Abstract
Major histocompatibility complex (MHC) polymorphism is thought to be driven by antagonistic coevolution between pathogens and hosts, mediated through either overdominance or frequency-dependent selection. However, investigations under natural conditions are still rare for endangered mammals which often exhibit depleted variation, and the mechanism of selection underlying the maintenance of characteristics remains a considerable debate. In this study, 87 wild giant pandas were used to investigate MHC variation associated with parasite load. With the knowledge of the MHC profile provided by the genomic data of the giant panda, seven DRB1, seven DQA1 and eight DQA2 alleles were identified at each single locus. Positive selection evidenced by a significantly higher number of non-synonymous substitutions per non-synonymous codon site relative to synonymous substitutions per synonymous codon site could only be detected at the DRB1 locus, which leads to the speculation that DRB1 may have a more important role in dealing with parasite infection for pandas. Coprological analyses revealed that 55.17% of individuals exhibited infection with 1–2 helminthes and 95.3% of infected pandas carried Baylisascaris shroederi. Using a generalized linear model, we found that Aime-DRB1*10 was significantly associated with parasite infection, but no resistant alleles could be detected. MHC heterozygosity of the pandas was found to be uncorrelated with the infection status or the infection intensity. These results suggested that the possible selection mechanisms in extant wild pandas may be frequency dependent rather than being determined by overdominance selection. Our findings could guide the candidate selection for the ongoing reintroduction or translocation of pandas.
Introduction
Genes of the major histocompatibility complex (MHC), with their extremely high levels of polymorphism, represent classic examples for the selective advantage of genetic diversity (Hughes and Yeager, 1998). MHC encodes glycoproteins that recognize foreign peptides for the subsequent presentation to immune cells. Viable MHC enables the wide recognition and binding of pathogens by activating an antigen-specific immune response, and in turn, determines the disease and parasite resistance of an organism (Sommer, 2005). However, the mechanism of parasite-driven selection underlying the maintenance of diverse functions and characteristics at MHC (overdominance or heterozygote advantage versus frequency-dependent selection) remains a considerable debate (Sommer, 2005; Piertney and Oliver, 2006).
The overdominance hypothesis predicts that individuals that are heterozygous for key MHC exons (especially those which encode antigen binding residues) are expected to show lower levels of infection than those individuals that are homozygous. It is believed that the possession of two alleles provides genetic ‘insurance' against infection in an environment where a diverse range of organisms can challenge the immune system of the host (Doherty and Zinkernagel, 1975). The most convincing evidence for the heterozygote advantage was shown by McClelland et al. (2003) by using co-infections with multiple pathogens in MHC-congenic mice with reciprocal resistance/susceptibility profiles. A superiority of MHC heterozygotes to homozygotes against multiple pathogens could be observed. More recently, MHC heterozygote advantage was also evidenced by significantly higher viral infection in MHC homozygotes than their heterozygotes counterparts in Chinook salmon (Oncorhynchus tshawytscha) (Evans and Neff, 2009). In contrast, the frequency-dependent selection predicts that local disease outbreaks select for or against specific alleles that confer resistance or susceptibility to the host, respectively. Here, individuals possessing a particular allele or alleles are expected to show a relationship between the presence and absence of that allele and infection level (Takahata and Nei, 1990). A growing body of empirical evidences for associations between particular alleles and resistance/susceptibility to infection have been suggested to be consistent with negative frequency-dependent selection driving the maintenance of polymorphism in MHC genes (Piertney and Oliver, 2006). MHC class II DQA genes were found to be associated with Puumala virus infection in the bank vole (Myodes glareolus) (Deter et al., 2008); and it was revealed that MHC heterozygotes were simultaneously co-infected by fewer parasite types than homozygotes in the water vole (Arvicola terrestris). This was the first demonstration that MHC heterozygotes have an advantage against multi-parasite infections in a natural population (Oliver et al., 2009). ‘Good' MHC genes were also evidenced by the association of MHC class II DRB alleles with ectoparasitism resistance in the neotropical lesser bulldog bat (Noctilio albiventris) (Schad et al., 2012). These studies showing the interaction between MHC and parasite infection, however, have been mostly limited in small or experimental animals.
Endangered species generally exhibits lower levels of genetic variation than common species, especially at the MHC (Marsden et al., 2009). Reduced MHC variability was suggested to increase the risk of diseases infection (Sommer, 2005), and direct links between MHC polymorphism and diversity in immune defense have been confirmed (Messaoudi et al., 2002). With the rising concerns about infectious diseases in biological conservation, the importance of adaptive genetic variation at MHC in the conservation and evolutionary ecology has been addressed (Sommer, 2005). Nevertheless, little evidence has been accumulated about the association between fitness characteristics in terms of parasite resistance/susceptibility and MHC variability in wild large endangered mammals. In addition, for numerically scarce species, detecting the selection within MHC associated with resistance/susceptibility to diseases will be statistically problematic, partly because of a lack of power to detect small-scale effects using a system possessing high levels of polymorphism and multiple loci with considerable number of alleles at each locus (Radwan et al., 2010).
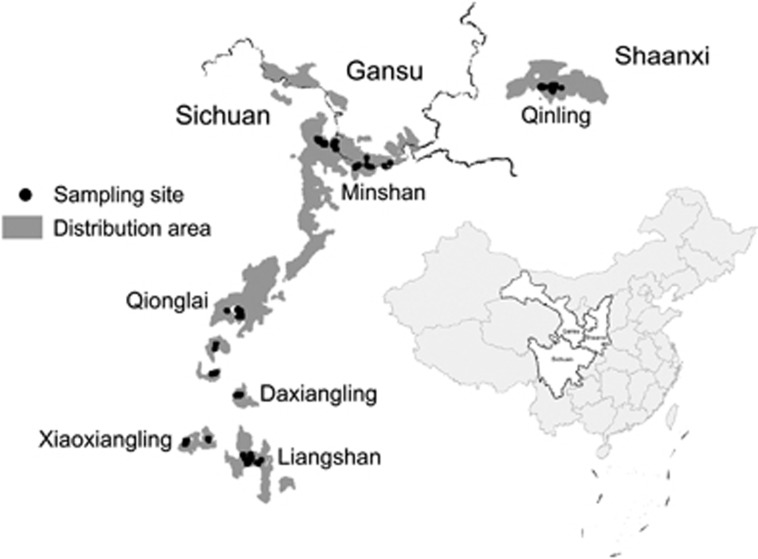
Giant pandas (Ailuropoda melanoleuca) are distributed in very small numbers, with habitats fragmented to six isolated mountain areas including Qinling, Minshan, Qionglai, Liangshan, Daxiangling and Xiaoxiangling mountains in China (Figure 1). Wild pandas are basically solitary animals that only occasionally interact with each other except during their short reproductive season (Schaller et al., 1985). Within past years, much progress has been made in research employing neutral markers such as mtDNA and microsatellites, which are very informative for exploring the evolutionary history of extant pandas. Moderate to relative high levels of diversity have been detected using these neutral markers in the giant panda, and a consistent anthropogenic pressure has been revealed in the recent population decline (Zhang et al., 2007; Qi et al., 2012; Wei et al., 2012; Zhao et al., 2013). Population fragmentation and demographic isolation were also shown to be significant in affecting dispersal and gene flow in wild pandas (Hu et al., 2010; Zhu et al., 2010; Wei et al., 2012). Nevertheless, direct information on contemporary selection processes involving the interaction of individuals with their environment can only be provided by genes under selection (Sommer, 2005). Though previous studies have shown evidence of low diversity but positive selection at MHC DRB and DQA genes in the giant pandas (Wan et al., 2006; Zhu et al., 2007; Chen et al., 2010), possible associations of these MHC variations with parasite infection were not explored to date.
Figure 1.
Distribution of wild pandas in six mountains in China.
Of the diseases known to affect wild pandas, parasitosis (especially Baylisascaris shroederi) causes the greatest morbidity and mortality in the field (Zhang and Wei, 2006), and physical examination of rescued pandas also revealed that most giant pandas are often heavily parasitized by B. shroederi (Hu, 2001). These parasites usually infest the intestines of giant pandas, which can cause intestinal obstruction, inflammation and death (Zhang and Wei, 2006). Due to giant pandas' behaviour of defecating while feeding, many food or water resources may be possibly polluted by eggs of B. shroederi (Zhang and Wei, 2006). These eggs can enter dormancy or diapause during adverse environmental conditions and develop into free-living infective stages rapidly during suitable conditions, specifically infecting other giant pandas (or cubs during reproductive seasons), or more frequently, infect the current host without an intermediate host (Wu et al., 1985, 1987). Given the profound effect parasites may have on their hosts and their potential to shape genetic variation, assessment of MHC polymorphism of endangered pandas, incorporating such parasitosis caused by B. shroederi, is in great need.
The aims of the present study were (1) to explore associations between MHC genetic diversity and parasite load in natural populations of giant pandas and (2) to determine which selective mechanisms may be acting on the MHC in the presence of parasites in this species.
Materials and methods
Samples
DNA extracts were used from the previous study that focused on parasite infection based on the individual identification by using giant panda-specific microsatellite genotyping (Zhan et al., 2006) in wild pandas across the six mountain habitats in China (see Zhang et al., 2011 for details). A total of 224 faecal samples were collected and 126 individuals were determined (Zhang et al., 2011). A subsample of 87 individuals could be amplified successfully at these MHC loci and was used in this study (Table 1).
Table 1. Sample sizes (N), number of alleles (L), allelic richness (Rs) and expected heterozygosities (H exp) for three MHC loci in five populations of the giant panda.
| Mountain | N |
DRB1 |
DQA1 |
DQA2 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | Rs | Hexp | Hobs | L | Rs | Hexp | Hobs | L | Rs | Hexp | Hobs | ||
| Qinling (QinL) | 32 | 3 | 2.219 | 0.514 | 0.500 | 5 | 2.656 | 0.535 | 0.375 | 4 | 3.046 | 0.392* | 0.156 |
| Minshan (MS) | 14 | 6 | 5.047 | 0.712* | 0.286 | 5 | 3.789 | 0.444 | 0.385 | 2 | 1.967 | 0.260* | 0.000 |
| Qionglai (QioL) | 25 | 5 | 4.069 | 0.702* | 0.320 | 6 | 4.906 | 0.687* | 0.222 | 5 | 3.655 | 0.613* | 0.333 |
| Liangshan (LS) | 8 | 4 | 4.000 | 0.648* | 0.375 | 3 | 3.000 | 0.648* | 0.500 | 1.992 | 0.219 | 0.000 | |
| Xiaoxiangling (XXL) | 8 | 4 | 3.876 | 0.451* | 0.111 | 3 | 3.000 | 0.663* | 0.714 | 2 | 0.245 | 0.000 | |
| Total | 87 (26 ♀ 57 ♂)a | ||||||||||||
Abbreviation: MHC, major histocompatibility complex.
With four gender unknown. *stands for significantly (P<0.05) lower levels of observed heterozygosity than expected.
Parasitic screening
For parasitological study, 30 g of faeces (including the outer layer) was crushed and stored in vials of 10% neutral-buffered formalin when sampling. The faecal samples were screened for helminth eggs by using a modification of the McMaster sedimentation and floatation techniques (Zhang et al., 2011). Samples were transferred into a 500-ml bottle containing 250 ml distilled water, and then the mixture was filtered using a 750-mm aperture wire mesh followed by a 370-mm aperture wire mesh to remove bamboo stem fragments left in faeces. The filtrate supernatant was siphoned off after 30 min standing and the faecal sediment was concentrated to 30 ml. Three 1-ml samples were examined microscopically using a modified McMaster slide. If no eggs or larvae were detected after the third 1-ml sample, then we used flotation techniques to examine the remainder of the sample following (Aguirre et al. 1998). Helminthes were assigned to morphotypes according to size and morphological characteristics (Zhang et al., 2011).
Two measures of parasite load were used: (1) prevalence (P) of parasites was determined by dividing the number of positive individuals by the total number of individuals examined; and (2) intensity (I) was defined as the number of conspecific parasites per gram (FEC, faecal egg count), with uninfected samples being excluded. Calculations of prevalence and intensity were performed using Quantitative Parasitology 3.0 (Rózsa et al., 2000), with 95% confidence intervals estimated by bootstrap.
MHC genotyping
To design MHC class II DRB-specific primers, extensive BLAST searches were carried out in the giant panda genome database. The DNA sequence of the second exon of dog DRB gene (NM_001014768) was used to perform BLAST (blastn) to panda genome sequence downloaded from EnSembl (http://www.ensembl.org/info/data/ftp/index.html). Five hits were obtained in five different scaffolds. We ignored one scaffold because its length is only 164 bp and cannot give any more gene information. Of the other four hits, three have annotation items in panda gene annotation gtf file, which were named DRB1, 2 and 4. For the last unannotated hit, the approach of manual check combined with de novo gene prediction was applied to obtain one five-exon gene sequence, which was named DRB3. As a result, four similar genes were found, which were named sequentially in the order of their orthologues in the DLA and HLA as DRB1, DRB2, DRB3 and DRB4. We designed DRB1-specific primers (DRB1F: 5′-AAGGGCGAGTGCTACTTCAC-3′ DRB1R: 5′-CCGGATGAGTCTGTCTCACA-3′) with upstream end located in the exon2 and downstream end anchored in the adjacent intron of DRB1 exon2, which led to successful amplification of partial exon2 and its downstream partial intron (288 bp in total). After alignment with DRB1 exon2 (256 bp) of the giant panda genome, intron parts (60 bp) were removed in the following analyses. All samples were also genotyped at the MHC class II DQA locus using previously published primer pairs DQAup and DQAdown (Zhu et al., 2007).
PCRs were performed in a total volume of 30 μl containing 0.75 U HotStarTaq polymerase (Qiagen, Shanghai, China), 1 × PCR buffer, 60 μM of each dNTP, 10 pM of each primer, 1 × BSA (Promega, Beijing, China) and ∼10 ng of genomic DNA. Each sample was amplified twice independently for the loci studied. Purified PCR fragments were cloned using the pMD18-T vector system (Takara, Dalian, China), and then transformed into Escherichia coli competent cells (Tiangen, Beijing, China). At least six positive clones per individual were selected at random for sequencing analysis. Sequencing reactions were carried out at Beijing Genomics Institute (BGI) using AB 3730xl DNA Analyzer (Applied Biosystem, Foster City, CA, USA). Since up to four unique sequences at DQA gene could be detected from certain individuals, six more positive clones at DQA per individual were added for sequencing. Sequences from at least three identical clones were defined as an allele (Kennedy et al., 2002). Direct sequencing of uncloned PCR products was also used to check for agreement with polymorphic sites of cloned sequences. Those alleles found only once were verified by extra typing of that individual.
To validate whether the assayed MHC loci are expressed, we obtained RNA from the spleen of a captive individual which died naturally. Immediately after excision, the tissues were flash frozen in liquid nitrogen, and then stored at −80 °C. RNA was extracted from about 10 mg of the homogenized tissue sample with the RNeasy kit (Qiagen) including the DNase treatment step (Promega). cDNA was obtained using the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, Pittsburgh, PA, USA) in 20 μl reactions containing 6 μl template RNA, 1 μl Oligo (dT)18 primer (0.5 μg μl−1), 4 μl reaction buffer (5 × ), 1 μl RiboLock RNase inhibitor (20 U μl−1), 2 μl dNTP (10 mM), 1 μl RevertAid H Minus M-MuLV reverse Transcriptase (200 U μl−1) and 6 μl of RNase-free water. The reaction was incubated at 42 °C for 60 min. Additional primer combinations located in the DRB1 exon2 and the second exon regions of the two DQA loci were designed (DRBF2: 5′-TTCACCAACGGGACGGAG-3′ DRBR2: 5′-GCTGCTGCTCCATGAAGTC-3′ DQAF1: 5′-GCTTCCGATGGCATAAATGT-3′ DQAR1: 5′-GCAGCGGTATAGTTGGAACG-3′) and used in the following analysis of DRB and DQA expression, respectively. PCR products from cDNA were cloned and sequenced. At least 30 positive clones for each primer pair were sequenced.
Alleles at each locus are denominated according to the nomenclature suggested by Klein et al. (1990).
Data analyses
MHC sequences were aligned using MEGA 5 (Tamura et al., 2011) and sequence variations were characterized using DnaSP v5 (Librado and Rozas, 2009). Average pairwise nucleotide distances (Kimura 2-parameter model) and Poisson-corrected amino-acid distances were computed in MEGA 5 (Tamura et al., 2011) using the Nei-Gojobori method with the Jukes-Cantor correction for multiple substitutions (Nei and Gojobori, 1986). Standard errors of estimates were obtained through 1000 bootstrap replications.
Positive selection evidenced by a significantly higher number of non-synonymous substitutions per non-synonymous codon site (dN) relative to synonymous substitutions per synonymous codon site (dS) was tested. A Z-test implemented in MEGA 5 (Tamura et al., 2011) was performed to compare dN with dS at all codons, antigen binding sites (ABS) and non-ABS. We also tested for effects of ABS and non-ABS on dN/dS in a generalized linear model (GLM) including locus as a factor. Here, the pairwise rather than the overall dN/dS ratios were used in the model. The locations of putative ABS were inferred according to Reche and Reinherz (2003).
Phylogenetic relationships among DRB and DQA sequences were reconstructed using HLA alleles as outgroups, respectively. DRB and DQA sequences (AY895155-AY895161; EF554075-EF4080; GQ496165-GQ496181) from previous papers (Wan et al., 2006; Zhu et al., 2007; Chen et al., 2010) were included in phylogenetic analyses.
The best-fitting models of sequence evolution were chosen on the basis of the AIC criterion using jModelTest 0.1.1 (Posada, 2008). The best-fitting model at DQA loci was Kimura 2-parameter plus Gamma (K80+G). Phylogenetic relationships were estimated using maximum likelihood and neighbor joining methods, respectively. Maximum likelihood analysis was performed using PHYML 3.0 (Guindon and Gascuel, 2003), and neighbor joining tree was constructed in Phylip 3.69 (Felsenstein, 2009). The robustness of each obtained tree topology was tested with 100 bootstrap replicates.
Estimates of allelic richness (Rs) per locus and population were calculated with FSTAT 2.9.3.2 (Goudet, 2001). Rs is a measure of the number of alleles independent of sample size, hence making it appropriate to be compared between different population sizes. Allele frequencies, gene diversity and observed and expected heterozygosity were estimated with the program GENETIX 4.05 (Belkhir et al., 2004). Differences in allele frequencies between mountain populations and deviation from Hardy–Weinberg equilibrium were assessed using GENEPOP 4.0 (Rousset, 2008).
Effects of sampling area, host gender and MHC heterozygosity were fitted using GLM. Compensating for the overdispersion, prevalence was defined as a binary response (infected and uninfected) in each model using a quasibinomial error structure and a logit link function, and infection intensity was analyzed by quasi-Poisson error rather than Poisson error to refit the models (Kloch et al., 2010). On the basis of deletion tests, factors were retained in the model only if it caused a significant increase in deviance when it was removed from the current model. The change in deviance was assessed by F-test, respectively, with degrees of freedom equal to the number of factors (or levels of a factor) dropped (Crawley, 2007). To analyze the effect of individual alleles, they were included singly in the GLM with the other factors. We preselected the high frequency alleles that occurred in >5% of individuals as predictors. All analyses were performed in R 2.11.1 (R Development Core Team, 2011). The sequential Bonferroni procedure was applied where appropriate to keep the type I error levels at α⩽0.05 (Rice, 1989).
Results
Parasite load
We identified five distinct types of helminth eggs: three nematode morphotypes, one trematode morphotype and 1 strongy. Among the examined individuals, 55.17% (48/87) exhibited infection with 1–2 helminthes, and the remaining 44.83% did not show any helminth infection. The majority of infected pandas (95.3%) carried B. shroederi, whereas the other four helminthes occurred in <10% of the pandas.
MHC variability
Seven unique DRB1 alleles were identified with sequences deposited in GenBank (Aime-DRB1*08-14, accession numbers: JF518949–JF518955). Among the DRB1 alleles, 29 of 228 nucleotides were variable. The alleles differed between 1 and 22 (27.4%) nucleotide positions, and pairwise comparisons of amino-acid sequences revealed up to 12 (42.9%) positions were variable (Figure 2). Both nucleotide and amino-acid distances were higher in the putative ABS than in non-ABS within the DRB1 locus (Table 2). At the DRB1 locus, dN was significantly higher than dS at ABS; however, no such significances were detected within the non-ABS or all nucleotides (Table 3).
Figure 2.
Amino-acid alignment of 7 DRB1 alleles (a) and 15 DQA alleles (b). Sign ‘*' above the sequences represents putative ABS according to Reche and Reinherz (2003).
Table 2. The average nucleotide and amino-acid distances among the giant panda alleles from Aime-DRB1, Aime-DQA1, Aime-DQA2 and all Aime-DQA.
|
K2P nucleotide distance |
Poisson-corrected amino-acid distance |
||||
|---|---|---|---|---|---|
| All sites | ABS | Non-ABS | All sites | ABS | Non-ABS |
| Aime-DRB1 | |||||
| 5.7 (1.0) | 20.7 (4.9) | 2.1 (0.7) | 10.5 (2.5) | 34.4 (9.5) | 4.8 (1.9) |
| Aime-DQA1 | |||||
| 2.2 (0.7) | 7.5 (2.9) | 0.6 (0.4) | 4.0 (1.6) | 16.5 (7.4) | 0.5 (0.5) |
| Aime-DQA2 | |||||
| 2.3 (0.7) | 9.0 (3.1) | 0.4 (0.3) | 4.5 (1.7) | 19.2 (7.6) | 0.4 (0.4) |
| All Aime-DQA | |||||
| 3.4 (0.8) | 12.2 (3.6) | 1.0 (0.5) | 6.4 (2.1) | 25.3 (9.7) | 1.4 (1.0) |
Nucleotide distances are corrected for multiple substitutions with the Kimura 2-parameter model (K2P), and amino-acid distances are corrected using expectations from the Poisson distribution. ABS, putative antigen binding sites as determined in Reche and Reinherz (2003). Distances are given as percentages per site with standard errors (in parenthesis) based on 1000 bootstrap replicates.
Table 3. The estimation of non-synonymous (d N) and synonymous (d S) substitutions for antigen binding sites (ABS) and non-antigen binding sites (non-ABS) at MHC loci, with standard errors obtained through 1000 bootstrap replicates.
| Locus | Position | N | dN | dS | dN/dS | P |
|---|---|---|---|---|---|---|
| Aime-DRB1 | ABS | 17 | 0.252±0.079 | 0.085±0.054 | 2.965 | 0.043 |
| Non-ABS | 59 | 0.021±0.008 | 0.020±0.014 | 1.050 | 0.939 | |
| All | 76 | 0.065±0.016 | 0.034±0.016 | 1.912 | 0.137 | |
| Aime-DQA1 | ABS | 18 | 0.081±0.037 | 0.054±0.079 | 1.500 | 0.755 |
| Non-ABS | 58 | 0.002±0.002 | 0.018±0.014 | 0.111 | 0.265 | |
| All | 76 | 0.021±0.009 | 0.026±0.015 | 0.808 | 0.931 | |
| Aime-DQA2 | ABS | 18 | 0.099±0.042 | 0.048±0.068 | 2.063 | 0.551 |
| Non-ABS | 58 | 0.002±0.002 | 0.010±0.011 | 0.200 | 0.420 | |
| All | 76 | 0.025±0.010 | 0.017±0.012 | 1.471 | 0.644 | |
| Aime-DQA (DQA1 and DQA2) | ABS | 18 | 0.124±0.047 | 0.111±0.098 | 1.117 | 0.909 |
| Non-ABS | 58 | 0.002±0.002 | 0.010±0.010 | 0.200 | 0.409 | |
| All | 76 | 0.033±0.012 | 0.038±0.019 | 0.868 | 0.809 |
Abbreviation: MHC, major histocompatibility complex.
N is the number of codon, P is the probability that dN and dS are different by Z-test. Significant P-values (P<0.05) are typed in bold.
ABS, putative antigen binding sites as determined in Reche and Reinherz (2003).
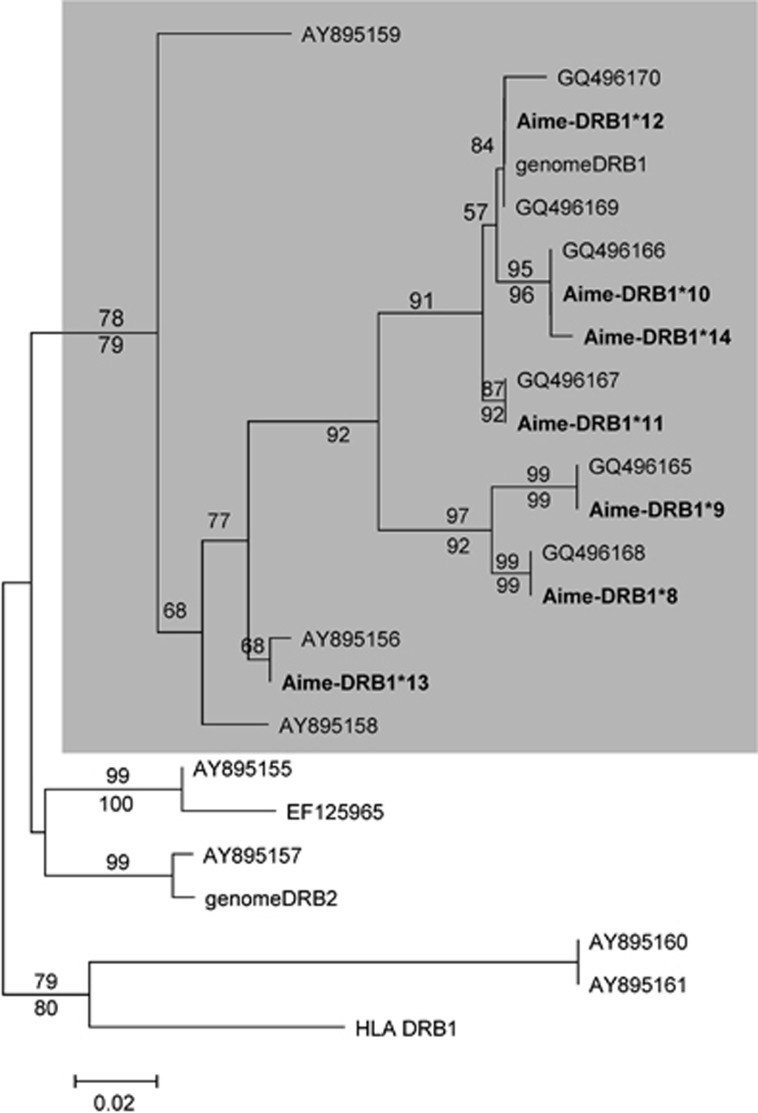
Phylogenetic analysis distinguished two well-supported clusters (Figure 3), and seven DRB alleles identified in this study were clustered together with panda genome DRB1, suggesting that these alleles are from one single locus.
Figure 3.
Phylogenetic relationships of the giant panda MHC class II DRB alleles. Kimura 2-parameter model was used with 100 bootstrap replicates. Bootstrap values above 50% from the neighbor joining analyses and above 70% from the maximum likelihood analyses are shown below and above the branches, respectively. The following sequence was included as an outgroup: Homo sapiens (HLA-DRB1, NM_002124). GenomeDRB1 and genomeDRB2 are from the giant panda genomic data. AY895155–AY895161, GQ496165–GQ496170 and EF125965 are from Wan et al. (2006), Chen et al. (2010) and Zeng et al. (2007), respectively.
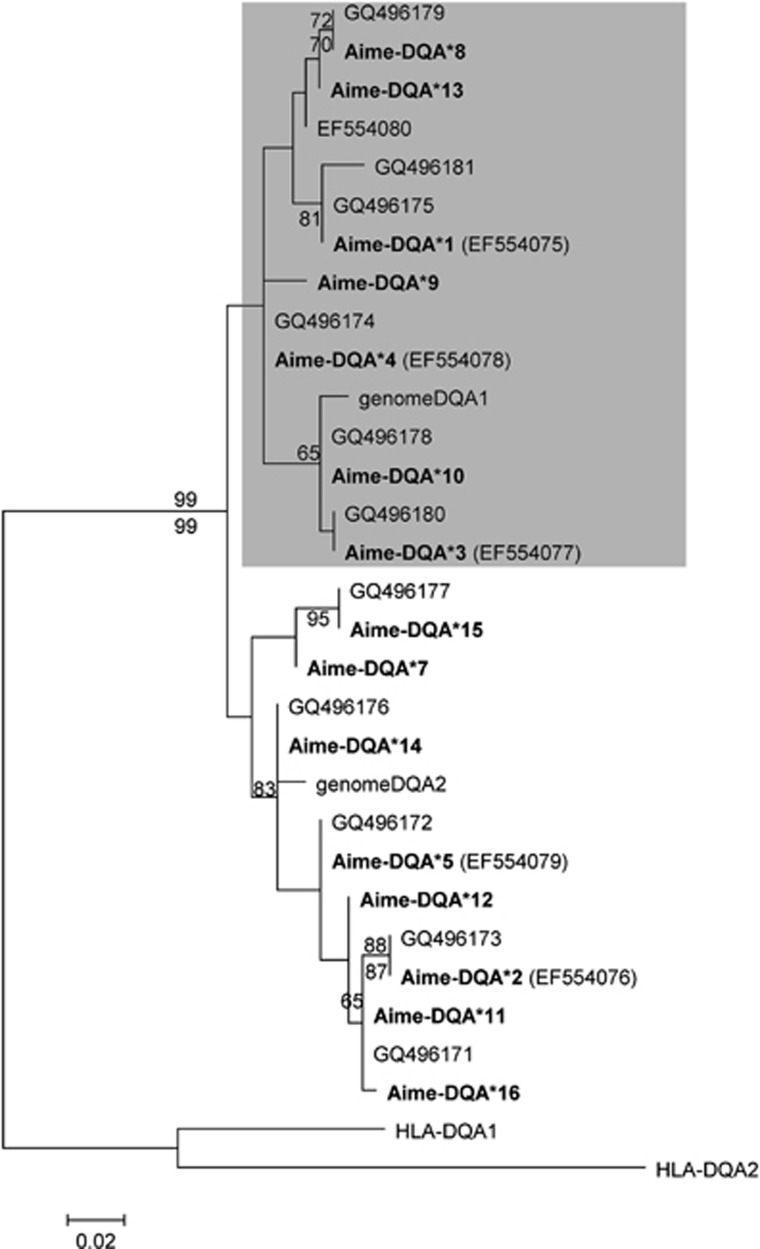
Fifteen DQA sequences (Aime-DQA*1-5 and Aime-DQA*7-16) were identified with new ones deposited in GenBank (Aime-DQA*7-16, accession numbers: JF518939–JF518948). However, up to four unique sequences could be detected from certain individuals, implying the existence of at least two loci in the DQA gene. The existence of two DQA loci was further verified using BLAST search with Aime-DQA*7 (JF518939) as a query in the giant panda genome sequence. Phylogenetic analysis distinguished 15 DQA alleles to two well-supported clusters (Figure 4). Within each cluster, no more than two different sequences were detected in a single individual, which indicates that alleles from different clusters belong to two different loci. The average nucleotide and amino-acid distances at putative ABS were much higher than those at non-ABS, no matter if they were from the DQA1 locus, DQA2 or all DQA alleles (Table 2). Nevertheless, for codons in the putative ABS, the dN/dS ratio is greater than one, suggesting positive selection, though the difference was not statistically significant within DQA1, DQA2 or overall DQA fragments (Table 3). Positive selection could not be detected for non-ABS codons or for all the codons (Table 3).
Figure 4.
Phylogenetic relationships of the giant panda MHC class II DQA alleles. Kimura 2-parameter model was used with bootstrap supported based on 100 bootstrap replicates. Bootstrap values above 50% from the neighbor joining analyses and above 70% from the maximum likelihood analyses are shown below and above the branches, respectively. GenomeDQA1 and genomeDQA2 are from the genomic data. The following sequences were included as outgroups: Homo sapiens (HLA-DQA1, NM_002122) and Homo sapiens (HLA-DQA2, NM_020056).
To examine for common effects of ABS/non-ABS on dN/dS ratio including locus as a predictor, pairwise dN/dS ratios were calculated for sites (ABS, non-ABS and all sites) at the respective DRB1, DQA1 and DQA2 locus. It revealed that dN/dS ratios were significantly associated with sites (χ2=85.365, d.f.=2, P=0.000) and loci (χ2=8.680, d.f.=2, P=0.006). dN/dS ratios at ABS were higher than those at non-ABS and all sites (P=0.000), while no significant differences could be observed for dN/dS between non-ABS and all sites (P=0.030, insignificant after Bonferroni correction). dN/dS ratios at the DRB1 locus were significantly higher than at both DQA loci (P=0.002), with no differences between DQA1 and DQA2 (P=0.035, insignificant after Bonferroni correction).
Analysis of PCR products obtained from spleen mRNA showed that DRB1 and both of DQA loci were expressed.
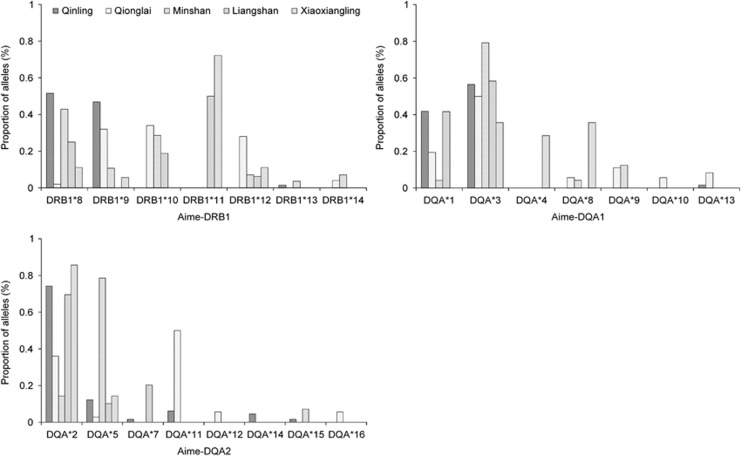
In general, some alleles (for example, Aime-DRB1*8, Aime-DQA*2, Aime-DQA*3 and Aime-DQA*5) were dominant and could be detected from all mountains, whereas others were observed at very low frequencies and only existed in specific populations, for example, Aime-DQA*10, Aime-DQA*12 and Aime-DQA*16 (Figure 5). The number of alleles among populations was similar, ranging from 3 to 6 at DRB1, from 5 to 6 at DQA1 and from 2 to 5 at DQA2. The number of alleles among populations did not increase in proportion to sample size, as was evident from estimate of Rs. For instance, the highest Rs value for DRB1 was recorded for MS, which contained 14 individuals, whereas the Rs values of QinL and QioL with 32 and 25 individuals, respectively, were lower (Table 1).
Figure 5.
DRB1, DQA1 and DQA2 allele frequencies in Qinling, Qionglai, Minshan, Liangshan and Xiaoxiangling mountains.
The estimation of heterozygosity (Table 1) showed that all populations exhibited lower levels of observed heterozygosity than expected.
Associations between MHC alleles and parasite load
Five Aime-DRB1 (Aime-DRB1*8∼Aime-DRB1*12), three Aime-DQA1 (Aime-DQA*1, 3 and 8) and three Aime-DQA2 alleles (Aime-DQA*2, 5 and 11) were dominant, associated with a prevalence of >5%, whereas the remaining were rarely represented in the samples and thus were not included in the following association analyses.
Mean intensity of parasite varied among mountains (P=0.014, insignificant after Bonferroni correction), and a marginally significant difference in the prevalence could be detected (P=0.082, Table 4). No significant effect of the host gender on the parasite prevalence or intensity was identified.
Table 4. Results of generalized linear models (GLMs) for examining the associations of MHC class II alleles on parasite prevalence and infection intensity.
|
Prevalence |
Intensity |
|||||
|---|---|---|---|---|---|---|
| Δdeviance | d.f. | P-value | Δdeviance | d.f. | P-value | |
| Mountain | 8.27 | 4 | 0.082 | 1400.6 | 4 | 0.014 |
| Host gender | 0.006 | 1 | 0.940 | 153.5 | 1 | 0.230 |
| DRB1 | ||||||
| Aime-DRB1*8 | 0.207 | 1 | 0.649 | 57.108 | 1 | 0.460 |
| Aime-DRB1*9 | 0.517 | 1 | 0.472 | 73.859 | 1 | 0.398 |
| Aime-DRB1*10 | 3.268 | 1 | 0.071 | 1074.6 | 1 | <0.001 |
| Aime-DRB1*11 | 0.810 | 1 | 0.368 | 1.479 | 1 | 0.906 |
| Aime-DRB1*12 | 0.455 | 1 | 0.500 | 478.74 | 1 | 0.020 |
| Heterozygous | 0.721 | 1 | 0.471 | 240.670 | 1 | 0.144 |
| DQA1 | ||||||
| Aime-DQA*1 | 0.075 | 1 | 0.784 | 405.04 | 1 | 0.052 |
| Aime-DQA*3 | 1.988 | 1 | 0.159 | 663.51 | 1 | 0.010 |
| Aime-DQA*8 | 0.035 | 1 | 0.852 | 16.883 | 1 | 0.711 |
| Heterozygous | 2.409 | 1 | 0.121 | 366.55 | 1 | 0.064 |
| DQA2 | ||||||
| Aime-DQA*2 | 0.254 | 1 | 0.615 | 222.02 | 1 | 0.116 |
| Aime-DQA*5 | 2.941 | 1 | 0.086 | 96.621 | 1 | 0.315 |
| Aime-DQA*11 | 2.008 | 1 | 0.157 | 273.32 | 1 | 0.087 |
| Heterozygous | 4.625 | 1 | 0.045 | 262.73 | 1 | 0.085 |
Abbreviation: MHC, major histocompatibility complex.
Terms significant after the Bonferroni correction for the number of tested alleles are given in bold.
Effects of sampling area, host gender and heterozygous at the respective locus on parasite prevalence and intensity were fitted using GLMs, and then individual alleles that occurred in >5% were included singly in the GLMs with the other factors.
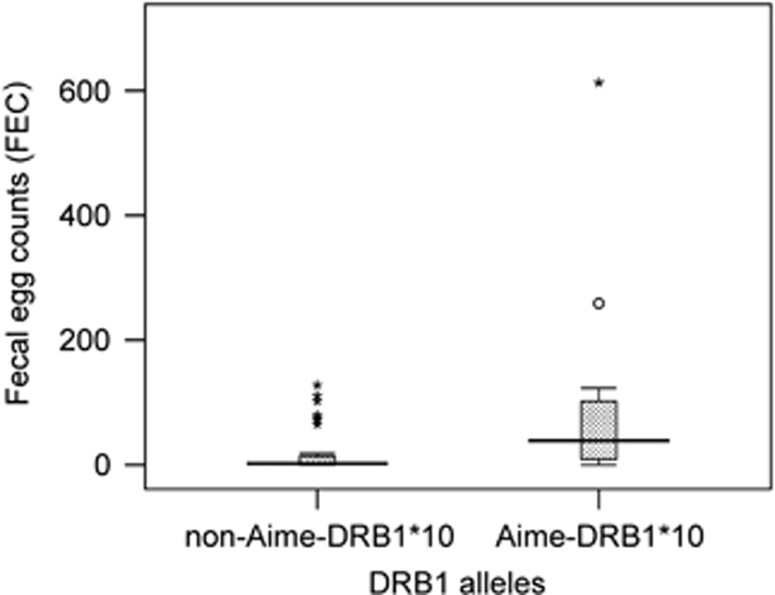
Associations were observed between specific alleles and the parasite load. The allele Aime-DRB1*10 occurred more frequently in infected individuals with marginal significance (P=0.071, Bonferroni not significant). However, the FEC values also differed with the allele configuration. Individuals carrying Aime-DRB1*10 exposed significantly higher FECs than individuals without the allele (Figure 6, P<0.001, Bonferroni significant). However, analyses with genotype or heterozygosity as predictors failed to show significant results associated with both prevalence and intensity at any MHC locus (Table 4).
Figure 6.
FEC values of individuals with and without Aime-DRB1*10 allele.
Discussion
Parasite infection in wild pandas
Giant pandas are suffering from B. shroederi infection and most of those pandas found dead in the wild have been heavily infected with B. shroederi (Hu, 2001). In this study, we documented that most of infected pandas were only parasitized by B. shroederi. Generally, a species that lives in social groups, as dense populations, or on a diverse diet may be especially vulnerable to parasite infection (Altizer et al., 2003; Petric et al., 2011). Pandas are known to be solitary in the wild and almost exclusively feed on bamboo, which may account for the limited species diversity of parasite fauna on the giant panda. On the other hand, it has been suggested that population density generally correlates with parasite abundance across species (Altizer et al., 2007). Wild pandas have been demonstrated to suffer from demographic decreases due to deforestation and habitat fragmentation for hundreds of years (Zhu et al., 2010), which might also lead to scarce parasite species observed in the giant panda.
MHC variability in the giant panda
It has been indicated that genetic diversity is much lower in endangered species, which is also the case with MHC genes (Radwan et al., 2010). In this study and previous studies (Wan et al., 2006; Zhu et al., 2007), a relatively low number of MHC class II alleles were observed in giant pandas. However, the sequences of alleles were divergent, especially at the ABS. Species with limited or absence of MHC polymorphism might exhibit higher levels of divergence among MHC alleles. The Australian bush rat (Rattus fuscipes greyii) has only one to two alleles in RT1.Ba (DQA homologue) in thirteen island populations but with high divergence (Seddon and Baverstock, 1999). An extreme variance of three MHC alleles in the Arabian oryx (Oryx leucoryx) was identified (Hedrick et al., 2000). Although limited DRB1 alleles were found in wild pandas, there was still a clear indication for selection processes acting on the DRB1 locus. Analyses of variation among the seven Aime-DRB1 sequences revealed compelling evidence for selection favouring functional diversity. Variation in terms of amino-acid differences occurred disproportionately in the ABS compared with the non-ABS, and the relative rate of dN was significantly higher than that of dS for ABS but not for non-ABS codons, which is characteristic of balancing selection acting to maintain functional polymorphism and was reported in a number of MHC studies (Piertney and Oliver, 2006). However, positive selection tests for DQA indicated that no strong positive selection exists in wild pandas, at least in the analyzed fragments. Consistent with positive selection at DQA can only be implied with a high rate of non-synonymous substitutions in the ABS of the gene. MHC class II genes are predominantly involved in monitoring the extracellular environment by presenting peptides mainly derived from parasites to the T cells (Sommer, 2005), which leads to the speculation that DRB1 may have a more important role than DQA loci in dealing with parasites infections of giant pandas.
Some DRB1 sequences found in this study are identical to DRB3 alleles in Chen et al. (2010), which indicated that DRB1 in our study and DRB3 in Chen et al. (2010) may be from the same locus. However, as shown in Figure 3, some DRB alleles found in Wan et al. (2006) are similar to DRB2 but divergent from DRB1.
DQA1 and DQA2 loci could be detected simultaneously using DQAup and DQAdown, inconsistent with the former study (Zhu et al., 2007) in which only one single DQA locus (Aime-DQA*1-6, accession numbers: EF554075–EF554080) was described using the same primer set. The existence of two DQA loci in the giant panda was evidenced by PCR result where up to four sequences could be amplified from many individuals and phylogenetic analysis, and was also supported by the genomic data and the work based on a bacterial artificial chromosome clone and sequencing (Chen et al., 2010). Incomplete knowledge of the number of loci that are actually being simultaneously PCR amplified may lead to problematic population genetic parameter estimation directly, or dubious observation of associations between MHC and parasite load (Piertney and Oliver, 2006). Every effort should be made to clarify that single expressed products are used for MHC genes, which are composed of different numbers of locus, high variability of alleles within a locus, and the similarity of alleles across loci, to ensure that the subsequent analysis based on genomic DNA is appropriate (Piertney, 2003; Piertney and Oliver, 2006).
MHC loci had significantly lower than expected heterozygosity in wild pandas for many studied populations. Multiple explanations for heterozygote deficiency have been proposed, among which the three most common explanations are the presence of null alleles, discontinuities in gene flow, or inbreeding and population decline (Allendorf and Luikart, 2007; Hagell et al., 2013). In our study, heterozygote deficiency was observed at most localities and all studied MHC loci instead of population or locus specific, which suggests that discontinuities in gene flow among isolated populations may explain the loss of heterozygotes in wild pandas. We did not consider null alleles to be an issue in this study, as all loci have low diversity or violate Hardy–Weinberg equilibrium and most sampling sites are out of Hardy–Weinberg equilibrium. It is unlikely that null alleles would persist in multiple loci. Furthermore, DRB1 and DQA loci were characterized based on the knowledge of gene structure from panda genome data and gene expression assays, while the utilization of the intron sequences flanking the exon 2 to obtain DRB1 genes could also assist the specific amplification of MHC genes, as suggested in Canal et al. (2010).
Association between MHC and parasite load in free-living pandas
We found that Aime-DRB1*10 was significantly associated with susceptibility to parasites. The allele Aime-DRB1*10 occurred more frequently in infected individuals and in individuals with high FEC values. The overall frequency of Aime-DRB1*10 was relatively high (15.9%) and varied significantly among mountain habitats. Notably, Aime-DRB1*10 was not found in Qinling and Xiaoxiangling mountains. Consistent with this finding, the mean intensity of B. shroederi in Qinling mountains was relatively lower than those in Minshan, Qionglai and Liangshan mountains, though no significant variation in B. shroederi prevalence among the five mountains could be observed (Zhang et al., 2011). The Xiaoxiangling population is the most isolated and the smallest population of the giant panda (State Forestry Administration, 2006), which may explain the absence of Aime-DRB1*10 therein.
We did not find any alleles related to the resistance to parasite infection in wild pandas. One potential reason is that the sample size may limit the power needed to detect significant associations in the analyses. Statistically, attempting to link MHC alleles to parasite resistance in natural populations with adequate statistical power is difficult to harness due to relative low sample sizes combined with high numbers of alleles at low frequencies (Hill, 1998; Piertney and Oliver, 2006). Another possibility is that alleles with capability of resistance to antigen produced by B. shroederi might be lost in the giant panda. For threatened species, depletion of variation at MHC genes is thought to compromise the ability of populations to respond to pathogen assault (Hedrick, 2001; Radwan et al., 2010). It is possible that MHC variants capable of presenting antigens of a given pathogen may no longer be present in the threatened population, making endangered species more susceptible to extinction (Radwan et al., 2010). In an examination of variation at DRB locus in the European bison (Bison bonasus), which has undergone an extreme bottleneck, no significant association between DRB alleles or genotypes and posthitis could be found, which leads to speculations that conferring resistance to posthitis may have been lost in the European bison (Radwan et al., 2007). Inference made here is from the spatial variation in the relationship between resistance and MHC. It would be better to have analyses of spatial-temporal changes in parasite infection and MHC variation combined with utilization of experimental challenges in the future. However, it should be noted that the possible reason of the findings that heterozygosity was uncorrelated with parasite infection in wild pandas may be related to the effect of deviation from Hardy–Weinberg equilibrium due to significant heterozygote deficiency observed across all studied loci.
Our finding of association between MHC alleles and parasite load in wild pandas would support the negative frequency-dependent selection hypothesis. The ‘susceptible' allele Aime-DRB1*10 was found to be common in most of the studied populations but no ‘resistant' alleles could be detected. Moreover, heterozygosity was found to be uncorrelated with parasite infection in wild pandas. A project of translocating pandas from other areas or reintroduction from the captivity has been implemented (Zhu et al., 2010) and will continue to help wild pandas recover, especially for those small or isolated populations which are experiencing drastic reductions and habitat loss. Our findings implicate that giant pandas not carrying Aime-DRB1*10 should be good candidates for future translocation and reintroduction.
Acknowledgments
We thank Professor Michael Bruford for comments on an earlier version of the manuscript. We appreciate the help of Dr Shixia Xu from Nanjing Normal University in data analyses. We thank BGI Shenzhen for providing gene de novo annotation results of panda genome. We are very grateful to the Sichuan, Gansu, Shaanxi Forestry Departments and Foping, Changqing, Wanglang, Tangjiahe, Dafengding, Fengtongzhai, Daxiangling, Yele, Liziping and Baishuijiang nature reserves for help with sampling. This research was supported by National Natural Science Foundation of China (grant 30970428; grant 30770319) and Key Program of Knowledge Innovation Program of Chinese Academy of Sciences (KSCX2-EW-Z-4).
The authors declare no conflict of interest.
References
- Aguirre DH, Viñabal AE, Gaido AB. (1998). Comparación de tres técnicas coprológicas para el diagnóstico de Fasciola hepática en rumiantes. Vet Arg 15: 421–4427. [Google Scholar]
- Allendorf FW, Luikart G. (2007) Conservation and the Genetics of Populations. Blackwell: Malden. [Google Scholar]
- Altizer S, Nunn CL, Lindenfors P. (2007). Do threatened hosts have fewer parasites? A comparative study in primates. J Anim Ecol 76: 304–314. [DOI] [PubMed] [Google Scholar]
- Altizer S, Nunn CL, Thrall PH, Gittleman JL, Antonovics J, Cunningham AA et al. (2003). Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu Rev Ecol Evol Syst 34: 517–547. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. (2004) GENETIX 4.05, Logiciel Sous Windows TM Pour la Génétique Des Populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II: Montpellier, France.
- Canal D, Alcaide M, Anmarkrud JA, Potti J. (2010). Towards the simplification of MHC typing protocols: targeting classical MHC class II genes in a passerine, the pied flycatcher Ficedula hypoleuca. BMC Res Notes 3: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Zhang YY, Zhang HM, Ge YF, Wan QH, Fang SG. (2010). Natural selection coupled with intragenic recombination shapes diversity patterns in the major histocompatibility complex class II genes of the giant panda. J Exp Zool B Mol Dev Evol 314: 208–223. [DOI] [PubMed] [Google Scholar]
- Crawley M. (2007) The R Book. Wiley and Sons: New Jersey. [Google Scholar]
- Deter J, Bryja J, Chaval Y, Galan M, Henttonen H, Laakkonen J et al. (2008). Association between the DQA MHC class II gene and Puumala virus infection in Myodes glareolus, the bank vole. Infect Genet Evol 8: 450–458. [DOI] [PubMed] [Google Scholar]
- Doherty PC, Zinkernagel RM. (1975). Enhanced immunological surveillance in mice heterozygous at H-2 gene complex. Nature 256: 50–52. [DOI] [PubMed] [Google Scholar]
- Evans ML, Neff BD. (2009). Major histocompatibility complex heterozygote advantage and widespread bacterial infections in populations of Chinook salmon (Oncorhynchus tshawytscha). Mol Ecol 18: 4716–4729. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (2009) PHYLIP (Phylogeny Inference Package), v3.69. Distributed by the Author. Department of Genome Sciences, University of Washington: Seattle.
- Goudet J. (2001) FSTAT, a program to estimate and test gene diversities and fixation Indices (Version 2.9.3). Available from http://www2.unil.ch/popgen/softwares/fstat.htm. Updated from: Goudet J, 1995. FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86: 485–486.
- Guindon S, Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Hagell S, Whipple AV, Chambers CL. (2013). Population genetic patterns among social groups of the endangered Central American spider monkey (Ateles geoffroyi) in a human-dominated landscape. Ecol Evol 3: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. (2001). Conservation genetics: where are we now? Trends Ecol Evol 16: 629–636. [Google Scholar]
- Hedrick PW, Parker KM, Gutierrez-Espeleta GA, Rattink A, Lievers K. (2000). Major histocompatibility complex variation in the Arabian oryx. Evolution 54: 2145–2151. [DOI] [PubMed] [Google Scholar]
- Hill AVS. (1998). The immunogenetics of human infectious diseases. Annu Rev Immunol 16: 593–617. [DOI] [PubMed] [Google Scholar]
- Hu JC. (2001) Research on the Giant Panda. Shanghai Science and Technology Education Press: Shanghai. [Google Scholar]
- Hu YB, Qi DW, Wang HJ, Wei FW. (2010). Genetic evidence of recent population contraction in the southernmost population of giant pandas. Genetica 138: 1297–1306. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Yeager M. (1998). Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet 32: 415–435. [DOI] [PubMed] [Google Scholar]
- Kennedy LJ, Ryvar R, Gaskell RM, Addie DD, Willoughby K, Carter SD et al. (2002). Sequence analysis of MHC DRB alleles in domestic cats from the United Kingdom. Immunogenetics 54: 348–352. [DOI] [PubMed] [Google Scholar]
- Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER et al. (1990). Nomenclature for the major histocompatibility complexes of different species-a proposal. Immunogenetics 31: 217–219. [DOI] [PubMed] [Google Scholar]
- Kloch A, Babik W, Bajer A, Sinski E, Radwan J. (2010). Effects of an MHC-DRB genotype and allele number on the load of gut parasites in the bank vole Myodes glareolus. Mol Ecol 19: 255–265. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Marsden CD, Mable BK, Woodroffe R, Rasmussen GSA, Cleaveland S, McNutt JW et al. (2009). Highly endangered African wild dogs (Lycaon pictus) lack variation at the major histocompatibility complex. J Hered 100: S54–S65. [Google Scholar]
- McClelland EE, Penn DJ, Potts WK. (2003). Major histocompatibility complex heterozygote superiority during coinfection. Infect Immun 71: 2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi I, Guevara-Patino JA, Dyall R, LeMaoult J, Nikolich-Zugich J. (2002). Direct link between MHC polymorphism, T cell avidity, and diversity in immune defense. Science 298: 1797–1800. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Oliver MK, Telfer S, Piertney SB. (2009). Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris). Proc R Soc B 276: 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petric M, Mladineo I, Sifner SK. (2011). Insight into the short-finned Squid Illex Coindetii (Cephalopoda: Ommastrephidae) feeding ecology: is there a link between helminth parasites and food composition? J Parasitol 97: 55–62. [DOI] [PubMed] [Google Scholar]
- Piertney SB. (2003). Major histocompatibility complex B-LB gene variation in red grouse Lagopus lagopus scoticus. Wildlife Biol 9: 251–259. [Google Scholar]
- Piertney SB, Oliver MK. (2006). The evolutionary ecology of the major histocompatibility complex. Heredity 96: 7–21. [DOI] [PubMed] [Google Scholar]
- Posada D. (2008). jModelTest: Phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- Qi DW, Hu YB, Gu XD, Yang XY, Yang G, Wei FW. (2012). Quantifying landscape linkages among giant panda subpopulations in regional scale conservation. Integr Zool 7: 165–174. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2011) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Radwan J, Demiaszkiewicz AW, Kowalczyk R, Lachowicz J, Kawalko A, Wojcik JM et al. (2010). An evaluation of two potential risk factors, MHC diversity and host density, for infection by an invasive nematode Ashworthius sidemi in endangered European bison (Bison bonasus). Biol Conserv 143: 2049–2053. [Google Scholar]
- Radwan J, Kawalko A, Wojcik JM, Babik W. (2007). MHC-DRB3 variation in a free-living population of the European bison, Bison bonasus. Mol Ecol 16: 531–540. [DOI] [PubMed] [Google Scholar]
- Reche PA, Reinherz EL. (2003). Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J Mol Biol 331: 623–641. [DOI] [PubMed] [Google Scholar]
- Rice WR. (1989). Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Rousset F. (2008). GENEPOP ' 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8: 103–106. [DOI] [PubMed] [Google Scholar]
- Rózsa L, Reiczigel J, Majoros G. (2000). Quantifying parasites in samples of hosts. J Parasitol 86: 228–232. [DOI] [PubMed] [Google Scholar]
- Schad J, Dechmann DK, Voigt CC, Sommer S. (2012). Evidence for the 'good genes' model: association of MHC class II DRB alleles with ectoparasitism and reproductive state in the neotropical lesser bulldog bat, Noctilio albiventris. PLoS ONE 7: e37101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GB, Hu JC, Pan WS, Zhu J. (1985) The Giant Panda of Wolong. The University of Chicago Press: Chicago. [Google Scholar]
- Seddon JM, Baverstock PR. (1999). Variation on islands: major histocompatibility complex (Mhc) polymorphism in populations of the Australian bush rat. Mol Ecol 8: 2071–2079. [DOI] [PubMed] [Google Scholar]
- Sommer S. (2005). The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State Forestry Administration. (2006) The 3rd National Survey Report on Giant Panda in China. Science Press: Beijing. [Google Scholar]
- Takahata N, Nei M. (1990). Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124: 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan QH, Zhu L, Wu H, Fang SG. (2006). Major histocompatibility complex class II variation in the giant panda (Ailuropoda melanoleuca). Mol Ecol 15: 2441–2450. [DOI] [PubMed] [Google Scholar]
- Wei FW, Hu YB, Zhu LF, Bruford MW, Zhan XJ, Zhang L. (2012). Black and white and read all over: the past, present and future of giant panda genetics. Mol Ecol 21: 5660–5674. [DOI] [PubMed] [Google Scholar]
- Wu J, Jiang YK, He GZ, Wu GQ, Zhang DH, Hu HG. (1985). Life history of Ascaris schroederi. Chinese Vet Sci 6: 21–23. [Google Scholar]
- Wu J, Jiang YK, Wu GQ, He GZ, Zhang DH, Hu HG. (1987). Investigation on resistance of Ascaris schroederi eggs. Chinese J Vet Med 13: 7–9. [Google Scholar]
- Zeng CJ, Pan HJ, Gong SB, Yu JQ, Wan QH, Fang SG. (2007). Giant panda BAC library construction and assembly of a 650-kb contig spanning major histocompatibility complex class II region. BMC Genomics 8: 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XJ, Li M, Zhang ZJ, Goossens B, Chen YP, Wang HJ et al. (2006). Molecular censusing doubles giant panda population estimate in a key nature reserve. Curr Biol 16: R451–R452. [DOI] [PubMed] [Google Scholar]
- Zhang BW, Li M, Zhang ZJ, Goossens B, Zhu LF, Zhang SN et al. (2007). Genetic viability and population history of the giant panda, putting an end to the "evolutionary dead end"? Mol Biol Evol 24: 1801–1810. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yang XY, Wu H, Gu XD, Hu YB, Wei FW. (2011). The parasites of giant pandas: individual-based measurement in wild animals. J Wildlife Dis 47: 164–171. [DOI] [PubMed] [Google Scholar]
- Zhang ZH, Wei FW. (2006) Giant Panda Ex-situ Conservation Theory and Practice. Science Press: Beijing. [Google Scholar]
- Zhao SC, Zheng PP, Dong SS, Zhan XJ, Wu Q, Guo XS et al. (2013). Whole-genome sequencing of giant pandas provides insights into demographic history and local adaptation. Nat Genet 45: 67–71. [DOI] [PubMed] [Google Scholar]
- Zhu L, Ruan XD, Ge YF, Wan QH, Fang SG. (2007). Low major histocompatibility complex class II DQA diversity in the giant panda (Ailuropoda melanoleuca). BMC Genet 8: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LF, Zhan XJ, Wu H, Zhang SN, Meng T, Bruford MW et al. (2010). Conservation implications of drastic reductions in the smallest and most isolated populations of giant pandas. Conserv Biol 24: 1299–1306. [DOI] [PubMed] [Google Scholar]