Abstract
Mussels of the genus Mytilus have been used to assess the circumglacial phylogeography of the intertidal zone. These mussels are representative components of the intertidal zone and have rapidly evolving mitochondrial DNA, suitable for high resolution phylogeographic analyses. In Europe, the three Mytilus species currently share mitochondrial haplotypes, owing to the cases of extensive genetic introgression. Genetic diversity of Mytilus edulis, Mytilus trossulus and Mytilus galloprovincialis was studied using a 900-bp long part of the most variable fragment of the control region from one of their two mitochondrial genomes. To this end, 985 specimens were sampled along the European coasts, at sites ranging from the Black Sea to the White Sea. The relevant DNA fragments were amplified, sequenced and analyzed. Contrary to the earlier findings, our coalescence and nested cladistics results show that only a single M. edulis glacial refugium existed in the Atlantic. Despite that, the species survived the glaciation retaining much of its diversity. Unsurprisingly, M. galloprovincialis survived in the Mediterranean Sea. In a relatively short time period, around the climatic optimum at 10 ky ago, the species underwent rapid expansion coupled with population differentiation. Following the expansion, further contemporary gene flow between populations was limited.
Introduction
Sea mussels of the Mytilus edulis species complex, which consists of three subspecies: M. edulis, Mytilus trossulus, and Mytilus galloprovincialis, are widespread in northern and southern hemispheres (Gérard et al., 2008). All three taxa occur in the coastal water ecosystems surrounding Europe (Gosling, 1992). In Europe, M. trossulus prefers lower salinity waters: it has been identified in the Baltic Sea (Väinölä and Hvilsom, 1991), in Norwegian fjords (Ridgway and Nævdal, 2004), in Scotland (Beaumont et al., 2008), and in the Barents Sea (Väinölä and Strelkov, 2011). Single individuals possessing M. trossulus alleles were found in the Netherlands (Śmietanka et al., 2004). M. edulis ranges from the White Sea and the Barents Sea in the north, through the Atlantic coastal waters to southern France in the south. The range of M. galloprovincialis extends from the Azov and the Black Sea to the British Isles. In the areas where two subspecies coexist, hybridization has been observed. Two well-documented hybridization areas between M. edulis and M. trossulus exist in European waters. The first one extends from the Danish Straits into the Baltic Sea up to the Aland Islands, and the second has recently been discovered in Scotland (Beaumont et al., 2008). A strong, unidirectional genetic introgression from M. edulis to M. trossulus has been observed in the Baltic Sea area (Kijewski et al., 2006). The extreme effect is visible at the mitochondrial DNA (mtDNA) level, for the native M. trossulus mtDNA was replaced by M. edulis mitochondrial genome (Rawson and Hilbish, 1998). With regards to the M. edulis and M. galloprovincialis contact zone, the hybridization between both subspecies has been observed along the Atlantic coasts from Spain to Ireland (Gosling et al., 2008), with particularly extensive introgression of M. edulis mtDNA in Atlantic M. galloprovincialis (Quesada et al., 1998).
Mytilus mussels have an unusual mode of mtDNA transmission termed doubly uniparental inheritance. Under doubly uniparental inheritance, females transmit their mtDNA (F genome) to all their progeny, just as in the regular strict maternal inheritance. However, contrary to strict maternal inheritance, males also transmit their mtDNA exclusively to their sons resulting in all males being heteroplasmic for two, divergent (up to 30% in Mytilus) mitochondrial genomes: one F genome from their mothers and the second M genome from their fathers (Skibinski et al., 1994; Zouros et al., 1994). In rare cases, the sequence divergence between both genomes in male individuals may be lowered following the masculinization event in which typical M genome is replaced by the F genome. Such events reset the divergence between both lineages (Hoeh et al., 1997). This seems to be the case for Mytilus mussels from the Baltic Sea where the typical M genome is present only at a very low frequency (Śmietanka et al., 2004) and, in most of the male mussels from the Baltic Sea, highly divergent M genome has been replaced by one of the recently masculinized recombinant genomes (Burzyński et al., 2006).
The higher substitution rate in the M lineage in comparison with the F lineage is usually observed (Stewart et al., 1996). However, the resulting higher polymorphism of the M genome does not reveal a finer population structure, and a more pronounced differentiation was noticed for the F than for the M data (Śmietanka et al., 2009). It is generally accepted that the most rapidly evolving part of the mitochondrial genome is the non-coding D-loop region believed to be the control region (CR) of mitochondrial replication and transcription. Therefore, to resolve the phylogeographic structure and demographic history of European Mytilus mussels at higher resolution, the CR seems to be a suitable marker (Cao et al., 2004; Song et al., 2013). Here we report the analysis of its polymorphism in mussels sampled in coastal waters around Europe from the White Sea to the Sea of Azov. Our focus is on this most variable part of the F genome as the seemingly more variable M genome is also more prone to periodic sweeps (Śmietanka et al., 2009). Therefore, the F genome should be better suited for studies of the phylogeographic history of the species.
Materials and methods
Sample collection
Mussels of the genus Mytilus were collected from 23 localities along the European coastline (Figure 1). Taxonomic identification of the studied mussels was carried out previously using three nuclear markers: Me 15/16, EF-bis, and ITS (Kijewski et al., 2011). Three samples were collected from the Baltic Sea: Askö near Stockholm (ASK), Gulf of Gdansk (GDA), and Mecklenburg Bight (MEB); two samples were obtained in the north of Europe from the Onega Bay, White Sea (ONE) and Barents Sea (BAR); one of Iceland (ICE) and Ireland (IRE); three samples were from North Sea: Tjärno (TJA), Balgzand (BAL), and Westerschelde (WES); two samples from English Channel: Somme(SOM), and Seine(SEI); four samples were collected from Biscay Bay: Loire (LOI): Ile de Ré (IDR), Bidasoa (BID), and Mundaka (MUN); and two from Atlantic coast of Spain: Vigo (VIG), and Punta Camarinal (CAM). Mediterranean Mytilus mussels were sampled from Gerona (GER), Banyuls sur Mer (BAN) and Gulf of Oristano (ORI); one sample was from Odessa, Black Sea (ODE) and one from Crime, Azov Sea (AZO). Samples were collected in years 2003–2004, with the exception of AZO which was described previously (Śmietanka et al., 2004). Approximately 35–55 specimens were taken per sample, with the exception of the Barents Sea sample (10 individuals). All samples were stored at −70 °C or in 96% ethanol before DNA extraction.
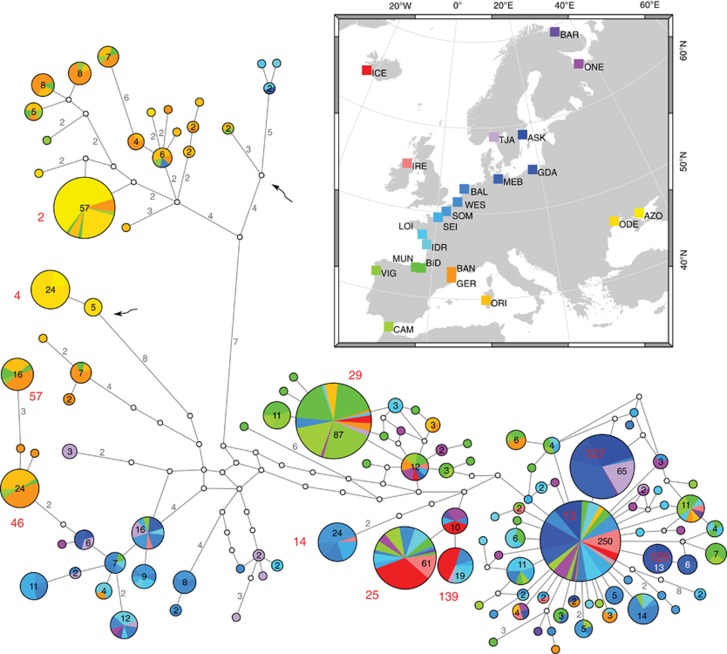
Figure 1.
Minimum Spanning Network of all obtained haplotypes. Each circle represents either single haplotype or a group of closely related haplotypes obtained by the star contraction procedure, with an area proportional to the number of observed individuals bearing the haplotype. This number is additionally given as the label inside the circles. Singleton haplotypes are not labeled. Two clades with sequences from potentially masculinized genomes are indicated by arrows. The most frequent haplotypes (>10 occurrences) are labeled in red font. The same labels can be found in Figure 3 and Supplementary Figure S1. Small, open circles represent median vectors inferred by the algorithm. They were used by alternative connections in the original network, which were removed by the Stainer procedure. Numbers on the lines connecting haplotypes indicate the number of mutational steps along each connection. Single step connections are not labeled. The location of all samples is color-coded and illustrated in the map (inset).
The studied DNA region
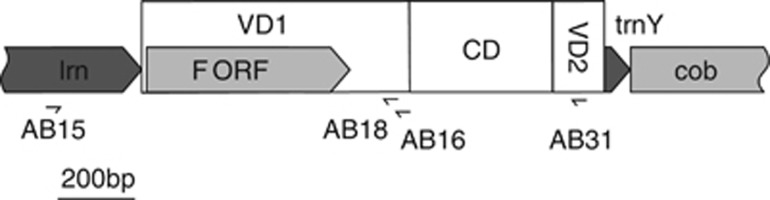
The marker selected for this study has some very attractive characteristics: it is located in the by far most variable part of the mitochondrial genome, and it is surrounded by very conservative regions, well suitable for primer design. We targeted a region spanning the end of lrn and the first variable domain of the CR in the F genome (VD1, Cao et al., 2004). Amplification was performed using the pair of universal primers AB15–AB16 (Filipowicz et al., 2008; Figure 2). The lengths of the AB15–AB16 products were approximately 900 base pairs (bp) for the F genome and 770 bp for the M genome, so these could easily be distinguished. However, several reports have claimed that recombination is possible, either within the region or in its vicinity (Burzyński et al., 2006; Venetis et al., 2007; Filipowicz et al., 2008), possibly changing the transmission route and consequently making the genome evolving faster. Certain precautions had to be taken to ensure that this did not affect our data in an unpredictable way. We took into account all the available information regarding the CR sequence of the recombinant/masculinized genomes. All the described recombination events involved the acquisition of M-like VD1 sequences into the otherwise F-like genome, but the original VD1F part was sometimes retained intact. Our primers AB15–AB16 would easily amplify from such a retained VD1F. This would, however, have been noticed, as also the original F genome sequence would be amplified, resulting in mixed signals in the sequencing reaction. Moreover, the haplotypes having such structures can be detected with repeat-specific PCR (AB16–AB32 primer pair, as described by Burzyński et al., 2006). Our survey showed that such genomes are present in a limited set of localities only, such as the Baltic Sea, the Black Sea and, to a much lesser extent, Mediterranean Sea and the Gulf of Biscay (Filipowicz et al., 2008). If either of the conditions occurred: suspicious sequencing results or suspicious location, the alternative primer set was used, with the F-specific reverse primer located at VD2 (AB31) instead of the universal AB16 primer (Filipowicz et al., 2008). We did not use this routinely for all samples because the AB15–AB16 reaction has the advantage of also detecting the highly divergent M genome. In no case such heteroplasmic individuals (usually confirmed as males) gave any indication of the presence of yet another F-like genome; they were heteroplasmic for two genomes only. Despite all efforts, two groups of haplotypes in the final data set most likely possess mosaic CR structures. This was positively verified by the AB16–AB32 PCR and the lack of any other amplifiable VD1F sequence in the individuals in question. Because there was no indication that such genomes are masculinized, as they were either present in females or in males heteroplasmic for the highly divergent M genome, we decided not to remove these sequences but to bear in mind that they may exhibit some unusual properties. They form two well-defined clades, indicated by arrows in Figure 1. The smaller clade contains two singletons from the Bay of Biscay and two doublets found in GDA, VIG and ORI. The sequences from this clade are very similar to the Baltic masculinized haplotypes from the 11a/15 group (Burzyński et al., 2006) but were found mostly in females. The larger, well-defined clade groups the sequences found mainly in the Black Sea. These are very similar to the haplotype described as a masculinized genome C (Venetis et al., 2007) but can be found in various heteroplasmic settings—both with the highly divergent M genome and with regular, recombination-free F genome as well as in homoplasmic females (Filipowicz et al., 2008). Here we included the sequences only if they were the only source of VD1F sequence in the analyzed individual, that excludes the obvious cases of males heteroplasmic for the regular F genome and the recombinant one. We tried to get only the regular F genome from such individuals, which was easily accomplished. Nevertheless, it is possible that some of the sequences from the two mentioned clades (there are 29 sequences in the ‘Black Sea' clade and 6 sequences in the ‘Baltic' clade) do represent genomes experiencing different evolutionary forces.
Figure 2.
Genetic map of the studied mtDNA region. RNA genes are shown in dark gray, protein (including putative) coding genes are in light gray. The location of primers used is visualized as well as the three major domains of the CR: two variable domains (VD1 and VD2) and the conserved domain (CD).
DNA extraction, amplification and sequencing
DNA was isolated from a small piece of the mantle tissue using the CTAB (hexadecyltrimethyl ammonium bromide) method, as described previously (Śmietanka et al., 2013). Obtained PCR products were separated by 1% agarose gel electrophoresis in a 0.5 × TBE buffer and visualized, after ethidium bromide staining, in ultraviolet light. In all cases where the M genome product was present additionally to the F genome product, supplementary F-specific primer AB18 was used instead of AB16 in sequencing. Sequencing was performed in both directions, after shrimp alkaline phosphatase and exonuclease I treatment of the PCR products (Werle et al., 1994), using the BigDye terminator cycle sequencing method. An ABI 3730 automatic sequencer was used to resolve reaction products (Macrogen, Seoul, Korea).
Bioinformatic analysis
Sequence assembly was facilitated by the Gap4 software from Staden Package version 1.7.0 (Staden et al., 2001). The consensuses were extracted and trimmed to the same range, then aligned with ClustalX version 1.83 (Thompson et al., 1997). In order to identify the signals of potential recombination that could interfere with the phylogenetic analyses, the RDP suite of programs was applied with the default settings (Martin et al., 2005). As no recombination was detected (P>0.05 for all methods), all sequences were used in further analyses. There was some length polymorphism associated with various indels, including one, relatively long, 36-bp indel in the central part of the studied region. In 72 cases, we observed single insertion (two repeats), and in 7 cases there was a double insertion (three tandemly repeated copies of the sequence motifs). These variants were detected primarily, but not exclusively, in the Mediterranean M. galloprovincialis. In one individual from the Black Sea (ODE7), the whole 36-bp long fragment was missing (zero repeats). We tested whether the repeat number is phylogenetically informative. To this end, we compared two data sets: the one with the indel removed and the second one with the indel coded as the number of repeats in a separate, ordered data partition. Both data sets were analyzed in MrBayes. The trees obtained without indel information had slightly better support for bipartitions, therefore we concluded that the repeats do not carry the useful phylogenetic signal. Consequently, all phylogenetic analyses were run on the data set with the 36-bp indel removed.
For each studied sample, standard diversity indices such as the number of segregating sites (S), the number of haplotypes (h), θ per site, haplotype diversity (hd) and nucleotide diversity (π) were calculated using DnaSP version 5.10 (Librado and Rozas, 2009). To evaluate the genetic structure, the hierarchical AMOVA (analysis of molecular variance) was performed in ARLEQUIN version 3.5.1.3 (Excoffier and Lischer, 2010). Variance components: ΦST, ΦSC, ΦCT and corresponding P-values were assessed by 10 000 permutations of the original data matrix following the Bonferroni correction (Rice, 1989). Additionally, the matrix of pairwise ΦST values and the absolute number of migrants exchanged between sampling sites were calculated in ARLEQUIN as Nm=(1−ΦST)/2ΦST. Slatkin genetic distances were calculated from ΦST, the resulting matrix was imported into MEGA5 (Tamura et al., 2011) and used to construct the neighbor-joining tree, illustrating genetic affinities among samples.
Phylogenetic relationships were reconstructed using Bayesian method using MrBayes, version 3.2.1 (Ronquist and Huelsenbeck, 2003). Model selection was done by Bayes Factor comparison. The marginal model likelihoods were estimated based on the harmonic means of the likelihood values using the Monte Carlo Markov chain samples. The GTR+I+Γ (general time reversible model with a proportion of invariable sites and a gamma-shaped distribution of rates across sites, nst=6, rates=invgamma) model was selected, but the tree topology was quite robust to model selection. It is essential that the Monte Carlo Markov chains were run with good mixing for sufficiently large number of generations to obtain meaningful results. Standard measures were taken to ensure that this is the case: each analysis was run with long (>1 million generations) burn-in, in multiple replicates (four runs, with four chains each), ensuring that all runs converged at the same solution. Each run lasted at least 20 million generations.
Haplotype networks are generally better suited than phylogenetic hierarchical trees to asses relationships within species. To this end, relationships among the observed haplotypes were assessed by constructing median-joining networks (Bandelt et al., 1999) using Network software version 4.6.1.1 (http://www.fluxus-engineering.com). In order to clarify connections in the network, star contraction procedure was applied before network calculation (Forster et al., 1996). Different settings for the homoplasy level parameter, ɛ, were tested, and ɛ=30 was eventually used. In order to account for differences in substitution rates, the weight of 1 was applied for transitions and 2 for transversions. The network was filtered by the MP procedure to remove uninformative, alternative branches (Polzin and Daneshmand, 2003).
The recently described F open reading frame (Breton et al., 2011) is located in the center of the studied region of mtDNA (Figure 2). The presence of a protein coding gene could have an impact on the analyses, as most of them assume that the scored variation is neutral. The dN/dS ratio in the F open reading frame was 0.72, much higher than typically observed for mitochondrial protein-coding genes (Śmietanka et al., 2009). However, when checked for the signatures of positive selection using methods applied previously to similar data (Śmietanka et al., 2010), it did not show any consistent and significant signals. Therefore, even if the F open reading frame does code a protein, it seems to evolve under much more relaxed pressure than other mitochondrial genes, and its presence should not influence the outcome of analyses relying on marker neutrality.
Nested cladistics analysis (nested clade phylogeographic analysis (NCPA); Templeton, 1998) was performed as follows. First, the network of haplotypes was estimated by statistical parsimony, using the TCS software (Clement et al., 2000). To improve the power of NCPA, additional criteria were used to resolve the obtained network (Templeton, 1998). One major cycle was resolved, and two missing links were added manually in the TCS. The final network still had a few small, local cycles, but overall, was consistent with both alternative clustering approaches: with the Minimum Spanning Network generated by the Network software and with the phylogenetic trees. Then, the procedure implemented in ANeCA (Panchal, 2007) was used to generate the input file for the GeoDis software version 2.6 (Posada et al., 2000). Finally, GeoDis was run on the modified data set, and the inference of past phylogeographic events was performed manually, using the newest inference key. The geographic distances between sampling sites were estimated manually, by computing shortest along the shore distance in GoogleEarth. The same matrix of geographic distances was used to perform Mantel test in ARLEQUIN.
Isolation-with-migration (IM) model was applied to the data using both the IM (Hey and Nielsen, 2004) and IMa2 software (Hey, 2010). Both implementations use Monte Carlo Markov chain approach, hence the convergence assessment and the effective size of the sampled data set are of critical importance. To ensure the correctness of the analysis, each was run in triplicate, with long burn-in phase of at least 107 generations, as described previously (Śmietanka et al., 2013). As single locus data could not possibly provide enough resolution to solve the full multipopulation model of IMa2, each pair of samples was compared separately in IM, following (Riginos and Henzler, 2008). To reduce the number of estimated parameters, several groups of samples were combined, as suggested by the results of AMOVA analysis and preliminary pairwise IM runs of geographically close samples. The resulting matrices of divergence times were used in UPGMA reconstructions of the chronograms relating both species populations in MEGA5 (Tamura et al., 2011). To illustrate the phylogeographic history, these trees were used in GeoPhylobuilder (Kidd and Liu, 2008) and projected over relevant maps in ArcGIS (ESRI, Redlands, CA, USA). To convert the time units used by IM/IMa2 to calendar years, evolutionary rate had to be estimated. To this end, the procedure described previously was used (Śmietanka et al., 2009; Śmietanka et al., 2013). The most common haplogroup data were subject to mismatch analysis in ARLEQUIN, and then following the assumption that its expansion started at the onset of the current interglacial, the absolute evolutionary rate was estimated.
Results
Relationships between haplotypes
The 985 obtained sequences of the F genome fragment came from Mytilus mussels comprising all three European species sharing common mitochondrial haplotypes. Standard diversity indices (Table 1) showed quite uniform distribution of the polymorphisms. Despite relatively low overall nucleotide diversity (1.5%), both the haplotype diversity (0.96) and the number of segregating sites (262) were high, consistent with substantial genetic variation in European Mytilus mussels. The nucleotide diversity (π) in the Mediterranean Mytilus (GER, BAN and ORI) was approximately twice as high as in the Atlantic samples, hinting at older diversity. A haplotype network was calculated to assess relationships within the whole data set (Figure 1). The generated network is remarkably well resolved, with only few cycles left. The cycles do not constitute major uncertainties, at least in relationships between the three main haplogroups. Despite the very high overall number, only a limited fraction of haplotypes, mostly located at the local centers of the network, has appreciable frequencies. This is a pattern characteristic for a population experiencing rapid growth following relatively recent bottleneck. Nonrandom geographic distribution of haplotypes is also clearly visible, with intriguing differences between major haplogroups. The inferred phylogenetic tree (Supplementary Figure S1) built with unique haplotype sequences also shows the three distinct clades, consistent with the results obtained earlier (Śmietanka et al., 2009), but with a much better resolution: several well-supported subclades are now clearly visible in all three major clades. Moreover, the deeper subdivisions within clades F2 and F3 than within F1 clade are now evident.
Table 1. Standard genetic diversity indices for all analyzed samples of European Mytilus populations.
| Sample | N | S | h | θ | hd | π (JC) |
|---|---|---|---|---|---|---|
| ASK | 50 | 8 | 9 | 0.002 | 0.67 | 0.001 |
| GDA | 50 | 43 | 13 | 0.013 | 0.73 | 0.004 |
| MEB | 55 | 31 | 19 | 0.009 | 0.86 | 0.005 |
| ONE | 37 | 42 | 22 | 0.014 | 0.92 | 0.006 |
| BAR | 10 | 22 | 8 | 0.011 | 0.93 | 0.007 |
| ICE | 45 | 20 | 19 | 0.007 | 0.86 | 0.004 |
| IRE | 40 | 51 | 27 | 0.016 | 0.95 | 0.006 |
| TJA | 35 | 39 | 17 | 0.014 | 0.90 | 0.011 |
| BAL | 40 | 62 | 26 | 0.020 | 0.94 | 0.012 |
| WES | 53 | 59 | 29 | 0.018 | 0.96 | 0.010 |
| SOM | 50 | 39 | 21 | 0.012 | 0.88 | 0.009 |
| SEI | 40 | 46 | 24 | 0.016 | 0.93 | 0.009 |
| LOI | 42 | 63 | 27 | 0.021 | 0.92 | 0.009 |
| IDR | 49 | 66 | 33 | 0.024 | 0.97 | 0.012 |
| BID | 50 | 72 | 34 | 0.023 | 0.97 | 0.012 |
| MUN | 50 | 64 | 30 | 0.020 | 0.94 | 0.009 |
| VIG | 46 | 80 | 34 | 0.025 | 0.95 | 0.012 |
| CAM | 43 | 69 | 28 | 0.022 | 0.94 | 0.010 |
| GER | 30 | 64 | 17 | 0.022 | 0.94 | 0.024 |
| BAN | 40 | 56 | 20 | 0.019 | 0.95 | 0.024 |
| ORI | 52 | 72 | 24 | 0.022 | 0.94 | 0.023 |
| ODE | 40 | 40 | 10 | 0.012 | 0.81 | 0.018 |
| AZO | 38 | 36 | 10 | 0.011 | 0.53 | 0.007 |
| Total data | 985 | 262 | 334 | 0.052 | 0.96 | 0.015 |
Abbreviations: h, number of haplotypes; hd, haplotype diversity; N, number of individuals; S, number of segregating sites; θ, theta per site; π, nucleotide diversity per site, with Jukes and Cantor correction.
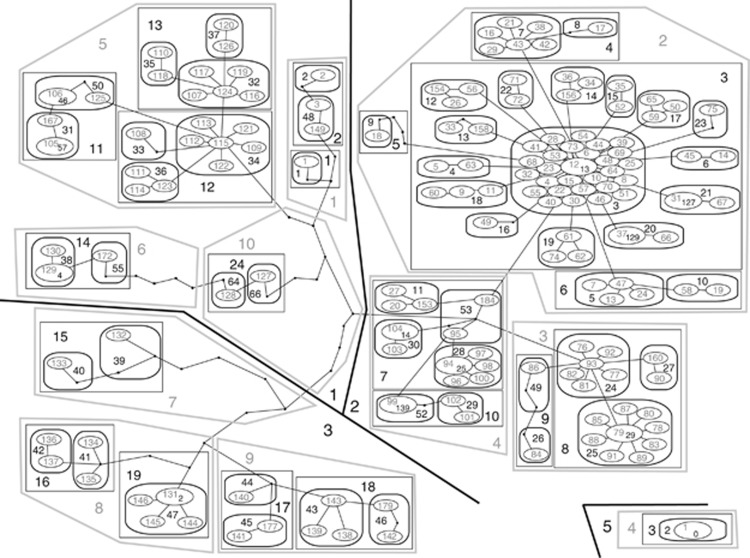
The haplotype-based approach to the inference of past phylogeographic events (NCPA) was attempted. The semi-automatically designed, six-level set of the nested clades (Figure 3) was quite complex, as expected for the observed number of different haplotypes. Nevertheless, the agreement between the NCPA clustering and the earlier mentioned results of phylogenetic and median-joining analyses was quite good. Nesting clade 5–1 corresponded with clade F2, visible also in the lower left part of the Minimum Spanning Network in Figure 1. Nesting clade 5–2 corresponded with the phylogenetic clade F1 (network lower right) and clade 5–3 with the clade F3 (network upper left). Unfortunately, NCPA analysis in GeoDis provided only few significant inferences (Table 2). Many of the inferences constitute ‘continuous range expansions', particularly several level 4 clades exhibited this. The clades expanding their ranges constituted the two major, primarily Mediterranean, parts of the 5–3 clade (4–8 and 4–9), represented in Figure 1 by the upper leftmost branch connected with the rest of the network via a single four-substitution long link. Clearly, the inferred expansion must have been from the Mediterranean Sea to the Atlantic in these cases. The other interesting range-expanding clades are 4–1 and 4–5. They are both parts of the 5–1 clade but have strikingly different geographic distribution. The clade 4–5 constitutes the majority of the 5–1 clade, with disjoint geographic distribution of its nested clades: clade 3–11 is found in M. galloprovincialis, primarily in the Mediterranean Sea, whereas clades 3–12 and 3–13 are almost exclusively from M. edulis inhabiting the vicinity of the North Sea. With the given relationship of nested clades (3–11 is at the tip, whereas 3–12 is not), the direction of the apparent range expansion of clade 4–5 must have been from the Atlantic to the Mediterranean Sea. Interestingly, the inference for the whole clade 5–1, after the inclusion of the most geographically and genetically disjoint clades 4–10 and 4–6 (they include the haplogroups dominating the Black Sea), suggests past fragmentation followed by range expansion. Other inferences are limited to rather small, local clades.
Figure 3.
Nested cladogram designed with the ANeCA software. The clade level is indicated by its graphic appearance, and clade numbers are given directly. In a few cases, additional numbers are present within the ovals representing 1-level clades: these are the labels of the most frequent haplotypes (zero-level clades).
Table 2. Nested cladistics inference of population history.
| Clade | Chain of inference | Outcome |
|---|---|---|
| 1–12 | 1-2-3-4 NO | Restricted dispersal by distance |
| 1–120 | 1-2-11-12 NO | Continuous range expansion |
| 2–3 | 1-2-3-5-6-7-8 YES | Restricted gene flow/dispersal but with some long-distance dispersal over intermediate areas not occupied by the species; or past gene flow followed by extinction of intermediate populations |
| 2–25 | 1-2-3-4 NO | Restricted dispersal by distance |
| 2–28 | 1-2-11-12 NO | Continuous range expansion |
| 3–6 | 1-2-3-4 NO | Restricted dispersal by distance |
| 3–7 | 1-2-3-4 NO | Restricted dispersal by distance |
| 3–10 | 1-2-3-5-6-7-8 YES | Insufficient genetic resolution to discriminate between range expansion/colonization and restricted dispersal/gene flow |
| 3–12 | 1-2-3-4 NO | Restricted dispersal by distance |
| 3–13 | 1-2-3-4 NO | Restricted dispersal by distance |
| 4–1 | 1-2-11-12 NO | Continuous range expansion |
| 4–5 | 1-2-11-12 NO | Continuous range expansion |
| 4–8 | 1-2-11-12 NO | Continuous range expansion |
| 4–9 | 1-2-11-12 NO | Continuous range expansion |
| 5–1 | 1-2-11-12-13 YES | Long-distance colonization, possibly coupled with subsequent fragmentation or past fragmentation followed by range expansion |
| 5–2 | 1-2-11-12 NO | Continuous range expansion |
| 5–3 | 1-2-3-4 NO | Restricted dispersal by distance |
| Total | 1-2-3-5-6-7-8 YES | Restricted gene flow/dispersal but with some long-distance dispersal over intermediate areas not occupied by the species; or past gene flow followed by extinction of intermediate populations |
Grouping of samples
With this high-resolution genetic data, we hoped to see finer resolution of the genetic structure of the studied populations. Population pairwise ΦST comparisons are presented in Supplementary Table S1. Most of the comparisons showed significant differentiation. At the first glance, the increasing ΦST values were correlated with the longer geographical distances between compared sampling sites. The formal comparison of the genetic distances (Slatkin distance) with the physical (geographic) distance (Mantel test, Mantel, 1967) confirmed that the effect of isolation by distance is highly significant (P<0.0001) and quite strong (coefficient of determination R2=60%). In some cases, however, the distance was not so important: there was no genetic differentiation between Atlantic mussels on the Iberian Peninsula, from Bidasoa (BID) to Punta Camarinal (CAM), separated by >1700 km. On the other hand, significant differentiation was observed between Baltic mussels from Askö (ASK) and from the Gulf of Gdansk (GDA), separated only by approximately 500 km. In addition to the four mentioned samples of the Atlantic M. galloprovincialis from the Iberian Peninsula, there was also obviously no differentiation of the three Mediterranean M. galloprovincialis samples (BAN, GER and ORI). Overall, in only 18 out of the 253 comparisons the ΦST was non-significant, indicating highly structured genetic composition of European Mytilus. Still, the real number of populations must be somewhat smaller than the number of samples. To find the best grouping of samples into natural population units, the matrix of genetic distances was used and the neighbor-joining algorithm was applied to construct the population tree (Supplementary Figure S2). The major split into three groups of samples was visible, with distinguishable Mediterranean, Iberian and Atlantic areas. However, the split between Black Sea and Mediterranean Sea samples was much more pronounced than any of the possible intra-Atlantic splits we hoped to identify. The groups suggested by the graph were then tested by hierarchical AMOVA (Table 3). Subtle changes were introduced in sample grouping, but they did not substantially affect the results. The primary source of genetic variation (64% on average) came from the intra-population level. Still, the differentiation among groups of populations constituted about 31% of observed diversity, while it was only ca. 5% among populations within groups, confirming that the approach was largely correct. The overall fixation index was quite high, exceeding 30% in all cases. The remaining significant differentiation observed in many pairwise intra-group comparisons suggested that even a finer genetic structure is present.
Table 3. Hierarchical AMOVA with four data clusterings given under the table.
| Data set (n) | Va (%) | ΦCT | Vb (%) | ΦSC | Vc (%) | ΦST |
|---|---|---|---|---|---|---|
| 4 | 33.32 | 0.333 | 5.35 | 0.080 | 61.34 | 0.387 |
| 51 | 30.28 | 0.303 | 4.37 | 0.063 | 65.35 | 0.347 |
| 52 | 30.84 | 0.308 | 4.79 | 0.069 | 64.37 | 0.356 |
| 12 | 31.13 | 0.311 | −0.12 | −0.002 | 69.00 | 0.310 |
Abbreviations: AMOVA, analysis of molecular variance; n, number of groups, V, variance components: a—among groups; b—among populations within groups; c—within populations, ΦCT, ΦSC, ΦST, fixation indices.
P<0.001 in all cases.
4—{ONE, BAR, ICE, IRE, TJA, BAL, WES, SOM, SEI, LOI, IDR, MEB, GDA, ASK} {BID, MUN, VIG, CAM} {GER, BAN, ORI} {ODE, AZO}.
51—{ONE, BAR, ICE, IRE, TJA, BAL, WES, SOM, SEI, LOI, IDR} {MEB, GDA, ASK} {BID, MUN, VIG, CAM} {GER, BAN, ORI} {ODE, AZO}.
52—{ONE, BAR, ICE, IRE, TJA, BAL, WES, SOM, SEI, LOI, IDR, MEB} {GDA, ASK} {BID, MUN, VIG, CAM} {GER, BAN, ORI} {ODE, AZO}.
12—{ONE, BAR} {ICE} {IRE} {TJA} {BAL, WES, SOM, SEI} {LOI, IDR} {MEB, GDA} {ASK} {BID, MUN, VIG, CAM} {GER, BAN, ORI} {ODE} {AZO}.
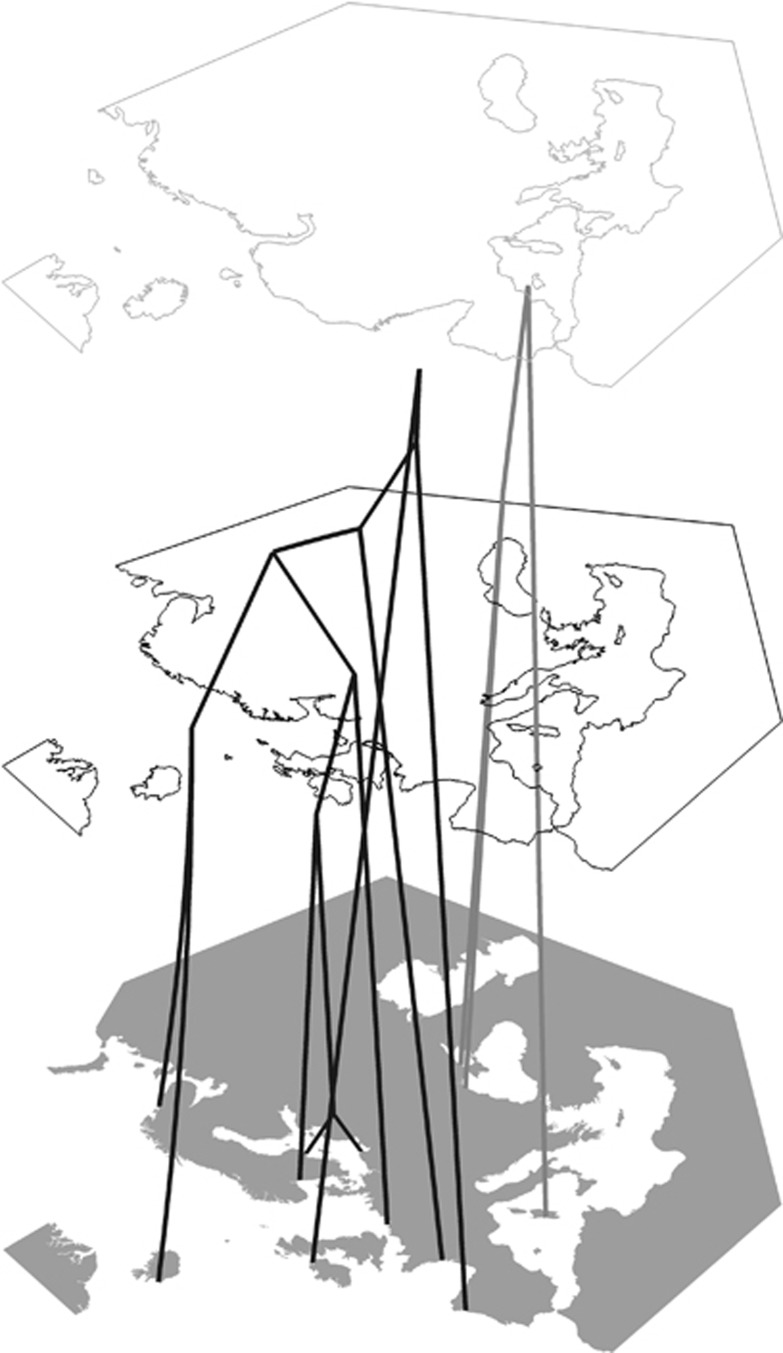
To put the observed phylogeography into historical perspective, we tried to fit the IMM model to the data using the IMa2 (Hey, 2010). The program was run in all pairwise comparisons first. These preliminary IMa2 results suggested that there may be large differences in effective population sizes between samples, a feature which could affect the estimated indices of differentiation. To account for this, older software implementing population split parameter s was used (IM, Nielsen and Wakeley, 2001). Indeed, the populations showing anomalously long terminal branches: ASK, AZO and ICE (Supplementary Figure S2) were scored in this analysis as having very low s (<0.02), and thus their genetic composition could be affected by a recent founder effect. Therefore, these samples were not combined with their closest relatives not to introduce sampling bias. The preliminary results were used to cluster samples: the pairs for which either very high migrations in both directions were inferred or the ones with divergence times not differing significantly from zero were combined. The resulted grouping of samples into 12 populations was checked in AMOVA (Table 3, last row), confirming that the proposed groups were indeed quite homogenous (no variation was observed among samples within groups). Then, all groups were compared pairwise by running full model in IMa2 and IM. Surprisingly, in a few comparisons only a significant migration was found, and usually the migration parameter m was small even in those cases (Table 4). Most of the samples seemed to be well isolated, and the inferred time of their differentiation was similar. Unfortunately, the full model has too many parameters to estimate with just one marker. To focus on the time of divergence, we ran the limited model with no migration in all pairwise comparisons. The resulting matrix of times since divergence was then used to construct the chronogram. This was done separately for the Atlantic and Mediterranean samples as they were the only ones connected by very high and significant migration rates. Due to the violation of the no-migration assumption, putting them together would lead to a highly distorted tree. The chronograms were subsequently put into geographic context (Figure 4). To convert the mutational time units into calendar years, the estimate of the substitution rate for the studied DNA fragment was needed. To this end, the mismatch analysis of the most abundant group of haplotypes was used. We assumed that the onset of its expansion corresponded with the end of last glacial maximum (LGM) at 18 kya (Clark et al., 2009). The mismatch analysis (Supplementary Figure S3) showed that the data fit the expansion model remarkably well (sum of square deviations<0.0005) and allowed a quite reliable estimate of the per locus substitution rate (4.4 × 10−5), which was then used to convert the chronogram units into calendar years. Two schematic paleomaps were created based on the sea level changes (Clark et al., 2009): one represented by a 150-m bathymetric contour as a proxy of the shoreline at the LGM and the second one represented by a 30-m bathymetric contour as a proxy of the shoreline at the climatic optimum, ca. 10 kya. The vertical scaling of the chronograms has been synchronized with the placement of the paleomaps (Figure 4).
Table 4. The indices inferred by IMa2 for population pairs with at least one significantly >0 migration rate: migration rates (m) in both directions and effective population sizes (q=4 Nμ) for contemporary (0 and 1) and ancestral (A) populations.
| Sites | qA | q0 | q1 | m0 | m1 | Nm0 | Nm1 | |
|---|---|---|---|---|---|---|---|---|
| TJA | bas | 123.47 | 33.75 | 40.58 | 1.65 | 0.22 | 95.03 | 5.39 |
| ICE | nor | 24.17 | 249.17 | 17.50 | 0.01 | 0.09 | 0.01 | 16.43 |
| hol | nor | 304.27 | 185.47 | 46.75 | 0.02 | 0.01 | 6.33 | 0.44 |
| bas | hol | 47.50 | 299.17 | 62.50 | 0.01 | 0.05 | 0.01 | 7.71 |
| TJA | hol | 55.83 | 302.50 | 57.50 | 0.01 | 0.02 | 9.04 | 3.95 |
| ICE | TJA | 22.50 | 42.50 | 4.17 | 0.01 | 0.25 | 0.01 | 5.28 |
| ibe | nor | 619.33 | 171.33 | 66.00 | 0.01 | 0.01 | 6.73 | 0.01 |
| IRL | hol | 189.17 | 305.83 | 42.50 | 0.01 | 0.12 | 1.24 | 13.32 |
| TJA | nor | 47.50 | 154.17 | 27.50 | 0.01 | 0.14 | 9.89 | 3.00 |
| ibe | med | 1265.67 | 50.33 | 84.33 | 0.44 | 0.80 | 19.98 | 19.98 |
| bas | nor | 42.50 | 204.17 | 37.50 | 0.07 | 0.09 | 3.02 | 13.65 |
| hol | frs | 310.83 | 349.17 | 74.17 | 0.04 | 0.01 | 7.84 | 1.45 |
Number of migrants received per generation by first and second column population is shown as Nm0 and Nm1, respectively.
Samples were grouped as shown in the last row of Table 3, nor=ONE+BAR, hol=BAL+WES+SOM+SEI, frs=LOI+IDR, bas=MEB+GDA, ibe=BID+MUN+VIG+CAM, med=GER+BAN+ORI.
Figure 4.
Two chronograms of Atlantic (black) and Mediterranean (gray) populations, based on coalescence estimates of their times of divergence. The vertical scale corresponds to time before present; the middle paleomap represents 10 kya, the top paleomap represents the LGM, at ca. 18 kya.
Discussion
The observed high level of genetic polymorphism is remarkable but not unexpected: the marine bivalves are known for their extremely large effective population sizes (Bazin et al., 2006), and consequently, population-level genetic polymorphism is expected to be substantial. However, the relatively low connectivity and strong population structuring seems to contradict the expectations: other mussel species, such as Mytilus californianus, do not show any population structuring over relatively long geographic distances (Ort and Pogson, 2007). The apparently different situation observed in European Mytilus populations was noted in a number of studies and was usually attributed to its unique history, including recent range changes, hybridization events and introgression (Quesada et al., 1998; Śmietanka et al., 2004; Stuckas et al., 2009; Kijewski et al., 2011). However, in the earlier studies the limits of techniques used: limited sampling, short sequences or RFLP data only precluded high resolution phylogeographic analysis. What can we say about the phylogeographic history of European Mytilus based on our higher resolution mitochondrial data set? Coalescence-based approaches seem to indicate that all Atlantic populations are separated by a similar amount of time, comparable to the time of the onset of the expansion of a major mitochondrial lineage. If we assume that this event took place at the end of the LGM, then all Atlantic Mytilus populations must have survived the last glaciation in a single refugium. The substitution rate required for that is not much different than usually assumed for neutral mitochondrial markers in Mytilus—in the order of 10−8 substitutions per site per year. This is comparable to the rates used by Wares and Cunningham (2001). The most likely location of the refugium (Figure 4) would correspond to the potential refugium number 4 in Maggs et al. (2008), to the north of the Bay of Biscay, in accordance with recent modeling of late Pleistocene species distribution (Waltari and Hickerson, 2013). The population of M. galloprovincialis survived the LGM in the Mediterranean Sea. There are indications of high gene flow between current Atlantic and Mediterranean populations of M. galloprovincialis. The haplogroup currently dominating Atlantic M. galloprovincialis population is of M. edulis origin and diverged from the most of other common haplogroups very recently, most likely shortly before the postglacial expansion, this is also supported by NCPA analysis showing expansion of the respective clades. Based on coalescence results, we must conclude that this expansion constitutes, in fact, a very recent and prominent gene flow event between Mediterranean and Atlantic. The long history of Mytilus populations inferred by Wares and Cunningham (2001) is not necessarily in contrast with our finding of a very sharp coalescence into one refugium within the last 10 ky. Apparently, the surviving population was very diverse and retained much of its ancestral genetic polymorphism, a feature that could have affected the earlier estimates. The most striking example of this is the NCPA clade 5–1 whose expansion must have occurred long before the last glacial cycle, the most recent common ancestor of this group was dated at ca. 0.3 MYA (Śmietanka et al., 2010). Yet this haplogroup is more differentiated in the Atlantic than in any other location (Mediterranean and Black Seas). The potential existence of the third refugium in the west Atlantic remains controversial. Most species critically examined indicated recolonization from Europe rather than persistence of the species in America throughout the last glaciation (Maggs et al., 2008; Ilves et al., 2010). However, the results obtained for M. edulis seem to indicate that it could have survived also on the west coasts of the Atlantic (Wares and Cunningham, 2001; Riginos and Henzler, 2008; Waltari and Hickerson, 2013). These results were, however, primarily driven by the reciprocal monophyly of the mitochondrial M marker. The M genome of the Atlantic populations diverged ca. 0.35 MYA, clearly long before the last glaciation event (Śmietanka et al., 2010). To account for this apparent discrepancy, the possibility of selective sweeps in the F lineage was considered by Ilves et al. (2010), in effect favoring the persistence hypothesis. An alternative, more likely scenario, under which the sweep occurred in both M genome lineages, effectively fixing the preexisting diversity, would favor the alternative, colonization hypothesis. Our data bring only a little to this discussion as our focus is on European populations, and we did not sample any populations from the west Atlantic. Moreover, the choice of the marker precluded us from direct comparison with the existing data set of Riginos and Henzler (2008). However, the identity of some haplogroups can be traced based on available full genome sequences, and hence the overall observed patterns can be compared. It seems like the limited sampling of European mussels did bias their conclusions to some extent: the diversity was underestimated and some haplogroups considered exclusively American can be found in Europe. Moreover, the inferred direction of contemporary gene flow: from America to Iceland (Riginos and Henzler, 2008) and from Iceland to Europe (Table 4) is most likely associated with the clades 2–28 and 3–7, strongly suggesting that the private haplotypes found in the west Atlantic are very closely related to the European haplotypes, and hence the time estimates for transatlantic split given by Riginos and Henzler (2008) may be overestimated. Further research involving more markers is needed to clarify this, but our data strongly support a single Atlantic refugium hypothesis.
Data archiving
DNA sequences: GenBank (Accession numbers KF631456–KF632440). Voucher and locality information has been included in GenBank records.
Alignments of DNA sequences, input file for GeoDis, formatted MSN from Network software, and output files from IM and IMa2 are available from the Dryad Digital Repository: doi:10.5061/dryad.3kn08.
Acknowledgments
This work was supported by the EU Grant BioCoMBE, contract number EVK3-CT-2002-00072 and by the Polish supporting Grant KBN 31/E-45/5.PR UE to RW. Computer intense analyses were run on supercomputers of the Academic Computer Centre (TASK) in Gdańsk, as well as using the PLGRID (www.plgrid.pl) infrastructure.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Bandelt HJ, Forster P, Röhl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Bazin E, Glémin S, Galtier N. (2006). Population size does not influence mitochondrial genetic diversity in animals. Science 312: 570–572. [DOI] [PubMed] [Google Scholar]
- Beaumont A, Hawkins M, Doig F, Davies I, Snow M. (2008). Three species of Mytilus and their hybrids identified in a Scottish Loch: natives, relicts and invaders? J Exp Mar Biol Ecol 367: 100–110. [Google Scholar]
- Breton S, Ghiselli F, Passamonti M, Milani L, Stewart DT, Hoeh WR. (2011). Evidence for a fourteenth mtDNA-encoded protein in the female-transmitted mtDNA of marine Mussels (Bivalvia: Mytilidae). PLoS One 6: e19365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyński A, Zbawicka M, Skibinski DOF, Wenne R. (2006). Doubly uniparental inheritance is associated with high polymorphism for rearranged and recombinant control region haplotypes in Baltic Mytilus trossulus. Genetics 174: 1081–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Kenchington E, Zouros E, Rodakis GC. (2004). Evidence that the large noncoding sequence is the main control region of maternally and paternally transmitted mitochondrial genomes of the marine mussel (Mytilus spp.). Genetics 167: 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B et al. (2009). The last glacial maximum. Science 325: 710–714. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. (2000). TCS: a computer program to estimate gene genealogies. Mol Ecol 9: 1657–1659. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Filipowicz M, Burzyński A, Śmietanka B, Wenne R. (2008). Recombination in mitochondrial DNA of European mussels Mytilus. J Mol Evol 67: 377–388. [DOI] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt HJ. (1996). Origin and evolution of native American mtDNA variation: a reappraisal. Am J Hum Genet 59: 935–945. [PMC free article] [PubMed] [Google Scholar]
- Gosling E, Doherty S, Howley N. (2008). Genetic characterization of hybrid mussel (Mytilus) populations on Irish coasts. J Mar Biol Assoc UK 88: 341–346. [Google Scholar]
- Gosling EM. (1992). Genetics of Mytilus. In: Gosling EM, (ed). The Mussels Mytilus: Ecology, Physiology, Genetics and Culture. Elsevier: The Netherlands. [Google Scholar]
- Gérard K, Bierne N, Borsa P, Chenuil A, Féral J. (2008). Pleistocene separation of mitochondrial lineages of Mytilus spp. mussels from Northern and Southern Hemispheres and strong genetic differentiation among southern populations. Mol Phylogenet Evol 49: 84–91. [DOI] [PubMed] [Google Scholar]
- Hey J. (2010). Isolation with migration models for more than two populations. Mol Biol Evol 27: 905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. (2004). Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeh WR, Stewart DT, Saavedra C, Sutherland BW, Zouros E. (1997). Phylogenetic evidence for role-reversals of gender-associated mitochondrial DNA in Mytilus (Bivalvia: Mytilidae). Mol Biol Evol 14: 959–967. [DOI] [PubMed] [Google Scholar]
- Ilves KL, Huang W, Wares JP, Hickerson MJ. (2010). Colonization and/or mitochondrial selective sweeps across the North Atlantic intertidal assemblage revealed by multi-taxa approximate Bayesian computation. Mol Ecol 19: 4505–4519. [DOI] [PubMed] [Google Scholar]
- Kidd D, Liu X. (2008). Geophylobuilder 1.0: an arcgis extension for creating ‘geophylogenies'. Mol Ecol Res 8: 88–91. [DOI] [PubMed] [Google Scholar]
- Kijewski T, Śmietanka B, Zbawicka M, Gosling E, Hummel H, Wenne R. (2011). Distribution of Mytilus taxa in European coastal areas as inferred from molecular markers. J Sea Res 65: 224–234. [Google Scholar]
- Kijewski TK, Zbawicka M, Väinölä R, Wenne R. (2006). Introgression and mitochondrial DNA heteroplasmy in the Baltic populations of mussels Mytilus trossulus and M. edulis. Mar Biol 149: 1373–1385. [Google Scholar]
- Librado P, Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Maggs CA, Castilho R, Foltz D, Henzler C, Jolly MT, Kelly J et al. (2008). Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 89: S108–S122. [DOI] [PubMed] [Google Scholar]
- Mantel N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res 27: 209–220. [PubMed] [Google Scholar]
- Martin DP, Williamson C, Posada D. (2005). RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21: 260–262. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J. (2001). Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort BS, Pogson GH. (2007). Molecular population genetics of the male and female mitochondrial DNA molecules of the California sea mussel, Mytilus californianus. Genetics 177: 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal M. (2007). The automation of nested clade phylogeographic analysis. Bioinformatics 23: 509–510. [DOI] [PubMed] [Google Scholar]
- Polzin T, Daneshmand SV. (2003). On Steiner trees and minimum spanning trees in hypergraphs. Operations Res Lett 31: 12–20. [Google Scholar]
- Posada D, Crandall KA, Templeton AR. (2000). GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Mol Ecol 9: 487–488. [DOI] [PubMed] [Google Scholar]
- Quesada H, Gallagher C, Skibinski DAG, Skibinski DOF. (1998). Patterns of polymorphism and gene flow of gender-associated mitochondrial DNA lineages in European mussel populations. Mol Ecol 7: 1041–1051. [Google Scholar]
- Rawson PD, Hilbish TJ. (1998). Asymmetric introgression of mitochondrial DNA among European population of blue mussels (Mytilus spp.). Evolution 52: 100–108. [DOI] [PubMed] [Google Scholar]
- Rice WR. (1989). Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Ridgway G, Nævdal G. (2004). Genotypes of Mytilus from waters of different salinity around Bergen, Norway. Helgol Mar Res 58: 104–109. [Google Scholar]
- Riginos C, Henzler CM. (2008). Patterns of mtDNA diversity in North Atlantic populations of the mussel Mytilus edulis. Mar Biol 155: 399–412. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Skibinski DOF, Gallagher C, Beynon CM. (1994). Mitochondrial DNA inheritance. Nature 368: 817–818. [DOI] [PubMed] [Google Scholar]
- Śmietanka B, Burzyński A, Wenne R. (2009). Molecular population genetics of male and female mitochondrial genomes in European mussels Mytilus. Mar Biol 156: 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śmietanka B, Burzyński A, Wenne R. (2010). Comparative genomics of marine mussels (Mytilus spp.) gender associated mtDNA: rapidly evolving atp8. J Mol Evol 71: 385–400. [DOI] [PubMed] [Google Scholar]
- Śmietanka B, Zbawicka M, Sańko T, Wenne R, Burzyński A. (2013). Molecular population genetics of male and female mitochondrial genomes in subarctic Mytilus trossulus. Mar Biol 160: 1709–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śmietanka B, Zbawicka M, Wołowicz M, Wenne R. (2004). Mitochondrial DNA lineages in the European populations of mussels (Mytilus spp.). Mar Biol 146: 79–92. [Google Scholar]
- Song N, Jia N, Yanagimoto T, Lin L, Gao T. (2013). Genetic differentiation of Trachurus japonicus from the Northwestern Pacific based on the mitochondrial DNA control region. Mitochondrial DNA 24: 705–712. [DOI] [PubMed] [Google Scholar]
- Staden R, Judge DP, Bonfield JK. (2001). Sequence assembly and finishing methods. In: Baxevanis AD, Ouellette BFF, (eds). Bioinformatics. A Practical Guide to the Analysis of Genes and Proteins. Wiley: New York, NY, USA. [DOI] [PubMed] [Google Scholar]
- Stewart DT, Kenchington ER, Singh RK, Zouros E. (1996). Degree of selective constraint as an explanation of the different rates of evolution of gender-specific mitochondrial DNA lineages in the mussel Mytilus. Genetics 143: 1349–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckas H, Stoof K, Quesada H, Tiedemann R. (2009). Evolutionary implications of discordant clines across the Baltic Mytilus hybrid zone (Mytilus edulis and Mytilus trossulus). Heredity 103: 146–156. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton A. (1998). Nested clade analyses of phylogeographic data: testing hypotheses about gene flow and population history. Mol Ecol 7: 381–397. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetis C, Theologidis I, Zouros E, Rodakis GC. (2007). A mitochondrial genome with a reversed transmission route in the Mediterranean mussel Mytilus galloprovincialis. Gene 406: 79–90. [DOI] [PubMed] [Google Scholar]
- Väinölä R, Hvilsom M. (1991). Genetic divergence and a hybrid zone between Baltic and North Sea Mytilus populations (Mytilidae: Mollusca). Biol J Linn Soc Lond 43: 127–148. [Google Scholar]
- Väinölä R, Strelkov P. (2011). Mytilus trossulus in Northern Europe. Mar Biol 158: 817–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltari E, Hickerson MJ. (2013). Late Pleistocene species distribution modelling of North Atlantic intertidal invertebrates. J Biogeogr 40: 249–260. [Google Scholar]
- Wares JP, Cunningham C. (2001). Phylogeography and historical ecology of the North Atlantic intertidal. Evolution 55: 2455–2469. [DOI] [PubMed] [Google Scholar]
- Werle E, Schneider C, Renner M, Volker M, Fiehn W. (1994). Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res 22: 4354–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros E, Ball AO, Saavedra C, Freeman KR. (1994). Mitochondrial DNA inheritance. Nature 368: 818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.