The role that autophagy pathways play in cancer, as a suppressor of tumor development or a driver of oncogenic transformation, remains a topic of debate and controversy. In some instances, there are indicative links between mutations that drive cancers and regulation of autophagy. For example, activation of the PI3K/AKT/mTOR pathway is well recognized in many different tumor types—a result often of activating mutations in PI3K, AKT or mTOR, or loss of function mutations in the negative regulators, PTEN, TSC1 and TSC2. PI3K signaling is also activated by a number of receptor tyrosine kinases, including EGFR, HER2, c-Kit and PDGFRA. There is little doubt that PI3K pathway activation suppresses autophagy. However, activation of PI3K pathways has other direct tumor-promoting capabilities, such as promoting cell growth, translation and proliferation. Thus while the frequency of activation of PI3K/AKT/mTOR signaling in cancers raises the very distinct possibility that repression of autophagic flux contributes to oncogenic transformation, it does not prove it.
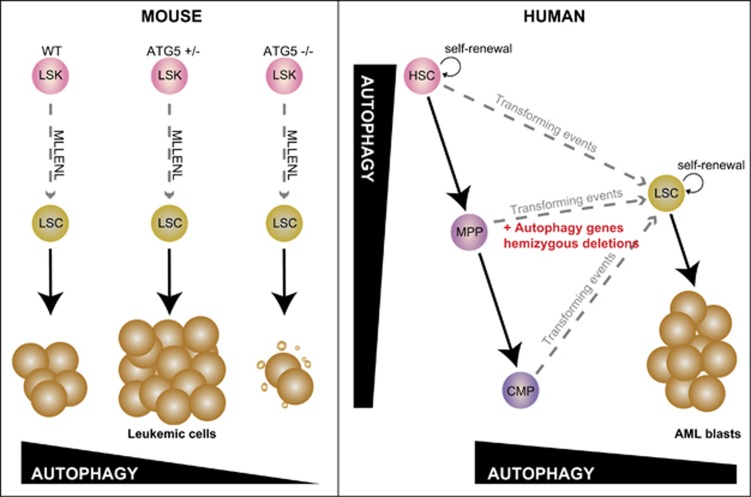
Studying genes that are uniquely involved in the regulation of autophagy can more directly establish how autophagic pathways contribute to cancer initiation or progression. In Cell Death and Discovery, the paper from Watson et al.1 provide both direct and circumstantial evidence that diminished autophagic flux results in the development of a myeloproliferative state and accelerates the progress of acute myeloid leukemia (AML) in a mouse model of the disease driven by an oncogenic fusion gene involving the mixed lineage leukemia (MLL) histone methyl transferase (KMT2A). The data they present make another strong case that diminished autophagy accelerates the development of AML (Figure 1).
Figure 1.
In mouse models of MLL-driven leukemia (left panel), Watson et al.1 show heterozygous deletion of Atg5 gene increases leukemogeneisis, whereas loss of both alleles is deleterious for leukemic cells. In human hematopoiesis (right panel), the more differentiated myeloid hematopoietic cells represses autophagic flux. However, in human leukemic cells, there is copy-number loss of chromosomal regions that encode a number of autophagy genes, and this occurs more frequently than might be expected by chance alone. That raises the possibility that that decreased autophagy flux, as a result of haploinsufficiency of autophagy genes, might contribute to the development of acute myeloid leukemia
Watson and colleagues noted that haematopoietic stem and progenitor cells have higher autophagic flux than mature hematopoietic cells, whilst AML blasts tend to have reduced flux. This, together with the fact that a number of autophagic genes exist (along, of course, with many other genes), in genomic regions that are subject to copy-number loss in AML, hint at an association between diminished autophagy and the development of AML.
The most intriguing and direct results come from the conditional Atg5−/− knockout mice. ATG5 (and ATG7) are involved in the assembly of the assembly of autophagic vesicles. Atg5, conjugated to Atg12, are together required for the lipidation of microtubule-associated protein light chain 3 (LC3), a key readout of autophagic flux. When Atg5 is deleted from hematopoietic cells, Watson and colleagues found that mice develop a myeloproliferative disease, a phenotype very similar to that of the Atg7-deficient animals they had previously described.2 At least within the time frame of the current study, these mice did not develop a florid myeloid leukemia, although infiltrating myeloid cells were observed in a range of tissues. The phenotypic similarities between the Atg5−/− and Atg7−/− mice clearly establishes the importance of autophagic vesicle assembly in normal hematopoiesis, and shows that disruption of this process is sufficient to initiate a myeloproliferative disorder in mice. Watson et al. extended these observations to models of AML. Heterozygous deletion of Atg5 was sufficient to reduce the latency of AML in the MLL-ENL model, such that all mice transplanted with MLL-ENL-Atg5+/− leukemic cells were dead before any control mice had begun show symptoms. This striking acceleration of this disease constitutes strong genetic evidence that reduced autophagic functions, perhaps as a result of somatic copy-number loss, can drive the progression of AML. In these experiments the autophagy gene Atg5 is acting as tumor suppressor, and haploinsufficiency is enough to remove this tumor suppressor activity. Even if Atg5 (or Atg7) mutations are rarely described in AML (or rarely identified in sequence databases such as COSMIC3), the findings of this paper are entirely consistent with a study from Qu et al.4 that shows that beclin-1 is also a haploinsufficient tumor-suppressor gene. Both studies support the hypothesis that reduced autophagic activity contributes to the oncogenic functions of more established mutations in cancer, including in the PI3K/AKT/mTor pathway. Formal proof of this waits further experimental testing and detailed functional analyses.
Like any good paper, this one poses as many questions as are answered. In particular, the potential therapeutic consequences of these observations remain wide open. It is not yet clear if the effects of ATG5 loss on MLL-ENL-driven leukemogenesis will be recapitulated when other MLL-fusion proteins (e.g., MLL-AF9) are the oncogenic drivers or indeed if ATG5 affects AML initiated by alterations in other AML driver genes (e.g., DNMT3a, IDH1/2, p53). Certainly there are indications that ATG5 can regulate the onset and development of other malignancies such as colorectal cancer in preclinical models5 so its plausible that the effects of hemizygous Atg5 deletion described by Watson and colleagues may apply to other tumor types.
The investigators could not study MLL-ENL transformed cells that lacked both copies of Atg5, because of excessive death in these leukemic cells. It would be interesting to know how these cells died and whether any of the few remaining leukemic cells could still cause leukemia in the recipient animals if they were transplanted. As dead leukemic cells are the ultimate goal of antileukemic treatment, it might turn out be that inhibition of autophagy in AML, even recognizing that Atg5 (and Atg7) deletion drives abnormal myeloproliferation, has therapeutic potential. Indeed, an exciting possibility is that AML that have heterozygous deletions of autophagy genes, may be particularly susceptible to autophagy inhibitors such as chloroquine. An alternative approach, inverting this logic, would be to try and restore autophagy with PI3K/mTOR inhibitors. One might predict that this would apply in instances where autophagic genes are not deleted, but the pathway is inhibited by other mechanisms. More generally, the models described by Watson et al. provide a direct means to test how disruption of autophagy pathways contribute to the response to chemotherapy. Although the deletion of Atg5 resulted in a more rapid onset of AML, it would still be intriguing to determine whether the deletion of this (or other) autophagic genes also make these tumors less sensitive to standard chemotherapy. Could it be that the haploinsufficiency for autophagy genes in AML is a biomarker of drug resistance?
Another interesting therapeutic avenue arising from these results is the use of glycolysis inhibitors. The authors show that the elevated proliferation in MLL-ENL Atg5+/− cells is accompanied by higher glycolytic metabolism. Interestingly, the inhibition of glycolysis has reported to have a synergistic effect with mTOR inhibitor in AML cell lines.6
Watson and colleagues have provided another line of evidence that reinforces the dual function of autophagy in regulating cell death and cell survival. The complete blockade of autophagy resulted in the death of leukemic cells, while a reduction of this pathway increased their proliferation and significantly reduced the latency of the disease. This duality is one of the reasons why the role of autophagy in cancer progression and resistance is complicated, making the rational design of a targeted clinical approach somewhat challenging. Indeed, this study offers alternate clinical options for AML that will need to be tested in preclinical settings. However, the experimental approach of developing malignancy models on genetic backgrounds with targeted lesions in specific autophagic genes looks to be a very fruitful line of enquiry. Using genetic models to enhance our understanding of the roles autophagic genes have in specific cancer models will be the basis on which therapeutic regimens that modulate autophagy will be built.
The authors declare no conflict of interest.
References
- Watson AS et al. Cell Death Discovery 2015; 1: 15008. [DOI] [PMC free article] [PubMed]
- Mortensen M et al. J Exp Med. 2011; 208: 455–467. [DOI] [PMC free article] [PubMed]
- Forbes SA et al. Nucleic Acids Res 2015; 43(Database issue): D805–D811. [DOI] [PMC free article] [PubMed]
- Qu X et al. J Clin Invest 2003; 112: 1809–1820. [DOI] [PMC free article] [PubMed]
- Wang L et al. Cancer Biol Ther 2015; 16: 383–391. [DOI] [PMC free article] [PubMed]
- Akers LJ et al. Leuk Res. 2011; 35: 814–820. [DOI] [PMC free article] [PubMed]