The prognostication of patients with chronic lymphocytic leukemia (CLL) currently relies on both clinical and biological parameters (Figure 1). The prime example concerns the TP53 gene, whereby inactivation of TP53, resulting from either a mutation or chromosome 17p deletion, is associated with a short time to progression, an early need for treatment, and an overall dismal outcome.1,2 The presence of TP53 aberrations is also a strong predictor of treatment response, as patients carrying such lesions respond poorly to standard chemoimmunotherapy (CIT) (i.e. fludarabine, cyclophosphamide, and rituximab).3 Although TP53 abnormalities are infrequent at diagnosis (5%–10%), they are found in 40%–50% of advanced or therapy-refractory cases, hence underscoring the need to re-assess TP53 gene status as the disease progresses and clones evolve.2,4,5
Figure 1.
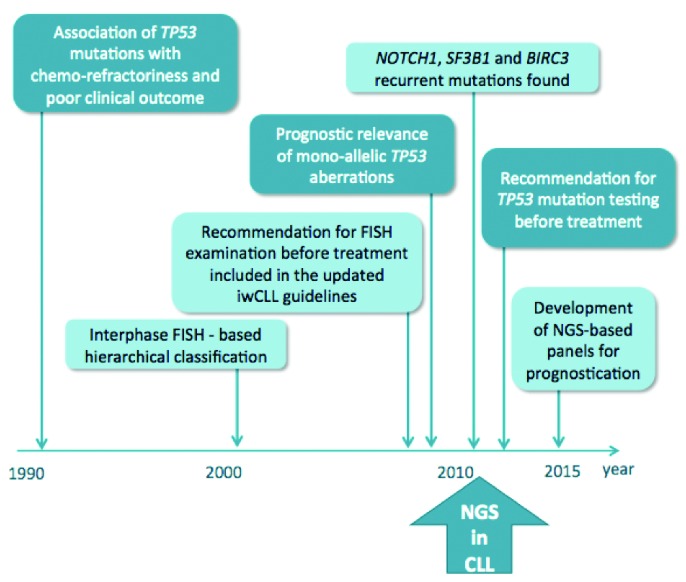
Gene and chromosomal defects in the management of chronic lymphocytic leukemia. Chronology of the introduction of clinically relevant chromosomal aberrations and gene mutations into clinical routine practice. FISH: fluorescence in situ hybridization; NGS: next generation sequencing.
TP53 inactivation in CLL was identified more than 20 years ago in the context of advanced disease, chemo-refractoriness and poor clinical outcome. However, almost a decade passed before the analysis of TP53 defects by fluorescence in situ hybridization (FISH) was introduced into routine clinical practice, spurred on by the establishment of the Döhner hierarchical classification of cytogenetic abnormalities.1 In this model, patient stratification was based on the presence of recurrent chromosomal aberrations (17p deletion, including the TP53 locus, 11q deletion, including the ATM locus, trisomy 12 and 13q deletion), with patients carrying del(17p) having the shortest survival.1 More recently, studies have shown that approximately 80% of patients harboring del(17p) also carry a mutation on the second allele of the TP53 gene.4,6,7 Conversely, only approximately 60% of patients with TP53 mutations carry del(17p); nevertheless, studies have revealed that both biallelic and monoallelic defects (sole mutation or sole deletion) convey an equally poor prognosis. The poor prognosis conveyed by TP53 aberrations is mainly due to refractoriness to CIT treatment.4–8
The advent of agents acting independently of the p53 pathway looks promising for the treatment of CLL patients carrying TP53 aberrations. Recently, two inhibitors of B-cell receptor (BCR) signaling, namely ibrutinib [inhibitor of Bruton’s tyrosine kinase (BTK)], and idelalisib [inhibitor of phosphatidylinositol 3-kinase delta (PI3Kδ)] have been approved by both the Food and Drug administration (FDA) and the European Medicines Agency (EMA) for the first-line treatment of cases harboring del(17p)/TP53 mutations or the treatment of CLL cases refractory to CIT or unfit for CIT.9,10 Additional small molecules targeting other pathophysiological processes are set to be approved, e.g. the BH3 mimetic ABT199 (Venetoclax), with preliminary results indicating efficacy in del(17p)/TP53 mutated CLL cases.11
These new therapeutic options have also accelerated the need to analyze del(17p)/TP53 mutations prior to therapy, and the European Research Initiative on CLL (ERIC) (www.ericll.org) has been at the forefront of this field by: i) formulating recommendations on the mutational analysis of TP53 in CLL, including guidance about when and in whom del(17p)/TP53 mutations should be investigated;5 ii) establishing the European CLL TP53 Network, aimed at promoting and advancing the analysis of TP53 gene aberrations across the medical community; iii) launching a certification system with external quality control for laboratories assessing TP53 mutations; and finally, iv) arranging dedicated educational workshops (the 1st ERIC workshop on TP53 analysis in CLL was held in October 2015 in Brno, the Czech Republic).
While many laboratories currently utilize Sanger sequencing for TP53 analysis, other laboratories use pre-screening techniques such as denaturing high performance liquid chromatography (DHPLC) or functional analysis of separated allele in yeast (FASAY), in combination with Sanger sequencing.5 Technological progress over the last few years has led to the introduction of next generation sequencing (NGS), which will increasingly be applied to clinical diagnostics in the near future.12 In addition to monitoring the clonal population, targeted NGS, in which selected genes, e.g. TP53, are analyzed, now allows minor clones to be detected, even those below 1% variant allelic frequency (VAF), thus facilitating the study of clonal evolution. This is extremely important since resistance to CIT has been shown to be associated with the expansion of minor CLL clones carrying TP53 defects, which are already present but remain undetected at initiation of therapy, whereas the acquisition of novel TP53 defects in untreated patients is only rarely observed.13,14 In additon to cytotoxic therapy, other factors must be involved in clonal selection since TP53-mutated subclones persist at low allelic burden in some cases despite having received several lines of therapy.13–15
Next generation sequencing, specifically whole-genome sequencing (WGS) and whole-exome sequencing (WES), has been invaluable for deciphering the molecular heterogeneity of CLL. It is now known that CLL is characterized by a relatively well-defined set of recurrent mutations in addition to TP53, e.g. SF3B1, NOTCH1, ATM, BIRC3, NFKBIE and MYD88, as well as the known cytogenetic lesions.16 Although the molecular mechanisms by which some of the newly identified genetic lesions correlate with clinical aggressiveness are not fully understood, the fact that mutations often cluster in evolutionarily conserved hotspots or functional domains strongly suggests they have a role in the pathogenesis of CLL. In addition, the enrichment of some of these alterations in patients requiring treatment or refractory to therapy suggests a contributing role.16 In fact, it has recently been shown that CLL cases co-expressing mutations of TP53, NOTCH1 and BIRC3 have a poorer outcome than cases carrying only TP53 mutations.17 In line with this, studies have shown that mutations within the aformentioned genes may refine the prognostic value of the Döhner hierarchical classification.1,18,19 However, it is important to note that none of the recently identified mutations have yet been incorporated into clinical practice. Furthermore, while subclonal TP53 mutations may infer a poor prognosis, data regarding subclonal mutations within other recurrently mutated genes remain limited. Nevertheless, a few key studies have provided some insight into the intratumoral composition of mutations and their (sub)clonal behavior, and have demonstrated significant dynamic changes as the disease progresses, as well as the selection of subclones conveying poor prognosis.13,14,20,21 Thus, as we prepare for an era of tailored therapy, we need not only to appreciate the effect of mutations on response to treatment, but also the clinical impact of subclones harboring specific mutations.
While WGS and WES remain primarily a discovery tool in biomedical cancer research, targeted NGS, in which a selected number of genes are sequenced, is emerging as a much more efficient means to identify genetic variants and contains many attractive features, including the possibility to: i) detect mutations present at low VAFs due to the high sensitivity achievable; ii) analyze all coding exons within a gene, irrespective of size; and iii) custom design a gene panel and screen several genes with known or predicted clinical implications in multiple patient samples simultaneously.12–14 In addition, technology is now evolving such that multiple types of lesions, i.e. single nucleotide variants, insertions, deletions and chromosomal rearrangements, can be analyzed in a single sequencing run; a task that cannot be achieved by any other stand-alone test.
Although it is likely that NGS will change molecular diagnostics in CLL, the routine implementation of NGS is in its infancy, and there is still a long way to go before we consider NGS not just as a discovery tool for research, but also as a clinically useful prognostic tool. To begin with, standard analytical methods must be established and checked for accuracy and reproducibility. To help in this transition, ERIC has recently initiated a multi-center project to harmonize the analysis of clinically relevant genes in CLL, including TP53, using NGS (Figure 2). This initiative aims to reach a consensus on which genomic variants are clinically relevant in CLL and thereafter rigorously assess various gene panel technologies and bioinformatic approaches across the collaborative sites. These studies may help to set the stage for the wider adoption of targeted NGS gene panels in clinical practice by not only identifying the strengths and weaknesses of various technologies, but also by defining quality metrics that will improve accuracy and ensure consistency and reproducibility. Prior to incorporating NGS within a clinical setting, criteria should be harmonized at an international level, and these should include: i) the determination of performance specifications for NGS, such as ascertaining cut-off values for low frequency (subclonal) variants; ii) dealing with regions harboring a high GC content; iii) distinguishing between mean depth of coverage and uniformity of coverage; and iv) the standardization and evaluation of NGS at a clinical level.
Figure 2.
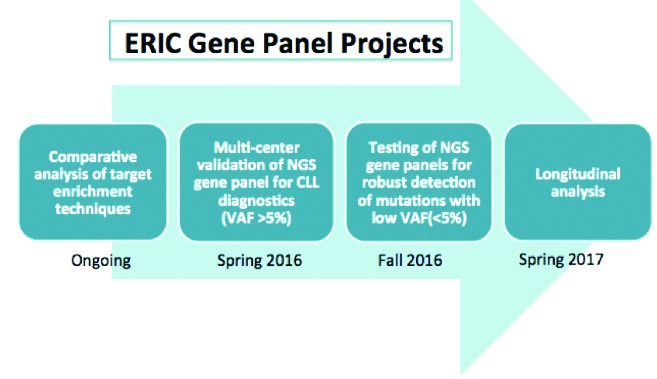
Ongoing and planned ERIC Gene Panel Projects. NGS: next generation sequencing; VAF: variant allelic frequency.
In conclusion, incredible progress has been made in CLL genomics during the last few years, largely due to the rise of NGS technology. The ERIC initiatives set out here will hopefully help in the clinical deployment of NGS as both a research and a clinical tool for CLL. An important caveat is that validity and utility differ between research and clinical settings, with crucial issues relating to: the cost per test, the time to actionable result, the possibility of establishing automated interpretation of the tests, and the actual clinical significance of each mutation. With regards to the latter, identifying mutations is only the first step, while determining the clinical significance of novel mutations can be a lengthy process, and elucidating which variants are clinically actionable or have clinical validity in CLL requires substantial research and prospective studies. Another important issue is the difference in capability of larger academic institutes/hospitals as compared to community or regional healthcare facilities. Therefore, a model has to be devised to cater for both; one such option is the provision of NGS at specialized, referral centers.
Despite the challenges, NGS is (and will) change many aspects of modern medicine, and while its widespread implementation may take several years, the journey should be viewed with both hope and as an iterative process that will continually incorporate new findings while discarding the irrelevant.
Acknowledgments
This work was supported in part by research projects CEITEC CZ.1.05/1.1.00/02.68, H2020 Twinning (MEDGENET/2016–2018/no.692298) and MZ CR projects AZV 15-31834A and AZV 15-30015A; the Swedish Cancer Society, the Swedish Research Council, Selander’s Foundation and Lion’s Cancer Research Foundation, Uppsala, Sweden; H2020 “AEGLE, An analytics framework for integrated and personalized healthcare services in Europe”, by the EU; the Special Program in Molecular Clinical Oncology, 5 × 1000 No. #9965 and 10007, AIRC, Milan Italy, Ricerca Finalizzata RF-2010-2318823 and RF-2011-02349712, Ministero della Salute, Rome, Italy; Else Kröner-Forschungskolleg Ulm. The authors would like to thank all the participants of the 1st TP53 ERIC workshop held in Brno, in October 2015, for the lively and constructive discussion on the present and future analysis of TP53 in CLL.
Footnotes
Financial and other disclosures provided by the author using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are available with the full text of this paper at www.haematologica.org.
References
- 1.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. [DOI] [PubMed] [Google Scholar]
- 2.Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10(1):37–50. [DOI] [PubMed] [Google Scholar]
- 3.Stilgenbauer S, Schnaiter A, Paschka P, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123(21):3247–3254. [DOI] [PubMed] [Google Scholar]
- 4.Zenz T, Eichhorst B, Busch R, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28(29):4473–4479. [DOI] [PubMed] [Google Scholar]
- 5.Pospisilova S, Gonzalez D, Malcikova J, et al. ERIC recommendations on TP53 mutation analysis in chronic lymphocytic leukemia. Leukemia. 2012;26(7):1458–1461. [DOI] [PubMed] [Google Scholar]
- 6.Malcikova J, Smardova J, Rocnova L, et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: selection, impact on survival, and response to DNA damage. Blood. 2009;114(26):5307–5314. [DOI] [PubMed] [Google Scholar]
- 7.Rossi D, Cerri M, Deambrogi C, et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res. 2009;15(3):995–1004. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez D, Martinez P, Wade R, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29(16):2223–2229. [DOI] [PubMed] [Google Scholar]
- 9.Brown JR, Byrd JC, Coutre SE, et al. Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110delta, for relapsed/refractory chronic lymphocytic leukemia. Blood. 2014;123(22):3390–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd JC, Brown JR, O’Brien S, et al. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N Engl J Med. 2014;371(3):213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stilgenbauer S, Eichhorst B, Schetelig J, et al. Venetoclax (ABT-199/GDC-0199) Monotherapy Induces Deep Remissions, Including Complete Remission and Undetectable MRD, in Ultra-High Risk Relapsed/Refractory Chronic Lymphocytic Leukemia with 17p Deletion: Results of the Pivotal International Phase 2 Study. American Society of Hematology; 2015; Orlando, Florida. [Google Scholar]
- 12.Sutton LA, Ljungstrom V, Mansouri L, et al. Targeted next-generation sequencing in chronic lymphocytic leukemia: a high-throughput yet tailored approach will facilitate implementation in a clinical setting. Haematologica. 2015;100(3):370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi D, Khiabanian H, Spina V, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123(14):2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malcikova J, Stano-Kozubik K, Tichy B, et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia. 2015;29(4):877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutton LA, Rosenquist R. Deciphering the molecular landscape in chronic lymphocytic leukemia: time frame of disease evolution. Haematologica. 2015;100(1):7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guieze R, Robbe P, Clifford R, et al. Presence of multiple recurrent mutations confers poor trial outcome of relapsed/refractory CLL. Blood. 2015;126(18):2110–2117. [DOI] [PubMed] [Google Scholar]
- 18.Rossi D, Rasi S, Spina V, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121(8):1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baliakas P, Hadzidimitriou A, Sutton LA, et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29(2):329–336. [DOI] [PubMed] [Google Scholar]
- 20.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4): 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuh A, Becq J, Humphray S, et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012;120(20):4191–4196. [DOI] [PubMed] [Google Scholar]


