Abstract
For many highly mobile species, the marine environment presents few obvious barriers to gene flow. Even so, there is considerable diversity within and among species, referred to by some as the ‘marine speciation paradox'. The recent and diverse radiation of delphinid cetaceans (dolphins) represents a good example of this. Delphinids are capable of extensive dispersion and yet many show fine-scale genetic differentiation among populations. Proposed mechanisms include the division and isolation of populations based on habitat dependence and resource specializations, and habitat release or changing dispersal corridors during glacial cycles. Here we use a phylogenomic approach to investigate the origin of differentiated sympatric populations of killer whales (Orcinus orca). Killer whales show strong specialization on prey choice in populations of stable matrifocal social groups (ecotypes), associated with genetic and phenotypic differentiation. Our data suggest evolution in sympatry among populations of resource specialists.
Introduction
In the marine environment, connectivity is facilitated by the lack of physical barriers across large distances and yet considerable diversity has evolved within and among species (Palumbi, 1994; Bierne et al., 2003). Delphinid species provide a good study system for investigating this paradox due to their recent radiation, great diversity and the taxonomic complexities of many lineages within the group (Steeman et al., 2009; Moura et al., 2013). Although capable of extensive dispersion (Stevick et al., 2002), many cetacean species show fine-scale genetic differentiation among populations (Hoelzel, 2009). In some cases there is a correlation between population structure and apparent habitat boundaries, as for the bottlenose dolphin (Tursiops truncatus) populations in European waters (Natoli et al., 2005) or with resource specializations as for the killer whale (Orcinus orca) populations in the North Pacific (Hoelzel et al., 2007). Environmental cycles releasing habitat or opening/closing dispersal corridors may also influence the evolution of population structure in these species (Amaral et al., 2012; Moura et al., 2013). For killer whales, some well-studied populations show strong resource specializations based on consistent prey choice (ecotypes) within stable, matrifocal social groups (pods), together with genetic and phenotypic differentiation (Hoelzel et al., 1998; Pitman and Ensor, 2003; Hoelzel et al., 2007; Morin et al., 2010). A key question is whether or not differentiation has occurred in sympatry through ecologically based divergent selection with the potential to lead to sympatric speciation.
In this study, we generate the first multilocus phylogeny based on nuclear DNA for this genus, providing an important test of earlier inference based on mitochondrial DNA (mtDNA) trees (Hoelzel et al., 1998; Pitman and Ensor, 2003; Morin et al., 2010). We compared high-resolution phylogenetic reconstructions for mtDNA (alignment length of 4370 bp) with nuclear sequence phylogenies, built from restriction-associated DNA (RAD) fragments (see methods) consisting of a total alignment of 1 730 328 bp, with 5191 bp being variable among the killer whale samples. The earlier studies based on mtDNA (based on both Control Region and whole-mitogenome studies; for example, Hoelzel et al. (1998) and Morin et al. (2010)) showed that a lineage comprised of the marine-mammal-eating populations in the North Pacific (known as ‘transients') branched from the most basal node. A later study based on mtDNA proposed that a North Atlantic population was derived from ancestral North Pacific lineages, perhaps during an opening in the northwest passage during the last (Eemian) interglacial (Foote et al., 2011a). The authors further hypothesized that two fish-eating populations (known as ‘residents' and ‘offshores') represent a later re-invasion of the North Pacific back from the North Atlantic, establishing secondary contact and sympatry between the different ecotype populations (Foote et al., 2011a).
An alternative interpretation is that the diversity and distribution of mtDNA haplotypes have been impacted by historical demographic events (Hoelzel et al., 2002), and therefore do not fully reflect the true pattern of phylogeography. The single gene tree represented by mtDNA can also be impacted by simple stochasticity and historical introgression. The mtDNA phylogenies show good support for some lineages that are consistent with geography or ecotype. However, branches are shallow, with the most distinct haplotypes differentiated by only 0.56% (consistent with a loss of diversity during a bottleneck event, as indicated by both mtDNA and nuclear genomic data; Hoelzel et al., 2002; Moura et al., 2014). To help resolve ambiguities that may have arisen from the analysis of a single gene tree, we generated a phylogenomic analysis and undertook biogeographic analyses comparing inference from the mtDNA and nuclear DNA data. We test the hypothesis that differentiation between ecotypes evolved in sympatry within the North Pacific.
Materials and methods
DNA samples were obtained from archives available from previous studies (Hoelzel et al., 2007), and their number and provenance is provided in Supplementary Table S1. We further included new samples obtained from Marion Island (Southern Ocean), representing an Antarctic lineage (see Results). Sampling design was based on the inclusion of multiple geographic populations and ecotypes. Marion Island samples were obtained as biopsies (see similar protocol in Hoelzel et al. (2007)) from a population of known individuals (Reisinger et al., 2011). Fieldwork at Marion Island was permitted by the Prince Edward Islands Management Committee and procedures approved by the University of Pretoria's Animal Use and Care Committee (EC023-10). Sample number and ecotypes included are described in Supplementary Table 1. For the North Atlantic, we include samples from Iceland and the UK, representing both of the main mtDNA lineages identified previously for this region (Foote et al., 2011b).
Nuclear data
Nuclear genome-wide sequence data was obtained through RAD sequencing. The RAD sequencing protocol was modified from the version described by Baird et al. (2008) as follows. To reduce the requirements for high levels (30–50%) of the Illumina-supplied control phiX library (Illumina, San Diego, CA, USA), the adapter from which the forward read commences (p5 adapter) was modified such that a pool of four adapters was employed during the initial ligation to the NotI-digested DNA. These four adapters allow the start of the forward sequencing read to be staggered, ensuring the complexity of reads was greater over the first five bases and therefore improving the ability of the HiSeq instrument control software to differentiate between the sequencing clusters (see similar approach in Fadrosh et al. (2014)). In addition, a 5′ biotin modification in this adapter design allowed for specific selection of adapter-ligated sequences. Further, the 8 bp barcodes were added within the p7 adapter region during the PCR amplification step. The index read is performed separately as per any standard Illumina TruSeq library and demultiplexing performed using CASAVA (Illumina), instead of using the start of the forward reads as a barcode. To determine the success of this approach, an initial pool of five libraries generated using both the modified and the Baird et al. (2008) approach were sequenced on two separate 2 × 150 MiSeq runs without the presence of phiX (Illumina).
Genomic DNA (500 ng–1 μg) was digested to completion overnight at 37 °C with 1–2 μl NotI HF restriction enzyme (R3189L, New England Biolabs, Ipswich, MA, USA, 20 000 U ml−1). The complementary adapter sequences were annealed together by mixing the individual compatible oligonucleotides at 10 mM in annealing buffer (100 mM Tris, pH 7.5, 500 mM NaCl, 10 mM EDTA). The four adapters were mixed in equimolar amounts. One μl of 100 nM adapter mix was used to ligate to NotI fragments (from initial starting amount of 500 ng and in a volume of 34 μl) using NEBnext Quick Ligation module (New England Biolabs E6056L). Adapter-ligated fragments were sheared to an average size of 500 bp using a Covaris S2 sonicator (Covaris, Wobern, MA, USA) and selected after mixing the sample with strepavidin magnetic beads (Dynabeads M-280 Streptavidin cat no11205D, Life Technologies, Grand Island, NY, USA). Fragmented DNA was A-tailed (NEBNext dA-Tailing Module cat no E6053L) to make it blunt ended. DNA on beads was ligated to a universal p7 sequence adapter. A series of 47 amplification primers were designed with 8 bp barcodes to enable subsequent multiplexing of samples for a single lane of sequencing. A single barcoded primer and a universal primer were used to amplify each sample. Cycling conditions were 98 °C for 30 s followed by 12–14 cycles at 98 °C for 10 s, 60 °C for 30 s and 72 °C for 30 s followed by an extension at 72 °C for 5 min and 4 °C hold. Samples were purified with AMPure XP (Beckman Coulter, Brea, CA, USA) (1:1) and beads washed with 80% ethanol. After drying the beads, samples were resuspended in 22 μl of 10 mM Tris, pH 7.5. Samples were assessed for quantity (Qubit high sensitivity kit—Life Technologies) and quality (Agilent Bioanalyser 2100, Palo Alto, CA, USA). A fragment size distribution (‘smear') analysis was performed for each sample between 400 and 600 bp and this value was used to normalize the samples for multiplexing. The pooled samples were size selected on a 1.5% Pippin prep cassette (Sage Scientific, Beverly, MA, USA). The recovered library pools were assessed by quantitative PCR (Kapa, Wilmington, MA, USA) for quantification. Sequencing was performed as 2 × 100 bp paired-end reads on five lanes of the Illumina HiSeq 2000 using v3 chemistry. For further details see Supplementary Methods.
Trimmed short reads were mapped against bottlenose dolphin genome version 1.68 (which does not include mitochondrial DNA sequences; only version 1.72 and higher include this information) using Burrows-Wheeler Aligner short read mapper (Li and Durbin, 2009). Genotypes were called using a multisample Bayesian algorithm as implemented in the Unified Genotyper module (DePristo et al., 2011) from the Genome Analysis Toolkit software package (McKenna et al., 2010), with a minimum preliminary quality score filter set to 10. The resulting VCF file was processed to remove all positions with average coverage below 20 using VCFtools (Danecek et al., 2011), so that the final filtering is at a minimum mapping quality of Q20. All positions with indels were also removed, as were positions for which at least a single individual did not pass the set filters (that is, all positions with missing data were removed). The resulting VCF file was converted into a fasta file using a custom perl script.
mtDNA
Data from Morin et al. (2010) were used to identify the most informative regions of mtDNA in retrieving the same cetacean topology as from full mitogenomes. A set of 10 primers was designed to target this region using standard PCR and Sanger sequencing (Supplementary Table 2), resulting in a sequence 4370 bp long. PCR reactions were set up using 1 × Taq buffer (Promega, Madison, WI, USA), 0.2 mM deoxynucleotide triphosphates and varying concentrations of Mg+, primers and Taq (Supplementary Table 2). Thermocycling conditions were: one initial denaturation step at 95 °C for 2 min, followed by 45 cycles of denaturation at 95 °C for 30 s, annealing at varying temperatures (Supplementary Table 2) for 30 s, extension at 72 °C for 1 min and a final extension step at 72 °C for 10 min. Sequences were obtained from five Marion Island samples, and one North Atlantic sample obtained in the UK to match the range of lineages represented in the nuclear phylogeny. Corresponding sequences from the other ecotypes were retrieved from Morin et al. (2010), and a bottlenose dolphin sequence was used as an outgroup from Moura et al. (2013).
Phylogenetic analysis
The adequacy of using Marion Island samples as representative of Antarctic ecotypes was assessed by inferring a phylogenetic tree based on the same 4370 bp comparing Marion Island with sequences representative of Antarctic ecotypes from Morin et al. (2010). Nuclear phylogenetic trees were based on contigs up to 1028 bp in length (with 90% of the contig length range within ±100 bp of the 196 bp mode) built using MRBAYES (Ronquist and Huelsenbeck, 2003) under the GTR+G model of evolution (after similar RAD-based phylogenetic reconstructions in Wagner et al. (2012)). This model allows for rate variation along the sequence, and is therefore appropriate for concatenated alignments such as the one used here. Trials were also run using the GTR+I+G model, and no difference in topology found (data not shown). Two separate runs were started for each of four independent chains, three of them heated, and runs were considered to have achieved convergence if effective sample size values were all over 200, the PSRF+ statistic was close to 1, further confirmed by visual inspection of the log-likelihood plots for both runs. For the mtDNA trees, the best fit model of evolution was determined using TOPALI (Milne et al., 2009). The initial assessment of the Marion Island phylogenetic position based on mtDNA was run for 10 000 000 iterations, with the first 25% iterations discarded as burn-in. For the main mtDNA tree, MRBAYES was run for 12 000 000 iterations, with the first 25% iterations discarded as burn-in.
To assess the bias created by sites potentially under positive selection, all variable positions were extracted using the software SEAVIEW (Gouy et al., 2009), and converted into GenePop format using a custom perl script. Signal for selection was investigated using the FST outlier method implemented in LOSITAN (Antao et al., 2008). Mean neutral FST was calculated using the infinite alleles model, and assuming nine demes of size 10, following the different a priori-defined populations (based on the results obtained in Hoelzel et al. (2007) and Parsons et al. (2013)): Marion Island, North Atlantic, North Pacific offshores, Alaskan residents, Southern residents, Alaska transients, California transients, Bering Sea and Russia. Although some sample sizes were small per putative population, this is more likely to artificially inflate FST, generating false outliers (which would be conservative in this case). An initial run to remove potential selected loci was done to calculate the baseline mean neutral FST, which was estimated using the bisection algorithm over repeated simulations (Antao et al., 2008). A total of 50 000 simulations were run, with a false discovery rate of 0.1. Sites identified as being under positive selection by the LOSITAN algorithm were then removed from the full RAD alignment, and a new phylogenetic tree was constructed based on the shorter sequence. In both, the full data set and in the trimmed data set, MRBAYES was run for 1 000 000 iterations with the first 25% iterations discarded as burn-in.
Given the known biases that GC-rich regions might impose on phylogenetic reconstruction (Romiguier et al., 2013), the RAD data set was further divided between GC- and AT-rich regions. Reads mapped to consecutive reference positions with a gap of <20 bp were assembled into contigs, for which GC content was calculated. Contigs were then pooled into GC-rich and AT-rich alignments based on a 50% GC content threshold. MRBAYES was then run for 10 000 000 iterations (with 25% burn-in) for the full alignment where the evolutionary parameters were estimated independently (using the GTR+G model as described above) for two partitions defined according to GC content. Romiguier et al. (2013) found that for placental mammals, the AT-rich regions were ‘better at retrieving well-supported, consensual nodes', therefore we also constructed a tree using the same methods based only on the AT-rich contigs. Because the enzyme chosen for the RAD library construction (NotI) is GC rich, the proportion of AT-rich contigs was relatively small (191 544 bp, 1490 of which were variable).
Further, to assess the effect of concatenating different genomic locations in a single alignment, the CAT-GTR model (see Lartillot and Philippe, 2004) implemented with the software PHYLOBAYES (Lartillot et al., 2009) was used in the full alignment, but considering only variable sites. We focused on variable sites because the software PHYLOBAYES cannot accommodate the full-sequence input file. However, for an evolution model based on site heterogeneity, this should not affect the topology significantly, although it can be expected to affect branch length. The program was run for 437 000 cycles with 50 000 burn-in, with trees recorded every 1000 cycles. Convergence of the run was assessed through checking effective sample size values and the stability of the log-likelihood plots after burn-in.
Reconstruction of ancestral distributions and dating analysis
To estimate phylogeographic patterns, we applied different ancestral distribution reconstruction methods as applied in the software RASP (Yu et al., 2013), for both mtDNA and RAD trees. Phylogenetic trees for this analysis were obtained by building a 50% majority consensus tree in RASP from all the phylogenetic trees retained after burn-in in the MRBAYES analysis. Three distributional ranges were considered, Southern Ocean (Marion Island), North Atlantic (Iceland and UK) and North Pacific (offshores, transients, residents, Russia and Bering Sea). Bottlenose dolphin was used as an outgroup and defined as occurring in all three areas, and therefore uninformative. Statistical Dispersal-Vicariance Analysis (S-Diva) is a parsimony-based method that minimizes the number of dispersal and extinction events in a tree (Ronquist, 1997). The maximum number of areas per node was set to 3, and with the ‘Allow reconstruction' option enabled. Uncertainty was assessed using the S-Diva value (Yu et al., 2010) based on all the post-burn-in trees inferred by MRBAYES (see above). In addition, the Bayesian Binary (BB) Markov chain Monte Carlo method was also implemented, which uses a full hierarchical Bayesian approach to quantify uncertainty in the reconstruction of ancestral distributions (Ronquist, 2004). The maximum number of areas per node was set to 3, and the root distribution was set to null, given that the outgroup used has a wider distribution than the three considered for the ingroup. Analysis was run with 10 chains, 9 of which were heated, for 1 000 000 iterations with 10 000 burn-in.
Dated phylogenies were obtained using BEAST (Drummond et al., 2012) by applying a strict clock under a Yule speciation model. Given the lack of robust and unambiguous calibration points to determine mutation rate in killer whales, our objective was only to gain an idea of the temporal range of possible splitting times using credible mutation rates from the literature (Dornburg et al., 2012; Moura et al., 2013). For the mtDNA tree, we used a rate of 0.03 substitutions per site per million years (Moura et al., 2013), whereas for the RAD tree we used a rate of 0.0011 substitutions per site per million years estimated for Odontocetes (Dornburg et al., 2012).
Results
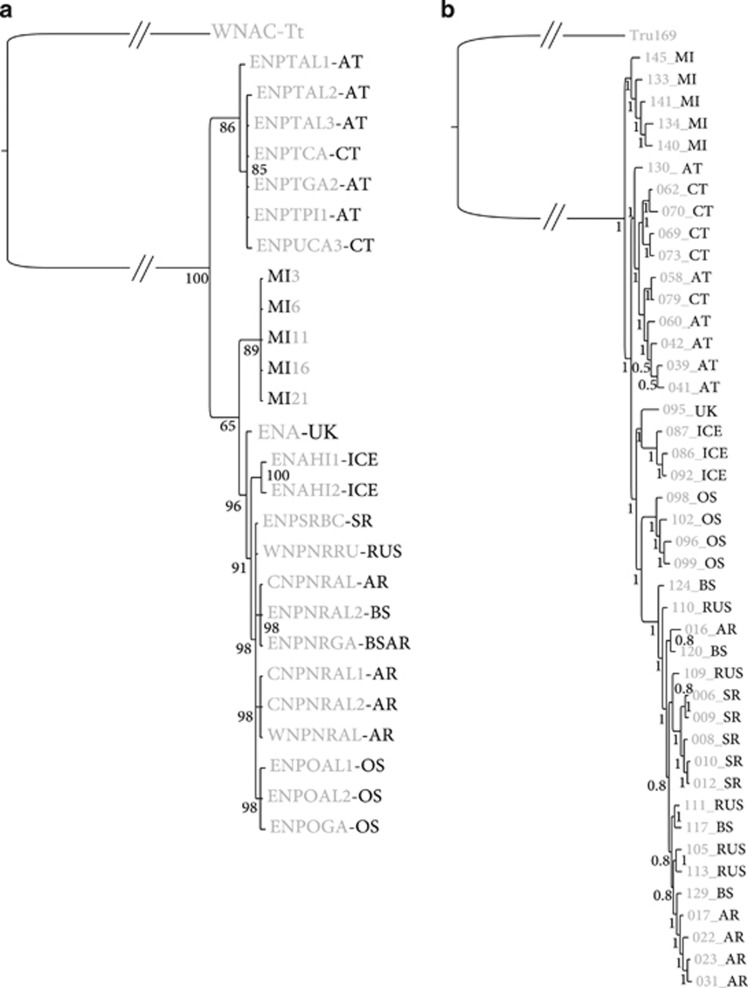
Our mtDNA phylogeny (based on sufficient sequence data to recapitulate the topology of the published mitogenome tree; see Methods) was confirmed to provide the same structure and similar inference (Figures 1 and 2) as reported in the earlier studies (Hoelzel et al., 1998; Morin et al., 2010; Foote et al., 2011a). A Southern Ocean population is represented in our tree using samples from Marion Island, which group tightly with the ‘type B' Antarctic lineage haplotypes (Supplementary Figure 1a).
Figure 1.
Bayesian phylogenetic trees of killer whale ecotypes for (a) mitochondrial DNA and (b) nuclear DNA obtained through RAD-associated sequencing. Both trees were inferred using MRBAYES software. AR, Alaskan residents; AT, Alaskan transients; BS, Bering Sea; CT, Californain transients; ICE, Iceland; MI, Marion Island; OS, offshores; RUS, Russian residents; SR, Southern residents.
Figure 2.
Phylogeographical reconstruction of killer whale ancestral distributions and dispersal patterns based on (A) nuclear DNA obtained through RAD-associated sequencing and (B) mitochondrial DNA. Inference was done in RASP software, using the BB Markov chain Monte Carlo method. Node numbers next to nodes refer to numbers given in Tables 1 and 2 and in Supplementary Figure 2. Numbers within some nodes refer to paths in map figures.
Reconstruction of the geographical distribution of ancestral nodes based on our mtDNA tree showed some inconsistencies between S-DIVA and the BB method (Table 1), although both methods suggest colonization of the North Atlantic followed by a later dispersal event from the North Atlantic back to the North Pacific, consistent with the earlier study (Foote et al., 2011a). However, there is some indication that the initial dispersal into the North Atlantic is more likely via the Antarctic from this analysis (Figure 2; Supplementary Figure 2), rather than over the pole (as suggested earlier; Foote et al., 2011a).
Table 1. Assignment probability for the reconstruction of ancestral distributions using the software RASP, for key nodes of interest in the mitochondrial phylogeny (Figure 2).
| Method | Region | Node 52 | Node 45 | Node 44 | Node 43 | Node 42 | Node 32 | Node 51 |
|---|---|---|---|---|---|---|---|---|
| S-Diva | SO | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NA | 0 | 0 | 47.34 | 0 | 0 | 100 | 0 | |
| NP | 32.88 | 0 | 0 | 0 | 100 | 0 | 100 | |
| SO\NA | 8.25 | 31.11 | 0 | 0 | 0 | 0 | 0 | |
| SO\NP | 23.18 | 33.68 | 0 | 0 | 0 | 0 | 0 | |
| NA\NP | 14.13 | 1.19 | 52.66 | 100 | 0 | 0 | 0 | |
| SO\NA\NP | 21.56 | 34.02 | 0 | 0 | 0 | 0 | 0 | |
| Bayesian Binary | SO | 8.53 | 43.11 | 0.49 | 0.10 | 0 | 0 | 0.01 |
| NA | 6.09 | 24.73 | 90.75 | 80.49 | 0.40 | 99.27 | 0.02 | |
| NP | 68.88 | 14.04 | 1.61 | 4.90 | 92.23 | 0 | 98.4 | |
| SO\NA | 0.77 | 9.00 | 1.65 | 0.29 | 0 | 0.31 | 0 | |
| SO\NP | 8.72 | 5.11 | 5.37 | 0.02 | 0.08 | 0 | 0.37 | |
| NA\NP | 6.22 | 2.93 | 0.03 | 14.15 | 7.29 | 0.59 | 1.19 | |
| SO\NA\NP | 0.79 | 1.07 | 0.10 | 0.05 | 0.01 | 0 | 0 |
Highest value for each test is shown in bold text for each node.
The nuclear data generate a well-supported tree (Figure 1), although the overall level of divergence remains low (0.07% at the deepest node, Hasegawa, Kishino and Yano model based on a distance matrix constructed using GENEIOUS). The killer whale short reads from the RAD sequencing have been deposited in NCBI Genbank in BioProject PRJNA236163. Analysis of the nuclear data using LOSITAN revealed the presence of 365 single nucleotide polymorphism outliers for positive selection, but removal of these positions did not alter the topology (see Supplementary Figure 1b), so all loci were retained for further analyses.
The topology recovered for the nuclear phylogeny using the full alignment differed from the mtDNA tree in several key respects (Figure 1). Southern Ocean haplotypes that were nested well within North Pacific lineages in the mtDNA tree now branch from the most basal node, whereas North Atlantic samples and ‘offshores' from the North Pacific now form reciprocally monophyletic lineages (Figure 1). The 'resident' and 'offshore' fish-eating ecotypes are more clearly delineated into separate lineages, and the North Pacific ‘residents' form a broad lineage with incomplete lineage sorting among regional populations. The topology of the nuclear tree was robust to partitioning with respect to GC content and to the reconstruction employing the heterogeneous CAT-GTR evolution model, with the exception that for the latter analysis, offshores and North Atlantic haplotypes were not as clearly separated into a bifurcating relationship (Supplementary Figure 2). The AT-rich tree (Supplementary Figure 2) again supported the broader topology, but the ‘offshore' group clustered with the ‘transients'. The observed discordance between the nuclear and mtDNA phylogenies has been noted earlier in the North Pacific (Pilot et al., 2010) and among North Atlantic ecotypes (Foote et al., 2009, 2013) based on comparisons between mtDNA control region sequences and microsatellite DNA genotypes.
Reconstruction of the geographical distribution of ancestral nodes also recovered a phylogeographic scenario from the nuclear tree that is distinct from that obtained from the mtDNA data (Figure 2, Table 2). As the biogeographic inference was the same for the nuclear tree reconstructions based on the full data set without partitioning, for the partitioned tree based on GC content, for the AT-rich tree and for the CAT-GTR tree (data not shown), we report on the analyses of the full data set as presented in Figure 1. Both S-DIVA and BB suggested that killer whales expanded from the Southern Ocean into the North Pacific, with North Atlantic ecotypes diverging from North Pacific lineages, and the divergence between North Pacific ecotypes occurring locally in sympatry (Figure 2; Supplementary Figure 3). Ancestry in the Southern Oceans is consistent with the present day abundance of killer whales in the region, and the relative stability of that habitat over the course of the Quaternary (Francois et al., 1997; Latimer and Filippelli, 2001). Inference about dispersal and vicariance from the BB model is shown in Figure 2. From the S-DIVA model based on the nuclear phylogeny, North Atlantic ecotypes diverged from North Pacific lineages by dispersal (at ‘2' in Figure 2a), whereas the node separating the Southern Oceans from other regions suggests vicariance (at ‘1' in Figure 2a). For the mtDNA reconstruction based on S-DIVA, the inference is the same as for the BB model.
Table 2. Assignment probability for the reconstruction of ancestral distributions using the software RASP, for key nodes of interest in the nuclear phylogeny (Figure 2).
| Method | Region | Node 86 | Node 81 | Node 80 | Node 76 | Node 85 | Node 54 | Node 79 |
|---|---|---|---|---|---|---|---|---|
| S-Diva | SO | 0 | 0 | 0 | 0 | 100 | 0 | 0 |
| NA | 0 | 0 | 0 | 0 | 0 | 0 | 100 | |
| NP | 0 | 100 | 0 | 100 | 0 | 100 | 0 | |
| SO\NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| SO\NP | 100 | 0 | 0 | 0 | 0 | 0 | 0 | |
| NA\NP | 0 | 0 | 100 | 0 | 0 | 0 | 0 | |
| SO\NA\NP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Bayesian Binary | SO | 48.45 | 1.21 | 0.17 | 0 | 98.96 | 0 | 0.02 |
| NA | 1.21 | 1.03 | 4.57 | 0.08 | 0.01 | 0.01 | 96.02 | |
| NP | 29.86 | 93.27 | 85.63 | 98 | 0.08 | 99.49 | 0.55 | |
| SO\NA | 0.77 | 0.03 | 0.02 | 0 | 0.09 | 0 | 0.13 | |
| SO\NP | 18.93 | 2.39 | 0.34 | 0.07 | 0.85 | 0.12 | 0 | |
| NA\NP | 0.47 | 2.03 | 9.24 | 1.73 | 0 | 0.36 | 3.27 | |
| SO\NA\NP | 0.30 | 0.05 | 0.04 | 0 | 0 | 0 | 0 |
Highest value for each test is shown in bold text for each node.
Discussion
In this study, we generate a phylogeny for the genus Orcinus based on a large number of nuclear DNA loci. The topology of the nuclear tree was consistent even after partitioning for GC content and testing alternative evolution models. The CAT-GTR tree based only on variable sites showed greater depth (as expected) and poorer resolution of the North Atlantic and offshore lineages, but retained the key aspects of topology seen in the other tree reconstructions, in particular the position of the Southern Ocean samples from Marion Island. The nuclear trees were based on relatively short, dispersed sequences, but several evolution models that account for rate variation across the sequence were applied and the trees consistently showed the same overall topology. The AT-rich tree again agreed with the overall topology, but grouped the offshores into the same lineage as the transients, a result that is consistent with inference from microsatellite DNA loci in Pilot et al. (2010).
When comparing the nuclear and mtDNA trees, the main differences were associated with the position of the Marion Island lineage, and the strength of support for the offshores as a lineage distinct from the North Pacific residents. Biogeographic analyses suggested a relatively uncomplicated pattern for the establishment of populations, compared with the mtDNA tree. For the nuclear tree, the pattern was consistent with the division of extant North Pacific populations within the North Pacific and without the need for a period of allopatric divergence in the North Atlantic. Allopatric or parapatric differentiation within the North Pacific is possible, but published data suggest that both local specialization and geographic distance reduce gene flow in a similar way. In particular, sympatric ecotype populations show levels of differentiation comparable to that found between populations of the resident ecotype either side of the North Pacific, and there is evidence for isolation by distance within an ecotype (Hoelzel et al., 2007). It may be that prey choice changes temporal and spatial patterns of habitat use enough to minimize interactions among specialist groups, thereby reducing gene flow without requiring a period of physical isolation. The extensive ranging capabilities of this species also makes allopatric or parapatric boundaries on their own seem less likely drivers within an ocean basin than resource specializations.
Earlier studies indicated ongoing gene flow between North Pacific ecotypes, and suggested that gene flow was generally male mediated during temporary encounters between matrifocal pods (Hoelzel et al., 2007; Pilot et al., 2010). However, key distinguishing features of the nuclear phylogeny could not be explained by male-mediated gene flow following secondary contact. The scenario implicit in the mtDNA phylogeny indicates isolation of a fish-eating form in the North Atlantic, derived from North Pacific ‘transient' ancestors, and the re-invasion of this form into the North Pacific, now represented by the residents and offshores (which share similar mtDNA haplotypes). However, secondary contact could not explain why the Southern Ocean ecotype branches from the most basal node in the nuclear phylogeny, or why offshores and residents show greater divergence at nuclear loci. Instead, the implication is that the mtDNA phylogeny is distorted by historical demography (possibly in conjunction with a bottleneck event; Hoelzel et al., 2002; Moura et al., 2014) or other stochastic factors.
The nuclear data suggest North Pacific ancestry of at least some North Atlantic populations, similar to what was proposed based on mtDNA data (Foote et al., 2011a). If movement was across the pole, this could only have happened during interglacial periods when there may have been an open passage. Using a fixed rate clock and a published average substitution rate for the Odontocete nuclear genome (Dornburg et al., 2012), the node defining the separation of the North Atlantic lineage from the North Pacific falls within the Eemian interglacial (∼155 kya; Supplementary Figure 1c). However, the mutation rate applied was derived from relatively deep phylogenetic calibrations. As has been established in numerous publications for mtDNA (see review in Ho et al. (2007)), calibrating for more recent events may require the use of a higher mutation rate, typically at least an order of magnitude higher for mtDNA. The correct rate to apply is not known in this case, but an order of magnitude increase would still allow for transfer during an interglacial, just before the beginning of the Holocene (∼16 kya).
Although sampling was not inclusive of all populations on a global scale, two key aspects of the nuclear phylogeny indicate that inference about differentiation in sympatry is not due to incomplete taxon sampling. First, the North Pacific transient form does not branch from the ancestral node in this tree (a result that further sampling is unlikely to change), and second, the transient and resident types remain reciprocally monophyletic, with the node distinguishing the North Atlantic and North Pacific resident lineages apparently younger than the node that separates them from the transient lineage (Supplementary Figure 1c). Together, these factors indicate that transients and residents most likely share ancestry in the North Pacific, and additional details about the relationship among unsampled populations from other parts of the world should not affect this interpretation. The possibility of populations or species differentiating in sympatry has remained controversial, although there are some instances that are now generally accepted (see Bolnick and Fitzpatrick, 2007). In general, most models invoke strong disruptive ecological selection (for example, in association with differential resource use) together with high initial levels of phenotypic polymorphism, and strong mating preferences (Gavrilets, 2004). Ultimately, this process may promote ecological speciation (see Nosil (2012) for various examples), and the possibility of incipient ecological speciation based on the cultural transmission of foraging specializations has been raised previously for the killer whale (for example, Hoelzel et al. (2002) and Riesch et al. (2012)).
Killer whales feed on a wide variety of prey, however, this diversity results from a range of local specializations on relatively few prey species (de Bruyn et al., 2013). These local populations of resource specialists are often genetically differentiated, but as indicated earlier, differentiation between populations of the same ecotype is also seen, and reflects a pattern of isolation by distance (Hoelzel et al., 2007). Ecotypes may also exhibit differences in social structure, morphology, behavior and vocal signatures (see for review de Bruyn et al. (2013)). In the North Pacific, the resident and transient ecotypes occupy largely sympatric distribution ranges (Ford et al., 2000), but specialize on very different prey resources (fish and marine mammals, respectively; Ford et al., 1998; Krahn et al., 2007), are genetically differentiated (Hoelzel et al., 1998, 2002, 2007), and exhibit different social organization (Ford et al., 2000), mating systems (Pilot et al., 2010) and vocal behavior (Yurk et al., 2002; Deecke et al., 2005). Less is known about the ‘offshore' ecotype, however, our data indicate that we need to consider their differentiation in sympatry as well. Krahn et al. (2007) and Dahlheim et al. (2008) found that ‘offshore' killer whales feed on fish resources (possibly with some overlap with residents including halibut—Jones, 2006—but also distinct prey; Krahn et al., 2007), and sighting data indicate a largely but not exclusively pelagic distribution, (likely overlapping with both 'transient' and 'resident' ecotypes in some regions; Dahlheim et al., 2008), whereas the residents are more dependent on coastal resources. The average group size is larger and adult body size smaller for offshores than for either residents or transients, but data are based on just 59 sightings over 30 years (Dahlheim et al., 2008). Re-sightings of photographically identified pods revealed the potential for very large scale movement (>4000 km), greater than that so far conclusively documented for the other regional ecotypes (Dahlheim et al., 2008).
The first nuclear phylogenetic division within the North Pacific was between transients and offshores, followed by an apparently later division between offshores and residents. An earlier division between fish-eating and marine-mammal-eating ecotypes in pelagic waters is reasonable if the nearshore habitat was unavailable at that time (under ice). Differences in dispersal range, social behavior and prey choice between transients and offshores (Yurk et al., 2002) may have reinforced isolation. We suggest that dependence on learned behavior, likely transferred within social groups by tradition, serves to isolate populations of resource specialists, as discussed previously (Hoelzel et al., 2007). This may lead to local adaptation through disruptive selection and differentiation by drift among populations whose foraging behavior determines different spatial and temporal patterns of dispersion (for example, Hoelzel et al. (2007) and Riesch et al. (2012)). The apparent conflict between ease of connectivity among these populations and their genetic differentiation may be explained by these processes. At the same time, when habitats change (as during the interglacial warming periods), changing resources may require changes in foraging strategies, and different foraging strategies that do not also lead to physical or temporal isolation need not lead to genetic differentiation (Hoelzel et al., 2007; de Bruyn et al., 2013). A recent study based on isotopic markers suggesting specialization among North Atlantic groups not clearly differentiated for nuclear or mtDNA markers (Foote et al., 2013) may be an example. Our data for the North Pacific suggest that in this case, life history and behavioral changes associated with resource use led to lineage differentiation between ecotypes, and the potential for incipient speciation.
Data archiving
RAD sequence data are provided at Genbank under accessions SRX564829-SRX564955 in Bioproject PRJNA236163. Mitochondrial DNA sequences are available under Genbank accession numbers KM016850–KM016879.
Acknowledgments
We thank Howard Gray for providing primer sequences for the amplification of mitochondrial DNA, and Charlene Janse van Rensburg and Colin Nicholson for labwork associated with DNA extraction and archiving. This study was funded by the Natural Environment Research Council UK (grant number NE/014443/1). We thank the South African Department of Environmental Affairs for providing logistical support within the South African National Antarctic Programme and the Department of Science and Technology (administered through the South African National Research Foundation) for funding the marine mammal monitoring programme at Marion Island.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Amaral AR, Beheregaray LB, Bilgmann K, Freitas L, Robertson KM, Sequeira M et al. (2012). Influences of past climatic changes on historical population structure and demography of a cosmopolitan marine predator, the common dolphin (genus Delphinus). Mol Ecol 9: 4854–4871. [DOI] [PubMed] [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. (2008). LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics 9: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA et al. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Bonhomme F, David P. (2003). Habitat preference and the marine-speciation paradox. Proc R Soc Lond B Biol Sci 270: 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick DI, Fitzpatrick BM. (2007). Sympatric speciation: models and empirical evidence. Annu Rev Ecol Evol Syst 38: 459–487. [Google Scholar]
- Dahlheim ME, Schulman-Janiger A, Black N, Ternullo R, Ellifrit D, Balcomb Iii KC. (2008). Eastern temperate North Pacific offshore killer whales (Orcinus orca): occurrence, movements, and insights into feeding ecology. Mar Mammal Sci 24: 719–729. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011). The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyn PJN, Tosh CA, Terauds A. (2013). Killer whale ecotypes: is there a global model? Biol Rev 88: 62–80. [DOI] [PubMed] [Google Scholar]
- Deecke VB, Ford JKB, Slater PJB. (2005). The vocal behaviour of mammal-eating killer whales: communicating with costly calls. Anim Behav 69: 395–405. [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornburg A, Brandley MC, McGowen MR, Near TJ. (2012). Relaxed clocks and inferences of heterogeneous patterns of nucleotide substitution and divergence time estimates across whales and dolphins (Mammalia: Cetacea). Mol Biol Evol 29: 721–736. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J. (2014). An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, Morin PA, Durban JW, Willerslev E, Orlando L, Gilbert MTP. (2011. a). Out of the Pacific and back again: insights into the matrilineal history of Pacific killer whale ecotypes. PLoS One 6: e24980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, Vilstrup JT, de Stephanis R, Verborgh P, Abel Nielsen SC, Deaville R et al. (2011. b). Genetic differentiation among North Atlantic killer whale populations. Mol Ecol 20: 629–641. [DOI] [PubMed] [Google Scholar]
- Foote AD, Newton J, Avila-Arcos MC, Kampmann ML, Samaniego JA, Post K et al. (2013). Tracking niche variation over millennial timescales in sympatric killer whale lineages. Proc R Soc Lond B Biol Sci 280: 20131481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, Newton J, Piertney SB, Willerslev E, Gilbert MTP. (2009). Ecological, morphological and genetic divergence of sympatric North Atlantic killer whale populations. Mol Ecol 18: 5207–5217. [DOI] [PubMed] [Google Scholar]
- Ford JKB, Ellis GM, Balcomb KC. (2000) Killer Whales: The Natural History and Genealogy of Orcinus orca in British Columbia and Washington State 3rd edn UBC Press: Vancouver, BC, Canada. [Google Scholar]
- Ford JKB, Ellis GM, Barrett-Lennard LG, Morton AB, Palm RS, Balcomb KC III. (1998). Dietary specialization in two sympatric populations of killer whales (Orcinus orca) in coastal British Columbia and adjacent waters. Can J Zool 76: 1456–1471. [Google Scholar]
- Francois R, Altabet MA, Yu E-F, Sigman DM, Bacon MP, Frank M et al. (1997). Contribution of Southern Ocean surface-water stratification to low atmospheric CO2 concentrations during the last glacial period. Nature 389: 929–935. [Google Scholar]
- Gavrilets S. (2004) Fitness Landscapes and the Origin of Species. Princeton University Press: Princeton, NJ, USA. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. (2009). SeaView Version 4: a Multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Kolokotronis SO, Allaby RG. (2007). Elevated substitution rates estimated from ancient DNA sequences. Biol Lett 3: 702–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzel AR. (2009). Evolution of population structure in marine mammals. In: Bertorelle G, Bruford MW, Hauffe HC, Rizzoli A, Vernesi C, (eds) Population Genetics for Animal Conservation. Cambridge University Press: Cambridge, UK. [Google Scholar]
- Hoelzel AR, Dahlheim M, Stern SJ. (1998). Low genetic variation among killer whales (Orcinus orca) in the Eastern North Pacific and genetic differentiation between foraging specialists. J Hered 89: 121–128. [DOI] [PubMed] [Google Scholar]
- Hoelzel AR, Hey J, Dahlheim ME, Nicholson C, Burkanov V, Black N. (2007). Evolution of population structure in a highly social top predator, the killer whale. Mol Biol Evol 24: 1407–1415. [DOI] [PubMed] [Google Scholar]
- Hoelzel AR, Natoli A, Dahlheim ME, Olavarria C, Baird RW, Black NA. (2002). Low worldwide genetic diversity in the killer whale (Orcinus orca): implications for demographic history. Proc R Soc Lond B Biol Sci 269: 1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones IM. (2006). A Northeast Pacific offshore killer whale (Orcinus orca) feeding on a pacific halibut (Hippoglossus stenolepis). Mar Mammal Sci 22: 198–200. [Google Scholar]
- Krahn MM, Herman DP, Matkin CO, Durban JW, Barrett-Lennard L, Burrows DG et al. (2007). Use of chemical tracers in assessing the diet and foraging regions of eastern North Pacific killer whales. Mar Environ Res 63: 91–114. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. (2004). A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol 21: 1095–1109. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. (2009). PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288. [DOI] [PubMed] [Google Scholar]
- Latimer JC, Filippelli GM. (2001). Terrigenous input and paleoproductivity in the Southern Ocean. Paleoceanography 16: 627–643. [Google Scholar]
- Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF et al. (2009). TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25: 126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PA, Archer FI, Foote AD, Vilstrup J, Allen EE, Wade P et al. (2010). Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res 20: 908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura AE, Janse van Rensburg C, Pilot M, Tehrani A, Best PB, Thornton M et al. (2014). Killer whale nuclear genome and mtDNA reveals widespread population bottleneck during the last glacial maximum. Mol Biol Evol 31: 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura AE, Nielsen SCA, Vilstrup JT, Moreno-Mayar JV, Gilbert MTP, Gray H et al. (2013). Recent diversification of a marine genus (Tursiops spp.) tracks habitat preference and environmental change. Syst Biol 62: 865–877. [DOI] [PubMed] [Google Scholar]
- Natoli A, Birkun A, Aguilar A, Lopez A, Hoelzel AR. (2005). Habitat structure and the dispersal of male and female bottlenose dolphins (Tursiops truncatus). Proc R Soc Lond B Biol Sci 272: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P. (2012) Ecological Speciation. Oxford University Press:. Oxford, UK, pp 304. [Google Scholar]
- Palumbi SR. (1994). Genetic divergence, reproductive isolation, and marine speciation. Annu Rev Ecol Syst 25: 547–572. [Google Scholar]
- Parsons KM, Durban JW, Burdin AM, Burkanov VN, Pitman RL, Barlow J et al. (2013). Geographic patterns of genetic differentiation among killer whales in the northern North Pacific. J Hered 104: 737–754. [DOI] [PubMed] [Google Scholar]
- Pilot M, Dahlheim ME, Hoelzel AR. (2010). Social cohesion among kin, gene flow without dispersal and the evolution of population genetic structure in the killer whale (Orcinus orca). J Evol Biol 23: 20–31. [DOI] [PubMed] [Google Scholar]
- Pitman RL, Ensor P. (2003). Three forms of killer whales in Antarctic waters. J Cetacean Res Manage 5: 131–139. [Google Scholar]
- Reisinger RR, de Bruyn PJN, Bester MN. (2011). Abundance estimates of killer whales at -subantarctic Marion Island. Aquatic Biol 12: 177–185. [Google Scholar]
- Riesch R, Barrett-Lennard LG, Ellis GM, Ford JKB, Deeke VB. (2012). Cultural traditions and the evolution of reproductive isolation: ecological speciation in killer whales? Biol J Linn Soc 106: 1–17. [Google Scholar]
- Romiguier J, Ranwez V, Delsuc F, Galtier N, Douzery EJP. (2013). Less is more in mammalian phylogenomics: AT-rich genes minimize tree conflicts and unravel the root of placental mammals. Mol Biol Evol 30: 2134–2144. [DOI] [PubMed] [Google Scholar]
- Ronquist F. (1997). Dispersal-Vicariance analysis: a new approach to the quantification of historical biogeography. Syst Biol 46: 195–203. [Google Scholar]
- Ronquist F. (2004). Bayesian inference of character evolution. Trends Ecol Evol 19: 475–481. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Steeman ME, Hebsgaard MB, Fordyce RE, Ho SY, Rabosky DL, Nielsen R et al. (2009). Radiation of extant cetaceans driven by restructuring of the oceans. Syst Biol 58: 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevick PT, McConnell BJ, Hammond PS. (2002). Patterns of movement. In: Hoelzel AR, (ed) Marine Mammal Biology: an Evolutionary Approach. Blackwell Science: Oxford, UK. pp 185–216. [Google Scholar]
- Wagner CE, Keller I, Wittwer S, Selz OM, Mwaiko S, Greuter L et al. (2012). Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in Lake Victoria cichlid adaptive radiation. Mol Ecol 22: 787–798. [DOI] [PubMed] [Google Scholar]
- Yu Y, Harris AJ, He X-J. (2013) RASP (Reconstruct Ancestral State in Phylogenies) 2.1 beta. Available at: http://mnhscueducn/soft/blog/RASP. [DOI] [PubMed]
- Yu Y, Harris AJ, He X. (2010). S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Mol Phylogenet Evol 56: 848–850. [DOI] [PubMed] [Google Scholar]
- Yurk H, Barrett-Lennard L, Ford JKB, Matkin CO. (2002). Cultural transmission within maternal lineages: vocal clans in resident killer whales in southern Alaska. Anim Behav 63: 1103–1119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.