Abstract
Erythropoietin is essential for the production of mature erythroid cells, promoting both proliferation and survival. Whether erythropoietin and other cytokines can influence lineage commitment of hematopoietic stem and progenitor cells is of significant interest. To study lineage restriction of the common myeloid progenitor to the megakaryocyte/erythroid progenitor of peripheral blood CD34+ cells, we have shown that the cell surface protein CD36 identifies the earliest lineage restricted megakaryocyte/erythroid progenitor. Using this marker and carboxyfluorescein succinimidyl ester to track cell divisions in vitro, we have developed a mathematical model that accurately predicts population dynamics of erythroid culture. Parameters derived from the modeling of cultures without added erythropoietin indicate that the rate of lineage restriction is not affected by erythropoietin. By contrast, megakaryocyte/erythroid progenitor proliferation is sensitive to erythropoietin from the time that CD36 first appears at the cell surface. These results shed new light on the role of erythropoietin in erythropoiesis and provide a powerful tool for further study of hematopoietic progenitor lineage restriction and erythropoiesis.
Introduction
The question of whether extrinsic signals, in particular cytokines, have a deterministic role in lineage commitment of hematopoietic stem and progenitor cells has been much debated. It is difficult to distinguish a definite role in commitment from the well-characterized roles of cytokines in promoting proliferation and survival. Recent work has provided strong evidence that lineage commitment of granulocyte/monocyte progenitors (GMP) to granulocytes or macrophages can be specified by the appropriate colony stimulating factor (CSF)1–3 but it is less clear whether cytokines can control the lineage restriction of less mature stem and progenitor cells.4,5 Common myeloid progenitors (CMPs) are immature hematopoietic progenitors that become restricted to either the erythroid lineage as megakaryocyte/erythroid progenitors (MEP), giving rise to red blood cells and megakaryocytes/ platelets, or the myeloid lineage as GMP, giving rise to neutrophils, eosinophils, mast cells and macrophages. Mutations in regulators of lineage commitment from CMP are often found in leukemia,6–9 highlighting the importance of understanding this key regulatory event.
The erythroid master regulator erythropoietin (epo) is fundamental to the control of both homeostatic and stress erythropoiesis.10 Erythroid cell proliferation is dependent on epo during the early S phase of the second colony forming unit-erythroid (CFU-E) division, coincident with the appearance of the erythroid marker glycophorin A (GPA).11,12 Epo is also required for erythroid precursor cell survival through signal transducer and activator of transcription (Stat-5) mediated activation of the apoptotic inhibitor B-cell lymphoma-extra large (Bcl-XL).13 However, the role of epo in controlling proliferation, maturation and survival of the earliest erythroid progenitors is not completely understood. In vivo, epo is essential from the CFU-E stage onward as mutants defective for epo or epo receptor do not produce mature red cells.14 Such mutants are able to produce both early and late erythroid progenitors, burst forming unit-erythroid (BFU-E) and CFU-E, respectively, suggesting that epo signaling is not absolutely required for erythroid commitment. However, a recent study has suggested that high levels of epo can bias commitment toward the erythroid lineage over the myeloid lineage both in vivo and in vitro.15 To clarify these apparently contradictory findings on the role of epo in lineage commitment and erythropoiesis, we have determined the effect of epo on the kinetics of proliferation and lineage restriction during the maturation from CMP to MEP.
Population dynamics of hematopoietic culture is a complex output generated by the interplay of proliferation, maturation and cell death. Proliferation and cell death can be followed directly using the carboxyfluorescein succinimidyl ester (CFSE) division-tracking dye and Annexin V/propidium iodide staining, respectively, but maturation associated with lineage restriction can only be analyzed indirectly as a measure of the number of mature cells that arise in culture which cannot be accounted for by proliferation or death. To disentangle the individual contributions of these different cell behaviors, we have developed a mathematical model of the early stages of erythroid culture during which cells become committed to the megakaryocyte/erythroid lineage.
Mathematical models have been developed as tools to assist in the analysis of population dynamics during hematopoiesis,16–19 and to determine the transcription factor regulatory interactions that control hematopoietic differentiation pathways.20 To understand how proliferation integrates with lineage commitment and impacts on the overall population dynamics, a number of mathematical approaches for cell division have also been proposed, taking advantage of CFSE division-tracking. In particular, the Smith-Martin model that takes into account progression of cells through the cell cycle, has been successfully used to predict and model population dynamics of in vitro erythropoiesis.21–25
We have combined immunophenotyping, proliferation analysis and cytokine dependence to refine the analysis of early erythroid culture of CD34+ cells isolated from human peripheral blood (PB). Isolated PB CD34+ cells are CMP, and the appearance of detectable surface levels of the erythroid marker CD36 is the earliest identifier of progenitors restricted to the megakaryocyte/erythroid lineage.26 Interestingly, CMP give rise to MEP independently of cell division in erythroid culture and most of the starting CMP become lineage-restricted prior to the onset of proliferation. We have developed an adaptation of the Smith-Martin mathematical model that describes the population dynamics of erythroid culture during lineage restriction from CMP to MEP. The model suggests that cells become responsive to epo as soon as they are committed to the megakaryocyte/erythroid lineage, but epo does not control lineage restriction.
Methods
Antibodies and reagents
Antibodies used were CD34-BV421, CD36-APC, CD90-PE, CD123-PE/Cy5, CD38-APC/Cy7, CD135-PE and CD45RA-BV421 (Biolegend) and CD36-PE, CD45-APC/Vio770, CD235 (GPA)-APC, CD61-APC/Vio770 (Miltenyi Biotec). AnnexinV-BV421 (Biolegend) was used for analysis of dying cells. Monastrol (Merck Chemicals Ltd, UK) was used at 100 μM to block cells in M phase of the cell cycle.
Erythroid culture
Peripheral blood mononuclear cells (PBMCs) were isolated from leukocyte cones by density purification over Histopaque (Sigma) from healthy donors with informed consent. Isolated cells were cultured in erythroid medium (EM); Stem Span (Stem Cell Technologies) + 10 ng/mL stem cell factor (SCF) (for Day 0 to Day 4) and 50 ng/mL (for Day 5 to Day 11), 40 ng/mL insulin-like growth factor1 (IGF-1), 1 ng/mL interleukin-3 (IL-3), 1 μM dexamethasone and with or without 2 U/mL epo (details in Online Supplementary Methods). Research was reviewed and approved by Southmead and Bristol Research Ethics Committee Centre (08/H0102/26 and 12/SW/0199, respectively).
Cell cycle analysis
K562 cells were grown in Iscove modified Dulbecco medium + 20% fetal calf serum, fixed in ice cold 70% ethanol and rehydrated in phosphate buffered saline (PBS). Fixed cells were stained in PBS (Sigma) + 0.1% Triton X-100 + 10 μg/mL propidium iodide + 100 mg/mL RNase and analyzed on the MACSQuant VYB flow cytometer.
Flow cytometry and fluorescence assisted cell sorting
Flow cytometry and fluorescence assisted cell sorting (FACS) was used to analyze cells stained with anti-human-specific antibodies on the MACSQuant and post-acquisition analysis was carried out with FlowJo v.7.6.5 for proliferation analysis and FlowJo v.X0.7 for all other analysis (details in Online Supplementary Methods).
CFSE tracking
CD34+ cells were labeled on the day of isolation with CFSE (Biolegend) at 2 μM in PBS for 15 min at 37°C. Excess CFSE was quenched by incubating in Stem span + 10% fetal calf serum for 5 min at 37°C. Cells were washed in PBS and transferred into EM at a density of 2×104 cells/mL in multi-well dishes so that each time point was taken from a separate well. For extended cultures beyond Day 4, cells were fed by 1:2 partial medium change into EM with 100 ng/mL SCF.
Results
CD36 expression marks megakaryocyte/erythroid restricted progenitors
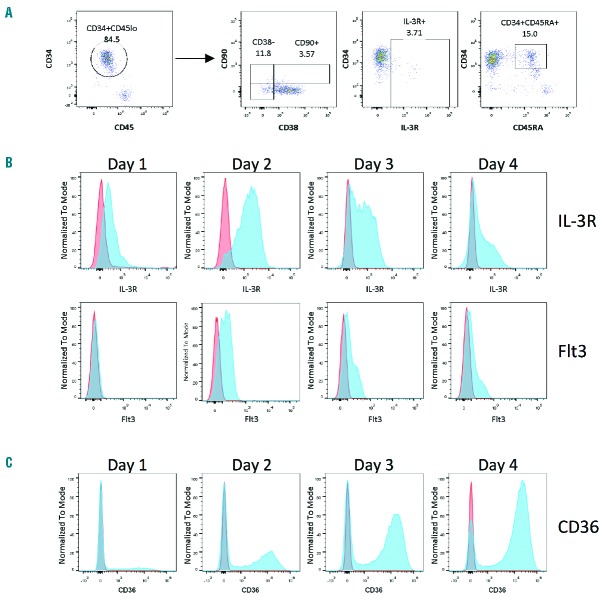
At the time of isolation, CD34+ cells from human PBMNC, are 85% CD34+CD38+IL3Rα−CD45RA−CD90−CD45lo (Figure 1A), previously defined as CMP27,28 and 15% of CD34+ cells are CD34+CD45lo IL−3Rlo CD38+CD90− CD45RA+, GMP as previously described,29,30 which do not persist in erythroid culture (Online Supplementary Figure S1). CD34+ cells cultured in serum-free medium supplemented with appropriate cytokines including SCF, IL-3 and epo mature into erythroid cells. The appearance of erythroid precursors and erythroblasts is easily detected by the appearance of the erythroid marker GPA (Ter119) at Day 7 to Day 10 of culture, depending on conditions. To identify the less mature erythroid progenitors, MEP, at the point of lineage restriction from CMP, prior to GPA expression, we examined the change during culture of the previously defined markers interleukin-3 receptor (IL-3R/CD123) and Fms-like tyrosine kinase 3 (Flt-3/CD135), which are present at low levels on CMP and GMP and absent on MEP isolated from human bone marrow and cord blood.27,31,32 Surface levels of both markers increase over the first two days in culture and then decrease to Day 4 on the bulk population, so they do not provide useful markers for early lineage restriction in erythroid culture (Figure 1B). The erythroid marker CD36 was originally identified as a cell surface marker of erythroblasts33 and later shown to be detectable prior to the appearance of the erythroid differentiation marker GPA.34 At isolation, PB CD34+ cells do not express CD36, but give rise to CD34+CD36+ cells within the first few days of culture (Figure 1C). To determine whether surface expression of CD36 marks the lineage restriction event, CD34+ cells were cultured for 2–4 days and then separated by FACS into CD34+CD36− and CD34+CD36+ populations. CD34+CD36− cells give rise to both erythroid (burst forming unit-erythroid, BFU-E) and myeloid [colony forming unit-granulocyte/monocyte (CFU-GM)] colonies in methylcellulose, indicating these cells are the CMP population. CD34+CD36+ cells give rise only to BFU-E (Figure 2A). BFU-E colonies contain small numbers of megakaryocytes (Figure 2B) consistent with the assignment of CD34+CD36+ cells as bipotent MEPs. This is further supported by the finding that CD34+CD36+ cells are CD61lo (Figure 2C, population I,) reflecting megakaryocyte potential.36
Figure 1.
Immunophenotyping of peripheral blood (PB) CD34+ cells in erythroid culture. (A) Surface expression of CD34, CD45, CD38, IL-3R (CD123), CD45RA and CD90 on PB CD34+ cells on the day of isolation. All populations were gated first on Propidium iodide incorporation and scatter properties. Cells were gated for CD34+ and CD45lo (left panel) prior to analysis of CD38 and CD90. IL-3R and CD45RA were analyzed on the total live population. Gates were set with isotype controls. (B) Surface expression of IL-3R and Flt3 on CD34+ cells cultured in EM for four days. Stained cells in blue and isotype controls in orange. Expression of both markers increases in the total population during the first three days of culture and then decreases. (C) Surface expression of CD36 over four days of culture demonstrating that CD36− and CD36+ cells comprise distinct populations. Stained cells in blue and isotype controls in orange.
Figure 2.
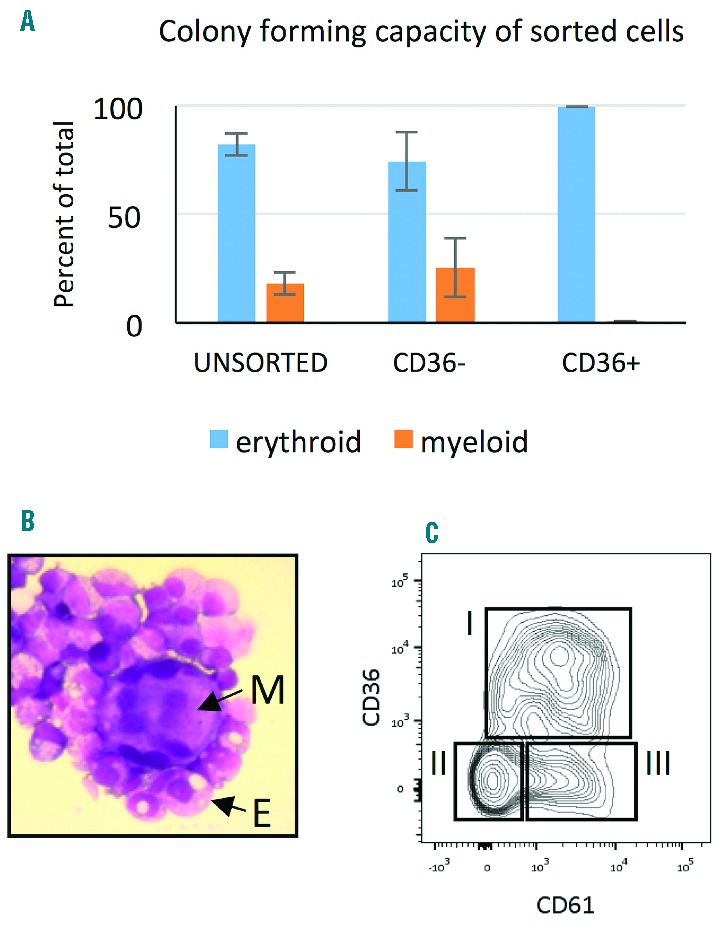
Surface expression of CD36 marks the transition from CMP to MEP in erythroid culture. (A) CD34+ cells cultured in EM for two or four days were sorted by FACS into CD34+CD36− and CD34+CD36+ populations. Sorted cells were cultured in methylcellulose for two weeks and colonies enumerated. Results are from 3 independent experiments, 2 tests per experiment. Error bars are standard deviation. (B) Cytospin from an individual erythroid colony stained with May Grünwald/Giemsa showing a megakaryocyte (M) with multiple nuclei and surrounding erythroblasts (E). (C) The megakaryocyte marker CD61 is absent on cells at isolation (data not shown) and heterogeneous at Day 2 with low expression on CD36+ MEP (population I, 46% CD34+CD36+CD61lo/−) and largely absent on CD36− CMP (population II, 37% CD34+CD36− CD61−). There is a small population of CD61− cells (population III, 17% CD34+CD36+CD61−) which may be megakaryocyte progenitors directly descended from CMP as previously described for mouse multipotent progenitors.47
Maturation of CMP to MEP is independent of cell division
Previous work had suggested that PB CD34+ cells divided asymmetrically dependent on unequal distribution of the determinants Notch and Numb.37 One possible explanation for the observed population dynamics of erythroid culture, in particular the retention of CMP concurrent with MEP production (Online Supplementary Figure S2), is that CMP might divide asymmetrically to produce 1 MEP and 1 CMP. To test this, we first examined whether maturation of CMP to MEP was coincident with cell division by following proliferation of PB CD34+ cells by CFSE tracking. Cells were CFSE-labeled at the time of isolation and the number of cells in each generation was determined separately for CMP and MEP by gating on CD36. After 24 h in culture, all cells in both CMP and MEP populations remain in generation 0 [G(0)] and do not progress to G(1) until the second day of culture, indicating a lag phase for both CMP and MEP from the time of isolation to the initiation of proliferation between Days 1 and 2 (Figure 3A, top panels). Maturation of CMP to MEP begins immediately, as is clear from the emergence of CD36+ MEP on Day 1, before the appearance of G(1) cells (Figure 3B). This indicates that maturation does not require cell division, at least for G(0) CMP, and rules out the possibility that MEP arise from CMP by asymmetric division. This was confirmed by demonstrating that inhibiting cell cycle progression with Monastrol does not block maturation of CD34+ cells to MEP (Online Supplementary Figure S3).
Figure 3.
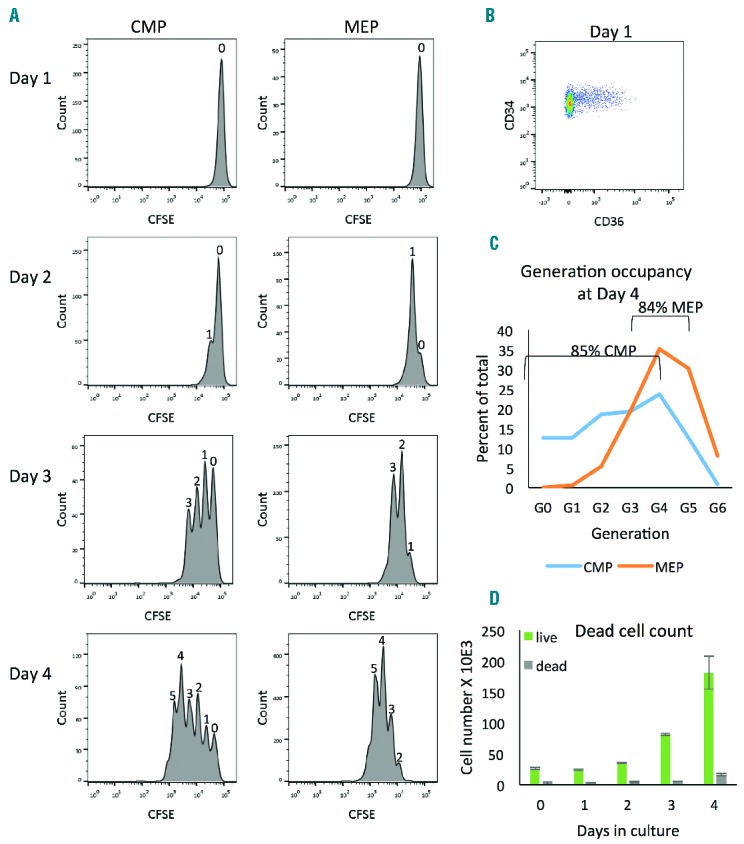
CFSE tracking highlights key features of early erythroid culture. CD34+ cells were labeled with CFSE on the day of isolation and cultured in EM for four days. (A) Cells were gated on CD34+CD36− (CMP) or CD34+CD36+ (MEP) and generation occupancy was determined by flow cytometry. Numbers above peaks indicate generation. After one day in culture, cells have not divided and all cells are in G(0), (top panels). Proliferation begins for CMP and MEP between Day 1 and Day 2 as demonstrated by the appearance of a peak at half the mean fluorescence intensity (MFI) of the original peak (G(0) MFI=63000 for G(0) and MFI=34000 for G(1)). This generation analysis was done manually in FlowJo vX0.7. (B) CD36 staining on Day 1 of the same cells as shown in the left panels demonstrates the appearance of CD36+ cells prior to the onset of proliferation. (C) Generational occupancy at Day 4 shows that MEP proliferation is more synchronized than CMP with 84% MEP within 3 generations, G(3–5), and 85% CMP evenly distributed in 5 generations, G(0–4). (D) Cell death is insignificant during the first four days of culture as measured by propidium iodide staining to identify dead cells (gray) as compared to live cells (green). Error bars are standard deviation.
By Day 2, 75% MEP and 24% CMP have undergone a first division and are in G(1) (Figure 3A). Overall, CMP proliferation is heterogeneous with an even distribution of cells over 5 generations from G(0–4). By contrast, all of the MEP cells divide within the first three days as indicated by the absence of cells in G(0) and, by Day 4, 84% MEP are found in 3 generations G(3–5) with a distinct peak (35%) in G(4), suggesting a largely synchronously proliferating population (Figure 3C).
To determine whether cell death has a significant effect on population dynamics, dying and dead cells were quantified over a 4-day time course by Annexin V staining and Propidium Iodide incorporation, respectively. The total number of dead cells was insignificant compared to live cells (Figure 3D). Annexin V staining was undetectable.
Mathematical model of population dynamics of erythroid culture
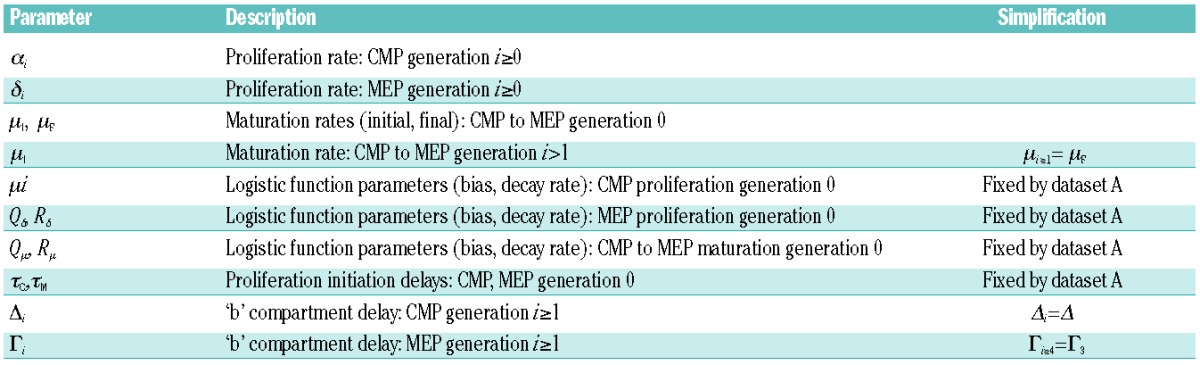
Understanding the role of extrinsic factors on CMP to MEP maturation and progenitor proliferation requires a means of measuring the relative parameters independently, which is not possible using population dynamics alone. Instead, we have developed a mathematical model that simulates the population dynamics, in terms of CMP and MEP numbers, dependent on a set of parameters (Table 1) derived from our experimental data, which include total cell number, CMP:MEP ratio and cell division by CFSE tracking with sampling every eight hours during the first four days of culture
Table 1.
Parameters of the mathematical model. Symbols and physiological meaning of the parameters used in the full (37 parameter) mathematical model, and (column 3) their values in the simplified (20 parameter) model.
Our model is an adaptation of the Smith-Martin model. It separates the cell cycle into two distinct compartments ‘a’ and ‘b’, representing the G1 (‘a’) and the combined S, G2 and M phases (‘b’) of the cell cycle. We incorporated maturation of CMP to MEP, as shown schematically in Figure 4A. Cells originate in the ‘a’ compartment, with their transition governed by a stochastic process. This gives rise to rates αi and δi for generation i CMP and MEP cells, respectively, transitioning to the ‘b’ compartment, and rate mi for generation i CMP maturing to generation i MEP. Cells remain in the ‘b’ compartment for a fixed time (Di and Gi for CMP and MEP, respectively) before they divide and then re-enter the ‘a’ compartment as a next generation cell.
Figure 4.
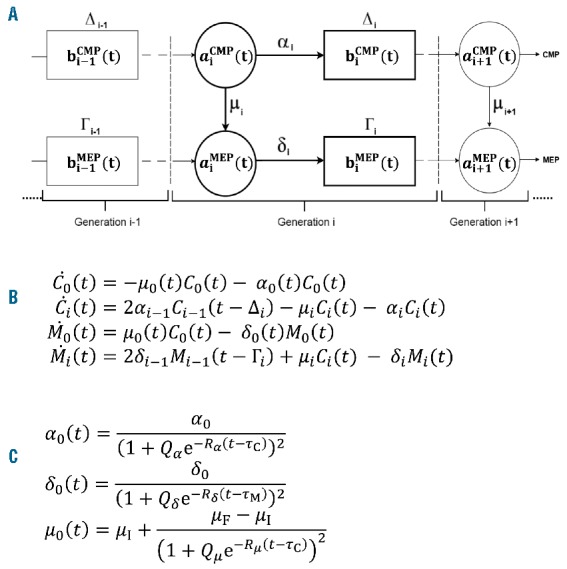
Mathematical model of the system. (A) Schematic diagram of erythroid cell culture. The cell cycle is modelled as a 2-stage process, composed of compartments ‘a’ approximating the G1 phase of the cell cycle, and ‘b’ approximating the S/G2/M phases. CMP and MEP both originate in the ‘a’ compartment. Their transition is governed by a stochastic process, with rates 훼푖 and 훿푖 for generation 푖 CMP and MEP cells, respectively, to transition to the ‘b’ compartment, and a rate 휇푖 of maturing (transitioning) from CMP into same-generation MEP cells. Cells remain in the ‘b’ compartment for a fixed time (∆푖 and Γ푖 for CMP and MEP, respectively) before they divide and then re-enter the ‘a’ compartment as a next generation cell. (B) The size of the CMP and MEP populations in each generation at each time point are given by 4 delay differential equations (DDEs) where (푡) and (푡) are the generational populations of CMP and MEP, respectively. (C) G(0) proliferation and maturation rates are time-dependent, taking the form of generalized logistic functions, where 푃훼 is the maximum proliferation rate for G(0) CMP, 푃훿 is the maximum proliferation rate for G(0) MEP, and 휇I and 휇F are the initial and final maturation rates for G(0). The parameters 푄훼,,,푅훿,푄휇, 푅휇, 휏C and 휏M control the dynamics of the generalized logistic functions and were optimized for. Equations were solved using MATLAB® solver dde23.48
In order to simplify the model whilst maintaining the key dynamics that relate to lineage restriction, we made the following assumptions based on data presented in Figure 3: 1) death rate plays an insignificant role over the time course; 2) cell maturation initiates prior to the onset of CMP and MEP proliferation and, thus, does not require transition through the cell cycle; 3) GMP are excluded from the analysis as this population does not contribute significantly to the population dynamics in the first four days of culture (Online Supplementary Figure S1). The CMP and MEP populations in each generation evolve according to a set of coupled delay differential equations (Figure 4B). To account for the observation that maturation initiates prior to the onset of proliferation (Figure 3B), we allowed the first generation proliferation and maturation rates to be time dependent. Constant rates for proliferation and maturation in G(0) do not provide a good qualitative or quantitative match to data, while piecewise constant rates (a single step function for each rate) substantially improve the fit. To maintain physiological plausibility, we model G(0) rates as smoothed step functions, in the form of generalized logistic functions (Figure 4C).
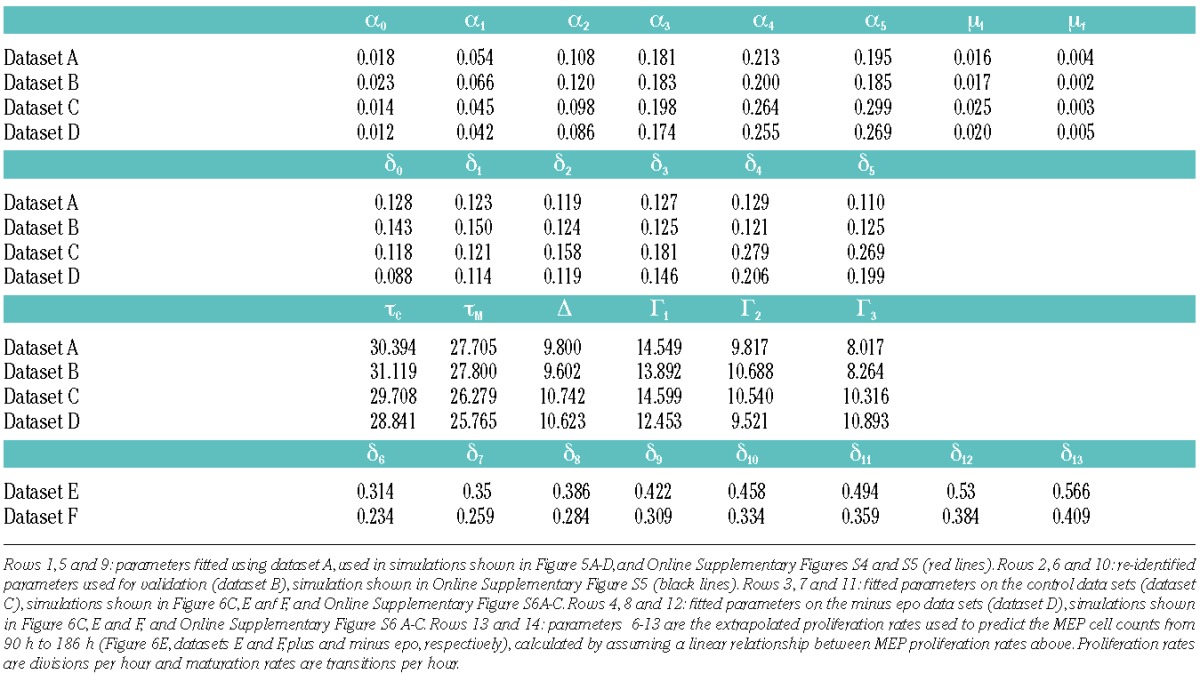
We then used optimization routines to find the set of parameters that minimizes the root mean square error (RMSE) to the experimental data (CFSE tracking) (see Online Supplementary Methods). The system was fitted to experimental data (Online Supplementary Table S1, dataset A) initially in the most general case, where all 37 parameters are generation dependent (Online Supplementary Table S2). This provided insight as to the system behavior and allowed us to make simplifications with no significant increase in RMSE (see Online Supplementary Methods), and reduce the system to 20 parameters: 12 proliferation rates (α(0–5), δ(0–5)), 2 maturation rates (μI, μF), 4 compartmental delays (Δ, Γ(1–3)), and 2 proliferation initiation delays (τC, τM).
We observed very good quantitative agreement between model and experiment for all generations (Figure 5A and B and Online Supplementary Figure S4A and B). To quantify the experimental error, standard deviation was calculated and normalized over total cell counts (shaded regions in Figure 5B). The standard deviation found within the experimental data was 222.44 (3.07%) for CMP, and 279.46 (2.75%) for MEP. The RMSE error between experimental data and model predictions for optimal parameter values (Figure 5B) was 101.02 (1.38%) for CMP and 138.29 (1.85%) for MEP. Table 2 shows the rates (divisions per hour) and delays (hours) obtained from the optimization algorithm.
Figure 5.

Fitted model simulations provide proliferation and maturation parameters and recapitulate population dynamics. (A) Fitted model simulations (solid lines) of CMP (left panel) and MEP (right panel) cell numbers in each generation. Experimental data (dataset A) are dashed lines, ± standard error (shaded regions). (B) Model simulation of total CMP (left panel) and MEP (right panel) cell numbers using model derived parameters (solid lines) together with experimental data (dashed lines) ± standard error (shaded regions). (C) Proliferation rates dataset A for CMP (α) in blue, and MEP (δ) in orange, as divisions per hour over the first 5 generations. (D) Changes for dataset A in maturation rate (m0) of G(0) CMP in the first 90 h of culture expressed as CMP to MEP transitions per hour.
Table 2.
Parameter values of the mathematical model.
The model was validated using a second, independent experimental data set (dataset B) in two different ways (Online Supplementary Methods and Online Supplementary Figure S5), and indicated high confidence in the model and parameter values. Further in silico experiments were conducted on an extensive set of simulated data, confirming robustness of the parameter identification methods used (Online Supplementary Methods and Online Supplementary Table S3).
The identified parameters for dataset A (Table 2) show that the average MEP proliferation rate (δ) is 0.12 divisions per hour, and that this rate does not vary greatly from the onset of proliferation to Day 4 (Figure 5C). The rate of proliferation of CMP cells is initially 10% of the proliferation rate of MEP, and increases with each generation such that CMP transit from G(5) to G(6) is approximately twice the average MEP rate (Figure 5C). The maturation rate for G(0) (μ0(t)) decreases over time, from 0.016 to 0.004 transitions per hour (Figure 5D). Importantly, the maturation rate, even at the beginning of the culture when it is at its highest, is an order of magnitude smaller than the MEP proliferation rate. These parameters show important features of the relative contributions of proliferation and maturation to overall population dynamics at different stages in the culture: while maturation plays a crucial role at the beginning, proliferation is the main effector of later culture stages.
The role of epo in maturation from CMP to MEP
To investigate a possible role for epo in the earliest stages of erythropoiesis in vitro, CD34+ cells were isolated and cultured under standard conditions or in the absence of epo. After one day in culture, prior to the onset of proliferation, 20% CMP have matured to MEP under both conditions (Figure 6A), suggesting that CMP to MEP transition during the first 24 h is not responsive to epo in G(0). However, over nine days in culture, there is a dramatic difference in the number of MEP in control and minus epo cultures and by Day 9 there is over 20 times more MEP produced in cultures with epo (Figure 6B).
Figure 6.
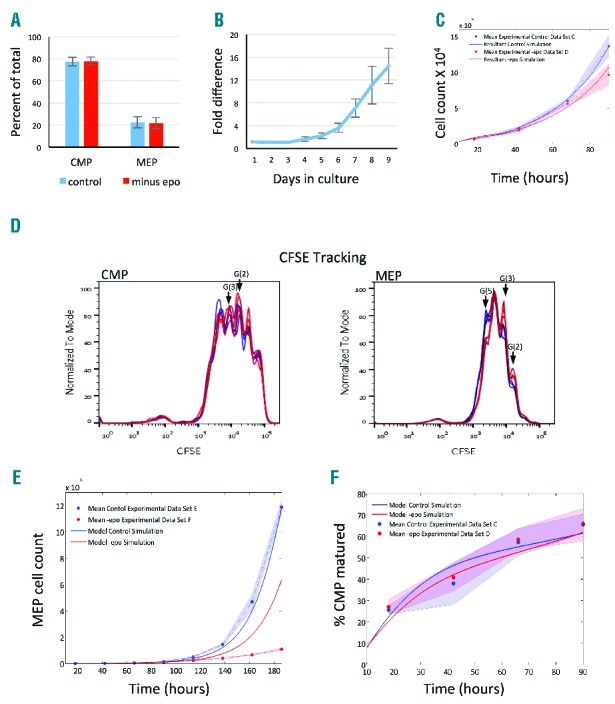
MEP proliferation rate is epo-sensitive but CMP maturation is independent of epo. (A) Cell numbers for CMP and MEP after one day in control (blue) and minus epo (red) culture ± standard deviation. Data are from 4 experiments, 3 samples per experiment. (B) Relative difference in MEP numbers in control and minus epo cultures (fold difference = N. control/N. minus epo MEP ± standard deviation) from 3 experiments. (C) Model fitting simulations for control (blue) and minus epo (red) cultures compared to experimental data from datasets C and D (dots). Shaded regions represent standard error in the experimental data. (D) CFSE in CMP and MEP after four days in control culture (blue) or minus epo (red). Traces from 3 samples are overlaid. Generations 2, 3 and 5 are labeled. (E) Extrapolation of MEP cell counts from datasets E and F to Day 8 (186 h) using best-fit parameters in Table 2 for both control and minus epo populations. Additional proliferation rates (훿6−13) predicted by assuming a linear relationship between the optimized proliferation rates, found from the Day 0–4 fitting for G(0) to G(5) MEP, allowing us to extrapolate forward for G(6) onwards. (F) Maturation over four days expressed as percentage of CMP matured from experimental data (circles) and model predictions (lines). Experimental values were calculated as the number of CMP taking into account cell division. CMP matured = N. CMP in G(0) + sum over 6 generations N. CMP G(i)/2i. Model predicted maturation was tracked from all generations of CMP throughout the simulations.
The lower MEP production in the absence of epo could be due to defects in maturation, proliferation, or both. To resolve the individual effects of epo on maturation and proliferation, we used the model to fit parameters from generational CMP and MEP counts of 4-day cultures with and without epo (datasets C and D). The results are shown in Figure 6C and Online Supplementary Figure S6A and B. Parameter fitting shows that the proliferation rate of MEP (δ) is 21% lower in the absence of epo, whereas there is less than 10% difference in proliferation rates of CMP (α) (Table 2). The traces from CFSE tracking also support this result in that a distinct delay in generation progression for MEP is detectable in cultures without epo (Figure 6D). Also, CFSE tracking shows no significant effect of epo on CMP generation occupancy (Figure 6D); this result, taken together with insignificant change in proliferation rate of CMP (Table 2), indicates that CMP proliferation is not epo-sensitive. Model validation, using fitted parameters for δ to reproduce MEP amplification in 2 additional experimental datasets (Online Supplementary Table S1, datasets E and F), further supports this result (Online Supplementary Figure S6C).
We have used the parameters calculated from 4-day cultures with and without epo to simulate MEP population expansion to Day 8 (Table 2 and Figure 6E, datasets E and F). For control cultures, there is good agreement between the predicted cell numbers and experimental data. For cultures without epo, there is good agreement for the first 5–6 days (up to time 130 h in Figure 6E), and then the experimental data show a significantly slower proliferation than predicted. This suggests that there is a change in epo sensitivity between Days 5 and 6, after which MEP proliferate very slowly without epo.
The maturation rate for G(0) (μ0(t) (Table 2) is not significantly different with or without epo (datasets C and D). Although the final maturation rate is slightly higher in the absence of epo, this cannot account for the decrease in MEP formation, suggesting that epo effects on population dynamics and MEP production are not mediated by altered maturation rates. To determine whether epo confers an erythroid bias, we used CFSE tracking to quantify CMP maturation and showed that epo does not affect the total number of CMP that mature to MEP (Figure 6F). These results provide strong evidence that neither the rate nor the bias of the CMP to MEP transition is influenced by epo.
Discussion
Understanding the fundamental mechanisms controlling lineage commitment, proliferation and survival during erythropoiesis is becoming an achievable goal thanks to the advent of novel high throughput technologies to explore the transcriptome,39–42 proteome43 and epigenome.44,45 In order to maximize the specificity of the information afforded by these techniques, it is essential to start with well-defined populations. A recent report provided an excellent protocol to identify populations at distinct stages of erythroid differentiation,46 but early erythroid culture has proved more challenging. To achieve this, it is necessary to measure the transition event under experimental conditions where proliferation and survival can be controlled and/or measured simultaneously. We identified CD36 as a cell surface marker that distinguishes CMP and MEP as distinct populations.
Identification of CD36 as an MEP marker provides a powerful tool to investigate the influence of extrinsic factors on lineage restriction of CMP to MEP. Epo is essential for steady-state and stress erythropoiesis and is used therapeutically to increase red cell production in clinical anemia. It is of considerable interest, therefore, to understand the precise role of epo in hematopoiesis. There is a significant body of work demonstrating that an essential role of epo is to promote proliferation of erythroid progenitors and survival of differentiating erythroblasts.47,48 This raises the question of whether epo acts on less mature progenitors. Early work from Lodish et al.14 showed that epo and the epo receptor are not required to generate early erythroid-specified progenitors in vivo, suggesting that epo does not have an instructive role in early hematopoiesis. On the other hand, a more recent study15 showed that early progenitors are responsive to epo, and that high levels of epo suppress GMP formation through transcriptional reprogramming, both in vivo and in vitro. Interestingly, this study showed that although there is a relative increase in erythroid progenitors compared to non-erythroid progenitors, the absolute number of erythroid progenitors is not affected by high levels of epo in vivo. Our results using mathematical modeling correspond well with these studies, demonstrating that the time-dependent maturation rate of CMP is not affected by epo. Importantly, equal numbers of CMP convert to MEP with and without epo, further supporting the idea that epo does not bias CMP commitment toward the erythroid lineage. Population dynamics of cord blood CD34+ cells in erythroid culture show similar dynamics to PB CD34+ cells (Online Supplementary Figure S7) suggesting that these results are likely to be applicable to CMP from other sources. This provides validation for the proposed model in demonstrating consistency with in vivo data.
It is important to note that, while our results clearly indicate that the MEP population is homogeneous, the CMP population behaves heterogeneously in terms of proliferation. It is, therefore, possible that there is an earlier restricted progenitor within the CMP population that is indistinguishable from other CMP using known markers. Developing a transcriptomic fingerprint of CD34+CD36+ cells would allow us to investigate this possibility using single cell analysis applied to the CMP population.
Although it is well established that epo is required for proliferation of late erythroid progenitors, a possible influence of epo on earlier progenitors has not been described. To test this, we determined the progenitor proliferation rates using the model. This showed that, from the initial stages of megakaryocyte/erythroid lineage commitment, cells are epo responsive and proliferation is slower in the absence of epo. By disentangling the independent contributions of proliferation and maturation to population dynamics, the model allowed a high-resolution quantification of proliferation rate that revealed the sensitivity of MEP to epo. This means of quantification will be highly beneficial to the efforts to amplify erythroid cells in culture for the production of red cells for clinical transfusion; a 4–5 day culture significantly decreases the cost and simplifies screening of changes in culture conditions as compared to end point analysis from 2–3 week cultures.
Model simulations using a proliferation rate derived from the first four days of culture and extrapolating to Day 8 show that the derived proliferation rate faithfully reproduces the experimental data. This confirms the power of the model to describe erythroid culture. Interestingly, simulating MEP amplification to Day 8 in the absence of epo using the proliferation rate derived from the first four days of culture shows a marked change after Day 5. This suggests that there are two MEP stages with different epo sensitivity: an early stage (Days 1–4) with a small increase in proliferation rate in response to epo and a later stage (Days 4–8), which is increasingly dependent on epo for proliferation. The increased resolution of cell behavior parameters given by the mathematical model provides a valuable tool to further investigate both the mechanism of lineage restriction and proliferation behavior of hematopoietic progenitors in culture.
Acknowledgments
The authors would like to thank Dr Ashley Toye and members of his group for critical reading of the manuscript, and anonymous reviewers for constructive comments that improved the manuscript.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/101/3/286
Funding
The work was supported by funding from the National Institute for Health Research program grant RP-PG-0310-1004-AMT and from the EPSRC Doctoral Training Programme (DW).
References
- 1.Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325(5937):217–218. [DOI] [PubMed] [Google Scholar]
- 2.Sarrazin S, Mossadegh-Keller N, Fukao T, et al. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell. 2009;138(2): 300–313. [DOI] [PubMed] [Google Scholar]
- 3.Mossadegh-Keller N, Sarrazin S, Kandalla PK, et al. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 2013;497(7448):239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolff L, Humeniuk R. Concise review: erythroid versus myeloid lineage commitment: regulating the master regulators. Stem Cells. 2013;31(7):1237–1244. [DOI] [PubMed] [Google Scholar]
- 5.Sarrazin S, Sieweke M. Integration of cytokine and transcription factor signals in hematopoietic stem cell commitment. Semin Immunol. 2011;23(5):326–334. [DOI] [PubMed] [Google Scholar]
- 6.Dore LC, Crispino JD. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118(2): 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wechsler J, Greene M, McDevitt MA, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32(1):148–152. [DOI] [PubMed] [Google Scholar]
- 8.Mueller BU, Pabst T, Osato M, et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 2002;100(3):998–1007. [DOI] [PubMed] [Google Scholar]
- 9.Steidl U, Steidl C, Ebralidze A, et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest. 2007;117(9):2611–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koulnis M, Porpiglia E, Hidalgo D, Socolovsky M. Erythropoiesis: from molecular pathways to system properties. Adv Exp Med Biol. 2014;844:37–58. [DOI] [PubMed] [Google Scholar]
- 11.Landschulz KT, Boyer SH, Noyes AN, Rogers OC, Frelin LP. Onset of erythropoietin response in murine erythroid colony-forming units: assignment to early S-phase in a specific cell generation. Blood. 1992;79(10):2749–2758. [PubMed] [Google Scholar]
- 12.Socolovsky M, Murrell M, Liu Y, Pop R, Porpiglia E, Levchenko A. Negative autoreg-ulation by FAS mediates robust fetal erythropoiesis. PLoS Biol. 2007;5(10):e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction. Cell. 1999;98(2):181–191. [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Liu X, Jaenisch R, Lodish HF. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 1995;83(1):59–67. [DOI] [PubMed] [Google Scholar]
- 15.Grover A, Mancini E, Moore S, et al. Erythropoietin guides multipotent hematopoietic progenitor cells toward an erythroid fate. J Exp Med. 2014;211(2): 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colijn C, Mackey MC. A mathematical model of hematopoiesis–I. Periodic chronic myelogenous leukemia. J Theor Biol. 2005; 237(2):117–132. [DOI] [PubMed] [Google Scholar]
- 17.Ostby I, Rusten LS, Kvalheim G, Grottum P. A mathematical model for reconstitution of granulopoiesis after high dose chemotherapy with autologous stem cell transplantation. J Math Biol. 2003;47(2):101–136. [DOI] [PubMed] [Google Scholar]
- 18.Stiehl T, Ho AD, Marciniak-Czochra A. The impact of CD34+ cell dose on engraftment after SCTs: personalized estimates based on mathematical modeling. Bone Marrow Transplant. 2014;49(1):30–37. [DOI] [PubMed] [Google Scholar]
- 19.Manesso E, Teles J, Bryder D, Peterson C. Dynamical modelling of haematopoiesis: an integrated view over the system in homeostasis and under perturbation. J R Soc Interface. 2013;10(80):20120817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glauche I, Cross M, Loeffler M, Roeder I. Lineage specification of hematopoietic stem cells: mathematical modeling and biological implications. Stem Cells. 2007;25(7):1791–1799. [DOI] [PubMed] [Google Scholar]
- 21.Banks HT, Thompson WC. Mathematical Models of Dividing Cell Populations: Application to CFSE Data. Math Model Nat Phenom. 2012;7(5):24–52. [Google Scholar]
- 22.Nordon RE, Nakamura M, Ramirez C, Odell R. Analysis of growth kinetics by division tracking. Immunol Cell Biol. 1999;77(6):523–529. [DOI] [PubMed] [Google Scholar]
- 23.Smith JA, Martin L. Do cells cycle? Proc Natl Acad Sci USA 1973;70(4):1263–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akbarian V, Wang W, Audet J. Measurement of generation-dependent proliferation rates and death rates during mouse erythroid progenitor cell differentiation. Cytometry A. 2012;81(5):382–389. [DOI] [PubMed] [Google Scholar]
- 25.Nordon RE, Ko KH, Odell R, Schroeder T. Multi-type branching models to describe cell differentiation programs. J Theor Biol. 2011; 277(1):7–18. [DOI] [PubMed] [Google Scholar]
- 26.Heideveld E, Masiello F, Marra M, et al. CD14+ cells from peripheral blood positively regulate hematopoietic stem and progenitor cell survival resulting in increased erythroid yield. Haematologica. 2015;100(11): 1396–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci USA. 2002; 99(18):11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baum CM, Weissman IL, Tsukamoto AS, Buckle AM, Peault B. Isolation of a candidate human hematopoietic stem-cell population. Proc Natl Acad Sci USA. 1992;89(7):2804–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edvardsson L, Dykes J, Olofsson T. Isolation and characterization of human myeloid progenitor populations–TpoR as discriminator between common myeloid and megakaryocyte/erythroid progenitors. Exp Hematol. 2006;34(5):599–609. [DOI] [PubMed] [Google Scholar]
- 30.Olthof SG, Fatrai S, Drayer AL, Tyl MR, Vellenga E, Schuringa JJ. Downregulation of signal transducer and activator of transcription 5 (STAT5) in CD34+ cells promotes megakaryocytic development, whereas activation of STAT5 drives erythropoiesis. Stem Cells. 2008;26(7):1732–1742. [DOI] [PubMed] [Google Scholar]
- 31.Huang S, Chen Z, Yu JF, et al. Correlation between IL-3 receptor expression and growth potential of human CD34+ hematopoietic cells from different tissues. Stem Cells. 1999;17(5):265–272. [DOI] [PubMed] [Google Scholar]
- 32.Rappold I, Ziegler BL, Kohler I, et al. Functional and phenotypic characterization of cord blood and bone marrow subsets expressing FLT3 (CD135) receptor tyrosine kinase. Blood. 1997;90(1):111–125. [PubMed] [Google Scholar]
- 33.Edelman P, Vinci G, Villeval JL, et al. A monoclonal antibody against an erythrocyte ontogenic antigen identifies fetal and adult erythroid progenitors. Blood. 1986;67(1):56–63. [PubMed] [Google Scholar]
- 34.Tirelli V, Ghinassi B, Migliaccio AR, et al. Phenotypic definition of the progenitor cells with erythroid differentiation potential present in human adult blood. Stem Cells Int. 2011;2011:602483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasko JE, O’Flaherty E, Begley CG. Mpl ligand (MGDF) alone and in combination with stem cell factor (SCF) promotes proliferation and survival of human megakaryocyte, erythroid and granulocyte/macrophage progenitors. Stem Cells. 1997;15(1):33–42. [DOI] [PubMed] [Google Scholar]
- 36.Nteliopoulos G, Gordon MY. Protein segregation between dividing hematopoietic progenitor cells in the determination of the symmetry/asymmetry of cell division. Stem Cells Dev. 2012;21(14):2565–2580. [DOI] [PubMed] [Google Scholar]
- 37.May G, Soneji S, Tipping AJ, et al. Dynamic analysis of gene expression and genomewide transcription factor binding during lineage specification of multipotent progenitors. Cell Stem Cell. 2013;13(6):754–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gazit R, Garrison BS, Rao TN, et al. Transcriptome analysis identifies regulators of hematopoietic stem and progenitor cells. Stem Cell Reports. 2013;1(3):266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pimkin M, Kossenkov AV, Mishra T, et al. Divergent functions of hematopoietic transcription factors in lineage priming and differentiation during erythromegakary-opoiesis. Genome Res. 2014;24(12):1932–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin HL, Kim JS, Kim YJ, Kim SJ, Broxmeyer HE, Kim KS. Dynamic expression of specific miRNAs during erythroid differentiation of human embryonic stem cells. Mol Cells. 2012;34(2):177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cadeco S, Williamson AJ, Whetton AD. The use of proteomics for systematic analysis of normal and transformed hematopoietic stem cells. Curr Pharm Des. 2012;18(13): 1730–1750. [DOI] [PubMed] [Google Scholar]
- 42.Ji H, Ehrlich LI, Seita J, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467(7313):338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madzo J, Liu H, Rodriguez A, et al. Hydroxymethylation at gene regulatory regions directs stem/early progenitor cell commitment during erythropoiesis. Cell Rep. 2014;6(1):231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu J, Liu J, Xue F, et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood. 2013;121(16): 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silva M, Grillot D, Benito A, Richard C, Nunez G, Fernandez-Luna JL. Erythropoietin can promote erythroid progenitor survival by repressing apoptosis through Bcl-XL and Bcl-2. Blood. 1996;88(5):1576–1582. [PubMed] [Google Scholar]
- 46.Migliaccio G, Migliaccio AR, Adamson JW. In vitro differentiation of human granulocyte/macrophage and erythroid progenitors: comparative analysis of the influence of recombinant human erythropoietin, G-CSF, GM-CSF, and IL-3 in serum-supplemented and serum-deprived cultures. Blood. 1988;72(1):248–256. [PubMed] [Google Scholar]
- 47.Suda J, Suda T, Ogawa M. Analysis of differentiation of mouse hemopoietic stem cells in culture by sequential replating of paired progenitors. Blood. 1984;64(2):393–399. [PubMed] [Google Scholar]
- 48.The MathWorks I. MATLAB 8.0 and Statistics Toolbox 8.1. Natick, Massachusetts, United States, 2013. [Google Scholar]