Abstract
After cessation of lactation, involution of the mouse mammary gland proceeds in two distinct phases, a reversible and an irreversible one, which leads to the death and removal of alveolar cells. Cell death is preceded by the loss of STAT5 activity, which abrogates cell differentiation and gain of STAT3 activity. Despite early observations implicating BCL2 (B cell lymphoma 2) family proteins in this process, recent evidence suggests that STAT3-controlled cathepsin activity is most critical for cell death at the early stage of involution. Somewhat surprisingly, this cell death associates with but does not depend on the activation of pro-apoptotic effector caspases. However, transgenic overexpression of BCL2, that blocks caspase activation, delays involution while conditional deletion of BclX accelerates this process, suggesting that BCL2 family proteins are needed for the effective execution of involution. Here, we report on the transcriptional induction of multiple pro-apoptotic BCL2 family proteins of the ‘BH3-only' subgroup during involution and the rate-limiting role of BIM in this process. Loss of Bim delayed epithelial cell clearance during involution after forced weaning in mice, whereas the absence of related Bmf had minor and loss of Bad or Noxa no impact on this process. Consistent with a contribution of BCL2 family proteins to the second wave of cell death during involution, loss of Bim reduced the number of apoptotic cells in this irreversible phase. Notably, the expression changes observed within the BCL2 family did not depend on STAT3 signalling, in line with its initiating role early in the process, but rather appear to result from relief of repression by STAT5. Our findings support the existence of a signalling circuitry regulating the irreversible phase of involution in mice by engaging BH3-only protein-driven mitochondrial apoptosis.
Apoptosis is an important physiological mechanism in the development and maintenance of tissue architecture in multicellular organisms. Cells with limited lifespan need to be removed and potentially dangerous cells are kept in check by this mechanism to prevent pathologies like autoimmunity or cancer. Most cell death by apoptosis is mediated via the intrinsic or mitochondrial pathway that is controlled by pro- and anti-apoptotic proteins of the BCL2 (B cell lymphoma 2) family.1, 2 A well-known example for apoptosis involved in tissue remodelling is the programmed cell death observed in the process of involution, which promotes the deletion and clearance of secretory epithelial cells in the lactating mammary gland after weaning. During this process, a virgin-like tissue structure is re-established.3, 4, 5
During pregnancy, sex hormones such as progesterone and prolactin induce a massive change in the cellular composition of the mammary gland, driving the development of epithelial structures, including ducts and alveoli, forming an effective milk-producing organ securing survival of the newborn. Upon cessation of lactation, the large number of secretory alveolar epithelial cells in the mammary gland are no longer needed and removed.3 The initial triggers for the induction of cell death during involution have been intensely investigated and involve the gain of STAT3 and the loss of STAT5 activity upon cessation of hormone signalling.5, 6 Glucocorticoids,7 transforming growth factor-β 3 (TGFbeta3)8 and leukaemia inhibitory factor (LIF),9 which control the activity of additional transcription factors, including the glucocorticoid receptor, SMA and MAD-related proteins (SMADs) or CCAAT/enhancer-binding proteins (C/EBPs)10 also help to orchestrate this complex process. In addition, enzymes such as matrix metalloproteinases11 or other factors usually secreted into the lumen, like insulin-like growth factor-binding protein 5 (IGFBP-5), contribute to remodelling.12 For example, IGFBP-5 negatively regulates the activity of the growth hormone by binding to the survival factor insulin-like growth factor I (IGF-I), and thereby mitigates IGF-receptor signalling and its coupled PI3K/AKT signalling network that prevents cell death and, when hyperactivated, can effectively delay involution.13, 14
STAT3 has been shown to have a central role in involution as its loss in the mammary gland causes significantly delayed this process.15, 16 In normal physiology, STAT3 activity appears to be counteracted by the suppressor of cytokine signalling 3 (SOCS3), as its tissue-specific deletion triggers increased STAT3 activity and precocious involution, associated with increased activation of c-MYC and its pro-apoptotic effectors E2F1, BAX and p53.10, 17 These observations suggested the involvement of the apoptotic cell death machinery downstream of STAT3 activation for involution. However, STAT3 also represses PI3K signalling by changing the regulatory subset composition of the active kinase complex thereby reducing its activity, and by upregulating the lysosomal cathepsins (B and L) while repressing one of their most potent inhibitors, SPI2A.18, 19 These proteins can be involved in cell death after lysosomal membrane permeabilization (LMP) and pharmacological inhibition of cathepsins was shown to delay involution in vivo suggesting that caspase-independent cell death suffices to initiate the involution process and subsequent remodelling of the mammary gland.20, 21
Despite this intriguing observation, ample data actually support the involvement of the BCL2 family in mammary gland morphogenesis. Involution can be divided into two main phases in mice. The first phase is reversible and accompanied by the accumulation of dying cells in the lumen of ducts that also stain positive for caspase-3.5, 18 The second phase that starts after 48 h can no longer be reversed and involves several proteases, including cathepsins, but besides this, also mitochondria-controlled caspase activity, most notably that of caspase-7.20, 22 Involvement of mitochondrial apoptosis is further supported by studies demonstrating that transgenic BCL2 under the control of the whey acidic protein (WAP) promoter delays involution in mice,23 whereas WAP-Cre mediated loss of BclX accelerates it, a phenotype that was not prevented by co-deletion of Bax,20 suggesting redundancy with Bak. Furthermore, early studies revealed that pro-apoptotic BAK and the BH3-only protein BAD (Bcl-2 antagonist of cell death) were both upregulated during the late phase of lactation and during involution, whereas the anti-apoptotic members BCL2 and BCLW were found repressed, suggesting that their downregulation enables the induction of apoptosis.24 At the end of the late phase of involution, anti-apoptotic BCL2-like proteins reappeared to balance the rheostat in the remodelled tissue.24
Surprisingly, signals that lead to changes in BCL2 family expression remain poorly defined, similar to the role of most pro-apoptotic BH3-only proteins, including BIM, BMF, PMAIP/NOXA or PUMA/BBC3, not known at the time of these early studies. Prolactin stimulation, for example, induces BCL2 mRNA levels in mammary epithelium,25 whereas cessation of hormone signalling leads to a downregulation of AKT activity.26 The AKT pathway, repressed upon STAT3 activation, is known to delay involution upon hyperactivation.14, 18 This might be achieved by phosphorylation of BAD that inactivates its pro-apoptotic function.27 The observed upregulation of BAD protein during involution further suggests a role for this BH3-only protein in tissue remodelling.24 However, the consequences of the lack of hormone or AKT signalling on the expression or activity of other BH3-only proteins remain undefined. Of note, the AKT signalling pathway represses expression of BH3-only proteins BIM, BMF and PUMA in lymphocytes upon cytokine- stimulation.28, 29 PUMA and NOXA are also key effectors of p53 that has been implicated to synergize with STAT3 in involution.10, 30, 31 BIM, together with BMF, is reportedly involved in initial stages of the mammary gland development as they contribute to the clearing of ducts and terminal end buds.32, 33, 34 Interestingly, both proteins can interact with microtubules or actin filaments, respectively, by binding to dynein light-chain molecules.35, 36 Of note, the shedding of epithelial cells into the lumen of ducts, as noted during early involution, triggers a special form of cell death called anoikis that associates with the rearrangement of the cytoskeleton and that is believed to involve BIM and/or BMF.37
Taken together, a rate-limiting role of the BCL2 protein network in involution appears plausible, either as direct activator of mitochondrial apoptosis and/or as a modulator of LMP activity, as recently discussed.38 Hence, we investigated the role of different BH3-only proteins in mammary gland remodelling during involution.
Results
BCL2 family protein expression is highly dynamic during mammary gland morphogenesis
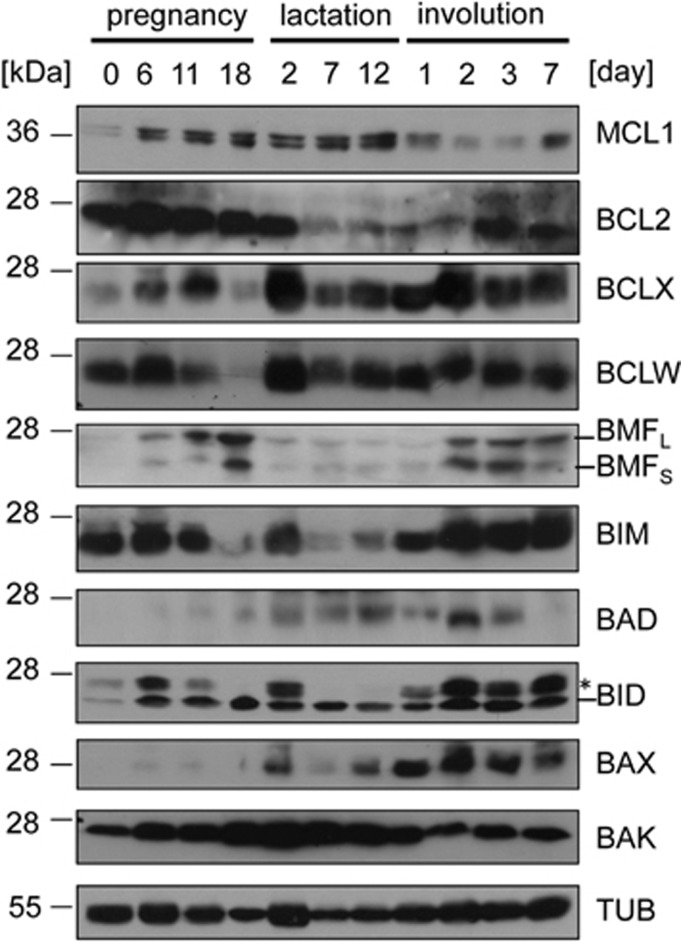
First, the relative expression levels of individual BCL2 family proteins in protein lysates isolated from mammary tisue of virgin 8-week-old non-parous C57BL/6 mice, as well as glands from animals sacrificed at different stages of pregnancy, lactation or involution were investigated. Most BCL2 proteins analysed showed dynamic changes in their expression levels (Figure 1). Anti-apoptotic BCLX (Bcl-2-related protein X) or BCLW were downregulated towards the end of pregnancy but appeared rather stable during lactation and involution while BCL2 was low during lactation, increasing again later in involution. MCL1 levels dropped early upon forced weaning but similar to BCL2 recovered later (Figure 1).
Figure 1.
Dynamic regulation of BCL2 family protein expression in the mammary gland. Mammary gland tissue was isolated at the indicated developmental stages and protein lysates (40 μg/lane) were size fractionated by SDS-PAGE and transferred to nitrocellulose membranes that were sequentially probed, or when necessary, stripped and reprobed with the listed primary antibodies. Anti-tubulin served to control for protein loading. Displayed are representative results from two independent experiments yielding similar results. *indicates a nonspecific signal; BMFS and BMFL represent individual isoforms generated from different translation initiation sites53
Most notably, the levels of the BH3-only proteins BIM and BMF were strongly elevated, albeit with different kinetics during the involution phase, where most cell death occurs, but expressed at rather low levels during lactation. Levels of BAD and BID showed a comparatively minor induction (Figure 1). NOXA and PUMA were not detectable by western analysis with the antibodies used. Of note, all these expression changes were detectable before or at 48 h after weaning where the switch from the reversible to irreversible involution phase occurs. Interestingly, expression of multi-domain pro-apoptotic Bak seemed to increase during pregnancy and lactation, but was actually found lower during involution, whereas the levels of Bax behaved in an opposite way (Figure 1).
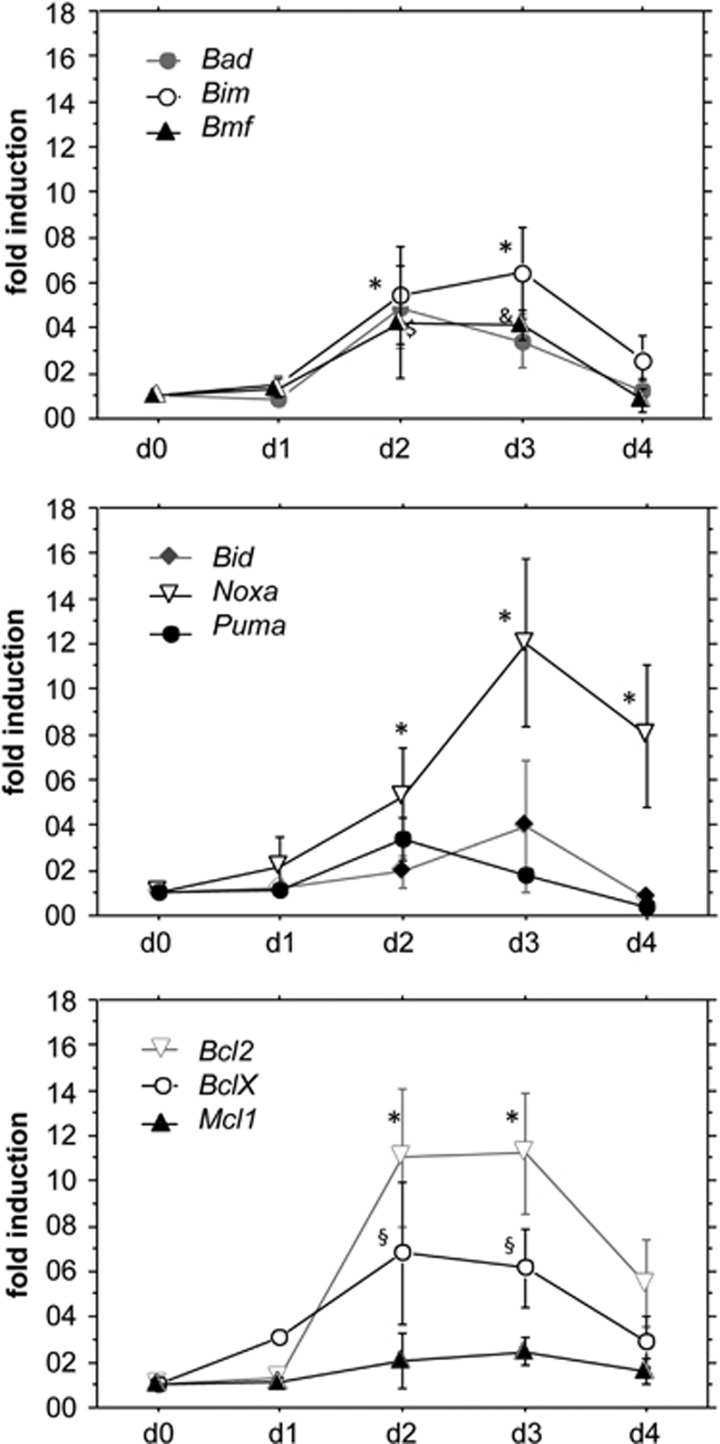
Next, it was assessed whether the changes during involution were due to increased target gene transcription or rather a consequence of post-translational modifications. Therefore, glands were isolated after 12 days of lactation or additional 1, 2, 3 or 4 days after forced weaning to trigger involution. Relative expression levels were normalized to mRNA levels detected on day 12 of lactation (involution d0). This analysis revealed strong and significant induction of Bim and Noxa mRNA on day 2 and day 3, less pronounced increases for Bmf and Bad on either day 2 and/or day 3 but no significant changes in Puma and Bid transcripts (Figure 2). Induction of Noxa was of particular interest, as Mcl1 mRNA levels did not change while protein transiently decreased, suggesting that the induction of Noxa might promote the degradation of MCL1 during involution (Figure 2). Despite a significant increase in the levels of BclX mRNA, the protein appeared rather stable (Figures 1 and 2). Consistent with the observed protein re-expression during involution, Bcl2 mRNA also peaked on day 2 (Figure 2) indicating that the increase of BH3-only proteins is compensated again at later stages of involution to re-establish tissue homeostasis.
Figure 2.
Differential regulation of Bcl-2 family mRNA expression during involution. qRT-PCR analysis was performed on RNA samples extracted from C57BL/6 mammary glands isolated after 12 days of lactation (day 0) or 1, 2, 3 or 4 days of involution triggered by forced weaning. Symbols represent means ±S.E. of n=3–4 individual specimens per time point analysed. Expression levels of the indicated target mRNAs were normalized to actin and relative expression on day 0 (i.e., 12 days of lactation) was set to 1.0. Statistically significant differences between day 0 and day 2, day 3 or day 4 of involution, as assessed by paired Student's t-test (P<0.05) are indicated by * for Bim, $ for Bmf and § for Bad
Loss of Bim delays late-stage involution of the mouse mammary gland
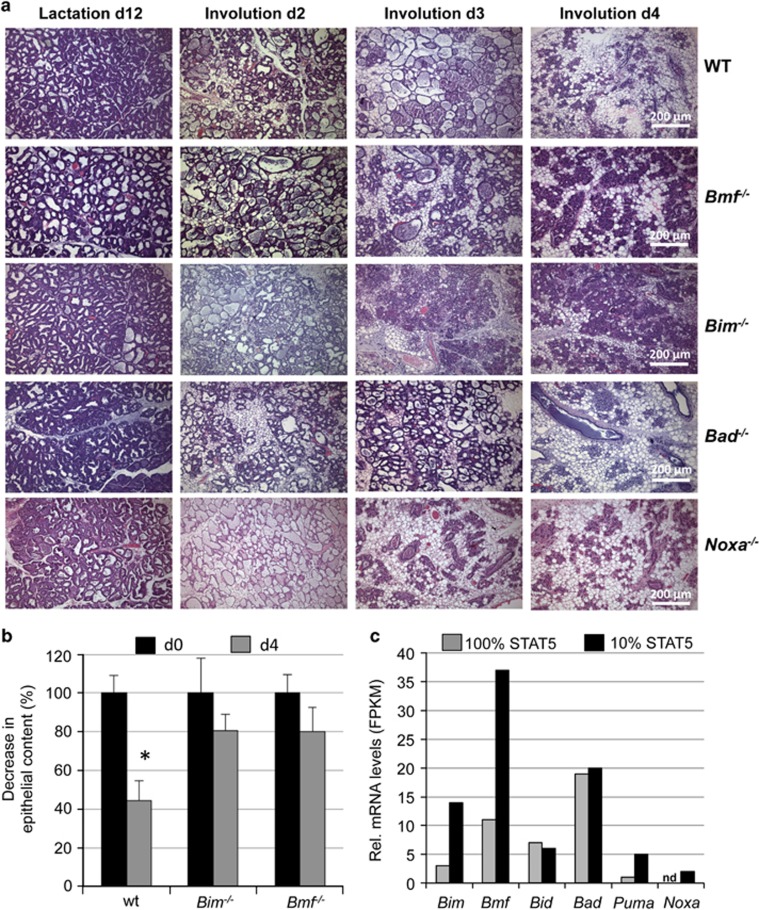
As Bim, Bmf, Bad and Noxa mRNA and/or protein were found increased during involution, the actual contribution of individual BH3-only proteins to tissue remodelling was investigated during the first 4 days after forced weaning. Mammary glands of single-parous mice were isolated for histological analysis at different time points after forced weaning. At early stages (day 2), the absence of none of these proteins appeared to impact on involution. On day 4, wild-type, Bad- or Noxa-deficient mice had cleared most of the epithelial structures, an observation that was in contrast to findings made in glands from Bim−/− and Bmf−/− animals (Figure 3a). These tissues still contained residual alveolar structures and the number of epithelial cells appeared higher in haematoxylin and eosin (H&E) sections by genotype-blinded visual inspection (Figure 3a). This finding was corroborated by automated image analysis on multiple sections from individual mammary glands derived from Bim- or Bmf-deficient mice on day 0 versus day 4, suggesting redundant roles for these proteins in this process (Figure 3b; Supplementary Figure 1).
Figure 3.
Impact of BH3-only protein loss on mammary gland remodelling in involution. (a) Representative images of H&E-stained mammary gland #2, 3 or #4, five sections from mice of the indicated genotypes per time point, after forced weaning. 3–6 individual mice/genotype were analysed for each time point. (b) Quantification of epithelial cell loss during involution using ImageJ-Fiji software, as described in the methods section. Bars represent means ±S.E.M. of glands from 4–6 animals per genotype. *indicate statistically significant differences between days 0 and 4 as assessed by paired Student's t-test (P<0.05). (c) Relative mRNA expression levels are displayed as Fragments Per Kilobase of exon per Million reads (FPKM) of individual BH3-only proteins assessed by RNA-seq analysis comparing epithelial cells from MMTV-Cre+ wt (100% STAT5 activity) or Stat5af/f Stat5b+/− mice (<10% STAT5 activity)40
Bim is induced during involution in mammary epithelial cells
A key feature of the first stage of involution is the sharp drop of STAT5 activity and concomitant reduction of genes that are activated by STAT5, such as those controlling differentiation of mammary epithelium. STAT5 can also suppress expression of genes, such as Bcl6,39 and it is feasible that Bim may fall into this category. In search for evidence that Bim- or other BH3-only protein-encoding genes may be induced selectively in mammary epithelium during involution as a result of decreased STAT5 activity, or in infiltrating phagocytes, we analysed RNA-seq data from mice lacking about 90% of that activity due to the absence of both Stat5a and one Stat5b allele.40 This analysis revealed an approximately threefold increase in Bim and Bmf mRNA levels in Stat5-deficient epithelium but no or minor changes in the relative expression levels of Bid, Bad, Puma or Noxa (Figure 3c). These findings suggested that Bim and Bmf are under negative regulation of STAT5 during lactation, in line with data from western blots (Figure 2). Indeed, analysis of CHIP-seq data40 demonstrated strong direct binding of STAT5A to a GAS motif in exon 4 of the Bim gene (Supplementary Figure 2). No binding was detected at GAS motifs in the promoter region or in introns. Strong STAT5A binding was also detected at a GAS motif in the 3′ exon of the Bmf gene as well as in intronic sequences. Some of these did not coincide with GAS motifs suggesting indirect binding through other transcription factors. Bid expression did not change in the Stat5-deficient mammary tissue and no STAT5A binding was observed in the 3′ exon. No STAT5A binding was detected in other BH3-only protein-encoding gene loci (data not shown).
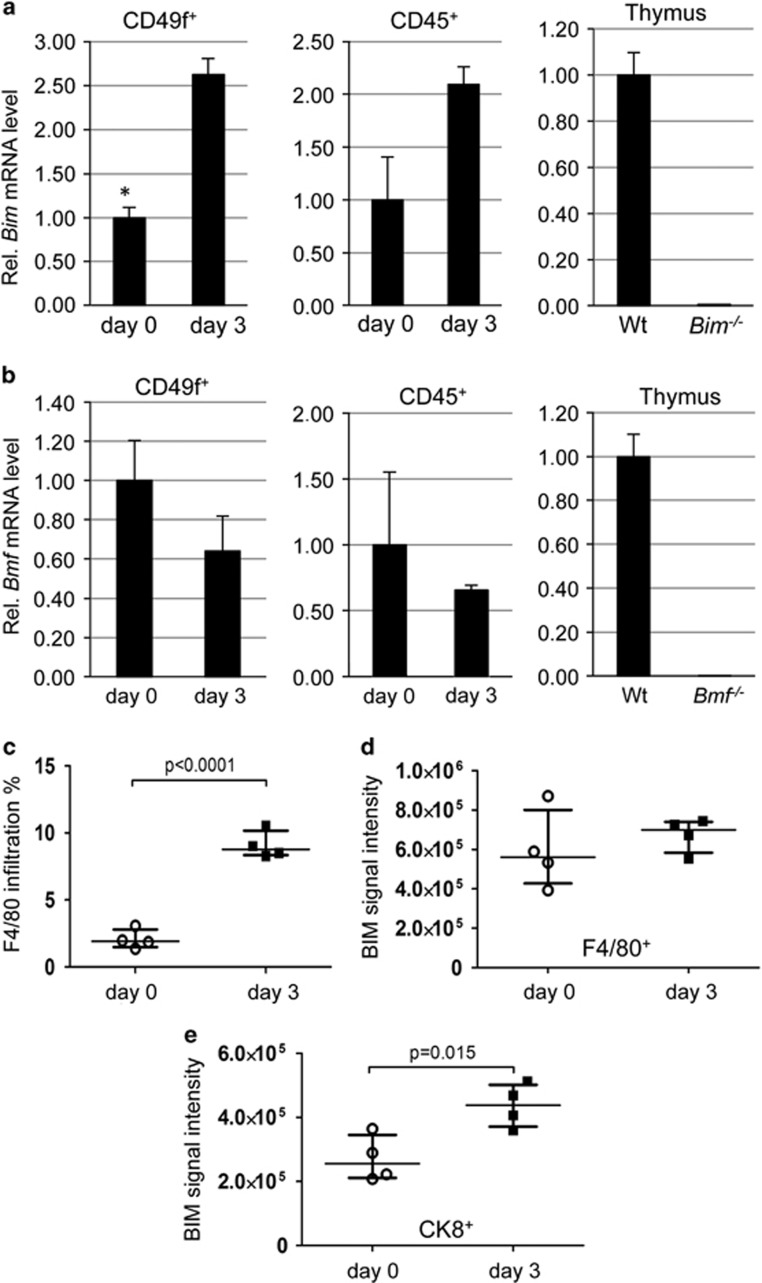
To address the question whether Bim-induction occurs selectively in epithelial cells during involution and/or infiltrating macrophages, needed to clear dying cells, we isolated CD45+ haematopoietic and CD49f+CD45− epithelial cells by FACS sorting from the lactating or involuting mammary tissue (Supplementary Figure 3). qRT-PCR analysis on isolated RNA revealed a significant induction of Bim transcripts in the epithelial fraction while no such effect was detected for Bmf. Neither Bim nor Bmf mRNA was found increased in CD45+ haematopoietic cells, albeit Bim mRNA levels showed a trend (Figures 4a and b). To scrutinize these findings, we also performed immunofluorescence analysis for BIM (please note that available BMF antibodies are not suitable for IF, AV unpublished). This analysis revealed (i) an increase in the number of BIM-expressing infiltrating F4/80+ macrophages on day 3 of involution (Figure 4c) and (ii) low-level expression of BIM in cytokeratin 8 (CK8)+ epithelial cells that increased on day 3, as assessed by image quantitation (Figure 4e). In contrast, the mean fluorescence intensity per macrophage did not increase during involution (Figure 4d, Supplementary Figure 4).
Figure 4.
Increased Bim mRNA and protein in mammary epithelium during involution. qRT-PCR analysis of (a) Bim or (b) Bmf mRNA in sorted CD45+ haematopoietic or CD49f+CD45− epithelial cells isolated on day 0 or day 3 of involution. Bars represent means ±S.D. of two independent experiments performed in duplicates. RNA derived from wt, Bim- or Bmf-deficient thymocytes was used as reference. (c) Quantification of macrophage infiltration during involution (see also Supplementary Figure 4) (d and e) Quantification of relative BIM expression levels as assessed by immunofluorescence analysis shown in Supplementary Figure 4. Bars represent means ±S.D. Four IF-images per gland, derived from two mice per genotype were analysed (see also Supplementary Figure 4 for representative images and image quantification procedure). *indicates statistically significant differences between days 0 and 3 as assessed by paired Student's t-test (P<0.05)
Together these findings demonstrate that Bim expression is induced in mammary epithelial cells during involution and suggest that the Bim gene is a direct negative target of STAT5 during lactation, similar to the well-studied Bcl6 gene. Furthermore, infiltrating macrophages do contribute to the observed increase in BIM levels noted in western blots (Figure 1). Opposite to the effects observed by western with total mammary gland extracts, Bmf mRNA levels in isolated populations of epithelial cells and macrophages did not increase during involution. This may indicate an increase of protein levels by post-translational modifications and/or in a cell type not analysed individually, for example, adipocytes or stromal fibroblasts. Alternatively, Bmf may have been induced only in a transient manner in epithelial cells.
Loss of Bim delays apoptosis during mammary gland involution
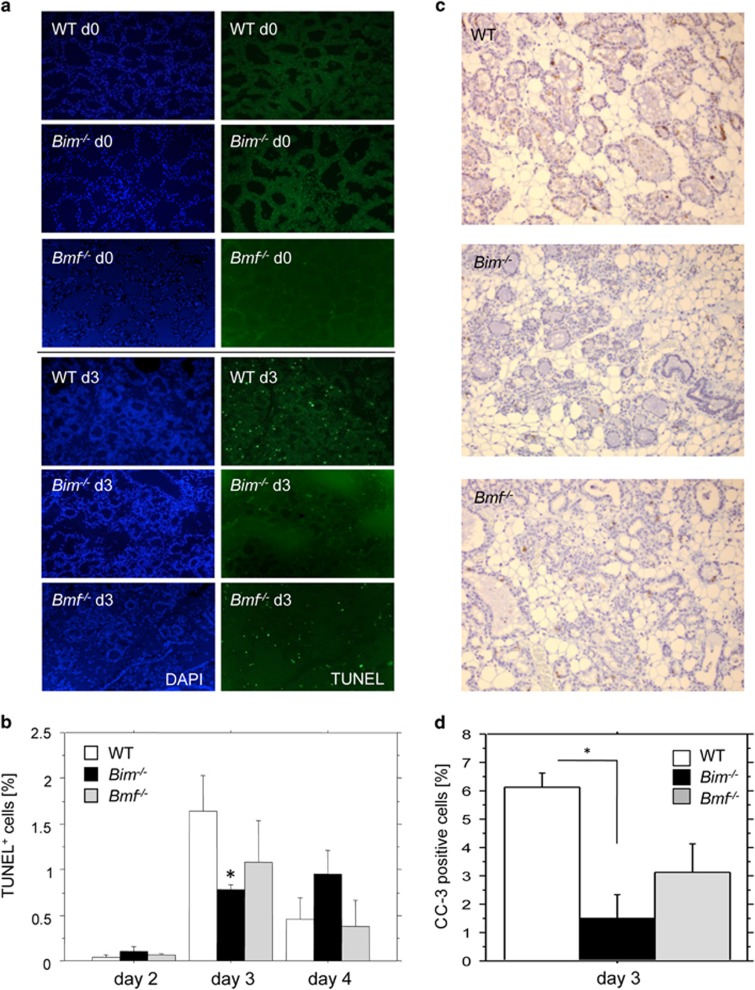
To confirm that the delayed clearance of Bim-deficient glands was indeed due to reduced apoptosis, we performed terminal transferase-mediated dUTP nick end labelling method (TUNEL) staining on paraffin sections from mammary glands isolated during involution to quantify cell death in relation to the total number of epithelial cells (Figure 5a). This analysis revealed that in the absence of BIM, the number of TUNEL-positive cells was significantly decreased on day 3 of involution, when compared with wild-type sections, confirming our findings made in H&E stainings from Bim−/− mice (Figure 3). In addition, although not statistically significant, there was a trend towards a reduction in the number of TUNEL-positive cells on day 3 in Bmf−/− mice (Figure 3). To back up this finding and to assess the possible involvement of BIM and BMF in more detail, we also analysed caspase-3 activation in the wild-type, Bim- and Bmf-deficient mammary glands on involution day 2 and 3. The ratio of caspase-3-positive cells per total cells was quantified and statistically evaluated. This analysis revealed a clear difference in the number of epithelial cells with active caspase-3 on day 3 of involution in the absence of BIM and a trend when BMF was lacking, albeit a statistical significant difference was not observed in the latter (Figure 5d). Taken together, our analyses suggest a rate-limiting role for BIM in the induction of cell death in the second phase of involution, whereas the BH3-only proteins BMF, BAD or NOXA appear either redundant with BIM or dispensable.
Figure 5.
Loss of Bim delays apoptosis during involution. (a) Representative examples of immunofluorescence-based TUNEL analysis performed on mammary gland #4 sections from tissue isolated on day 0 or day 3 of involution from wt, Bmf−/− or Bim−/− mice. (b) Quantification of samples shown in a. Bars represent means ±S.E. of n=3 mice per day and genotype analysed. (c) Representative examples of immunohistochemistry-based analysis of caspase-3 activation performed on mammary gland #4 sections from tissue isolated on day 0 or day 3 of involution from wt, Bim−/− or Bmf−/− mice. (d) Quantification of samples shown in c. Bars represent means ±S.E.M. of n=3 mice per day and genotype analysed. *indicates a statistically significant difference between wt and Bim-deficient mice, as assessed by unpaired Student's t-test (P<0.05)
Changes in the BCL2 rheostat do not depend on STAT3
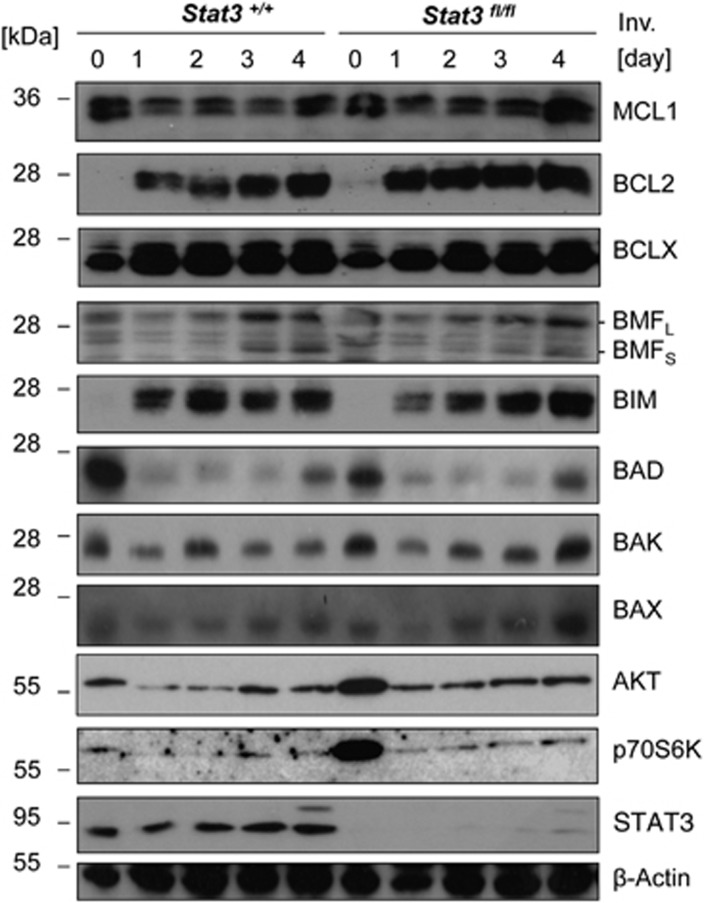
Loss of Stat3 in mammary epithelium strongly delays the initiating events of involution and this has been mechanistically linked to defective repression of SPI2 expression, a potent inhibitor of cathepsins released from lysosomes at early phases of involution.19 To address if STAT3 is also critical for the induction of BH3-only proteins, in particular BIM, to facilitate the second wave of involution, we analysed BCL2 family protein expression in mammary glands derived from mice conditionally deleted for STAT3 in the secretory epithelium (BLG-Cre;Stat3f/f) during lactation. This analysis failed to reveal any substantial differences in the expression levels of BCL2 proteins caused by the absence of STAT3, in line with the idea that the increase of BH3-only proteins is a delayed event in the involution process, uncoupled form the initiating STAT3-dependent signalling cascade (Figure 6) and regulated by an independent signalling circuitry. The drop in AKT level and activity noted during involution appeared largely unaffected by STAT3 and hence may constitute or contribute to such a signal.
Figure 6.
BCL2 protein family regulation during involution does not depend on STAT3. Mammary gland tissue was isolated at the indicated times after forced weaning on day 10 from Stat3f/f and Stat3f/f BLG-Cre mice. Protein lysates (40 μg/lane) were size fractionated by SDS-PAGE and transferred to nitrocellulose membranes which were sequentially probed, or when necessary, stripped and reprobed with the listed primary antibodies. Reprobing with anti-tubulin served to control for comparable protein loading
Discussion
This study reports a rate-liming role for the BH3-only protein BIM during the irreversible phase of involution in the mouse mammary gland. Previous studies have shown a role for BIM in the clearing of developing ducts and end buds of the mammary gland32, 33 but none of them have addressed the role of BIM or other BH3-only proteins by genetic means in involution. Early studies using transgenic mice overexpressing BCL2 under control of the WAP-gene promoter suggested a role for BCL2 proteins in this process as these animals showed reduced cell death during involution.23 Similarly, conditional ablation of BclX caused enhanced loss of epithelia in the mammary gland during involution20 and temporary downregulation of pro-survival BCL2 and BCLW, paralleled by an induction of BAK and BAD, were also considered to contribute to this process.24 Together, these findings consolidated the idea that involution is regulated by the intrinsic apoptosis pathway and the BCL2 family.24
However, this concept was severely challenged by more recent findings demonstrating that cathepsins, released from lysosomes early in involution, are critical for the remodelling of the mammary gland into a virgin-like structure, whereas effector caspases, whose activation is usually controlled by BCL2 family proteins, were largely dispensable.19 Interestingly, although Caspase-3 and -6 are clearly active during early involution, mice lacking Casp-3 and -6 showed normal involution phenotypes, suggesting that processing of effectors observed early on is secondary to lysosomal membrane permeabilization (LMP). In contrast, Caspase-7 was only found activated during the second wave of involution from day 3 on,19 opening the possibility that BCL2 proteins may contribute to this second irreversible stage. Consistent with this hypothesis, we observed the induction of multiple BCL2 family proteins at the mRNA and/or protein level during involution. Most strikingly, the induction of BH3-only proteins such as BIM and BMF was paralleled by a drop in MCL1 levels and the re-expression of BCL2, indicating that identical developmental cues co-regulate pro- and anti-apoptotic BCL2 family proteins during this process to ensure tissue homeostasis at the end of involution. A critical regulator here seems to be STAT5 that appears to repress BH3-only proteins during lactation, possibly by direct interaction with the respective gene loci, as suggested by RNA-seq and ChIP-seq analysis. Most interestingly, loss of STAT5 signalling also promotes the death of pro-B cells that rely on MCL1 for survival and it is tempting to speculate that this is mediated by BIM induction, known to limit the lifespan of developing B cells.41
Although other BH3-only proteins were also upregulated, either at the protein or mRNA level during involution, that is, Bad and Noxa, respectively, we noted that neither of them was essential for cell death after weaning or significantly increased upon near complete Stat5-deletion in epithelial cells. A proof for a rate-limiting role in this process was only found for Bim with signs for auxiliary roles for Bmf, because in their absence clearance of epithelium during involution was reduced, correlating with a reduced number of TUNEL positivity and lower numbers of cells staining positive for active caspase-3. Cross-compensation in Bim- and Bmf-mutant mice by increased expression of the remaining BH3-only protein may contribute to the rather mild phenotypes observed, but this remains mere speculation at the moment. In contrast, lack of Bad or Noxa did not cause a delay in the removal of epithelial structures (Figure 3). Of note, as the increase of Noxa mRNA was accompanied by the downregulation of MCL1 protein, this suggests that the levels of this pro-survival protein may need to be reduced by NOXA-mediated proteasomal degradation42 for involution to occur. However, as loss of Noxa did not delay involution, we assume that it may act in concert with other BH3-only proteins to promote epithelial cell death, most likely BIM. On the basis of our findings, we assume further that there is potential redundancy between BIM and BMF during involution, as noted also in lymphocyte death.43 However, this could not be directly addressed, as mice lacking both genes show vaginal atresia, rendering them infertile.44, 45 It would, however, be of interest to explore possible redundancies between BIM and NOXA in this process in future studies, as these two proteins show functional overlap, for example, in the control of NK-cell or granulocyte survival46, 47 as well as epithelial cells stalled in mitosis.48
In summary, upregulation of the pro-apoptotic Bim, Bad, Bmf and Noxa mRNA and/or protein at a time where levels of BCL2 and MCL1 transiently declined suggests that this assists successful and timely involution. Notably, none of these changes in protein levels was controlled by STAT3 signalling. The minor differences between our analyses performed in wild-type mice and those carrying a floxed Stat3 allele, for example, on the induction of BAD or the kinetics and amplitude of BMF, may be explained by differences in genetic background (C57BL6 versus CBAxC57BL6) and/or the time point of forced weaning (day 12 versus day 10).
The signals involved in activating the BCL2 controlled cell death pathway remain to be defined in detail. One possible cue, next to cessation of hormone signalling, may be the release of TGFβ from macrophages engulfing dying epithelial cells.49 This can contribute to the suppression of inflammation, on one hand, but may induce transcription of pro-death genes in the epithelium priming them to cell death. Consistent with this hypothesis, both, Bim and Bmf are established TGFβ targets in breast cancer cell lines50 and T cells.51 As macrophages that engulf apoptotic cells produce TGFβ to suppress immune responses to cells dying by apoptosis, it seems plausible that this may contribute to increases in BIM expression in epithelial cells driving a feed-forward loop. Another cue may be increased expression of IGFBP-5, a STAT3 target8 and a negative regulator of IGF-I signalling that may contribute to a reduction of AKT-mediated survival stimuli. STAT3-dependent expression of PI3K p55α and p50α regulatory subunits at the onset of involution may affect survival signalling and, consistently, p55α/p50α double-mutant mice show a delay in involution.18, 52 Reduced levels of AKT, however, may also allow for increased BIM expression, as documented in lymphocytes after cytokine deprivation, and possibly also BMF, shown to increase upon PI3K inhibition in MEF.53 Of note, STAT5 positively controls AKT1 expression in response to prolactin and overexpression of STAT5 delays involution correlating with sustained AKT activity.54
However, as our analysis was based on the use of total-body knockout mutants lacking the relevant genes in all cells, it remains to be determined in detail to what extent the effect caused by loss of Bim is due to effects on mammary epithelium versus infiltrating macrophages. Hence, future analysis of compound mutant mice and mice with conditional alleles will be required to fully elucidate the contribution of the individual BH3-only proteins in mammary gland morphogenesis at the cellular level.
Materials and Methods
Mice
All mice used in this study were maintained on C57BL/6 genetic background. The generation and genotyping of Noxa-, Bad-,Bim- and Bmf-deficient mice has been previously described.30, 55, 56, 57 Deletion of Stat3 in the mammary gland was achieved using BLG-Cre delete mice that were on a mixed genetic background (CBAxC57BL/6).19
Involution
Mammary tissue from virgin, pregnant or lactating animals was isolated and processed after killing, as previously described. Tissue was then ground to powder under liquid nitrogen as described.58, 59 Involution was induced in single-parous mice by forced weaning at day 12 of lactation, representing day 0 in our analysis. Gland #4, including the inguinal lymph node, was taken for histological analysis. The second gland #4, without the lymph node, and glands #2 and #3 were isolated for protein or RNA isolation (see below). The tissue was snap frozen in liquid nitrogen, or placed in tubes with 4% PFA (see below) for further processing.
Preparation of histological sections and image quantification
Glands were fixed in 4% PFA/PBS after several washes in excess PBS to remove residual milk. Tissues were processed according to standard procedures, cut in 3-μm sections and stained in H&E or were processed for TUNEL staining by immunofluorescence or active caspase-3 detection by immunohistochemistry (see below).
Epithelial area of wt, Bim−/− and Bmf−/− H&E-stained slides of involuting mammary glands (day 0 versus day 4) was quantified using the following custom-made macro and ImageJ-Fiji: run (‘8-bit'); make Rectangle (837, 615, 1011, 756); run (‘Crop'); set AutoThreshold (‘Default'); //run (‘Threshold...'); set Threshold (0, 129); run (‘Measure').
Cell sorting
Wild-type females were killed on day 12 of lactation (day 0) or on day 3 after forced weaning. Mammary glands #2–5 were minced and incubated for 2 h at 37 °C, 5% CO2 in DIG1 mix (5 ml RPMI1640 (PAA), 24 μl liberase (Roche, Mannheim, Germany) 100 μl hyaluronidase 10 mg/ml (Sigma-Aldrich, Munich, Germany)), washed in excess PBS/10%FCS twice and incubated in red blood cell lysis buffer, 7 min on ice and 3 min on RT, washed again twice followed by digestion for 2 min in trypsin, washed twice and 2 min in DNase1 (20 ng/ml) containing buffer (Krebs-Henseleit Buffer 98 ml, Hepes (stock solution 1 M) 1.3 ml, d-Glucose (stock solution 1M) 500 μl, Gentamycin (stock solution 50 mg/ml) 500 μl, Penicillin/Streptomycin (10000 U/ml) 100x5 ml, pH=7.4 (sterile filter) at 37 °C. Cells were then stained with anti-CD45-PE (Biolegend, London, UK: clone 30-F11) and anti-CD49f-FITC (Biolegend: clone GoH3) for 30' at 4 °C; DAPI was used for dead cell exclusion.
Immunofluorescence microscopy
Mammary gland tissue on days 0 and 3 involution were embedded in OCT Compound (Sakura, Alphen aan den Rijn, Netherlands) and snap frozen in liquid nitrogen. Tissue samples were cut in 10-μm thick slices and mounted onto silanized microscopy slides. Tissue samples were stored at −80 °C till further processing. For the immunoflourescence staining, slides were air dried at room temperature, fixed in acetone at −20 °C, blocked with 10% horse serum in PBS and incubated for 1 h at room temperature with antibodies diluted in the blocking solution. The following antibodies were utilized: anti-F4/80 (clone BM8, conjugated with Alexa488, BioLegend), anti-CD11b (M1/70, Alexa488, BioLegend), anti-cytokeratin 8 (E432, MERCK, Darmstadt, Germany) and rat anti-BIM (3C5, ENZO), anti-rat IgG (polyclonal, Cy3, Life Technologies, Vienna, Austria), anti-mouse IgG (polyclonal, Alexa488, Life Technologies). DNA was counterstained with DAPI dissolved in mounting medium (Dako, Glostrup, Denmark). Photos were taken with the Axio Imager fluorescence microscope with a × 70 lenses (Zeiss) as 16-bit tiff images. Images were further processed with the ImageJ software. In brief, the background was subtracted (rolling ball algorithm), the contrast adjusted and images overlaid. For the measurement of CD11b, BIM and F4/80 expression, images were binarized (IsoData algorithm) and the fraction of positive area per field was calculated with the Measure function. For the determination of BIM expression in F4/80-positive and CK8-positive cells, F4/80 and CK8 images were binarized as above, converted to masks (positive pixels =0) and subtracted from BIM images. BIM signal density was measured in the resulting images representing BIM positivity in F4/80+ and CK8+ regions. BIM expression in the particular cell type was presented as mean and total BIM signal intensity divided by F4/80+ and CK8+ area respectively. The scheme of image calculations is depicted in Supplementary Figures 5a and b.
Caspase-3 and TUNEL staining
Mammary gland tissue embedded in paraffin was cut at 3 μm, deparaffinized and rehydrated. Antigens were retrieved in 10 mM citrate buffer, pH 6.0, at 98 °C for 20 min. The slides were blocked with 5% horse serum for 1 h at room temperature. Primary anti-cleaved caspase-3 antibody (clone 5A1, Cell Signaling, Leiden, Netherlands) was used at a 1:400 dilution. Immunoreactivity was detected using the Dako REAL EnVision Detection System (Dako). Five random regions were imaged per section in a blinded manner and the nuclei of six random alveoles were counted, equalling 30 alveoles per mouse. Tissues were counterstained with haematoxylin. The number of caspase-3-positive cells within these alveoles was assessed in a blinded manner and the percentage per section was calculated. For visualization of apoptotic cells via the in situ TUNEL (Roche), rehydrated slides of tissue sections on cover slips were treated with 0.1% Triton-X-100/0.1% sodium citrate buffer and dehydrated by graded series of alcohol and chloroform. The TUNEL reaction was carried out in a humidified chamber for 1 h at 37 °C using recombinant terminal transferase and FITC-dUTP (Roche), using the protocol provided by Roche. Counterstaining of nuclei was done with DAPI (Sigma-Aldrich, St. Louis, MO, USA). For evaluation, three randomly taken images were analysed from each tissue sample in a blinded manner. The numbers of nuclei staining with DAPI ± FITC-dUTP+ were counted to calculate the percentage of TUNEL+ cells per tissue sample.
Real-time quantitative PCR
RNA was extracted from tissue powder using TRIzol (Invitrogen, Paisley, UK) according to the protocol provided by the manufacturer. cDNA was prepared using Omniscript RT-kit (Qiagen, Hilden, Germany). Primers used for the RT-PCR: Bad 5′-GGA CTT ATC AGC CGA AGC AG-3′ and 5′-GCT CAA ACT CTG GGA TCT GG-3′ Bcl-2 5′-CTGGCATCTTCTCCTTCCAG-3′ and 5′-GACGGTAGCGACGAGAGAAG-3′ BclX 5′-TTCGGGATGGAGTAAACTGG-3′ and 5′-TGGATCCAAGGCTCTAGGTG-3′ Bid 5′-CTG CCT GTG CAA GCT TAC TG-3′ and 5′-GTC TGG CAA TGT TGT GGA TG-3′ Bim 5′-GAGATACGGATTGCACAGGA-3′ and 5′-TCAGCCTCGCGGTAATCATT-3′ Bmf 5′-CCCATAAGCCAGGAAGACAA-3′ and 5′-AGGGAGAGGAAGCCTGTAGC-3′ Mcl1 5′-TAA CAA ACT GGG GCA GGA TT-3′ and 5′-GTC CCG TTT CGT CCT TAC AA-3′ Noxa 5′-CCC ACT CCT GGG AAA GTA CA-3′ and 5′-AAT CCC TTC AGC CCT TGA TT-3′ Puma 5′-CAA GAA GAG CAG CAT CGA CA-3′ and 5′-TAG TTG GGC TCC ATT TCT GG-3′ and actin 5′-ACTGGGACGACATGGAGAAG-3′ and 5′-GGGGTGTTGAAGGTCTCAAA-3′ was used as housekeeping gene. Real-time quantitative PCR was run on a realplex2, Mastercycler, ep gradient S from Eppendorf using SYBR Green (Biozym Diagnostics, Vienna, Austria). The results were normalized to β-Actin expression and evaluated using the –ΔΔCt relative quantification method.
Cell lysis and immunoblotting
Tissue powder was lysed in ONYX buffer (20 mM TrisHCl pH 7.5, 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 1% Triton-X-100, 10% glycerol, protease inhibitor (Roche Complete Protease Inhibitor Cocktail Tablets) for 30 min at 4 °C on a rocking platform. Lysates were cleared by centrifugation for 5 min at 13 000 r.p.m. at 4 °C in a microfuge. Proteins were separated by SDS-PAGE using 14% Tris-Glycine polyacrylamide gels. After semidry transfer onto nitrocellulose (Hybond N, GE Healthcare, Vienna, Austria), the membranes were probed sequentially with the following primary antibodies purchased from Rockland, BD, Cell Signalling (Leiden, Netherlands), ENZO Life Science or Sigma-Aldrich: BIM 3C5 (rat anti-human/mouse/rat/monkey/dog), BMF 17A9 (rat anti-mouse/rat), BAD CS9292 (rabbit anti-mouse), BID 2D1 (rat anti-mouse/human), BAK ALX-210-002 (rabbit anti-human/mouse/rat), BAX CS2772 (rabbit anti-human/mouse/rat/monkey), BCLX (54H6) CS2764 (rabbit anti-human/rat/mouse/monkey), BCLW 13F9 (rat anti-human/mouse/rat/monkey/dog), BCL2 3F11 (hamster anti-mouse), MCL1 600-401-394S (rabbit anti-mouse), cleaved Caspase-3 5A1E CS9664 (rabbit anti-human/mouse/rat/monkey/dog), STAT3 79D7 CS4904 (rabbit anti-human/mouse/rat/monkey), pan-AKT CS4491 (rabbit anti-human/mouse/rat), pS70S6 kinase (T389) CS9234 (rabbit anti-human/mouse/rat), α-Tubulin DM1A (mouse anti-mouse/rat/human) from Sigma-Aldrich. HRP (horseradish peroxidase)-labelled secondary antibodies were used in combination with ECL detection reagents (GE Healthcare).
ChIP-seq and RNA-seq analyses
STAT5A (GSM1005189) and H3K4me3 (GSM1005192) ChIP-seq data from mammary tissue were downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/) and mapped to the mouse reference genome mm9 using bowtie aligner. For visualization, HOMER and IGV were used. GAS motifs were identified by IGV. Genome-wide mRNA levels from wild-type mammary tissue and tissue carrying only one copy of Stat5b had been determined by RNA-seq.40
Statistics
Results were expressed as mean ±S.E. of three or more separate experiments, unless indicated otherwise. Paired or unpaired Student's t-test was used for statistical analysis. A P-value <0.05 was considered statistically significant.
Acknowledgments
We thank A Strasser, L Huber and D Huang for mice and/or reagents; I Gaggl for mouse genotyping, C Soratroi and S Faserl for technical assistance, K Rossi and B Rieder for animal care. This work was supported by grants from the ‘Tiroler Krebshilfe' to FB, as well as the graduate school Molecular Cell Biology and Oncology (MCBO) funded by the Austrian Science Fund (FWF; W1101) and the Integrated Center for Research and Therapy (IFTZ) of the Medical University Innsbruck. FS is recipient of a DOC fellowship of the Austrian Academy of Science (ÖAW). SO and LH were funded by the intramural research program (IRP) of the NIDDK/NIH. SO is a member of the NIH/Dankook Graduate Partnership Program.
Glossary
- Bad
Bcl-2 antagonist of cell death
- BclX
Bcl-2-related protein X
- Bcl-2
B-cell lymphoma 2
- BH
Bcl-2 homology domain
- Bid
Bcl-2 interacting domain death agonist
- Bim
Bcl-2 interacting mediator of cell death
- PUMA
p53-upregulated modulator of apoptosis
- AKT
v-AKT murine thymoma viral oncogene homologue 1
- Stat
signal transducer and activator of transcription
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by P Bouillet
Supplementary Material
References
- Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 2013; 15: 49–63. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell 2010; 37: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol 2005; 6: 715–725. [DOI] [PubMed] [Google Scholar]
- Watson CJ, Kreuzaler PA. Remodeling mechanisms of the mammary gland during involution. Int J Dev Biol 2011; 55: 757–762. [DOI] [PubMed] [Google Scholar]
- Li M, Liu X, Robinson G, Bar-Peled U, Wagner KU, Young WS et al. Mammary-derived signals activate programmed cell death during the first stage of mammary gland involution. Proc Natl Acad Sci USA 1997; 94: 3425–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K et al. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of stat3. Genes Dev 1999; 13: 2604–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Marti A, Jehn B, Altermatt HJ, Chicaiza G, Jaggi R. Glucocorticoid and progesterone inhibit involution and programmed cell death in the mouse mammary gland. J Cell Biol 1995; 131: 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AV, Pollard JW. Transforming growth factor beta3 induces cell death during the first stage of mammary gland involution. Development 2000; 127: 3107–3118. [DOI] [PubMed] [Google Scholar]
- Kritikou EA, Sharkey A, Abell K, Came PJ, Anderson E, Clarkson RW et al. A dual, non-redundant, role for LIF as a regulator of development and STAT3-mediated cell death in mammary gland. Development 2003; 130: 3459–3468. [DOI] [PubMed] [Google Scholar]
- Sutherland KD, Vaillant F, Alexander WS, Wintermantel TM, Forrest NC, Holroyd SL et al. c-myc as a mediator of accelerated apoptosis and involution in mammary glands lacking Socs3. EMBO J 2006; 25: 5805–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint DJ, Boutinaud M, Whitelaw CB, Allan GJ, Kolb AF. Prolactin inhibits cell loss and decreases matrix metalloproteinase expression in the involuting mouse mammary gland but fails to prevent cell loss in the mammary glands of mice expressing IGFBP-5 as a mammary transgene. J Mol Endocrinol 2006; 36: 435–448. [DOI] [PubMed] [Google Scholar]
- Tonner E, Barber MC, Travers MT, Logan A, Flint DJ. Hormonal control of insulin-like growth factor-binding protein-5 production in the involuting mammary gland of the rat. Endocrinology 1997; 138: 5101–5107. [DOI] [PubMed] [Google Scholar]
- Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev 2002; 23: 824–854. [DOI] [PubMed] [Google Scholar]
- Schwertfeger KL, Richert MM, Anderson SM. Mammary gland involution is delayed by activated Akt in transgenic mice. Mol Endocrinol 2001; 15: 867–881. [DOI] [PubMed] [Google Scholar]
- Chapman RS, Lourenco P, Tonner E, Flint D, Selbert S, Takeda K et al. The role of Stat3 in apoptosis and mammary gland involution. Conditional deletion of Stat3. Adv Exp Med Biol 2000; 480: 129–138. [DOI] [PubMed] [Google Scholar]
- Humphreys RC, Bierie B, Zhao L, Raz R, Levy D, Hennighausen L. Deletion of Stat3 blocks mammary gland involution and extends functional competence of the secretory epithelium in the absence of lactogenic stimuli. Endocrinology 2002; 143: 3641–3650. [DOI] [PubMed] [Google Scholar]
- Robinson GW, Pacher-Zavisin M, Zhu BM, Yoshimura A, Hennighausen L. Socs 3 modulates the activity of the transcription factor Stat3 in mammary tissue and controls alveolar homeostasis. Dev Dyn 2007; 236: 654–661. [DOI] [PubMed] [Google Scholar]
- Abell K, Bilancio A, Clarkson RW, Tiffen PG, Altaparmakov AI, Burdon TG et al. Stat3-induced apoptosis requires a molecular switch in PI(3)K subunit composition. Nat Cell Biol 2005; 7: 392–398. [DOI] [PubMed] [Google Scholar]
- Kreuzaler PA, Staniszewska AD, Li W, Omidvar N, Kedjouar B, Turkson J et al. Stat3 controls lysosomal-mediated cell death in vivo. Nat Cell Biol 2011; 13: 303–309. [DOI] [PubMed] [Google Scholar]
- Walton KD, Wagner KU, Rucker EB3rd, Shillingford JM, Miyoshi K, Hennighausen L. Conditional deletion of the bcl-x gene from mouse mammary epithelium results in accelerated apoptosis during involution but does not compromise cell function during lactation. Mech Dev 2001; 109: 281–293. [DOI] [PubMed] [Google Scholar]
- Schorr K, Li M, Bar-Peled U, Lewis A, Heredia A, Lewis B et al. Gain of Bcl-2 is more potent than bax loss in regulating mammary epithelial cell survival in vivo. Cancer Res 1999; 59: 2541–2545. [PubMed] [Google Scholar]
- Watson CJ, Kreuzaler PA. The role of cathepsins in involution and breast cancer. J Mammary Gland Biol Neoplasia 2009; 14: 171–179. [DOI] [PubMed] [Google Scholar]
- Jager R, Herzer U, Schenkel J, Weiher H. Overexpression of Bcl-2 inhibits alveolar cell apoptosis during involution and accelerates c-myc-induced tumorigenesis of the mammary gland in transgenic mice. Oncogene 1997; 15: 1787–1795. [DOI] [PubMed] [Google Scholar]
- Metcalfe AD, Gilmore A, Klinowska T, Oliver J, Valentijn AJ, Brown R et al. Developmental regulation of Bcl-2 family protein expression in the involuting mammary gland. J Cell Sci 1999; 112(Pt 11): 1771–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff MA, Buckley DJ, Krumenacker JS, Reed JC, Miyashita T, Buckley AR. Rapid modulation of the apoptosis regulatory genes, bcl-2 and bax by prolactin in rat Nb2 lymphoma cells. Endocrinology 1996; 137: 5456–5462. [DOI] [PubMed] [Google Scholar]
- Strange R, Metcalfe T, Thackray L, Dang M. Apoptosis in normal and neoplastic mammary gland development. Microsc Res Tech 2001; 52: 171–181. [DOI] [PubMed] [Google Scholar]
- Danial NN. BAD: undertaker by night, candyman by day. Oncogene 2008; 27(Suppl 1): S53–S70. [DOI] [PubMed] [Google Scholar]
- You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J Exp Med 2006; 203: 1657–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon JD, Labi V, Egle A, Villunger A. Bim and Bmf in tissue homeostasis and malignant disease. Oncogene 2008; 27(Suppl 1): S41–S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036–1038. [DOI] [PubMed] [Google Scholar]
- Matthews JR, Clarke AR. p53 mediates a default programme of mammary gland involution in the absence of STAT3. Oncogene 2005; 24: 3083–3090. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Mailleux AA, Overholtzer M, Carroll JS, Solimini NL, Lightcap ES et al. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci USA 2007; 104: 3787–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailleux AA, Overholtzer M, Schmelzle T, Bouillet P, Strasser A, Brugge JS. BIM regulates apoptosis during mammary ductal morphogenesis, and its absence reveals alternative cell death mechanisms. Dev Cell 2007; 12: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J et al. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol 2003; 5: 733–740. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Huang DCS, O'Reilly LA, King SM, Strasser A. The pro-apoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 1999; 3: 287–296. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Villunger A, O'Reilly LA, Beaumont JG, Coultas L, Cheney RE et al. Bmf: a pro-apoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science 2001; 293: 1829–1832. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Strasser A. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ 2002; 9: 505–512. [DOI] [PubMed] [Google Scholar]
- Repnik U, Turk B. Lysosomal-mitochondrial cross-talk during cell death. Mitochondrion 2010; 10: 662–669. [DOI] [PubMed] [Google Scholar]
- Tran TH, Utama FE, Lin J, Yang N, Sjolund AB, Ryder A et al. Prolactin inhibits BCL6 expression in breast cancer through a Stat5a-dependent mechanism. Cancer Res 2010; 70: 1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji D, Kang K, Robinson GW, Hennighausen L. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res 2013; 41: 1622–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol 2010; 11: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, Huang DC et al. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc Natl Acad Sci USA 2007; 104: 6217–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woess C, Tuzlak S, Labi V, Drach M, Bertele D, Schneider P et al. Combined loss of the BH3-only proteins Bim and Bmf restores B-cell development and function in TACI-Ig transgenic mice. Cell Death Differ 2015. [DOI] [PMC free article] [PubMed]
- Hubner A, Barrett T, Flavell RA, Davis RJ. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol Cell 2008; 30: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labi V, Woess C, Tuzlak S, Erlacher M, Bouillet P, Strasser A et al. Deregulated cell death and lymphocyte homeostasis cause premature lethality in mice lacking the BH3-only proteins Bim and Bmf. Blood 2014; 123: 2652–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington ND, Puthalakath H, Gunn P, Naik E, Michalak EM, Smyth MJ et al. Interleukin 15-mediated survival of natural killer cells is determined by interactions among Bim, Noxa and Mcl-1. Nat Immunol 2007; 8: 856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschnek S, Vier J, Gautam S, Frankenberg T, Rangelova S, Eitz-Ferrer P et al. Molecular analysis of neutrophil spontaneous apoptosis reveals a strong role for the pro-apoptotic BH3-only protein Noxa. Cell Death Differ 2011; 18: 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschka MD, Soratroi C, Kirschnek S, Häcker G, Hilbe R, Geley S et al. The NOXA–MCL1–BIM axis defines lifespan on extended mitotic arrest. Nat Commun 2015; 6: 6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanders KC, Wakefield LM. Transforming growth factor-(beta)s and mammary gland involution; functional roles and implications for cancer progression. J Mammary Gland Biol Neoplasia. 2009; 14: 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramjaun AR, Tomlinson S, Eddaoudi A. Downward J: Upregulation of two BH3-only proteins, Bmf and Bim, during TGF beta-induced apoptosis. Oncogene 2007; 26: 970–981. [DOI] [PubMed] [Google Scholar]
- Tischner D, Wiegers GJ, Fiegl H, Drach M, Villunger A. Mutual antagonism of TGF-beta and Interleukin-2 in cell survival and lineage commitment of induced regulatory T cells. Cell Death Differ 2012; 19: 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensa S, Neoh K, Resemann HK, Kreuzaler PA, Abell K, Clarke NJ et al. The PI3K regulatory subunits p55alpha and p50alpha regulate cell death in vivo. Cell Death Differ 2014; 21: 1442–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grespi F, Soratroi C, Krumschnabel G, Sohm B, Ploner C, Geley S et al. BH3-only protein Bmf mediates apoptosis upon inhibition of CAP-dependent protein synthesis. Cell Death Differ 2010; 17: 1672–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer BA, Sakamoto K, Schmidt JW, Triplett AA, Moriggl R, Wagner KU. Stat5 promotes survival of mammary epithelial cells through transcriptional activation of a distinct promoter in Akt1. Mol Cell Biol 2010; 30: 2957–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P, Metcalf D, Huang DCS, Tarlinton DM, Kay TWH, Köntgen F et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 1999; 286: 1735–1738. [DOI] [PubMed] [Google Scholar]
- Labi V, Erlacher M, Kiessling S, Manzl C, Frenzel A, O'Reilly L et al. Loss of the BH3-only protein Bmf impairs B cell homeostasis and accelerates gamma irradiation-induced thymic lymphoma development. J Exp Med 2008; 205: 641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger AM, Zha J, Harada H, Datta SR, Danial NN, Gilmore AP et al. Bad-deficient mice develop diffuse large B cell lymphoma. Proc Natl Acad Sci USA 2003; 100: 9324–9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannesdottir L, Tymoszuk P, Parajuli N, Wasmer MH, Philipp S, Daschil N et al. Lapatinib and doxorubicin enhance the Stat1-dependent antitumor immune response. Eur J Immunol 2013; 43: 2718–2729. [DOI] [PubMed] [Google Scholar]
- Tonko-Geymayer S, Goupille O, Tonko M, Soratroi C, Yoshimura A, Streuli C et al. Regulation and function of the cytokine-inducible SH-2 domain proteins, CIS and SOCS3, in mammary epithelial cells. Mol Endocrinol 2002; 16: 1680–1695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.