding depression in fecundity and inbred line extinction in the bulb mite, Rhizoglyphus robini. Heredity 90: 371–376. [DOI] [PubMed] [Google Scholar]
- Roff DA. (2002). Inbreeding depression: tests of the overdominance and partial dominance hypotheses. Evolution 56: 768–775. [DOI] [PubMed] [Google Scholar]
- Saito Y, Sahara K, Mori K. (2000). Inbreeding depression by recessive deleterious genes affecting female fecundity of a haplo-diploid mite. J Evol Biol 13: 668–678. [Google Scholar]
- Simmons FH, Crow JF. (1977). Mutations affecting fitness in Drosophila populations. Annu Rev Genet 11: 49–78. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. (1995). Biometry. The Principles and Practice of Statistics in Biological Research. Freeman and Company: New York, NY, USA. [Google Scholar]
- Swindell WR, Bouzat JL. (2006. a). Reduced inbreeding depression due to historical inbreeding in Drosophila melanogaster: evidence for purging. J Evol Biol 19: 1257–1264. [DOI] [PubMed] [Google Scholar]
- Swindell WR, Bouzat JL. (2006. b). Selection and inbreeding depression: effects of inbreeding rate and inbreeding environment. Evolution 60: 1014–1022. [PubMed] [Google Scholar]
- Szövenyi P, Ricca M, Hock Z, Shaw JA, Shimizu KK, Wagner A. (2013). Selection is no more efficient in haploid than in diploid life stages of an angiosperm and a moss. Mol Biol Evol 30: 1929–1939. [DOI] [PubMed] [Google Scholar]
- Wang J, Hill WG, Charlesworth D, Charlesworth B. (1999). Dynamics of inbreeding depression due to deleterious mutations in small populations: mutation parameters and inbreeding rate. Genet Res 74: 165–178. [DOI] [PubMed] [Google Scholar]
- Werren JH. (1993). The evolution of inbreeding in haplodiploid organisms. in: Thornhill NW (ed.) The Natural History of Inbreeding and Outbreeding. Theoretical and Empirical Perspectives. The University of Chicago Press: Chicago, IL, USA. [Google Scholar]
- Willis JH. (1999). The role of genes of large effect on inbreeding depression in Mimulus guttatus. Evolution 53: 1678–1691. [DOI] [PubMed] [Google Scholar]