Abstract
Inter-specific hybridization may be especially detrimental when one species is extremely rare and the other is abundant owing to the potential for genetic swamping. The Cuban crocodile (Crocodylus rhombifer) is a critically endangered island endemic largely restricted to Zapata Swamp, where it is sympatric with the widespread American crocodile (C. acutus). An on-island, C. rhombifer captive breeding program is underway with the goals of maintaining taxonomic integrity and providing a source of individuals for reintroduction, but its conservation value is limited by lack of genetic information. Here we collected mtDNA haplotypic and nuclear genotypic data from wild and captive C. rhombifer and C. acutus in Cuba to: (1) investigate the degree of inter-specific hybridization in natural (in situ) and captive (ex situ) populations; (2) quantify the extent, distribution and in situ representation of genetic variation ex situ; and (3) reconstruct founder relatedness to inform management. We found high levels of hybridization in the wild (49.1%) and captivity (16.1%), and additional evidence for a cryptic lineage of C. acutus in the Antilles. We detected marginally higher observed heterozygosity and allelic diversity ex situ relative to the wild population, with captive C. rhombifer exhibiting over twice the frequency of private alleles. Although mean relatedness was high in captivity, we identified 37 genetically important individuals that possessed individual mean kinship (MK) values lower than the population MK. Overall, these results will guide long-term conservation management of Cuban crocodiles for maintaining the genetic integrity and viability of this species of high global conservation value.
Introduction
Interbreeding of genetically differentiated populations, independent of their evolutionary trajectory, has had a key role in the evolution of both animals (Dowling and Secor, 1997) and plants (Stebbins, 1950). Nevertheless, despite wide recognition of hybridization as part of the evolutionary process mediating adaptation and speciation, it remains an important issue in species conservation. Hybridization caused by human activities has resulted in significant threats to many taxa by both replacement and genetic mixing (Rodriguez et al., 2011). For that reason, it is generally desirable to implement management practices that avoid anthropogenically driven hybridization with some exceptions (Allendorf et al., 2001).
Hybridization has been widely reported among members of the genus Crocodylus, both in the wild and in captivity, with several examples primarily due to anthropogenic causes (FitzSimmons et al., 2002; Rodriguez et al., 2011). From a conservation perspective, hybridization is particularly concerning when one species is extremely rare and the other is more abundant (Rhymer and Simberloff, 1996; Allendorf et al., 2001). In terms of New World crocodiles, there is evidence of hybridization involving two range-restricted species, the Cuban crocodile (C. rhombifer; Varona, 1966; Weaver et al., 2008; Milián-García et al., 2011) and Morelet's crocodile (C. moreletii; Hekkala, 2004; Ray et al., 2004; Cedeño-Vazquez et al., 2008; Rodriguez et al., 2008; Machkour-M'Rabet et al., 2009) with overlapping populations of the American crocodile (C. acutus), that is more broadly distributed in North America, South America and the Caribbean (Ramos et al., 2004; Thorbjarnarson et al., 2006).
Crocodylus rhombifer is endemic to the main island of Cuba, largely restricted to Zapata Swamp (Matanzas Province), where it is sympatric with C. acutus; in most other coastal areas in Cuba, including Birama Swamp, C. acutus is found in allopatry (Soberón, 2009). A previous study involving captive individuals revealed two distinct mitochondrial DNA (mtDNA) lineages originally characterized as C. rhombifer-α and C. rhombifer-β, with the latter exhibiting C. acutus morphology (Weaver et al., 2008). The authors proposed that C. rhombifer-β may constitute a previously unidentified lineage of New World Crocodylus, but suggested that reference samples from wild populations in Cuba were necessary to directly test this hypothesis (Weaver et al., 2008). A subsequent study involving wild populations of Cuban Crocodylus, including C. rhombifer from Zapata Swamp, an allopatric population of C. acutus in Birama Swamp and exemplars of mainland C. acutus, likewise revealed a split between Cuban C. acutus (equivalent to C. rhombifer-β from Weaver et al., 2008) and mainland C. acutus, both of which were genetically and morphologically distinct from C. rhombifer (Milián-García et al., 2011). Both of these previous studies also revealed evidence for low levels of hybridization between C. rhombifer and C. acutus in captivity (Weaver et al., 2008) and in the wild (Milián-García et al., 2011), yet they were not equipped to explicitly characterize patterns of in situ hybridization and population history within Cuban Crocodylus, as neither included representative samplings of C. rhombifer and C. acutus where their ranges overlap in Zapata Swamp.
The Cuban crocodile is listed as critically endangered (IUCN, 2013) owing to the effects of illegal hunting, habitat modifications, small population size and limited knowledge of species biology. Compounding these issues is the serious potential threat of loss of the Cuban crocodile's genomic identity through hybridization with the American crocodile (Targarona et al., 2008).
Ex situ management in the form of captive breeding constitutes one important conservation strategy for preserving the genetic identity of this Cuban endemic species. The primary breeding facility is located in Zapata Swamp, originally founded in 1959 with hundreds of individuals of both species (C. rhombifer and C. acutus) caught in the surrounding area. In 1974, the program began focusing on maintaining the genetic identity of C. rhombifer using an arbitrary set of morphological characters to remove suspected C. acutus and C. acutus–C. rhombifer hybrids. To date, the captive breeding program in Zapata Swamp has increased its population to 4086 individuals divided into multiple age categories, including neonates (0–1 years), juveniles (>1–6 years), replacement (subadults) and adults.
Although Cuba's in-country captive population in Zapata Swamp is considered the most important source for C. rhombifer, the lack of genetic information is impeding the establishment of effective long-term conservation management plans. In general, there are three genetic issues of special concern in captive breeding programs: (1) loss of genetic variability; (2) inbreeding depression; and (3) adaptation to the captive environment (Lynch and O'Hely, 2001; Frankham et al., 2002). A fourth genetic threat also applies to C. rhombifer in terms of the potential negative effects of hybridization on genomic and taxonomic integrity. To date, the degree of hybridization that has occurred in the Zapata Swamp captive population has not been evaluated.
Given the critically endangered listing of C. rhombifer (IUCN, 2013), a more thorough understanding of population history and in situ hybridization with C. acutus is necessary for fully evaluating the status and informing recovery. In addition, the Zapata Swamp captive population represents an important hedge against extinction, yet the lack of genetic management may further undermine future conservation goals. Here we used mtDNA haplotypic and microsatellite genotypic data to: (1) investigate patterns of hybridization and introgression between C. rhombifer and C. acutus both in the wild and captivity; (2) assess the degree to which captive C. rhombifer is representative of the gene pool of the extant wild population; and (3) estimate levels of relatedness and inbreeding among C. rhombifer breeders to inform management recommendations based on a strategy of minimizing mean kinship (MK).
Materials and Methods
Sampling
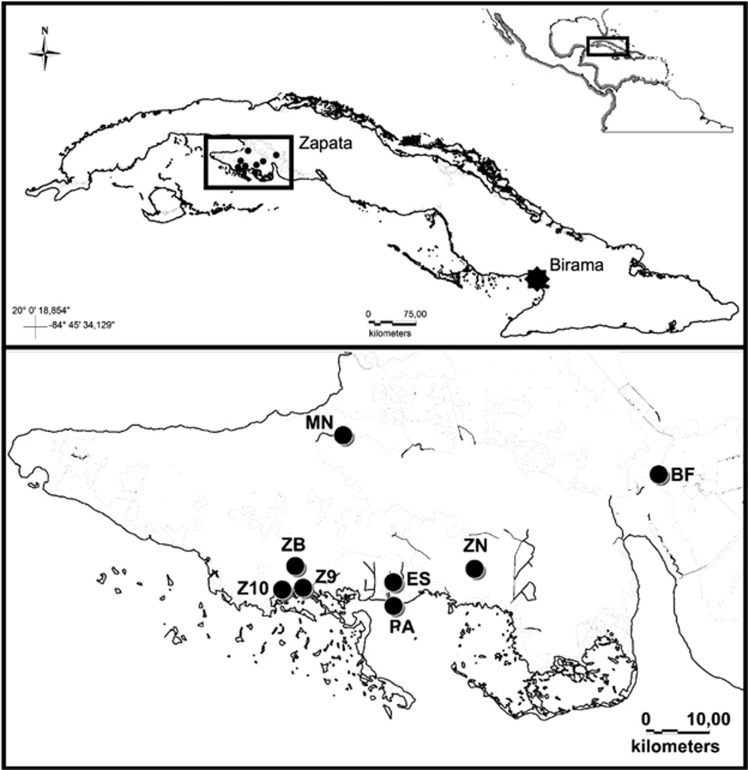
A total of 227 individuals were sampled from wild populations in Zapata Swamp (n=67) and Birama Swamp (n=23), as well as the ex situ population at the Zapata Swamp Captive Breeding Farm (n=137) between 2007 and 2012, including exemplars sampled in 2000 and 2002 (Figure 1; Supplementary Table S1). Each sample consisted of a piece of scale clipped from the animal's tail. Individuals were morphologically classified as C. rhombifer, C. acutus or as suspected hybrids based on 26 external characters (Ramos Targarona, 2013).
Figure 1.
Sampling of C. rhombifer and C. acutus, including specific localities within Cuba. Sites within Zapata Swamp (inset) are indicated by black circles, while Birama Swamp is shown with a black star. Sampling localities in Zapata Swamp are as follows: Breeding Farm (BF), Estamento (ES), Maneadero (MN), Punta Arena (PA), Zanja del 9 (Z9), Zanja del 10 (Z10), Zona del Brigadista (ZB), and Zona Novo (ZN).
Samples from the wild were taken from adults, sub-adults and neonates from different nests to avoid underestimation of genetic variability. Individuals sampled in the wild Zapata Swamp population were morphologically classified as C. rhombifer (n=42), C. acutus (n=14) or suspected hybrids (n=11) as described above and distributed across seven collecting sites (Estamento, Maneadero, Punta Arena, Zona del Brigadista, Zanja del 9, Zanja del 10 and Zona Novo; Figure 1; Supplementary Table S1). The 23 samples from Birama Swamp were all wild C. acutus from the Wildlife Refuge Monte Cabaniguán, where they are allopatric relative to C. rhombifer (Figure 1). Precise geographic coordinates were withheld to protect these threatened wild populations. Regarding the ex situ population, only adults and sub-adults were included. All samples were collected and transported in accordance with an agreement established between the Faculty of Biology at the University of Havana and the National Enterprise for the Protection of Flora and Fauna in Cuba and CITES permits C0001166 and C0001455.
DNA extraction, sequencing and genotyping
Total DNA was isolated from ethanol-preserved tail scale tissue using the NucleoSpin kit for DNA extraction (Macherey-Nagel, Düren, Germany) following the manufacturer's instructions. A ~458-base-pair segment of the mitochondrial genome (mtDNA), including the tRNAPro-tRNAPhe-D-loop region was amplified as a single fragment for all wild individuals (n=90) using primers drL15459 (5′-AGGAAAGCGCTGGCCTTGTAA-3′) and CR2HA (5′-GGGGCCACTAAAAACTGGGGGGA-3′) (Weaver et al., 2008). PCRs were carried out on an ABI Veriti thermal cycler in 25 μl reactions, containing: 20–50 ng of DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, 0.5 μM of each primer and 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA, USA). Cycling conditions were as follows: 94 °C (2 min), 33 cycles of 94 °C (30 s), 58 °C (30 s), 72 °C (45 s), and a final extension of 72 °C (7 min). All PCR products were purified by ExoSAP-IT (USB Products, Santa Clara, CA, USA) and sequenced using an ABI 3130XL Genetic Analyzer (Applied Biosystems).
All samples were genotyped at nine polymorphic microsatellite loci (Cj16, Cj18, Cj20, Cj35, Cj109, Cj119, Cj127, Cj131, CU5-123) using published primers (FitzSimmons et al., 2000; Dever and Densmore, 2001). All forward primers were 5′-tailed with an M13 sequence (5′-TCCCAGTCACGACGT-3′) to facilitate automated genotyping. Specifically, the M13-tailed forward primer was used in combination with an M13 primer of the same sequence 5′-labeled with one of four fluorescent dyes (6-FAM, VIC, NED, PET), effectively incorporating the label into the resulting PCR amplicon (Schuelke, 2000). In addition, reverse primers were modified following Brownstein et al., (1996) to improve genotyping. All PCRs were carried out on an ABI Veriti thermal cycler in 12.5 μl reactions containing: ~20–50 ng of DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, 0.08 μM of the M13-tailed forward primer, 0.8 μM of each of the reverse primer and the M13 fluorescent dye-labeled primer, and 0.5 U of AmpliTaq DNA polymerase (Applied Biosystems). Reaction conditions for all primers were as follows: 95 °C (2 min), 8 cycles of 95 °C (30 s), 59 °C (30 s), 72 °C (45 s), followed by 32 cycles of 95 °C (30 s), 51 °C (30 s), 72 °C (45 sec), and a final extension of 72 °C (10 min). Fragments were separated on an ABI 3130XL Genetic Analyzer and scored using GENEMAPPER 4.0 (Applied Biosystems).
Haplotypic variation
MtDNA sequences were edited and aligned in Sequencher 4.5 (Gene Codes, Ann Arbor, MI, USA). Haplotypic (h) and nucleotide (π) diversity (Nei, 1987) estimates were calculated based on mtDNA sequences as executed in DNASP v5 (Librado and Rozas, 2009). A haplotype network was generated using statistical parsimony as implemented in TCS (Clement et al., 2000).
To examine observed haplotypic variation recovered in Cuban Crocodylus within the context of all New World crocodiles, we reconstructed a Bayesian haplotype tree. Previously published overlapping mtDNA sequences were downloaded from Genbank representing C. rhombifer-α (GU064608; Rodriguez et al., 2011), C. acutus haplotypes published to date, including β (EU034586; Weaver et al., 2008) and Ca1-Ca8 (AY568314, GU064603, AY568312, AY568310, AY568311, GU086321, AY341530, GU064607; Rodriguez et al., 2011), as well as exemplars from vouchered specimens (Oaks, 2011) of C. intermedius (JF315382), C. moreletii (JF315380), C. niloticus (JF315356) and C. porosus (JF315369). Sequences were unambiguously aligned using MUSCLE as implemented in GENEIOUS 4.6 (Biomatters, Ltd., San Francisco, CA, USA) employing default settings. The Bayesian haplotype tree was reconstructed using MRBAYES 3.1 (Ronquist and Huelsenbeck, 2003) assuming the HKY+I+G model of nucleotide substitution as selected according to the Akaike information criterion as implemented in MODELTEST (Posada and Crandall, 1998). The saltwater crocodile (C. porosus; JF315369) was used as an outgroup to root the tree. The analysis ran four simultaneous chains for 2.0 × 106 total generations, each using a random tree as a starting point, the default heating scheme and saving a tree every 100 generations for a total 20 000 trees. The first 2000 trees were discarded as burn-in samples and the remaining 18 000 trees were used to construct a majority-rule consensus tree and derive posterior probability values.
Genotypic variation
The genotypic data set was examined for the presence of null alleles using MICROCHECKER (Van Oosterhout et al., 2004). Deviation from Hardy–Weinberg equilibrium was assessed using exact tests, as implemented in GENEPOP 3.3 (Raymond and Rousset, 1995; Rousset, 2008). Linkage disequilibrium was investigated for all pairs of loci using GENEPOP 3.3 (Raymond and Rousset, 1995; Rousset, 2008). Type I error rates for tests of linkage disequilibrium and departure from Hardy–Weinberg equilibrium were corrected for multiple comparisons using the sequential Bonferroni's procedure (Rice, 1989).
Allelic richness, number of private alleles, observed (Ho) and expected heterozygosity (He) were calculated at each locus for each site using GENALEX 6.5 (Peakall and Smouse, 2006). Estimates of allelic richness and frequency of private alleles were also corrected for sample size using HP-RARE (Kalinowski, 2005). Levels of genetic differentiation among groups were estimated by pairwise comparisons of the unbiased estimator, θ (Weir and Cockerham, 1984), as calculated in FSTAT (Goudet, 2001). In addition, the model-based clustering method implemented in STRUCTURE 2.3.4 (Pritchard et al., 2000) was used to infer population structure and to identify potential hybrids within the sampled wild populations. Run length was set to 1 000 000 MCMC (Markov chain Monte Carlo) replicates after a burn-in period of 500 000 using correlated allele frequencies under a straight admixture model. We varied the number of clusters (K) from 1 to 8, with 20 replicates for each value of K. The most likely number of clusters was determined by plotting the log probability of the data (ln Pr(X|K) (Pritchard et al., 2000) across the range of K values tested and selecting the K where the value of ln Pr(X|K) plateaued as suggested in the STRUCTURE manual. We also employed the ΔK method (Evanno et al., 2005) as calculated in STRUCTURE HARVESTER (Earl and vonHoldt, 2011). Results for the identified optimal values of K were summarized using CLUMPP (Jakobsson and Rosenberg, 2007) and used to estimate average cluster membership coefficients for each individual. The threshold membership coefficient for purebred identification was 0.05, similar to a previous study of hybridization between Cuban and American crocodiles (Weaver et al., 2008). In this case, any value between 0.0 and ⩽0.05 or ⩾0.95 and 1.0 was identified as purebred C. rhombifer and C. acutus, respectively. All other individuals were deemed to be of mixed ancestry. Final bar plots were constructed using DISTRUCT (Rosenberg, 2004).
In situ representation and relatedness of C. rhombifer in captivity
We used STRUCTURE 2.3.4 (Pritchard et al., 2000) to identify individuals of mixed ancestry within the Zapata Swamp captive population. Run length was set to 1 000 000 MCMC replicates after a burn-in period of 500 000 using correlated allele frequencies and prior population information, the latter corresponding to the distinct clusters identified in the analysis of the wild populations of C. rhombifer and C. acutus. As above, the threshold membership coefficient for purebred identification was ⩽0.05; all other individuals were deemed to be of mixed ancestry.
Pairwise relatedness was calculated among all non-hybrid individuals according to the method of Queller and Goodnight (1989) in iREL (Gonçalves da Silva and Russello, 2011). The observed distribution of pairwise relatedness were plotted with those of 1000 simulated dyads for each of the four relatedness categories (unrelated, half sibs, full sibs and parent–offspring) also using iREL (Gonçalves da Silva and Russello, 2011). The individual and overall inbreeding coefficients with 95% confidence intervals were calculated using the estimator of Ritland (1996) as implemented in COANCESTRY (Wang, 2011). Individual kinship coefficients were calculated in SPAGeDi 1.3 (Hardy and Vekemans, 2002) and used to calculate individual and population MK.
Results
Haplotypic variation
For the wild individuals (n=90), we recovered 10 polymorphic sites across the 458-base-pair mtDNA fragment, constituting four haplotypes (Figure 2). Overall haplotypic and nucleotide diversities were 0.485 and 0.00905, respectively. The most common haplotype was identical to the α haplotype previously described (Weaver et al., 2008) and was shared among C. rhombifer (n=26), C. acutus (n=5) and hybrids (n=26) in Zapata Swamp (Figures 2 and 3). The other shared haplotype was identical to the β haplotype (Weaver et al., 2008), differing by nine steps from the α haplotype, and was only detected in morphologically and genetically identified C. acutus (Figures 2 and 3). Two new haplotypes were detected in the current study, one in a single C. rhombifer individual (haplotype D; Figures 2 and 3) and the other in a single C. acutus individual in Zapata Swamp (haplotype C; Figures 2 and 3).
Figure 2.
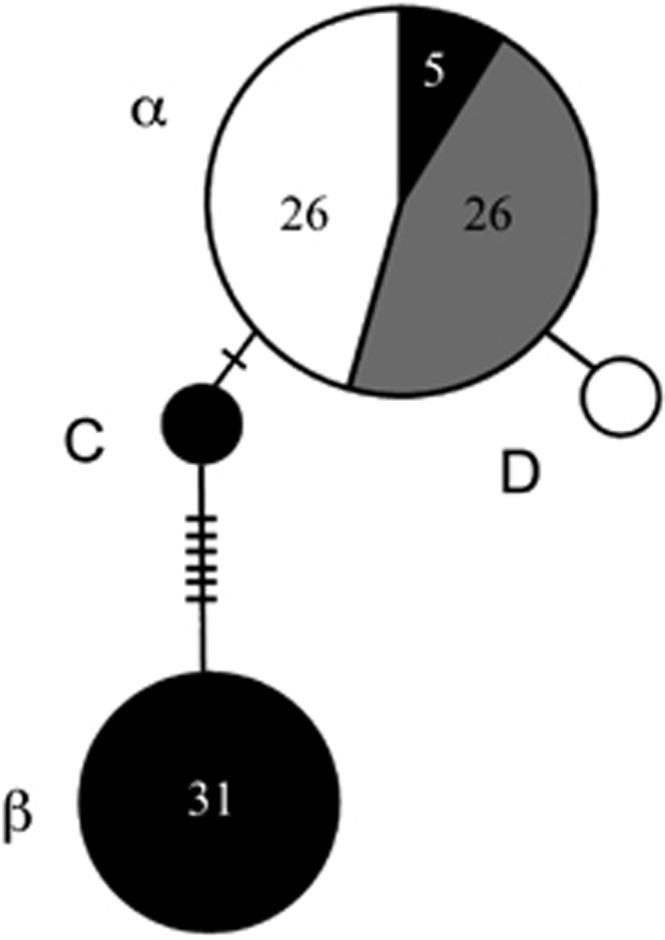
Haplotype network based on mtDNA control region data for wild Cuban Crocodylus. Shading indicates proportion of C. acutus (black), C. rhombifer (white) and putative hybrids (gray) possessing a given haplotype, as genetically identified by the STRUCTURE analysis (also see Figure 3, lower panel). The numbers of individuals possessing a haplotype is indicated inside the circles except for singletons. Inferred but unsampled haplotypes are indicated by slashes.
Figure 3.
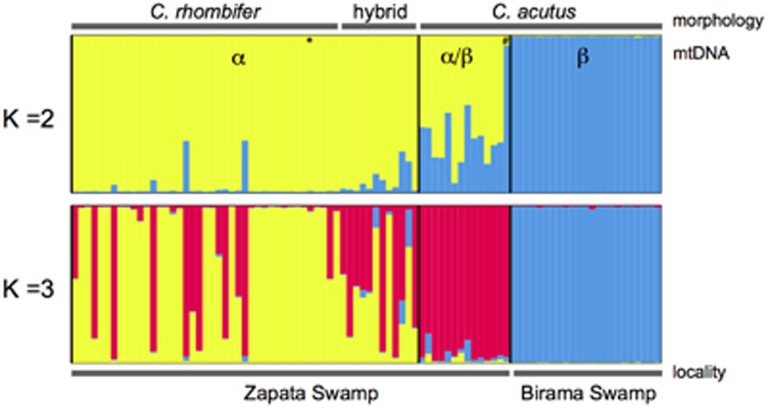
STRUCTURE bar plots depicting the model-based clustering results for inferred K=2 and K=3. Inferred genetic clusters are displayed as different colours. Each bar on the x axis represents an individual, with the y axis displaying the proportion of membership in each genetic cluster. Morphological identification as C. rhombifer, C. acutus or hybrid is indicated at the top, and sampling locality (Zapata Swamp or Birama Swamp) at the bottom. Recovered mtDNA haplotypes, including α and β, are superimposed on the plot, with black vertical lines indicating the boundaries. Individuals possessing rare haplotypes are indicated by * for haplotype D and # for haplotype C. A full color version of this figure is available at the Heredity journal online.
As detected in previous studies (Weaver et al., 2008; Milián-García et al., 2011), Cuban Crocodylus form a monophyletic group, relative to all other New World crocodiles analyzed, including exemplars from mainland C. acutus, rendering the latter species paraphyletic (Supplementary Figure S1). Within the Cuban Crocodylus clade, there is a well-supported split between the α and β haplotypes (posterior probability=1.00), with new haplotype D strongly clustering with α (posterior probability=0.91), and new haplotype C showing an affinity with β, although the latter is not well supported (Supplementary Figure S1).
Genotypic variation and degree of hybridization
In the microsatellite data, there was no evidence for null/false alleles in any population. There were two instances of evidence for linkage disequilibrium among pairs of loci following Bonferroni correction (Cj127/Cj131 in C. acutus from Birama Swamp; Cj127/Cj131 in Zapata Swamp captive population). There were two instances of deviation from Hardy–Weinberg equilibrium (Cj109 in C. acutus from Birama Swamp; Cj119 in Zapata Swamp captive population) once hybrids were removed (see below). As no consistent patterns of deviations from Hardy–Weinberg equilibrium or linkage equilibrium were detected across sites, downstream analyses were based on genotypic data at all nine microsatellite loci.
STRUCTURE analyses revealed two clusters in the data set (ΔK2=1081.28; Figure 3 and Supplementary Figure S2; Supplementary Table S2), generally corresponding to C. acutus and C. rhombifer, with a high degree of admixture in Zapata Swamp across morphologically identified C. rhombifer, C. acutus and hybrids (Figure 3). All Birama Swamp wild individuals were morphologically and genetically identified as C. acutus (Figure 3).
STRUCTURE analyses further provided evidence for three clusters in the data (ΔK3 =207.98; Figure 3 and Supplementary Figure S2; Supplementary Tables S1 and S2), corresponding to two distinct populations of C. acutus in Zapata and Birama Swamps, respectively, and a highly admixed group that consisted of morphological C. rhombifer individuals and hybrids (Figure 3). In the latter case, 26 of the 53 individuals (49.1%) were C. rhombifer × C. acutus hybrids. All genetic hybrids possessed the C. rhombifer-α mtDNA haplotype (Figure 3; Supplementary Table S1). Given the biologically meaningful pattern that emerges under a K=3 in light of morphology and geography, we proceeded with the identification of hybrids and subsequent analyses of the wild and captive populations under this framework as recommended by Pritchard et al. (2000).
With hybrids removed, rarefied allelic richness within wild populations ranged from 2.7 in C. acutus from Birama Swamp to 5.1 in C. acutus from Zapata Swamp (Table 1). The highest levels of observed heterozygosity were exhibited by C. acutus (0.62) and C. rhombifer (0.49) populations in Zapata Swamp (Table 1), with comparatively lower levels found in C. acutus from Birama Swamp (0.44; Table 1). The C. acutus wild populations exhibited 14 and 12 private alleles for Zapata and Birama Swamps, respectively, while C. rhombifer from Zapata Swamp possessed 13 (Table 1 and see footnote). Significant population differentiation was recovered across all pairwise comparisons among wild populations (Table 2).
Table 1. Haplotypic and genotypic variation within populations, including sample size (N), the number of haplotypes (N haplotypes), haplotypic diversity (h), allelic richness (A), rarified allelic richness (AR), observed heterozygosity (Ho) and expected heterozygosity (He), and the number of private alleles (PA) with the multi-locus average private allele frequency following rarefaction in parentheses.
| Species | Population | Na |
Mitochondrial DNA |
Microsatellites |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Nhaplotypes | h | A | AR | HO | HE | PAb | |||
| C. acutus | Zapata | 14 | 3 | 0.60 | 5.1 | 5.1 | 0.62 | 0.64 | 11 (1.6) |
| C. acutus | Birama | 23 | 1 | 0.00 | 3.0 | 2.7 | 0.44 | 0.42 | 12 (1.1) |
| C. rhombifer | Zapata | 27 | 2 | 0.09 | 4.1 | 3.6 | 0.49 | 0.54 | 2 (0.3) |
| C. rhombifer | Captive | 113 | ND | ND | 5.6 | 3.6 | 0.53 | 0.56 | 15 (0.7) |
Abbreviation: ND, not detected.
Sample sizes used for calculating summary statistics included only purebred individuals after hybrids removed.
When captive C. rhombifer is removed, private alleles among wild populations include C. rhombifer-Zapata (n=13), C. acutus-Zapata (n=14) and C. acutus-Birama (n=12).
Table 2. Matrix of θ (Weir and Cockerham, 1984) and P-values (below diagonal) for all pairwise comparisons of sampled populations of C. rhombifer and C. acutus, including from Zapata Swamp (ZS), Birama Swamp (BS) and the Zapata Swamp captive population (Captive).
| Population | C. rhombifer (ZS) | C. acutus (ZS) | C. acutus (BS) | Captive |
|---|---|---|---|---|
| C. rhombifer (ZS) | 0.3009 | 0.4695 | 0.0182 | |
| C. acutus (ZS) | <0.01** | 0.2606 | 0.3128 | |
| C. acutus (BS) | <0.01** | <0.01** | 0.4549 | |
| Captive | <0.01** | <0.01** | <0.01** |
**Significance at P⩽0.01.
In situ representation and relatedness of C. rhombifer in captivity
Twenty-two out of the 137 (16.1%) individuals sampled from the Zapata Swamp captive population were genetically classified as hybrids. Two captive individuals (C22012 and CB036) were genetically assigned to C. acutus, while the remainder (n=113) were assigned to C. rhombifer. The captive C. rhombifer exhibited comparable levels of rarified allelic richness (3.6) and observed heterozygosity (0.53) relative to the wild population in Zapata Swamp (Table 1). Fifteen of the 50 detected alleles in captive C. rhombifer were private, with over twice the frequency than that detected in the wild population following rarefaction (Table 1).
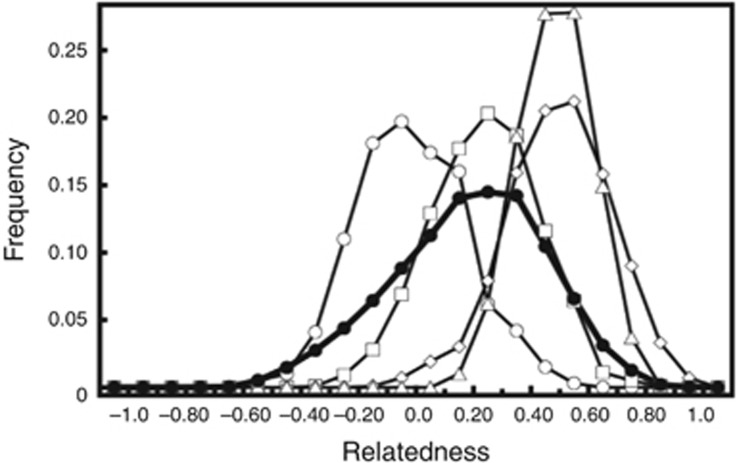
Mean pairwise relatedness (rxyQG) among the 113 C. rhombifer in captivity was 0.18. The observed distribution of rxyQG largely overlapped with the expected distribution for half-siblings (Figure 4). Average inbreeding coefficients were 0.086, with individual inbreeding coefficients ranging from −0.200 to 3.22 (Table 3). Population MK was −0.00501, with individual MKs ranging from −0.02909 to 0.00024 (Table 3). Thirty-seven of the 113 individuals exhibited MK values lower than the average MK for the captive population (Table 3).
Figure 4.
Observed distribution of pairwise relatedness (●) for captive C. rhombifer at the Zapata Swamp Breeding Farm plotted against expected distributions for each of the following relationship categories: non-related (○); half-siblings (□); full sibling (◊); and parent–offspring (△).
Table 3. Genetically important males (♂) and females (♀) within the Zapata Swamp captive population arranged in ascending order based on mean kinship (MK) with associated inbreeding coefficients (F) (Ritland, 1996); all high priority individuals with MK lesser than the population MK (−0.00501) are shown in bold.
| ID | Morphology | MK | F | Sex | ID | Morphology | MK | F | Sex | ID | Morphology | MK | F | Sex |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C22083 | rhombifer | −0.02909 | 3.2234 | ♀ | C22065 | rhombifer | −0.00489 | −0.0211 | ♂ | CB017 | rhombifer | −0.00248 | 0.0215 | ♀ |
| CB044 | rhombifer | −0.02407 | 0.4165 | ♀ | C22077 | rhombifer | −0.00488 | −0.0477 | ♀ | C22016 | rhombifer | −0.00240 | 0.3913 | ♀ |
| C22014 | rhombifer | −0.01891 | 0.0541 | ♂ | C22024 | rhombifer | −0.00484 | 0.1290 | ♀ | CB021 | rhombifer | −0.00238 | 0.1899 | − |
| C22069 | rhombifer | −0.01733 | −0.0936 | ♀ | CB012 | rhombifer | −0.00457 | −0.0465 | ♀ | CB004 | rhombifer | −0.00229 | 0.1049 | ♀ |
| C22055 | rhombifer | −0.01689 | 1.7682 | ♀ | C22085 | rhombifer | −0.00441 | −0.1223 | ♀ | CB013 | rhombifer | −0.00224 | −0.0503 | ♀ |
| C22018 | rhombifer | −0.01685 | 0.0166 | ♀ | CB006 | rhombifer | −0.00421 | 0.1464 | ♀ | C22063 | rhombifer | −0.00221 | 0.0269 | ♀ |
| CB003 | rhombifer | −0.01556 | 0.0288 | ♀ | C22023 | rhombifer | −0.00408 | 0.0381 | − | C22009 | rhombifer | −0.00221 | 0.0928 | ♀ |
| C22017 | rhombifer | −0.01434 | −0.1005 | ♀ | C22087 | rhombifer | −0.00399 | 0.0791 | ♀ | C22091 | rhombifer | −0.00214 | 0.1523 | ♀ |
| C22005 | rhombifer | −0.01365 | −0.0397 | ♀ | C22111 | rhombifer | −0.00385 | 0.1466 | ♀ | C22086 | rhombifer | −0.00213 | 0.0387 | ♂ |
| C22021 | rhombifer | −0.01198 | −0.1256 | ♂ | C22071A | rhombifer | −0.00381 | 0.1142 | − | C22112 | rhombifer | −0.00211 | −0.0231 | ♀ |
| CB027 | rhombifer | −0.01151 | −0.0985 | ♀ | C22107 | rhombifer | −0.00380 | 0.0646 | ♂ | C22015 | rhombifer | −0.00209 | 0.1697 | − |
| C22052 | rhombifer | −0.01148 | −0.1628 | ♀ | C22075 | rhombifer | −0.00377 | 0.0308 | ♀ | C22054 | rhombifer | −0.00202 | 0.0321 | ♀ |
| CB047 | rhombifer | −0.01014 | 0.0516 | ♂ | CB029 | rhombifer | −0.00355 | 0.0651 | ♀ | C22084 | rhombifer | −0.00202 | 0.0486 | ♀ |
| C22076 | rhombifer | −0.00902 | 0.0190 | ♀ | CB008 | rhombifer | −0.00348 | 0.3101 | ♀ | CB038 | rhombifer | −0.00201 | 0.1053 | ♀ |
| CB009 | rhombifer | −0.00894 | −0.0458 | ♂ | C22066 | rhombifer | −0.00346 | −0.0812 | ♀ | C22103 | rhombifer | −0.00197 | −0.0081 | ♀ |
| C22081 | rhombifer | −0.00846 | −0.0656 | ♀ | CB043 | rhombifer | −0.00344 | 0.3268 | ♀ | C22097 | rhombifer | −0.00190 | 0.1154 | ♂ |
| C22008 | rhombifer | −0.00830 | 0.4910 | ♀ | CB005 | rhombifer | −0.00324 | −0.1391 | ♀ | C22104 | rhombifer | −0.00189 | −0.0739 | ♀ |
| CB026 | rhombifer | −0.00797 | 0.0990 | ♀ | C22022 | rhombifer | −0.00319 | 0.0272 | ♀ | CB010 | rhombifer | −0.00187 | −0.0704 | ♀ |
| CB049 | rhombifer | −0.00733 | 0.1245 | ♂ | C22110 | rhombifer | −0.00313 | −0.0120 | ♀ | C22007 | rhombifer | −0.00187 | −0.0805 | ♂ |
| C22002 | rhombifer | −0.00731 | −0.1749 | ♀ | C22072 | rhombifer | −0.00312 | −0.0301 | ♂ | C22113 | rhombifer | −0.00186 | 0.0200 | ♀ |
| C22011 | rhombifer | −0.00729 | 0.3707 | ♀ | C22071B | rhombifer | −0.00311 | 0.1581 | — | C22060 | rhombifer | −0.00175 | 0.0768 | ♀ |
| C22094 | rhombifer | −0.00722 | 0.5451 | ♂ | C22099 | rhombifer | −0.00309 | 0.0977 | ♂ | CB041 | rhombifer | −0.00148 | −0.0847 | ♂ |
| C22098 | rhombifer | −0.00717 | −0.1592 | ♀ | CB050 | rhombifer | −0.00308 | 0.2986 | ♀ | C22004 | rhombifer | −0.00145 | −0.0977 | ♀ |
| C22096 | rhombifer | −0.00688 | 0.1144 | ♀ | C22082 | rhombifer | −0.00308 | −0.0982 | ♀ | CB019 | rhombifer | −0.00145 | 0.0683 | ♀ |
| C22053 | rhombifer | −0.00686 | −0.1222 | ♀ | C22073 | rhombifer | −0.00304 | −0.0412 | ♀ | C22068 | rhombifer | −0.00141 | −0.1103 | ♀ |
| CB011 | rhombifer | −0.00681 | −0.0590 | ♂ | C22062 | rhombifer | −0.00294 | 0.0271 | ♀ | C22058 | rhombifer | −0.00139 | −0.1391 | ♀ |
| C22064 | rhombifer | −0.00646 | −0.0104 | ♀ | CB024 | rhombifer | −0.00286 | −0.0335 | ♀ | CB048 | rhombifer | −0.00124 | 0.0741 | ♀ |
| CB032 | rhombifer | −0.00638 | 0.5503 | ♂ | C22095 | rhombifer | −0.00282 | 0.1159 | ♀ | CB020 | rhombifer | −0.00120 | 0.1785 | ♂ |
| C22061 | rhombifer | −0.00599 | −0.1592 | ♂ | CB031 | rhombifer | −0.00275 | 0.1711 | ♂ | C22026 | rhombifer | −0.00115 | −0.200 | ♂ |
| C22092 | rhombifer | −0.00585 | −0.0864 | ♀ | C22056 | rhombifer | −0.00272 | −0.0146 | ♂ | CB002 | rhombifer | −0.00114 | 0.0351 | ♀ |
| CB030 | rhombifer | −0.00568 | 0.0051 | ♂ | C22109 | rhombifer | −0.00264 | −0.0243 | ♂ | C22051 | rhombifer | −0.00099 | −0.0058 | ♀ |
| C22088 | rhombifer | −0.00550 | 0.1676 | ♀ | CB045 | rhombifer | −0.00263 | −0.0049 | ♀ | C22079 | rhombifer | −0.00085 | −0.1997 | ♀ |
| C22059 | rhombifer | −0.00534 | 0.0066 | ♀ | C22108 | rhombifer | −0.00262 | −0.0029 | ♂ | C22006 | rhombifer | −0.00083 | −0.1967 | ♀ |
| C22020 | rhombifer | −0.00529 | 0.4282 | ♀ | C22078 | rhombifer | −0.00258 | 0.0680 | ♂ | C22090 | rhombifer | −0.00076 | −0.1592 | ♀ |
| C22100 | rhombifer | −0.00520 | 0.4897 | ♀ | C22089 | rhombifer | −0.00258 | −0.0068 | ♀ | C22074 | rhombifer | −0.00052 | −0.1022 | ♂ |
| C22003 | rhombifer | −0.00518 | 0.4013 | ♂ | CB022 | rhombifer | −0.00256 | −0.0562 | ♂ | C22070 | rhombifer | −0.00052 | −0.1625 | ♀ |
| CB023 | rhombifer | −0.00514 | 0.1245 | ♀ | CB028 | rhombifer | −0.00256 | 0.0365 | ♀ | CB015 | rhombifer | 0.00024 | −0.1628 | ♀ |
| C22106 | rhombifer | −0.00490 | 0.0961 | ♂ | C22114 | rhombifer | −0.00252 | −0.0092 | ♀ |
Discussion
Population history and taxonomic status
New World crocodiles have a complex history that is continuing to be unraveled by the growing body of research dedicated to this topic (Dever and Densmore, 2001; Hekkala, 2004; Ray et al., 2004; Cedeño-Vazquez et al., 2008; Rodriguez et al., 2008; Weaver et al., 2008; Machkour-M'Rabet et al., 2009; Meredith et al., 2011; Milián-García et al., 2011; Oaks, 2011; Rodriguez et al., 2011). It has been previously recognized that the lineages endemic to Cuba are unique, corresponding to the morphologically divergent Cuban crocodile (C. rhombifer) and a cryptic lineage morphologically similar to the American crocodile (β) (Weaver et al., 2008; Milián-García et al., 2011), most likely the result of mitochondrial capture (Rodriguez et al., 2011). Our results provide additional insights, for the first time analyzing wild populations of Cuban C. acutus and C. rhombifer both in sympatry and allopatry within their native range. We found significant differentiation between non-hybrids of the endemic C. rhombifer and C. acutus both where their ranges overlap in Zapata Swamp and where they do not (Birama Swamp). Interestingly, C. acutus populations in Zapata and Birama Swamps also exhibited significant differentiation from each other (Table 2), possessed a high degree of private alleles (Table 1) and were reconstructed as distinct clusters in the STRUCTURE analysis (Figure 3). Moreover, whereas the Birama Swamp C. acutus was fixed for the β haplotype, morphological C. acutus in Zapata Swamp exhibited a mixture of both the α and β haplotypes, as well as a novel haplotype (C) (Figure 2). These patterns are consistent with the phylogeographic scenario of Milián-García et al. (2011), which proposed multiple colonization events of Cuba by the ancestors of modern-day C. rhombifer ~2 mya followed by a second event >0.35–0.22 mya by the ancestor of Cuban C. acutus. This reconstruction further suggests that ancient hybridization occurred and then ceased before 0.22 mya, accounting for the reconstructions of three distinct lineages, including C. rhombifer, Cuban C. acutus and mainland C. acutus (Milián-García et al., 2011). Here, our results indicate that hybridization has not ceased in Zapata Swamp; rather the distinctiveness of Zapata Swamp C. acutus may be more indicative of long-term, unidirectional hybridization (see below) that accounts for the distribution of both the α and β mtDNA haplotypes, continued morphological distinctiveness and significant differentiation from allopatric conspecifics in Birama Swamp.
These patterns present challenges for taxonomy, as is the case for other crocodilians where cryptic diversity has been detected (Eaton et al., 2009; Hekkala et al., 2011) or where hybridization occurs (for example, C. moreletii x C. acutus) (Hekkala, 2004; Ray et al., 2004; Cedeño-Vazquez et al., 2008; Rodriguez et al., 2008; Machkour-M'Rabet et al., 2009). Combined with the results of previous studies (Weaver et al., 2008; Milián-García et al., 2011), we found evidence for a cryptic lineage of American crocodile currently found in Cuba and other localities in the Antilles. Moreover, the persistent hybridization between C. rhombifer and C. acutus in Zapata Swamp (see below) violate the criterion for species status under the biological species concept (Mayr, 1942), as is the case in other parts of the American crocodile range where it overlaps with other species (for example, C. moreletii). Yet, the detectable genetic, morphological, ecological and ethological characteristics exhibited by C. rhombifer (Cuvier, 1807) still suggest that it is a unique lineage. Our ability to fully evaluate the taxonomic status of C. rhombifer and the cryptic Antillean lineage of C. acutus is limited by our sampling of the American crocodile, which was restricted to their Cuban distribution, as well as the limited number of loci (mtDNA and microsatellite) providing character data. Expanded population and character (morphological and molecular) samplings are required to more definitely disentangle the relationships between these distinct lineages and for placing them within the overall context of range-wide variation exhibited by the broadly distributed C. acutus.
Hybridization between Cuban and American crocodiles
Hybridization has long been recognized as having an important role in the diversification of both plants (Stebbins, 1950) and animals (Dowling and Secor, 1997) yet becomes a conservation concern when anthropogenic factors directly cause or exacerbate the frequency of the events. Complexities arise, however, with regard to detecting hybridization, quantifying introgression and inferring underlying causes (Allendorf et al., 2001). Hybridization and introgression are especially concerning for conservation when it involves the potential for genetic swamping of a rare species by a more abundant one, the frequency of which may be largely underappreciated (Rhymer and Simberloff, 1996). Crocodilians present additional challenges, where inter-specific hybridization is common (FitzSimmons et al., 2002; Machkour-M'Rabet et al., 2009; Rodriguez et al., 2011) and, in the case of the Cuban and American crocodiles, where evidence exists for both ancient and contemporary hybridization (this study; Weaver et al., 2008; Milián-García et al., 2011).
In this study, we detected a high degree of hybridization within wild (49.1% (26/53)) and captive (16.1% (22/137)) C. rhombifer populations in which most genetic hybrids (57.7% (15/26)) were misclassified on the basis of morphology. These results add to the growing list of studies documenting interspecific hybridization between members of the genus Crocodylus, many of which also found high misclassification rates when using morphology alone (Dever and Densmore, 2001; Hekkala, 2004; Cedeño-Vazquez et al., 2008; Rodriguez et al., 2008; Weaver et al., 2008; Machkour-M'Rabet et al., 2009; Rodriguez et al., 2011).
In addition, our microsatellite data revealed a substantial underestimation of the frequency of interspecific hybridization between Cuban Crocodylus reported in previous studies (Weaver et al., 2008; Milián-García et al., 2011), most likely due to the use of non-Cuban C. acutus specimens as a reference. We found that the C. acutus population in Zapata Swamp is genetically differentiated from conspecifics in Birama Swamp (FST=0.2606, P<0.01; Table 2) to a similar degree as to sympatric C. rhombifer (FST=0.3009, P<0.01; Table 2). Given these patterns and the potential extent of admixture within Zapata Swamp, clustering analyses conventionally used in assessing the presence and degree of hybridization (for example, STRUCTURE; Pritchard et al., 2000) may not infer multiple clusters with admixture but rather lump individuals into a single cluster relative to additional highly differentiated populations, such as when using as sole reference the allopatric population in Birama Swamp (Milián-García et al., 2011) or mainland populations (Weaver et al., 2008).
Ex situ genetic representation of the wild C. rhombifer population
From a genetic perspective, the management goals of any captive breeding program may be achieved by avoiding a decrease in the gene diversity of the founders. This diversity is defined relative to the source population from which the founders were drawn and represents just a subset of its gene diversity (Lacy, 1995). In the present study, we found slightly higher levels of observed heterozygosity and average number of alleles per locus in captivity relative to the wild C. rhombifer population (Table 1), including a large number of private alleles. These differences in the extent and distribution of in situ and ex situ genetic variation could be indicative of gene variants retained in the captive population that are now extirpated in the wild, as has been found in other critically endangered species (Henry et al., 2009). Alternatively, this pattern may have been influenced by long-term introgression with C. acutus in captivity dating back to the early years of the breeding program. Since establishment of the Zapata Swamp captive population in 1959, the population size has increased to 4086 individuals from a few hundred wild-caught founders of unknown identity. Over this time period, it is certainly feasible that generations of backcrossing may yield morphologically and genetically (mtDNA and nDNA) diagnosed C. rhombifer, yet still have some increased genetic variation from past hybridization with C. acutus. Additional studies involving genome-wide scans would be better equipped to tease apart these non-mutually exclusive interpretations.
Inbreeding and relatedness among captive C. rhombifer
Inbreeding has demonstrated negative effects on both reproduction and survival, leading to decreased reproductive fitness and increased extinction risk (Lacy, 1995; Frankham, 2005). In the on-island, captive population of C. rhombifer, we estimated a mean inbreeding coefficient >0 (0.086), and detected a high level of mean relatedness (0.18), on the order of half-siblings. This outcome was expected, however, considering the following: (1) the Zapata Swamp captive population has been effectively a closed population; with few exceptions, the current individuals were born in captivity dating back up to 10 generations; (2) the population was initially founded with hundreds of individuals with uncertain ancestry, yet the source was still a small wild population, which increases the probability of founders being close relatives; (3) due to the polygynous mating system of this species, the breeding pens typically contain one male with three or four females, immediately lowering the effective population size.
These challenges to maintaining a viable and representative captive C. rhombifer population can be somewhat mitigated by the use of genetic evidence for determining mate pairings based on a strategy of minimizing MK. Empirical and theoretical studies have shown that breeding based on minimizing MK helps maximize gene diversity and allows the propagation of founder alleles with lowest frequency (Lacy, 1995; Frankham et al., 2002). Prior to the current study, the captive breeding program of C. rhombifer in Zapata Swamp has proceeded in the absence of genetic data, relying solely on morphological, ethological, demographic and logistic information for implementing short-term management plans. The results presented here now provide an important genetic baseline for informing management decisions. Specifically, 37 out of the 113 non-hybrid individuals (32.4%) were highlighted as genetically important for management purposes, exhibiting values of MK lower than the overall MK in the captive population (Table 3). This collection of genetically important individuals included 11 males and 26 females, a reasonable sex ratio given the polygynous mating system of C. rhombifer, providing flexibility in implementing a scientifically informed breeding program.
Implications for in situ/ex situ conservation
The results of our study provide genetic data that will be integrated with morphological, ethological and demographic information together with logistical considerations for designing long-term management plans for C. rhombifer, a charismatic and critically endangered member of the Cuban herpetofauna. This work constitutes a first step in establishing an appropriate source population for prospective reintroduction and recovery plans in the wild. Also, it contributes to maintaining the genetic integrity of the Cuban crocodile ex situ, allowing the detection of hybrids and identification of genetically important individuals for implementing a breeding strategy based on minimizing MK. Overall, this work contributes to the first formal attempt in Cuba to establish a scientifically managed in situ/ex situ conservation program guided by genetic information.
Just as important, we now have reference data for evaluating the genetic identity of C. rhombifer outside of Cuba. For example, some specimens assigned to the Cuban crocodile abroad have been used in the establishment of phylogenetic hypotheses for the still unresolved genus Crocodylus (Meredith et al., 2011). The primary identification for these specimens was originally based on morphological characters that have proven to be insufficient in most hybridization scenarios involving Crocodylus spp. Moreover, the historical legacy of exporting C. rhombifer for intentional hybridization with other species (for example, C. siamensis) (FitzSimmons et al., 2002) for the purpose of improving stocks for the skin trade may further confound the genetic integrity of Cuban crocodiles abroad.
Similar to the results for C. rhombifer, the genetic data collected for C. acutus in Zapata and Birama Swamps now provide critical reference data for further characterizing cryptic diversity within mainland and Antillean American crocodiles, the resolution of which will likely directly inform conservation prioritization.
Data Archiving
Mitochondrial haplotypes are available in Genbank: accession numbers KM577701 and KM577702. Microsatellite data are available from the Dryad Digital Repository: 10.5061/dryad.r86n4.
Acknowledgments
We would like to dedicate this work to the memory of Juan Caiman (John Thorbjarnarson) for his invaluable contribution to the study of crocodilians in Cuba and abroad. We would particularly like to acknowledge the work of Roberto Carlos Frias Soler and the breeders at the Zapata Swamp Breeding Farm, as well as Paulo Bouza Rodríguez, Francisco Medina Tejera and Andrés Arencibia Estenoz, and Jose Álvarez Lemus for helping with essential logistics. We are grateful to George Amato, Lucia Reis Ribeiro and three anonymous reviewers for feedback on a previous version of this manuscript, David Rodriguez for providing C. acutus reference sequences and Roberto Rodríguez Soberón for thoughtful discussions regarding crocodilian conservation. The Canadian Department of Foreign Affairs and International Trade (DFAIT) and Canadian Bureau for International Education (CBIE) provided funding through the Emerging Leaders in the Americas Program (ELAP) that supported collaborations and student exchange between the University of British Columbia and the University of Havana (YMG). Research funding was provided by the Wildlife Conservation Society (nos 20100618 and 20110707), the Rufford Foundation (Small Grant for Nature Conservation nos 11409-1 and 15172-2), and the International Foundation for Science (no. D 5354).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001). The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16: 613–622. [Google Scholar]
- Brownstein MJ, Carpten JD, Smith JR (1996). Modulation of non-templated nucleotide addition by Taq DNA polymerase: Primer modifications that facilitate genotyping. Biotechniques 20: 1004–1010. [DOI] [PubMed] [Google Scholar]
- Cedeño-Vazquez JR, Rodriguez D, Calme Sophie, Perran Ross J, Densmore L III, Thorbjarnarson J (2008). Hybridization between Crocodylus acutus and Crocodylus moreletii in the Yucatan Peninsula: I. Evidence from mitochondrial DNA and morphology. J Exp Zool A Ecol Genet Physiol 309: 661–673. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada M, Crandall KA (2000). TCS: a computer program to estimate gene genealogies. Mol Ecol 9: 1657–1660. [DOI] [PubMed] [Google Scholar]
- Cuvier G (1807). Sur les différentes espèces de crocodiles vivants et sur leurs caractères distinctifs. Annls Mus Hist Nat Paris 10: 8–66. [Google Scholar]
- Dever JA, Densmore LD (2001). Microsatellites in Morelet's crocodile (Crocodylus moreletii) and their utility in addressing crocodilian population genetics questions. J Herpetol 35: 541–544. [Google Scholar]
- Dowling TE, Secor CL (1997). The role of hybridization and introgression in the diversification of animals. Annu Rev Ecol Syst 28: 593–619. [Google Scholar]
- Earl DA, vonHoldt BM (2011). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resourc 4: 359–361. [Google Scholar]
- Eaton MJ, Martin A, Thorbjarnarson J, Amato G (2009). Species-level diversification of African dwarf crocodiles (Genus Osteolaemus): a geographic and phylogenetic perspective. Mol Phylogenet Evol 50: 496–506. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- FitzSimmons NN, Buchan JC, Lam PV, Polet G, Hung TT, Thang NQ et al. (2002). Identification of purebred Crocodylus siamensis for reintroduction in Vietnam. J Exp Zool A Ecol Genet Physiol 294: 373–381. [DOI] [PubMed] [Google Scholar]
- FitzSimmons NN, Tanksley S, Forstner MRJ, Loius EE, Daglish R, Gratten J et al. (2000). Microsatellite markers for Crocodylus: new genetic tools for population genetics, mating system studies and forensics. In: Grigg GC, Seebacher F, Franklin CE (eds). Crocodilian Biology and Evolution. Surrey Beatty & Sons: Chipping Norton. pp 51–57. [Google Scholar]
- Frankham R (2005). Genetics and extinction. Biol Conserv 126: 131–140. [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA (2002). Introduction to Conservation Genetics. Cambridge University Press: Cambridge, MA, USA. [Google Scholar]
- Gonçalves da Silva A, Russello MA (2011). iREL: software for implementing pairwise relatedness estimators and evaluating their performance. Conserv Genet Resourc 3: 69–71. [Google Scholar]
- Goudet J (2001). FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html.
- Hardy OJ, Vekemans X (2002). SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620. [Google Scholar]
- Hekkala E, Shirley MH, Amato G, Austin JD, Charter S, Thorbjarnarson J et al. (2011). An ancient icon reveals new mysteries: mummy DNA resurrects a cryptic species within the Nile crocodile. Mol Ecol 20: 4199–4215. [DOI] [PubMed] [Google Scholar]
- Hekkala ER (2004). Conservation genetics at the species boundary: case studies from African and Caribbean crocodiles (Genus: Crocodylus). PhD thesis Columbia University: New York, New York, USA. [Google Scholar]
- Henry P, Miquelle D, Sugimoto T, McCullough DR, Caccone A, Russello MA (2009). In situ population structure and ex situ representation of the endangered Amur tiger. Mol Ecol 18: 3173–3184. [DOI] [PubMed] [Google Scholar]
- IUCN (2013). Red List of Threatened Species http://www.iucnredlist.org.
- Jakobsson M, Rosenberg NA (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST (2005). HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5: 187–189. [Google Scholar]
- Lacy RC (1995). Clarification of genetic terms and their use in the management of captive populations. Zoo Biol 14: 565–578. [Google Scholar]
- Librado P, Rozas J (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lynch M, O'Hely M (2001). Captive breeding and the genetic fitness of natural populations. Conserv Genet 2: 363–378. [Google Scholar]
- Machkour-M'Rabet S, Henaut Y, Charruau P, Gevrey M, Winterton P, Legal L (2009). Between introgression events and fragmentation, islands are the last refuge for the American crocodile in Caribbean Mexico. Mar Biol 156: 1321–1333. [Google Scholar]
- Mayr E (1942). Systematics and the Origin of Species. Columbia University Press: New York, NY, USA. [Google Scholar]
- Meredith RW, Hekkala ER, Amato G, Gatesy J (2011). A phylogenetic hypothesis for Crocodylus (Crocodylia) based on mitochondrial DNA: evidence for a trans-Atlantic voyage from Africa to the New World. Mol Phylogenet Evol 60: 183–191. [DOI] [PubMed] [Google Scholar]
- Milián-García Y, Venegas-Anaya M, Frías-Soler R, Crawford AJ, Ramos-Targarona R, Rodríguez-Soberón R et al. (2011). Evolutionary history of Cuban Crocodiles Crocodylus rhombifer and Crocodylus acutus inferred from multilocus markers. J Exp Zool A Ecol Genet Physiol 315: 358–375. [DOI] [PubMed] [Google Scholar]
- Nei M (1987). Molecular Evolutionary Genetics. Columbia University Press: New York, NY, USA. [Google Scholar]
- Oaks JR (2011). A time-calibrated species tree of Crocodylia reveals a recent radiation of the true crocodiles. Evolution 65: 3285–3297. [DOI] [PubMed] [Google Scholar]
- Peakall ROD, Smouse PE (2006). genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller DC, Goodnight KF (1989). Estimating relatedness using genetic markers. Evolution 43: 258–275. [DOI] [PubMed] [Google Scholar]
- Ramos R, de Buffrenil V, Ross J (2004). Current status of the Cuban crocodile, Crocodylus rhombifer, in the wild. In: Crocodiles, Proceedings of the 12th Working Meeting of the Crocodile Specialist Group. IUCN: Gland, Switzerland. pp 113–140. [Google Scholar]
- Ramos Targarona R (2013) Ecología y conservación del cocodrilo cubano (Crocodylus rhombifer) en la ‘Ciénaga de Zapata', Cuba. PhD thesis, Universidad de Alicante, España and Universidad de Pinar del Río, Cuba.
- Ray DA, Dever JA, Platt SG, Rainwater TR, Finger AG, McMurry ST et al. (2004). Low levels of nucleotide diversity in Crocodylus moreletii and evidence of hybridization with C. acutus. Conserv Genet 5: 449–462. [Google Scholar]
- Raymond M, Rousset F (1995). GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J Hered 86: 248–249. [Google Scholar]
- Rhymer JM, Simberloff D (1996). Extinction by hybridization and introgression. Annu Rev Ecol Evol Syst 27: 83–109. [Google Scholar]
- Rice WR (1989). Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Ritland K (1996). Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res 67: 175–185. [Google Scholar]
- Rodriguez D, Cedeño-Vazquez JR, Forstner MRJ, Densmore LD III (2008). Hybridization between Crocodylus acutus and Crocodylus moreletii in the Yucatan Peninsula: II. Evidence from microsatellites. J Exp Zool A Ecol Genet Physiol 309: 674–686. [DOI] [PubMed] [Google Scholar]
- Rodriguez D, Forstner MRJ, Moler PE, Wasilewski JA, Cherkiss MS, Densmore LD III (2011). Effect of human-mediated migration and hybridization on the recovery of the American crocodile in Florida (USA). Conserv Genet 12: 449–459. [Google Scholar]
- Ronquist F, Huelsenbeck JP (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA (2004). DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4: 137–138. [Google Scholar]
- Rousset F (2008). GENEPOP'007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resourc 8: 103–106. [DOI] [PubMed] [Google Scholar]
- Schuelke M (2000). An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18: 233–234. [DOI] [PubMed] [Google Scholar]
- Soberón RAR (2009) Situación, estrategia de conservación y uso sostenible de Crocodylus acutus en Cuba. PhD thesis, Universidad de Alicante, Alicante, Spain.
- Stebbins GL (1950). Variation and Evolution in Plants. Columbia University Press. [Google Scholar]
- Targarona R, Soberón R, Cotayo L, Alonso Tabet M, Thorbjarnarson J. (2008). Red List of Theattened Species. IUCN. [Google Scholar]
- Thorbjarnarson J, Mazzotti F, Sanderson E, Buitrago F, Lazcano M, Minkowski K et al. (2006). Regional habitat conservation priorities for the American crocodile. Biol Conserv 128: 25–36. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004). Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535–538. [Google Scholar]
- Varona LS (1966). Notas sobre los crocodilidos de Cuba y una descripcion de una nueva especie del Pleistoceno. Poeyana 16: 1–34. [Google Scholar]
- Wang J (2011). COANCESTRY: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resourc 11: 141–145. [DOI] [PubMed] [Google Scholar]
- Weaver JP, Rodriguez D, Venegas-Anaya M, Cedeño-Vazquez JR, Forstner M, Densmore L III (2008). Genetic characterization of captive Cuban crocodiles (Crocodylus rhombifer) and evidence of hybridization with the American crocodile (Crocodylus acutus). J Exp Zool A Ecol Genet Physiol 309: 649–660. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC (1984). Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.