Abstract
Nanoparticle probes enable implementation of advanced on-surface assay formats, but impose often underappreciated size-associated constraints, in particular on assay kinetics and sensitivity. Here, we highlight substantially slower diffusion-limited assay kinetics due to the rapid development of a nanoprobe depletion layer next to the surface, which static incubation and mixing of bulk solution employed in conventional assay setups often fail to disrupt. In contrast, cyclic solution draining and replenishing (CDR) yields reaction-limited assay kinetics irrespective of the probe size. Using common surface bioassays, ELISA and immunofluorescence, we show that this conceptually distinct approach effectively “erases” size-dependent diffusion constraints, providing a straightforward route to rapid on-surface bioassays employing bulky probes and procedures involving multiple labeling cycles, such as multi-cycle single-cell molecular profiling. For proof-of-concept, we demonstrate that the assay time can be shortened from hours to minutes with the same probe concentration and, at a typical incubation time, comparable target labeling can be achieved with up to 8 times lower nanoprobe concentration. We expect our findings to enable realization of novel assay formats and stimulate development of rapid on-surface bioassays with nanoparticle probes.
Keywords: nanoparticle probes, diffusion limitation, on-surface bioassays, assay kinetics, molecular profiling
1. Introduction
On-surface bioassays have proven essential for biomedical research, drug discovery, and clinical diagnostics, with a number of enzyme-linked immunosorbent assays (ELISAs), Western blots, DNA microarrays, and cell and tissue staining procedures (such as immunofluorescence, IF, and immunohistochemistry, IHC) performed on a daily basis. Exciting technological advances have been made toward improving the throughput and multiplexing capacity of these methods.[1] In particular, novel nanoparticle-based probes[2] are being developed for exploiting previously unattainable functionalities to address continuously expanding demands of new research and clinical applications. For example, surface-enhanced Raman scattering nanoparticle probes now offer a highly sensitive tool for biomolecular detection and imaging,[3] fluorescent nanoparticles enable multiplexed molecular imaging via conventional fluorescence microscopy,[4-6] and recently introduced polymeric rare earth metal-containing bioconjugates promise an unprecedented level of multiplexing via mass-spectrometry imaging techniques.[7] However, majority of on-surface assays face the same fundamental limitation in assay kinetics and sensitivity due to slow probe diffusion from the bulk solution to surface-immobilized targets. Remaining largely underappreciated, size-associated diffusion constraints might substantially diminish or completely obliterate the added functionality expected from the incorporation of nanoparticle probes in bioassays.
Mass transfer limitation is typically experienced when the probe binding kinetics greatly exceeds probe diffusion from solution to the surface, which creates a depletion layer next to the surface and exposes the specimen to a substantially lower effective probe concentration[8] that free probes in bulk solution need to fill via diffusion (Figure 1a). Given the relatively fast binding kinetics of most common target-probe systems, such as antigen-antibody[9] and DNA-DNA’[10] pairing, mass transfer limitation might be expected even in on-surface assays utilizing relatively small molecular probes. For example, IgG probe depletion layer forms around microspheres in suspension within 1 min of incubation, taking over an hour for the assay to reach steady state.[11] Mass transfer effect amplifies dramatically for larger nanoparticle probes, as diffusion constant inversely relates to particle size by Stokes-Einstein equation, while the time it takes for reaction to switch to a diffusion control is directly proportional to diffusion constant:[11]
| (1) |
| (2) |
where D is diffusion constant, kb is the Boltzmann constant, η is the fluid viscosity, T is absolute temperature, r is probe hydrodynamic radius, τ is time to reaching diffusion control, kf is intrinsic forward binding rate, and Γ0 is the surface concentration of reactive sites. As a result, assays proceed at a substantially slower speed, often requiring multi-hour incubation steps and presenting a major technical hurdle for time-sensitive applications, such as intraoperative rapid diagnosis, and advanced multi-step procedures, such as multi-cycle staining methodologies increasingly employed for in situ single-cell molecular profiling.[6,12,13]
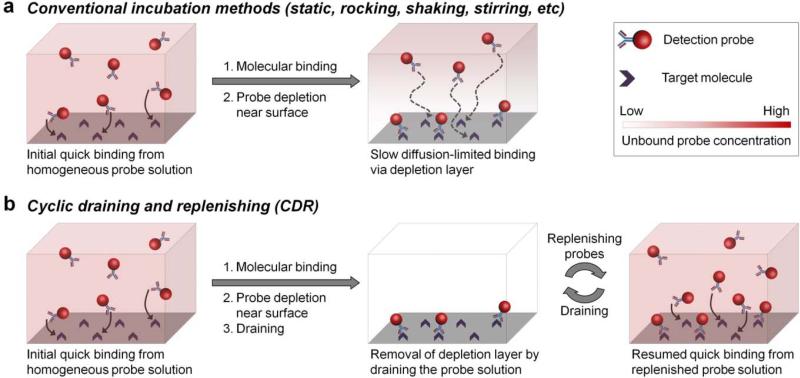
Figure 1.
Schematic illustration of the cyclic draining-replenishing (CDR) technology. (a) Conventional surface-based assays typically employ static incubation for probe binding to surface-immobilized targets. As a result of fast binding kinetics and slow diffusion, probes get rapidly depleted from the volume adjacent to the surface, yielding a substantially lower effective probe concentration and limiting further target labeling kinetics by slow mass transfer from the bulk solution. Improving bulk fluid exchange by rocking, stirring, and shaking has little effect on depletion layer near the surface. (b) CDR method enables rapid target labeling by eliminating the probe diffusion limitation. The probe depletion layer is quickly removed by complete draining of the staining solution, rather than speeding up probe diffusion in the solution. Subsequent refilling of the surface with the same bulk solution keeps the probe concentration near the surface constant and equivalent to bulk solution, thus maintaining fast target labeling kinetics.
Mild shaking routinely employed with on-surface assays typically achieves probe mixing within the bulk solution, but fails to eliminate the depletion layer due to the lack of efficient fluid movement next to the surface, thus offering little improvement in assay performance. Similar challenges are faced by efforts in assay miniaturization and automation with microfluidic devices,[14] where laminar flow offers poor mixing, quickly developing a diffusion-limited probe depletion layer.[15] A number of techniques have attempted to address this issue. Some examples include introduction of turbulence,[16] oscillatory flow,[17] and rapid liquid exchange and recirculation[18] for improved reagent mixing in microfluidic devices and microchambers, use of electric field for active probe enrichment at the surface,[19] and perfusion of samples through porous membranes for enhanced interaction between the surface and reagents.[20] Diffusion constraints have also been recognized with cell and tissue staining applications, where microfluidic processors,[21] ultrasound mixing,[22] microwave treatment,[23] and automated instruments[24] have been used to speed-up the process. Yet, often marginal benefits achieved at an expense of substantially increased complexity with strictly application-specific setups still limit use of these methodologies in routine practice.
Here, we explore the extent of diffusion constraints on the assay kinetics with nanoparticle probes and report a conceptually distinct methodology of cyclic draining-replenishing (CDR) to completely bypass diffusion limitation regardless of the probe size. CDR directly addresses the need for efficient molecular mixing at the surface by repeatedly draining all liquid from the specimen to strip the surface of its probe depletion layer and then refilling the surface with a freshly mixed probe solution, consistently exposing the surface to bulk probe concentration and reusing the same probe solution through multiple cycles (Figure 1b). In a simple implementation of this concept broadly accessible to biomedical laboratories (Supporting Information Figure S1, Movie S1), a molecular target or specimen is immobilized on the plate surface (usually bottom of the well), covered by the probe solution, and sealed with Parafilm. The sealed plate is then affixed onto a laboratory rotator and inverted at the rate of 8 rpm, creating a continuous flow of solution from the well bottom onto the walls, Parafilm cover, and then back to the well bottom, thus driving the probe solution from and to the surface-bound specimen and mixing the solution in the process. Complete solution drainage by gravity and surface tension ensures efficient molecular mixing directly at the specimen surface. At the same time, it is unlikely that any target degradation occurs during the CDR procedure, as effects of drying on the target antigen were only observed if the sample was dried for over 5 minutes prior to replenishing the solution (Supporting Information Figure S2). To demonstrate the concept, we show that both ELISA and IF can be achieved within minutes without sacrificing data quality. Furthermore, we highlight the utility of CDR methodology for rapid single-cell molecular profiling by staining multiple targets in multiple cycles, making this high-content molecular analysis a highly practical approach for the first time.
2. Results and Discussion
2.1. Kinetics of rapid ELISA diagnostics
To explore the effect of mass transfer on the overall assay kinetics and quantitatively evaluate the performance of CDR methodology in a diffusion-limited assay setting, a simple ELISA was established by immobilizing a molecular target (mouse IgG) onto the bottom surface of a microplate at an optimal concentration as determined from standard curves for each probe (Supporting Information Figure S3). Two types of probes routinely employed in ELISA were tested – relatively small monomeric horseradish peroxidase (HRP)-linked antibody (Ab) bioconjugate (mono-HRP, 8-10 nm in size) and larger polymeric HRP-Ab cluster (poly-HRP, ~100 nm) as an ultrasensitive nanoprobe. The CDR incubation process was benchmarked against the well-established conditions, static incubation on a bench and rotary shaking on a shaker. Nonspecific binding was checked by either using rabbit IgG as a target together with anti-mouse HRP-Ab detection or using anti-rabbit HRP-Ab probe on mouse IgG-coated plates.
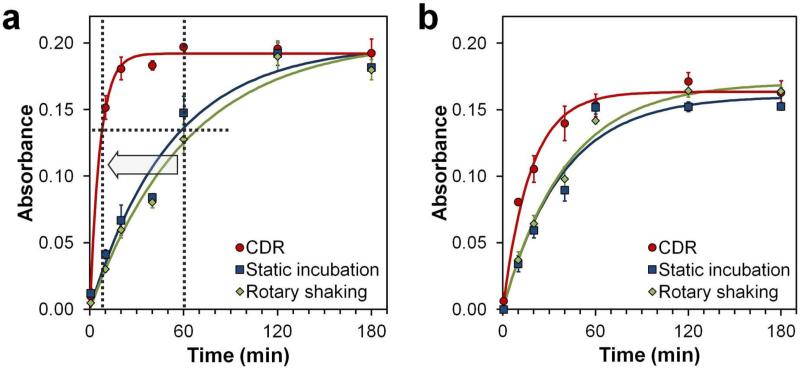
Assay kinetics was characterized by the increase in HRP substrate absorbance at 450 nm with respect to incubation time (10-180 min), and the data was fitted with an exponential growth curve modeled after a typical bimolecular association-dissociation reaction.[25] Under the conventional static and rotary shaking conditions, both Poly-HRP and Mono-HRP probes exhibited only initial fast binding, which then slowly leveled off as incubation time increased. In contrast, CDR produced a sustained fast binding, quickly reaching equilibrium (Figure 2). In comparison to a typical ELISA incubation time of ca. 1 hour, when a major proportion of signal build-up happens without necessarily reaching equilibrium, CDR method produced the same signal intensity within 7 minutes of incubation with Poly-HRP nanoprobes, thus yielding an approximately one order of magnitude increase in assay speed (Figure 2a), while retaining assay specificity (Supporting Information Figure S4). It should be noted, nonetheless, that all methods yielded similar maximum steady-state labeling intensities, confirming that only the diffusion component was eliminated by CDR, while keeping the antibody-antigen binding parameters unperturbed. It is also noteworthy that smaller mono-HRP probes, which already exhibited faster binding kinetics under static incubation, showed further increase in binding speed under CDR (Figure 2b), confirming the presence of mass transfer limitation even with relatively small mono-HRP-Ab bioconjugates.
Figure 2.
Kinetics of rapid ELISA diagnostics. (a) Assay performed with Poly-HRP probes exhibited slow mass transfer-limited kinetics under static incubation (t½ = 36 min) and rotary shaking (t½ = 43 min) conditions, but demonstrated a dramatically improved performance with CDR (t½ = 5 min). Compared to standard ELISA protocols where incubation typically takes 1 h, CDR reaches the same staining intensity in 7 minutes. (b) Smaller mono-HRP probes exhibited faster kinetics under static incubation (t½ = 26 min) and rotary shaking (t½ = 28 min) conditions, featuring further improvement in assay speed with CDR (t½ = 12 min). Solution absorbance at 450 nm with respect to assay time is shown and fitted by an exponential curve. Background absorbance by the TMB substrate alone was subtracted from all measurements. Error bars represent one standard deviation from triplicate assays.
Importantly, CDR-based assay with Poly-HRP probes proceeded with a binding half-time of just 5 min, despite probes being over 100 nm in size. Considering typical kinetic parameters for antibody-antigen binding (kon = 5×105 M−1 s−1 and koff = 5×10−4 s−1),[9] labeling half-time of 2-20 min can be estimated for 10-0.1 nM antibody concentration (i.e., an order of magnitude above and below KD) and no mass transfer limitation. The half-time value of 5 min directly falls within this range, suggesting that the assay proceeds in reaction-limited regime under CDR incubation, while being heavily diffusion-limited under static and rotary shaking conditions.
It follows that the phenomenon of enhanced target labeling is directly associated with fluid draining from the surface rather than bulk fluid movement. The general perception that performing ELISA (or other surface-based bioassays) on a shaker or rocker would help with reagent mixing and improve binding kinetics is not supported by our data, which instead shows that static and shaking tests produce quite similar binding kinetics profiles and highlights the lack of additional molecular mixing at the surface via rotary shaking. In contrast, CDR eliminates diffusion limitation under a range of rotation rates, as long as efficient draining is achieved (Supporting Information Figure S5).
2.2. Rapid IF staining with antibody-QDot nanoprobes
The utility of CDR approach for rapid immunofluorescence assays with nanoprobes was evaluated using fluorescent quantum dot (QDot)-antibody conjugates. QDot technology represents a promising tool for variety of sensing and imaging applications, and it has solved the multiplexing limitation often experienced with organic dyes. However, having a hydrodynamic size of ~20 nm, this advanced class of fluorescent imaging probes is expected to experience major diffusion constraints. Indeed, 2-3 hour incubation steps are typical with QDot-based IF.[5,6,26]
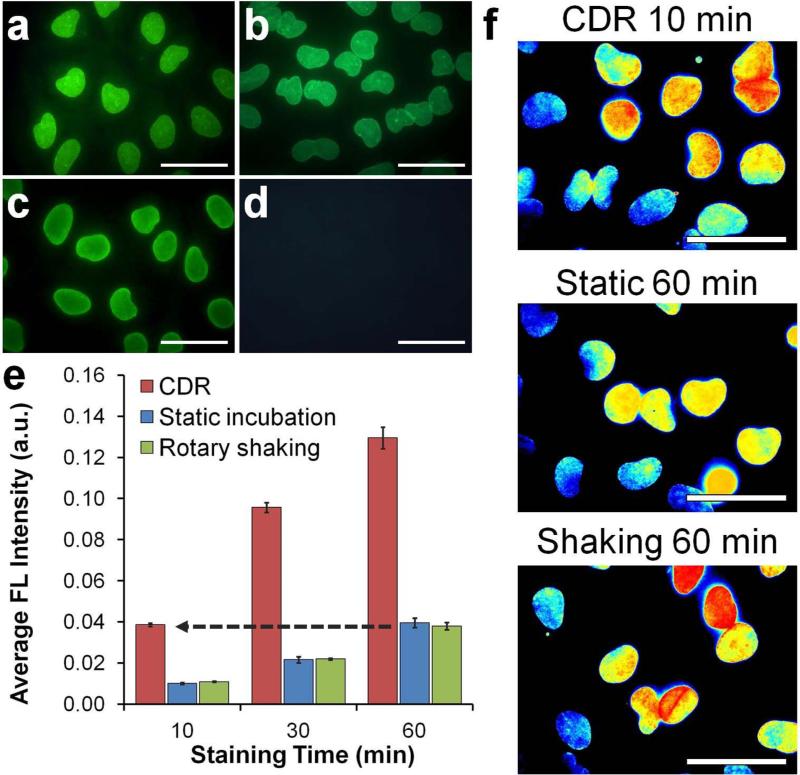
Studies were performed on HeLa cells that were grown directly inside the wells of a glass-bottom 6-well plate, fixed with formaldehyde, and permeabilized with detergents to allow probe access to intracellular targets. Lamin A, a component of nuclear envelope, was used as a target due to its characteristic staining pattern, ease of quantitative analysis of staining intensity, and intracellular localization. The staining specificity was confirmed by incubating cells with anti-Lamin A QDot-Ab and dye-labeled Ab probes under static conditions and comparing results to a conventional 2-step staining procedure performed with unmodified primary antibodies and secondary Ab-QDot bioconjugates (Figure 3a-d).
Figure 3.
Rapid immunofluorescence staining with QDot probes. (a-d) Characterization of QDot-1'Ab and dye-labeled 1'Ab probes targeting Lamin A. One-step immunofluorescence images obtained with (a) QDot-1’Ab and (b) dye-labeled 1’Ab probes produced staining patterns consistent with the nuclear membrane localization of Lamin A and (c,d) results obtained with QDot-2'Ab in a conventional two-step staining procedure (positive Lamin A staining is shown in (c) and control lacking 1’Ab incubation in (d)). Scale bar, 50 μm. (e) Quantitative evaluation of staining intensity with respect to staining time achieved via CDR, static incubation, and rotary shaking techniques. Notably, CDR achieved comparable staining intensity 6 times faster than conventional methods, producing detectable signal within the first 10 minutes of staining. Error bars represent one standard deviation of an average Lamin A staining intensity from four different fields of view. (f) Representative cell staining intensity maps obtained after 10-min CDR in comparison to 60-min static and rotary shaking incubation. All images were normalized and color-coded with a heat map for direct comparison of staining pattern and intensities. Scale bar, 50 μm.
The evolution of staining intensity under CDR, static incubation, and rotary shaking was monitored over a period of 8 h. Consistent with the expectation of slow diffusion-limited kinetics, IF staining indeed approached a steady-state intensity level only after 8 hours of incubation under static and shaking conditions (Supporting Information Figure S6). At the same time, CDR efficiently reduced the binding time toward saturation to 2 hours. More importantly, under practical IF staining applications, it took just 10 min to yield the same level of fluorescence signals of 1-hour conventional IF incubation (Figure 3e,f).
Taking advantage of the substantial enhancement of binding kinetics with CDR, we proceeded to explore two applications previously inaccessible with conventional IF methods: background-free monitoring of staining evolution and high-content single-cell molecular profiling in practical time frame (hours rather than a week).
2.3. Background-free monitoring of staining evolution
Strong background fluorescence originating from the bulk high-concentration probe solution often interferes with in situ IF staining applications, requiring specimen washing prior to imaging and, thus, precluding real-time monitoring of staining evolution and increasing assay time. Reducing the probe concentration might be beneficial in this situation (which also helps to reduce costs of expensive biological reagents), but it would necessarily slow-down staining kinetics under diffusion-limited conditions. In this regard, we hypothesized that elimination of probe diffusion limitation should yield comparable staining intensity with lower probe concentration due to improved assay kinetics.
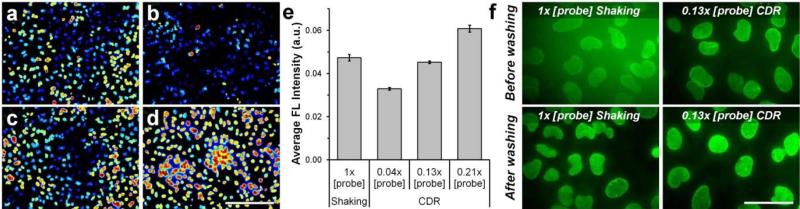
To test this hypothesis we first compared staining intensities obtained after 1-h CDR with 0.3 nM, 0.9 nM, and 1.5 nM QDot-Ab probes to a conventional 1-h staining with 7 nM QDots. Qualitative evaluation of representative normalized images (Figure 4a-d) and quantitative analysis of average staining intensities (Figure 4e) confirmed that similar labeling intensity could be achieved after 1-h incubation with either 7 nM probes under rotary shaking or 0.9 nM probes under CDR. Therefore, the CDR methodology enabled an approximately 8-fold reduction in probe concentration without any loss in sensitivity.
Figure 4.
Background-free immunofluorescence staining via CDR. (a-d) Representative fluorescence intensity maps obtained after a 1-hour incubation with (a) 7 nM QDot-1'Ab (1× [probe]) under rotary shaking and (b) 0.3 nM (0.04× [probe]), (c) 0.9 nM (0.13× [probe]), and (d) 1.5 nM (0.21× [probe]) QDot-1'Ab using CDR procedure. Images were normalized and color-coded with a heat map. Scale bar, 250 μm. (e) Average fluorescence intensities of Lamin A staining achieved after a 1-hour incubation under rotary shaking with 1× [probe] and CDR with 0.04×, 0.13×, and 0.21× QDot-1'Ab concentration. Consistent with qualitative observations in (a-d), quantitative analysis demonstrated that comparable staining could be obtained with ~8-times lower probe concentration via CDR methodology. Error bars represent one standard deviation of an average Lamin A staining intensity from four different fields of view. (f) A strong background fluorescence originating from the QDots in 1× [probe] bulk solution used for staining under rotary shaking required extensive specimen washing prior to imaging. At the same time, 0.13× [probe] solution employed with CDR methodology featured nearly no background, enabling real-time monitoring of staining evolution. Scale bar, 50 μm.
We then tested whether such reduction of probe concentration below 1 nM was sufficient in order to eliminate the undesirable fluorescence background. Fixed HeLa cells were incubated with QDot-Ab probes at 7 nM under rotary shaking or 0.9 nM under CDR and imaged prior to and after washing (Figure 4f). As expected, 7 nM QDot solution produced a strong fluorescence background, which masked specific target labeling and had to be removed by washing. In contrast, 0.9 nM QDots featured nearly no background, while producing comparable nuclear membrane staining via CDR. Thus, we confirmed that CDR could be used for background-free monitoring of staining evolution and, potentially, could be applied for efficient capture of low-abundance targets.
2.4. Enabling practical multicycle IF staining
A number of recently developed multicycle IF staining methods have captivated bioengineers and biologists because of their enormous potential in comprehensive in situ molecular analysis of single cells.[6,12] Subsets of molecular targets in the same cells are stained with antibody-fluorescent reporter conjugates in multiple cycles with 2-10 targets in each cycle (depending on the availability of spectrally distinctive fluorophores). One such technology, recently reported by our group, is based on QDot probes that enable simultaneous 5-10 color labeling during each cycle (in comparison to organic dyes that are typically limited to 3 colors).[6,27] Repeated staining of subgroups of antigens potentially allows examination of over 100 molecular targets in individual cells with the resolution of optical imaging. However, a drawback of this powerful technology for real-world research and clinical applications is the slow assay speed, as each staining cycle takes approximately 4 hours, largely due to the long diffusion-limited incubations including blocking, staining, destaining, and all the washing steps in between. When 10 cycles are performed at this speed, it takes 40 hours (whole week of work time) to complete the experiment. Therefore, enhancement of assay kinetics 5-10 times without the loss of sensitivity and capacity for quantitative analysis is highly desirable and will significantly enhance the practicality of virtually all the sequential cyclic staining technologies.[6,12]
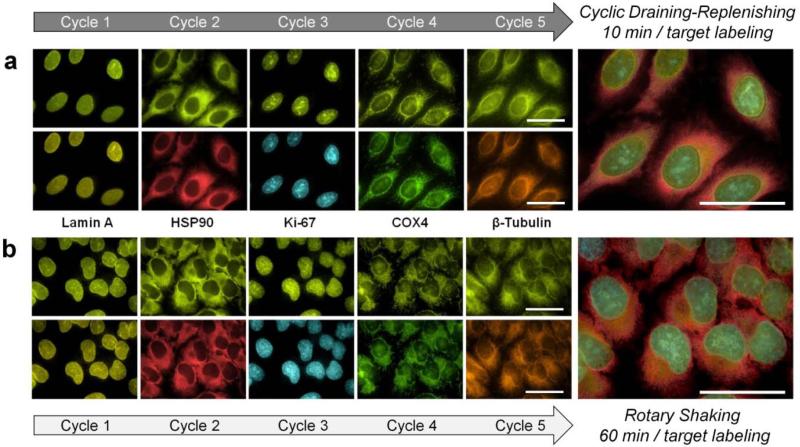
To demonstrate the concept, five intracellular targets (Lamin A, HSP90, Ki-67, Cox-4 and β-tubulin) were labeled with same-color QDot probes through 5 sequential staining / imaging / regeneration cycles. QDot-Ab bioconjugates were applied to cells either for 10 min via CDR or 1 h under conventional rotary shaking. Fluorescence microscopy with hyperspectral imaging was employed for imaging and quantitative analysis of staining intensity. In complete consistency with the rapid IF results shown above, shortened probe incubation through CDR produced accurate staining patterns (Figure 5a) comparable to those produced via a regular multicycle procedure with 1-h QDot incubation steps (Figure 5b). Importantly, quantitative analysis indicated that the fast CDR procedure yielded staining intensity levels very similar to those obtained with reference static incubation (Supporting Information Figure S7), producing accurate molecular expression profile at a substantially reduced assay time.
Figure 5.
Rapid multicycle immunofluorescence staining. Five sequential cycles of single-color staining and imaging were performed on the same cell subpopulation for multiplexed detection of five molecular targets (Lamin A, HSP90, Ki-67, Cox-4 and β-tubulin) using self- assembled QDot-SpA-Ab probes. During each cycle, labeling was done via either a 10-minute CDR (a) or 1-hour rotary shaking (b) incubation. Both methods produced highly specific staining patterns with no carry-over fluorescence, build-up of background fluorescence, or cross-staining between cycles, yielding accurate 5-target imaging and analysis. However, CDR methodology enabled a substantial reduction in the overall assay time due to a dramatically improved labeling kinetics. Composite 5-target images were generated from false-colored, aligned, and cropped images of individual targets. Scale bar, 50 μm.
3. Conclusions
On-surface assays have become an indispensable tool for numerous bio-analytical and imaging applications, with nanoparticle probes expanding assay capabilities and enabling novel assay formats. However, most of these techniques are strictly limited by the slow probe diffusion from the bulk solution to the surface, which has proven virtually inevitable with conventional assay setups. Diffusion limitation is highly detrimental to advanced multi-step procedures, such as powerful multi-cycle IF techniques for single-cell molecular profiling,[27,28] and is undesirable in single-step procedures when the assay time is of essence, such as in intraoperative[29] and infectious disease[30,31] diagnostics.
The cyclic draining-replenishing (CDR) technology reported here completely eliminates the chronic and fundamental problem of diffusion limitation, achieving efficient molecular mixing at the surface and “erasing” the effect of the probe size in a simple and affordable setup easily adaptable by a wide range of biomedical laboratories. Through proof-of-concept studies, we have demonstrated that ELISA with ~100 nm poly-HRP nanoprobes could be performed in a much faster reaction-limited regime, and IF cell staining with QDots could be achieved in under 10 min incubation with CDR. Further, a substantial enhancement in assay kinetics has rendered multi-step and multi-cycle high-content molecular analysis technologies practically useful for the first time.
We anticipate CDR technology to be incorporated within existing on-surface assays and become an enabling component of novel ultra-rapid technologies for high-content scientific analysis and fast clinical diagnostics. The concept of CDR could also inspire a broad spectrum of applications in basic and clinical research (e.g., cell transfection and fast detection of time-sensitive diseases, such as infectious and cardiovascular diseases)[31,32] and industrial fields (e.g., on-surface catalysis).[33] Finally, elimination of at-surface molecular depletion might prove instrumental for applications requiring efficient capture of low-abundance targets onto a surface, which might take hours or days under mass transfer-limited condition.
4. Experimental Section
Surface-based bioassay setup
Glass-bottom 6-well plates (In Vitro Scientific) were used for cell staining studies, and plastic 6-well plates (BD Biosciences) were used for enzyme-linked immunosorbent assay (ELISA). Static incubation was done by resting the plate on a lab bench. Incubation under shaking was done by placing the plate on a rotary shaker (Thermo Scientific). CDR procedure was performed using a Labquake rotator (Barnstead Thermolyne) by sealing individual wells of 6-well plates with Parafilm M (Bemis Flexible Packaging), fixing plates onto the rotator, and rotating at the speed of 8 rpm.
Probe preparation
Dye-labeled antibodies and QDot-1'Ab probes targeting Lamin A were prepared following the protocol provided by the manufacturer. QDot-SpA probes were prepared following the protocol described elsewhere.[27] Detailed probe preparation protocols are available in SI Methods.
ELISA diagnostics
Mouse and Rabbit IgG (Sigma-Aldrich) were used as model targets for ELISA experiments. IgG was immobilized onto 6-well plastic plates (BD Biosciences) and blocked by bovine serum albumin. Target labeling was done via either CDR or conventional methods with Poly-HRP (Pierce Goat Anti-Mouse or Goat Anti-Rabbit Poly-HRP, Thermo Scientific) and Mono-HRP (Goat Anti-Mouse or Goat Anti-Rabbit IgG (whole molecule)-HRP, Sigma-Aldrich). HRP abundance was assessed by adding 1-Step Turbo TMB-ELISA substrate (Thermo Scientific) and measuring solution absorbance at 450 nm using Infinite M 200 plate reader (Tecan).
Immunofluorescence cell staining
Human cervical cancer cell line HeLa (ATCC) was used for immunofluorescence staining experiments. Cells were grown in glass-bottom 6-well plates (In Vitro Scientific) and processed following the protocol described elsewhere.[27] Prior to staining cells were blocked with 2% BSA/0.1% casein (Novagen) in 1×TBS. For a two-step immunofluorescence, cells were first incubated with polyclonal rabbit anti-Lamin A IgG (Sigma-Aldrich) and then with QDot-2'Ab (Qdot 565 goat F(ab')2 anti-rabbit conjugates, Invitrogen). For a one-step IF, cells were incubated with 7 nM QDot-1'Ab probes or 5 μg/ml dye-labeled 1'Abs targeting Lamin A via either CDR or conventional (static incubation or rotary shaking) methods. For low-background staining, 0.3 nM, 0.9 nM, and 1.5 nM QDot-1'Ab solutions were used separately for staining via CDR method. Fluorescence imaging was done immediately following staining.
Multicycle immunofluorescence staining
Multicycle IF staining was performed in 5 single-plex labeling cycles in a sequential manner as described elsewhere[27] to obtain multiplexed composite images of five model targets: Lamin A, HSP90, Ki-67, Cox-4 and β-tubulin. Each staining cycle consisted of (i) pre-blocking, (ii) staining, (iii) imaging, and (iv) de-staining, where staining of each target was done with pre-assembled QDot-SpA-1'Ab probes for 1 hour under rotary shaking or 10 minutes under CDR. Following imaging, cells were de-stained for 15 min, washed with TBS and blocking buffer, and blocked for 1 hour (as part of next staining cycle). Note that although CDR is applicable to all the incubation steps in immunostaining for an overall shortened assay time, it was applied solely to QDot incubation step in this proof-of-concept study, because this is the only step generating detectable signal for quantitative assessment of CDR performance. It is expected that other steps, such as blocking, washing, and regeneration, can be sped-up under CDR in a similar manner.
Immunofluorescence imaging and signal analysis
Cell imaging was done with IX-71 inverted fluorescence microscope (Olympus). True-color images were captured with QColor5 camera (Olympus) for qualitative evaluation of staining patterns. Quantitative analysis was performed on images captured with a hyperspectral imaging camera (Nuance, CRI, now Advanced Molecular Vision) using Nuance image analysis software and following procedure described previously.[27] For sequential staining, a permanent reference point was marked on the bottom of the well to aid in finding the same cell subset for each imaging cycle. All images were acquired with cells attached to the coverslip bottom of the well and immersed in TBS (for QDot probes) or Vectashield mounting medium (Vector Laboratories, for dye-labeled antibody probes). Detailed image acquisition and signal analysis procedure is described in SI Methods.
Supplementary Material
Acknowledgements
This work was supported in part by NIH (R01CA131797, R21CA192985) and the Department of Bioengineering at the University of Washington. P.Z. thanks the National Cancer Institute for a T32 fellowship (T32CA138312). JWL thanks the Howard Hughes Medical Institute for the international student scholarship. We are also grateful to Dr. Barry Lutz for fruitful discussions and valuable comments of the manuscript.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Mitchell P. Nature Biotechnol. 2002;20:225. doi: 10.1038/nbt0302-225. [DOI] [PubMed] [Google Scholar]
- 2.Howes PD, Chandrawati R, Stevens MM. Science. 2014;346:1247390. doi: 10.1126/science.1247390. [DOI] [PubMed] [Google Scholar]
- 3.a Cao YC, Jin R, Mirkin CA. Science. 2002;297:1536. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]; b Cao YC, Jin R, Nam JM, Thaxton CS, Mirkin CA. J. Am. Chem. Soc. 2003;125:14676. doi: 10.1021/ja0366235. [DOI] [PubMed] [Google Scholar]; c Liu Z, Tabakman S, Sherlock S, Li X, Chen Z, Jiang K, Fan S, Dai H. Nano Res. 2010;3:222. doi: 10.1007/s12274-010-1025-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Chan YH, Ye F, Gallina ME, Zhang X, Jin Y, Wu IC, Chiu DT. J. Am. Chem. Soc. 2012;134:7309. doi: 10.1021/ja3022973. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wu C, Schneider T, Zeigler M, Yu J, Schiro PG, Burnham DR, McNeill JD, Chiu DT. J. Am. Chem. Soc. 2010;132:15410. doi: 10.1021/ja107196s. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Yezhelyev MV, Al-Hajj A, Morris C, Marcus AI, Liu T, Lewis M, Cohen C, Zrazhevskiy P, Simons JW, Rogatko A, Nie S, Gao X, O'Regan RM. Adv. Mater. 2007;19:3146. [Google Scholar]
- 5.Xing Y, Chaudry Q, Shen C, Kong KY, Zhau HE, Chung LW, Petros JA, O'Regan RM, Yezhelyev MV, Simons JW, Wang MD, Nie S. Nature Protoc. 2007;2:1152. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- 6.Zrazhevskiy P, Gao X. Nature Commun. 2013;4:1619. doi: 10.1038/ncomms2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y, Nolan GP. Nature Med. 2014;20:436. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, Varga Z, Wild PJ, Gunther D, Bodenmiller B. Nature Methods. 2014;11:417. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 8.a Nygren H, Stenberg M. Immunology. 1989;66:321. [PMC free article] [PubMed] [Google Scholar]; b Stenberg M, Stiblert L, Nygren H. J. Theor. Biol. 1986;120:129. doi: 10.1016/s0022-5193(86)80169-2. [DOI] [PubMed] [Google Scholar]; c Kusnezow W, Syagailo YV, Ruffer S, Klenin K, Sebald W, Hoheisel JD, Gauer C, Goychuk I. Proteomics. 2006;6:794. doi: 10.1002/pmic.200500149. [DOI] [PubMed] [Google Scholar]; d Squires TM, Messinger RJ, Manalis SR. Nature Biotechnol. 2008;26:417. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 9.Canziani GA, Klakamp S, Myszka DG. Anal. Biochem. 2004;325:301. doi: 10.1016/j.ab.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Pappaert K, Van Hummelen P, Vanderhoeven J, Baron GV, Desmet G. Chem. Eng. Sci. 2003;58:4921. doi: 10.1016/s0021-9673(03)00715-5. [DOI] [PubMed] [Google Scholar]
- 11.Stenberg M, Nygren H, Immunol J. Methods. 1988;113:3. doi: 10.1016/0022-1759(88)90376-6. [DOI] [PubMed] [Google Scholar]
- 12.a Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, Hollman D, Kamath V, Kaanumalle S, Kenny K, Larsen M, Lazare M, Li Q, Lowes C, McCulloch CC, McDonough E, Montalto MC, Pang Z, Rittscher J, Santamaria-Pang A, Sarachan BD, Seel ML, Seppo A, Shaikh K, Sui Y, Zhang J, Ginty F. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11982. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Micheva KD, Smith SJ. Neuron. 2007;55:25. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Bockelmann R, Malykh Y, Gollnick H, Friedenberger M, Bode M, Dress AW. Nature Biotechnol. 2006;24:1270. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 13.Stack EC, Wang C, Roman KA, Hoyt CC. Methods. 2014;70:46. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 14.a Ng AH, Uddayasankar U, Wheeler AR. Anal. Bioanal. Chem. 2010;397:991. doi: 10.1007/s00216-010-3678-8. [DOI] [PubMed] [Google Scholar]; b Wang L, Li PC. Anal. Chim. Acta. 2011;687:12. doi: 10.1016/j.aca.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 15.a Glaser RW. Anal. Biochem. 1993;213:152. doi: 10.1006/abio.1993.1399. [DOI] [PubMed] [Google Scholar]; b Karlsson R, Roos H, Fagerstam L, Persson B. Methods. 1994;6:99. [Google Scholar]
- 16.a Golden JP, Floyd-Smith TM, Mott DR, Ligler FS. Biosens. Bioelectron. 2007;22:2763. doi: 10.1016/j.bios.2006.12.017. [DOI] [PubMed] [Google Scholar]; b Yoon SK, Fichtl GW, Kenis PJ. Lab Chip. 2006;6:1516. doi: 10.1039/b609289f. [DOI] [PubMed] [Google Scholar]; c Morales-Narvaez E, Guix M, Medina-Sanchez M, Mayorga-Martinez CC, Merkoci A. Small. 2014;10:2542. doi: 10.1002/smll.201303068. [DOI] [PubMed] [Google Scholar]
- 17.a Liu J, Williams BA, Gwirtz RM, Wold BJ, Quake S. Angew. Chem. Int. Ed. Engl. 2006;45:3618. doi: 10.1002/anie.200503830. [DOI] [PubMed] [Google Scholar]; b Liu J, Williams BA, Gwirtz RM, Wold BJ, Quake S. Angew. Chem. 2006;118:3700. doi: 10.1002/anie.200503830. [DOI] [PubMed] [Google Scholar]; c Rupp J, Schmidt M, Munch S, Cavalar M, Steller U, Steigert J, Stumber M, Dorrer C, Rothacher P, Zengerle R, Daub M. Lab Chip. 2012;12:1384. doi: 10.1039/c2lc21110f. [DOI] [PubMed] [Google Scholar]; d Liu RH, Lenigk R, Druyor-Sanchez RL, Yang J, Grodzinski P. Anal. Chem. 2003;75:1911. doi: 10.1021/ac026267t. [DOI] [PubMed] [Google Scholar]; e Hertzsch JM, Sturman R, Wiggins S. Small. 2007;3:202. doi: 10.1002/smll.200600361. [DOI] [PubMed] [Google Scholar]; f Vanderhoeven J, Pappaert K, Dutta B, Van Hummelen P, Desmet G. Anal. Chem. 2005;77:4474. doi: 10.1021/ac0502091. [DOI] [PubMed] [Google Scholar]
- 18.a Erickson D, Liu X, Krull U, Li D. Anal. Chem. 2004;76:7269. doi: 10.1021/ac049396d. [DOI] [PubMed] [Google Scholar]; b Wei C-W, Cheng J-Y, Huang C-T, Yen M-H, Young T-H. Nucleic Acids Res. 2005;33:e78. doi: 10.1093/nar/gni078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a Morozov VN, Groves S, Turell MJ, Bailey C. J. Am. Chem. Soc. 2007;129:12628. doi: 10.1021/ja075069m. [DOI] [PubMed] [Google Scholar]; b Sosnowski RG, Tu E, Butler WF, O'Connell JP, Heller MJ. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1119. doi: 10.1073/pnas.94.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a Cheek BJ, Steel AB, Torres MP, Yu YY, Yang H. Anal. Chem. 2001;73:5777. doi: 10.1021/ac0108616. [DOI] [PubMed] [Google Scholar]; b Xu Y, Bao G. Anal. Chem. 2003;75:5345. doi: 10.1021/ac034613g. [DOI] [PubMed] [Google Scholar]
- 21.Ciftlik AT, Lehr HA, Gijs MA. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5363. doi: 10.1073/pnas.1211273110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatta H, Tsuneyama K, Kondo T, Takano Y. J. Histochem. Cytochem. 2010;58:421. doi: 10.1369/jhc.2010.955096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a Long DJ, 2nd, Buggs C. J. Mol. Histol. 2008;39:1. doi: 10.1007/s10735-007-9093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kumada T, Tsuneyama K, Hatta H, Ishizawa S, Takano Y. Mod. Pathol. 2004;17:1141. doi: 10.1038/modpathol.3800165. [DOI] [PubMed] [Google Scholar]
- 24.Chang KH, Finn DT, Lee D, Bhawan J, Dallal GE, Rogers GS. J. Am. Acad. Dermatol. 2011;64:107. doi: 10.1016/j.jaad.2010.02.055. [DOI] [PubMed] [Google Scholar]
- 25.Hulme EC, Trevethick MA. Br. J. Pharmacol. 2010;161:1219. doi: 10.1111/j.1476-5381.2009.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Lau SK, Varma VA, Moffitt RA, Caldwell M, Liu T, Young AN, Petros JA, Osunkoya AO, Krogstad T, Leyland-Jones B, Wang MD, Nie S. ACS Nano. 2010;4:2755. doi: 10.1021/nn100213v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zrazhevskiy P, True LD, Gao X. Nature Protoc. 2013;8:1852. doi: 10.1038/nprot.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedenberger M, Bode M, Krusche A, Schubert W. Nature Protoc. 2007;2:2285. doi: 10.1038/nprot.2007.320. [DOI] [PubMed] [Google Scholar]
- 29.a Gal AA, Cagle PT. JAMA. 2005;294:3135. doi: 10.1001/jama.294.24.3135. [DOI] [PubMed] [Google Scholar]; b Balog J, Sasi-Szabo L, Kinross J, Lewis MR, Muirhead LJ, Veselkov K, Mirnezami R, Dezso B, Damjanovich L, Darzi A, Nicholson JK, Takats Z. Sci. Transl. Med. 2013;5:194ra93. doi: 10.1126/scitranslmed.3005623. [DOI] [PubMed] [Google Scholar]
- 30.a Diekema DJ, Pfaller MA. Clin. Infect. Dis. 2013;56:1614. doi: 10.1093/cid/cit038. [DOI] [PubMed] [Google Scholar]; b Liesenfeld O, Lehman L, Hunfeld KP, Kost G. Eur. J. Microbiol. Immunol. (Bp) 2014;4:1. doi: 10.1556/EuJMI.4.2014.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caliendo AM, Gilbert DN, Ginocchio CC, Hanson KE, May L, Quinn TC, Tenover FC, Alland D, Blaschke AJ, Bonomo RA, Carroll KC, Ferraro MJ, Hirschhorn LR, Joseph WP, Karchmer T, MacIntyre AT, Reller LB, Jackson AF. Clin. Infect. Dis. 2013;57(Suppl 3):S139. doi: 10.1093/cid/cit578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.a Luo D, Saltzman WM. Nature Biotechnol. 2000;18:893. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]; b Daubert MA, Jeremias A. Vasc. Health Risk Manag. 2010;6:691. doi: 10.2147/vhrm.s5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a Lee AF, Bennett JA, Manayil JC, Wilson K. Chem. Soc. Rev. 2014;43:7887. doi: 10.1039/c4cs00189c. [DOI] [PubMed] [Google Scholar]; b Naik SN, Goud VV, Rout PK, Dalai AK. Renew. Sust. Energ. Rev. 2010;14:578. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.