Abstract
Understanding the genetic architecture of host resistance is key for understanding the evolution of host–parasite interactions. Evolutionary models often assume simple genetics based on few loci and strong epistasis. It is unknown, however, whether these assumptions apply to natural populations. Using a quantitative trait loci (QTL) approach, we explore the genetic architecture of resistance in the crustacean Daphnia magna to two of its natural parasites: the horizontally transmitted bacterium Pasteuria ramosa and the horizontally and vertically transmitted microsporidium Hamiltosporidium tvaerminnensis. These two systems have become models for studies on the evolution of host–parasite interactions. In the QTL panel used here, Daphnia's resistance to P. ramosa is controlled by a single major QTL (which explains 50% of the observed variation). Resistance to H. tvaerminnensis horizontal infections shows a signature of a quantitative trait based in multiple loci with weak epistatic interactions (together explaining 38% variation). Resistance to H. tvaerminnensis vertical infections, however, shows only one QTL (explaining 13.5% variance) that colocalizes with one of the QTLs for horizontal infections. QTLs for resistance to Pasteuria and Hamiltosporidium do not colocalize. We conclude that the genetics of resistance in D. magna are drastically different for these two parasites. Furthermore, we infer that based on these and earlier results, the mechanisms of coevolution differ strongly for the two host–parasite systems. Only the Pasteuria–Daphnia system is expected to follow the negative frequency-dependent selection (Red Queen) model. How coevolution works in the Hamiltosporidium–Daphnia system remains unclear.
Introduction
In natural populations, genetic polymorphisms among hosts for disease-related traits are widespread and prominent (Klein, 1990; Bergelson et al., 2001; Flainik and Du Pasquier, 2004), generating intensive discussion about the processes maintaining them. Several population genetic models have been put forward, linking this genetic diversity to host–parasite coevolution, both within and among populations connected by geneflow. Most models are based either on the idea of beneficial mutations spreading in the populations (selective sweep models) or on standing genetic variation maintained by negative frequency-dependent selection (NFDS, the Red Queen models) (Woolhouse et al., 2002; Ebert, 2008; Lively, 2010). The genetic assumptions underlying these models are very different. Models based on novel beneficial mutations make no assumption about the underlying genetics. Beneficial mutations, no matter what the selection coefficient and where they occur in the genome, may spread and reach fixation. Several mutations may sweep through a sexual population simultaneously. If selective sweeps lead to fixation of a variant, genetic polymorphisms are transient. In contrast, coevolution by NFDS operates under a rather strict set of underlying genetic assumptions. They depend on one locus or a few loci with strong effects and, for multilocus systems, on epistatic interactions among loci. In theory, the NFDS model can sustain genetic polymorphisms endlessly (Clarke, 1976).
As we still know little about the genetic architecture of host resistance and parasite virulence, it is difficult to judge which genetic model best explains phenotypic observations of coevolutionary dynamics (Edmunds and Alstad, 1978; Lively et al., 2004; Morgan et al., 2005; Laine, 2006; Decaestecker et al., 2007; Jokela et al., 2009; Morran et al., 2011; Kerstes et al., 2012). A review of quantitative trait locus studies in host–parasite systems suggested that host resistance is often influenced by a few loci with rather strong effects and epistatic interactions (Wilfert and Schmid-Hempel, 2008). This evidence—together with the general notion that disease-related traits show high levels of polymorphisms in natural populations—has been used to emphasize the role of NFDS as a key mechanism of coevolution (references as before; for reviews, see (Thompson, 2005; Schmid-Hempel, 2011)). Other studies, however, stress the role of selective sweeps for host and parasite adaptation (Buckling and Rainey, 2002; Jiggins, 2003; Little et al., 2004; Nash et al., 2005; Obbard et al., 2011). To understand which model best explains coevolution in any given system requires, however, we must clarify the genetic architecture of host resistance. Here we aim to do this for D. magna and two of its naturally coevolving parasites—the bacterium P. ramosa (Metchnikoff, 1888) and the microsporidium H. tvaerminnensis (Haag et al., 2011) (formerly called Octosporea bayeri). We explore the genetic architecture of resistance to these parasites and link our findings to what is known about coevolution in these systems.
D. magna and its parasites are frequently used as model systems for the study of natural diseases (Green, 1974; Ebert, 2008; Auld, 2014). Phenotypic studies have suggested specific coevolution between Daphnia and its antagonists. The pathogenic bacterium P. ramosa transmits exclusively horizontally via environmentally resistant endospores released from dead hosts. It sterilizes its host, killing it after about 50 days. Studies of sediment cores have revealed fast evolutionary dynamics between host and parasite suggestive of a Red Queen-like coevolution (Decaestecker et al., 2007) and genetic crosses revealed strong host–parasite genetic interactions based on few loci and strong epistasis (Little et al., 2006; Luijckx et al., 2012; Luijckx et al., 2013), making it a strong candidate for coevolution by NFDS (Ebert, 2008). Nonetheless, although we understand relatively well Daphnia resistance to Pasteuria, we have lacked knowledge about this system's underlying genomic architecture.
The other parasite studied here, the microsporidium H. tvaerminnensis, infects mainly the fat body and ovaries of D. magna, reducing life span, fecundity and competitive ability (Bieger and Ebert, 2009; Ben-Ami et al., 2011). This parasite is transmitted via mixed modes, that is, both vertically (mother to offspring) and horizontally (through water-borne environmental spores). H. tvaerminnensis seems to coevolve with its D. magna host, as revealed by studies on local adaptation and transmission in relation to host sexual reproduction (Altermatt et al., 2007; Ebert et al., 2007; Ebert, 2008). Experimental evolution further suggests that the host adapts rapidly to cope with the parasite, but at a cost in the absence of infections (Zbinden et al., 2008). Nothing is known about the genetics of disease-related traits in this system.
These Daphnia–parasite systems are particularly suited for quantitative trait loci (QTL) studies, because the hosts can be crossed sexually, but also kept clonally, allowing us to produce QTL panels starting with individual females collected in the field and to maintain recombinant lines as clones for many generations. This clonality also allows us to replicate genotypes, which reduces non-genetic effects, including variation in maternal effects. The QTL F2 panel used in this study originates with the crossing of a Finnish D. magna clone, susceptible to both P. ramosa and H. tvaerminnensis, and a German D. magna clone resistant to both parasites. This F2 panel is kept clonally in the laboratory and has previously been used to map other D. magna traits, such as recessive deleterious mutants, induction of males and production of resting eggs (Routtu et al., 2010; Roulin et al., 2013). Here, our aim was to use this panel to elucidate the number of QTL in the host genome that contribute to resistance against P. ramosa and H. tvaerminnensis, test for interactions among QTLs (epistasis) and test for colocalization of QTLs. The results are interpreted with regard to the genetic architecture of host resistance and—in combination with previously published material—with regard to the mechanisms of coevolution between the host and its parasites.
Materials and methods
D. magna is a cyclic parthenogenetic planktonic crustacean found in standing fresh water in Eurasia, Africa and North America. Females reproduce primarily asexually, but can also reproduce sexually on environmental induction. Clonal lines can be maintained for many years on a diet of green algae. The two clones used in the QTL panel for this study are from a carp-breeding pond near Munich, Germany (clone ID: Iinb1; coordinates: 48°12′24′′N, 11°42′35′′E) and from a rock pool in southwest Finland near Tvärminne (clone ID: Xinb3; coordinates 59°49′58 N, 23°15′45E). For all experiments, animals were kept in artificial Daphnia medium (Klüttgen et al., 1994) at 20 °C and a light:dark cycle of 16:8 with the green algae Scenedesmus sp. as the only food.
The C19 clone of the Gram-positive bacterium P. ramosa used here was isolated from an infected D. magna collected from a pond in Gaarzerfeld in northern Germany and was characterized in an earlier study (Luijckx et al., 2011). Infection occurs when hosts are exposed to aqueous suspensions of parasite spores. We produced P. ramosa spores by exposing immature female D. magna individuals from a susceptible host clone to the parasite and letting the parasite grow for 40 days. The tissue of the infected hosts with mature spores was then homogenized in artificial Daphnia medium and the spore concentration was determined with a Neubauer-improved counting chamber.
An isolate of the microsporidium H. tvaerminnensis was obtained from an infected female D. magna collected from a rock pool on the island Ören in the Tvärminne archipelago in southwest Finland. About 50% of D. magna populations on the archipelago are infected with H. tvaerminnensis and all D. magna clones from the region appear susceptible; D. magna clones outside of Finland, however, are mostly resistant (Ebert, 2008) (Lange et al., submitted). Other host species or secondary hosts are not known for this parasite (Haag et al., 2011; Haag et al., 2013). H. tvaerminnensis is transmitted both horizontally (from environmental spores released by decaying hosts) and vertically (from mothers to offspring). Vertical transmission is 100% efficient to parthenogenetic (asexual) offspring, but only partially effective to sexually produced offspring (via resting eggs) (Ebert et al., 2007). Spores were produced from the vertically infected offspring of an infected mother. Infected hosts with mature spores were collected when 40 days old and spore suspensions were produced as described for Pasteuria.
The D. magna F2 panel
All experiments used the Basel D. magna QTL panel. This F2 panel is described in full detail in earlier publications (Routtu et al., 2010; Roulin et al., 2013) (summary at http://evolution.unibas.ch/ebert/research/qtl/index.htm). In short, two parental strains, Iinb1 and Xinb3 from Germany and Finland, respectively, were crossed to form an F1 hybrid clone. Iinb1, which does not easily reproduce sexually, was previously selfed one time and Xinb3 was selfed three times. The F1 hybrid clone was then selfed to produce the sexual offspring of the F2 generation, which were kept as clonal lines in the laboratory. The current study used 195 F2 clones. The clones of the F2 panel were previously genotyped for 109 VNTR markers (Routtu et al., 2010) and later for 1324 SNP markers (Routtu et al., submitted), which were used to construct genetic maps for the phenotypic traits (Routtu et al., 2010; Roulin et al., 2013).
Infection trial design
Clonal females from the F2 panel were raised individually in 100-ml jars filled with 90 ml artificial Daphnia medium and fed 3–10 million algal cells per day, accommodating the increasing needs of the growing animals in optimal conditions. Medium was changed every 4 days or after the females released a clutch. From each female, one daughter from the third clutch was randomly chosen for the infection trials. These offspring were placed individually in 100-ml jars filled with 20 ml artificial Daphnia medium. Three to five days after birth, each individual was exposed to 200 000 spores of either P. ramosa (6 replicates) or H. tvaerminnensis (3 replicates) or to a control suspension (3 replicates). This high spore dose was used, as it resulted in 100% infection rates of susceptible genotypes, thus reducing the chance of failed infection in susceptible hosts due to stochastic effects. We knew from previous studies that variation in resistance manifests itself most clearly in the presence/absence patterns for P. ramosa (Carius et al., 2001) and in spore counts for H. tvaerminnensis (Ebert, 2008; Roth et al., 2008). At the age of 40 days, Pasteuria exposed animals were killed to check for infection and H.tvaerminensis exposed animals (and controls) were homogenized to quantify the number of spores in their body using a cell counting chamber. To test for resistance to vertical transmission, we raised two offspring from the third clutch of all Daphnia, which had previously been exposed to H. tvaerminnensis (horizontally infection trial), until they were 40 days old and then quantified their spore loads. The presence of spores, in these animals, indicates successful vertical transmission.
Data handling and statistics
All statistics were calculated using the R statistical software version 2.9.2 (R Development Core Team, 2008). QTLs were analyzed with the R package R/qtl version 1.29-2 (9 September 2013) (Broman et al., 2003; Broman and Sen., 2009; http://www.rqtl.org/). A description of the structure and construction of the SNP array linkage map used can be found in Routtu et al. (2010), Roulin et al. (2013) and Routtu et al., submitted. For the QTL analysis, genome-wide significance level was established using 10 000 permutation tests with significant (α=0.05) and suggestive (α=0.1) LOD scores of 3.78 and 3.44, respectively. Analysis of variance was used to estimate the proportion of total variance explained by the fitted models. To find epistatic interactions, we ran scantwo (for two-dimensional scans) and further analyzed the candidates with fitqtl (for fitting a defined multiple-QTL model) (Broman et al., 2003; Broman and Sen, 2009). The minimum QTL effect size for our design was estimated with package qtlDesign (Sen et al., 2007). In all experiments, controls were uninfected and are therefore not further discussed. The final data set for resistance analysis included 195 F2 clones.
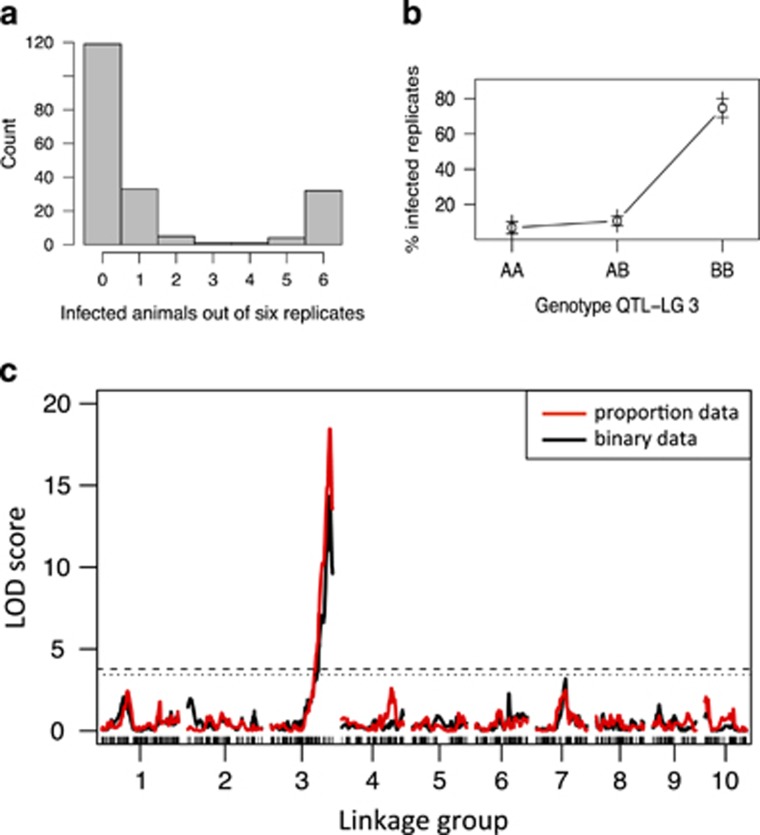
Analysis of Pasteuria infection data was carried out in two ways. First, data were treated as proportions of infected animals over number of replicates (n=6). Second, we used a binary model (infection status 0/1) for the QTL analysis as described by Broman et al. (2003). This is justified, because infection data of Pasteuria clones typically show strongly bimodal patterns, with either most replicates being infected or hardly any (see Figure 1a for this pattern in the current data set). For this, we designated clones as susceptible when three or more of the six replicates showed infection; otherwise, we designated them as resistant (binary classification on the clone level).
Figure 1.
Analysis of infection data for D. magna after exposure to P. ramosa. (a) Frequency distribution showing the number of infected replicates (of six total) for 195 F2 clones. (b) Effect plot (means±s.e.) for the three genotypes at scaffold00288_965 on lg 3 for the infected replicates. A alleles are from the resistant German parental clone; B alleles are from the susceptible Finnish parental clone. (c) QTL mapping for resistance against Pasteuria. Black solid lines indicate the analysis with the proportion data; the red line shows the analysis with binary coded data; dashed lines show the LOD threshold for significance (dashed line: P<0.05; dotted: P<0.1).
For the H. tvaerminnensis data, we log10(x+1) transformed counts of horizontally and vertically infected individuals to normalize spore count distribution and then calculated the means of this log-transformed data for the QTL analysis. We used Haley–Knott regression in the QTL analysis for robustness and speed (Haley and Knott, 1992). Although we expected that totally resistant females (judged from spore counts=0) in the horizontal infection assay would be unable to pass the infection to the next generation, we had some cases of zero counts in the horizontal infections but found spores in the offspring of these animals. This discrepancy is likely explained by the counting chambers' high detection threshold for spores (spores were counted in a sub-sample volume of 0.1 μl of the total volume of 100 μl). In addition, as has previously been observed, horizontal infections generally produce much lower spore counts than vertical infections (Vizoso and Ebert, 2004), making it more likely to miss an infection in horizontally infected animals (false negative) than in vertically infected animals. Therefore, we included all animals in the vertical infection analysis, disregarding the infection status of their mother. Repeating the vertical infection analysis after excluding replicates with zero spore counts in the horizontal infection trials did not change the QTL results for vertical infections.
Results
The estimated minimum effect size to detect a genetic signal with the QTL panel used here was 10.97% variance explained for significant (α=0.05) and 10.19% variance explained for suggestive (α=0.1) LOD scores. Thus, loci explaining less than about 10% of the total variation will not be picked up by our analysis.
Resistance against P. ramosa
The P. ramosa infection data revealed strong bimodality with most replicates of many clones being totally resistant or most replicates being infected (Figure 1a). We designated F2 clones ‘susceptible' if three or more of the six replicates were infected and as ‘resistant' otherwise. This resulted in 20.6% susceptible clones. Assuming that resistance is dominant and coded by a single locus with two alleles, the proportion of susceptible clones would be expected to be 25%, which is not significantly different from 20.6% (χ2=1.986, P=0.16, n=195). This single locus Mendelian segregation was also seen for Pasteuria resistance in a cross between two Finnish D. magna clones (Luijckx et al., 2012). The QTL analysis of these binary (0/1) infection data (clones being treated as susceptible or resistant) revealed a single major QTL that explains 50.04% of the total variation (Figure 1c and Table 1). No epistatic interactions were detected. A minor peak in the middle of linkage group (lg) 7 was far from significance. Running the QTL analysis with the Pasteuria infection data coded as proportions of infected replicates among all replicates revealed the same result with a single peak at the same place and 50.1% of the variance explained (Figure 1c and Table 1). No epistatic interactions were detected.
Table 1. Estimates of total LOD scores and total variation explained by genotype effects at the QTL sites (Q).
| Infection by | Mean square | LOD score | % Variance explained | Model |
|---|---|---|---|---|
| Pasteuria (proportions) | 245.00 | 29.44 | 50.10 | y~Q1 |
| Pasteuria (binary) | 7.816 | 29.53 | 50.04 | y~Q1 |
| Hamiltosporidium (horizontal infection) | 0.9574 | 20.43 | 38.276 | y~Q2+Q3+Q4+ Q5+Q6+Q2:Q4 |
| Hamiltosporidium (vertical infection) | 7.0873 | 6.154 | 13.526 | y~Q4 |
Abbreviation: QTL, quantitative trait loci.
Sites refer to the D. magna genome and are numbered continuously (compare with Table 2): Q1=scaffold00288_965, Q2=scaffold00773_975, Q3=scaffold02114_353, Q4=scaffold00606_2933, Q5=contig29113_349, Q6=contig14698_81. All four models are significant (P<0.0001). Total degrees of freedom were 194 in each case.
Resistance to H. tvaerminnensis
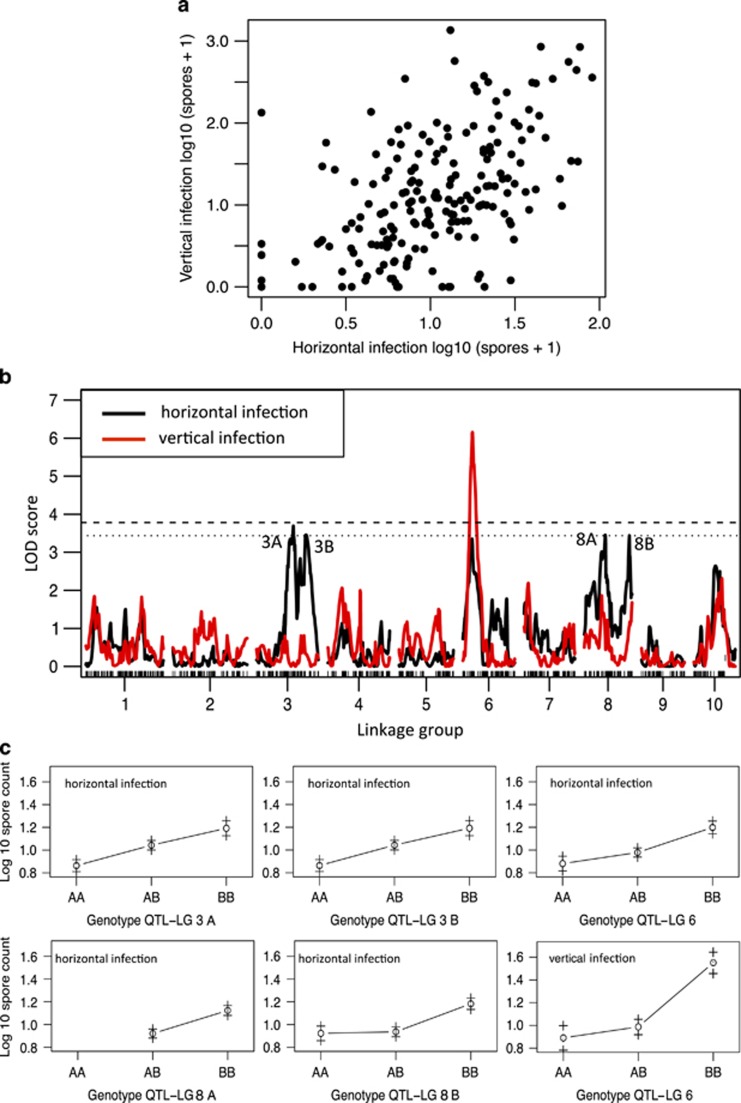
The distribution of the log-transformed spore counts for vertical and horizontal infections with H. tvaerminnensis were nearly perfectly normal, with only a few clones with mean spore counts of zero (Figure 2a). Horizontal and vertical infection spore counts were positively correlated (Spearman: r2=0.43, P=4.19e−10; Figure 2a).
Figure 2.
Analysis of infection data for D. magna after exposure to H. tvaerminnensis. (a) Scatter plot of mean clonal spore counts for vertically infected versus horizontally infected animals (both traits log10(spore count+1) transformed). (b) QTL mapping of H. tvaerminnensis log10(spore count+1) in horizontally infected (black line) and vertically infected (red line) hosts. Dashed lines show the LOD threshold for significance (dashed line: P<0.05; dotted: P<0.1). Five QTLs are detected in horizontally infected hosts. One QTL is detected for vertically infected hosts, colocalized with a QTL for horizontal infections on lg 6. (c) Effect plots (means±s.e.) for genotypes at the identified QTLs. A alleles are from the resistant German parental clone; B alleles from the susceptible Finnish parental clone. Multiple QTLs on lg 3 and lg 8 are indicated with letters A and B. The effect plot for marker contig29113_349 (lg 8A) has no AA genotypes shown, because we had only phenotypic data for a single AA genotype. This region of the map showed strong transmission ratio distortion due to an infertility allele (Routtu et al., 2010).
H. tvaerminnensis spore counts for horizontally infected Daphnia revealed five QTLs, approaching significance in lg 3, 6 and 8 (Figures 2b and c, and Table 1). These QTLs showed weak epistatic interactions with each other, but only the interaction between lg 3A and lg 6 approached significance (Table 1). No other epistatic interactions where detected. Together, these five QTLs and their interactions explained 38.3% of the observed variation (Table 1). H. tvaerminnensis spore counts from vertical infections produced a single significant peak colocating with a peak detected for horizontal infection spore counts on lg 6 and explaining 13.5% of the total variation (Figures 2b and c, and Table 1). Repeating this analysis with the few clones seemingly being resistant to horizontal infections (zero spore count in horizontal infection trials) being excluded from the vertical infection analysis did not change this picture.
Comparing QTL for the two parasites
The spore counts of H. tvaerminnensis were not correlated with susceptibility to P. ramosa (Spearman r=0.047 and r=0.074 for horizontal and vertical infections, respectively, P>0.2 in both cases), although resistance was correlated in the two parent clones, with the Finnish clone being highly susceptible to both parasites. Consistent with this, the effect plots for both parasites clearly show that the German parent (AA genotype) carries the alleles for resistance (Figure 2). None of the QTLs observed in the Hamiltosporidium analysis colocalized with the QTL for Pasteuria resistance at the end of lg 3.
Discussion
The aim of this study was to examine the genetic architecture of D. magna's resistance to two parasites in the context of previous work on host–parasite interactions and genetic polymorphisms for resistance in natural populations. Using a QTL panel based on a cross of a resistant host clone from Germany and a susceptible clone from Finland, we found contrasting patterns of resistance for the two parasites. A single major QTL was found for resistance against the bacterium Pasteuria, whereas five weak QTLs were observed for resistance against horizontal infections of the microsporidium H. tvaerminnensis and one QTL was found explaining resistance to vertical infections of H. tvaerminnensis. QTLs of the two parasites did not colocalize in the host genome. The QTLs for resistance against horizontal infections of H. tvaerminnensis showed only moderate evidence for epistasis.
The result of the QTL study shows that major QTL for resistance against Pasteuria segregates among the F2 clones and that this segregation is consistent with a single locus with Mendelian inheritance, with the German clone carrying a dominant resistance allele. Furthermore, the data show that variation for resistance is mostly determined by clone identity, as replicates within clones are highly consistent with each other, suggesting little non-genetic variation (Figure 1a). These results were not surprising: a previous study using genetic crosses had shown that D. magna's resistance against P. ramosa is likely based on a locus with Mendelian inheritance, without evidence for non-genetic effects (Luijckx et al., 2012). However, this previous study, using material from Finnish populations exclusively, revealed a second locus strongly linked to the first, coding for resistance against a different P. ramosa genotype (Luijckx et al., 2013). This second locus seemed not to segregate in our study's QTL F2 panel, as the parental clones and all F2 clones were resistant against this P. ramosa genotype (unpublished data). However, given that we started from only two parent genotypes and that we had only 195 F2 clones, it is likely that we did not capture all polymorphism with our panel design, as some polymorphisms might have been excluded by the choice of the parents and some minor QTL might have been under the detection threshold. Nevertheless, our results are consistent with what is known about the Pasteuria–Daphnia system, namely that resistance is strongly determined by one (more likely a few loci) in one region of the genome with a strong fitness effect, without evidence for loci explaining genetic variation in other parts of the genome or evidence for prominent non-genetic effects. These findings corroborate the suggestion that Daphnia and Pasteuria evolve by NFDS (Decaestecker et al., 2007). Cyclic coevolution caused by NFDS is most easily observed in models with one locus or few loci with strong effects: the fewer loci, the stronger the link between fitness and allelic variants (Otto and Nuismer, 2004).
The H. tvaerminnensis infection trials revealed a different pattern, however. The continuous spore count distribution across F2 clones suggests that multiple host loci with small effects contribute to resistance and segregate among the F2 clones. Indeed, several minor QTLs were identified for horizontal infections and one minor QTL was identified for vertical infections (Figure 2). Other QTLs might have been below the detection threshold, which is ~10% variation explained. The QTLs on lg 6 for horizontal and vertical infection have the highest LOD scores on the same marker, suggesting that the same locus might be responsible for this effect. However, horizontal infections seem to be influenced by more loci, perhaps suggesting that the process for horizontal infections is more complex. For example, genes that determine entry into the host or that modulate host behavior to regulate exposure to the spores may have a role in horizontal, but not vertical, infection processes, while genes responsible for within host transmission and growth might affect both types of infections. The QTL for spore counts after vertical infection is also higher than any QTL for horizontal infections, possibly because less noise is produced by the segregation of alleles at other loci, or because the vertical infection process is less influenced by non-genetic effects.
Although coevolution for the D. magna–Pasteuria system is likely based on NFDS, this seems unlikely for the D. magna–Hamiltosporidium system. We can only speculate about the coevolutionary processes at work in this system. Previous work demonstrated genetic variation for resistance to Hamiltosporidium (Ebert, 2008), local parasite adaptation (Altermatt et al., 2007) and that transmission to sexual eggs is more impaired if the host outbreeds than in case of selfing (Vizoso et al., 2005; Ebert et al., 2007). The current study suggests that the genetic variation underlying these findings is of a quantitative nature, with several minor QTLs contributing to it. Therefore, coevolution is unlikely to be driven by NFDS.
A study by Zbinden et al. (2008) suggests that the dynamic nature of the metapopulation may be the driving force for shaping resistance polymorphisms in the D. magna–H. tvaerminnensis system. Experimental evolution in the field suggested that the hosts in parasitized populations adapted to the parasite and that this adaptation was costly in the parasite's absence. Spore counts and infection assays were not conducted. This metapopulation is highly dynamic, with an average of 16% of all D. magna populations going extinct each year, although this is balanced by recolonizations (Pajunen and Pajunen, 2003). At any moment, ~50% of D. magna populations are infected by H. tvaerminnensis (Ebert et al., 2001). Most newly colonized pools are free of the parasite but will become infected with a high likelihood in the following years (Ebert et al., 2001). Thus, the evolution of resistance may progress in the following way: a Daphnia population loses resistance after colonizing a pool without the parasite, but then adapts to the parasite after the parasite colonized the host population. This form of evolution is not limited to a specific genetic architecture. A complex, quantitative genetic trait architecture, as revealed by our QTL study, would suffice.
No evidence for colocalization of defence genes
In our QTL panel, Daphnia's resistance to the two parasites is based on different underlying genetics, with different phenotype distributions, different QTLs strengths and no colocalized QTLs. Therefore, we cannot assume that common features of the Daphnia immune system (McTaggart et al., 2009) explain the observed variation. This is also consistent with an experimental evolution study in the presence and absence of Hamiltosporidium (Zbinden et al., 2008), where no correlation between resistance to Pasteuria and Hamiltosporidium was found. Still, certain aspects of the immune system could act on both parasite systems during an infection, even if these seem not to contribute to variation in resistance.
Fine mapping of resistance genes
As Pasteuria resistance shows a single, strong QTL peak, its resistance locus could feasibly be fine mapped. Indeed, the single strong QTL and evidence for Mendelian segregation indicate the presence of a single locus, and the strength of the QTL and the mostly consistent infection results within clones suggest weak environmental effects, making fine mapping an attractive prospect. Currently, however, the region of interest is over 1 MB long, with ~200 annotated genes, many without known functions; thus, it would be difficult to select candidate genes in the region of interest. We are thus developing markers for a recombination breakpoint analysis to narrow the region down. In contrast, fine mapping the loci responsible for H. tvaerminnensis spore count variation does not seem promising. Multiple minor peaks, epistatic interactions and variation for spore counts within clones make it difficult to link phenotypes with genotypes. The single peak for vertical infections offers a better option for fine mapping, but with only 13.8% of the variance explained by this peak, it would still be difficult. In addition, the regions in the D. magna genome (version 2.4), where the QTLs for H. tvaerminnensis spore counts are located are poorly assembled, further complicating the prospect of fine mapping these QTLs. With the current version of the D. magna genome (version 2.4 of the D. magna genome consortium) still divided into about 50 000 contigs and scaffolds, progress is limited.
Steps of the infection process
P. ramosa's infection process can be subdivided into a number of steps: encounter, spore activation, attachment, penetration of the host cuticle and within-host steps (Duneau et al., 2011; Hall and Ebert, 2012). The host's interference with any of these steps could contribute to host resistance. Our experimental design and knowledge of the system allow us to pinpoint the step that explains most of variance in the current study The encounter step, for example, can be ruled out, as our experimental design treated all hosts with a suspension of finely suspended spores in the water. Spore activation can also be excluded, as it shows no genetic or environmental variation (Duneau et al., 2011). The attachment step—during which the parasite attaches to the lining of the host esophagus—is the best candidate to explain most of the here observed variation. Attachment has been shown to be invariable with regard to environmental conditions and has been shown to be a binomial trait, with Daphnia clones either allowing attachment or not (Duneau et al., 2011; Luijckx et al., 2011). If attachment fails, hosts do not become infected. We believe our results here mainly reflect variation in the host locus for Pasteuria attachment. In the next step, the attached parasite needs to penetrate the host body wall. If the host moults before Pasteuria completed penetration into the body cavity, it is able to shed its carapace with the parasite and remain uninfected (Duneau and Ebert, 2012). At 20 °C, if the host moults within ~12 h after attachment, the likelihood of infection is strongly reduced. The moulting step may therefore explain cases where only some of the six host clone replicates became infected (Figure 1). In the final step, the parasite is exposed to the host's immune system, which sometimes clears the infection, again explaining the failure of some replicates to became infected (Ben-Ami et al., 2010; Hall and Ebert, 2012), although this is not common under good environmental conditions (Ben-Ami et al., 2010; Hall and Ebert, 2012).
The stepwise infection model cannot explain why some host clones had only one or two infected replicates. It may be that the high spore dose (200 000 spores per host) used in this study enabled infection by another mechanism for some individuals of clones that do not allow attachment otherwise. Other points of entry into the host have been observed (Duneau, personal communication) but were not monitored in the current study. However, these alternative infection routes may only occur rarely and might be overlooked in large-scale experiments such as the QTL analysis conducted here. Another possible explanation is that a second parasite genotype (mutation or contamination) was present in our material but did not lead to high infection rates due to its low abundance. In any case, the low number of infections observed in some clones seemed not to interfere with the discovery of the major QTLs for resistance in these systems. Coding the data in binary form or using the proportion of infected replicates as dependent variables resulted in the same outcome (Table 1 and Figure 1).
Conclusions
What factors determine the type of resistance genetic architecture at work? A crucial difference between the Pasteuria and the Hamiltosporidium system is the barrier defence (the attachment step with a genetically determined yes/no infection response) that governs the overall infection success for Pasteuria. The genetics of this attachment step fall in line perfectly with the assumptions of Red Queen models (Luijckx et al., 2013). This is not the case for microsporidian infections, which infect the host by shooting—in a harpoon-like manner—a tube into the host tissue and forcefully injecting their cytoplasm directly into the host cell. Thus, no barrier defence seems to exist against microsporidians (Wittner and Weiss, 1999) and the host's degree of resistance seems determined by genetic polymorphisms at the level of its within-host cell immune system, which is likely much more complex than the apparently simple genetics of the Pasteuria attachment step (Auld et al. 2010). Although Pasteuria resistance is also influenced by the host's immune system (Hall and Ebert, 2012), the immune system's overall role in explaining variation in resistance is much less because it encounters only those parasites that were able to pass the barrier defence, that is, that attached to and penetrated into the host. Thus, we speculate that barrier defences early in the infection process may play a crucial role in shaping the distribution of genetic variation for resistance in a given system. Barrier defences are also at the heart of gene-for-gene models in plant–pathogen interactions, which have been suggested to have an important role in Red Queen-like coevolutionary dynamics (Frank, 1993; Thrall et al., 2012).
Data archiving
The SNP data for the genetic map are archived on Dryad (doi:10.5061/dryad.2260t).
Table 2. Infection traits and their corresponding linkage group (lg).
| Trait | lg | Marker_SNP nucleotide | Sequence (5′–3′) |
|---|---|---|---|
| Pasteuria infection | 3 | scaffold 00288_965 | TTGTTAAAGTCCATTGTaAGTGTTTAAGTAGCAAA |
| Hamiltosporidium horizontal infection | 3A | scaffold 00773_975 | ATCGAATCAGCATTTTaTCCTCCTAATTCAAC |
| Hamiltosporidium horizontal infection | 3B | scaffold 02114_353 | ACACTGTCGAAAAAAAAaTTTTGATTTCCCTTAAAC |
| Hamiltosporidium horizontal infection | 6 | scaffold 00606_2933 | TTGTAGATTTTACCGAaCTTTTATTCTTACATCAC |
| Hamiltosporidium horizontal infection | 8A | contig 29113_349 | CGATATATCTTTCAAAACaATCTATATGGATACAATC |
| Hamiltosporidium horizontal infection | 8B | contig 14698_81 | TGGCGATAAAGGACaTTGAATCAAAGGACG |
| Hamiltosporidium vertical infection | 6 | scaffold 00606_2933 | TTGTAGATTTTACCGAaCTTTTATTCTTACATCAC |
Acknowledgments
We thank Jürgen Hottinger and Urs Stiefel for laboratory support. Suzanne Zweizig improved the language of the manuscript. Anne Roulin, Andrea Kaufmann, Gilberto Bento, David Preiswerk, Roberto Arbore, Cesar Metzger and other members of the host-symbiont group at the Zoological Institute of Basel University provided helpful comments on the manuscript. This work was supported by the Swiss National Science Foundation.
The authors declare no conflict of interest.
References
- Altermatt F, Hottinger JW, Ebert D (2007). Parasites promote host gene flow in a metapopulation. Evol Ecol 21: 561–575. [Google Scholar]
- Auld SKJR (2014). Immunology and immunity. In: Smirnov NN (ed). The Physiology of the Cladocera. Academic Press: Amsterdam, pp 219–234. [Google Scholar]
- Auld SKJR, Scholefield JA, Little TJ (2010). Genetic variation in the cellular response of Daphnia magna (Crustacea: Cladocera) to its bacterial parasite. Proc R Soc B 277: 3291–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami F, Ebert D, Regoes RR (2010). Pathogen dose infectivity curves as a method to analyze the distribution of host susceptibility: A quantitative assessment of maternal effects after food stress and pathogen exposure. Am Nat 175: 106–115. [DOI] [PubMed] [Google Scholar]
- Ben-Ami F, Rigaud T, Ebert D (2011). The expression of virulence during double infections by different parasites with conflicting host exploitation and transmission strategies. J Evol Biol 24: 1307–1316. [DOI] [PubMed] [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian DC (2001). Evolutionary dynamics of plant R-genes. Science 292: 2281–2285. [DOI] [PubMed] [Google Scholar]
- Bieger A, Ebert D (2009). Expression of parasite virulence at different host population densities under natural conditions. Oecologia 160: 247–255. [DOI] [PubMed] [Google Scholar]
- Broman KW, Sen Ś (2009). A Guide to QTL Mapping with R/qtl. Springer: New York. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Buckling A, Rainey PB (2002). Antagonistic coevolution between a bacterium and a bacteriophage. Proc R Soc B 269: 931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carius HJ, Little TJ, Ebert D (2001). Genetic variation in a host-parasite association: Potential for coevolution and frequency-dependent selection. Evolution 55: 1136–1145. [DOI] [PubMed] [Google Scholar]
- Clarke BC (1976). The ecological relationship of host-parasite relationships. In: Taylor AER, Muller RM (eds). Genetic Aspects of Host-Parasite Relationships. Blackwell: Oxford, pp 87–104. [Google Scholar]
- Decaestecker E, Gaba S, Raeymaekers JAM, Stoks R, Van Kerckhoven L, Ebert D et al. (2007). Host-parasite ‘Red Queen' dynamics archived in pond sediment. Nature 450: 870–873. [DOI] [PubMed] [Google Scholar]
- Duneau D, Ebert D (2012). The role of moulting in parasite defence. Proc R Soc B 279: 3049–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duneau D, Luijckx P, Ben-Ami F, Laforsch C, Ebert D (2011). Resolving the infection process reveals striking differences in the contribution of environment, genetics and phylogeny to host-parasite interactions. BMC Biol 9: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D (2008). Host-parasite coevolution: Insights from the Daphnia-parasite model system. Curr Opin Mircobiol 11: 290–301. [DOI] [PubMed] [Google Scholar]
- Ebert D, Altermatt F, Lass S (2007). A short term benefit for outcrossing in a Daphnia metapopulation in relation to parasitism. J R Soc Interface 4: 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D, Hottinger JW, Pajunen VI (2001). Temporal and spatial dynamics of parasites in a Daphnia metapopulation: Which factors explain parasite richness? Ecology 82: 3417–3434. [Google Scholar]
- Edmunds GF, Alstad DN (1978). Coevolution in insect herbivores and conifers. Science 199: 941–945. [DOI] [PubMed] [Google Scholar]
- Flainik MF, Du Pasquier L (2004). Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol 25: 640–644. [DOI] [PubMed] [Google Scholar]
- Frank SA (1993). Coevolutionary genetics of plants and pathogens. Evol Ecol 7: 45–75. [Google Scholar]
- Green J (1974). Parasites and epibionts of Cladocera. Trans Zool Soc Lond 32: 417–515. [Google Scholar]
- Haag KL, Larsson JIR, Refardt D, Ebert D (2011). Cytological and molecular description of Hamiltosporidium tvaerminnensis gen. et sp nov., a microsporidian parasite of Daphnia magna, and establishment of Hamiltosporidium magnivora comb. nov. Parasitology 138: 447–462. [DOI] [PubMed] [Google Scholar]
- Haag KL, Traunecker E, Ebert D (2013). Single-nucleotide polymorphisms of two closely related microsporidian parasites suggest a clonal population expansion after the last glaciation. Mol Ecol 22: 314–326. [DOI] [PubMed] [Google Scholar]
- Haley CS, Knott SA (1992). A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324. [DOI] [PubMed] [Google Scholar]
- Hall MD, Ebert D (2012). Disentangling the influence of parasite genotype, host genotype and maternal environment on different stages of bacterial infection in Daphnia magna. Proc R Soc B 279: 3176–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM (2003). Male-killing Wolbachia and mitochondrial DNA: Selective sweeps, hybrid introgression and parasite population dynamics. Genetics 164: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokela J, Dybdahl MF, Lively CM (2009). The maintenance of sex, clonal dynamics, and host-parasite coevolution in a mixed population of sexual and asexual snails. Am Nat 174: S43–S53. [DOI] [PubMed] [Google Scholar]
- Kerstes NAG, Berenos C, Schmid-Hempel P, Wegner KM (2012). Antagonistic experimental coevolution with a parasite increases host recombination frequency. BMC Evol Biol 12: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J (1990). Immunology. Blackwell: Oxford. [Google Scholar]
- Klüttgen B, Dülmer U, Engels M, Ratte HT (1994). ADaM, an artificial freshwater for the culture of zooplankton. Water Res 28: 743–746. [Google Scholar]
- Laine AL (2006). Evolution of host resistance: looking for coevolutionary hotspots at small spatial scales. Proc R Soc B 273: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little TJ, Colbourne JK, Crease TJ (2004). Molecular evolution of Daphnia immunity genes: Polymorphism in a gram-negative binding protein gene and an alpha-2-macroglobulin gene. J Mol Evol 59: 498–506. [DOI] [PubMed] [Google Scholar]
- Little TJ, Watt K, Ebert D (2006). Parasite-host specificity: Experimental studies on the basis of parasite adaptation. Evolution 60: 31–38. [PubMed] [Google Scholar]
- Lively CM (2010). A review of Red Queen models for the persistence of obligate sexual reproduction. J Hered 101: S13–S20. [DOI] [PubMed] [Google Scholar]
- Lively CM, Dybdahl ME, Jokela J, Osnas EE, Delph LE (2004). Host sex and local adaptation by parasites in a snail-trematode interaction. Am Nat 164: S6–S18. [DOI] [PubMed] [Google Scholar]
- Luijckx P, Ben-Ami F, Mouton L, Du Pasquier L, Ebert D (2011). Cloning of the unculturable parasite Pasteuria ramosa and its Daphnia host reveals extreme genotype-genotype interactions. Ecol Lett 14: 125–131. [DOI] [PubMed] [Google Scholar]
- Luijckx P, Fienberg H, Duneau D, Ebert D (2012). Resistance to a bacterial parasite in the crustacean Daphnia magna shows Mendelian segregation with dominance. Heredity 108: 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijckx P, Fienberg H, Duneau D, Ebert D (2013). A matching-allele model explains host resistance to parasites. Curr Biol 23: 1085–1088. [DOI] [PubMed] [Google Scholar]
- McTaggart SJ, Conlon C, Colbourne JK, Blaxter ML, Little TJ (2009). The components of the Daphnia pulex immune system as revealed by complete genome sequencing. BMC Genomics 10: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metchnikoff ME (1888). Pasteuria ramosa un représentant des bactéries a division longitudinale. Annales de L'Institut Pasteur (Paris) 2: 165–170. [Google Scholar]
- Morgan AD, Gandon S, Buckling A (2005). The effect of migration on local adaptation in a coevolving host-parasite system. Nature 437: 253–256. [DOI] [PubMed] [Google Scholar]
- Morran LT, Schmidt OG, Gelarden IA, Parrish RC, Lively CM (2011). Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science 333: 216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash D, Nair S, Mayxay M, Newton PN, Guthmann JP, Nosten F et al. (2005). Selection strength and hitchhiking around two anti-malarial resistance genes. Proc R Soc B 272: 1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Jiggins FM, Bradshaw NJ, Little TJ (2011). Recent and recurrent selective sweeps of the antiviral RNAi gene Argonaute-2 in three species of Drosophila. Mol Biol Evol 28: 1043–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Nuismer SL (2004). Species interactions and the evolution of sex. Science 304: 1018–1020. [DOI] [PubMed] [Google Scholar]
- Pajunen VI, Pajunen I (2003). Long-term dynamics in rock pool Daphnia metapopulations. Ecography 26: 731–738. [Google Scholar]
- R Development Core Team (2008). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. http://www.R-project.org. [Google Scholar]
- Roth O, Ebert D, Vizoso DB, Bieger A, Lass S (2008). Male-biased sex-ratio distortion caused by Octosporea bayeri, a vertically and horizontally-transmitted parasite of Daphnia magna. Int J Parasit 38: 969–979. [DOI] [PubMed] [Google Scholar]
- Roulin AC, Routtu J, Hall MD, Janicke T, Colson I, Haag CR et al. (2013). Local adaptation of sex induction in a facultative sexual crustacean: insights from QTL mapping and natural populations of Daphnia magna. Mol Ecol 22: 3567–3579. [DOI] [PubMed] [Google Scholar]
- Routtu J, Jansen B, Colson I, De Meester L, Ebert D (2010). The first-generation Daphnia magna linkage map. BMC Genomics 11: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel P (2011). Evolutionary Parasitology. Oxford University Press: Oxford, UK. [Google Scholar]
- Sen S, Satagopan JM, Broman KW, Churchill GA (2007). R/qtlDesign: Inbred line cross experimental design. Mammalian Genome 18: 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN (2005). The Geographic Mosaic of Coevolution. University of Chicago Press: Chicago, USA. [Google Scholar]
- Thrall PH, Laine AL, Ravensdale M, Nemri A, Dodds PN, Barrett LG et al. (2012). Rapid genetic change underpins antagonistic coevolution in a natural host-pathogen metapopulation. Ecol Lett 15: 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizoso DB, Ebert D (2004). Within-host dynamics of a microsporidium with horizontal and vertical transmission: Octosporea bayeri in Daphnia magna. Parasitology 128: 31–38. [DOI] [PubMed] [Google Scholar]
- Vizoso DB, Lass S, Ebert D (2005). Different mechanisms of transmission of the microsporidium Octosporea bayeri: a cocktail of solutions for the problem of parasite permanence. Parasitology 130: 501–509. [DOI] [PubMed] [Google Scholar]
- Wilfert L, Schmid-Hempel P (2008). The genetic architecture of susceptibility to parasites. BMC Evol Biol 8: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittner M, Weiss LM (1999). The Microsporidia and Microsporidiosis. ASM Press: Washington, D.C. [Google Scholar]
- Woolhouse MEJ, Webster JP, Domingo E, Charlesworth B, Levin BR (2002). Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet 32: 569–577. [DOI] [PubMed] [Google Scholar]
- Zbinden M, Haag CR, Ebert D (2008). Experimental evolution of field populations of Daphnia magna in response to parasite treatment. J Evol Biol 21: 1088–1078. [DOI] [PubMed] [Google Scholar]