Abstract
The recombination suppression models of chromosomal speciation posit that chromosomal rearrangements act as partial barriers to gene flow allowing these regions to accumulate genetic incompatibilities, thus contributing to the divergence of populations. Empirical and theoretical studies exploring the requirements of these models have mostly focused on the role of inversions. Here, the recombination landscape of heterozygosity for Robertsonian (Rb) fusions is investigated in the house mouse. Laboratory-bred F1 males and females between highly differentiated races from Tunisia (Rb: 2n=22, Standard, St: 2n=40) were produced in which all Rb fusions are present as trivalents in meiosis. Recombination patterns were determined by the analysis of chiasmata and compared with previous data on the Tunisian parental mice. A comparative analysis was performed on wild-caught male mice spanning the hybrid zone between two Italian races (2n=40, 2n=22). The results showed that the chiasma characteristics of both male and female Tunisian F1 and Italian hybrids clearly differed from those of Rb and St mice. Not only was the mean chiasma number (CN) intermediate between those of the parental mice in both geographic samples, but the distribution of chiasmata along the chromosomal arms of the F1 showed a distinct mosaic pattern. In short, the proximal region in the F1 exhibited a reduced CN similar to that observed in homozygous Rb, whereas distal regions more closely matched those in St mice. These results suggest that Rb rearrangements (homozygous or heterozygous) reduce recombination in the proximal regions of the chromosomes supporting their potential role in recombination-mediated speciation models.
Introduction
Historically, chromosomal rearrangements have long been recognized as a major driving force promoting divergence (White, 1978), but it was only recently that the role of rearrangements as recombination modifiers has been recognized and formalized in the recombination suppression models of chromosomal speciation (Noor et al., 2001; Rieseberg, 2001; Navarro and Barton, 2003; Ayala and Coluzzi, 2005). Thus, the emphasis is no longer on the decrease in fitness of chromosomal hybrids, but on the suppression of recombination between the rearranged chromosomes, resulting in a partial barrier to gene flow between populations. The association with speciation events lies in the tight linkage between the rearrangements and reproductive isolation or locally adaptive genes owing to the suppression of recombination in chromosomal heterozygotes. Thus, rearrangements can contribute to the persistence of reproductive isolation genes in the face of gene flow for a longer time than if they were absent (Noor et al., 2001), and extend the action of linked isolation genes over larger genomic regions than if no rearrangements were present (Rieseberg, 2001; Butlin, 2005). The emergence of the recombination suppression models stimulated a considerable amount of both empirical (for example, Lowry and Willis, 2010; Ayala et al., 2011; MacGaugh and Noor, 2012; Farré et al., 2013) and theoretical (for example, Kirkpatrick and Barton, 2006; Feder and Nosil, 2009; Faria and Navarro, 2010) studies exploring the operational conditions, mechanisms and processes involved. Most, if not all, of these analyses have focused on inversions and reciprocal translocations, whereas the most widespread rearrangement, Robertsonian (Rb) fusion/fission, has received considerably less attention (King, 1993).
The aim of the present study is to assess the effect of heterozygosity for Rb fusions on recombination patterns. These rearrangements consist of the joining of two nonhomologous acrocentric chromosomes by the centromere to form one biarmed chromosome. The house mouse, Mus musculus domesticus, was chosen as the biological model for several reasons. First, extensive chromosomal diversity through fixation of Rb fusions occurs throughout its distribution and all chromosomes, except the sex pair, are involved in Rb fusions in wild populations (Piálek et al., 2005). Second, underdominance levels of simple Rb heterozygotes, that is, carrying trivalents formed by the pairing of Rb fusions with the two homologous acrocentrics, are well documented in this subspecies. In particular, the data indicate that heterozygosity for a single-Rb fusion has a limited effect on hybrid fitness (Hauffe and Searle, 1998; Sans-Fuentes et al., 2010); in this case, there will be almost no constraint on its fixation probability, but conversely, its contribution to postmating isolation will be extremely reduced unless it is associated with changes in recombination pattern. Finally, several studies have indicated a reduction in crossover rates in wild homozygous Rb vs standard mice in the proximal centromeric regions (Bidau et al., 2001, Castiglia and Capanna, 2002; Dumas and Britton-Davidian, 2002; Merico et al., 2003, 2013). Analyses of recombination rates in wild Rb heterozygotes have also been assessed but involve, in most cases, polymorphic individuals, that is, carrying variable numbers of Rb bivalents and trivalents, making it difficult to disentangle the effect of each meiotic type of configuration (Wallace et al., 1992; Bidau et al., 2001; Castiglia and Capanna, 2002; Capilla et al., 2014). In contrast, extensive analyses of recombination patterns in single-Rb heterozygotes have been performed by genetic assays in crosses between different laboratory strains (Davisson and Akeson, 1993). Recombination suppression was recorded in almost all of the Rb fusions tested, but the extent of the suppression varied between chromosomes and crosses, suggesting an influence of the genetic background on this trait. The present analysis is performed on male and female laboratory-bred F1 hybrids (2n=31) between two chromosomal races of the house mouse (2n=40 and 2n=22) from Tunisia, as well as wild-caught mice spanning the hybrid zone between two North Italian races (2n=40 and 2n=22). In both cases, structural genomic differences exist between races, as all chromosomes except two pairs (a small acrocentric pair and the sex chromosomes) are involved in Rb fusions. The Tunisian F1 hybrids exhibit a homogeneous meiotic architecture in which all Rb chromosomes are present as trivalents, whereas the number of Rb fusions (heterozygous and homozygous) was variable in the Italian mice. Although recent cytogenetic estimates of recombination rely on the analysis of MLH1 foci (mismatch repair protein) on synaptonemal complexes (Anderson et al., 1999), recombination was assessed in the present study by meiotic chiasma analyses permitting direct comparisons with previously published data (Dumas and Britton-Davidian, 2002). The effects of Rb heterozygosity on recombination patterns were identified by comparing the chiasma-based assessment of the Tunisian F1 mice with those previously recorded for the Tunisian parental standard and Rb races from which they originated (Dumas and Britton-Davidian, 2002). An estimate of variation in recombination rates was approached by comparison with the data from the Italian samples. From this, we infer the extent and genomic distribution of the barrier to gene flow between chromosomal races and discuss the relevance of Rb rearrangements to reproductive isolation and divergence processes.
Materials and methods
Samples
Rb heterozygotes (F1; 2n=31) were generated from eight reciprocal crosses between the two previously studied wild-derived chromosomal strains of house mice from Tunisia (Dumas and Britton-Davidian, 2002), one carrying the standard (St) 2n=40 karyotype and the other (Rb) 2n=22, resulting from nine pairs of Rb fusions: Rb(1.11), Rb(2.16), Rb(3.12), Rb(4.6), Rb(5.14), Rb(7.18), Rb(8.9), Rb(10.17), Rb(13.15) (Ould Brahim et al., 2005). These strains originated from three localities in Tunisia where these races interact through hybrid zones (Chatti et al., 1999): Djemmal and Monastir (2n=22) and Kairouan (2n=40). A total of 33 F1 individuals (10 males, 23 females) yielding a total of 162 cells (104 for males and 58 for females) were analyzed. The North Italian sample consisted of 17 male mice from 11 different localities captured along a transect between Cremona and Mantova during a field trip in 2000 (Table 1). The diploid number varied from 2n=22 to 2n=40. The Rb fusions belong to the Cremona race (ICRE, see Piálek et al., 2005): Rb(1.6), Rb(2.8), Rb(3.4), Rb(5.15), Rb(7.18), Rb(9.14), Rb(10.12), Rb(11.13), Rb(16.17). An additional three laboratory-bred F1 individuals from two reciprocal crosses between the Italian 2n=22 and 2n=40 mice were analyzed.
Table 1. Chromosomal analysis of the wild-caught and laboratory-bred F1 Italian mice.
| Type | N | 2n | Rb fusions | Cells | |
|---|---|---|---|---|---|
| Homozygous | Heterozygous | ||||
| Rb | 5 | 22 | 1.6, 2.8, 3.4, 5.15, 7.18 | 50 | |
| 9.14, 10.12, 11.13,16.17 | |||||
| Wild | 1 | 25 | 1.6, 2.8, 3.4, 5.15, 9.14, 11.13 | 7.18, 10.12, 16.17 | 10 |
| hybrids | 1 | 26 | 5.15, 7.18, 9.14, 10.12, 16.17 | 1.6, 2.8, 3.4, 11.13 | 36 |
| 1 | 35 | 10.12 | 3.4, 9.14, 16.17 | 8 | |
| 1 | 37 | 1.6, 3.4, 16.17 | 10 | ||
| 1 | 38 | 5.15 9.14 | 3 | ||
| 1 | 39 | 10.12 | 10 | ||
| 1 | 39 | 3.4 | 10 | ||
| Sub-total | 87 | ||||
| F1 | 3 | 31 | 1.6, 2.8, 3.4, 5.15, 7.18 | 25 | |
| 9.14, 10.12, 11.13,16.17 | |||||
| St | 5 | 40 | 50 | ||
| Total | 202 | ||||
Abbreviations: Cells, number of cells analyzed per individual or type of mouse; N, number of mice; 2n, diploid number.
Rb fusions in a homozygous or heterozygous state.
Chromosomal preparations
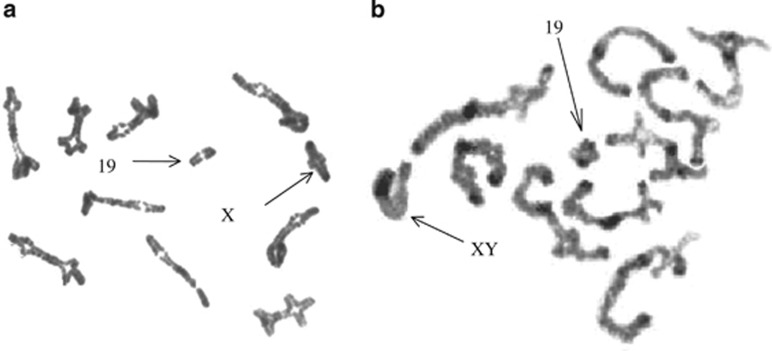
All mice were killed by cervical dislocation, the Tunisian specimens between 88 and 554 days postpartum, and the wild Italian ones after capture. Testes were removed from males and meiotic chromosome preparations were obtained using the air-drying method. At least 10 spermatocytes at the metaphase I stage were examined per male. Ovaries were extracted from females and cultured for 4 h in an incubator at 37 °C to obtain metaphase I chromosomes. The meiosis preparations were stained using a slightly modified C-banding protocol in order to identify the centromeres (Figure 1). The karyotypes of the Italian mice were prepared from bone marrow cells by the air-drying procedure followed by G-banding identification of the chromosomes involved in the Rb fusions. All procedures are detailed in Dumas and Britton-Davidian (2002). Observations were performed using a Zeiss Axiophot microscope at × 1250 magnification and analyzed and archived with the Genevision system (Applied Imaging, Santa Clara, CA, USA).
Figure 1.
Metaphase I plates of F1 Tunisian mice. (a) female F1; (b) male F1.
Chiasma analysis
The meiotic metaphases of F1 hybrids presented a diploid number of 2n=31 consisting of two bivalents (the sex chromosomes and autosome pair 19), and nine trivalents formed by the pairing of Rb fusions with their homologous acrocentrics (Figure 1). The Italian wild-caught sample showed meiotic configurations that depended on their karyotype composition (Table 1): 20 (2n=40) or 11 bivalents (2n=22), 1–4 trivalents (25<2n<39).
Chiasma number (CN) and distribution were recorded per chromosomal arm for each bivalent and trivalent. As the chromosomes were not individually identified, the chiasma counts were totalled over all chromosomes in each cell and averaged per individual. As the sex bivalents differed between males and females, they were excluded from the analyses. Thus, all values of mean CN and distribution refer to the autosomal complement (that is, 19 autosomal arms). However, the X bivalent and bivalent 19 could be morphologically identified in the metaphases of the Tunisian F1 (that is, the X bivalent is the largest and chromosome 19 the smallest), so their CN was determined individually and compared between karyotypes. Chiasmata were divided into two types: i) single, when only one chiasma was present on the arms and ii) multiple when more than one was observed.
Chiasma positions were measured using the option available in the Genevision software, which provided a distance from the centromere expressed as a percentage of the total length of the chromosomal arm. Two measurements were made, one along each chromatid, and averaged per chromosomal arm. Measurement error was estimated by an ANOVA, which was first calculated on a subsample of 120 arms in the F1 individuals (60 per sex), and then separately for the 30 shortest values and the 30 longest ones within each sex. Measurement error accounted for 0.53–1.76% of the total variance in the location of chiasmata with the short bivalents having the highest error fraction (range 1.04–1.76%). The overall low values of measurement error justified dividing chromosomal arms into 10% segments to estimate the mean distribution of chiasmata along the chromosomal arms. In addition, three regions were delimited with similar sizes as in Dumas and Britton-Davidian (2002): proximal (from 0–50% of the arm length), distal (50–90%) and terminal (90–100%). Moreover, the distances between multiple chiasmata occurring on one arm (named chiasma interference) as well as between the centromere and the most proximal chiasma (named centromeric interference) were recorded and averaged. The chiasma data (mean CN and distribution) for the Tunisian F1 males and females were compared with previously published data for the 2n=22 and 2n=40 parents (Dumas and Britton-Davidian, 2002), as were the wild-caught Italian mice.
Statistical tests
Preliminary investigations within sexes involved testing for variation among the Tunisian F1 according to geographic origin (Djemmal, Monastir) and direction of the cross (female Rb × male St and vice-versa) using non-parametric Mann–Whitney U tests, as well as according to age, using Spearman rank correlation tests. Mann–Whitney U tests were also used to compare variation in mean CN between sexes, between karyotypes, and between geographic regions; the data involved the laboratory-bred F1 and their 2n=22 and 2n=40 parents (Tunisian sample; Dumas and Britton-Davidian, 2002), as well as the Italian sample: wild-caught hybrids, laboratory-bred F1, and Rb and St mice. The same testing procedure was performed for the comparisons of mean CN among the three regions of the chromosomal arm and for individual chromosomes (specifically, bivalent 19 and the X chromosome pair in females; see above). The distribution of chiasmata along the 10 segments of the chromosomal arms was compared between samples using χ2- tests. The correlation between diploid number and mean CN in the Italian mice was tested using the Spearman rank correlation test. All tests were performed with Statistica 4.3 (StatSoft, Inc., Tulsa, OK, USA). Corrections for series of k=2–9 tests were made using the sequential Bonferroni tests (Dunn-Sidak method, see Sokal and Rohlf (1995), p. 241). The probability values provided are those corrected according to this method.
Results
Chiasma number
Tunisian sample
The preliminary tests in the F1 mice showed no significant effect of the geographic origin (MW: males: Z=−1.322, P>0.05; females: Z=−1.005, P>0.05), or type of cross (MW: males: Z=−0.685, P>0.05; females: Z=−1.642, P>0.05) on mean CN within sexes. Thus, for each sex, the data were pooled and labeled as F31 and M31 corresponding, respectively, to female and male F1. The correlation between age (days) and CN was calculated for F1 males (range: 88–114) and females (range: 339–554) and was not significant (Spearman: males: rho=0.1385, P=0.70; females: rho=0.3068, P=0.15).
Comparisons between sexes showed that the mean autosomal CN in the Tunisian F1 was significantly higher in females than in males (Table 2, Supplementary Table S1a; MW: P=8.8 × 10-5). Within sexes, the comparison of the F1 with both parental samples revealed that the mean CN of the F1 was intermediate between those of the parental strains, that is, lower than St mice, but higher than Rb individuals (Table 2). All values were significantly different except those between F1 and St females (Supplementary Table S1a).
Table 2. Mean CN values per sex, karyotype and geographic origin of the sample (the sex chromosomes are excluded).
| Origin | Sex |
Rb |
F1/HYB |
St |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | s.d. | N | Mean | s.d. | N | Mean | s.d. | N | ||
| Tunisia | F | 22.29 | 1.52 | 26 | 23.82 | 1.98 | 23 | 24.39 | 1.79 | 21 |
| M | 20.25 | 0.22 | 10 | 21.04 | 0.77 | 10 | 23.00 | 1.60 | 10 | |
| Italy | M | 20.78 | 0.55 | 5 | 21.87 | 0.40 | 10 | 22.16 | 0.53 | 5 |
Abbreviations: F, females; F1, Rb heterozygotes between the Tunisian Rb and St populations; HYB, F1 and hybrids between the Italian Rb and St populations; M, males; N, number of individuals analyzed; Rb, Rb homozygotes; St, standard populations/homozygotes.
The data for the Rb and St Tunisian mice are from Dumas and Britton-Davidian (2002).
Italian sample
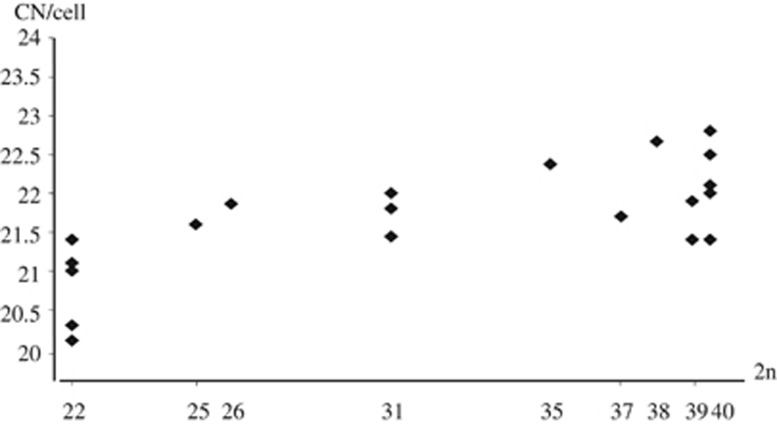
The distribution of mean CN among the Italian individuals showed a significant positive correlation between CN and diploid number (Spearman: rho=0.694, P=7 × 10−4; Figure 2). The Italian hybrid sample consisted of laboratory-bred F1 and wild-caught mice. The preliminary tests on the mean CN values between these mice showed that they were not significantly different (data not shown; MW: −0.247<Z<−0.612; 0.54<P<0.98). In the following tests, the data for Italian mice with intermediate diploid numbers were pooled (hereafter referred to as hybrids: HYB).
Figure 2.
Distribution of mean autosomal chiasma number per cell (CN) according to diploid number in the Italian wild-caught and F1 male mice. Rb=22, St=40, 31=laboratory-bred F1. Data for the Rb and St Tunisian mice are from Dumas and Britton-Davidian (2002).
The mean CN values in the Italian sample showed the same trend as in the Tunisian sample. The mean CN was lowest in the Rb mice, highest in the St individuals and intermediate in the hybrids (Table 2). These differences were significant except for the comparison between Italian St mice and hybrids (Supplementary Table S2).
Geographic variation
Only slight and nonsignificant differences in mean CN were observed between geographic regions within each karyotypic category (Rb, F1/HYB, St; Supplementary Table S2). This was further investigated by comparing CN levels among similar meiotic configurations (bivalents, trivalents), and among sexes, karyotypes and geographic regions (Table 3). The results showed that females had systematically more chiasmata than males for all meiotic types except bivalent 19 (MW: −7.950<Z<−5.286; P<0.001). No significant differences were observed between Tunisian and Italian males with one exception: the mean CN of metacentric bivalents of Rb mice were significantly higher in Italian than in Tunisian mice (Table 3; MW: Z=−6.684; P<0.001). Within the Italian sample, the only significant difference was recorded for metacentric bivalents, the CN of which was lower in hybrids than in Rb mice (Table 3; MW; Z=−3.350; P<0.01).
Table 3. Mean CN values per sex and meiotic configuration in the Tunisian and Italian samples.
| Sex |
Bivalent 19 |
Acrocentrics |
Trivalents |
Metacentrics |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | s.d. | Cells | Mean | s.d. | n | Mean | s.d. | n | Mean | s.d. | n | |||
| Tunisia | M | Rb | 1 | 0 | 229 | 1.069 | 0.062 | 4122 | ||||||
| F1 | 1.039 | 0.050 | 104 | 1.111 | 0.073 | 1872 | ||||||||
| St | 1.013 | 0.032 | 195 | 1.22(1.21*) | 0.088 | 3510 | ||||||||
| F | Rb | 1.024 | 0.075 | 55 | 1.181 | 0.095 | 990 | |||||||
| F1 | 1.081 | 0.227 | 58 | 1.263 | 0.12 | 1044 | ||||||||
| St | 1.16 | 0.321 | 51 | 1.29 | 0.109 | 918 | ||||||||
| Italy | M | Rb | 1 | 0 | 50 | 1.154 | 0.087 | 900 | ||||||
| HYB | 1.011 | 0.024 | 73 | 1.191 | 0.09 | 562 | 1.175 | 0.161 | 1018 | 1.112 | 0.136 | 436 | ||
| St | 50 | 1.166* | 0.095 | 950 | ||||||||||
Abbreviations: Acrocentrics, all autosomes except bivalent 19 (*these values include bivalent 19); Cells, number of cells analyzed; F1, laboratory-bred F1 between the Tunisian Rb and St populations; HYB, laboratory-bred F1 and wild hybrids between the Italian Rb and St populations; n, number of chromosomal arms analyzed; Rb, Rb homozygotes; St, Standard homozygotes.
The data for the Rb and St Tunisian mice are from Dumas and Britton-Davidian (2002).
Chiasma distribution
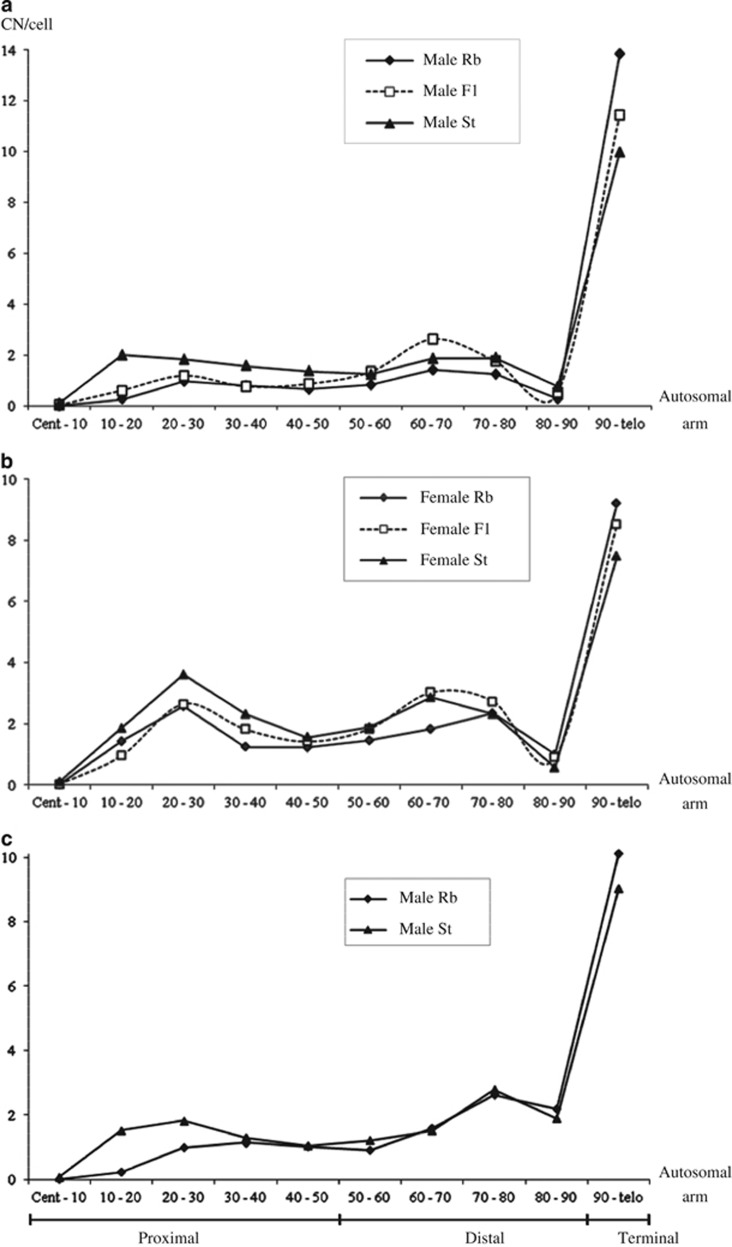
The distribution of chiasmata along the chromosomal arms was determined for the Tunisian F1 mice as well as the Italian Rb and St individuals (Figure 3). The Italian hybrids were not included in this analysis owing to their variable karyotype. Within the Tunisian F1, all tests that compared the overall and regional distribution of total chiasmata, as well as the distribution by chiasma type (single and multiple), were significant between sexes (Supplementary Table S1a–c). Comparisons within sexes between the F1 and the parental karyotypes showed significant differences in the overall distribution of chiasmata, whether total chiasmata were considered, or separated into single or multiple (Supplementary Table S1c; Figure 3; Supplementary Figure S1). The regional distribution of total chiasmata highlighted a particular mosaic pattern. In both male and female F1, the localization of chiasmata in the proximal regions closely matched that in Rb mice, whereas the distal chiasmata followed the same pattern as that in St individuals—the CN in the terminal regions being intermediate between the parental values (for details, see Figures 3a and b). This distinctive pattern in the F1 was supported by the significance values of the comparative tests (Supplementary Table S1b). Although the overall distribution of chiasmata differed between the Italian Rb and St mice for all chiasma types (χ2: total chiasmata=69.4; single=41.7; multiple=42.0; P<0.01), the regional distribution was not significantly different between karyotypes (MW: proximal: Z=−2.41; distal: Z=−0.522; terminal: Z=−2.627; all P>0.05; see Figure 3c); this may be due to the small sample size (50 cells), or to the extent of the region defined as proximal (50%). Comparison within karyotypes from different geographic regions showed that the regional distribution of mean CN in Rb and St mice differed only with respect to distal and terminal chiasmata. This was due to a higher number of single-distal chiasmata in the Italian mice than the Tunisian mice (data not shown; MW: 2.817<Z<3.065; P<0.05). The opposite was observed for the terminal chiasmata among the Rb mice (data not shown; MW: Z=−3.065; P<0.05).
Figure 3.
Mean per cell distribution of chiasmata along 10% segments of the autosomal arms in Tunisian (a) males and (b) females (data for the Rb and St Tunisian mice are are from Dumas and Britton-Davidian, 2002), and (c) Rb and St Italian wild-caught males. The three regional distributions are indicated: proximal (0–50%), distal (50–90%), terminal (90–100%). Cent: centromere; Telo: telomere.
Individual bivalents
The XY bivalents invariably presented only one terminal chiasma in all males (data not shown). Tests comparing chiasma patterns of the X bivalent were performed for Tunisian females carrying different karyotypes (Supplementary Table S1). No significant differences in overall CN were found between F1 and St (respective means: 1.42±0.38 and 1.57±0.42; MW: P=0.169), or Rb mice (mean: 1.24±0.35; MW: P=0.077). Results on chiasma distribution along the X bivalent of the F1 indicated that the overall patterns were similar to those of the parental mice (Supplementary Table S1c; MW: all tests P-values >0.05), except for the distribution of single chiasmata, which differed significantly between F1 and the St mice (χ2: P=0.001). Finally, comparisons involving bivalent pair 19 (the only autosomal bivalent common to all karyotypes) revealed no differences between sexes or chromosomal types (Table 3); in fact, all mean CN values did not significantly differ from one chiasma per bivalent (MW: 0.054<Z<0.843; all P>0.05).
Interference distances
Mean values of chiasma interference, that is, the distance between multiple chiasmata occurring on one arm, and of centromere interference, that is, the distance between the centromere and the most proximal chiasma, were calculated for the Tunisian sample (Table 4). Within sexes, the interference distances in the Tunisian F1 hybrids were generally intermediate between those of the parental races. All values were significantly different with the exception of the centromere-to-proximal component of multiple chiasmata and the chiasma interference distance between Rb and F1 females (Supplementary Table S3). Significant between-sex differences were observed in the Tunisian F1 as (i) distances between chiasmata were significantly higher in males than in females and (ii) the centromere-to-proximal distances in multiple chiasmata were higher in females than in males (Table 4; Supplementary Table S3). This differential pattern was not observed for centromere-to-single-chiasma distances, as males always presented significantly higher distance values than females (Table 4; Supplementary Table S3).
Table 4. Mean values (standard deviation) expressed in % of chromosomal arm length for different interference distances in Tunisian Rb, F1 and St females (F) and males (M).
| Interference distance |
Rb |
F1 |
St |
|||
|---|---|---|---|---|---|---|
| F | M | F | M | F | M | |
| Between chiasmata | 69.91 (12.87) | 67.18 (12.68) | 71.39 (9.54) | 74.90 (9.12) | 73.38 (11.58) | 75.57 (11.65) |
| Centromere to multiple proximal chiasmata | 27.25 (11.52) | 32.27 (12.30) | 27.67 (8.87) | 24.76 (7.88) | 25.23 (9.39) | 23.51 (11.02) |
| Centromere to single chiasma | 73.39 (26.68) | 87.76 (21.43) | 72.30 (21.34) | 83.13 (20.95) | 62.49 (23.37) | 74.25 (26.50) |
Abbreviations: F1, Rb heterozygotes between the Tunisian Rb and St populations; Rb, Rb homozygotes; St, Standard homozygotes.
Chiasma interference refers to the distance between two chiasmata on a chromosomal arm. Centromere interference refers to the distance between the centromere and a chiasma. Multiple proximal corresponds to the most proximal component of multiple chiasmata. Data for the Rb and St populations are from Dumas and Britton-Davidian (2002).
Discussion
Effect of Rb heterozygosity on recombination patterns
The present analysis provides data on the effects of genome-wide Rb heterozygosity on chiasma patterns in both male and female hybrids between highly differentiated chromosomal races of the house mouse. Neither the age of the mice, nor of the direction of the cross had an effect on recombination estimates. These results are consistent with most, but not all, previous observations (Koehler et al., 2002; but see Merico et al., 2003; Dumont and Payseur, 2011; Vrooman et al., 2014). Comparison of mean chiasma counts between Rb heterozygotes (F1/HYB) and the parental races showed that the mean CN in the former was intermediate, lower than in the St race, and higher than in homozygous Rb mice. Owing to disjunctional constraints of at least one crossover per chromosomal arm for proper segregation (Pardo-Manuel de Villena and Sapienza, 2001), the reduction in CN evidenced in F1 individuals was related to a decrease in the number of multiple chiasmata compared with standard individuals (Pardo-Manuel de Villena and Sapienza, 2001), decrease which was nevertheless less pronounced than that observed in homozygous Rb mice (Dumas and Britton-Davidian, 2002; Capilla et al., 2014).
More importantly, female and male F1 hybrids showed chiasma distribution patterns that were significantly different from those in the parental races. The proximal regions of meiotic chromosomes in F1 hybrids exhibited a reduction in mean CN similar to that observed in Rb homozygotes, whereas distal regions more closely matched values in St mice; telomeric regions showed a mean CN that was intermediate between the two parental karyotypes (Figure 3). As demonstrated in Rb mice (Dumas and Britton-Davidian, 2002), the reduction in mean CN along the proximal regions of Rb trivalents was related to an increase in centromere interference compared with St individuals (Table 4). However, the centromere interference in the F1 generally extended over a smaller chromosomal arm length than in Rb bivalents, and only a distal shift of chiasmata was observed, instead of the more terminal relocation evidenced in Rb mice (Figures 3a and b). The expansion in centromere interference observed in Rb fusions has recently been related to alterations in the epigenetic signatures of heterochromatinization (Capilla et al., 2014).
To our knowledge, few analyses on chiasma or MLH1 patterns have compared trivalent-carrying heterozygous wild mice with homozygous standard or Rb individuals, and those that have, involve either few cells (that is, 12 cells; Wallace et al., 1992), few Rb fusions (that is, 4; Bidau et al., 2001) or mostly polymorphic hybrid karyotypes (24<2n<39; Castiglia and Capanna, 2002; Capilla et al., 2014). The exception is the study by Merico et al. (2003) involving other North Italian races (2n=24, St mice) and their laboratory-bred F1 hybrids (2n=32 with eight trivalents). The trends revealed in all these studies mirror ours in that they detect (i) a negative correlation between CN/MLH1 foci and the number of Rb configurations in the genome (whether homozygous or heterozygous), and (ii) an intermediate recombination rate in Rb hybrids or trivalents compared with Rb and St mice. The only difference was recorded by Merico et al. (2003) who observed an even lower number of MLH1 foci in the North Italian F1 hybrids than in the Rb race. Although mean values varied between analyses, F1 hybrids/trivalents always exhibited the same distribution pattern: a pronounced decrease in recombination in the proximal regions of the chromosomes compared with St mice/St bivalents. Such modifications in the recombination landscape owing to Rb fusions appear to be a general feature of this type of rearrangement as they have also been observed in a diversity of organisms, that is, shrews, grasshoppers and fish (Borodin et al., 2008; Colombo, 2013; Ostberg et al., 2013). Thus, a growing number of studies demonstrate that heterozygosity for Rb rearrangements is associated with reduced recombination in the proximal regions of chromosomes.
In house mice, the intensity of the decrease in recombination rates in Rb fusions varied slightly among geographic regions and chromosomal races, irrespective of the method of analysis (chiasma or MLH1; range: 22.16<St<23.0; 20.1<Rb<22.28; the present study; Wallace et al., 1992; Bidau et al., 2001; Castiglia and Capanna, 2002; Dumas and Britton-Davidian, 2002; Merico et al., 2013). Exceptionally high levels were observed by Merico et al. (2003) in two North Italian races (Rb=24.58; St=25.13) that exceeded estimates described so far for the subspecies (Dumont and Payseur, 2011). A large part of the variation among Rb races most likely depends on the number of Rb fusions present, and on the chromosomes involved in the Rb fusions. In the present study, as chiasma values were averaged over all chromosomes, the inter-chromosomal variability could not be assessed. Such data require identification of meiotic chromosomes by specific probes such as FISH-BACs or whole-chromosome probes (Capilla et al., 2014). However, the diversity of rates among races with the same diploid number, as well as among St mice (see for example Tunisia, Italy), strongly suggests that the genetic background also contributes to the recombination landscape (Davisson and Akeson, 1993; Koehler et al., 2002), possibly through the fixation of variant alleles at the Prdm9 gene as suggested by Capilla et al. (2014).
Intriguingly, both Rb homozygotes and heterozygotes exhibit a similar decrease in proximal recombination rates. As the reduction in Rb heterozygotes is related to meiotic pairing defects in the centromeric region of trivalents (Davisson and Akeson, 1993), the same mechanism cannot apply to Rb bivalents that show conventional pairing patterns (Manterola et al., 2009). Such discrepancies require further assessment to determine the nature of the mechanisms involved in the modifications of recombination patterns (Dumas and Britton-Davidian, 2002; Colombo, 2013; Capilla et al., 2014).
Differences between sexes within hybrids were observed for both CN and localization. Female F1 specimens had higher CN values as well as a larger proportion of nonterminal vs terminal chiasmata than did F1 males. These are the first data available for wild female mice, although sex-biased chiasma patterns have been described in laboratory strains of house mice with a standard karyotype (Jagiello and Fang, 1987). Thus, sex differences in recombination patterns are maintained regardless of the chromosomal change in structure.
Effect of Rb heterozygosity on gene flow
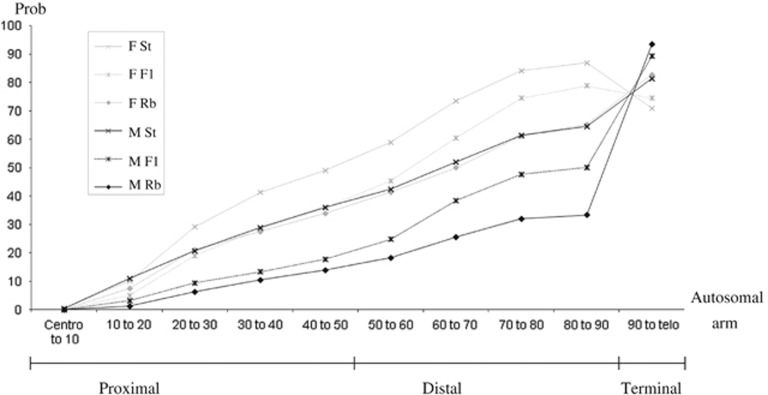
The recombination suppression models of speciation emphasize the role of rearrangements as modifiers of the recombination landscape rather than that of fitness (Navarro and Barton, 2003). These models are of particular interest in species such as the house mouse, in which heterozygosity for single-Rb fusions is associated with very low levels of aneuploidy (Hauffe and Searle, 1998; Sans-Fuentes et al., 2010). The present study provides experimental data on chiasma patterns in chromosomal male and female hybrids from which recombination rates can be inferred and by extension the expected patterns of gene flow between chromosomal races. The decrease in proximal recombination rates affects both rearranged homozygotes and heterozygotes. These results contrast with current models in which the suppression of recombination is restricted to chromosomal heterozygotes (inversions, reciprocal translocations; Rieseberg, 2001; Navarro and Barton, 2003). How do these unexpected results fit in with gene flow expectations? Gene flow between chromosomal races can only occur through hybrids, so it is a function of the rate of gene exchange between the Rb fusion and the homologous acrocentric chromosomes during meiosis. Thus, the recombination parameter relevant to gene flow is the probability that an allele move from one chromosomal type to the other. The nonrandom distribution of chiasmata along the chromosomal arms of trivalents suggests that the probability of exchange will be related to its degree of linkage to the centromere. This probability was computed by cumulating the mean number of chiasmata per chromosomal segment starting from the centromere, and by subtracting from these data the number of distal multiple chiasmata, as they restore the terminal fragment to its original chromosome (Figure 4). The results indicate that the probability for a gene to be included in a crossover event increases with its distance from the centromere, and that females will have higher exchange rates than males in all regions of the chromosomes except the telomeric ones.
Figure 4.
Cumulative probability of the occurrence of allelic exchanges during meiosis between homologous chromosomes relative to the location on the chromosomal arm (10% segments), the karyotype and the sex of the animal. The data are estimated from CN values and distribution of the Tunisian mice (F1: present study; Rb and St: Dumas and Britton-Davidian, 2002). Rb=Rb homozygotes; St=standard populations. The three regional distributions are indicated: proximal (0–50%), distal (50–90%), terminal (90–100%). Centro: centromere; Telo: telomere.
The gradient in recombination rate is expected to produce a similar gradient in the flow of genes from one race to the other through the hybrid zone (Feder and Nosil, 2009). Estimates of divergence between the Rb and St populations studied here (Tunisia and Italy) did not have the required resolution to test for differences in local recombination rates (that is, allozymes; Britton-Davidian, 1990; Saïd et al., 1999; Ould Brahim et al., 2005). However, several studies have addressed this question by measuring the level of divergence of mapped microsatellite markers between chromosomally differentiated house mice. In all cases, centromeric loci exhibited higher differentiation levels than distal ones, regardless of the type of meiotic heterozygosity of the hybrids, that is, complex chains or rings (Panithanarak et al., 2004; Förster et al., 2013; Gimenez et al., 2013 ) or trivalents (Franchini et al., 2010). Previous studies have indicated that recombination suppression extended up to 15–20 cM from the centromere depending on the chromosomes involved in the Rb fusion and the genetic background (Davisson and Akeson, 1993; Gimenez et al., 2013; Capilla et al., 2014). In the present analysis, the mean reduction in recombination affected the proximal 50% of the chromosome, but the intensity of the decrease appeared stronger in males than in females, and was more pronounced in the Tunisian than Italian mice. Conversely, divergence estimates in shrew populations differing by Rb fusions produced mixed results. In two parapatric species of shrews, molecular markers on rearranged chromosomes showed higher divergence levels than those on collinear chromosomes (Basset et al., 2006), whereas such differences were not observed among several chromosomal races of the common shrew (Horn et al., 2011). In addition to the possible lack of resolution due to the imprecise localization of the markers in shrews, these discrepancies highlight confounding difficulties in disentangling the relative contributions of reduced gene flow, ancestral polymorphism and selection at linked sites (Nachman and Payseur, 2012), particularly in closely related taxa (that is, races vs species).
In the studies on house mice cited above, the distribution of genetic divergence levels between chromosomal races closely matched the one predicted by the gradient in recombination. This concordance suggests that the recombinational effect of Rb fusions is sufficient to significantly affect the pattern of gene flow between chromosomally differentiated populations. However, using a simulation approach, Gimenez et al. (2013) concluded that hybrid dysgenesis alone could account for the differentiation pattern they observed. Hybrid breakdown and the proximal decrease in recombination are, to a large extent, both related to pairing disturbances during meiosis leading to germ cell death and aneuploidy due to malsegregation (King, 1993). Meiotic pairing of trivalents involves either no (asynapsis) or heterologous synapsis in the proximal regions, that is, respectively open and closed configurations (Davisson and Akeson, 1993; Manterola et al., 2009). These pairing alterations will lead to recombination suppression in this region, as crossing over requires homologously and timely synapsed segments. Although asynaptic cells are generally eliminated through apoptosis, a recent study demonstrated that 37% of such cells were able to bypass the apoptotic meiotic checkpoints enabling the meiocyte to develop into viable sperm (Manterola et al., 2009). Thus, all surviving meiocytes of Rb heterozygotes are expected to show reduced proximal recombination. Among these, a certain percentage (depending on the degree of heterozygosity) will be aneuploid and eliminated after fertilization. These two processes will influence gene flow differently. Euploid cells will lead to the divergence of genes in the proximal regions of the chromosomes involved in the Rb fusions, whereas the elimination of dysfunctional cells or aneuploid embryos will contribute to the divergence of genes throughout the genome. Thus, a differential effect on gene flow of hybrid breakdown vs reduced proximal recombination is expected. This suggests that (i) both recombination reduction and hybrid unfitness act synergistically to modify the gene flow landscape between chromosomal races of the house mouse as previously stated (Panithanarak et al., 2004; Franchini et al., 2010; Gimenez et al., 2013), and (ii) the effect of hybrid unfitness is expected to increase with the degree and type of heterozygosity. Combining simulation approaches with analyses, using high resolution genomic markers in wild populations, will undoubtedly assist in teasing apart the respective contribution of both factors, and further refine the expected patterns of divergence related to Rb fusions.
We therefore conclude that heterozygosity for Rb fusions in the house mouse may promote the divergence of proximal genomic regions. Although the role of centromeres as recombination suppressors has long been recognized, their relevance as promoters of divergence has generally been downscaled owing to their surrounding low gene density (Navarro and Barton, 2003; Faria and Navarro, 2010). What the present study clearly highlights is that Rb rearrangements, in fact, extend the suppression recombination effect of centromeres to a larger region of the chromosome, thus providing the opportunity for the divergence of proximally located genes. If these chromosomal regions harbor loci involved in postzygotic (that is, genetic incompatibilities) and/or prezygotic isolation (that is, assortative mating loci), divergence between the chromosomally differentiated taxa may lead to speciation (Butlin, 2005; Gimenez et al., 2013). Interestingly, several genes involved in mate choice discrimination and preferences have been localized in the house mouse; these pertain to the signaling and reception of odors (Smadja and Ganem, 2008; see Karn and Laukaitis, 2012): androgen-binding protein, major urinary protein and pheromone receptors in the vomeronasal organ (V1r and V2r multigene families). Several of these genes occupy sufficiently proximal positions (<15 cM; see for example the V1r receptor genes on chromosomes 4, 5 and 7, http://www.jax.org) to be affected by changes in recombination patterns in chromosomal heterozygotes. Further studies on the chromosomal distribution of genes involved in genetic incompatibilities are required to quantitatively support the role of Rb fusions in the development of reproductive isolation in the house mouse.
Data archiving
The data are not genotypes nor sequences and have thus not been archived.
Acknowledgments
We acknowledge the assistance of M Perriat-Sanguinet for help in the maintenance of mice. We express our very grateful thanks to C Smadja and T J Robinson for insightful comments on a previous version of the manuscript, to M Kirkpatrick for enlightening discussions, and to G Ganem for her unwavering support in the statistical analyses. This work was financed by a grant from ACC-SV7 ‘Systématique et Biodiversité' and recurrent funds from the Centre National de la Recherche Scientifique and the Université de Montpellier 2. This is publication N° ISEM-2014-099.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Anderson LK, Reeves A, Webb LM, Ashley T. (1999). Distribution of crossing-over on mouse synaptonemal complexes using immunofluorescent localization of Mlh1 protein. Genetics 151: 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D, Fontaine MC, Cohuet A, Fontenille D, Vitalis R, Simard F. (2011). Chromosomal inversions, natural selection and adaptation in the malaria vector Anopheles funestus. Mol Biol Evol 28: 745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala FJ, Coluzzi M. (2005). Chromosome speciation: humans, Drosophila and mosquitoes. Proc Natl Acad Sci 102: 6535–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basset P, Yannic G, Brünner H, Hausser J. (2006). Restricted gene flow at specific parts of the shrew genome in chromosomal hybrid zones. Evolution 60: 1718–1730. [PubMed] [Google Scholar]
- Bidau CJ, Gimenez MD, Palmer CL, Searle J. (2001). The effects of Robertsonian fusions on chiasma frequency and distribution in the house mouse (Mus musculus domesticus) from a hybrid zone in northern Scotland. Heredity 87: 305–313. [DOI] [PubMed] [Google Scholar]
- Borodin PM, Karamysheva TV, Belonogova NM, Torgasheva AA, Rubtsov NB, Searle JB. (2008). Recombination map of the common shrew, Sorex araneus (Eulipotyphla, Mammalia). Genetics 178: 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton-Davidian J. (1990). Genic differentiation in M. m. domesticus populations from Europe, the Middle East and North Africa: geographic patterns and colonization events. Biol J Linn Soc 41: 27–45. [Google Scholar]
- Butlin RK. (2005). Recombination and speciation. Mol Ecol 14: 2621–2635. [DOI] [PubMed] [Google Scholar]
- Capilla L, Medarde N, Alemany-Scmidt A, Oliver-Bonnet M, Ventura J, Ruiz-Herrera A. (2014). Genetic recombination variation in wild mice Robertsonian mice: on the role of chromosomal fusions and Prdm9 allelic background. Proc Roy Soc London B 281: 1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglia R, Capanna E. (2002). Chiasma repatterning across a chromosomal hybrid zone between chromosomal races of Mus musculus domesticus. Genetica 114: 35–40. [DOI] [PubMed] [Google Scholar]
- Chatti N, Ganem G, Benzekri K, Catalan J, Britton-Davidian J, Saïd K. (1999). Microgeographical distribution of two chromosomal races of house mice in Tunisia: pattern and origin of habitat partitioning. Proc Roy Soc London B 266: 1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo PC. (2013). Micro-evolution in grashoppers mediated by polymorphic Robertsonian translocations. J Insect Sci 13: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson MT, Akeson EC. (1993). Recombination suppression by heterozygous Robertsonian chromosomes in the mouse. Genetics 133: 649–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas D, Britton-Davidian J. (2002). Chromosomal rearrangements and evolution of recombination: comparison of chiasma distribution patterns in standard and Robertsonian populations of the house mouse. Genetics 162: 1355–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. (2011). Evolution of the genomic recombination rate in murid rodents. Genetics 187: 643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria R, Navarro A. (2010). Chromosomal speciation revisited: rearranging theory with pieces of evidence. TREE 25: 660–669. [DOI] [PubMed] [Google Scholar]
- Farré M, Micheletti D, Ruiz-herrera A. (2013). Recombination rates and genomic shuffling in human and chimpanzee - new twist in the chromosomal speciation theory. Mol Biol Evol 30: 853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Nosil P. (2009). Chromosomal inversions and species differences: when are genes affecting adaptive divergence and reproductive isolation expected to reside within inversions? Evolution 63: 3061–3075. [DOI] [PubMed] [Google Scholar]
- Förster DW, Mathias ML, Britton-Davidian J, Searle JB. (2013). Origin of the chromosomal radiation of Madeiran house mice: a microsatellite analysis of metacentric chromosomes. Heredity 110: 380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini P, Colangelo P, Solano E, Capanna E, Verheyen E, Castiglia R. (2010). Reduced gene flow at pericentromeric loci in a hybrid zone involving chromosomal races of the house mouse Mus musculus domesticus. Evolution 64: 2020–2032. [DOI] [PubMed] [Google Scholar]
- Gimenez MD, White TA, Hauffe HC, Panithanarak T, Searle JB. (2013). Understanding the basis of diminished gene flow between hybridizing chromosome races of the house mouse. Evolution 67: 1446–1462. [DOI] [PubMed] [Google Scholar]
- Hauffe HC, Searle JB. (1998). Chromosomal heterozygosity and fertility in house mice (Mus musculus domesticus) from Northern Italy. Genetics 150: 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn A, Basset P, Yannic G, Banaszek A, Borodin PM, Bulatova NS et al. (2011). Chromosomal rearrangements do not seem to affect the gene flow in hybrid zones between karyotypic races of the common shrew (Sorex araneus). Evolution 66: 882–889. [DOI] [PubMed] [Google Scholar]
- Jagiello G, Fang J-S. (1987). Observations on chiasmata in mouse diplotene oocytes and spermatocytes. Cytologia 52: 283–292. [Google Scholar]
- Karn RC, Laukaitis CM. (2012). The roles of gene duplication, gene conversion and positive selection in rodent Esp and Mup pheromone gene families with comparison to the Abp family. PloS ONE 7: e47697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. (1993). Species evolution. The role of chromosome change. Cambridge University Press: Cambridge. [Google Scholar]
- Kirkpatrick M, Barton N. (2006). Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KE, Cherry JP, Lynn A, Hunt PA, Hassold TJ. (2002). Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162: 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. (2010). A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biology 8: e10000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola M, Page J, Vasco C, Berrios S, Parra MT, Viera A et al. (2009). A high incidence of meiotic silencing of unsynapsed chromatin is not associated with substantial pachytene loss in heterozygous male mice carrying multiple Robertsonian translocations. PloS Genetics 5: e1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh SE, Noor MAF. (2012). Genomic impacts of chromosomal inversions in parapatric Drosophila species. Phil Trans Roy Soc B 367: 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico V, Giménez MD, Vasco C, Zuccotti M, Searle JB, Hauffe HC et al. (2013). Chromosomal speciation in mice: a cytogenetic analysis of recombination. Chrom Res 21: 523–533. [DOI] [PubMed] [Google Scholar]
- Merico V, Pigozzi MI, Esposito A, Merani MS, Garagna S. (2003). Meiotic recombination and spermatogenic impairment in Mus musculus domesticus carrying multiple simple Robertsonian translocations. Cytogenet Genome Res 103: 321–329. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Payseur BA. (2012). Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Phil Trans Roy Soc B 367: 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Barton NH. (2003). Accumulating postzygotic isolation genes in parapatry: A new twist on chromosomal speciation. Evolution 57: 447–459. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J. (2001). Chromosomal inversions and the reproductive isolation of species. Proc Nat Acad Sci 98: 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostberg CO, Hauser L, Pritchard VL, Garza JC, Naish KA. (2013). Chromosome rearrangements, recombination suppression, and limited segregation distortion in hybrids between Yellowstone cutthroat trout (Oncorhynchus clarkii bouvieri) and rainbow trout (O. mykiss). BMC Genomics 14: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ould Brahim I, Chatti N, Britton-Davidian J, Said K. (2005). Origin and evolution of the Robertsonian populations of the house mouse (Rodentia, Muridae) in Tunisia based on allozyme studies. Biol J Linn Soc 84: 515–521. [Google Scholar]
- Panithanarak T, Hauffe HC, Dallas JF, Glover A, Ward RG, Searle JB. (2004). Linkage-dependent gene flow in a house mouse chromosomal hybrid zone. Evolution 58: 184–192. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. (2001). Recombination is proportional to the number of chromosome arms in mammals. Mamm Genome 12: 318–322. [DOI] [PubMed] [Google Scholar]
- Piálek J, Hauffe HC, Searle JB. (2005). Chromosomal variation in the house mouse. Biol J Linn Soc 84: 535–563. [Google Scholar]
- Rieseberg LH. (2001). Chromosomal rearrangements and speciation. TREE 16: 351–358. [DOI] [PubMed] [Google Scholar]
- Saïd K, Auffray J-C, Boursot P, Britton-Davidian J. (1999). Is chromosomal speciation occurring in house mice in Tunisia? Biol J Linn Soc 68: 387–399. [Google Scholar]
- Sans-Fuentes M, Garcia-Valero J, Ventura J, Lopez-Fuster MJ. (2010). Spermatogenesis in the house mouse in a Robertsonian polymorphism zone. Reproduction 140: 569–581. [DOI] [PubMed] [Google Scholar]
- Smadja C, Ganem G. (2008). Divergence of odorant signals within and betwen the two European subspecies of the house mouse. Behav Ecol 19: 223–230. [Google Scholar]
- Sokal RR, Rohlf FJ. (1995) Biometry. Freeman: New York. [Google Scholar]
- Vrooman LA, Nagaoka SI, Hassold TJ, Hunt PA. (2014). Evidence for paternal age-related alterations in meiotic chromosome dynamics in the mouse. Genetics 196: 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BMN, Searle JB, Everett CA. (1992). Male meiosis and gametogenesis in wild house mice (Mus musculus domesticus) from a chromosomal hybrid zone; a comparison between ‘simple' Robertsonian heterozygotes and homozygotes. Cytogenet Cell Genet 61: 211–220. [DOI] [PubMed] [Google Scholar]
- White MJD. (1978) Modes of speciation Eds Freeman WH & co: San Francisco. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.