Abstract
Bacillus rubiinfantis sp. nov. strain mt2T is the type strain of B. rubiinfantis sp. nov., isolated from the fecal flora of a child with kwashiorkor in Niger. It is Gram-positive facultative anaerobic rod belonging to the Bacillaceae family. We describe the features of this organism alongside the complete genome sequence and annotation. The 4 311 083 bp long genome (one chromosome but no plasmid) contains 4028 protein-coding gene and 121 RNA genes including nine rRNA genes.
Keywords: Bacillus rubiinfantis, culturomics, genome, taxonogenomics
Introduction
Bacterial classification is currently based on phenotypic and genomic characteristics and the building of phylogenetic relationships using a comparison of the 16S ribosomal RNA [1], [2], [3]. Culturomics studies are based on the multiplication of culture conditions with a rapid identification method, which has allowed us to recently extend the gut microbiota repertoire [4], [5]. In parallel, a new concept of bacterial description, termed taxonogenomics, has recently been developed combining a proteomic description with the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) profile associated with a biochemical and genomic description of the new bacterial species [4]. To date, nine bacterial species have officially been recognized through taxonogenomics [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15].
The genus Bacillus was established in 1872 by Cohn and is composed of strictly aerobic and facultatively anaerobic rod-shaped bacteria that form heat-resisting endospores [16], [17], [18], [19]. It includes over 200 described species and subspecies belonging to the Firmicutes phylum (http://www.bacterio.net/bacillus.html). These bacteria are widely distributed in the environment (soil, water, air), plants, food and human clinical samples [20]. The most known pathogenic Bacillus species is Bacillus anthracis, but most Bacillus are considered to be harmless to humans [20].
Bacillus rubiinfantis strain mt2T (= Collection de souches de l’Unité des Rickettsies (CSUR) P1141 = Deutsche Sammlung von Mikroorganismen (DSM) 28615) is the type strain of B. rubiinfantis sp. nov. This bacterium is Gram positive, spore forming, facultative anaerobic and motile. It was isolated from the stool of a child living in Niamey, Niger, with severe acute malnutrition (kwashiorkor).
Material and Methods
Organism information: classification and features
A stool sample was collected from a 2-year-old girl living in Niamey, Niger, with severe acute malnutrition. Consent was obtained from the child's parents by the National Hospital of Niamey, and the study was approved by the Institut Fédératif de Recherche 48, Faculty of Medicine, Marseille, France (agreement 09-022). The patient did not receive any antibiotics at the time of sample collection; the fecal sample was stored at −80°C.
Strain identification
All the colonies were identified by MALDI-TOF, as described below. In the case of no matching spectra in the database, we further characterized the colonies using 16S rRNA sequencing, as previously described. If the 16S rRNA sequence similarity value was lower than 98.7%, we considered a new species without performing DNA-DNA hybridization as suggested by Stackebrandt and Ebers [21].
Phenotypic characteristics
The main phenotypic characteristics (i.e. Gram staining, motility, sporulation, catalase and oxidase test) were performed as previously described [22]. The chemical characteristics of the newly isolated strain were investigated using API 20NE, API ZYM and API 50CH strips (bioMérieux, Marcy l’Étoile, France). Growth temperatures were tested at 25°C, 30°C, 37°C, 45°C and 55°C. The growth of the strain was tested under anaerobic and microaerophilic conditions using GENbag anaer and GENbag microaer systems, respectively (bioMérieux) and under aerobic conditions with or without 5% CO2. Regarding electron microscopy, detection formvar coated grids were deposited on a 40 μL bacterial suspension drop and incubated during 30 minutes at 37°C. The grids were incubated for 10 seconds on ammonium molybdate 1%, dried on blotting paper and then observed with a Morgani 268D at an operating voltage of 60 kV.
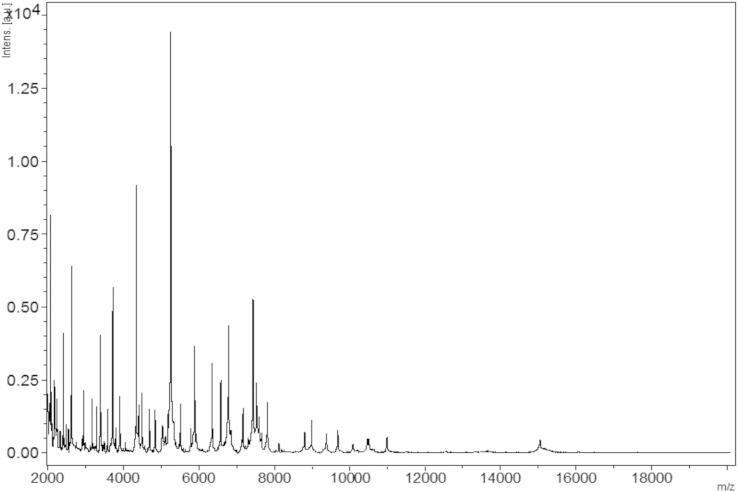
MALDI-TOF protein analysis was carried out as previously described [23] using a Microflex spectrometer (Brüker Daltonics, Leipzig, Germany). Twelve individual colonies were deposited on a MTP 96 MALDI-TOF target plate (Brüker). The 12 spectra were imported into the MALDI BioTyper software (version 2.0; Brüker) and analysed by standard pattern matching (with default parameter settings) against the main spectra of 6.252 bacteria, including 199 spectra from 104 validly named Bacillus species used as reference data in the BioTyper database. A score enabled the presumptive identification and discrimination of the tested species from those in a database: a score of >2 with a validated species enabled the identification at the species level, and a score of <1.7 did not enable any identification [23]. No significant score was obtained for strain mt2, thus suggesting that our isolate was not a member of any known species. The obtained reference spectrum for B. rubiinfantis strain mt2T (Fig. 4) was incremented in our database and compared to other members of the Bacillus genus. The observed differences are shown in gel view in Fig. 5.
Fig. 4.
Reference mass spectrum from B. rubiinfantis strain mt2T. Spectra from 12 individual colonies were compared and reference spectrum was generated.
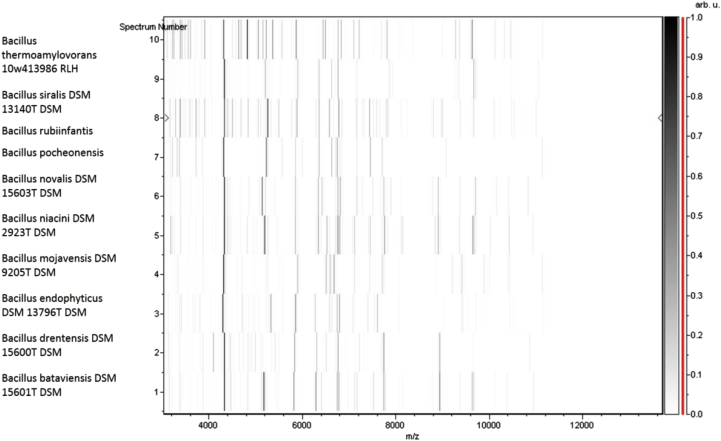
Fig. 5.
Gel view comparing B. rubiinfantis (= CSUR P1141 = DSM 28615) to other species within genus Bacillus. Gel view displays raw spectra of loaded spectrum files arranged in pseudo-gel-like look. X-axis records m/z value. Left y-axis displays running spectrum number originating from subsequent spectra loading. Peak intensity is expressed by greyscale scheme code. Color bar and right y-axis indicate relation between color peak; peak intensity in arbitrary units. Displayed species are indicated on left.
Growth conditions and genomic DNA preparation
B. rubiinfantis sp. nov., strain mt2T (= CSUR P1141 = DSM 28615), was grown on 5% sheep's blood–enriched Columbia agar (bioMérieux) at 37°C in aerobic atmosphere. Bacteria grown on three petri dishes were collected and resuspended in 4 × 100 μL of Tris-EDTA (TE) buffer. Then 200 μL of this suspension was diluted in 1 ml TE buffer for lysis treatment that included a 30- minute incubation with 2.5 μg/μL lysozyme at 37°C, followed by an overnight incubation with 20 μg/μL proteinase K at 37°C. Extracted DNA was then purified using 3 successive phenol–chloroform extractions and ethanol precipitations at −20°C overnight. After centrifugation, the DNA was resuspended in 160 μL TE buffer.
Genome sequencing and assembly
Genomic DNA of Bacillus rubiinfantis was sequenced on the MiSeq Technology (Illumina, San Diego, CA, USA) with the mate-pair strategy. The gDNA was barcoded in order to be mixed with 11 other projects with the Nextera Mate Pair sample prep kit (Illumina). gDNA was quantified by a Qubit assay with the high sensitivity kit (Life Technologies, Carlsbad, CA, USA) to 66.2 ng/μL. The mate-pair library was prepared with 1 μg of genomic DNA using the Nextera mate-pair Illumina guide. The genomic DNA sample was simultaneously fragmented and tagged with a mate-pair junction adapter. The pattern of the fragmentation was validated on an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with a DNA 7500 labchip. The DNA fragments ranged in size from 1 kb up to 11 kb, with an optimal size of 3.927 kb. No size selection was performed, and 505 ng of tagmented fragments were circularized. The circularized DNA was mechanically sheared to small fragments with an optimal at 597 bp on a Covaris S2 device in microtubes (Covaris, Woburn, MA, USA). The library profile was visualized on a High Sensitivity Bioanalyzer LabChip (Agilent), and the final concentration library was measured at 59.2 nmol/L. The libraries were normalized at 2 nM and pooled. After a denaturation step and dilution at 15 pM, the pool of libraries was loaded onto the reagent cartridge and then onto the instrument along with the flow cell. Automated cluster generation and sequencing run were performed in a single 39-hour run at 2 × 251 bp. Total information of 10.7 Gb was obtained from a 1283 K/mm2 cluster density, with a cluster passing quality control filters of 99% (24 072 208 clusters). Within this run, the index representation for Bacillus rubiinfantis was determined to 9.71%. The 2 053 904 paired reads were filtered according to the read qualities. These reads were trimmed, then assembled. A genome with 12 scaffolds and a size of 4.32 Mb was generated. The percentage GC was calculated at 40.0%.
Genome annotation and genome analysis
Open reading frames (ORFs) were predicted using Prodigal (http://prodigal.ornl.gov/) with default parameters, but the predicted ORFs were excluded if they spanned a sequencing gap region (contain N). The predicted bacterial protein sequences were searched against the Clusters of Orthologous Groups (COGs) using BLASTP (E-value 1e−03, coverage 0.7, identity 30%). If no match was found, it searched against the NR database using BLASTP with an E-value of 1e−03, coverage 0.7 and identity 30%, If the sequence lengths were smaller than 80 amino acids, we used an E-value of 1e−05. The tRNAScanSE tool [24] was used to find tRNA genes, whereas ribosomal RNAs were found by RNAmmer [25]. Lipoprotein signal peptides and the number of transmembrane helices were predicted using Phobius [26]. ORFans were identified if all the performed BLASTP did not give positive results (E value smaller than 1e−03 for ORFs with sequence size upper than 80 aa or E value smaller than 1e−05 for ORFs with sequence length smaller 80 aa). Such parameter thresholds have already been used in previous works to define ORFans.
For the genomic comparison, we compared our type strain Bacillus rubiinfantis strain mt2T to Bacillus bataviensis (GenBank accession number AJ542508.1), Bacillus infantis (AY904032.1), Bacillus endophyticus (AF295302.1), Bacillus pseudomycoides (AF013121.1), Bacillus thuringiensis (D16281.1), Bacillus amyloliquefaciens (AB006920.1), Bacillus mojavensis (AB021191.1), Bacillus thermoamylovorans (L27478.1) and Clostridium butyricum (AJ58420.1). Genomes were automatically retrieved from the 16S RNA tree using Xegen software (Phylopattern) [27]. For each selected genome, the complete genome sequence, proteome genome sequence and Orfeome genome sequence were retrieved from the File Transfer Protocol (FTP) of National Center for Biotechnology Information. All proteomes were analysed with proteinOrtho [28]. For each couple of genomes, a similarity score was then computed. This score is the mean value of nucleotide similarity between all couples of orthologues between the two genomes studied (AGIOS) [6]. An annotation of the entire proteome was performed to define the distribution of the functional classes of predicted genes according to the clusters of orthologous groups of proteins (using the same method as for the genome annotation).
Results
Phenotypic description
Strain mt2T (Table 1) was first isolated in January 2014 by a 21-day anaerobic preincubation in blood culture with sheep's blood and cultivation 5% sheep's blood–enriched Colombia agar (BioMérieux) in anaerobic atmosphere at 37°C.
Table 1.
Classification and general features of Bacillus rubiinfantis strain mt2
| Property | Term |
|---|---|
| Current classification | Domain: Bacteria |
| Phylum: Firmicutes | |
| Class: Bacilli | |
| Order: Bacillales | |
| Family: Bacillaceae | |
| Genus: Bacillus | |
| Species: Bacillus rubiinfantis | |
| Type strain: mt2 | |
| Gram stain | Positive |
| Cell shape | Rod |
| Motility | Motile |
| Sporulation | sporulating |
| Temperature range | Mesophilic |
| Optimum temperature | 37°C |
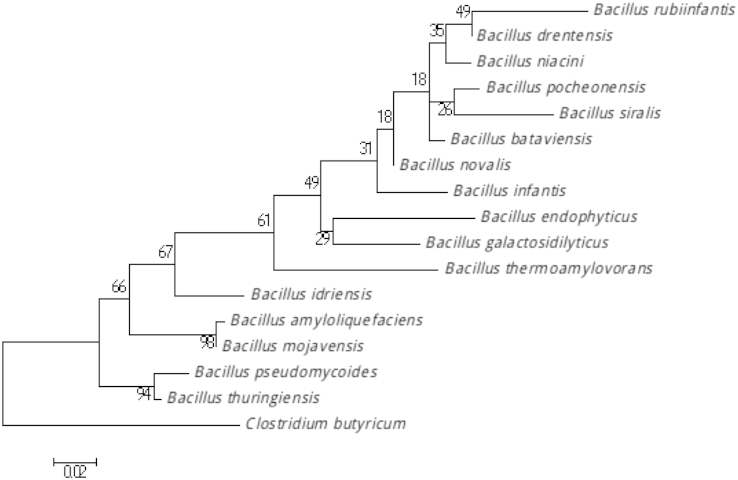
This strain showed a 96% nucleotide sequence similarity with B. bataviensis, the phylogenetically closest Bacillus species with a valid published name (Fig. 1). This similarity value is below the 16S rRNA threshold of 98.7% set by Stackebrandt and Ebers to delineate a new species without carrying out DNA-DNA hybridization [21].
Fig. 1.
Phylogenetic tree highlighting position of Bacillus rubiinfantis sp. nov. strain mt2T (= CSUR P1141 = DSM 28615) relative to other type strains within Bacillus genus. Strains and their corresponding GenBank accession numbers for 16S rRNA genes are (type = T): B. novalis strain SCTB 113, JN650278; B. drentensis DRG 4, HQ436340; B. niacini strain TSII-13, JN993716; B. bataviensis strain LMG21832, AJ542507; B. pocheonensis strain Gsoil 420, AB245377.1; B. infantis strain A-49, KC751070; B. siralis strain 171544, NR_028709.1; B. endophyticus strain 2DT, NR_025122.1; B. pseudomycoides strain BIHB 360, FJ859700.1; B. thuringiensis strain GMC 108, AB741470; B. idriensis strain SMC 4352–2, NR_043268.1; B. amyloliquefaciens strain BIHB 35, FJ859496 ; B. mojavensis strain BCRC 17124, EF433405.1 ; B. galactosidilyticus strain LMG 17892, NR_025580.1 ; B. thermoamylovorans strain DKP, NR_029151.1. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA6. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 1000 times to generate majority consensus tree. Bacteroides thetaiotaomicron strain ATCC 29148T (L16489) was used as outgroup. Scale bar = 1% nucleotide sequence divergence.
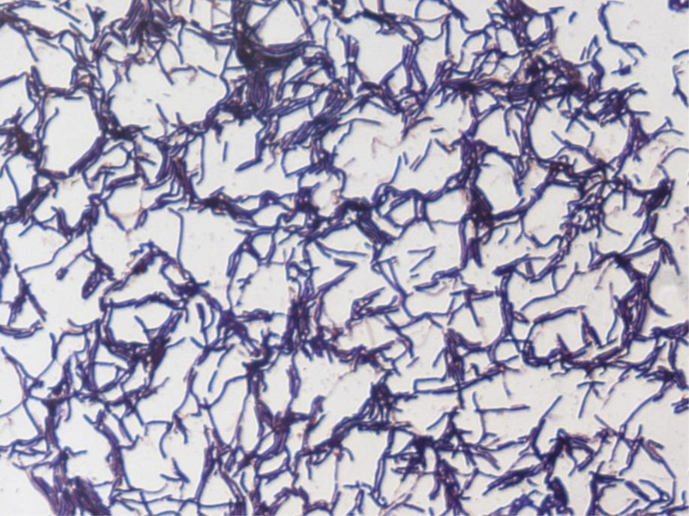
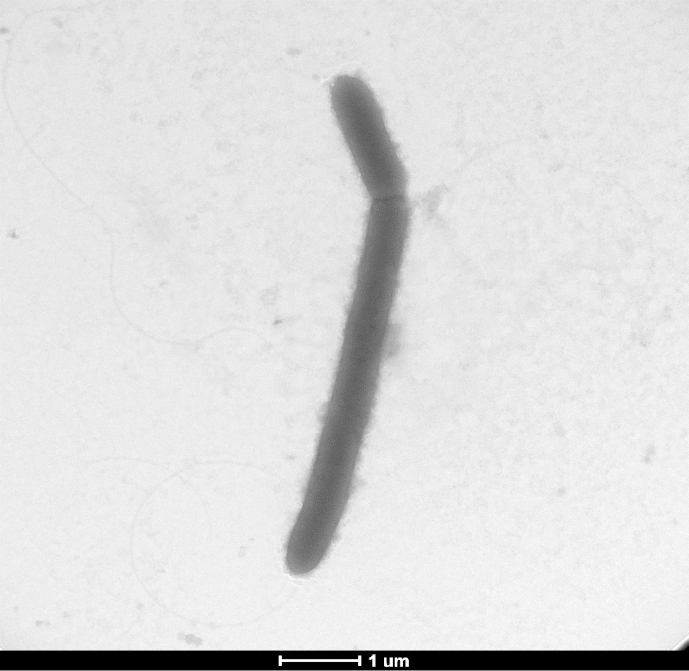
Various growth temperatures (25°C, 30°C, 37°C, 45°C and 56°C) were tested. Growth occurred between 25°C and 45°C on blood-enriched Columbia agar (bioMérieux), with the optimal growth being obtained at 37°C after 48 hours of incubation. Growth of the strain was tested under anaerobic and microaerophilic conditions using GENbag Anaer and GENbag microaer systems, respectively (bioMérieux), and under aerobic conditions, with or without 5% CO2. Optimal growth was achieved aerobically. Weak cell growth was observed under microaerophilic and anaerobic conditions. The motility test was positive, and the cells were sporulating. Colonies were translucent and 10 mm in diameter on blood-enriched Columbia agar. Cells were Gram-positive rods (Fig. 2). Under electron microscopy, the bacteria grown on agar had a mean diameter and length of 0.54 and 0.62 μm, respectively (Fig. 3).
Fig. 2.
Gram staining of B. rubiinfantis strain mt2T.
Fig. 3.
Transmission electron microscopy of B. rubiinfantis strain mt2T using Morgani 268D (Philips) at operating voltage of 60 kV. Scale bar = 1 μm
Strain mt2T exhibited catalase activity but was negative for oxidase. Using an API ZYM strip (BioMérieux), positive reactions were observed for esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, cystine arylamidase, α-chymotrypsin, acid phosphatase, naphtol-AS-BI-phosphohydrolase, α-galactosidase and β-galactosidase. Negative reactions were observed for lipase (C14), trypsin, alkaline phosphatase, β-glucuronidase, α-glucosidase, β-glucosidase, α-fucosidase, α-mannosidase and N-acetyl-β-glucosaminidase. Using an API 20 NE strip, positive reactions were obtained for l-arginine, urea, 4-nitrophenyl- βd-galactopyranoside and capric acid. All other reactions were negative.
Using an API 50 CH strip (BioMérieux), positive reactions were observed for d-galactose, d-glucose, d-fructose, d-mannose, N-acetylglucosamine, esculin ferric citrate, d-maltose, d-lactose, d-melibiose and d-trehalose. Negative reactions were observed glycerol, erythrol, d-arabinose, l-arabinose, d-ribose, l-xylose, d-xylose, d-adonitol, methyl-βd-xylopranoside, l-sorbose, l-rhamnose, dulcitol, inositol, d-mannitol, d-sorbitol, methyl-αd-mannopyranoside, methyl-αd-glucopyranoside, amygdaline, arbutin, salicin, d-cellobiose, d-maltose, d-saccharose, inulin, d-melezitose, d-raffinose, amidon, glycogen, gentiobiose, d-turanose, d-lyxose, d-tagatose, d-fucose, l-fucose, d-arabitol, l-arabitol, potassium gluconate, potassium 2-ketogluconate and potassium 5-ketogluconate.
Cells are susceptible to vancomycin, rifampicin, doxycycline, erythromycin, amoxicillin, nitroflurantoin, ciprofloxacin, ceftriaxone, penicillin G, gentamicin and imipenem but resistant to trimethoprim/ sulfamethoxazole and metronidazole. The differences exhibited by the comparison with other representatives of the genus Bacillus are detailed in Table 2.
Table 2.
Differential characteristics of Bacillus rubiinfantis strain mt2T, Bacillus novalis LMG 21837, Bacillus bataviensis LMG 21833, Bacillus drentensis LMG 21831, Bacillus niacini DSM 2923, Bacillus endophyticus UCM 5715, Bacillus siralis NCIMB 13601, Bacillus thermoamylovorans CNCM l 1378, Bacillus pocheonensis DSM 18135, Bacillus mojavensis NRRL B 14698 [31], [32], [33], [34], [35], [36], [37], [38], [39]
| Property | B. rubiinfantis | B. nolis | B. bataviensis | B. drentensis | B. niacini | B. endophyticus | B. siralis | B. thermo-amylovorans | B. pocheonensis | B. mojavensis |
|---|---|---|---|---|---|---|---|---|---|---|
| Cell diameter (μm) | 0.5–0.6 | 0.6–1.2 | 0.7–1.2 | 0.6–1.2 | 0.9–1.4 | 0.5–1.5 | 0.5–0.8 | 0.45–0.5 | 1.5–3 | 1–2 |
| Oxygen requirement | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic |
| Gram stain | + | + | + | + | + | + | + | + | + | + |
| Salt requirement | NA | NA | NA | NA | NA | + | + | NA | + | NA |
| Motility | + | + | + | + | − | − | NA | + | − | + |
| Endospore formation | + | + | + | + | + | + | + | − | + | + |
| Indole | − | − | − | − | + | − | NA | − | − | − |
| Production of: | ||||||||||
| Alkaline phosphatase | − | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Catalase | + | NA | NA | NA | + | + | + | + | + | + |
| Oxidase | − | NA | NA | NA | + | + | + | + | + | + |
| Nitrate reductase | − | + | + | +/− | + | − | + | − | + | + |
| Urease | + | − | − | − | − | − | NA | NA | − | − |
| β-Galactosidase | + | + | + | + | + | − | − | + | + | + |
| N-Acetyl-glucosamine | − | + | + | + | + | NA | NA | + | + | NA |
| Acid from: | ||||||||||
| l-Arabinose | − | − | − | − | − | + | − | − | + | + |
| Ribose | − | + | + | + | − | + | − | + | − | − |
| Mannose | + | + | + | + | + | + | − | + | + | + |
| Mannitol | − | + | + | − | − | + | − | − | − | + |
| Sucrose | −+ | − | + | + | + | + | − | + | − | + |
| d-Glucose | + | + | + | + | + | + | − | + | − | + |
| d-Fructose | + | + | + | + | + | NA | − | + | + | + |
| d-Maltose | − | + | + | + | + | + | − | + | + | + |
| d-Lactose | + | − | + | + | + | + | + | + | − | − |
| Habitat | Human gut | Soil | Soil | Soil | soil | Cotton plants | Grassland soil | Palm vine | Soil | soil |
NA: non-available data
Genome properties
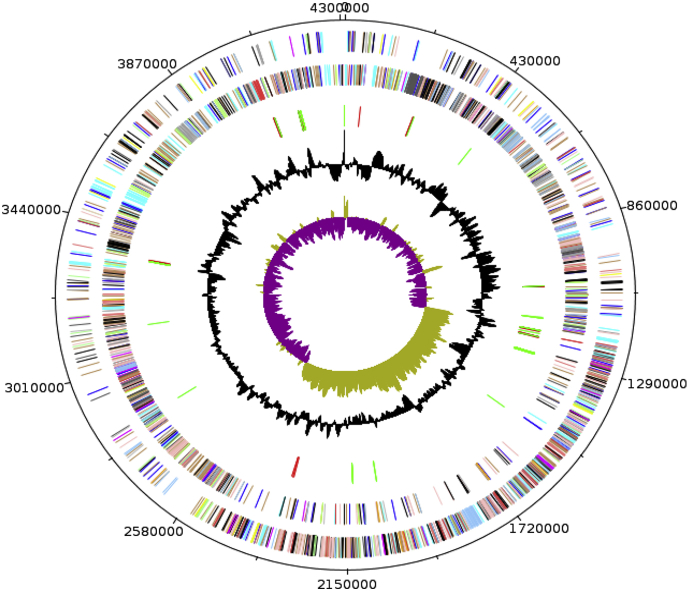
The genome of B. rubiinfantis strain mt2T is 4 311 083 bp long with a 40.0% G+C content (Fig. 6). Of the 4149 predicted genes, 4028 were protein-coding genes and 121 were RNAs. Out of these 121 rRNA genes, 11 are 16S rRNA genes, 9 are 23S rRNA, 11 are 5S rRNA and 90 predicted tRNA genes were identified in the genome. A total of 3026 genes (72.93%) were assigned a putative function. Two hundred two genes were identified as ORFans (5.04%). The remaining genes were annotated as hypothetical proteins. The properties and statistics of the genome are summarized in Table 3, Table 4. The distribution of genes into COGs functional categories is presented in Table 4.
Fig. 6.
Graphical circular map of chromosome. From outside to center: genes on forward strain colored by COGs categories (only gene assigned to COGs), RNA genes (Trnas green, rRNAs red), GC content and GC skew. COGs, Clusters of Orthologous Groups.
Table 3.
Nucleotide content and gene count levels of the genome
| Attribute | Genome (total) |
|
|---|---|---|
| Value | % of totala | |
| Size (bp) | 4 311 083 | 100 |
| G+C content (bp) | 1 724 433 | 40.0 |
| Coding region (bp) | 3 672 471 | 84.88 |
| Total genes | 4149 | 100 |
| RNA genes | 121 | 2.80 |
| Protein-coding genes | 4028 | 97.08 |
| Genes with function prediction | 3026 | 72.93 |
| Genes assigned to COGs | 2666 | 64.25 |
| Genes with peptide signals | 244 | 5.88 |
| Genes with transmembrane helices | 1045 | 25.18 |
| Genes with Pfam domains | 1405 | 34.02 |
| CRISPR repeats | 01 | 0.02 |
COGs, Clusters of Orthologous Groups.
Total is based on either size of genome (in base pairs) or total number of protein coding genes in annotated genome.
Table 4.
Number of genes associated with 25 general COGs functional categories
| Code | Value | % of totala | Description |
|---|---|---|---|
| J | 163 | 3.97 | Translation |
| A | 0 | 0 | RNA processing and modification |
| K | 265 | 5.34 | Transcription |
| L | 235 | 4.62 | Replication, recombination, repair |
| B | 1 | 0.02 | Chromatin structure and dynamics |
| D | 40 | 0.92 | Cell cycle control, mitosis, meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 65 | 1.46 | Defense mechanisms |
| T | 197 | 3.57 | Signal transduction mechanisms |
| M | 184 | 3.60 | Cell wall/membrane biogenesis |
| N | 60 | 1.24 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 46 | 0.97 | Intracellular trafficking and secretion |
| O | 122 | 2.73 | Posttranslational modification, protein turnover, chaperones |
| C | 211 | 4079 | Energy production and conversion |
| G | 183 | 3.85 | Carbohydrate transport and metabolism |
| E | 397 | 9.14 | Amino acid transport and metabolism |
| F | 91 | 2.09 | Nucleotide transport and metabolism |
| H | 107 | 2.46 | Coenzyme transport and metabolism |
| I | 133 | 3.15 | Lipid transport and metabolism |
| P | 218 | 4.87 | Inorganic ion transport and metabolism |
| Q | 81 | 1.79 | Secondary metabolites biosynthesis, transport, catabolism |
| R | 428 | 9.41 | General function prediction only |
| S | 291 | 6.41 | Function unknown |
| — | 3518 | 33.47 | Not in COGs |
COGs, Clusters of Orthologous Groups.
Total is based on total number of protein coding genes in annotated genome.
It was the 70th organism identified within the Bacillus genera. The genome's GenBank accession number is CCFE01000000 and consists of 12 scaffolds and 33 contigs.
Genome comparison
The draft genome sequence of Bacillus rubiinfantis is larger than those of Bacillus amyloliquefaciens, Bacillus mojavensis and Bacillus thermoamylovorans (4.01, 3.96 and 3.71) but smaller than those of Bacillus pseudomycoides, Bacillus bataviensis, Bacillus thuringiensis, Bacillus infantis, Bacillus endophyticus, Bacillus infantis and Clostridium butyricum (5.75, 5.37, 5.24, 5.1, 4.88 and 4.62 MB, respectively). The G+C content of Bacillus rubiinfantis is smaller than those of Bacillus amyloliquefaciens, Bacillus infantis and Bacillus mojavensis (46.1, 46.0 and 43.7) but larger than those of Bacillus bataviensis, Bacillus thermoamylovorans, Bacillus endophyticus, Bacillus pseudomycoides, Bacillus thuringiensis and Clostridium butyricum (39.6%, 37.3%, 36.4%, 35.4%, 35.4% and 28.8%, respectively). The gene content of Bacillus rubiinfantis is larger than those of Bacillus mojavensis, Bacillus amyloliquefaciens and Bacillus thermoamylovorans (4131, 4005 and 3458) but lower than that of Bacillus pseudomycoides, Bacillus thuringiensis, Bacillus bataviensis, Bacillus endophyticus, Bacillus infantis and Clostridium butyricum (5941, 5263, 5238, 4996, 4837 and 4231, respectively). To evaluate the genomic similarity among studied Bacillus strains, we determined two parameters, dDDH, Which exhibits a high correlation with DDH [29], [30], and AGIOS [6], which was designed to be independent from DDH (Table 5, Table 6).
Table 5.
Pairwise comparison of Bacillus rubiinfantis (upper right) with eight other species using GGDC, formula 2 (DDH estimates based on identities/HSP length)a
| B. rubiinfantis | B. amyloliquefaciens | B. bataviensis | B. endophyticus | B. mojavensis | B. infantis | B. pseudomycoides | B. thermoamylovorans | B. thuringiensis | C. butyricum | |
|---|---|---|---|---|---|---|---|---|---|---|
| B. rubiinfantis | 100% ± 00 | 2.54% ± 0.14 | 2.66% ± 0.23 | 2.54% ± 0.16 | 2.55% ± 0.14 | 2.57% ± 0.20 | 2.54% ± 0.16 | 2.54% ± 0.23 | 2.55% ± 0.14 | 2.53% ± 0.16 |
| B. amyloliquefaciens | 100% ± 00 | 2.54% ± 0.22 | 2.54% ± 0.20 | 3.01% ± 0.22 | 2.54% ± 0.20 | 2.53% ± 0.19 | 2.53% ± 0.24 | 2.54% ± 0.13 | 2.53% ± 0.18 | |
| B. bataveinsis | 100% ± 00 | 2.54% ± 0.21 | 2.54% ± 0.19 | 2.61% ± 0.20 | 2.54% ± 0.21 | 2.53% ± 0.24 | 2.55% ± 0.18 | 2.53% ± 0.17 | ||
| B. endophyticus | 100% ± 00 | 2.55% ± 0.18 | 2.54% ± 0.23 | 2.55% ± 0.22 | 2.53% ± 0.22 | 2.56% ± 0.20 | 2.53% ± 0.19 | |||
| B. mojavensis | 100% ± 00 | 2.54% ± 0.16 | 2.54% ± 0.16 | 2.53% ± 0.23 | 2.55% ± 0.12 | 2.53% ± 0.17 | ||||
| B. infantis | 100% ± 00 | 2.53% ± 0.19 | 2.53% ± 0.22 | 2.55% ± 0.18 | 2.52% ± 0.11 | |||||
| B. pseudomycoides | 100% ± 00 | 2.53% ± 0.19 | 3.00% ± 0.16 | 2.53% ± 0.18 | ||||||
| B. thermoamylovorans | 100% ± 00 | 2.53% ± 0.17 | 2.52% ± 0.21 | |||||||
| B. thuringiensis | 100% ± 00 | 2.54% ± 0.17 | ||||||||
| C. butyricum | 100% ± 00 |
GGDC, Genome-to-Genome Distance Calculator; DDH, DNA-DNA Hybridization; HSP, High-Scoring Pair.
Confidence intervals indicate inherent uncertainty in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size) These results are in accordance with 16S rRNA (Fig. 1) and phylogenomic analyses as well as GGDC results.
Table 6.
Numbers of orthologous protein shared between genomes (upper right) average percentage similarity of nucleotides corresponding to orthologous protein shared between genomes (lower left)
| B. amyloliquefaciens | B. bataviensis | B. endophyticus | B. mojavensis | B. infantis | B. pseudomycoides | B. rubiinfantis | B. thermoamylovorans | B. thuringiensis | C. butyricum | |
|---|---|---|---|---|---|---|---|---|---|---|
| B. amyloliquefaciens | 4022a | 1674 | 1864 | 2028 | 686 | 1642 | 1486 | 1442 | 1707 | 798 |
| B. bataviensis | 65.44 | 5265a | 1765 | 1653 | 878 | 1785 | 1850 | 1599 | 1807 | 854 |
| B. endophyticus | 65.43 | 66.90 | 5077a | 1849 | 749 | 1769 | 1547 | 1481 | 1839 | 833 |
| B. mojavensis | 77.91 | 65.86 | 66.25 | 4078a | 693 | 1637 | 1466 | 1413 | 1694 | 776 |
| B. infantis | 65.39 | 74.32 | 67.37 | 65.97 | 2152a | 744 | 740 | 633 | 757 | 391 |
| B. pseudomycoides | 64.79 | 67.00 | 68.06 | 65.55 | 67.10 | 5863a | 1556 | 1416 | 2212 | 868 |
| B. rubiinfantis | 65.31 | 73.95 | 66.82 | 65.78 | 72.33 | 67.19 | 4028a | 1532 | 1571 | 778 |
| B. thermoamylovorans | 64.64 | 67.30 | 66.27 | 65.10 | 67.67 | 66.64 | 67.41 | 3468a | 1445 | 785 |
| B. thuringiensis | 64.42 | 66.65 | 67.70 | 65.23 | 66.92 | 83.41 | 66.80 | 66.33 | 5271a | 879 |
| C. butyricum | 57.74 | 60.66 | 61.81 | 59.14 | 61.44 | 62.07 | 60.72 | 61.49 | 62.11 | 4148a |
Number of proteins per genome.
however, the distribution of genes into COGs categories was similar in all compared genomes (Fig. 7(. In addition, B. rubiinfantis shared 4022, 5265, 5077, 4078, 2152, 5863, 3468, 5271 and 4148 orthologous genes with B. amyloliquefaciens, B. bataviensis, B. endophyticus, B. mojavensis, B. infantis, B. pseudomycoides, B. thuringiensis, B. thermoamylovorans and Clostridium butyricum (Table 6). Among species with standing in nomenclature, AGIOS values ranged from 57.74 between Clostridium butyricum and B. amyloliquefaciens to 83.41% between B. thuringiensis and B. pseudomycoides.
Fig. 7.
Distribution of functional classes of predicted genes according to COGs. COGs, Clusters of Orthologous Groups.
Conclusion
On the basis of phenotypic, phylogenetic and genomic analyses, we formally propose the creation of Bacillus rubiinfantis sp. nov. that contains the strain mt2. This bacterial strain has been isolated from the fecal flora of a 2-year-old girl from Niamey, Niger, with kwashiorkor.
Taxonomic and nomenclatural proposals: description of Bacillus rubiinfantis strain mt2T sp. nov.
The cells are Gram-positive, sporulating and rod-shaped bacilli with a diameter of 0.5 μm. Colonies are translucent and 10 mm in diameter on 5% sheep's blood–enriched Columbia agar (bioMérieux) and are catalase positive and oxidase negative. Positive reactions are observed for esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, cystine arylamidase, α-chymotrypsin, acid phosphatase, naphtol-AS-BI-phosphohydrolase, α-galactosidase and β-galactosidase. Negative reactions are observed for lipase (C14), trypsin, nitrate reduction, alkaline phosphatase, β-glucuronidase, α-glucosidase, β-glucosidase, α-fucosidase, α-mannosidase and N-acetyl-β-glucosaminidase.
Cells are susceptible to vancomycin, rifampicin, doxycycline, erythromycin, amoxicillin, nitroflurantoin, ciprofloxacin, ceftriaxone, penicillin G, gentamicin and imipenem but resistant to trimethoprim/sulfamethoxazole and metronidazole.
The G+C content of the genome is 40.0%. The 16S rRNA gene sequence and whole-genome shotgun sequence of B. rubiinfantis strain mt2T are deposited in GenBank under accession numbers LK021113 and CCFE01000000, respectively. The type strain mt2T (= CSUR P1141 = DSM 28615) was isolated from the stool of a child living in Niamey, Niger, with kwashiorkor.
Acknowledgements
The authors thank the Xegen Company (http://www.xegen.fr/) for automating the genomic annotation process. This study was funded by the Mediterranée-Infection Foundation. We thank K. Griffiths for her English-language review.
Conflict of Interest
None declared.
References
- 1.Viale A.M., Arakaki A.K., Soncini F.C., Ferreyra R.G. Evolutionary relationships among eubacterial groups as inferred from GroEL (Chaperonin) sequences comparison. Int J Syst Bacteriol. 1944;44(3):527–533. doi: 10.1099/00207713-44-3-527. [DOI] [PubMed] [Google Scholar]
- 2.Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposals for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf M., Muller T., Dandekar T., Pollack J.D. Phylogeny of Firmicutes with special reference to Mycoplasma (Mollicutes) as inferred from phosphoglycerate kinase amino acid sequence data. Inst J Syst Evol Microbiol. 2004;54(Pt3):871–875. doi: 10.1099/ijs.0.02868-0. [DOI] [PubMed] [Google Scholar]
- 4.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015 Jan;28(1):237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 6.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi-Tamisier M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of new bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 7.Lagier J.C., El Karkouri K., Nguyen T.T., Armougom F., Raoult D., Fournier P.E. Non-contiguous finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand Genomic Sci. 2012;6:116–125. doi: 10.4056/sigs.2415480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagier J.C., Armougom F., Mishra A.K., Nguyen T.T., Raoult D., Fournier P.E. Non-contiguous finished genome sequence and description of Alistipes timonensis sp. nov. Stand Genomic Sci. 2012;6:315–324. doi: 10.4056/sigs.2685971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roux V., El Karkouri K., Lagier J.C., Robert C., Raoult D. Non-contiguous finished genome sequence and description of Kurthia massiliensis sp. nov. Stand Genomic Sci. 2012;7:221–232. doi: 10.4056/sigs.3206554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kokcha S., Ramasamy D., Lagier J.C., Robert C., Raoult D., Fournier P.E. Non-contiguous finished genome sequence and description of Brevibacterium senegalense sp. nov. Stand Genomic Sci. 2012;7:233–245. doi: 10.4056/sigs.3256677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramasamy D., Kokcha S., Lagier J.C., Nguyen T.-T., Raoult D., Fournier P.E. Genome sequence and description of Aeromicrobium massiliense sp. nov. Stand Genomic Sci. 2012;7:246–257. doi: 10.4056/sigs.3306717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagier J.C., Elkarkouri K., Rivet R., Couderc C., Raoult D., Fournier P.E. Non contiguous-finished genome sequence and description of Senegalemassilia anaerobia gen. nov., sp. nov. Stand Genomic Sci. 2013;7:343–356. doi: 10.4056/sigs.3246665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagier J.C., El Karkouri K., Mishra A.K., Robert C., Raoult D., Fournier P.E. Non contiguous-finished genome sequence and description of Enterobacter massiliensis sp. nov. Stand Genomic Sci. 2013;7:399–412. doi: 10.4056/sigs.3396830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugon P., Mishra A.K., Lagier J.C., Nguyen T.T., Couderc C., Raoult D. Non-contiguous finished genome sequence and description of Brevibacillus massiliensis sp. nov. Stand Genomic Sci. 2013;8:1–14. doi: 10.4056/sigs.3466975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagier J.C., Gimenez G., Robert C., Raoult D., Fournier P.E. Non-contiguous finished genome sequence and description of Herbaspirillum massiliense sp. nov. Stand Genomic Sci. 2012 Dec 19;7(2):200–209. doi: 10.4056/sigs.3086474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Priest F.G., Goodfellow M., Todd C. A numerical classification of the genus Bacillus. J Gen Microbiol. 1988;134:1847–1882. doi: 10.1099/00221287-134-7-1847. [DOI] [PubMed] [Google Scholar]
- 17.Fritze D. Taxonomy of the genus Bacillus and related genera: the aerobic endospore-forming bacteria. Physiopathology. 2004;94(11):1245–1248. doi: 10.1094/PHYTO.2004.94.11.1245. [DOI] [PubMed] [Google Scholar]
- 18.Gordon R.E., Haynes W.C., Pang C.H. The genus Bacillus. US Dep Agric Handh. 1973;427 [Google Scholar]
- 19.Goto K., Omura T., Hara Y., Sadaie Y. Application of the 16s rDNA sequence as an index for rapid identification of species in the genus Bacillus. J Gen Appl Microbiol. 2000;46(1):1–8. doi: 10.2323/jgam.46.1. [DOI] [PubMed] [Google Scholar]
- 20.http://www.bacterio.net/bacillus.html.
- 21.Prevot A.R. Dictionnaire des bactéries pathogens. In: Hauduroy P., Ehringer G., Guillot G., Magrou J., Prevot A.R., Rosset, Urbain A., editors. Masson; Paris: 1953. pp. p.1–692. [Google Scholar]
- 22.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 23.Lagier J.C., Edouard S., Pagnier I., Mediannikov O., Drancourt M., Raoult D. Current and past strategies for bacterial culture in clinical microbiology. Clin Microbiol Rev. 2015 Jan;28(1):208–236. doi: 10.1128/CMR.00110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prodigal (http://prodigal.ornl.gov/).
- 26.Lagesen K., Hallin P., Rodland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 28.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 29.Gouret P., Thompson J.D., Pontarotti P. PhyloPattern: regular expressions to identify complex patterns in phylogenetic trees. BMC Bioinformatics. 2009;10:298. doi: 10.1186/1471-2105-10-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lechner M., Findeib S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auch A.F., Von Jan M., Klenk H.P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyram J., Vanparys B., Logan N.A., Balcaen A., Rodriguez-Diaz M., Felske A. Bacillus novalis sp. nov., Bacillus vireti sp. nov.,Bacillus soli sp. nov., Bacillus bataviensis sp. nov., and Bacillus drentensis sp. nov., from the Drentse A grasslands. Int J Syst Evol Microbiol. 2004;54(1):47–57. doi: 10.1099/ijs.0.02723-0. [DOI] [PubMed] [Google Scholar]
- 34.Nagel M., Andreesen J.R. Bacillus niacini sp. nov., a nicotinate- metabolizing mesophile isolated from soil. Int J Syst Evol Microbiol. 1991;41(1):134–139. [Google Scholar]
- 35.Reva O.N., Smirnov V.V., Petterson B., Priest F.G. Bacillus endophyticus sp. nov., isolated from the inner tissues of cotton plants (Gossypium sp.) Int J Syst Evol Microbiol. 2002;52(1):101–107. doi: 10.1099/00207713-52-1-101. [DOI] [PubMed] [Google Scholar]
- 36.Petterson B., De Silva S.K., Uhlén M., Priest F.G. Bacillus siralis sp. nov., a novel species from silage with a higher order structural attribute in the 16S rRNA genes. Int J Syst Evol Microbiol. 2000;50(6):2181–2187. doi: 10.1099/00207713-50-6-2181. [DOI] [PubMed] [Google Scholar]
- 37.Combet-Blanc Y., Olivier B., Streicher C., Patel B.K., Dwivedi P.P., Pot B. Bacillus thermoamylovorans sp. nov., a moderately thermophilic and amylolytic bacterium. Int J Syst Evol Microbiol. 1995;45(1):9–16. doi: 10.1099/00207713-45-1-9. [DOI] [PubMed] [Google Scholar]
- 38.Ten L.N., Baek S.H., Im W.T., Larina L.L., Lee J.S., Oh H.M. Bacillus pocheonensis sp. nov., a moderately halotolerant, aerobic bacterium isolated from soil of a ginseng field. Int J Syst Evol Microbiol. 2007;57(11):2532–2537. doi: 10.1099/ijs.0.64491-0. [DOI] [PubMed] [Google Scholar]
- 39.Robert M.S., Nakamura L.K., Cohan F.M. Bacillus mojavensis sp. nov., distinguishable from Bacillus subtilis by sexual isolation, divergence in DNA sequence, and differences in fatty acid composition. Int J Syst Evol Microbiol. 1994;44(2):256–264. doi: 10.1099/00207713-44-2-256. [DOI] [PubMed] [Google Scholar]