Abstract
Over the last 15 years, protein acetylation has emerged as a globally important post-translational modification that fine-tunes major cellular processes in many life forms. This dynamic regulatory system is critical both for complex eukaryotic cells and for the viruses that infect them. HIV-1 accesses the host acetylation network by interacting with several key enzymes, thereby promoting infection at multiple steps during the viral life cycle. Inhibitors of host histone deacetylases and bromodomain-containing proteins are now being pursued as therapeutic strategies to enhance current antiretroviral treatment. As more acetylation-targeting compounds are reaching clinical trials, it is timely to review the role of reversible protein acetylation in HIV-infected CD4+ T cells.
Keywords: Post-translational modification, epigenetic regulation, virus–host Interactions, viral infection, therapeutic inhibitors, latency
Introduction
The survival and function of cells are critically dependent on their ability to rapidly integrate multiple, intersecting cell-signaling circuits. A key strategy for effectively regulating complex signals is the reversible post-translational modification (PTM) of proteins. Over 200 PTMs are known, many of which are highly conserved among a wide range of organisms (Jensen, 2006). Despite their ubiquitous presence, only a few PTMs have been comprehensively studied including acetylation of lysines. The global role of protein acetylation was initially underappreciated (Verdin & Ott, 2014). Originally found to reversibly modify lysines in the tails of histones, acetylation was thought to regulate gene expression primarily by altering the structural properties of the chromatin environment (Box 1). However, with the identification of acetylation-modifying enzymes and improvements in high-resolution mass spectrometry, it became clear that the regulation of cellular function by protein acetylation extends beyond the nucleus. Over 3600 novel acetyl-lysine sites have been identified in a broad range of human proteins in different subcellular compartments (Choudhary et al., 2009). Acetylation of these proteins has been linked to the regulation of diverse cellular pathways, including cell-cycle control, DNA damage response, cytoskeletal organization, and immune signaling (Spange et al., 2009; Shakespear et al., 2011).
Box 1.
Translating electrostatic and physical properties of acetylation into the regulation of cellular function. The reversible addition of an acetyl group to the ε-NH2 group of lysines modifies the electrostatic and physical properties of the target protein. The positive charge of the lysine is neutralized by the acetyl group, thereby altering hydrogen bonding capabilities through the formation of a circumferential hydrophobic milieu. At the level of the chromatin, this means that increased acetylation of histone tails will decrease the affinity of the histone for the negatively charged DNA strand. As a result, the chromatin environment becomes less compact, nucleosomes are remodeled, and gene transcription increases as a result of the greater accessibility of transcription factors. Furthermore, transcription factors themselves are often subject to cycles of acetylation and deacetylation. The acetylation of transcription factors modifies their DNA-binding capacity, protein stability, and nuclear translocation, thereby regulating their ability to activate target genes. Lysine acetylation can also alter cellular signaling pathways by modifying the dynamics of protein–protein interactions. Depending on the context, the addition of the acetyl group may create steric hindrance that prevents key interactions. At the same time, acetylated lysines can serve as docking sites for proteins that contain acetyl-lysine recognition domains (e.g. bromodomains), thereby facilitating transient interactions between proteins. Functionally, acetylation of enzymes can modify their catalytic activity through allosteric regulation, restricting access to substrates, or altering interactions with other critical regulatory proteins. Many metabolic enzymes appear to be governed by reversible acetylation—thereby not only ensuring rapid cellular responses to environmental cues, but also flexibly fine-tuning reaction rates. Another level of complexity involves functional crosstalk between acetylation and other PTMs. Because lysine residues are subject to additional PTMs—such as methylation, ubiquitylation, and sumoylation—acetylation can compete with PTMs at the same site. Moreover, by controlling the recruitment of specific enzymes, acetylation can influence PTMs at nearby sites.
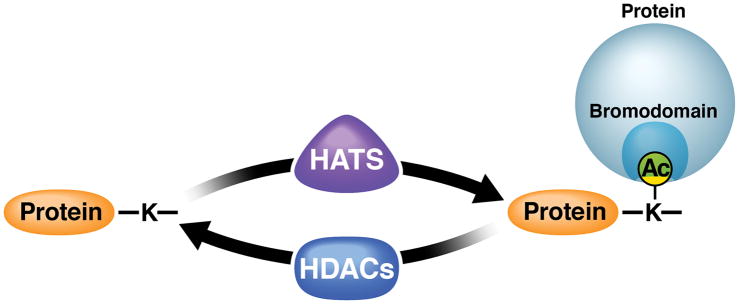
Histone acetyltransferases (HATs) are considered the “writers” of acetylation because they transfer an acetyl group from the cofactor, acetyl-coenzyme A, to the target lysine (Figure 1). At least 26 human HATs are known, nine of which are grouped into three major families based on similarities in their structure and sequence: (1) GNAT (Gcn5-related N-acetyltransferases), including PCAF and GCN5; (2) MYST (MOZ, Ybf2/Sas3, Sas2, TIP60), including HBO1; and (3) p300/CBP proteins (E1A-associated protein of 300kDa/CREB-binding protein) (Berndsen & Denu, 2008; Li et al., 2012).
Figure 1. The different players in the host acetylation network.
Histone acetyltransferases (HATs) transfer acetyl groups to target lysines in proteins while histone deacetylases (HDACs) remove them. Proteins containing bromodomains bind acetyl-lysines via a distinct structural binding pocket and recruit complexes relevant for the function of the acetylated protein.
The activity of HATs is counterbalanced by HDACs, which remove the acetyl groups and are therefore considered “erasers” (Figure 1). Thus far, 18 mammalian HDACs are known, which are categorized into three classes based on distinct catalytic characteristics. Class I and II HDACs (HDACs 1–11) use a Zn2+-dependent deacetylation mechanism and are inhibited by hydroxamic acids such as trichostatin A, vorinostat (SAHA), givinostat (ITF2357), and panobinostat (LBH589) (Shirakawa et al., 2013). Notably, class II HDACs shuttle between the nucleus and cytoplasm; a subset of these HDACs (class IIa) have little in vitro HDAC activity unless associated with the class 1 HDAC3/N-CoR complex (Jones et al., 2008). Class III HDACs are NAD+-dependent sirtuin deacetylases (SIRTs 1–7), which are found in the nucleus, cytoplasm, and mitochondria, and are not responsive to classical HDAC inhibitors (Houtkooper et al., 2012).
Besides HATs and HDACs, so-called reader proteins have been identified that contain protein domains which bind specifically to acetylated lysines (Figure 1). Best known and characterized are proteins containing bromodomains—conserved ~110–amino acid protein modules that form a deep hydrophobic cavity that specifically accommodates acetyl-lysine residues (Filippakopoulos & Knapp, 2014). The human genome is predicted to encode 46 bromodomain-containing proteins, which are usually epigenetic regulators; some of these proteins contain more than one bromodomain (Filippakopoulos et al., 2012).
The three groups of acetylation-associated proteins engage in regulatory crosstalk. Many HATs, including p300/CBP and GNAT enzymes, contain bromodomains. Since they can both write and recognize acetylation marks, HATs can be recruited to acetylated sites and promote spreading of the mark (Josling et al., 2012). Because class IIa HDACs have negligible intrinsic deacetylase activity, they might function as acetyl-lysine readers rather than erasers and recruit other chromatin-modifying enzymes to sites of transcription (Bradner et al., 2010). Moreover, HAT and HDAC activities are regulated by acetylation of the enzymes themselves. p300/CBP proteins bind and regulate the activity of several HDACs (e.g. HDAC1, HDAC6, and SIRT2) by directly acetylating lysines (Qiu et al., 2006; Han et al., 2008, 2009). Conversely, SIRT2 can regulate the autoacetylation of p300 and thereby modulate its ability to bind to transcription pre-initiation complexes (Black et al., 2006, 2008). HATs and HDACs not only regulate each other, but they are also intimately tied to the metabolism of cells through their cofactors acetyl-coenzyme A (HATs) and NAD+ (Class III HDACs). This regulatory crosstalk serves to maintain a dynamic equilibrium between the acetylation and deacetylation of specific substrates within cells and to rapidly translate environmental cues into shifts in complex cellular processes.
Viruses have evolved intricate strategies to usurp complex cellular processes in support of their own propagation. HIV-1 is a complex lentivirus that reverse transcribes its RNA genome into cDNA and integrates into the host chromatin of CD4+ T lymphocytes and macrophages. Through recruitment of the host transcriptional machinery, HIV promotes high-level transcription of its viral genome or becomes transcriptionally silenced in a subset of latently infected memory T cells (Ott et al., 2011).
Acetylation of the chromatin environment near the viral integration site can affect HIV transcription (Shirakawa et al., 2013). However, acetylation of non-histone proteins is also important in the viral life cycle. Multiple interactions exist between HIV and HATs, HDACs, and bromodomain-containing proteins encoded by the host. These interactions can alter the function of these epigenetic regulators, thereby disrupting the host acetylation network. Viral proteins, including the virally encoded integrase enzyme and transactivator of transcription (Tat), serve as substrates for cellular HATs and HDACs and require timely acetylation and deacetylation events for their proper function. In this review, we discuss the known mechanisms by which HIV taps into the host acetylation network as a basis for our understanding of how acetylation-targeting strategies interfere with the HIV life cycle.
HIV and the host acetylation machinery
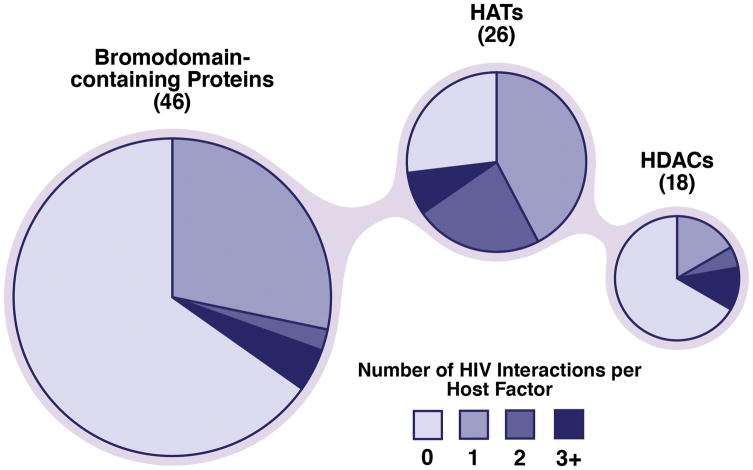
Recent system-wide mass spectrometry approaches identified a large number of interactions of acetylation-related proteins with HIV (Gautier et al., 2009; Fahey et al., 2011; Jäger et al., 2012), many of which have yet to be experimentally confirmed. Importantly, because these proteomic strategies rely on affinity purifications, it is unclear whether the interactions are direct or indirect via a larger protein complex. The majority of human HATs (19 of 26) interact with at least one HIV protein; some, such as p300 and p160, interact with up to five (Figure 2). As much as one third of human HDACs (6 of 18) have been formally identified as HIV interaction partners. Notable HDACs that can interact with up to three HIV proteins include HDAC1 (Tat, Vpr, and integrase) and HDAC6 (Tat, gp41, and gp120). Similarly, about one third of bromodomain-containing proteins (16 of 46) display HIV-binding potential. Interestingly, the majority of these interactions are made with HIV Tat (Tat binds to 11 HATs, 4 HDACs, and 14 bromodomain-containing proteins), underscoring the importance of reversible acetylation in the function and regulation of this accessory HIV protein. Below, we focus on confirmed interactions and modifications of critical regulators of the HIV life cycle.
Figure 2. Global interactions of HIV with the host acetylation network.
Recent unbiased interaction studies between HIV and host proteins have identified a high degree of interplay between HIV proteins and host acetylation factors: 19 of 26 HATs, 6 of 18 HDACs, and 16 of 46 bromodomain-containing proteins bind diverse HIV proteins. These data were determined with GPS-Prot (http://www.gpsprot.org), a web-based software tool that integrates HIV–host interaction datasets.
Acetylation of host and viral factors during HIV entry and integration
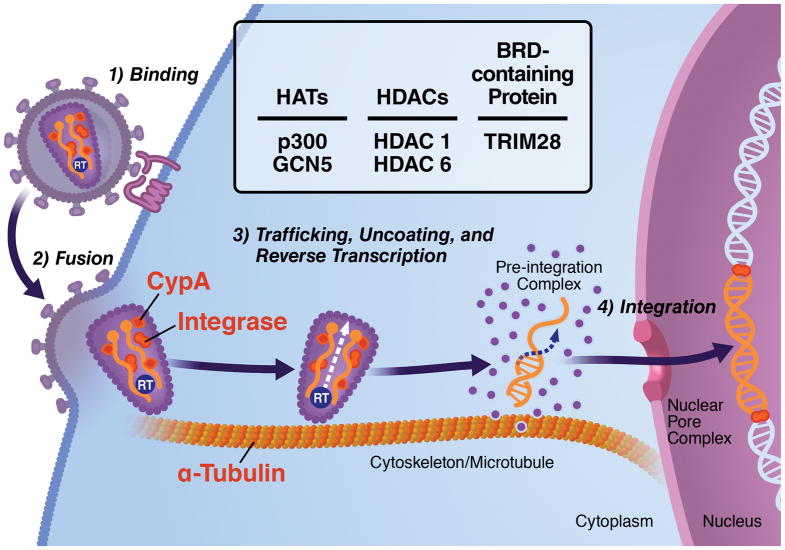
Although studies of the effect of acetylation on HIV replication have focused traditionally on transcriptional regulation, mounting evidence suggests that acetylation is critical in early steps of the viral life cycle. After HIV-1 attaches to the plasma membrane of the host cell, the viral envelope (Env) proteins gp120 and gp41 interact with the host receptor CD4 and one of two coreceptors—CXC chemokine receptor type 4 (CXCR4) and CC chemokine receptor type 5 (CCR5)—to facilitate viral fusion and entry (Figure 3) (Loetscher et al., 2000). Once within the cytoplasm, the viral nucleocapsid uses the host microtubule network to move toward the nuclear pore complex by manipulating cytoplasmic factors such as dynein (McDonald et al., 2002). During this time, the core of the HIV-1 particle progressively disassembles (viral uncoating), and the viral RNA genome is reverse transcribed by the viral reverse transcriptase enzyme to form a pre-integration complex with the proviral double-stranded cDNA at its center. The pre-integration complex, composed of viral and host factors, mediates the transport of double-stranded cDNA into the nucleus, where it integrates into the host chromatin with the assistance of host factors such as the lens epithelium-derived growth factor (LEDGF/p75) and the virally encoded integrase enzyme. During these early steps of HIV infection, several host and viral factors undergo reversible acetylation. Here, we will discuss how these early steps are regulated by the acetylation of α-tubulin, HIV-1 integrase, and cyclophilin A (Figure 3).
Figure 3. The role of reversible protein acetylation in early HIV infection.
The different steps of the early stages of the HIV life cycle are depicted with viral (integrase) and host (cyclophilin A (CypA) and α-tubulin) factors regulated by acetylation depicted in red. Engagement of HIV envelope protein gp120 with its host receptor CD4 induces α-tubulin acetylation and microtubule stabilization, a process required for successful fusion of the virus to the cell. Overexpression of HDAC6, which decreases α-tubulin acetylation, impairs virus–cell fusion and subsequent infection. Cellular CypA, a peptidylprolyl isomerase, is packaged into budding virions and regulates early steps of infection. Acetylation of CypA impairs its catalytic activity and disrupts its interaction with the HIV gag protein. The viral integrase enzyme is critical for the integration of proviral double-stranded cDNA into the host chromatin. Acetylation of the integrase enzyme by p300 enhances its affinity to genomic DNA and its strand transfer activity. It also recruits bromodomain-containing protein TRIM28 and its associated HDAC1 activity.
Microtubules are composed of α/β tubulin heteropolymers, which form key structures in cell division, vesicular trafficking, and multiple signaling pathways. These dynamic filaments are stabilized by acetylation of the α-tubulin polymer—a highly conserved mechanism that is increasingly recognized as a key factor in human health and disease (Piperno et al., 1987; Perdiz et al., 2011). As shown by Valenzuela-Fernández et al., the interaction between the HIV gp120 protein and the CD4 T-cell surface receptor induces α-tubulin acetylation, leading to microtubule stabilization necessary for fusion of the virus to the host cell (Valenzuela-Fernández et al., 2005). By altering the activity of HDAC6, one of two reported tubulin deacetylases (North et al., 2003)–either through overexpression of the wildtype protein or a dominant-negative mutant—the authors linked α-tubulin acetylation to early viral fusion (Figure 3).
Recently, the matrix region of HIV Gag was shown to recruit the EB1-binding protein Kif4 to the ends of microtubules, thereby regulating the formation of acetylated α-tubulin necessary for early stages of HIV-1 infection (Sabo et al., 2013). Occurring specifically at the postentry stage of infection, this recruitment affects nuclear import, initiation of reverse transcription, and viral cDNA synthesis. Human herpes virus 8 also induces microtubule acetylation during early stages of viral infection, pointing to a common phenomenon conserved among different viruses (Naranatt et al., 2004). However, the molecular mechanism by which HIV uses the host acetylation machinery to promote α-tubulin acetylation remains unclear. Although tubulin was one of the first non-histone proteins shown to undergo acetylation, the α-tubulin acetyltransferase αTAT-1 (or MEC17 in worms) was not identified until recently (Akella et al., 2010; Shida et al., 2010). Further studies are required to determine whether HIV regulates microtubule acetylation by directly recruiting αTAT-1, by inhibiting the α-tubulin deacetylases HDAC6 and SIRT2, or by alternative mechanisms.
Integration occurs within the large pre-integration nucleoprotein complex, which consists of the viral cDNA, viral proteins (integrase, matrix, Vpr, nucleocapsid, and reverse transcriptase), and several host factors. p300 can directly acetylate the viral integrase at three carboxy-terminal lysines: K264, K266, and K273 (Cereseto et al., 2005). Acetylation increases the affinity of integrase for genomic DNA and enhances strand transfer activity (Figure 3). Conversely, point mutations in acetylation sites or inhibition of p300 inhibited viral integration and replication (Ceresto et al., 2005). An additional report confirmed the acetylation of integrase by p300, but could not find a replication defect of point mutants when an untagged viral construct was used (Topper et al., 2007). A second integrase acetyltransferase, human GCN5, has partially overlapping specificity for integrase lysine residues, suggesting that integrase acetylation is more complex than originally assumed (Terreni et al., 2010).
The acetylated residues in HIV integrase are interaction sites for host bromodomain-containing proteins. Data from a tethered catalysis yeast two-hybrid screen identified host TRIM28 (also known as KAP1 and Tif-1B) as a bromodomain factor that binds preferentially to acetylated integrase (Allouch & Cereseto, 2009). TRIM28 recruits HDAC1, thus triggering deacetylation of HIV integrase and restricting viral integration (Allouch et al., 2011). This highlights the evolution of cellular mechanisms to counter infection by exploiting viral dependence on protein acetylation. Interestingly, HDAC1 was previously identified as a component of the integrase complex (Sorin et al., 2009). Here, the authors showed, using the yeast two-hybrid system, that integrase interacts with SAP18, a component of the cellular Sin3a/HDAC complex. They further showed that this complex, packaged into HIV-1 virions, is critical for postentry viral infection. In sum, these studies suggest that HDAC1 can regulate HIV infection either positively or negatively, depending on the context of viral interaction.
Cyclophilin A (CypA), a highly conserved host peptidyl-prolyl cis-trans isomerase, has complex functions in diverse cellular processes such as protein folding, signal transduction, and cell-cycle regulation. During HIV infection, CypA is recruited by the group-specific antigen (Gag) precursor polyprotein, which consists of important components of the HIV virion such as matrix and capsid proteins, and is packaged into budding virions (Figure 3) (Franke et al., 1994; Thali et al., 1994). After entering target cells, CypA is associated with multiple steps of early infection, including uncoating, reverse transcription, and nuclear trafficking (Fassati, 2012). These functions are regulated by acetylation (Lammers et al., 2010). The authors used a new in vitro system to generate large amounts of acetylated CypA protein using synthetically evolved acetyl-lysyl-tRNA synthetase/tRNACUA pair system in E. coli., which co-translationally directs the incorporation of acetyllysine into a target protein in response to specifically encoded amber codons (Neumann et al., 2008). They showed that acetylation of lysine 125 (K125) inhibits the catalytic activity of the CypA enzyme and disrupts its interactions with the HIV Gag protein. It is possible, although not shown, that the positive function of HDAC1 in early viral infection is associated with deacetylation of K125 in CypA by the co-packaged integrase/Sin3a/HDAC complex, thereby promoting optimal CypA activity (Sorin et al., 2009).
Reversible protein acetylation and HIV transcription
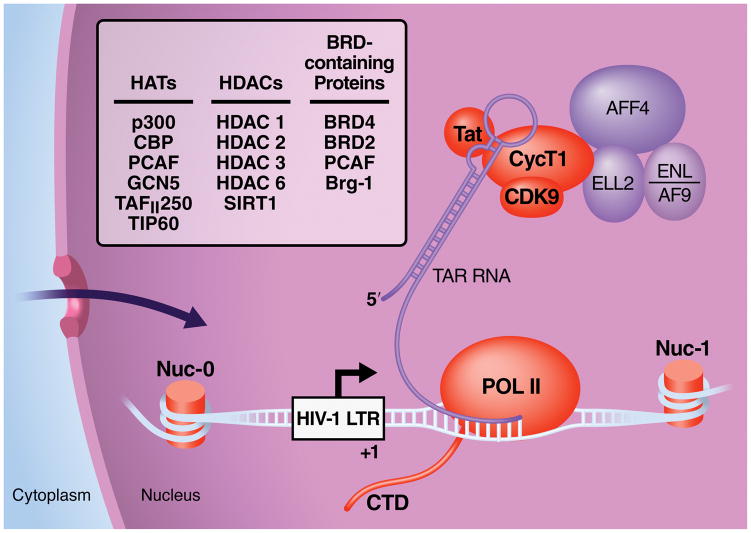
Once integrated into the host chromatin, the HIV genome, like a human protein coding gene, is subject to transcriptional regulation by the host RNA polymerase II (Pol II) enzyme. During the first phase of HIV-1 transcription, short incomplete viral transcripts accumulate that cannot support full viral replication (Kao et al., 1987). These incomplete transcripts results from pausing of the Pol II complex shortly after transcription starts. This elongation block is not unique to HIV and is found in many human genes (Core et al., 2008). To overcome it, HIV encodes the transcriptional transactivator Tat, an RNA-binding protein required for elongation of HIV transcription. Tat recruits a critical multicomponent host factor, the positive transcription elongation factor b (P-TEFb), to the 5′ extremities of elongating HIV transcripts, specifically to a conserved RNA stem-loop structure called TAR (Figure 4). The recruitment of P-TEFb to TAR promotes transcriptional elongation through its intrinsic serine/threonine kinase activity, enhancing the processivity of Pol II and dissociating negative elongation factors that physically obstruct transcription. Subsequent splicing of these elongated HIV RNA transcripts fuel the viral life cycle and give rise to novel Tat molecules which, in an autoregulatory loop, activate HIV transcription (Weinberger et al., 2005).
Figure 4. Regulation of HIV transcription by protein acetylation.
HIV transcription is closely associated with the host acetylation machinery and is currently a target for acetyllysine-targeting drug regimens. Viral (Tat) and host factors (histones in nucleosomes (nuc), the Tat cofactor P-TEFb, RNA polymerase II (POL II)) that are targets of acetylation are depicted in orange. HATs, HDACs and bromodomain (BRD)-containing factors associated with HIV transcription are listed in the box. Proteins in purple depict factors of the super elongation complex (SEC) recently identified as interacting with P-TEFb and Tat. See text for details.
Throughout this process, viral transcription is regulated at multiple levels by reversible acetylation (Figure 4). The integrated HIV provirus is fully chromatinized, controlling access of host transcription factors to the HIV promoter in the 5′ long-terminal repeat (LTR) (Verdin et al., 1993). This chromatin structure is under the control of HATs and HDACs, as first shown in studies in which the HDAC inhibitor trichostatin A potently remodeled the chromatin structure at the HIV LTR in cells (Van Lint et al., 1996) and in in vitro reactions (Sheridan et al., 1997). Since then, interest has grown to identify which HATs and HDACs play distinct roles in regulating HIV transcription. These efforts were recently comprehensively reviewed (Hakre et al., 2011; Shirakawa et al., 2013) Briefly, HDACs 1–3 are known to reside at the HIV LTR in cells with transcriptionally inactive (latent) HIV through interactions with various transcription factors, including YY1, LSF, CTIP2, CBP-1, NF-κB p50, c-myc and Sp1 (Shirakawa et al., 2013). In contrast, cellular HATs such as p300, CBP, PCAF, and GCN5 are recruited to the LTR by Tat and activating transcription factors such as NF-κB p65, AP-1, Myb, GR C/EBP, NFAT, Ets-1, LEF-1 and IRF (Hakre et al., 2011). Interestingly, increased histone acetylation during HIV activation appears to be associated with chromatin modifications during G2 arrest of the cell cycle—demonstrating the ability of HIV to manipulate critical cellular processes through the host acetylation network (Thierry et al., 2004).
Many transcription factors involved in recruiting HATs or HDACs to the HIV promoter are themselves targets of acetylation. For example, the p65 subunit of NF-κB is acetylated at multiple sites. One of which is K310, a target site of SIRT1 and SIRT2; this acetylation event is critical for full transcriptional activity of p65 (Yeung et al., 2004; Rothgiesser et al., 2010). The p50 subunit of NF-κB is also acetylated at multiple sites, including K431, K440, and K441; these acetylation events enhance the DNA-binding activity and transcriptional activity of the heterodimeric NF-κB complex (Furia et al., 2002; Deng et al., 2003). Similarly, acetylation of Sp1 at K703 increases affinity of the transcription factor for DNA (Ryu et al., 2003). Interestingly, both components of the Tat-associated P-TEFb complex, cyclin T1 and CDK9, also undergo reversible acetylation—a modification that alters their association with inhibitory ribonucleoprotein complexes and the kinase activity of CDK9 directly (Fu et al., 2007; Sabò et al., 2008; Cho et al., 2009, 2010). Three acetylation sites in the cyclin T1 subunit also serve to bind the second bromodomain of the double bromodomain and extraterminal domain (BET) protein BRD4, a process associated with activation of the P-TEFb complex (Schröder et al., 2012). Interestingly, eight lysines in the C-terminal domain of Pol II undergo reversible acetylation by p300 (Schröder et al., 2013). Acetylation of the Pol II C-terminal domain is specifically enriched downstream of polymerase-occupied gene promoters and is required for optimal activation of genes carrying paused Pol II. However, a direct connection with HIV has not yet been established (Figure 4).
Tat itself interacts with HAT enzymes including TIP60, PCAF, CBP, p300, TAFII250, and human GCN5 (Kamine et al., 1996; Yamamoto & Horikoshi, 1997; Benkirane et al., 1998; Hottiger & Nabel, 1998; Marzio et al., 1998; Weissman et al., 1998; Col et al., 2001). These interactions can target the individual enzymes to specific transcription factor complexes (e.g. TBP, TFII, NF-κB), recruit them to the HIV LTR, or modulate their catalytic activities (Caron et al., 2002). In certain cases, interactions between Tat and HATs promote extra-transcriptional effects, such as neuronal cell death (by disrupting neurotrophin signaling) or increased neoplasia (by impairing p53 tumor suppressor function) (Harrod et al., 2003; Wong et al., 2005). However, understanding the relationship between these individual interactions (i.e. temporal and spatial kinetics) within the context of the viral life cycle still remains a major challenge in the field.
Tat has at least two acetylation sites. Acetylation of lysine 28 (K28) by PCAF supports the cooperative interaction of Tat with its target RNA structure TAR and the P-TEFb cofactor, thereby promoting Pol II phosphorylation and efficient transcript elongation (Kiernan et al., 1999; D’Orso & Frankel, 2009). Acetylation of K50/51 by p300/CBP and human GCN5 terminates the P-TEFb-dependent step in Tat transactivation, mediates dissociation of Tat from TAR/P-TEFb, and recruits instead the PCAF HAT via the PCAF bromodomain (Kiernan et al., 1999; Ott et al., 1999; Deng et al., 2000; Dorr et al., 2002; Mujtaba et al., 2002; Kaehlcke et al., 2003). Other interactions of acetylated Tat with host bromodomain-containing proteins include its recruitment of Brg-1—a component of the SWI/SNF nucleosome remodeling complex—and the HAT and transcription initiation factor TAFII250 (Weissman et al., 1998; Mahmoudi et al., 2006). Bromodomain-containing protein BRD2 has also recently emerged as a potential regulator of the HIV LTR; this mechanism, however, appears to occur independently of Tat involvement (Boehm et al., 2013).
Tat also interacts with host HDACs, such as SIRT1 and HDAC6, to undergo lysine deacetylation (Figure 4) (Pagans et al., 2005; Huo et al., 2011). Deacetylation of Tat by SIRT1 is necessary for optimal transactivator function—supporting a model in which timely and balanced acetylation/deacetylation events are important to fully support Tat function during HIV transcription. Tat serves as a super-substrate for SIRT1, associating avidly with the SIRT1 HDAC domain and thereby preventing other substrates (e.g. p65 K310) from accessing the enzyme (Kwon et al., 2008). By effectively inhibiting SIRT1 activity on other substrates, Tat induces hyperacetylation of p65, rendering it more active and activating infected CD4+ T lymphocytes. Thus, Tat is not only a bona fide substrate and recruitment module for HATs, HDACs, and bromodomain-containing proteins, it also directly manipulates the activity of HATs and HDACs, resulting in reprogramming of infected T cells and manipulation of the infection rates of neighboring lymphocytes. Besides Tat, the accessory HIV protein Vpr also binds to p300/CBP HAT proteins and supports HIV transcription (Kino et al., 2002).
Acetylation during late stages of HIV infection
It remains to be determined whether acetylation also regulates the late stages of viral replication. However, it is clear that the changes in HIV entry, integration, and transcription described above will also indirectly alter the rates of virion assembly and budding. Furthermore, because stable microtubules are important for virion assembly and budding, it is likely that altering the acetylation of α-tubulin will also directly affect these later stages of the viral lifecycle (Jolly et al., 2007). Similarly, the co-packaging of integrase with the Sin3/HDAC complex into virions is likely associated with more widespread acetylation/deacetylation processes during assembly, budding, and maturation of HIV virions (Sorin et al., 2009). However, the exact nature of the host or viral factors critical for these late steps of the viral life cycle remains unclear at this stage.
Therapeutic manipulation of the acetylation network
These intricate interactions between HIV and host acetylation-associated processes make acetylation-targeting drugs ideal candidates to support current antiretroviral therapy (ART). ART potently inhibits actively replicating HIV, but cannot eradicate the virus from patients (Chun et al., 1997; Wong et al., 1997; Siliciano et al., 2003). The major barrier to curing HIV-1 remains the persistence of long-lived, resting CD4+ memory T cells harboring replication-competent but transcriptionally silenced proviruses (Chomont et al., 2009). These latent reservoirs are established early after infection, are resistant to ART, and trigger viral rebound after ART is stopped (Zhang et al., 1999; Fischer et al., 2004; Kaufmann et al., 2004; Lewin et al., 2008). One current approach is to “shock and kill” latently infected T cells with latency-reversing agents, forcing latent proviruses into active transcription under the protection of ART to eliminate them through the immune system or additional intervention.
HDAC inhibitors and reversal of HIV latency
Since early studies demonstrated that HDAC inhibitors modify the chromatin environment of the integrated provirus and potently activate HIV, considerable effort has focused on identifying HDACs that are important for maintaining the latent state (Hakre et al., 2011; Shirakawa et al., 2013). A growing library of small molecules that inhibit class I and II HDACs reactivate HIV within in vitro models of latent HIV infection; some of these compounds, previously approved for the treatment of cancer, have advanced into clinical trials (Sgarbanti & Battistini, 2013; Cillo et al., 2014; Falkenberg & Johnstone, 2014; Campbell et al., 2015). Compounds such as valporic acid, vorinostat, and givinostat showed success in increasing viral RNA levels in latently infected resting T cells from treated patients; however, the results were either not reproducible (Archin et al., 2009; Blazkova et al., 2012; Routy et al., 2012; Wei et al., 2014) or pointed to the finding that repeated intake of HDAC inhibitors desensitizes cells to their latency-reversing activities (Archin et al., 2010).
The findings from studies of select latency-reactivating agents are summarized in Table 1, which also gives the working concentrations for each of the compounds used in vitro or ex vivo and outlines their mechanism of action. A more extensive list of HDAC inhibitors used in vitro can be found in a recent review (Wightman et al., 2012). Various comparative studies indicate that panobinostat and romidepsin are most efficient at targeting class I HDACs (Rasmussen et al., 2013; Wei et al., 2014). Notably, panobinostat, decreased the size of the latent pool in patients in a phase I clinical trial (Rasmussen et al., 2014). Furthermore, prolonged treatment with romidepsin had robust latency-reversing activity in patient-derived cells and induced virion release in a clinical study in patients (Wei et al., 2014). It remains to be shown whether panobinostat or romidepsin treatment will effectively delay viral rebound in patients. A single-drug will likely prove insufficient to overcome latency in all cells, and thus a combination treatment targeting multiple stages of the life cycle may be required (Bullen et al., 2014).
Table 1. Inhibitors of HDACs, bromodomain-constaining proteins and HATs used as regulators of HIV transcription.
Summary of emerging or clinically relevant acetylation-based therapeutic strategies to combat HIV-1 latency by reactivation or sustained suppression of the HIV-1 LTR. Clinical trial information was obtained through https://clinicaltrials.gov/.
| Compound Concentration
|
Target
|
Effects on HIV-1+ Latent Cells
|
Status in Trial Trial ID
|
References
|
|---|---|---|---|---|
|
Romidepsin 0.1 nM-45nM |
Class I HDAC Inhibition |
|
On-going Phase I Trial NCT01933594 |
Bullen et al, 2014; Campbell et al, 2014; Wei et al, 2014; Bertino and Otterson, 2011 |
|
|
|
|
|
|
|
Panobinostat 5nM–30nM |
Pan-specific HDAC Inhibition |
|
Completed Phase I Trial NCT01680094 |
Bullen et al, 2014; Rasmussen et al, 2013; Rasmussen et al, 2014; Wei et al, 2014 |
|
|
|
|
|
|
|
Vorinostat 100nM–5μM |
Pan-specific HDAC Inhibition |
|
Completed Phase I Trial NCT1365065 On-going Phase II Trials NCT01933594 NCT01319383 |
Archin et al, 2014; Archin et al, 2012; Bullen et al, 2014; Cillo et al, 2014; Del Prete et al, 2014; Lucera et al, 2014; Elliott J, 2013 |
|
|
|
|
|
|
|
Valproic Acid
100μM–5mM |
Pan-specific HDAC Inhibition |
|
Terminated Phase I Trial NCT00289952 |
Archin et al, 2010; Bullen et al, 2014; Routy et al, 2012 |
|
|
|
|
|
|
|
Givinostat 30nM–250nM |
Class I and Class II HDAC Inhibition |
|
Completed Phase I Non-HIV Trials NCT00792467 NCT00570661 |
Rasmussen et al, 2013; Furlan et al, 2011; Matalon et al, 2010 |
|
|
|
|
|
|
|
JQ-1 100nM–10μM |
BET Protein Inhibition |
|
Tested in vitro and ex vivo | Bullen et al, 2014; Boehm et al, 2013; Bisgrove et al, 2007; Li et al, 2012; Filippakopoulos et al, 2010 |
|
|
|
|
|
|
|
I-BET, I-BET151 100nM–10uM |
BET Protein Inhibition |
|
Tested in vitro | Nicodeme et al, 2010; Boehm et al, 2013; Seal et al, 2012 |
|
|
|
|
|
|
|
MS417 50nM-5μM |
BET Protein Inhibition |
|
Tested in vitro | Boehm et al, 2013; Zhang et al, 2012 |
|
|
|
|
|
|
|
Compound 8 1 μM–140μM |
Distruption of PCAF/Tat Interaction |
|
Tested in vitro | Pan et al, 2007 |
|
|
|
|
|
|
|
LTK14 10μM–50μM |
p300 Inhibition |
|
Tested in vitro | Balasubramanyam et al, 2004a; Mantelingu et al, 2007 |
|
|
|
|
|
|
|
BPRHIV001 1 μM–140μM |
p300 Down-Regulation |
|
Tested in vitro | Lin et al, 2011 |
In addition, “shock” therapies like HDAC inhibitors may exert unwanted effects on the “kill” arm of the approach. Notably, HDAC inhibitor treatment caused defects in T-cell development and distorted CD8+ T cell activity, potentially diminishing the potential of these cells to effectively eliminate reactivated cells in patients (Shan et al., 2012; Tschismarov et al., 2014). Furthermore, treatment with vorinostat and panabinostat decreased interferon-γ production in primary activated CD8+ T cells, resulting in impaired elimination of HIV-Gag-positive CD4+ T cells in an in vitro model of HIV latency (Jones et al., 2014). Studies outside HIV also point to important roles of HDACs in the effector function of T cells and macrophages (Halili et al., 2010; Bagley et al., 2014; Cheng et al., 2014; Yan et al., 2014). For example, in conditional HDAC1 knockout mice, cytokine production in CD8+ T cells was enhanced, but the ability to fend off a viral challenge was decreased (Tschismarov et al., 2014). It remains to be tested whether the targeted inhibition of individual HDACs is effective in reactivating latent HIV while reducing unwanted effects on T-cell function (Archin et al., 2009; Barton et al., 2014; Klase et al., 2014).
Bromodomain inhibitors and HIV transcription
Recently, novel inhibitors of bromodomain-containing proteins, especially those targeting so-called BET (bromodomain and ET domain) proteins BRD2–4 and BRDT, have shown impressive effects in cancer, immunity, and contraception (French, 2012; Matzuk et al., 2012; Filippakopoulos & Knapp, 2014). These drugs occupy the binding pockets for acetyl-peptides in bromodomains and thereby displace BET proteins from chromatin or other binding partners (Shi & Vakoc, 2014). Much of the work characterizing these compounds, particularly the freely available compound JQ1, has focused on inhibiting BRD4, owing to its relevance in some malignant midline carcinomas (Filippakopoulos et al., 2010; French, 2012). Because BRD4 is a cofactor of the P-TEFb complex and competes with the HIV Tat protein for P-TEFb binding, BET inhibitors were also tested for the ability to reactivate latent HIV. Several BET bromodomain inhibitors, including JQ1, iBET151 and MS417, activate HIV transcription in cell culture models of latency (Table 1) (Banerjee et al., 2012; Li et al., 2012; Boehm, Conrad, et al., 2013), but their effect in patient-derived cells varies (Zhu et al., 2012; Shi & Vakoc, 2014). BET inhibitors do not synergize with HDAC inhibitors to activate HIV transcription, supporting the notion that both drugs target similar molecular pathways (Bartholomeeusen et al., 2012; Boehm et al., 2013; Loosveld et al., 2014). However, strong synergies exists with activators of the cellular protein kinase C pathway in cell culture (Li et al., 2012; Wang et al., 2012; Boehm et al., 2013; Jiang et al., 2014) and with monoclonal antibodies against HIV in humanized mouse models of HIV (Halper-Stromberg et al., 2014). Further studies are still required to better understand the molecular mechanisms of how BET inhibitors activate HIV transcription, which BET protein is targeted, and how best to combine these inhibitors in clinical trials.
HAT inhibitors and the permanent silencing of HIV transcription
Alternatives to the “shock and kill” approach, which include durable transcriptional silencing of latently infected cells, are so far less developed. Since HIV–HAT interactions have a central role in the activation of viral transcription, a structure-guided approach could be used to develop specific inhibitors against Tat–HAT interactions (Vendel & Lumb, 2004; Zeng et al., 2005; Pan et al., 2007). While compounds such as curcumin and isogarcinol can inhibit p300/PCAF HAT activity, they also demonstrate low specificity or high levels of toxicities in cell lines (Balasubramanyam et al., 2004a,b). Mantelingu and colleagues demonstrated that chemical manipulation of these naturally derived compounds could increase the specificity of binding to p300, and in the case of one of the derived compounds, LTK14, suppress viral reactivation in vitro (Mantelingu et al., 2007). Another, more recent study, has shown that coumarin derivitaves, specifically, BPRHIV001, also inhibits p300 activity and is able to suppress Tat mediated transcription in vitro (Lin et al., 2011). While these studies collectively highlight the importance of p300/CBP as a potential target in HIV treatment, it is unclear whether HAT inhibitors can effectively inhibit viral rebound from latency in vivo. Further investigation of the molecular role of HATs, HDACs, and bromodomain-containing proteins in the establishment and maintenance of latency are required to develop better and more targeted therapeutic interventions.
Conclusion and outlook
Protein acetylation, a highly conserved regulatory system in a broad range of organisms, can rapidly translate environment signals into critical cellular functions. Viruses such as HIV have evolved intricate strategies to manipulate this system to facilitate viral propagation at multiple steps of the viral life cycle. As a result, there is growing interest in using inhibitors of acetylation-associated proteins to disrupt these interactions. Despite an impressive body of work, much remains to be learned about the complex role of reversible acetylation during the HIV life cycle and within the immune system. The ability to continuously monitor acetyl-stoichiometric changes (i.e. frequency of each acetylated site within a cell)—on both a local and systemic scale—will be critical in assessing the biological significance of the effects of these modifications on different cellular processes. Two studies have begun to address this topic in Saccharomyces cerevisiae and Escherichia coli (Baeza et al., 2014; Weinert et al., 2014). These studies revealed that significant acetylation alterations occur in distinct subcellular compartments during specific cell-cycle phases or upon deletion of a particular HDAC. Since viruses operate in distinct host compartments at different time points, it will be interesting to use this technology to map acetylation dynamics in an infected cell during different phases of the viral life cycle. This knowledge will promote a more comprehensive understanding of the dynamics of host–virus interactions and highlight critical areas of interest for therapeutic intervention.
In addition, as new players are still continually being added to the acetylation network, novel hypotheses and opportunities for treating HIV will arise. Besides bromodomains, some tandem plant homeodomain zinc-finger proteins may also bind histones in an acetylation-specific manner (Zeng et al., 2010; Ali et al., 2012; Qiu et al., 2012). Furthermore, the tandem pleckstrin-homology domain of Rtt106, a yeast chaperone protein, binds acetylated histone H3 at lysine 56 (Su et al., 2012). Most relevant to HIV, the highly conserved YEATS domain, named for its five founding proteins (Yaf9, ENL, AF9, Taf14, and Sas5), binds acetyl-lysine residues, with a preference for acetylated histone H3 lysine 9 (Li et al., 2014). ENL and AF9 are both members of the so-called super elongation complex (SEC), which is associated with HIV Tat and P-TEFb and critically involved in their function during HIV transcription elongation (He et al., 2010; Sobhian et al., 2010). It remains to be determined whether these interactions are dependent on the acetylation status of these factors and can be affected by acetylation-targeting drugs.
Acknowledgments
We thank John Carroll and Giovanni Maki for assistance with graphics. We also thank members of the Ott laboratory for helpful discussions, Stephen Ordway for editorial and Veronica Fonseca for administrative assistance.
Footnotes
Declarations of interest
We gratefully acknowledge support from the NIH (R01AI083139 and U19 AI096113 CARE Collaboratory). Mark Y. Jeng is supported in part by the NSF Graduate Research Fellowship Grant 1144247. Ibraheem Ali is supported in part by NIH Training Grant 2 T32 IA 7334-26.
References
- Akella JS, Wloga D, Kim J, et al. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 2010;467:218–22. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Yan K, Lalonde M-E, et al. Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J Mol Biol. 2012;424:328–38. doi: 10.1016/j.jmb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allouch A, Cereseto A. Identification of cellular factors binding to acetylated HIV-1 integrase. Amino Acids. 2009;41:1137–45. doi: 10.1007/s00726-009-0444-3. [DOI] [PubMed] [Google Scholar]
- Allouch A, Di Primio C, Alpi E, et al. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe. 2011;9:484–95. doi: 10.1016/j.chom.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Archin NM, Bateson R, Tripathy MK, et al. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis. 2014;210:728–35. doi: 10.1093/infdis/jiu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Cheema M, Parker D, et al. Antiretroviral intensification and valproic acid lack sustained effect on residual HIV-1 viremia or resting CD4+ cell infection. PLoS One. 2010;5:e9390. doi: 10.1371/journal.pone.0009390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Espeseth A, Parker D, et al. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses. 2009;25:207–12. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Dowell JA, Smallegan MJ, et al. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem. 2014;289:21326–38. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley JA, Yan Z, Zhang W, et al. Double-bromo and extraterminal (BET) domain proteins regulate dendrite morphology and mechanosensory function. Genes Dev. 2014;28:1940–56. doi: 10.1101/gad.239962.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanyam K, Altaf M, Varier RA, et al. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem. 2004a;279:33716–26. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- Balasubramanyam K, Varier RA, Altaf M, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004b;279:51163–71. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- Banerjee C, Archin N, Michaels D, et al. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol. 2012;92:1147–54. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeeusen K, Xiang Y, Fujinaga K, et al. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J Biol Chem. 2012;287:36609–16. doi: 10.1074/jbc.M112.410746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton KM, Archin NM, Keedy KS, et al. Selective HDAC inhibition for the disruption of latent HIV-1 infection. PLoS One. 2014;9:e102684. doi: 10.1371/journal.pone.0102684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M, Chun RF, Xiao H, et al. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem. 1998;273:24898–905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- Berndsen CE, Denu JM. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol. 2008;18:682–89. doi: 10.1016/j.sbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertino EM, Otterson GA. Romidepsin: a novel histone deacetylase inhibitor for cancer. Expert Opin Investig Drugs. 2011;20:1151–58. doi: 10.1517/13543784.2011.594437. [DOI] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, et al. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A. 2007;104:13690–5. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Choi JE, Lombardo SR, et al. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–18. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Black JC, Mosley A, Kitada T, et al. The SIRT2 deacetylase regulates autoacetylation of p300. Mol Cell. 2008;32:449–55. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazkova J, Chun TW, Belay BW, et al. Effect of histone deacetylase inhibitors on HIV production in latently infected, resting CD4+ T cells from infected individuals receiving effective antiretroviral therapy. J Infect Dis. 2012;206:765–69. doi: 10.1093/infdis/jis412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm D, Calvanese V, Dar RD, et al. BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle. 2013;12:452–62. doi: 10.4161/cc.23309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm D, Conrad R, Ott M. Bromodomain proteins in HIV infection. Viruses. 2013;5:1571–86. doi: 10.3390/v5061571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, West N, Grachan ML, et al. Chemical phylogenetics of histone deacetylases. Nat Meth. 2010;6:238–43. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen CK, Laird GM, Durand CM, et al. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med. 2014;20:425–29. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, Bruckman RS, Chu Y-L, et al. Autophagy induction by histone deacetylase inhibitors inhibits HIV type 1. J Biol Chem. 2014;290:5028–40. doi: 10.1074/jbc.M114.605428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron CC, Col E, Khochbin S. The viral control of cellular acetylation signaling. Bioessays. 2002;25:58–65. doi: 10.1002/bies.10202. [DOI] [PubMed] [Google Scholar]
- Cereseto A, Manganaro L, Gutierrez MI, et al. Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 2005;24:3070–81. doi: 10.1038/sj.emboj.7600770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Lienlaf M, Wang HW, et al. A novel role for histone deacetylase 6 in the regulation of the tolerogenic STAT3/IL-10 pathway in APCs. J Immunol. 2014;193:2850–62. doi: 10.4049/jimmunol.1302778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Schroeder S, Kaehlcke K, et al. Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. EMBO J. 2009;28:1407–17. doi: 10.1038/emboj.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Schroeder S, Ott M. CYCLINg through transcription: posttranslational modifications of P-TEFb regulate transcription elongation. Cell Cycle. 2010;9:1697–1705. doi: 10.4161/cc.9.9.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chun TW, Stuyver L, Mizell SB, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–97. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillo AR, Sobolewski MD, Bosch RJ, et al. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2014;111:7078–83. doi: 10.1073/pnas.1402873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Col E, Caron C, Seigneurin-Berny D, et al. The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J Biol Chem. 2001;276:28179–84. doi: 10.1074/jbc.M101385200. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–18. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete GQ, Shoemaker R, Oswald K, et al. Effect of suberoylanilide hydroxamic acid (SAHA) administration on the residual virus pool in a model of combination antiretroviral therapy-mediated suppression in SIVmac239-infected Indian rhesus macaques. Antimicrob Agents Chemother. 2014;58:6790–806. doi: 10.1128/AAC.03746-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, de la Fuente C, Fu P, et al. Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones. Virology. 2000;277:278–95. doi: 10.1006/viro.2000.0593. [DOI] [PubMed] [Google Scholar]
- Deng W-G, Zhu Y, Wu KK. Up-regulation of p300 binding and p50 acetylation in tumor necrosis factor-alpha-induced cyclooxygenase-2 promoter activation. J Biol Chem. 2003;278:4770–77. doi: 10.1074/jbc.M209286200. [DOI] [PubMed] [Google Scholar]
- Dorr A, Kiermer V, Pedal A, et al. Transcriptional synergy between Tat and PCAF is dependent on the binding of acetylated Tat to the PCAF bromodomain. EMBO J. 2002;21:2715–23. doi: 10.1093/emboj/21.11.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orso I, Frankel AD. Tat acetylation modulates assembly of a viral-host RNA-protein transcription complex. Proc Natl Acad Sci U S A. 2009;106:3101–06. doi: 10.1073/pnas.0900012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J, Solomon A, Wightman F, et al. The safety and effect of multiple doses of vorinostat on HIV transcription in HIV+ patients receiving cART. 20th Conference on Retroviruses and Opportunistic Infections.2013. [Google Scholar]
- Fahey ME, Bennett MJ, Mahon C, et al. GPS-Prot: A web-based visualization platform for integrating host-pathogen interaction data. BMC Bioinformatics. 2011;12:298. doi: 10.1186/1471-2105-12-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13:673–91. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- Fassati A. Multiple roles of the capsid protein in the early steps of HIV-1 infection. Virus Res. 2012;170:15–24. doi: 10.1016/j.virusres.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–56. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–31. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Joos B, Hirschel B, et al. Cellular viral rebound after cessation of potent antiretroviral therapy predicted by levels of multiply spliced HIV-1 RNA encoding nef. J Infect Dis. 2004;190:1979–88. doi: 10.1086/425983. [DOI] [PubMed] [Google Scholar]
- Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–62. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- French CA. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol Mech Dis. 2012;7:247–65. doi: 10.1146/annurev-pathol-011811-132438. [DOI] [PubMed] [Google Scholar]
- Fu J, Yoon HG, Qin J, et al. Regulation of P-TEFb elongation complex activity by CDK9 acetylation. Mol Cell Biol. 2007;27:4641–51. doi: 10.1128/MCB.00857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furia B, Deng L, Wu K, et al. Enhancement of nuclear factor-kappa B acetylation by coactivator p300 and HIV-1 Tat proteins. J Biol Chem. 2002;277:4973–80. doi: 10.1074/jbc.M107848200. [DOI] [PubMed] [Google Scholar]
- Furlan A, Monzani V, Reznikov LL, et al. Pharmacokinetics, safety and inducible cytokine responses during a phase 1 trial of the oral histone deacetylase inhibitor ITF2357 (givinostat) Mol Med. 2011;17:353–62. doi: 10.2119/molmed.2011.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier VW, Gu L, O’Donoghue N, et al. In vitro nuclear interactome of the HIV-1 Tat protein. Retrovirology. 2009;6:47. doi: 10.1186/1742-4690-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakre S, Chavez L, Shirakawa K, et al. Epigenetic regulation of HIV latency. Curr Opin HIV AIDS. 2011;6:19–24. doi: 10.1097/COH.0b013e3283412384. [DOI] [PubMed] [Google Scholar]
- Halili MA, Andrews MR, Labzin LI, et al. Differential effects of selective HDAC inhibitors on macrophage inflammatory responses to the Toll-like receptor 4 agonist LPS. J Leukoc Biol. 2010;87:1103–14. doi: 10.1189/jlb.0509363. [DOI] [PubMed] [Google Scholar]
- Halper-Stromberg A, Lu C-L, Klein F, et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. 2014;158:989–99. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Jeong HM, Jin Y-H, et al. Acetylation of histone deacetylase 6 by p300 attenuates its deacetylase activity. Biochem Biophys Res Commun. 2009;383:88–92. doi: 10.1016/j.bbrc.2009.03.147. [DOI] [PubMed] [Google Scholar]
- Han Y, Jin Y-H, Kim Y-J, et al. Acetylation of Sirt2 by p300 attenuates its deacetylase activity. Biochem Biophys Res Commun. 2008;375:576–80. doi: 10.1016/j.bbrc.2008.08.042. [DOI] [PubMed] [Google Scholar]
- Harrod R, Nacsa J, Van Lint C, et al. Human immunodeficiency virus type-1 Tat/co-activator acetyltransferase interactions inhibit p53Lys-320 acetylation and p53-responsive transcription. J Biol Chem. 2003;278:12310–18. doi: 10.1074/jbc.M211167200. [DOI] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–38. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO, Nabel GJ. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998;72:8252–56. doi: 10.1128/jvi.72.10.8252-8256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–38. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L, Li D, Sun X, et al. Regulation of Tat acetylation and transactivation activity by the microtubule-associated deacetylase HDAC6. J Biol Chem. 2011;286:9280–86. doi: 10.1074/jbc.M110.208884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Cimermancic P, Gulbahce N, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–70. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- Jiang G, Mendes EA, Kaiser P, et al. Reactivation of HIV latency by a newly modified Ingenol derivative via protein kinase Cδ–NF-κB signaling. AIDS. 2014;28:1555–66. doi: 10.1097/QAD.0000000000000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Mitar I, Sattentau QJ. Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1. J Virol. 2007;81:5547–60. doi: 10.1128/JVI.01469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Altamura S, De Francesco R, et al. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett. 2008;18:1814–19. doi: 10.1016/j.bmcl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- Jones RB, O’Connor R, Mueller S, et al. Histone deacetylase inhibitors impair the elimination of HIV-infected cells by cytotoxic T-lymphocytes. PLoS Pathog. 2014;10:e1004287. doi: 10.1371/journal.ppat.1004287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josling GA, Selvarajah SA, Petter M, et al. The role of bromodomain proteins in regulating gene expression. Genes. 2012;3:320–43. doi: 10.3390/genes3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaehlcke K, Dorr A, Hetzer-Egger C, et al. Acetylation of Tat defines a cyclinT1-independent step in HIV transactivation. Mol Cell. 2003;12:167–76. doi: 10.1016/s1097-2765(03)00245-4. [DOI] [PubMed] [Google Scholar]
- Kamine J, Elangovan B, Subramanian T, et al. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–66. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, et al. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–93. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kaufmann DE, Lichterfeld M, Altfeld M, et al. Limited durability of viral control following treated acute HIV infection. Plos Med. 2004;1:e36. doi: 10.1371/journal.pmed.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan RE, Vanhulle C, Schiltz L, et al. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 1999;18:6106–18. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Gragerov A, Slobodskaya O, et al. Human immunodeficiency virus type 1 (HIV-1) accessory protein Vpr induces transcription of the HIV-1 and glucocorticoid-responsive promoters by binding directly to p300/CBP coactivators. J Virol. 2002;76:9724–34. doi: 10.1128/JVI.76.19.9724-9734.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z, Yedavalli VSRK, Houzet L, et al. Activation of HIV-1 from latent infection via synergy of RUNX1 inhibitor Ro5-3335 and SAHA. PLoS Pathog. 2014;10:e1003997. doi: 10.1371/journal.ppat.1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H-S, Brent MM, Getachew R, et al. Human immunodeficiency virus type 1 Tat protein inhibits the SIRT1 deacetylase and induces T cell hyperactivation. Cell Host Microbe. 2008;3:158–67. doi: 10.1016/j.chom.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers M, Neumann H, Chin JW, et al. Acetylation regulates cyclophilin A catalysis, immunosuppression and HIV isomerization. Nat Chem Biol. 2010;6:331–37. doi: 10.1038/nchembio.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin SR, Murray JM, Solomon A, et al. Virologic determinants of success after structured treatment interruptions of antiretrovirals in acute HIV-1 infection. J Acquir Immune Defic Syndr. 2008;47:140–7. [PubMed] [Google Scholar]
- Li T, Du Y, Wang L, et al. Characterization and prediction of lysine (K)-acetyl-transferase specific acetylation sites. Mol Cell Proteomics. 2012;11:M111 011080. doi: 10.1074/mcp.M111.011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wen H, Xi Y, et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell. 2014;159:558–71. doi: 10.1016/j.cell.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Guo J, Wu Y, et al. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 2012;41:277–87. doi: 10.1093/nar/gks976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P-H, Ke Y-Y, Su C-T, et al. Inhibition of HIV-1 Tat-mediated transcription by a coumarin derivative, BPRHIV001, through the Akt pathway. J Virol. 2011;85:9114–26. doi: 10.1128/JVI.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loetscher P, Moser B, Baggiolini M. Chemokines and their receptors in lymphocyte traffic and HIV infection. Adv Immunol. 2000;74:127–80. doi: 10.1016/s0065-2776(08)60910-4. [DOI] [PubMed] [Google Scholar]
- Loosveld M, Castellano R, Gon S, et al. Therapeutic targeting of c-Myc in T-cell acute lymphoblastic leukemia, T-ALL. Oncotarget. 2014;5:3168–72. doi: 10.18632/oncotarget.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucera MB, Tilton CA, Mao H, et al. The histone deacetylase inhibitor vorinostat (SAHA) increases the susceptibility of uninfected CD4+ T cells to HIV by increasing the kinetics and efficiency of postentry viral events. J Virol. 2014;88:10803–12. doi: 10.1128/JVI.00320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Parra M, Vries RGJ, et al. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J Biol Chem. 2006;281:19960–68. doi: 10.1074/jbc.M603336200. [DOI] [PubMed] [Google Scholar]
- Mantelingu K, Reddy BAA, Swaminathan V, et al. Specific inhibition of p300-HAT alters global gene expression and represses HIV replication. Chem Biol. 2007;14:645–57. doi: 10.1016/j.chembiol.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Marzio G, Tyagi M, Gutierrez MI, et al. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci U S A. 1998;95:13519–24. doi: 10.1073/pnas.95.23.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, McKeown MR, Filippakopoulos P, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–84. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, et al. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–52. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, et al. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol Cell. 2002;9:575–86. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
- Naranatt PP, Krishnan HH, Smith MS, et al. Kaposi’s sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J Virol. 2004;79:1191–1206. doi: 10.1128/JVI.79.2.1191-1206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Peak-Chew SY, Chin JW. Genetically encoding Nε-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–34. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–23. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, et al. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–44. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Ott M, Geyer M, Zhou Q. The control of HIV transcription: Keeping RNA polymerase II on track. Cell Host Microbe. 2011;10:426–35. doi: 10.1016/j.chom.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Schnolzer M, Garnica J, et al. Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr Biol. 1999;9:1489–92. doi: 10.1016/s0960-9822(00)80120-7. [DOI] [PubMed] [Google Scholar]
- Pagans S, Pedal A, North BJ, et al. SIRT1 regulates HIV transcription via Tat deacetylation. Plos Biol. 2005;3:e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Mezei M, Mujtaba S, et al. Structure-guided optimization of small molecules inhibiting human immunodeficiency virus 1 Tat association with the human coactivator p300/CREB binding protein-associated factor. J Med Chem. 2007;50:2285–88. doi: 10.1021/jm070014g. [DOI] [PubMed] [Google Scholar]
- Perdiz D, Mackeh R, Poüs C, et al. The ins and outs of tubulin acetylation: more than just a post-translational modification? Cell Signal. 2011;23:763–71. doi: 10.1016/j.cellsig.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Liu L, Zhao C, et al. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 2012;26:1376–91. doi: 10.1101/gad.188359.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Zhao Y, Becker M, et al. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol Cell. 2006;22:669–79. doi: 10.1016/j.molcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Rasmussen TA, Schmeltz Søgaard O, Brinkmann C, et al. Comparison of HDAC inhibitors in clinical development: effect on HIV production in latently infected cells and T-cell activation. Hum Vaccin Immunother. 2013;9:993–1001. doi: 10.4161/hv.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TA, Tolstrup M, Brinkmann CR, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV. 2014;1:e13–e21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- Rothgiesser KM, Erener S, Waibel S, et al. SIRT2 regulates NF-κB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci. 2010;123:4251–58. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
- Routy JP, Tremblay CL, Angel JB, et al. Valproic acid in association with highly active antiretroviral therapy for reducing systemic HIV-1 reservoirs: results from a multicentre randomized clinical study. HIV Med. 2012;13:291–96. doi: 10.1111/j.1468-1293.2011.00975.x. [DOI] [PubMed] [Google Scholar]
- Ryu H, Lee J, Olofsson BA, et al. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc Natl Acad Sci U S A. 2003;100:4281–86. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo Y, Walsh D, Barry DS, et al. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe. 2013;14:535–46. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabò A, Lusic M, Cereseto A, et al. Acetylation of conserved lysines in the catalytic core of cyclin-dependent kinase 9 inhibits kinase activity and regulates transcription. Mol Cell Biol. 2008;28:2201–12. doi: 10.1128/MCB.01557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder S, Cho S, Zeng L, et al. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J Biol Chem. 2012;287:1090–99. doi: 10.1074/jbc.M111.282855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder S, Herker E, Itzen F, et al. Acetylation of RNA polymerase II regulates growth-factor-induced gene transcription in mammalian cells. Mol Cell. 2013;52:314–24. doi: 10.1016/j.molcel.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal J, Lamotte Y, Donche F, et al. Identification of a novel series of BET family bromodomain inhibitors: binding mode and profile of I-BET151 (GSK1210151A) Bioorg Med Chem Lett. 2012;22:2968–72. doi: 10.1016/j.bmcl.2012.02.041. [DOI] [PubMed] [Google Scholar]
- Sgarbanti M, Battistini A. Therapeutics for HIV-1 reactivation from latency. Curr Opin Virol. 2013;3:394–401. doi: 10.1016/j.coviro.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Shakespear MR, Halili MA, Irvine KM, et al. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 2011;32:335–43. doi: 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Shan L, Deng K, Shroff NS, et al. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity. 2012;36:491–501. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan PL, Mayall TP, Verdin E, et al. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 1997;11:3327–40. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–36. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Cueva JG, Xu Z, et al. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci U S A. 2010;107:21517–22. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa K, Chavez L, Hakre S, et al. Reactivation of latent HIV by histone deacetylase inhibitors. Trends Microbiol. 2013;21:277–85. doi: 10.1016/j.tim.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–28. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–51. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin M, Cano J, Das S, et al. Recruitment of a SAP18-HDAC1 complex into HIV-1 virions and its requirement for viral replication. PLoS Pathog. 2009;5:e1000463. doi: 10.1371/journal.ppat.1000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spange S, Wagner T, Heinzel T, et al. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–98. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Su D, Hu Q, Li Q, et al. Structural basis for recognition of H3K56-acetylated histone H3–H4 by the chaperone Rtt106. Nature. 2012;482:104–7. doi: 10.1038/nature10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terreni M, Valentini P, Liverani V, et al. GCN5-dependent acetylation of HIV-1 integrase enhances viral integration. Retrovirology. 2010;7:18. doi: 10.1186/1742-4690-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Bukovsky A, Kondo E, et al. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–5. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- Thierry S, Marechal V, Rosenzwajg M, et al. Cell cycle arrest in G2 induces human immunodeficiency virus type 1 transcriptional activation through histone acetylation and recruitment of CBP, NF-kappaB, and c-Jun to the long terminal repeat promoter. J Virol. 2004;78:12198–206. doi: 10.1128/JVI.78.22.12198-12206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper M, Luo Y, Zhadina M, et al. Posttranslational acetylation of the human immunodeficiency virus type 1 integrase carboxyl-terminal domain is dispensable for viral replication. J Virol. 2007;81:3012–17. doi: 10.1128/JVI.02257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschismarov R, Firner S, Gil-Cruz C, et al. HDAC1 controls CD8+ T cell homeostasis and antiviral response. PLoS One. 2014;9:e110576. doi: 10.1371/journal.pone.0110576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Fernández A, Alvarez S, Gordon-Alonso M, et al. Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol Biol Cell. 2005;16:5445–54. doi: 10.1091/mbc.E05-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C, Emiliani S, Ott M, et al. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–20. [PMC free article] [PubMed] [Google Scholar]
- Vendel AC, Lumb KJ. NMR mapping of the HIV-1 Tat interaction surface of the KIX domain of the human coactivator CBP. Biochemistry. 2004;43:904–8. doi: 10.1021/bi035612l. [DOI] [PubMed] [Google Scholar]
- Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2014;16:258–64. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- Verdin E, Paras P, Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 1993;12:3249–59. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Li Q, Helfer CM, et al. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J Biol Chem. 2012;287:10738–52. doi: 10.1074/jbc.M111.323493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei DG, Chiang V, Fyne E, et al. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 2014;10:e1004071. doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger LS, Burnett JC, Toettcher JE, et al. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–82. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Weinert BT, Iesmantavicius V, Moustafa T, et al. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716–16. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman JD, Brown JA, Howcroft TK, et al. HIV-1 tat binds TAFII250 and represses TAFII250-dependent transcription of major histocompatibility class I genes. Proc Natl Acad Sci U S A. 1998;95:11601–6. doi: 10.1073/pnas.95.20.11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman F, Ellenberg P, Churchill M, et al. HDAC inhibitors in HIV. Immunol Cell Biol. 2012;90:47–54. doi: 10.1038/icb.2011.95. [DOI] [PubMed] [Google Scholar]
- Wong JK, Hezareh M, Günthard HF, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–95. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- Wong K, Sharma A, Awasthi S, et al. HIV-1 Tat interactions with p300 and PCAF transcriptional coactivators inhibit histone acetylation and neurotrophin signaling through CREB. J Biol Chem. 2005;280:9390–99. doi: 10.1074/jbc.M408643200. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–98. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- Yan B, Xie S, Liu Z, et al. HDAC6 deacetylase activity is critical for lipopolysaccharide-induced activation of macrophages. PLoS One. 2014;9:e110718. doi: 10.1371/journal.pone.0110718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Li J, Muller M, et al. Selective small molecules blocking HIV-1 Tat and coactivator PCAF association. J Am Chem Soc. 2005;127:2376–77. doi: 10.1021/ja044885g. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Li S, et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–62. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu R, Zhong Y, et al. Down-regulation of NF-κB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem. 2012;287:28840–51. doi: 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ramratnam B, Tenner-Racz K, et al. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N Engl J Med. 1999;340:1605–13. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- Zhu J, Gaiha GD, John SP, et al. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep. 2012;2:807–16. doi: 10.1016/j.celrep.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]