Abstract
The toxicity of polybrominated diphenyl ethers (PBDEs), flame retardant components, was characterized in offspring from Wistar Han dams exposed by gavage to a PBDE mixture (DE71) starting at gestation day 6 (GD 6) and continuing to weaning on postnatal day 21 (PND 21). Offspring from the dams began PBDE direct dosing by gavage at the same dose as their dam on PND 12 – PND 21, and then after weaning, 5 days/week for another 13 weeks. Liver samples were collected at PND 22 and week 13 for liver gene expression analysis (Affymetrix Rat Genome 230 2.0 Array). PBDE treatment induced 1,066 liver gene transcript changes in females and 1,200 transcriptional changes in males at PND 22 (false discovery rate (FDR) < 0.01), but only 263 liver transcriptional changes at 13 weeks in male rats (FDR <0.05). No significant differences in dose response were found between male and female pups. Transcript changes at PND 22 coded for proteins in xenobiotic, sterol and lipid metabolism, and cell cycle regulation, and overlapped rodent liver transcript patterns after high fat diet or phenobarbital exposure. These findings, along with the observed PBDE-induced liver hypertrophy and vacuolization, suggests that long-term PBDE exposures have potential to modify cell functions that contribute to metabolic disease and/or cancer susceptibilities.
Keywords: polybrominated diphenyl ethers, liver toxicity, lipid metabolism
Introduction
Polybrominated diphenyl ether (PBDE) flame retardant exposures are widespread in the United States (Schecter et al. 2010). The PBDEs have physicochemical properties similar to those of other persistent organic chemicals; they are hydrophobic and lipophilic, have a low vapor pressure, and a high log Kow (U. S. Environmental Protection Agency 2008a, b, c). There are up to 209 possible isomers of PBDEs. The most common PBDEs found in the environment include PBDE 47, 99, and 153 (U. S. Environmental Protection Agency 2010; World Health Organization 2003). The most prevalent PBDE congener found in human tissue is PBDE 47 (Petreas et al. 2003; Schecter, Colacino, Sjodin, Needham and Birnbaum 2010; Sjodin et al. 2004). PBDE congeners show long-range atmospheric transport, environmental persistence and bioaccumulation in various species (including humans) and are considered persistent organic pollutants (POP) (World Health Organization 2003).
Environmental exposures come from the use of PBDEs as flame retardants found in polyester foams and other household products. Leaching of PBDEs from these foams deposited at waste dumps may result in the release PBDE isomers into the environment (water, fish, seals, humans) (Hale et al. 2003; Hale et al. 2002; Hale et al. 2001). The U. S. EPA has recently conducted an exposure assessment for PBDEs and reports that exposure in children (average PBDE intake, 47.2 ng/kg/day) is greater than in adults (average PBDE intake, 7.1 ng/kg/day) (U. S. Environmental Protection Agency 2010). PBDE exposure to the infant may occur from mother’s milk (Schecter, Colacino, Sjodin, Needham and Birnbaum 2010; Schecter et al. 2006) and house dust (Frederiksen et al. 2010; Harrad et al. 2010; Johnson et al. 2010; Lorber 2008). PBDE exposure is associated with delayed time to pregnancy in women (Harley et al. 2010) and in altered learning parameters in children (Roze et al. 2009).
The European Union banned the use and sale of PBDEs in 2004 and the U. S. manufacturer of PBDEs voluntarily phased out its production in 2004 (U. S. Environmental Protection Agency 2009). Nevertheless, the potential for PBDE exposures remain a concern because of continued widespread occurrence of these chemicals in the environment, and the long half life of PBDEs in humans (up to 6 years) (U. S. Environmental Protection Agency 2010).
The biological effects of PBDEs in rodents include thyroid and liver toxicity, developmental neurotoxic effects, and reproductive toxicity (U. S. Environmental Protection Agency 2008a, b, c). PBDEs disrupt endocrine activity (Hamers et al. 2006; Legler and Brouwer 2003; Meerts et al. 2001), alter thyroid hormone levels (Darnerud et al. 2007; Ellis-Hutchings et al. 2006; Zhou et al. 2002), and induce liver cytochrome p450 levels (Birnbaum and Cohen Hubal 2006; Sanders et al. 2005; Zhou, Taylor, DeVito and Crofton 2002). PBDE-induced toxicities may increase the risk for the development of other disease (Ervin 2009); (Baker et al. 2007; Yanovski and Yanovski 2002). Exposures to PBDEs, nongenotoxic chemicals (Agency for Toxic Substances and Disease Registry 2004), may cause conditions characteristic of those seen in metabolic disease (Lim et al. 2008).
Many PBDE toxicities, including endocrine and liver toxicity, are similar across species (Harley, Marks, Chevrier, Bradman, Sjodin and Eskenazi 2010; Roze, Meijer, Bakker, Van Braeckel, Sauer and Bos 2009; U. S. Environmental Protection Agency 2008a, b, c, d). In this paper we analyze PBDE-induced rat liver toxicity and gene transcript changes to better understand the molecular basis for PBDE toxicities.
Methods and Materials
Chemical
DE-71 (technical pentabromodiphenyl oxide; CASs. No. 32534-81-9) was obtained from Great Lakes Chemical Corporation (West Lafayette, IN - Lot 2550OA30A). Components of this mixture included primarily the tetra through penta PBDEs and a small component of hexabromodiphenyl ethers. The molecular weight for tetrabrominated diphenyl ethers is 469.6, for pentabromated diphenyl ethers, 548.5, and for hexabromo diphenyl ethers, 627.4. The DE71 bulk chemical analysis (National Toxicology Program 2004; Sanders, Burka, Smith, Black, James and Cunningham 2005) showed the following composition: 2,2’,4,4’-tetrabromodiphenyl ether- BDE-47 (36%), 2,2’,4,4’,5-pentabromodiphenyl ether - BDE-99 (42%); 2,2’,4,4’,6-pentabromodiphenyl ether - BDE-100 (10%), 2,2’,4,4’,5,5’-hexabromodiphenyl ether - BDE-153 (3%), 2,2’,4,4’,5,6’-hexabromodiphenyl ether - BDE-154 (4%), and 2,2’,3,4,4’-pentabromodiphenyl ether - BDE-85 (2%). The remaining 3% consisted of several identified tri-heptaBDEs and some unidentified PBDEs (National Toxicology Program 2004). This flame retardant mixture also contained approximately 66 nanograms/gram of brominated dioxins or furans. Consequently, a dose of 50 mg/kg is a dose of approximately 3.3 nanograms/kg body weight of brominated dioxins or furans, a dose not expected to contribute to liver toxicity (National Toxicology Program 2004; Sanders, Burka, Smith, Black, James and Cunningham 2005).
Experimental Design
Time mated female Wistar/Han dams (CRL:WI (HAN)) were obtained from Charles River Laboratories (Raleigh, NC). Dams were dosed by gavage with 0 or 50 mg/kg DE71 in corn oil (dosing volume 5 ml/kg) from GD 6 through PND 21, 7 days per week. Pups were dosed by gavage at the same dose as their dam starting on PND 12 and continuing through PND 21 for 7 days per week until weaning. Weaning occurred on PND 21, which was designated as Day 1 of the 13-week portion of the study. On day one of the 13-week portion of the study dosing began on a 5 days/week schedule. Pups used for PND 22 toxicogenomic study were dosed through PND 21, the day prior to PND 22 liver sample collection. Gestational exposure at 50 mg/kg did not affect littering endpoints for number of dams delivering live litters, number of live or dead pups per litter, or body weights of pups during lactation (data not shown). The NIH 07 diet (Ziegler Brothers, Inc., Gardners, PA) was given to dams and pups throughout the pregnancy and gestation periods and until PND 22 at which time pups were given NTP-2000 diet (Ziegler Brothers, Inc., Gardners, PA). Tap water and NTP-2000 diet were available ad libitum (National Toxicology Program 2005a, b).
On PND 22, liver samples from five male and five female rat pups from the control and five male and five female rat pups from the 50 mg/kg group were collected for toxicogenomic analysis (each pup came from a different dam – total of ten dams/control group and ten dams/50 mg/kg group). Liver samples were collected from ten males and ten females from the control and ten males and ten females from the 50 mg/kg groups for histopathlogic analysis (total of twenty dams/control group and twenty dams/50 mg/kg group). Five male rat liver samples were randomly selected from each group for the 13 week toxicogenomic analysis. No females were sampled for toxicogenomic analysis at 13 weeks due to possible interference of estrus cycling with interpretation of toxicogenomic findings at this timepoint.
The care of animals on this study was according to NIH procedures as described in the “The U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals”, available from the Office of Laboratory Animal Welfare, National Institutes of Health, Department of Health and Human Services, RKLI, Suite 360, MSC 7982, 6705 Rockledge Drive, Bethesda, MD 20892-7982 or online at http://grants.nih.gov/grants/olaw/olaw.htm#pol”. The study protocols were approved by the local animal care and use committee.
Necropsy and collection of liver samples for toxcogenomic analysis
Animals at PND 22 and 13 weeks were euthanized with carbon dioxide. For liver collection, the abdominal cavity of each rat was opened, and the liver removed. A portion of the liver was minced on an ice-cold surface, and placed into RNAlater® (Ambion, Inc., Austin, TX) for incubation overnight at 4° C. The samples were then transferred to – 20° C for storage prior to RNA extraction.
Liver Histopathology
Sections of the livers from the week 13 male rats were fixed in 10% neutral buffer formalin, embedded in paraffin, sectioned at approximately 5 microns, and mounted on glass slides. After staining with hematoxylin and eosin, cover-slips were attached to the slides. Tissues were then evaluated microscopically at different magnifications by a board certified veterinary pathologist.
The grading criteria for hepatocellular hypertrophy was as follows: minimal severity was recorded when hepatocellular hypertrophy was observed but was present in <10% of the hepatocytes in the section; mild severity was recorded when ≥10% and <50% of the hepatocytes in the section were affected; moderate severity was recorded when ≥50% and <75% of the hepatocytes in the section were affected; and marked severity was recorded when ≥75% of the hepatocytes in the section were affected. Cytoplasmic vacuolization was graded independently of hepatocellular hypertrophy, and used the following grading scale: minimal severity was recorded when cytoplasmic vacuolization was observed but was present in <25% of the hepatocytes in the section; mild severity was recorded when ≥25% and <50% of the hepatocytes in the section were affected; moderate severity was recorded when ≥50% and <75% of the hepatocytes in the section were affected; and marked severity was recorded when ≥75% of the hepatocytes in the section were affected. Livers from control and treated rats were cryosectioned and stained with Oil Red O to identify the presence of lipids.
RNA preparation and microarray hybridization
A section of the liver was collected at necropsy and placed into RNAlater® (Ambion, Inc., Austin, TX). RNA was extracted from liver using the QIAGEN Rneasy® (QIAGEN, Valencia, CA). The RNA was analyzed for quantity and purity by UV analysis using the NanoDrop ND-1000 (NanoDropTechnologies, Wilmington, DE). Samples were concentrated using Microcon filters (Millipore, Billerica, MA). All samples were evaluated for RNA integrity by gel electrophoresis using the Flash Gel RNA cassette system (Lonza, Rockland, ME). Total RNA (50 ng) was used to synthesize double-stranded cDNA for each sample using Affymetrix GeneChipExpression 3’amplication two-cycle target labeling and control reagents (Affymetrix Inc. Santa Clara, CA). The cDNA served as a template to synthesize biotin-labeled antisense cRNA using an in vitro transcription (IVT) labeling kit. Labeled cRNA was fragmented and hybridized to the Affymetrix Rat Genome 230 2.0 Genechip® Array. Array hybridization, washing, and staining were performed according to the Affymetrix recommended protocol EuKGE_Ws2v5. The chips were scanned using an Affymetrix GeneChip Scanner 3000. Quality control measurements were evaluated to determine if the data derived from the arrays were of sufficient quality prior to comparisons for differential expression.
Microarray Data Analysis
Data normalization
Probe intensity data from all Rat Genome 230 version 2 Affymetrix GeneChip® arrays was read into the R software environment (http://www.R-project.org) directly from .CEL files using the R/affy package (Gautier et al. 2004). Probe-level data quality was assessed using image reconstruction, histograms of raw signal intensities, hierarchical clustering of samples and MvsA plots. Normalization was carried out using the robust multi-array average (RMA) method using all probe intensity data sets together (Irrizarry et al. 2003). Briefly, the RMA method adjusts the background of perfect match (PM) probes, applies a quantile normalization of the corrected PM values, and calculates final expression measures using the Tukey median polish algorithm.
Statistical assessment of differential gene expression
Analysis of variance (ANOVA) methods were used to statistically resolve gene expression differences using the R/maanova package (Wu et al. 2003). To find differentially expressed probe sets in response to dose, the data was subset into male pups, female pups, or male rats. Then the model,
| (1) |
was used to fit the log-transformed gene expression measures Yi, where µ is the mean for each array, DOSE is the dose effect (control, 50 mg/kg) and εi captures random error. To find sex-related effects in pups, the data was subset into 59 mg/kg dose or control pups and the model,
| (2) |
was used, where SEX is the sex effect (male, female). To find age-related effects in males, the model,
| (3) |
was used where AGE is the effect for age (PND 22, 13 weeks), DOSE is the dose effect (control, 50 mg/kg) and AGE:DOSE is the interaction effect between age and dose. Finally, to test for sex-dependent responses in dose response, the model,
| (4) |
was used where SEX:DOSE is the interaction effect between sex and dose. All statistical tests were performed using Fs, a modified F-statistic incorporating shrinkage estimates of variance components (Cui et al. 2005). P-values were calculated by permuting model residuals 1000 times and corrected using the p.adjust function in R using method = “BH”.
Gene ontology (GO) analyses
Over-represented classifications of genes were determined from statistical outcomes by testing for association with ‘biological process’ GO terms. Enrichment of pathway members among differentially expressed probe sets was found using the one-tailed Fisher exact test for 2 × 2 contingency tables. Mappings between Affymetrix probe sets, Entrez gene identifiers and GO terms are based on the R/rat2302.db package (www.bioconductor.org).
Comparison of gene transcript expression to human disease
Gene lists and the normalized gene expression estimates (see Equation 1–Equation 4 above) were compared against results in the literature using the NextBio Platform (Kupershmidt et al. 2010). The NextBio Platform is a web-based tool to search, discover, and share knowledge using correlation analyses that are able to integrate data across various gene expression platforms and experiments for public and proprietary data. These studies include data sets which have been published and are available in GEO.
Using the NextBio datasets, we compared PND 22 male pup liver transcript data (∼ 1200 differentially expressed transcripts; false discovery rate (FDR) < 0.01) to gene transcript patterns in other disease conditions. PBDE transcript patterns were compared to those in rats after phenobarbital exposure (Waterman et al. 2010). Because brominated chemicals induce colon cancers in rodents (Dunnick et al. 1997b), and to further examine the potential of brominated flame retardants to create colon disease susceptibilities we compared the PBDE transcript patterns to those in human colon cancer gene expression profiles (Ancona et al. 2006; Irizarry et al. 2009; Jorissen et al. 2008; Saaf et al. 2007).
Results
Polybrominated diphenyl ether exposures (DE71) at 50 mg/kg caused no treatment-related effects on survival of rats. Body weight of treated males was 7% greater than controls (p = 0.1328) and 14% lower than controls for females at week 13 (p = 0.0015) (Table 1). Treatment-related liver toxicity and increased liver weights occurred in both sexes (Table 1; Figure 1). At week 13, the liver toxicity consisted of hepatocyte hypertrophy characterized by enlarged hepatocytes with an increased amount of cytoplasm, enlarged nuclei, and pale eosinophilic and granular cytoplasm. The hepatocyte hypertrophy was primarily observed in the centrilobular and midzonal regions of the hepaticlobule (lesion not shown).
Table 1.
Treatment-related effects of polybrominated diphenyl ether, male rats, week 13
| Dose group (mg/kg) |
Males 0 |
Males 50 |
Females 0 |
Females 50 |
|---|---|---|---|---|
| Hepatocyte hypertrophy | 0/10 | 10/10 (3.1)a |
0/10 | 10/10 (2.9) |
| Hepatocyte vacuolization | 3/10 (1.0) |
10/10 (1.2) |
2/10 (1.0) |
4/10 (1.0) |
| Thyroid gland Follicle hypertrophy |
0/10 | 5/10 (1.2) |
0/10 | 5/10 (1.2) |
| Mean body wt (gms) | 403.4 | 431.2 | 245.6 | 213.3 |
| Mean liver wt (gms) | 13.7 ± 1.24 | 19.5 ±2.39 | 7.94± 0.58 | 9.28 ± 1.35 |
Lesions severity grade
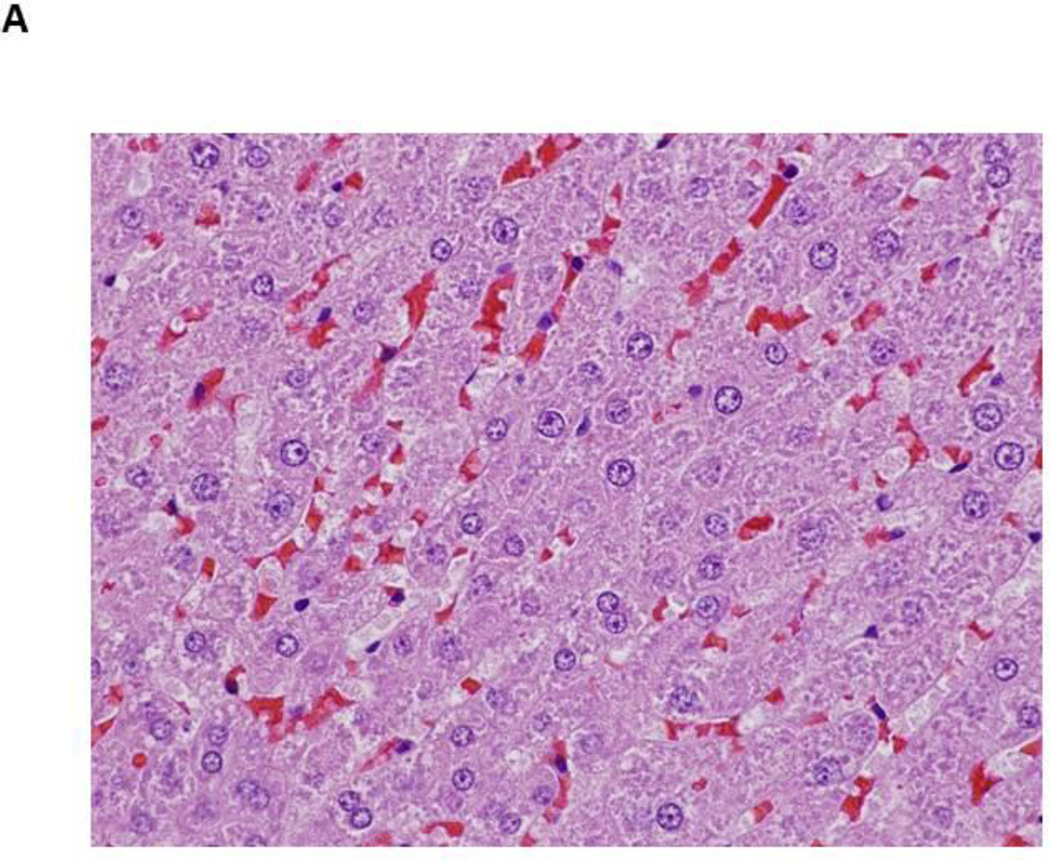
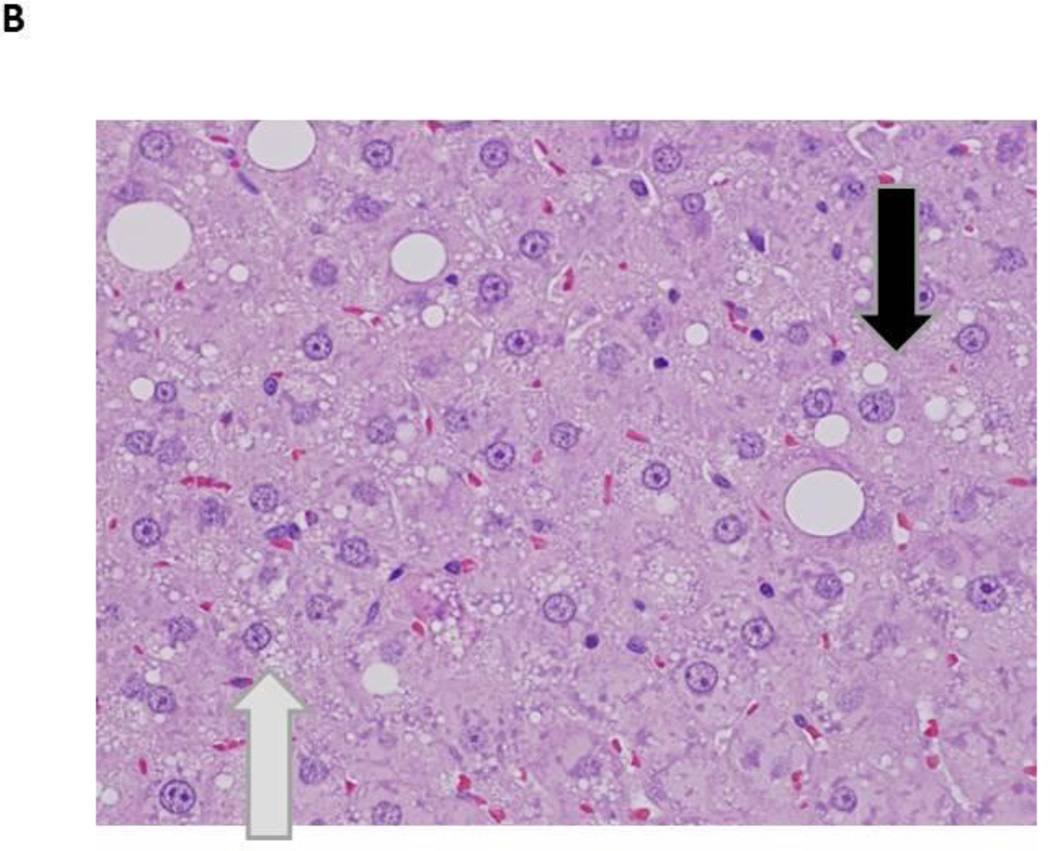
Figure 1.
Photomicrographs of control and PBDE treated livers at 13 weeks
- Male rat, control liver (H&E, 600X) – normal liver
- Male rat, 50 mg/kg (H&E, 600X) – microvesicular vacuolization of hepatocytes (blue arrow); macrovesicular vacuolization of hepatocytes (black arrow)
Hepatocyte cytoplasmic vacuolization was also observed at 13 weeks in treated rats and consisted of multiple, small, variably-sized vacuoles or large discrete, singular vacuoles within the cytoplasm of enlarged hepatocytes. Some hepatocytes contained a few scattered, small vacuoles within the cytoplasm and other heptocytes were distended with numerous small vacuoles in the cytoplasm (microvesicular vacuolization). In addition, scattered large, generally singular but occasionally multiple, clear vacuoles, compatible with macrovesicular vacuolization, were also noted in association with the microvesicular cytoplasmic vacuolization (Figure 1). Hepatocellular cytoplasmic vacuolization was most prominent in the periportal regions. Positive staining with Oil Red O indicated the vesicles within the cells from treated rats contained lipid (Figure 2).
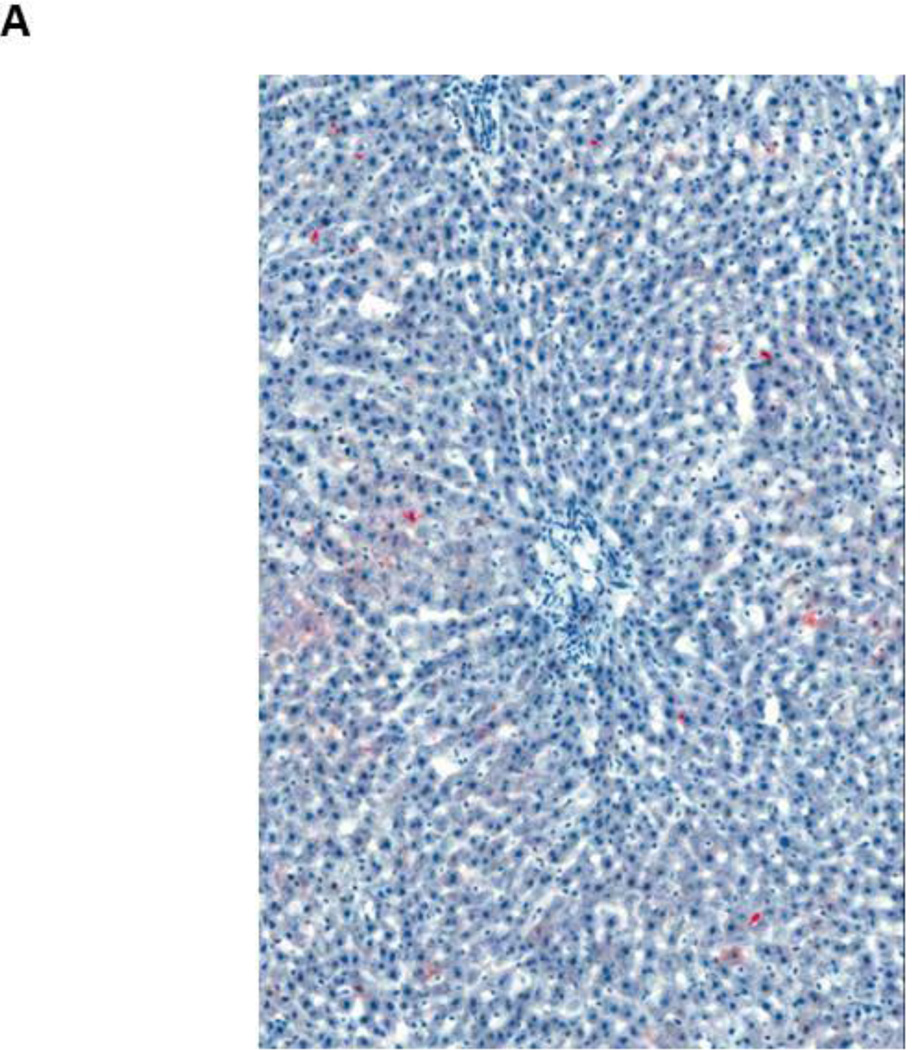
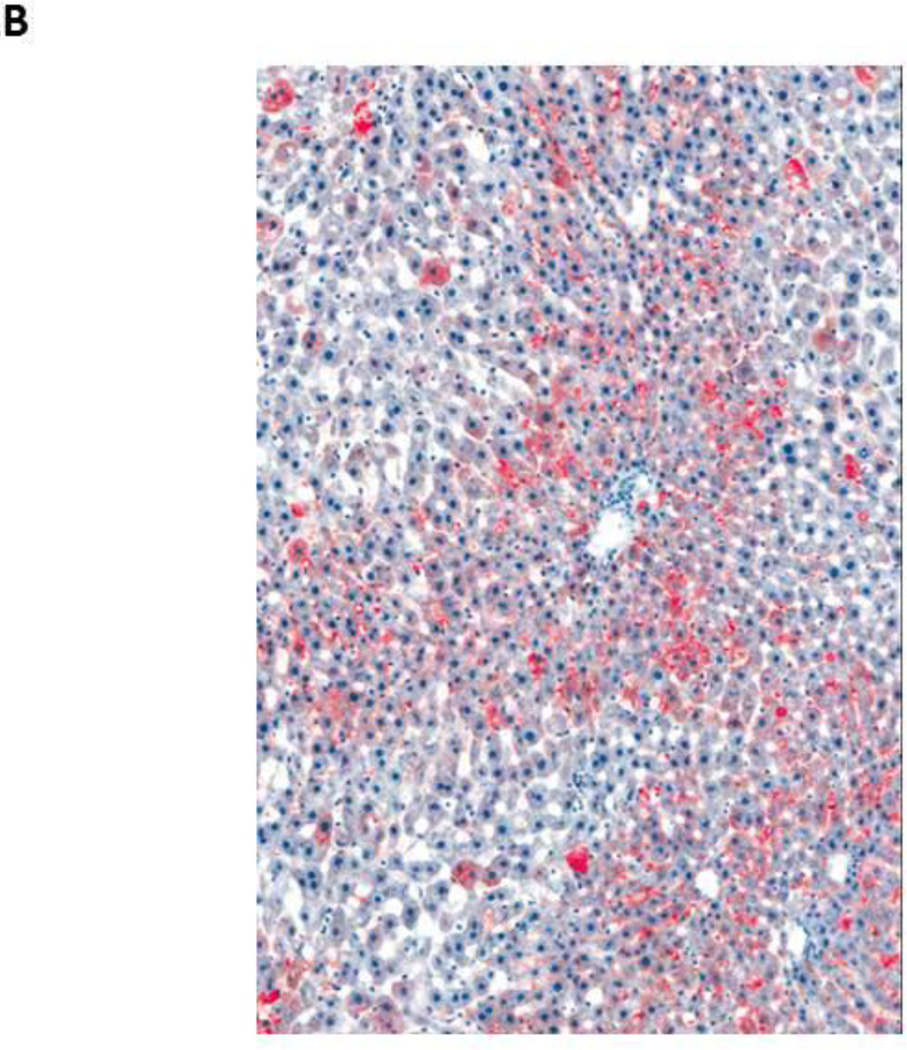
Figure 2.
Fat accumulation in PBDE treated liver at 13 weeks (Oil Red O staining of frozen liver sections)
- Male rat, control liver (10X) – normal liver
- Male rat, 50 mg/kg (10X) - Increase in the amount of fat in the liver of the treated rat, as evidenced by increased red stain (10X)
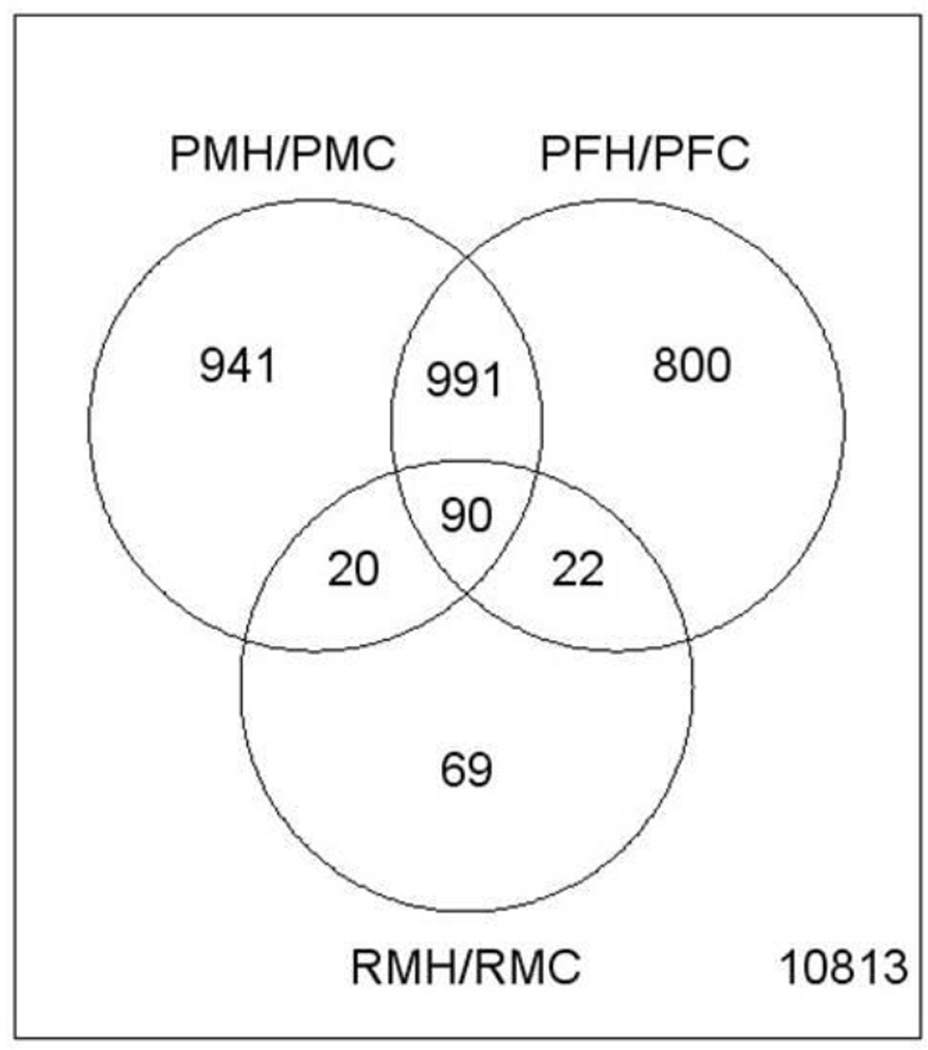
A total of 1,200 differentially expressed transcripts (DETs; control vs. treated animals) were found in male pups and 1,066 DETS were found in females, at PND 22 (FDR <0.01). The gene list size for 13 week male rats was too small to investigate at FDR < 0.01. However, at a FDR of 0.05 there were 263 DETs in male rats at week 13 (See Table 2 and Supplemental tables 1,2,3). Approximately 90 genes in common were found to be differentially expressed due to PBDE treatment (FDR <0.05) in PND 22 males, PND 22 females, and week 13 males (Figure 3; Supplement 4). Some of the transcripts that were upregulated at PND 22 and week 13 included aldh1a1, cyp1a1, abcc3, zshhx2, far4, cltb, abhd4, abhd4, ces2, zeint, fam134b, and vps26a. However, at PND 22 there were many more cytochrome P450s upregulated than at week 13.
Table 2.
Number of PBDE-induced significant liver gene transcripts
| Gene List Sizes for Given False Discovery Rate | ||||||
|---|---|---|---|---|---|---|
| 0.3 | 0.2 | 0.1 | 0.05 | 0.01 | 0.001 | |
| PBDE male pupa vs. male pup control | 9270 | 7015 | 4595 | 3055 | 1200 | 209 |
| PBDE female pupa vs. female pup control | 8178 | 6264 | 4113 | 2832 | 1066 | 109 |
| PBDE male ratb vs. male rat control | 3185 | 1748 | 611 | 263 | 22 | 0 |
| Male pup controla vs female pup control | 312 | 140 | 25 | 15 | 7 | 7 |
| PBDE male pupa vs. PBDE female pups | 39 | 26 | 19 | 11 | 9 | 5 |
| SEX-by-DOSE (pups) | 12 | 8 | 0 | 0 | 0 | 0 |
| AGE-by-DOSE (males) | 2923 | 1796 | 870 | 447 | 123 | 30 |
Figure 3.
Venn diagram of overlapping significant gene transcripts at PND 22 (males (PMH/PMC) and females (PFH/PFC)) and week 13 (males (RMH/RMC))
No sex-dependent differences in gene expression response to PBDEs were found at PND 22 (Table 2, FDR <0.01). Therefore, we used male PND 22 data to represent the PND 22 profile. With increasing time of PBDE exposures, an adaptive response to the PBDE liver toxicity was suggested by the larger number of DETs found at PND 22 (3,055 DETs FDR < 0.05) than at week 13. A total of 447 age-dependent changes in gene expression response to PBDEs were found here (FDR <0.05).
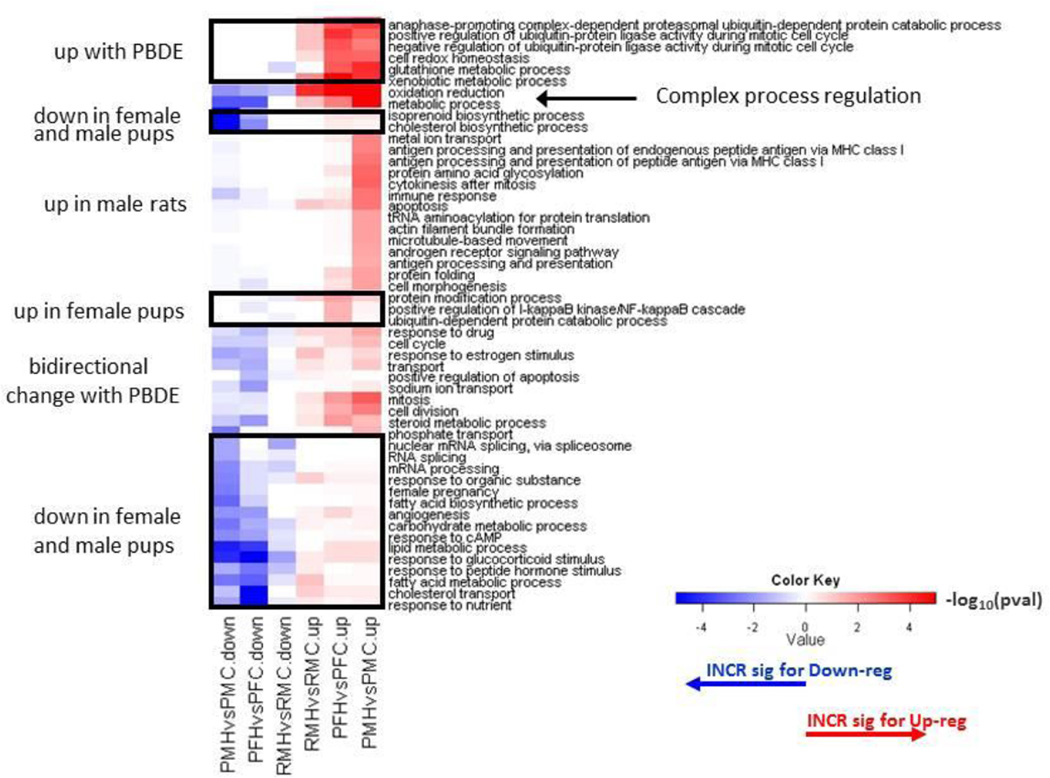
GO analysis (Supplement 5) indicated that gene transcript changes found at PND 22 (Supplement 1, 2) and week 13 (Supplement 3) are important in multiple cell functions, including oxidative and metabolic functions (Figure 4). In PND 22 male livers, there was upregulation of aldehyde dehyrogenases, aldo-keto reductases, glutathione S-transferases and carbony reductases (Table 3A). Alterations in transcripts for lipid metabolism were also significant at PND 22 (Table 3B). Many cytochrome P450 transcripts were increased in treated PND 22 pups (Table 3C). PBDE treatment induced alterations in cell cycle gene transcripts and in cancer disease pathways (Table 3D). Upregulation of the Abcc3 transcript (functions in metabolite conjugation) and glutathione S-transferase alpha 4 (Gsta4) occurred at both PND 22 and week 13.
Figure 4.
Clustered heat map of Biological Process Gene Ontology (GO) terms in upregulated (red) or downregulated (green) genes in comparison to PBDE treated (H) animals to controls (C). Differentially expressed genes for female and male pups (P) were determined by a false discovery rate (FDR) threshold of 0.01, while gene lists in 13 week old rats (R) were based on FDR < 0.05. Included processes have a p-value less than 0.05 and ≥5 differentially expressed genes in at least one comparison. Color key scale indicates the minus log10 p-value of GO term significance. Comparisons of female pups (PFHvsPFC), male pups (PMHvsPMC), 13 week old rats (RMHvsRMC).
Table 3.
Selected liver gene transcript changes in postnatal day 22 male pup livera
| AffyProbe Set ID |
Gene Symbol |
GeneName | Fold Change Male pups |
Fold change Female Pups |
|---|---|---|---|---|
|
Up regulation of genes combating oxidative stress |
||||
| “1387958_at” | “Akr1c18” | “aldo-keto reductase family 1, member C18” |
2.38 | 1.78 |
| “1371997_at” | “Akr1cl2” | “aldo-keto reductase family 1, member C-like 2” |
1.34 | 1.24 |
| “1398310_at” | “Akr1d1” | “aldo-keto reductase family 1, member D1 (delta 4–3-ketosteroid-5- beta-reductase)” |
1.42 | 1.16 |
| “1367843_at” | “Akr7a2” | “aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase)” |
1.39 | 1.46 |
| “1368121_at” | “Akr7a3” | “aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase)” |
3.04 | 2.40 |
| “1387022_at” | “Aldh1a1” | “aldehyde dehydrogenase 1 family, member A1” |
2.84 | 2.87 |
| “1368718_at” | “Aldh1a7” | “aldehyde dehydrogenase family 1, subfamily A7” |
2.78 | 2.65 |
| “1368130_at” | “Aldh3a1” | “aldehyde dehydrogenase 3 family, member A1” |
1.88 | 4.81 |
| “1368037_at” | “Cbr1” | “carbonyl reductase 1” | 1.43 | 1.52 |
| “1367774_at” | “Gsta3” | “glutathione S -transferase A3” |
1.30 | 1.22 |
| “1372297_at” | “Gsta4” | “glutathione S- transferase alpha 4” |
2.45 | 2.27 |
| “1386985_at” | “Gstm1” | “glutathione S- transferase mu 1” |
1.70 | 1.71 |
| “1370952_at” | “Gstm2” | “glutathione S- transferase mu 2” |
2.65 | 2.02 |
| “1369921_at” | “Gstm3” | “glutathione S- transferase mu 3” |
2.01 | 1.71 |
| “1375909_at” | “Gstm4” | “glutathione S- transferase mu 4” |
1.74 | 1.77 |
| “1368354_at” | “Gstt1” | “glutathione S- transferase theta 1” |
1.74 | 1.62 |
| “1371942_at” | “Gstt3” | “glutathione S- transferase, theta 3” |
1.96 | 1.73 |
| “1372599_at” | “Mgst2” | “microsomal glutathione S-transferase 2” |
1.45 | 1.46 |
|
Genes in lipid metabolism/ metabolic syndrome |
||||
| “1368126_at” | “Aacs” | “acetoacetyl-CoA synthetase” |
−3.51 | −2.70 |
| “1369455_at” | “Abcg5” | “ATP-binding cassette, sub-family G (WHITE), member 5” |
−4.57 | −4.24 |
| “1368718_at” | “Aldh1a7” | “aldehyde dehydrogenase family 1, subfamily A7” |
2.78 | 2.65 |
| “1369727_at” | “Apoa2” | “apolipoprotein A-II” | 3.64 | 3.79 |
| “1369011_at” | “Apoa5” | “apolipoprotein A-V” | −1.50 | −1.60 |
| “1393139_at” | “Apoc2” | “apolipoprotein C-II” | −1.23 | −1.11 |
| “1386901_at” | “Cd36” | “CD36 molecule (thrombospondin receptor)” |
1.66 | −1.06 |
| “1367689_a_at” | “Cd36” | “CD36 molecule (thrombospondin receptor)” |
1.26 | - |
| “1384341_at” | “Faah” | “fatty acid amide hydrolase” |
−1.52 | - |
| “1393751_at” | “Fabp12” | “fatty acid binding protein 12” |
−1.62 | - |
| “1370281_at” | “Fabp5” | “fatty acid binding protein 5, epidermal” |
−2.84 | - |
| “1370024_at” | “Fabp7” | “fatty acid binding protein 7, brain” |
−1.38 | - |
| “1368453_at” | “Fads2” | “fatty acid desaturase 2” | −2.97 | −1.94 |
| “1372476_at” | “Fads3” | “fatty acid desaturase 3” | 1.56 | - |
| “1367707_at” | “Fasn” | “fatty acid synthase” | −2.99 | - |
| “1367708_a_at” | “Fasn” | “fatty acid synthase” | −2.17 | - |
| “1373491_at” | “Gba” | “glucosidase, beta, acid” | 1.44 | 1.34 |
| “1370200_at” | “Glud1” | “glutamate dehydrogenase 1” |
−1.46 | - |
| “1373222_at” | “Hexa” | “hexosaminidase A” | 1.32 | - |
| “1375852_at” | “Hmgcr” | “3-hydroxy-3- methylglutaryl- Coenzyme A reductase” |
−1.70 | - |
| “1387848_at” | “Hmgcr” | “3-hydroxy-3- methylglutaryl- Coenzyme A reductase” |
−1.78 | - |
| “1367648_at” | “Igfbp2” | “insulin-like growth factor binding protein 2” |
−5.17 | −2.51 |
| “1367887_at” | “Lcat” | “lecithin cholesterol acyltransferase” |
−1.14 | - |
| “1381672_at” | “Ldlrap1” | “low density lipoprotein receptor adaptor protein 1” |
1.10 | - |
| “1386965_at” | “Lpl” | “lipoprotein lipase” | 7.13 | 4.71 |
| “1369150_at” | “Pdk4” | “pyruvate dehydrogenase kinase, isozyme 4” |
1.88 | - |
| “1386864_at” | “Pgam1” | “phosphoglycerate mutase 1 (brain)” |
1.25 | 1.26 |
| “1387803_at” | “Ppp2r2b” | “protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform” |
−4.83 | −6.34 |
| “1371143_at” | “Serpina7” | “serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7” |
−4.53 | −4.44 |
| “1387325_at” | “Slc27a5” | “solute carrier family 27 (fatty acid transporter), member 5” |
−1.56 | −1.49 |
| “1369636_at” | “Sord” | “sorbitol Dehydrogenase” |
1.29 | - |
| “1394786_at” | “Sorl1” | “sortilin-related receptor, LDLR class A repeats- containing” |
−1.44 | - |
| “1393933_at” | “Sorl1” | “sortilin-related receptor, LDLR class A repeats- containing” |
−1.44 | - |
| “1371104_at” | “Srebf1” | “sterol regulatory element binding transcription factor 1” |
−1.87 | - |
| “1371979_at” | “Srebf2” | “sterol regulatory element binding transcription factor 2” |
−1.31 | - |
| “1367938_at” | “Ugdh” | “UDP-glucose dehydrogenase” |
1.67 | 1.49 |
| “1389611_at” | “Vldlr” | “very low density lipoprotein receptor” |
−1.03 | 1.32 |
|
Cytochrome P450s |
||||
| 1369136_at | Cyp2a3a | cytochrome P450, family 2, subfamily A, polypeptide 3a |
37.40 | 6.84 |
| 1370269_at | Cyp1a1 | cytochrome P450, family 1, subfamily a, polypeptide 1 |
26.68 | 39.63 |
| 1370241_at | Cyp2c7 | cytochrome P450, family 2, subfamily c, polypeptide 7 |
23.68 | 9.17 |
| 1370495_s_at | Cyp2c13 | cytochrome P450 2c13 | 21.10 | - |
| 1387328_at | Cyp2c | cytochrome P450, subfamily IIC (mephenytoin 4- hydroxylase) |
6.99 | 2.45 |
| 1368990_at | Cyp1b1 | cytochrome P450, family 1, subfamily b, polypeptide 1 |
4.95 | 10.96 |
| 1370387_at | Cyp3a9 | cytochrome P450, family 3, subfamily a, polypeptide 9 |
2.95 | - |
| 1387243_at | Cyp1a2 | cytochrome P450, family 1, subfamily a, polypeptide 2 |
2.24 | 2.13 |
| 1387118_at | Cyp3a23/3a1 | cytochrome P450, family 3, subfamily a, polypeptide 23/polypeptide 1 |
1.97 | 2.03 |
| 1369424_at | Cyp2a2 | cytochrome P450, subfamily 2A, polypeptide 1 |
1.77 | - |
| 1387511_at | Cyp2a1 | cytochrome P450 IIA1 (hepatic steroid hydroxylase IIA1) gene |
1.64 | - |
| 1368155_at | Cyp2c12 | cytochrome P450, family 2, subfamily c, polypeptide 12 |
1.52 | - |
| 1387296_at | Cyp2j4 | cytochrome P450, family 2, subfamily J, polypeptide 4 |
1.46 | - |
| 1387296_at | Cyp2j4 | cytochrome P450, family 2, subfamily J, polypeptide 4 |
1.46 | - |
| 1387973_at | Cyp4f4 | cytochrome P450, family 4, subfamily f, polypeptide 4 |
1.28 | - |
| 1371142_at | Cyp2g1 | cytochrome P450, subfamily 2G, polypeptide 1 |
−1.03 | - |
| 1368458_at | Cyp7a1 | cytochrome P450, family 7, subfamily a, polypeptide 1 |
−2.30 | - |
| 1368435_at | Cyp8b1 | cytochrome P450, family 8, subfamily b, polypeptide 1 |
−2.93 | −4.20 |
| 1387123_at | Cyp17a1 | cytochrome P450, family 17, subfamily a, polypeptide 1 |
−6.16 | −5.23 |
| 1371076_at | Cyp2b | cytochrome P450, family 2b |
6.13 | 9.96 |
|
Cell cycle/cancer |
||||
| “1369698_at” | “Abcc3” | “ATP-binding cassette, sub-family C (CFTR/MRP), member 3” |
3.23 | 3.23 |
| “1379652_at” | “Ahctf1” | “AT hook containing transcription factor 1” |
−1.54 | - |
| “1379652_at” | “Ahctf1” | “AT hook containing transcription factor 1” |
−1.54 | - |
| “1369983_at” | “Ccl5” | “chemokine (C-C motif) ligand 5” |
2.21 | 2.02 |
| “1370345_at” | “Ccnb1” | “cyclin B1” | 1.94 | - |
| “1389566_at” | “Ccnb2” | “cyclin B2” | 1.78 | - |
| “1371643_at” | “Ccnd1” | “cyclin D1” | −2.13 | - |
| “1371150_at” | “Ccnd1” | “cyclin D1” | −2.28 | −1.94 |
| “1383075_at” | “Ccnd1” | “cyclin D1” | −2.33 | −2.1 |
| “1370810_at” | “Ccnd2” | “cyclin D2” | −1.42 | - |
| “1371953_at” | “Ccng2” | “cyclin G2” | 2.33 | - |
| “1380997_at” | “Ccnj” | “cyclin J” | −1.51 | - |
| “1374220_at” | “Ccny” | “cyclin Y” | 1.33 | - |
| “1372685_at” | “Cdkn3” | “cyclin-dependent kinase inhibitor 3” |
1.79 | - |
| “1387062_a_at” | “Chek1” | “CHK1 checkpoint homolog (S. pombe)” |
2.90 | 3.89 |
| “1367515_at” | “Cnot7” | “CCR4-NOT transcription complex, subunit 7” |
−1.31 | - |
| “1367515_at” | “Cnot7” | “CCR4-NOT transcription complex, subunit 7” |
−1.31 | - |
| “1368174_at” | “Egln3” | “EGL nine homolog 3 (C. elegans)” |
4.43 | 4.89 |
| “1373011_at” | “Fam134b” | “family with sequence similarity 134, member B” |
4.34 | 3.87 |
| “1368408_at” | “Grk5” | “G protein-coupled receptor kinase 5” |
3.28 | 2.95 |
| “1368549_at” | “Hbp1” | “HMG-box transcription factor 1” |
1.30 | - |
| “1368549_at” | “Hbp1” | “HMG-box transcription factor 1” |
1.30 | - |
| “1370728_at” | “Il13ra1” | “interleukin 13 receptor, alpha 1” |
1.73 | - |
| “1388711_at” | “Il13ra1” | “interleukin 13 receptor, alpha 1” |
1.47 | - |
| “1371170_a_at” | “Il1a” | “interleukin 1 alpha” | −1.37 | −1.59 |
| “1368592_at” | “Il1a” | “interleukin 1 alpha” | −1.75 | - |
| “1387394_at” | “Il2rb” | “interleukin 2 receptor, beta” |
1.50 | - |
| “1387935_at” | “Il3ra” | “interleukin 3 receptor, alpha” |
1.39 | 1.34 |
| “1386987_at” | “Il6ra” | “interleukin 6 receptor, alpha” |
−1.33 | - |
| “1373379_at” | “Irak1” | “interleukin-1 receptor- associated kinase 1” |
1.32 | - |
| “1380336_at” | “Irak3” | “interleukin-1 receptor- associated kinase 3” |
1.50 | - |
| “1368915_at” | “Kmo” | “kynurenine 3- Monooxygenase (kynurenine 3- hydroxylase)” |
−1.54 | −0.02 |
| “1371479_at” | “Mettl7a” | “methyltransferase like 7A” |
1.90 | 1.54 |
| “1384280_at” | “Nusap1” | “nucleolar and spindle associated protein 1” |
1.59 | - |
| “1392715_at” | “Ppargc1b” | “peroxisome proliferator-activated receptor gamma, coactivator 1 beta” |
−1.82 | - |
| “1370012_at” | “Ptgis” | “prostaglandin I2 (prostacyclin) synthase” |
−3.10 | −3.19 |
| “1398814_at” | “Rab11a” | “RAB11a, member RAS oncogene family” |
1.40 | 1.37 |
| “1389066_at” | “Rcan2” | “regulator of calcineurin 2” |
2.10 | 2.15 |
| “1378605_at” | “Sigirr” | “single immunoglobulin and toll-interleukin 1 receptor (TIR) domain” |
1.32 | - |
| “1379275_at” | “Snx10” | “sorting nexin 10” | 3.96 | 5.49 |
| “1371104_at” | “Srebf1” | “sterol regulatory element binding transcription factor 1” |
−1.87 | - |
| “1388426_at” | “Srebf1” | “sterol regulatory element binding transcription factor 1” |
−1.94 | - |
| “1367941_at” | “Tfam” | “transcription factor A, mitochondrial” |
1.04 | - |
| “1376668_at” | “Ttc39a” | “tetratricopeptide repeat domain 39A” |
8.84 | 14.87 |
False discovery rate < 0.01; p < .01 for all gene transcripts in this table (complete details in supplements 1 and 2). Most of these gene transcripts were not significantly changed in week13 male rat livers.
After scrutinizing the expression pattern of important genes (described above), PND 22 transcript data were subjected to a global bioinformatics analysis using NextBio software to compare the expression profiles obtained in this study to expression profiles found in the literature. PBDE liver gene transcript patterns correlated with other rodent studies on high fat diets including upregulation of cyp2c7, ces2, lpl, cyp2c, cyp2b2 (Almon et al. 2009; van Erk 2011). PBDE transcript patterns correlated with liver transcript patterns after phenobarbital exposure (rats) (Supplement 6), with 67 upregulated transcripts and 164 downregulated transcripts that overlapped those in phenobarbital treated rats including upregulation (>4 fold) of transcripts for P450, chemokine, carboxyesterase, and growth arrest activity (cyp1a1, cxcl9, ces2, zdhhc2, lpl, cyp2b2, rn71071, cyp1b1, egin3, and gadd45b).
PBDE transcript patterns were similar to some of the changes found in human cancers including colon cancers (Supplement 7). Transcripts upregulated in human colon cancer and after PBDE treatment included cyp2b6, fam134b, chek1, rcan2, ccng2, ccl5, and acacb.
Discussion
In utero/postnatal exposure of Wistar Han rats starting on GD 6 (start of organogenesis) to a mixture of low molecular weight polybrominated diphenyl ethers (PBDEs) resulted in liver toxicity characterized by hepatocyte hypertrophy and hepatocyte cytoplasmic vacuolization. This liver toxicity in the Wistar Han rat is similar to that observed in the F344/N rat after PBDE exposures (Dunnick and Nyska 2009). Hepatocellular vacuolization, as seen in PBDE rat livers, is often indicative of accumulation of fat deposits (Eustis et al. 1990). Non-alcoholic fatty liver is a hepatic manifestation of the metabolic syndrome (Lucero et al. 2010), and a common cause of liver enzyme elevation in humans and in rats (Stickel and Hellerbrand 2010).
The occurrence of hepatocellular hypertrophy and vacuolization may predispose rats to liver cancer. However, not all exposures that result in these liver histopathologic features lead to liver cancer (Maronpot et al. 2010). From a review of NTP chemical study findings at 90-days, only 45% of rat studies with chemical-induced liver hypertrophy went on to be rat liver carcinogens in 2-year cancer studies (Allen et al. 2004). Thus, there is a need for additional biomarkers to predict disease.
PBDE induced liver gene transcript changes that modify cell pathways and functions that, along with the liver toxicity, may lead to disease susceptibilities. This includes alterations in lipid and metabolic pathways. For instance at PND 22, PBDE increased the transcript for lipoprotein lipase 7-fold versus that in untreated liver. An increase in lipoprotein lipase activity has been associated with diabetes and metabolic diseases (Lopaschuk et al. 2010). PBDE decreased the cyp7a transcript which codes for an enzyme critical in metabolism of cholesterol to bile acids and in controlling hepatic triglyceride levels (Lee et al. 2010). The PBDE transcript patterns reported here, support other findings which show that PBDE exposures lead to increased body weights in rodents and alterations in adipocyte metabolism (Hoppe and Carey 2007; Suvorov and Takser 2010).
Thyroid homeostasis is important in preventing metabolic, reproductive, developmental and cardiac disease (Choksi et al. 2003; Jahnke et al. 2004; Miller et al. 2009; Pedrelli et al. 2010). PBDE-induced alterations in thyroid hormone levels (Dunnick and Nyska 2009; Richardson et al. 2008) may affect mitochondria biogenesis (Leigh-Brown et al. 2010) or alter mitochondria based TCA cycle and lipid distribution (Harper and Seifert 2008). Liver gene transcript analysis in the current study supports a finding of altered lipid metabolism. A PBDE-induced effect on lipid metabolism along with PBDE-induced endocrine effects may create susceptibilities to metabolic diseases.
The PBDE induced liver toxicity was accompanied by increases in liver P450 transcripts including those for cyp2a3a, cyp1a1 cyp1b1, cyp2c7, cyp2c13, cyt2c, cyp2b, coding for enzymes critical in metabolism of chemicals, drugs, fatty acids, and steroids (Yang et al. 2010). Up regulation of these cytochrome P450 liver enzymes has been found with other chemicals causing fatty liver (steatosis) (Lee et al. 2008; Lee, Kim, Kim, Kang, Kong and Lee 2010), and is associated with the development of carcinogenic events (Rendic and Guengerich 2010). An adaptive response to the PBDE liver toxicity was suggested by fewer significant liver gene transcript changes at week 13 than at PND 22, and induced liver transcripts for enzymes involved in decreasing aldehyde loads (Ellis 2007) and this may be part of an adaptive response to toxicity.
PBDE and dioxin exposures are both associated with increases in liver P450 enzyme levels (Boverhof et al. 2006a; Boverhof et al. 2006b; N’Jai et al. 2008; Szabo et al. 2009); (Sanders, Burka, Smith, Black, James and Cunningham 2005), and may involve activation of AH, PXR, or CAR receptors (Szabo, Richardson, Ross, Diliberto, Kodavanti and Birnbaum 2009). The PBDE mixture used in our studies contained a low level of brominated dioxins and furans that by themselves would not be expected to cause cancers (Dunnick and Nyska 2009), but in combination with the PBDEs may have synergistic carcinogenic effects. Whether that occurred in our studies would require further investigation.
Other studies report the PBDE exposures increase expression of genes coding for liver enzymes, including cyp2B, hepatic deiodinase 1 protein (D1) activities and cyp2b1, d1, and hepatic efflux transporter gene expression (mdr1, mrp2, mrp3) in rats at PND4, PND21, and PND60 (Szabo, Richardson, Ross, Diliberto, Kodavanti and Birnbaum 2009). Not all of these liver gene transcripts were upregulated in our PBDE studies, (although a different strain of rat (Long Evans rat) was used in the Szabo et al (2009) studies).
The liver gene expression profile generated by PBDE at PND 22 was similar to the transcriptional profile produced by phenobarbital in rodents ((Nesnow et al. 2009; Waterman, Currie, Cottrell, Dow, Wright, Waterfield and Griffin 2010), a chemical that induces liver tumors in rodents (Butler 1978; Imaida and Wang 1986; Rossi et al. 1977). The changes include transcript abundance increases for P450s, chemokines, carboxylesterases, and growth factor arrest (gadd45b) gene transcript levels, and were similar to liver gene transcript patterns found in human liver cancers (Frau et al. 2010; Tanaka and Arii 2010; Teoh 2009).
Liver genotoxic carcinogens are characterized by a specific liver fingerprint after 14 days or 13 weeks of exposure (upregulaton of mybl2, cyp1A1, and adam8 and downregulation of wwox, fhit, RGD1561899) (Auerbach et al. 2009). This “genotoxic chemical” fingerprint was not seen with PBDE exposure, except for the upregulation of cyp1A1. Based on these findings, the PBDE liver toxic mechanisms appear to differ from those of genotoxic liver toxins.
PBDE liver transcript patterns include those found in metabolic disease conditions. Metabolic disease conditions may contribute to the development of cancer (e.g. colorectal or uterine cancer) or heart disease particularly when occurring in combination with other underlying genetic conditions or environmental exposures (Lopaschuk, Ussher, Folmes, Jaswal and Stanley 2010).
PBDE liver gene transcript patterns overlapped those seen in human cancer (colon) including upregulation of cyclins (Koliadi et al. 2010) and other cell cycle controls (Stephens et al. 2011). In other studies, we reported that brominated chemicals may have a particular propensity to cause intestinal tumors (Dunnick et al. 1997a). Bromide radical formation in the intestine resulting in DNA damage (Ballmaier and Epe 2006) is one hypothesis for the cause of a brominated chemical carcinogenic response in the intestine, and additional studies would be needed to determine if this cancer occurs after long-term PBDE exposures.
PBDE treatment decreased expression of cyp17a1, a decrease that is associated with blocking androgen production (Hofland et al. 2010) and altering sex steroidogenesis (Canton et al. 2006). PBDE transcript patterns associated with alterations in the endocrine system are consistent with the ability of PBDEs to cause MCF7 cell proliferation (Darnerud 2008; Mercado-Feliciano and Bigsby 2008a, b; Song et al. 2008; Talsness et al. 2008). In addition, PBDE is an ER alpha and beta agonist in human T47D breast cancer cell lines (Meerts, Letcher, Hoving, Marsh, Bergman, Lemmen, van der Burg and Brouwer 2001).
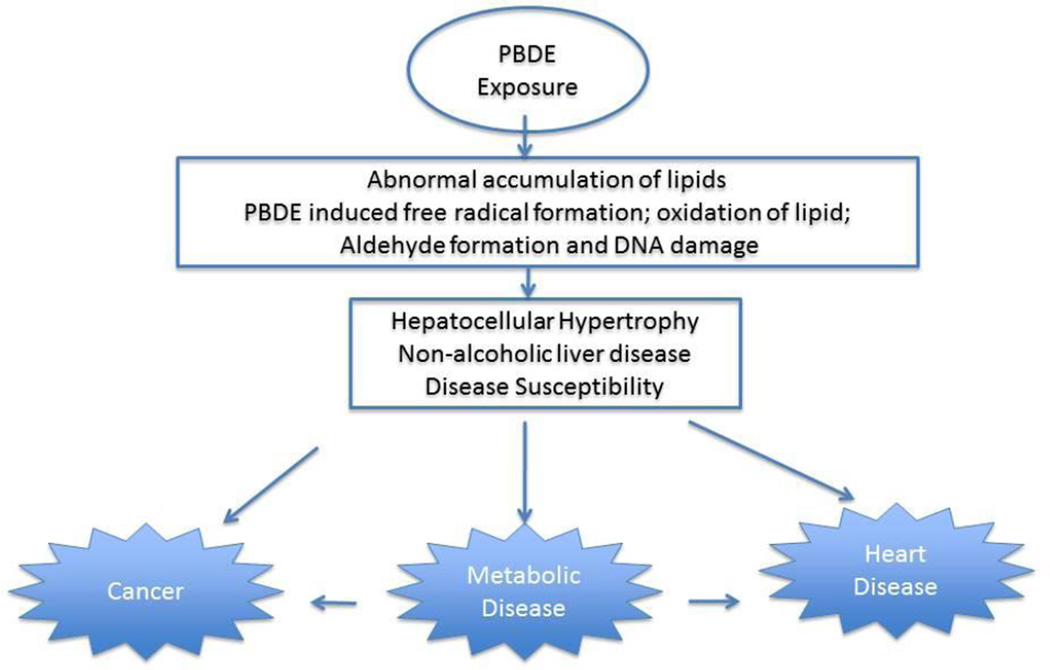
PBDE and its metabolites (e. g. 6-OH-BDE47) may increase reactive oxygen species leading to lipid peroxidation, aldehyde formation and DNA adduct formation (An et al. 2011; Jin et al. 2010). For these reasons, PBDE exposures could contribute to the develop of carcinogenesis processes as suggested by our findings (Figure 5).
Figure 5.
Steps in PBDE toxicity process
The liver gene transcript changes observed in this study suggest that PBDE exposures could lead to disease susceptibilities including those for metabolic/lipid disease (e.g. lipoprotein lipase (lpl)), cancer (e. g. cell cycle control transcripts (cyclins) or chemokine transcripts), and sex hormone synthesis (cyp17).
Supplementary Material
Supplement 1. Liver gene transcript changes – PND 22 males
Supplement 2. Liver gene transcript changes – PND 22 females
Supplement 3. Liver gene transcript changes – week 13 males
Supplement 4. PBDE significant genes at PND 22 and week 13
Supplement 5. GO Analysis
Supplement 6. PBDE PND 22 male liver gene transcripts versus phenobarbital gene transcript changes (developed from Nextbio database) 16
Supplement 7. PBDE PND 22 male liver gene transcripts versus human colon cancer genes (developed from Nextbio database)
Acknowledgments
This work was supported by the intramural program of the National Institute of Environmental Health Sciences (NIEHS), Research Triangle Park, NC. However, the statements, opinions or conclusions contained therein do not necessarily represent the statements, opinions or conclusions of NIEHS, NIH or the United States government. The in-life portion of this study was conducted under the supervision of Dr. C. Hébert, Southern Research Institute, under NIEHS contract NO1-ES-45516 and the microarray analysis under contract N01-ES-55536. Statistical support was provided under NIEHS contract ES 55547. We thank G. Kissling, A. Merrick, and M. Sanders, NIEHS, for their review of the manuscript.
Abbreviations in supplements
- PMH
PND 22 PBDE treated male pups
- PMC
PND 22 control male pups
- PFH
PND 22 PBDE treated female pups
- PFC
PND 22 control female pups
- RMH
week 13 PBDE treated males
- RMC
week 13 control males
References
- Agency for Toxic Substances and Disease Registry. Toxicological Profile for Polybrominated Biphenyls and Polybrominated Diphenyl Ethers (PBBs and PBDEs) 2004 http://www.atsdr.cdc.gov/toxprofiles/tp.asp?id=529&tid=94. [PubMed]
- Allen DG, Pearse G, Haseman JK, Maronpot RR. Prediction of rodent carcinogenesis: an evaluation of prechronic liver lesions as forecasters of liver tumors in NTP carcinogenicity studies. Toxicol Pathol. 2004;32:393–401. doi: 10.1080/01926230490440934. [DOI] [PubMed] [Google Scholar]
- Almon RR, DuBois DC, Lai W, Xue B, Nie J, Jusko WJ. Gene expression analysis of hepatic roles in cause and development of diabetes in Goto-Kakizaki rats. J Endocrinol. 2009;200:331–346. doi: 10.1677/JOE-08-0404. [DOI] [PubMed] [Google Scholar]
- An J, Li S, Zhong Y, Wang Y, Zhen K, Zhang X, Wu M, Yu Z, Sheng G, Fu J, Huang Y. The cytotoxic effects of synthetic 6-hydroxylated and 6-methoxylated polybrominated diphenyl ether 47 (BDE47) Environ Toxicol. 2011:26. doi: 10.1002/tox.20582. [DOI] [PubMed] [Google Scholar]
- Ancona N, Maglietta R, Piepoli A, D’Addabbo A, Cotugno R, Savino M, Liuni S, Carella M, Pesole G, Perri F. On the statistical assessment of classifiers using DNA microarray data. BMC Bioinformatics. 2006;7:387. doi: 10.1186/1471-2105-7-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Shah RR, Mav D, Smith CS, Walker NJ, Vallant MK, Boorman GA, Irwin RD. Predicting the hepatocarcinogenic potential of alkenylbenzene flavoring agents using toxicogenomics and machine learning. Toxicol Appl Pharmacol. 2009;243:300–314. doi: 10.1016/j.taap.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Baker JL, Olsen LW, Sorensen TI. Childhood body-mass index and the risk of coronary heart disease in adulthood. N Engl J Med. 2007;357:2329–2337. doi: 10.1056/NEJMoa072515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballmaier D, Epe B. DNA damage by bromate: mechanism and consequences. 2006 doi: 10.1016/j.tox.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Birnbaum LS, Cohen Hubal EA. Polybrominated diphenyl ethers: a case study for using biomonitoring data to address risk assessment questions. Environ Health Perspect. 2006;114:1770–1775. doi: 10.1289/ehp.9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Sharratt B, Chittim B, Harkema JR, Mendrick DL, Zacharewski TR. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol Sci. 2006a;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Kwekel JC, Humes DG, Burgoon LD, Zacharewski TR. Dioxin induces an estrogen-like, estrogen receptor-dependent gene expression response in the murine uterus. Mol Pharmacol. 2006b;69:1599–1606. doi: 10.1124/mol.105.019638. [DOI] [PubMed] [Google Scholar]
- Butler WH. Long-term effects of phenobarbitone-Na on male Fischer rats. Br J Cancer. 1978;37:418–423. doi: 10.1038/bjc.1978.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton RF, Sanderson JT, Nijmeijer S, Bergman A, Letcher RJ, van den Berg M. In vitro effects of brominated flame retardants and metabolites on CYP17 catalytic activity: a novel mechanism of action? Toxicol Appl Pharmacol. 2006;216:274–281. doi: 10.1016/j.taap.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Choksi NY, Jahnke GD, St Hilaire C, Shelby M. Role of thyroid hormones in human and laboratory animal reproductive health. Birth Defects Res B Dev Reprod Toxicol. 2003;68:479–491. doi: 10.1002/bdrb.10045. [DOI] [PubMed] [Google Scholar]
- Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- Darnerud PO. Brominated flame retardants as possible endocrine disrupters. Int J Androl. 2008;31:152–160. doi: 10.1111/j.1365-2605.2008.00869.x. [DOI] [PubMed] [Google Scholar]
- Darnerud PO, Aune M, Larsson L, Hallgren S. Plasma PBDE and thyroxine levels in rats exposed to Bromkal or BDE-47. Chemosphere. 2007;67:S386–S392. doi: 10.1016/j.chemosphere.2006.05.133. [DOI] [PubMed] [Google Scholar]
- Dunnick J, Heath J, Farnell D, Prejean J, Haseman J, Elwell MR. Carcinogenic activity of the flame retardant, 2,2-bis(bromomethyl)-1,3-propanediol in rodents, and comparison with the carcinogenicity of other NTP brominated chemicals. Toxicol Pathol. 1997a;25:541–548. doi: 10.1177/019262339702500602. [DOI] [PubMed] [Google Scholar]
- Dunnick JK, Heath JE, Farnell DR, Prejean JD, Haseman JK, Elwell MR. Carcinogenic activity of the flame retardant, 2,2-bis(bromomethyl)-1,3-propanediol in rodents, and comparison with the carcinogenicity of other NTP brominated chemicals. Toxicol Pathol. 1997b;25:541–548. doi: 10.1177/019262339702500602. [DOI] [PubMed] [Google Scholar]
- Dunnick JK, Nyska A. Characterization of liver toxicity in F344/N rats and B6C3F1 mice after exposure to a flame retardant containing lower molecular weight polybrominated diphenyl ethers. Exp Toxicol Pathol. 2009;61:1–12. doi: 10.1016/j.etp.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis EM. Reactive carbonyls and oxidative stress: potential for therapeutic intervention. Pharmacol Ther. 2007;115:13–24. doi: 10.1016/j.pharmthera.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Ellis-Hutchings RG, Cherr GN, Hanna LA, Keen CL. Polybrominated diphenyl ether (PBDE)-induced alterations in vitamin A and thyroid hormone concentrations in the rat during lactation and early postnatal development. Toxicol Appl Pharmacol. 2006;215:135–145. doi: 10.1016/j.taap.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Ervin RB. Prevalence of metabolic syndrome among adults 20 years of age and over, by sex, age, race and ethnicity, and body mass index: United States, 2003–2006. Natl Health Stat Report. 2009:1–7. [PubMed] [Google Scholar]
- Eustis SL, Boorman GA, Harada T, Popp JA. Liver. In: Boorman GA, Eustis SL, Elwell MR, Montgomery CA Jr, MacKenzie WF, editors. Pathology of the Fischer rat. New York: Academic Press, Inc; 1990. pp. 78–81. [Google Scholar]
- Frau M, Biasi F, Feo F, Pascale RM. Prognostic markers and putative therapeutic targets for hepatocellular carcinoma. Mol Aspects Med. 2010;31:179–193. doi: 10.1016/j.mam.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Frederiksen M, Thomsen C, Froshaug M, Vorkamp K, Thomsen M, Becher G, Knudsen LE. Polybrominated diphenyl ethers in paired samples of maternal and umbilical cord blood plasma and associations with house dust in a Danish cohort. Int J Hyg Environ Health. 2010 doi: 10.1016/j.ijheh.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Hale RC, Alaee M, Manchester-Neesvig JB, Stapleton HM, Ikonomou MG. Polybrominated diphenyl ether flame retardants in the North American environment. Environ Int. 2003;29:771–779. doi: 10.1016/S0160-4120(03)00113-2. [DOI] [PubMed] [Google Scholar]
- Hale RC, La Guardia MJ, Harvey E, Mainor TM. Potential role of fire retardant-treated polyurethane foam as a source of brominated diphenyl etheres to the U. S. environment. Chemosphere. 2002;46:729–735. doi: 10.1016/s0045-6535(01)00237-5. [DOI] [PubMed] [Google Scholar]
- Hale RC, La Guardia MJ, Harvey EP, Gaylor MO, Mainor TM, Duff WH. Persistant pollutants in land-applied sludges. Nature. 2001;412:140–141. doi: 10.1038/35084130. [DOI] [PubMed] [Google Scholar]
- Hamers T, Kamstra JH, Sonneveld E, Murk AJ, Kester MH, Andersson PL, Legler J, Brouwer A. In vitro profiling of the endocrine-disrupting potency of brominated flame retardants. Toxicol Sci. 2006;92:157–173. doi: 10.1093/toxsci/kfj187. [DOI] [PubMed] [Google Scholar]
- Harley KG, Marks AR, Chevrier J, Bradman A, Sjodin A, Eskenazi B. PBDE concentrations in women’s serum and fecundability. Environ Health Perspect. 2010;118:699–704. doi: 10.1289/ehp.0901450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper ME, Seifert EL. Thyroid hormone effects on mitochondrial energetics. Thyroid. 2008;18:145–156. doi: 10.1089/thy.2007.0250. [DOI] [PubMed] [Google Scholar]
- Harrad S, Goosey E, Desborough J, Abdallah MA, Roosens L, Covaci A. Dust from U.K. Primary School Classrooms and Daycare Centers: The Significance of Dust As a Pathway of Exposure of Young U.K. Children to Brominated Flame Retardants and Polychlorinated Biphenyls. Environ Sci Technol. 2010;44:4198–4202. doi: 10.1021/es100750s. [DOI] [PubMed] [Google Scholar]
- Hofland J, van Weerden WM, Dits NF, Steenbergen J, van Leenders GJ, Jenster G, Schroder FH, de Jong FH. Evidence of limited contributions for intratumoral steroidogenesis in prostate cancer. Cancer Res. 2010;70:1256–1264. doi: 10.1158/0008-5472.CAN-09-2092. [DOI] [PubMed] [Google Scholar]
- Hoppe AA, Carey GB. Polybrominated diphenyl ethers as endocrine disruptors of adipocyte metabolism. Obesity (Silver Spring) 2007;15:2942–2950. doi: 10.1038/oby.2007.351. [DOI] [PubMed] [Google Scholar]
- Imaida K, Wang CY. Effect of sodium phenobarbital and sodium saccharin in AIN-76A diet on carcinogenesis initiated with N-[4-(5-nitro-2-furyl)-2-thiazolyl]formamide and N,N-dibutylnitrosamine in male F344 rats. Cancer Res. 1986;46:6160–6164. [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke GD, Choksi NY, Moore JA, Shelby MD. Thyroid toxicants: assessing reproductive health effects. Environ Health Perspect. 2004;112:363–368. doi: 10.1289/ehp.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Yang F, Hui Y, Xu Y, Lu Y, Liu J. Cytotoxicity and apoptosis induction on RTG-2 cells of 2,2’,4,4’-tetrabromodiphenyl ether (BDE-47) and decabrominated diphenyl ether (BDE-209) Toxicol In Vitro. 2010;24:1190–1196. doi: 10.1016/j.tiv.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Johnson PI, Stapleton HM, Sjodin A, Meeker JD. Relationships between Polybrominated Diphenyl Ether Concentrations in House Dust and Serum. Environ Sci Technol. 2010 doi: 10.1021/es100697q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorissen RN, Lipton L, Gibbs P, Chapman M, Desai J, Jones IT, Yeatman TJ, East P, Tomlinson IP, Verspaget HW, Aaltonen LA, Kruhoffer M, Orntoft TF, Andersen CL, Sieber OM. DNA copy-number alterations underlie gene expression differences between microsatellite stable and unstable colorectal cancers. Clin Cancer Res. 2008;14:8061–8069. doi: 10.1158/1078-0432.CCR-08-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliadi A, Nilsson C, Holmqvist M, Holmberg L, de La Torre M, Warnberg F, Fjallskog ML. Cyclin B is an immunohistochemical proliferation marker which can predict for breast cancer death in low-risk node negative breast cancer. Acta Oncol. 2010;49:816–820. doi: 10.3109/02841861003691937. [DOI] [PubMed] [Google Scholar]
- Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, Shekar M, Wang H, Park J, Cui W, Wall GD, Wisotzkey R, Alag S, Akhtari S, Ronaghi M. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One. 2010:5. doi: 10.1371/journal.pone.0013066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Hong I, Kim M, Lee BH, Kim JH, Kang KS, Kim HL, Yoon BI, Chung H, Kong G, Lee MO. Gene expression profiles of murine fatty liver induced by the administration of methotrexate. Toxicology. 2008;249:75–84. doi: 10.1016/j.tox.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Lee MH, Kim JW, Kim JH, Kang KS, Kong G, Lee MO. Gene expression profiling of murine hepatic steatosis induced by tamoxifen. Toxicol Lett. 2010;199:416–424. doi: 10.1016/j.toxlet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Legler J, Brouwer A. Are brominated flame retardants endocrine disruptors? Environ Int. 2003;29:879–885. doi: 10.1016/S0160-4120(03)00104-1. [DOI] [PubMed] [Google Scholar]
- Leigh-Brown S, Enriquez JA, Odom DT. Nuclear transcription factors in mammalian mitochondria. Genome Biol. 2010;11:215. doi: 10.1186/gb-2010-11-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JS, Lee DH, Jacobs DR., Jr Association of brominated flame retardants with diabetes and metabolic syndrome in the U.S. population, 2003–2004. Diabetes Care. 2008;31:1802–1807. doi: 10.2337/dc08-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiol Rev. 2010;90:207–258. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- Lorber M. Exposure of Americans to polybrominated diphenyl ethers. J Expo Sci Environ Epidemiol. 2008;18:2–19. doi: 10.1038/sj.jes.7500572. [DOI] [PubMed] [Google Scholar]
- Lucero D, Zago V, Lopez GI, Graffigna M, Lopez GH, Fainboim H, Miksztowicz V, Rosso LG, Belli S, Levalle O, Berg G, Brites F, Wikinski R, Schreier L. Does non-alcoholic fatty liver impair alterations of plasma lipoproteins and associated factors in metabolic syndrome? Clin Chim Acta. 2010 doi: 10.1016/j.cca.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Maronpot R, Yoshizawa K, Nyska A, Harada T, Flake G, Muyeller G, Singh B, WArd J. Hepatic enzyme induction: histopathology. Toxicol Path in press. 2010 doi: 10.1177/0192623310373778. [DOI] [PubMed] [Google Scholar]
- Meerts IA, Letcher RJ, Hoving S, Marsh G, Bergman A, Lemmen JG, van der Burg B, Brouwer A. In vitro estrogenicity of polybrominated diphenyl ethers, hydroxylated PDBEs, and polybrominated bisphenol A compounds. Environ Health Perspect. 2001;109:399–407. doi: 10.1289/ehp.01109399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Feliciano M, Bigsby RM. Hydroxylated metabolites of the polybrominated diphenyl ether mixture DE-71 are weak estrogen receptor-alpha ligands. Environ Health Perspect. 2008a;116:1315–1321. doi: 10.1289/ehp.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Feliciano M, Bigsby RM. The polybrominated diphenyl ether mixture DE-71 is mildly estrogenic. Environ Health Perspect. 2008b;116:605–611. doi: 10.1289/ehp.10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Crofton KM, Rice DC, Zoeller RT. Thyroid-disrupting chemicals: interpreting upstream biomarkers of adverse outcomes. Environ Health Perspect. 2009;117:1033–1041. doi: 10.1289/ehp.0800247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N’Jai A, Boverhof DR, Dere E, Burgoon LD, Tan YS, Rowlands JC, Budinsky RA, Stebbins KE, Zacharewski TR. Comparative temporal toxicogenomic analysis of TCDD- and TCDF-mediated hepatic effects in immature female C57BL/6 mice. Toxicol Sci. 2008;103:285–297. doi: 10.1093/toxsci/kfn053. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program. Bulk chemical comprehensive analysis report - technical pentabromodiphenyl oxide. Battelle Report 7-173-BCCA-24, December 31, 2003. 2004 [Google Scholar]
- National Toxicology Program. Three-Month Toxicity Study of a Pentabromodiphenyl Oxide Mixture (DE-71) Administered by Gavage to B6C3FI Mice. Southern Research Institute Contract Report, Study number C20209, 11052.01.02 NTP Contract No. No1-ES-45516. 2005a [Google Scholar]
- National Toxicology Program. Three-Month Toxicity Study of a Pentabromodiphenyl Oxide Mixture (DE-71) Administered by Gavage to Fischer-344 Rats. Southern Research Institute Contract Report, Study number C20209, 11052.01.02 NTP Contract No. N01-ES-45516. 2005b [Google Scholar]
- Nesnow S, Ward W, Moore T, Ren H, Hester SD. Discrimination of tumorigenic triazole conazoles from phenobarbital by transcriptional analyses of mouse liver gene expression. Toxicol Sci. 2009;110:68–83. doi: 10.1093/toxsci/kfp076. [DOI] [PubMed] [Google Scholar]
- Pedrelli M, Pramfalk C, Parini P. Thyroid hormones and thyroid hormone receptors: Effects of thyromimetics on reverse cholesterol transport. World J Gastroenterol. 2010;16:5958–5964. doi: 10.3748/wjg.v16.i47.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreas M, She J, Brown FR, Winkler J, Windham G, Rogers E, Zhao G, Bhatia R, Charles MJ. High body burdens of 2,2’,4,4’-tetrabromodiphenyl ether (BDE-47) in California women. Environ Health Perspect. 2003;111:1175–1179. doi: 10.1289/ehp.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendic S, Guengerich FP. Update information on drug metabolism systems--2009, part I. Curr Drug Metab. 2010;11:4–84. doi: 10.2174/138920010791110908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson VM, Staskal DF, Ross DG, Diliberto JJ, DeVito MJ, Birnbaum LS. Possible mechanisms of thyroid hormone disruption in mice by BDE 47, a major polybrominated diphenyl ether congener. Toxicol Appl Pharmacol. 2008;226:244–250. doi: 10.1016/j.taap.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Rossi L, Ravera M, Repetti G, Santi L. Long-term administration of DDT or phenobarbital-Na in Wistar rats. Int J Cancer. 1977;19:179–185. doi: 10.1002/ijc.2910190207. [DOI] [PubMed] [Google Scholar]
- Roze E, Meijer L, Bakker A, Van Braeckel KN, Sauer PJ, Bos AF. Prenatal exposure to organohalogens, including brominated flame retardants, influences motor, cognitive, and behavioral performance at school age. Environ Health Perspect. 2009;117:1953–1958. doi: 10.1289/ehp.0901015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saaf AM, Halbleib JM, Chen X, Yuen ST, Leung SY, Nelson WJ, Brown PO. Parallels between global transcriptional programs of polarizing Caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer. Mol Biol Cell. 2007;18:4245–4260. doi: 10.1091/mbc.E07-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JM, Burka LT, Smith CS, Black W, James R, Cunningham ML. Differential expression of CYP1A, 2B, and 3A genes in the F344 rat following exposure to a polybrominated diphenyl ether mixture or individual components. Toxicol Sci. 2005;88:127–133. doi: 10.1093/toxsci/kfi288. [DOI] [PubMed] [Google Scholar]
- Schecter A, Colacino J, Sjodin A, Needham L, Birnbaum L. Partitioning of polybrominated diphenyl ethers (PBDEs) in serum and milk from the same mothers. Chemosphere. 2010;78:1279–1284. doi: 10.1016/j.chemosphere.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Schecter A, Papke O, Harris TR, Tung KC, Musumba A, Olson J, Birnbaum L. Polybrominated diphenyl ether (PBDE) levels in an expanded market basket survey of U.S. food and estimated PBDE dietary intake by age and sex. Environ Health Perspect. 2006;114:1515–15120. doi: 10.1289/ehp.9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjodin A, Jones RS, Focant JF, Lapeza C, Wang RY, McGahee EEI, Zhang Y, Turner WE, Slazyk B, Needham LL, Patterson DG., Jr Retrospective time trend study of polybrominate diphenyl ether and polybrominated and polychlorineated biphenyl levels in human serum form the United States. Environ Health Perspect. 2004;112:654–658. doi: 10.1289/ehp.112-1241957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, He Y, Murphy MB, Yeung LW, Yu RM, Lam MH, Lam PK, Hecker M, Giesy JP, Wu RS, Zhang W, Sheng G, Fu J. Effects of fifteen PBDE metabolites, DE71, DE79 and TBBPA on steroidogenesis in the H295R cell line. Chemosphere. 2008;71:1888–1894. doi: 10.1016/j.chemosphere.2008.01.032. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA, Campbell PJ. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickel F, Hellerbrand C. Non-alcoholic fatty liver disease as a risk factor for hepatocellular carcinoma: mechanisms and implications. Gut. 2010;59:1303–1307. doi: 10.1136/gut.2009.199661. [DOI] [PubMed] [Google Scholar]
- Suvorov A, Takser L. Global gene expression analysis in the livers of rat offspring perinatally exposed to low doses of 2,2’,4,4’-tetrabromodiphenyl ether. Environ Health Perspect. 2010;118:97–102. doi: 10.1289/ehp.0901031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo DT, Richardson VM, Ross DG, Diliberto JJ, Kodavanti PR, Birnbaum LS. Effects of perinatal PBDE exposure on hepatic phase I, phase II, phase III, and deiodinase 1 gene expression involved in thyroid hormone metabolism in male rat pups. Toxicol Sci. 2009;107:27–39. doi: 10.1093/toxsci/kfn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talsness CE, Kuriyama SN, Sterner-Kock A, Schnitker P, Grande SW, Shakibaei M, Andrade A, Grote K, Chahoud I. In utero and lactational exposures to low doses of polybrominated diphenyl ether-47 alter the reproductive system and thyroid gland of female rat offspring. Environ Health Perspect. 2008;116:308–314. doi: 10.1289/ehp.10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Arii S. Current status of molecularly targeted therapy for hepatocellular carcinoma: basic science. Int J Clin Oncol. 2010;15:235–241. doi: 10.1007/s10147-010-0083-4. [DOI] [PubMed] [Google Scholar]
- Teoh NC. Proliferative drive and liver carcinogenesis: too much of a good thing? J Gastroenterol Hepatol. 2009;24:1817–1825. doi: 10.1111/j.1440-1746.2009.06121.x. [DOI] [PubMed] [Google Scholar]
- U. S. Environmental Protection Agency. Toxicological review of 2,2’,4,4’-tetrabromodiphenyl ether (BDE-47) 2008a http://www.epa.gov/NCEA/iris/
- U. S. Environmental Protection Agency. Toxicological Review of 2,2’,4,4’,5,5’-hexabromodiphenyl ether (BDE-153) 2008b http://www.epa.gov/NCEA/iris/
- U. S. Environmental Protection Agency. Toxicological Review of 2,2’.4,4’,5-pentabromodiphenyl ether (BDE-99) 2008c http://www.epa.gov/NCEA/iris/
- U. S. Environmental Protection Agency. Toxicological Review of Decabromodiphenyl ether (BDE-209) 2008d http://www.epa.gov/NCEA/iris/
- U. S. Environmental Protection Agency. Chemical Hazard Review: Furniture Flame Retardancy Partnership: Environmental Profiles of Chemical Flame-Retardant Alternatives for Low-Density Polyurethane Foam. 2009 www.epa.gov/dfe 1.
- U. S. Environmental Protection Agency. An Exposure Assessment of Polybrominated Diphenyl Ethers National Center for Environmental Assessment Office of Research and Development U.S. Environmental Protection Agency Washington. 2010 http://cfpub.epa.gov/ncea/cfm/recordisplay.cfm?deid=210404#Download.
- van Erk MJ. Liver of humanized mice expressing ApoE on high fat wester diet plus PFOS. Nextbio Database - accessed June 2011. 2011 [Google Scholar]
- Waterman CL, Currie RA, Cottrell LA, Dow J, Wright J, Waterfield CJ, Griffin JL. An integrated functional genomic study of acute phenobarbital exposure in the rat. BMC Genomics. 2010;11:9. doi: 10.1186/1471-2164-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Health Risks Of Persistent Organic Pollutants From Long-Range Transboundary Air Pollution. 2003:199–212. [Google Scholar]
- Wu H, Kerr MK, ui XQ, Churchill GA. MAANOVA: A software package for the analysis of spotted cDNA microarray experiments. In: Parmgiani G, Garret ES, Irizarry RA, Zeger SL, editors. Analysis of gene expression data: an overview of methods and software. Springer; 2003. pp. 313–341. [Google Scholar]
- Yang X, Zhang B, Molony C, Chudin E, Hao K, Zhu J, Gaedigk A, Suver C, Zhong H, Leeder JS, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich RG, Slatter JG, Schadt EE, Kasarskis A, Lum PY. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 2010;20:1020–1036. doi: 10.1101/gr.103341.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovski SZ, Yanovski JA. Obesity. N Engl J Med. 2002;346:591–602. doi: 10.1056/NEJMra012586. [DOI] [PubMed] [Google Scholar]
- Zhou T, Taylor MM, DeVito MJ, Crofton KM. Developmental exposure to brominated diphenyl etheres results in thyroid hormone disruption. Toxicolog Sci. 2002;66:105–116. doi: 10.1093/toxsci/66.1.105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement 1. Liver gene transcript changes – PND 22 males
Supplement 2. Liver gene transcript changes – PND 22 females
Supplement 3. Liver gene transcript changes – week 13 males
Supplement 4. PBDE significant genes at PND 22 and week 13
Supplement 5. GO Analysis
Supplement 6. PBDE PND 22 male liver gene transcripts versus phenobarbital gene transcript changes (developed from Nextbio database) 16
Supplement 7. PBDE PND 22 male liver gene transcripts versus human colon cancer genes (developed from Nextbio database)