Abstract
Adaptation to low temperatures has been reasonably developed in the human species during the colonization of the Eurasian landmass subsequent to Out of Africa migrations of anatomically modern humans. In addition to morphological and cultural changes, also metabolic ones are supposed to have favored human isolation from cold and body heat production and this can be hypothesized also for most Neandertal and at least for some Denisovan populations, which lived in geographical areas that strongly experienced the last glacial period. Modulation of non-shivering thermogenesis, for which adipocytes belonging to the brown adipose tissue are the most specialized cells, might have driven these metabolic adaptations. To perform an exploratory analysis aimed at looking into this hypothesis, variation at 28 genes involved in such functional pathway was investigated in modern populations from different climate zones, as well as in Neandertal and Denisovan genomes. Patterns of variation at the LEPR gene, strongly related to increased heat dissipation by mitochondria, appeared to have been shaped by positive selection in modern East Asians, but not in Europeans. Moreover, a single potentially cold-adapted LEPR allele, different from the supposed adaptive one identified in Homo sapiens, was found also in Neandertal and Denisovan genomes. These findings suggest that independent mechanisms for cold adaptations might have been developed in different non-African human groups, as well as that the evolution of possible enhanced thermal efficiency in Neandertals and in some Denisovan populations has plausibly entailed significant changes also in other functional pathways than in the examined one.
Introduction
Adaptation to lower temperatures than those characterizing tropical and hot desert regions has been reasonably developed in the human species within the last 100 000 years, during the colonization processes of the Eurasian landmass subsequent to Out of Africa migrations of anatomically modern humans. The millennial settlement of certain human groups in temperate and boreal environments has shaped their morphological and cultural features, but also metabolic adaptations are supposed to have favored their isolation from cold and body heat production (Leonard et al., 2002).
This picture can be further extended to Neandertals and, perhaps, to some populations of Denisovans, as both paleoanthropological remains and genetic data suggest that their appearance and extinction took place in Late Pleistocene, mostly in geographical areas that strongly experienced the last glacial period (Lalueza-Fox and Gilbert, 2011) and for which low average temperatures have been estimated (Skrzypek et al., 2011). Accordingly, it can be hypothesized that also most Neandertal populations, and plausibly some Denisovan ones, have coped with significantly lower temperatures with respect to those experienced by their ancestors, and for a time interval long enough to adapt to them. Neandertals especially exhibited morphological traits that probably improved their thermal efficiency (Holliday, 1997) or that can be interpreted as a consequence of worse blood circulation due to cold-caused vasoconstriction (Steegmann, 2007). Moreover, their supposed massive muscular apparatus and hunter-gatherer subsistence strategies suggest that their lifestyle could be similar to that of modern populations from circumpolar regions, being characterized, at least at upper latitudes, by a diet rich in fat and meat proteins (El Zaatari et al., 2011), able to satisfy high calorie needs (that is, 4000–7000 kcal per day) (Snodgrass and William, 2009). Such features might have improved cold adaptation in this archaic species, being probably coupled with the high basal metabolic rate that is generally correlated with low temperatures in modern populations (Leonard et al., 2002).
In particular, metabolic adaptations to not tropical environments might be driven by modulation of thermogenesis and thermoregulation processes, which are complex functional pathways controlled by sympathetic signals of hypothalamus produced in response to cold exposure. This signaling induces the release of free fatty acids from triglycerides, activating thermogenin, which uncouples oxidative phosphorylation and ATP synthesis, leading to dissipation of the energy produced within mitochondria as heat (Wijers et al., 2008). Adipocytes belonging to the brown adipose tissue (BAT) are the most specialized cells in such non-shivering thermogenesis, according to a greater amount of mitochondria with respect to white fat adipocytes and to higher concentration of thermogenin in their inner membrane (Richard and Picard, 2011). In fact, BAT is especially abundant in hibernating mammals, but it has been found also in some apes, mainly in Macaca mulatta, one of the primates with the broadest geographical distribution beyond the human species. Moreover, although until few years ago the evidence of BAT presence in Homo sapiens was only limited to neonates, it has been recently observed also in adults (Cypess et al., 2009; Virtanen et al., 2009), appearing to be more metabolically active in response to cold exposure (Saito et al., 2009; van Marken Lichtenbelt et al., 2009).
Therefore, genes involved in BAT metabolism, storage and neogenesis reasonably constitute one of the main pathways responsible for thermogenesis and thermoregulation also in the human lineage. Genetic variation at these genes has thus the potential to represent one of the keys for an enhanced thermal efficiency and for metabolic adaptations to low temperatures in both modern and archaic humans.
To perform an exploratory analysis aimed at looking into this hypothesis, patterns of variation with potentially functional impact at 28 BAT genes were investigated in populations from different climate zones, as well as in Neandertal and Denisovan genomes. This approach promises to contribute to shed new light on the evolution of mechanisms involved in human body heat production and in those of our ancestors.
Materials and methods
Investigated genomic regions
A panel of 28 genes (Table 1) directly involved in BAT metabolism, or with potential regulatory functions on it, was selected by accurate literature survey and by exploring protein–protein interactions reported on the String database (http://string-db.org/). The panel includes also genes with still unknown biological functions, but associated with fat accumulation and body mass index according to results of genome-wide association studies reported on the Catalog of Published Genome-Wide Association Studies (www.genome.gov/gwastudies).
Table 1. Panel of investigated genes.
| Gene | Chr | Role | Referencesa |
|---|---|---|---|
| ADRA1A | 8 | G-protein-coupled receptor | Strosberg (1993) |
| ADRB3 | 8 | G-protein-coupled receptor | Bea et al. (2010) |
| CIDEA | 18 | Suppresses UCP1 activity | Puri et al. (2008) |
| CREB1 | 9 | Induces DIO2 and PGC-1 | Iourgenko et al. (2003) |
| DIO2 | 14 | Increases UCP1 expression | de Jesus et al. (2001) |
| FTO | 16 | Associated with BMI | Frayling et al. (2007) |
| HOXC4 | 12 | Involved in BAT differentiation | Gesta et al. (2007) |
| HOXA1 | 7 | Involved in BAT differentiation | Gesta et al. (2007) |
| LIPE | 19 | Hormone-sensitive lipase | Hoffstedt et al. (2001) |
| LEP | 7 | Regulates fat storage | Myers (2004) |
| LEPR | 1 | Regulates fat storage | Cinti et al. (1997) |
| NRF1 | 7 | Regulates mitochondrial biogenesis | Sawada et al. (2010) |
| NRIP1 | 21 | Regulates adipocyte differentiation | Seale et al. (2007) |
| PLIN1 | 15 | Coats lipid droplets of adipocytes/prevents LIPE action | Blanchette-Mackie et al. (1995) |
| PLIN2 | 9 | Coats lipid droplets of adipocytes/prevents LIPE action | Blanchette-Mackie et al. (1995) |
| PLIN3 | 19 | Coats lipid droplets of adipocytes/prevents LIPE action | Blanchette-Mackie et al. (1995) |
| PLIN5 | 19 | Coats lipid droplets of adipocytes/prevents LIPE action | Blanchette-Mackie et al. (1995) |
| PPARG | 3 | Regulates adipocyte differentiation | Ristow et al. (1998) |
| PPARGC1A | 4 | Regulates adipocyte differentiation | Wolf (2009) |
| PPARGC1B | 5 | Regulates adipocyte differentiation | Ristow et al. (1998) |
| PRDM16 | 1 | Induces BAT differentiation | Seale et al. (2008) |
| PRKAR1A | 17 | Involved in regulation of lipid/glucose metabolism | McKnight et al. (1998) |
| PRKAR2A | 3 | Involved in regulation of lipid/glucose metabolism | McKnight et al. (1998) |
| PRKAR1B | 7 | Involved in regulation of lipid/glucose metabolism | McKnight et al. (1998) |
| PRKAR2B | Involved in regulation of lipid/glucose metabolism | McKnight et al. (1998) | |
| UCP1 | 4 | Uncoupling protein | Cannon and Nedergaard (2004) |
| UCP2 | 11 | Uncoupling protein | Kovacs et al. (2005) |
| UCP3 | 11 | Uncoupling protein | Vidal-Puig et al. (2007) |
Abbreviations: BMI, body mass index; Chr, chromosome.
Listed in the Supplementary References file.
Exploited data sets
Sequence data from the 50 × and 30 × coverage Neandertal and Denisovan genomes aligned to hg19 human reference sequence were retrieved at http://cdna.eva.mpg.de/neandertal/altai/ and at https://bioinf.eva.mpg.de/download/HighCoverageDenisovaGenome/DenHC_catalog/, respectively. Reads covering the exons of the selected genes were extracted with ad hoc developed Phyton scripts and used to search for nucleotide substitutions.
As regards to modern populations, single-nucleotide polymorphisms (SNPs) located in the genomic regions under consideration were retrieved from those identified in the 1092 individuals belonging to the 14 human groups included in the 1000 Genomes Project phase I data set and confirmed also by the inspection of high-coverage exome alignments (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/phase1/).
SNPs identification
Bcftools utilities of the SAMtools package (Li et al., 2009) were used to call variants on collected reads from BAM archaic genomes by applying the same parameters setting used to identify 1000 Genomes Project SNPs (1000 Genomes Project Consortium et al., 2012), in order to obtain perfectly comparable modern and archaic data sets.
Variants identified respectively in the Neandertal and Denisovan genomes were searched for in the modern data set, and also genomic positions related to modern SNPs were cross-checked in the archaic genomes. Accordingly, we obtained a data set of 406 SNPs already annotated in the human species according to the dbSNP Build 137 and for which the ancestral alleles have been inferred by means of comparison with the chimpanzee genome.
Summary statistics and population structure analyses on modern variation
The complete data set of SNPs was used to compute summary statistics, such as nucleotide diversity (π), mean expected heterozygosity across loci (MEH) and number of polymorphic sites (S), for each present-day human group, by using the Arlequin package v.3.5.1.2.
Moreover, this data set was used to perform several population structure analyses (Supplementary Materials and Methods) aimed at contextualizing archaic variation into the modern one. For this purpose, the data set was filtered using PLINK v.1.07 (Purcell et al., 2007) to remove singletons and to prune SNPs according to their linkage disequilibrium (LD) levels, in the attempt to avoid LD effects on multivariate and admixture analyses, as well as on the adopted model-based clustering method. Loci in LD were removed by using a sliding windows approach. Windows of 50 SNPs, with LD being calculated between each possible pair of SNPs, were used and one of a pair of SNPs was removed if pairwise genotypic correlation (r2) was >0.1. Each window was subsequently shifted 10 SNPs forward and the same procedure was repeated, obtaining a pruned subset of 244 SNPs in approximate linkage equilibrium with each other.
Population groups differentiation analyses
Patterns of differentiation among clusters of genetically homogeneous populations pointed out by structure analyses (Supplementary Materials and Methods) were investigated by grouping samples according to their predominant ancestry component, such as African (AFR), European (EUR), East Asian (EAS) and Latin American (AMR) ones. Detection of the most differentiated genes among those under investigation was carried out by computing Fst index for each of the 406 identified SNPs, after removing those monomorphic in both the compared groups and by using the R package pegas. A χ2-test was also performed using PLINK v.1.07 to compare allele frequencies between samples and, according to the high correlation observed between χ2 and Fst values, it was used to provide an additional evidence of statistical significance related to the differentiation level of each SNP. Adjusted p-values for the Bonferroni correction were calculated for obtained asymptotic p-values using the R package multtest, in order to account for the adopted multiple testing procedures.
To identify the most plausible targets of selection, SNPs were divided into bins according to their global minor allele frequency and ranked within each bin on the base of their Fst values. Variants scoring in the top 1% of Fst distribution in each bin were then retained for subsequent analyses. This approach enabled to take into account also SNPs with moderate Fst, but representing potential outliers among those belonging to the same frequency interval.
Positions of the identified candidate SNPs within empirical genome-wide Fst distributions were then investigated to definitively identify genes with unusual high differentiation with respect to genomic patterns. In particular, six Fst genome-wide distributions were obtained by comparing the four continental groups of 1000 Genomes Project populations by means of an ad hoc developed Perl script and SNPs scoring in their 99th percentiles were considered as significant outliers.
Neutrality tests on candidate loci
Values of Integrated Haplotype Score (iHS) calculated on the HapMap Phase II and HGDP data sets were retrieved for the best candidate loci identified by differentiation analysis using Haplotter (http://haplotter.uchicago.edu/) and HGDP Selection Browser (http://hgdp.uchicago.edu/cgi-bin/gbrowse/HGDP/) web applications.
Full-gene sequence data for these loci were also retrieved from the 1000 Genomes Project phase I data set and used to compute several summary and neutrality statistics. In particular, the DnaSP package v.5.10 was used to calculate the average number of pairwise differences (Pi), the Watterson's nucleotide diversity estimator (θW), Tajima's D, Fu and Li's D* and F*, as well as Fay and Wu's H, on 10 kb non-overlapping sliding windows. The Fastsimcoal package v.2 (Excoffier et al., 2013) was used to perform 10 000 coalescent simulations conditioned on the local recombination rate and on the demographic models described by Gravel et al. (2011). The above-mentioned statistics were then calculated on simulated data sets with the DnaSP package v.5.10 and used to generate distributions of values under a neutral model of evolution. Estimates for each statistic were thus compared with obtained distributions in one-tailed tests and their significance resulted as the portion of coalescent simulations carrying more extreme values than the observed ones. Adjusted p-values for the adaptive Benjamini and Hochberg (ABH) procedure (Benjamini et al., 2006) were computed for obtained p-values using the R package multtest, in order to control the false discovery rate at α=0.05.
LD, haplotype and network analyses
Sequence data for a genomic interval covering the whole best candidate gene, as well as 100 kb upstream and downstream regions, were retrieved from the 1000 Genomes Project phase I data set and used to calculate pairwise LD for each possible SNP pair with PLINK v.1.07.
SNPs in high LD (r2>0.95) with the most promising candidate variant were used to reconstruct haplotypes using the Bayesian algorithm implemented in PHASE v.2.1. Evolutionary relationships of inferred haplotypes were then visualized by means of a median joining network using the Network package v.4.6.1.1 (http://www.fluxusengigeering.com).
Scripts used in the present study are available upon request to the corresponding author.
Results
Sequence variability of BAT genes
A total of 406 SNPs annotated according to the dbSNP Build 137 were observed in the modern data set (Supplementary Table 1), with derived alleles of 12 of them being found also in archaic species. In particular, both Neandertal and Denisovan genomes showed derived alleles for PLIN1 rs6496589 and PPARGC1B rs7732671, as well as for PLIN1 rs2304796, which were observed at high and moderate frequency (⩾76%, ⩾84%, up to 34%, respectively) in all modern populations; for PLIN1 rs150128694 that was present only in few AFR individuals (1%) and for LEPR rs1137101 that was highly represented in EAS (87%). Derived alleles of other six SNPs were observed only in the Neandertal genome, with LEPR rs6413506 being present only in few AFR individuals (4%), PPARG rs3856806 and PPARGC1B rs45520937, rs45588534 and rs45543631 being found at worldwide moderate frequency (up to 20%) and NRF1 rs3735006 being observed at low frequency (up to 7%) in all modern populations. Finally, the derived allele of PPARG rs41516544, which was present only in few AFR individuals (2%), was found in heterozygous state in the Denisovan genome.
Summary statistics and population structure for modern variation
Summary statistics of sequence variability for modern populations are reported on Supplementary Table 2 and acronyms of each population are repeated also in the legend of Supplementary Figure 1. As expected, nucleotide diversity was higher in ethnic groups with a predominant AFR ancestry with respect to not admixed Eurasian populations, being related to considerably larger percentages of polymorphic loci. Nevertheless, the highest heterozygosity values were found in EAS and northern EUR populations, with the exception of an unusual peak in Iberian populations from Spain (IBS) that could be explained by its high heterogeneity due to the inclusion of individuals from different populations belonging to the Iberian Peninsula, coupled with a small sample size.
Significant Fst genetic distances were computed for the great majority of pairwise population comparisons with few exceptions (Han Chinese from Beijing, CHB vs Han Chinese from Southern China, CHS, P=0.714; Colombians from Medellin, CLM vs Puerto Ricans, PUR, P=0.188; Utah residents with Northern and Western European ancestry, CEU vs British from England and Scotland, GBR, P=0.238 and most of comparisons involving IBS). The obtained matrix of pairwise distances (Supplementary Table 3) was graphically represented by means of a metric multidimensional scaling (MDS) that revealed an appreciable pattern of geographical structure for the observed variation, pointing out the presence of four clusters roughly corresponding to continental groups of populations (Supplementary Figure 2). Again, the sole exception was represented by the IBS sample that appeared to be more closely related to CLM and PUR than to other EUR. This pattern was statistically supported by analysis of the molecular variance (AMOVA) results (Supplementary Table 4) which showed significant, but relatively low, differentiation among worldwide populations (FST=0.075, P<0.001) and, when samples were subdivided into geographically based clusters, moderate and significant among-groups component of variance (FCT=0.081, P<0.001) that only slightly decreased when IBS were clustered with AMR populations, as suggested by MDS plot (FCT=0.079, P<0.001).
Contextualizing archaic variation into the modern one
In order to put archaic profiles into the context of modern variation, further analyses of population structure were performed including also Neandertal and Denisovan data and using a subset of 244 SNPs in approximate linkage equilibrium with each other.
The model with the best predictive accuracy for admixture analysis inferred three ancestral groups, roughly corresponding to population clusters of predominant AFR, EUR and EAS ancestry, with EUR and AMR populations being assigned to the same cluster (Supplementary Figure 1). At K=3 archaic genes showed a predominant AFR-like ancestry component (62.8%) and a more limited EAS-like one (37.2%). Such signatures were observable at both low and high resolution (Supplementary Figure 3), with the latter basically showing moderate percentages (23.9% at K=2 and 38.7% at K=4) and disappearing at K=5.
Discriminant analysis of principal components (DAPC) was then applied to 20 of the principal components (PCs) computed for modern data and archaic samples were represented as ‘supplementary individuals'. The AFR cluster showed the lowest mean membership probability (0.898), with the smallest single values related to people with African ancestry from Southwest United States (ASW) 29.5% of which were assigned to the EUR/AMR cluster, whereas values for EAS and EUR/AMR clusters were noticeably higher (0.951 and 0.966, respectively).
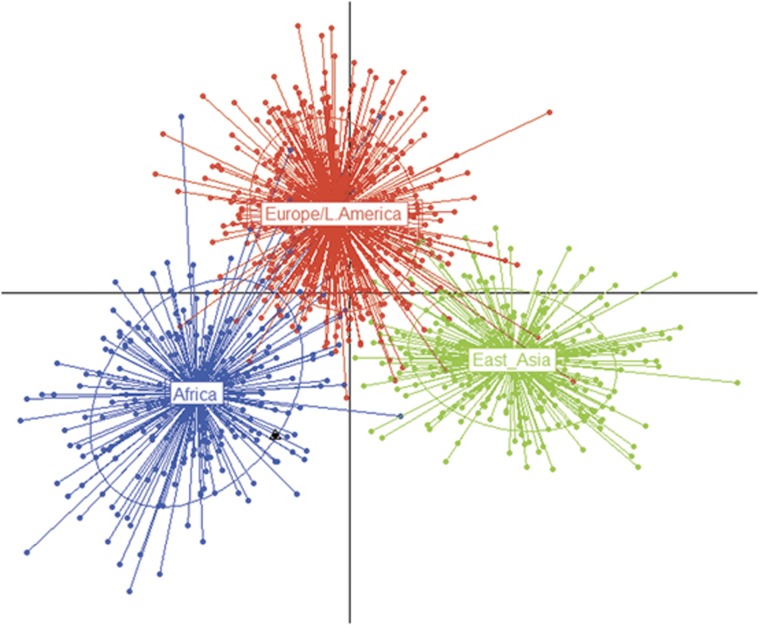
Archaic samples were again predicted to be essentially part of the AFR cluster (Figure 1), showing an AFR membership probability of 0.989, with respect to 0.001 and 0.009 for EAS and EUR/AMR components, respectively.
Figure 1.
First and second PCs of DAPC. Inferred population clusters are indicated by colors and ellipses, which model 95% of the corresponding variability. Archaic samples are represented as ‘supplementary individuals' and are overlapping, being indicated by a black dot and a white triangle.
A model-based clustering analysis finally confirmed the presence of three clusters that can be approximated as ‘AFR' (C1), ‘EUR/AMR' (C2) and ‘EAS' (C3) ones (Supplementary Figure 4). In particular, archaic samples, 94% of the examined AFR individuals, 5.3% of the EUR, 7.2% of the AMR and 0.7% of the EAS ones composed cluster C1. C2 included 67% of the EUR samples and 63% of the AMR ones, as well as 4.1% and 1.1% of AFR and EAS individuals, respectively. C3 was composed by 98% of the examined EAS samples, 30% of the AMR, 28% of the EUR and 2% of the AFR ones.
Differentiation of modern population groups
Detection of the most differentiated genes among groups of genetically homogeneous populations identified by structure analyses was obtained by computing Fst index and the highly correlated χ2-test (for example, r=0.992, P<0.0001 for AFR–EAS comparison) for each SNP. Variants scoring in both the top 1% Fst tail of each frequency bin and 99th percentiles of computed Fst genome-wide distributions were then pointed out as unusually differentiated loci.
Outlier SNPs that might have exerted an adaptive cold-related role according to high frequency of their derived alleles out of Africa were identified only in comparisons involving EAS. In fact, highly differentiated alleles in AFR–AMR and EUR–AMR comparisons showed only low to moderate frequencies in non-Africans (up to 0.392 in AMR and 0.339 in EAS for PRKAR2B rs75385144 and FTO rs2287142, respectively) (Table 2).
Table 2. Outlier SNPs scoring in 99th percentiles of Fst bins and genome-wide distributions obtained by comparing modern continental groups of populations.
| Comp | Chr | SNP | MA | F_AFR | F_EUR | F_EAS | F_AMR | Adj_p | Fst | Gene | FE |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AFR/EAS | 1 | rs1137100 | G | 0.128 | 0.289 | 0.811 | 0.265 | 6.85E-107 | 0.64 | LEPR | M |
| AFR/AMR | 7 | rs75385144 | T | 0.022 | 0.338 | 0.367 | 0.392 | 1.26E-41 | 0.37 | PRKAR2B | S |
| EUR/EAS | 1 | rs1137100 | G | 0.128 | 0.289 | 0.811 | 0.265 | 8.98E-76 | 0.43 | LEPR | M |
| 1 | rs1805096 | G | 0.573 | 0.600 | 0.126 | 0.536 | 2.15E-65 | 0.38 | LEPR | S | |
| EUR/AMR | 16 | rs2287142 | A | 0.004 | 0.003 | 0.339 | 0.102 | 1.79E-14 | 0.13 | FTO | I |
| EAS/AMR | 1 | rs1137100 | G | 0.128 | 0.289 | 0.811 | 0.265 | 2.04E-59 | 0.47 | LEPR | M |
| 1 | rs1137101 | Ga | 0.539 | 0.496 | 0.867 | 0.423 | 1.95E-44 | 0.37 | LEPR | M |
Abbreviations: Adj_p, p-values for χ2-test adjusted for Bonferroni correction; Chr, chromosome; Comp, populations comparison; F_, allele frequency; FE, SNP functional effect; I, intronic; M, missense; MA, minor allele; S, synonymous.
No examined SNPs scored in the 99th percentile of AFR/EUR Fst genome-wide distribution.
Derived allele observed also in archaic genomes.
Accordingly, LEPR turned out to be the most promising candidate gene to have been subjected to natural selection, presenting three variants (rs1137100, rs1137101 and rs1805096) which highly differentiate EAS from all the remaining groups and from AMR and EUR, respectively (Table 2). In particular, the rs1137100-derived allele showed very low frequency in AFR (0.128), moderate frequency in EUR (0.289) and AMR (0.265), as well as high frequency in EAS (0.811). The rs1137101-derived allele, which was observed also in archaic genomes, instead showed an even higher frequency in EAS (0.867), but being considerably represented also in the remaining populations (Table 2). On the contrary, the rs1805096-derived allele was common in all the examined groups, with the exception of EAS, probably as a consequence of its appreciable LD (r2=0.76) with the rs1137101 ancestral one.
LEPR neutrality tests and haplotype analyses
To further investigate putative signatures of natural selection on the LEPR gene, patterns of nucleotide diversity at 10 kb non-overlapping windows covering the related 220 858 bp genomic region were evaluated.
Despite the low to moderate Pi and θW values obtained for each population group, in line with the small number of segregating sites, no exceptionally reduced nucleotide diversities were observed according to p-values computed by means of coalescent simulations for 10 kb genomic windows encompassing the three candidate LEPR variants (Table 3).
Table 3. Summary statistics and neutrality tests for candidate LEPR windows.
| Win | Sample | N | S | Pi ( × 10-4) | θW ( × 10-4) | D | F* | D* | H |
|---|---|---|---|---|---|---|---|---|---|
| rs1137100 | AFR | 492 | 84 | 12.2 (0.545) | 12.4 (0.565) | −0.063 (0.536) | −1.357 (0.076) | −2.184 (0.019) | 2.795 (0.572) |
| EAS | 572 | 79 | 11.4 (0.592) | 11.4 (0.518) | 0.006 (0.602) | −3.049 (0.002*) | −5.056 (0.0007*) | 2.081 (0.486) | |
| EUR | 758 | 72 | 11.6 (0.622) | 11.2 (0.442) | 0.121 (0.607) | −2.4367 (0.010) | −3.991 (0.017) | 1.063 (0.449) | |
| AMR | 362 | 58 | 11.5 (0.857) | 8.9 (0.108) | 0.813 (0.864) | 0.993 (0.884) | 2.623 (0.946) | 1.138 (0.457) | |
| rs1137101 | AFR | 492 | 76 | 13.8 (0.805) | 11.218 (0.169) | 0.668 (0.808) | −1.969 (0.025) | −3.864 (0.014) | −0.251 (0.316) |
| EAS | 572 | 70 | 12.5 (0.808) | 10.1 (0.177) | 0.663 (0.811) | −2.379 (0.011) | −4.5134 (0.010) | −5.107 (0.141) | |
| EUR | 758 | 57 | 14.3 (0.950) | 8.8 (0.985) | 1.740 (0.954) | −0.679 (0.263) | −2.568 (0.014) | −0.417 (0.311) | |
| AMR | 362 | 47 | 14.1 (0.947) | 7.3 (0.990) | 2.623 (0.946) | 1.926 (0.939) | 0.739 (0.846) | 0.513 (0.396) | |
| rs1805096 | AFR | 492 | 76 | 9.1 (0.286) | 11.3 (0.879) | −0.546 (0.289) | −2.685 (0.013) | −3.864 (0.010) | −0.797 (0.333) |
| EAS | 572 | 62 | 8.1 (0.454) | 9.0 (0.737) | −0.275 (0.441) | −2.342 (0.011) | −3.497 (0.013) | −4.738 (0.139) | |
| EUR | 758 | 55 | 8.6 (0.580) | 8.8 (0.613) | 0.011 (0.588) | −2.129 (0.024) | −3.300 (0.010) | −1.437 (0.259) | |
| AMR | 362 | 57 | 8.7 (0.551) | 8.8 (0.528) | −0.024 (0.563) | −0.770 (0.215) | −1.176 (0.140) | −1.217 (0.107) |
Abbreviations: AFR, Africans; AMR, Latin Americans; D, Tajima's D; D*, Fu and Li's D* EAS, East Asians; EUR, Europeans; F*, Fu and Li's F* H, Fay and Wu's H; N, n° of chromosomes; Pi, average number of pairwise differences; S, n° of segregating sites; Win, 10 kb window encompassing the candidate SNP; θW, Watterson's nucleotide diversity estimator.
P-values obtained by 10 000 coalescent simulations are reported in brackets.
*Significant after ABH correction for multiple testing.
After controlling the false discovery rate, significantly and largely negative Fu and Li's D* and F* values (−5.056, P=0.0007, adjusted P=0.041 and −3.049, P=0.002, adjusted P=0.044, respectively) were instead obtained for EAS, as concerns the window encompassing rs1137100, suggesting a remarkable departure of allele frequency distribution from patterns expected under neutrality for variation surrounding this SNP. In particular, significance of Fu and Li's D* and F* estimates, but not of Tajima's D, pointed to an excess of singletons in EAS rather than of simply low frequency variants. In fact, in the rs1137100 window they were actually more represented in this group with respect to other ones, accounting for 41% of total segregating sites.
Patterns of LD were then explored in a genomic region extending 100 kb upstream and downstream with respect to the LEPR gene and leading to the identification of 10 intronic SNPs in high LD (r2⩾0.95) with rs1137100. These variants were used to reconstruct LEPR haplotypes characterizing modern populations and their evolutionary relationships were visualized by means of a median joining network (Figure 2). Out of a total of 20 inferred haplotypes, only two (that is, H1 and H20) overall accounted for 96% of the sampled chromosomes, pointing to the unlikely adaptive function of the remaining ones. Both of these common haplotypes were cosmopolitan ones, with H20 carrying the ancestral allele for each SNP and representing the predominant haplotype in all continental groups (frequency exceeding 58%), but EAS. On the contrary, H1 carried the derived allele for each SNP and showed moderate frequency in all examined groups (up to 36%), with the exception of EAS, in which it accounted for about 60% of chromosomes, thus having potentially exerted an adaptive role.
Figure 2.
Median joining network of haplotypes carrying LEPR rs1137100 and SNPs in high LD with it (r2⩾0.95). AFR are displayed in blue, EAS in green, EUR in red and AMR in yellow. Nodes are proportional to haplotype frequencies, whereas branch lengths are proportional to the number of variants occurred in the sequences.
As mentioned above, the remaining haplotypes were extremely rare, showing frequencies lower than 1–2% in all continental groups. Among them, six (that is, H2, H3, H6, H12, H14, H16) carried the rs1137100-derived allele, but being located on two different branches of the observed topology. In particular, H3 appeared to be the haplotype from which H2, H1 and H6 have originated as one-, two- and three-step derivatives, whereas H12, H14 and H16 resulted more closely related to the ancestral haplotype H20 (Figure 2). Accordingly, in addition to rs1137100, other variants turned out to be recurrent in the reconstructed topology, probably as a consequence of recombination or gene conversion events instead of representing actual homoplasies.
Discussion
In the last decade, studies aimed at exploring the genetic bases of human adaptation have moved from the investigation of few well-characterized adaptive traits, among others lactase persistence in adulthood, light skin pigmentation in Europeans and high-altitude adaptation, to genome-wide scans able to detect signatures of natural selection scattered along the entire human genome.
This approach has succeeded in pointing out a number of genes showing variation patterns consistent with departures from a neutral model of evolution and most of these loci have been proved to have an essential role in several metabolism functions (Hancock et al., 2008; Hancock et al., 2011; Hernandez et al., 2011; Klimentidis et al., 2011; Grossman et al., 2013). However, leading to the identification of large candidate regions and, in most cases, lacking of a priori hypotheses based on the knowledge of potentially adaptive traits, these studies have encountered considerable difficulties in pinpointing actual adaptive variants (Grossman et al., 2010).
That being so, fine-mapping studies focused on candidate regions highlighted by genome-wide surveys and addressing to well-defined biological questions are still fundamental to deepen the knowledge about the adaptive events occurred during our evolutionary history.
According to this rationale, the present study investigated sequence variability with a potential functional impact on highly selected genes involved in BAT metabolism, storage and neogenesis. This enabled the comparison of modern patterns of variation at different climate zones, as well as of modern and archaic sequences, related to one of the main pathways responsible for non-shivering thermogenesis, in search for signatures of metabolic adaptations to low temperatures in modern and archaic humans.
Although the highest genetic diversity was found in AFR populations, in accordance with the known landscape of human genetic variation (Li et al., 2008), heterozygosity turned out to be systematically larger in EAS and northern EUR groups (Supplementary Table 2), emphasizing that a considerable fraction of derived alleles at the examined loci reached remarkable frequencies out of Africa.
In fact, despite only variation at the exons of BAT genes was explored, results from both AMOVA (Supplementary Table 4) and population structure analyses (Figure 1) confirmed the presence of an appreciable geographical apportionment of genetic diversity, which roughly reflects divergence of populations according to continental clusters.
Structure analyses also enabled to set archaic variation into the landscape of modern one. For instance, admixture analysis inferred the presence of three hypothetical ancestral populations corresponding to clusters of predominant AFR, EUR and EAS ancestry (Supplementary Figure 1), also highlighting that archaic samples show a predominant AFR-like component and a moderate EAS-like one. This pattern was confirmed by results from both DAPC and cluster analyses, for which archaic sequences lie within the bulk of AFR samples (Figure 1) and in a cluster showing extremely low percentages of EUR and EAS subjects (Supplementary Figure 4).
These findings appear to be consistent with the supposed Neandertal and Denisovan positions within the phylogenetic tree of our ancestry (Green et al., 2010; Reich et al., 2010), and the limited sharing of BAT-derived alleles between archaic and modern genomes (2.9%), especially non-AFR ones, may also suggest the presence of completely different mechanisms of cold adaptation in these species.
To identify the best candidate genes that might be subject to natural selection in modern non-AFR populations, thus being potentially involved in their metabolic adaptation to low temperatures, differentiation among genetically homogenous population groups pointed out by structure analyses was investigated. In fact, significantly increased levels of differentiation in, or close to, genes are long expected to contribute a first hint about the action of natural selection (Barreiro et al., 2008).
The obtained picture of population differentiation highlighted significant outlier SNPs with respect to computed genome-wide Fst distributions for the LEPR, PRKAR2B and FTO genes (Table 2). However, PRKAR2B and FTO resulted unusually differentiated only between AFR–AMR and EUR–AMR groups and for two non-functional SNPs that show moderate frequencies out of Africa. Accordingly, SNPs that have plausibly exerted an adaptive role in non-tropical environments appear to be restricted to EAS and to be located on the LEPR gene (rs1137100, rs1137101 and rs1805096). This suggests that diverse mechanisms for independent cold adaptations in different non-AFR groups plausibly occurred, as observed for the evolution of light skin color (Norton et al., 2007).
LEPR thus turned out to be the best candidate selected gene, showing considerably higher frequencies of rs1137100- and rs1137101-derived alleles in EAS (0.811 and 0.867, respectively) with respect to the remaining population groups and an opposite pattern for the rs1805096-derived allele (Table 2).
Nevertheless, differentiation results are not sufficient at all to claim for signatures of positive selection and the detected high divergence levels between continental groups could be simply due to the diverse demographic histories of examined populations, rather than to the differential action of selective pressures on them. In fact, findings obtained by differentiation analysis are only partially in line with those reported by previous genome-wide scans. For instance, a study aimed at detecting signatures of selection by investigating correlations between allele frequencies and climate variables found strong correlations between the frequency of some LEPR variants and winter severity (Hancock et al., 2008). However, subsequent analyses exploiting both similar and innovative-related approaches did not identify this gene as a reliable target of selection (Hancock et al., 2011; Raj et al., 2013). Although this may be due to differences in the examined populations, in the sets of investigated SNPs per gene, as well as in possible adopted allele frequency thresholds, this can also point to the potential role of other evolutionary forces, rather than selection, in conditioning LEPR outlier position with respect to average levels of genomic differentiation.
To reject this possibility, we further investigated putative footprints of selection on the most promising LEPR SNPs by using both genotyping and sequence data. For instance, iHS values were retrieved for the obtained candidates from the HapMap Phase II database, showing extreme results only in EAS groups for rs1137100 (iHS=−2.915) and rs1137101 (iHS=−2.939), as well as a barely significant value for rs1805096 (iHS=2.088). Negative scores for rs1137100 and rs1137101 suggest that positive selection might have acted on their derived alleles, whereas the ancestral allele of rs1805096 appears to be the putative target of selection, even if this could be most likely explained by its LD with the rs1137101-derived allele. A significant value for the whole LEPR gene (empirical P=0.003), which lies within the top 1% tail of overall iHS scores in EAS (Voight et al., 2006), was also observed and iHS computed on HGDP data confirmed this pattern, according to a maximum score of −2.939 and to remarkable long strings of consecutive SNPs with extreme values (Li et al., 2008).
A sliding windows approach was then used to compute several diversity and neutrality statistics on full LEPR sequence data retrieved from the 1000 Genome Project, leading to the identification of a significant departure of allele frequency distribution from a neutral model of evolution in EAS. In particular, an actually significant excess of singletons was observed only in the 10 kb genomic interval centered on rs1137100 (Fu and Li's D*=−5.056, adjusted P=0.041 and Fu and Li's F*=−3.049, adjusted P=0.044), pinpointing this SNP as the most plausible candidate adaptive variant.
Exploration of LD patterns along 100 kb upstream and downstream LEPR regions also enabled the reconstruction of haplotype structure related to rs1137100 and to 10 intronic SNPs in high LD with it (Figure 2). The haplotype that carries derived alleles for each SNP (H1) was moderately represented in all continental groups, reaching instead high frequency (about 60%) in EAS. On the contrary, other haplotypes which include the rs1137100-derived allele were observed at extremely low frequencies, even in EAS, suggesting that the adaptive role of this SNP has been mainly exerted at the population level in the context of the H1 genetic background. Nevertheless, SNPs in high LD with rs1137100 are located on LEPR introns and no impact of their derived alleles on protein expression or splicing patterns is reported in the literature and public databases. Accordingly, rs1137100 appears to be the sole adaptive LEPR SNP and its distribution on rare haplotypes located in divergent branches of the reconstructed topology seems to be plausibly due to recombination or gene conversion events. It can be thus hypothesized that a strong selective pressure has acted on rs1137100 during early evolutionary history of EAS populations, having probably decreased in strength in more recent times, thus allowing recombination to erode patterns of LD linked to the selective sweep and to heavily influence the observed haplotype structure.
That being so, several complementary approaches pointed out variation at the LEPR gene as the sole among those investigated that could have realistically contributed to metabolic adaptation of EAS populations to not tropical climates. This gene encodes for the receptor responsible for the binding of leptin in hypothalamic regions and for the release of noradrenalin, whose targets are the ADRB3 and ADRA1A genes, which initiate signaling pathways related to fatty acid processing in adipose tissue. Accordingly, it mediates regulation of satiety and energy balance by modulating alimentary behavior, fat storage and glucose metabolism, also allowing leptin to inhibit insulin secretion (Myers, 2004). It also has a role in increasing norepinephrine turnover in BAT, mainly by enhancing its sympathetic nerve activity (Haynes et al., 1997).
In particular, LEPR SNPs showing some signatures of selection in EAS have been reported to be involved in specific metabolic patterns and/or disorders. For instance, rs1137100 is responsible for a non-synonymous substitution (K109R) that is found to be associated with increased respiratory quotient (that is, increased basal metabolic rate) (Loos et al., 2006), which is consistent with its important impact on non-shivering thermogenesis. Its derived allele was also reported to affect glucose tolerance and insulin response, being found to be associated with fasting insulin levels and insulin response in individuals with impaired glucose tolerance, suggesting that it can have a role in types II diabetes pathogenesis (Wauters et al., 2001). In fact, it reduces insulin inhibition by leptin, leading to the failure of insulin levels regulation and to its increased release, thus accelerating glucose intake and basal metabolic rate (that is, favoring heat dissipation), but also potentially leading to a condition of insulin resistance. Accordingly, this variant has the potential to be deleterious in hot climates, in which it has indeed maintained a low frequency, becoming progressively advantageous in colder climates due to the related increase in basal metabolic rate, and thus in heat dissipation that is the base of non-shivering thermogenesis.
Although the action of positive selection on rs1137101 in modern humans can be hypothesized only according to genotyping data, being questioned by results from sequence ones (Fu and Li's D*=−4.5134, adjusted P=0.068 and Fu and Li's F*=−2.379, adjusted P=0.069), it represents a very interesting SNP as its derived allele was the sole modern one with a proved functional impact to be observed also in archaic genomes. In particular, it is responsible for a non-synonymous substitution (Q223R) that is found to be associated with increased insulin release, body weight, body mass index and risk of obesity in several populations, especially in those from Pacific Islands, and whose impact has been supposed to strongly depend on other genetic and/or environmental factors (Furusawa et al., 2010). Although it represents the sole potentially cold-adapted allele shared between modern and archaic genomes, examination of single Denisovan and Neandertal individuals prevent to draw reliable conclusions about its actual impact on cold-adaptation of these species. In particular, we have no possibility to test whether this SNP was actually overrepresented in archaic populations from cold regions (for example, Siberia from which the examined samples derive) with respect to southern ones. In fact, reaching considerable frequency also in modern AFR (0.539) and being present in both the archaic species, rs1137101 could be supposed to have anciently arisen in the human lineage and this may explain its finding also in the sole until now sequenced Denisovan sample, irrespective to its potential role in increased thermogenesis and to the supposed sub-tropical Southern East Asian origin of Denisovans (Meyer et al., 2012).
Accordingly, the hypothesis that some Neandertal and Denisovan populations have adapted to cold environments through changes on other functional pathways than those implicated in modern humans appears to be the most plausible one.
That being so, in line with recent findings highlighted by transcriptomic analyses carried out on non-human mammals, disentangling the impact of selection also on pathways involved in shivering thermogenesis may represent a complementary and promising approach to investigate human adaptation to low temperatures. In fact, Cheviron et al. (2012) have proposed that enhanced capacity to use lipids as primary metabolic fuel source has been selected in high-altitude deer mice through significant changes in the expression of genes involved in fatty-acid oxidation and oxidative phosphorylation. This mechanism has been thus demonstrated to contribute to increase shivering thermogenesis, being also suggested to be more widespread in cold-adapted mammals than previously expected.
In conclusion, modifications in the BAT pathway that are supposed to contribute to the enhancement of heat dissipation by mitochondria have been detected in modern EAS populations, being only speculated for the Neandertal and Denisovan ones, but representing promising loci for follow-up functional studies. In fact, variation surrounding LEPR rs1137100 seems to have been actually shaped by positive selection in EAS, whereas only the potentially cold-adapted LEPR rs1137101 has been observed in archaic species. This suggests that both convergent evolution of modern and archaic increased thermogenesis mediated by the BAT pathway or the introgression of related archaic cold-adapted alleles into modern genomes have been unlikely occurred.
Moreover, adaptation to temperate climates appears to have been differently achieved in diverse modern human groups. Unfortunately, the exploited data set lacks populations from very cold environments, so that observed patterns can be linked only to overall adaptations occurred within broad geographical ranges. Although this limits the potential to identify signatures of specific local adaptions, the absence of signatures of selection for BAT genes at EUR groups might be due also to the fact that their adaptation entails a limited role for these genes. This suggests that other pathways should be considered in further studies (for example, those more strictly related to glucose metabolism or to shivering thermogenesis) or that, in addition to the few expected hard sweeps occurred in certain groups as modern humans Out of Africa migrations (Hernandez et al., 2011), adaptation to not tropical environments may involve also relatively weak selection on individual alleles (that is, polygenic adaptation and/or soft sweeps), in which multiple variants sweep simultaneously without necessarily generating large differences in allele frequencies between populations.
Data archiving
This article does not report new empirical data or software. The exploited data were already deposited at public databases (see the Materials and Methods section).
Acknowledgments
This work was supported by the RFO 2009-2010 grant to DL, by the Italian Ministry for University and Research (MIUR) PRIN 2009 grant 009WXT45Y to RC, by the MIUR PRIN 2010-2011 grant 20108XYHJS to PLM, by the EU RTD Framework Program COST BMBS Action TD1101 and by the MIUR PON grant PON01_02249 to RC. The authors would like to thank Luca Pagani for having provided a script to compute empirical genome-wide Fst distributions on the 1000 Genomes Project data set and the reviewers for their helpful suggestions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. (2008). Natural selection has driven population differentiation in modern humans. Nat Genet 40: 340–345. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Krieger AM, Yekutieli D. (2006). Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93: 491–507. [Google Scholar]
- Cheviron ZA, Bachman GC, Connaty AD, McClelland GB, Storz JF. (2012). Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proc Natl Acad Sci USA 109: 8635–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB et al. (2009). Identification and importance of brown adipose tissue in adult humans. New Engl J Med 360: 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Zaatari S, Grine FE, Ungar PS, Hublin JJ. (2011). Ecogeographic variation in Neandertal dietary habits: evidence from occlusal molar microwear texture analysis. J Hum Evol 61: 411–424. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Dupanloup I, Huerta-Sánchez E, Foll M. (2013). Robust demographic inference from genomic and SNP data. PLoS Genet 9: e1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Naka I, Yamauchi T, Natsuhara K, Kimura R, Nakazawa M et al. (2010). The Q223R polymorphism in LEPR is associated with obesity in Pacific Islanders. Hum Genet 127: 287–294. [DOI] [PubMed] [Google Scholar]
- Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM et al. (2012). An integrated map of genetic variation from 1092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG et al. (2011). Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci USA 108: 11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M et al. (2010). A draft sequence of the Neandertal genome. Science 328: 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A et al. (2013). Identifying recent adaptations in large-scale genomic data. Cell 152: 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Shlyakhter I, Karlsson EK, Byrne EH, Morales S, Frieden G et al. (2010). A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327: 883–886. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Alkorta-Aranburu G, Beall CM, Gebremedhin A, Sukernik R et al. (2011). Adaptations to climate-mediated selective pressures in humans. PLoS Genet 7: e1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Gordon AS, Eshel G, Pritchard JK, Coop G et al. (2008). Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet 4: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes WG, Morgan DA, Walsh SA, Mark AL, Sivitz WI. (1997). Receptormediated regional sympathetic nerve activation by leptin. J Clin Invest 100: 270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Kelley JL, Elyashiv E, Melton SC, Auton A, McVean G et al. (2011). Classic selective sweeps were rare in recent human evolution. Science 331: 920–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday TW. (1997). Postcranial evidence of cold adaptation in European Neandertals. Am J Phys Anthropol 104: 245–258. [DOI] [PubMed] [Google Scholar]
- Klimentidis YC, Abrams M, Wang J, Fernandez JR, Allison DB. (2011). Natural selection at genomic regions associated with obesity and type-2 diabetes: East Asians and sub-Saharan Africans exhibit high levels of differentiation at type-2 diabetes regions. Hum Genet 129: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalueza-Fox C, Gilbert MT. (2011). Paleogenomics of archaic hominins. Curr Biol 21: R1002–R1009. [DOI] [PubMed] [Google Scholar]
- Leonard WR, Sorensen MV, Galloway VA, Spencer GJ, Mosher MJ, Osipova L et al. (2002). Climatic influences on basal metabolic rates among circumpolar populations. Am J Hum Biol 14: 609–620. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S et al. (2008). Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104. [DOI] [PubMed] [Google Scholar]
- Loos RJ, Rankinen T, Chagnon Y, Tremblay A, Perusse L, Bouchard C. (2006). Polymorphisms in the leptin and leptin receptor genes in relation to resting metabolic rate and respiratory quotient in the Quebec Family Study. Int J Obes (Lond) 30: 183–190. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S et al. (2012). A high-coverage genome sequence from an archaic Denisovan individual. Science 338: 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M. (2004). Leptin receptor signaling and the regulation of mammalian physiology. Recent Prog Horm Res 59: 287–304. [DOI] [PubMed] [Google Scholar]
- Norton H, Kittles R, Parra E, McKeigue P, Mao X, Cheng K et al. (2007). Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol 24: 710–722. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj SM, Pagani L, Gallego Romero I, Kivisild T, Amos W. (2013). A general linear model-based approach for inferring selection to climate. BMC Genet 14: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY et al. (2010). Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468: 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard D, Picard F. (2011). Brown fat biology and thermogenesis. Front Biosci 16: 1233–1260. [DOI] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J et al. (2009). High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58: 1526–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek G, Wisniewski A, Grierson PF. (2011). How cold was it for Neanderthals moving to Central Europe during warm phases of the last glaciation? Quat Sci Rev 30: 481–487. [Google Scholar]
- Snodgrass JJ, William RL. (2009). Neandertal energetics revisited: insights into population dynamics and life history evolution. PaleoAnthropology 2009: 220–237. [Google Scholar]
- Steegmann AT. (2007). Human cold adaptation: an unfinished agenda. J Hum Biol 19: 218–227. [DOI] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND et al. (2009). Cold-activated brown adipose tissue in healthy men. New Engl J Med 360: 1500–1508. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T et al. (2009). Functional brown adipose tissue in healthy adults. N Engl J Med 360: 1518–1525. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. (2006). A map of recent positive selection in the human genome. PLoS Biol 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauters M, Mertens I, Rankinen T, Chagnon M, Bouchard C, Van Gaal L. (2001). Leptin receptor gene polymorphisms are associated with insulin in obese women with impaired glucose tolerance. J Clin Endocrinol Metab 86: 3227–3232. [DOI] [PubMed] [Google Scholar]
- Wijers SL, Schrauwen P, Saris WH, van Marken Lichtenbelt WD. (2008). Human skeletal muscle mitochondrial uncoupling is associated with cold induced adaptive thermogenesis. PLoS One 3: e1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.