Abstract
Healthy first-degree relatives with heredity of type 2 diabetes (FH+) are known to have metabolic inflexibility compared with subjects without heredity for diabetes (FH−). In this study, we aimed to test the hypothesis that FH+ individuals have an impaired response to exercise compared with FH−. Sixteen FH+ and 19 FH− insulin-sensitive men similar in age, peak oxygen consumption (V̇o2 peak), and body mass index completed an exercise intervention with heart rate monitored during exercise for 7 mo. Before and after the exercise intervention, the participants underwent a physical examination and tests for glucose tolerance and exercise capacity, and muscle biopsies were taken for expression analysis. The participants attended, on average, 39 training sessions during the intervention and spent 18.8 MJ on exercise. V̇o2 peak/kg increased by 14%, and the participants lost 1.2 kg of weight and 3 cm waist circumference. Given that the FH+ group expended 61% more energy during the intervention, we used regression analysis to analyze the response in the FH+ and FH− groups separately. Exercise volume had a significant effect on V̇o2 peak, weight, and waist circumference in the FH− group, but not in the FH+ group. After exercise, expression of genes involved in metabolism, oxidative phosphorylation, and cellular respiration increased more in the FH− compared with the FH+ group. This suggests that healthy, insulin-sensitive FH+ and FH− participants with similar age, V̇o2 peak, and body mass index may respond differently to an exercise intervention. The FH+ background might limit muscle adaptation to exercise, which may contribute to the increased susceptibility to type 2 diabetes in FH+ individuals.
Keywords: type 2 diabetes, exercise intervention, expression analysis, genetic predisposition, muscle
type 2 diabetes is a disease characterized by peripheral insulin resistance and failure of the pancreatic β-cells to compensate for the increased insulin need. Despite the identification of many loci linked to type 2 diabetes, they explain <20% of the observed heredity (9). Notably, individuals with a first-degree family history of type 2 diabetes (FH+) have a threefold higher risk of developing the disease compared with those individuals without a history (FH−) (19). FH+ individuals have reduced basal energy expenditure and decreased insulin sensitivity long before developing signs of clinical diabetes (8). FH+ individuals are further characterized by metabolic inflexibility (32), having lower carbohydrate oxidation in response to insulin and lower fat oxidation during a high-fat diet (10, 40).
These metabolic differences have been proposed to be dependent on primary defects in mitochondria (37), muscle fiber composition (22), lack of physical activity (41), and/or a sedentary lifestyle (11). Skeletal muscle is the main organ for insulin-mediated glucose disposal (4, 29), and regular exercise can prevent or postpone the development of type 2 diabetes in high-risk populations (5, 17, 18). Exercise increases insulin sensitivity (29), reducing the need for glucose-lowering drugs (2, 28), and is further known to increase skeletal muscle mitochondrial content and function in both nondiabetic and diabetic subjects (26). Interestingly, fat oxidation is related to insulin sensitivity in FH+ subjects (15). Fat oxidation does not increase after weight loss induced by low-calorie diet (3), but increases after an exercise intervention (7) in diabetic patients and overweight subjects. However, despite exercise intervention, differences in muscle ATPase activity (13), and glucose metabolism (25) remain in diabetic subjects, indicating that metabolic perturbations are difficult to reverse completely with exercise.
Our laboratory has previously reported decreased expression of mitochondrial genes and genes involved in fatty acid metabolism in skeletal muscle in insulin-sensitive FH+ individuals compared with FH− of a similar age, body mass index (BMI), and peak oxygen consumption (V̇o2 peak) (6). In the present study, we aimed to test the hypothesis that a FH+ background impairs the response to an exercise intervention compared with FH− controls.
MATERIALS AND METHODS
Study cohort.
Fifty sedentary men (age 30-45 yr) were recruited, 24 with (FH+) and 26 without a first-degree relative with type 2 diabetes (FH−), as described earlier (6, 33). Briefly, before enrollment, all subjects underwent a physical examination, a 75-g oral glucose tolerance test (OGTT), and a maximal exercise test. The FH+ and FH− groups were similar in age, BMI, V̇o2 peak, habitual activity, and glucose tolerance and insulin sensitivity (all with normal fasting glucose, OGTT, and homeostatic model assessment-insulin resistance). The study was approved by the local ethics committee at Lund University, and written, informed consent was obtained from all participants. Clinical characteristics of the study participants are given in Table 1. Of the included individuals, 17 FH+ and 19 FH− individuals completed the training program. The 14 participants not able to complete the study reported lack of time as the main reason for noncompliance (n = 12), in addition to minor injuries (n = 1) and moving from the area (n = 1). One FH+ individual was excluded from the analysis due to being >4 SD from the rest of the group in weight change. The FH− group included one and the FH+ group included two active smokers. Actiheart devices were used by 17 of the participants (9 FH+, 8 FH−) for 5 days to monitor physical activity level before the initiation of the intervention, as described earlier (6).
Table 1.
The clinical characteristics of the participants at the initiation of the study
| All |
FH− |
FH+ |
|||||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | MWU P value | |
| Age, yr | 38.1 | 4.3 | 37.8 | 4.7 | 38.4 | 3.9 | 0.683 |
| Weight, kg | 93.7 | 11.4 | 94.8 | 10.2 | 92.4 | 12.8 | 0.271 |
| BMI, kg/m2 | 28.7 | 2.9 | 29.0 | 2.9 | 28.4 | 2.8 | 0.367 |
| Waist circumference, cm | 99.6 | 8.0 | 101.0 | 7.8 | 98.1 | 8.2 | 0.161 |
| Hip circumference, cm | 105.0 | 5.6 | 106.1 | 4.9 | 103.8 | 6.3 | 0.082 |
| Waist/hip ratio | 0.95 | 0.04 | 0.95 | 0.05 | 0.95 | 0.04 | 0.832 |
| FFM, % | 76.7 | 5.3 | 75.7 | 4.6 | 78.0 | 5.8 | 0.172 |
| V̇o2peak/kg, ml·kg−1·min−1 | 31.4 | 4.7 | 30.5 | 4.7 | 32.5 | 4.6 | 0.286 |
| V̇o2peak/FFM, ml·kg FFM−1·min−1 | 41.0 | 5.0 | 40.4 | 5.3 | 41.7 | 4.6 | 0.481 |
| Fasting plasma glucose, mM | 4.3 | 0.5 | 4.3 | 0.6 | 4.3 | 0.4 | 0.707 |
| 2-h Glucose tolerance, mM | 5.8 | 1.3 | 5.8 | 1.4 | 5.8 | 1.2 | 0.883 |
| HbA1c, % | 4.3 | 0.3 | 4.3 | 0.3 | 4.3 | 0.4 | 0.781 |
| Fasting insulin, mU/ml | 7.5 | 3.2 | 7.4 | 3.0 | 7.7 | 3.6 | 1.000 |
| HOMA-IR ratio | 1.44 | 0.65 | 1.43 | 0.65 | 1.45 | 0.67 | 1.000 |
| HOMA-β, % | 309.8 | 376.2 | 354.4 | 483.6 | 242.9 | 175.5 | 0.708 |
| mtDNA/nDNA ratio | 2,509 | 727 | 2,443 | 835 | 2,587 | 590 | 0.735 |
The P value describes the significance of difference between the participants without and with first-degree heredity of type 2 diabetes (FH− and FH+ groups, respectively) using the Mann-Whitney U-test (MWU). BMI, body mass index; FFM, fat-free mass; V̇o2peak, peak oxygen consumption; HbA1c, glycosylated hemoglobin; HOMA-IR, homeostasis model assessment of insulin resistance; HOMA-β, homeostasis model of β-cell function; mtDNA, mitochondrial DNA; nDNA, nuclear DNA.
Exercise intervention.
The exercise intervention lasted 216 days (SD 26 days) and has been described earlier (30, 31). Briefly, the subjects were offered combined supervised group training 3 times per week for the entire intervention. One session of 1-h spinning class and two sessions of 1-h aerobics class were given weekly. During the spinning class, a specially designed cycle ergometer (Pulse, Pulse Fitness, Congleton, UK) was used, and the sessions were led by a certified instructor. The workload was individually adjusted by braking the wheel during cycling. The aerobics classes were conducted to music and led by certified instructors. The classes included a warm-up period (10 min), rhythmic aerobic training (15 min) mixed with strength exercises for arm, leg, abdominal, and back muscles (15 min), cool-down, and stretching exercises (10 min). Strength training was performed with the subject's own body weight as resistance, such as push-ups, crunches, sit-ups, and back extensions. Heart rate was monitored continuously during the spinning and aerobics classes by a Polar belt (Polar Fitness F1, POLAR), which was used to calculate exercise intensity and total energy expenditure, using the flex-heart rate method (16). In this method, we use the relation between the heart rate and the intensity of the training. The exercise volume (reported in MJ) was calculated by multiplying the intensity (in W) with time spent on exercise. The relative intensity of exercise was measured with heart rate reserve (HRR), in which the intensity is described in percentage of individual maximal working capacity. The participants were told to adhere to their normal diets during the study.
Muscle biopsy and sampling.
Muscle biopsies were taken before and after the exercise intervention from the right vastus lateralis muscle under local anesthesia (lidocaine 1%), using a 6-mm Bergström needle (Stille). The participants were instructed not to perform any vigorous exercise within 48 h before the biopsy and to fast from 10 PM the previous day. RNA was isolated using the RNeasy Fibrous Tissue Kit (Qiagen). Concentration and purity were measured using a NanoDrop ND-1000 spectrophotometer (ratio of 260- to 280-nm absorbance > 1.8 and ratio of 260- to 230-nm absorbance > 1.0) (NanoDrop Technologies, Wilmington, DE). No major signs of degradation were observed using agarose gel electrophoresis and Experion DNA 1K gel chips (Bio-Rad). Mitochondrial DNA content (mtDNA) was quantified using quantitative PCR, comparing the mitochondrial 16S and ND6 genes with the nuclear RNaseP gene. Plasma glucose was measured with the hexokinase method (Beckman Syncron CX System Chemistry), insulin with ELISA (Dako), and HbA1c was measured with HPLC (Mono S, GE Healthcare), as described earlier (6).
Expression analysis.
Synthesis of biotin-labeled cRNA and hybridization to the Affymetrix Custom Array NuGO-Hs1a520180 GeneChip (http://www.nugo.org) were performed according to the manufacturer's recommendation. The GeneChip contains 23,941 probe sets for interrogation. Images were analyzed using the GeneChip Operating System (Affymetrix) software. We used ENTREZ custom chip definition files (http://brainarray.mbni.med.umich.edu) for regrouping the individual probes into consistent probe sets and remap them to the sets of genes for Affymetrix Custom Array (NuGO-Hs1a520180), which resulted in a total of 16,313 genes. The data were filtered based on the MAS5.0 present/absent calls, which classifies each gene as expressed above background (present call) or not (marginal or absent call). The expression data were normalized using robust multiarray average (12). Data were analyzed for FH+ and FH− separately due to the difference in amount of exercise performed. A regression model was used to identify expression changes dependent on exercise volume. Genes with P < 0.05 were considered for further analysis. False discovery rate analysis was made in R (27) using the q-value package (36) with a false discovery rate acceptance of < 0.05.
Incremental maximal exercise test.
An incremental maximal exercise test was performed before and after the exercise intervention on a bicycle ergometer (Marquette Hellige Medical Systems 900 ERG, Milwaukee, WI). The initial workload was 50 W and increased by 15 W/min; pedaling rate was maintained at 60 rpm using both visual feedback and verbal encouragement. O2 consumption, CO2 production, and ventilation were measured breath by breath (Oxycon Mobile, Jaeger, Hoechberg, Germany). Heart rate was continuously monitored throughout the test (Polar T 61, POLAR, Oulu, Finland). The subjects were encouraged to exercise to exhaustion, and V̇o2 peak was reached when the respiratory exchange ratio exceeded 1.10. The gas sensors were calibrated before each test with a certified gas mixture, and air flow was calibrated using a calibration syringe.
Statistics.
Differences between the groups were analyzed using nonparametric Mann-Whitney U-tests and Wilcoxon rank test, and P < 0.05 was considered statistically significant. Linear regression was used with starting value and exercise volume as predictors of the end value. For comparison of gene expression on the pathway level, the mean centroid method was used, estimating the mean expression of all genes in a pathway (20). Data were analyzed with SPSS 19.0 (SPSS, Chicago, IL) and Graphpad 5 (Graphpad software, La Jolla, CA). For pathway analysis, Webgestalt (42) was used, utilizing the Gene Ontology (GO) (1) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (14) annotation databases.
RESULTS
Exercise increase V̇o2 peak and decrease body weight and waist circumference.
During the 216 days of the exercise intervention, the average participant attended 39 training sessions of 1-h duration (range 11–107 sessions), expending 18.8 MJ by exercise (range 5.3–42.1 MJ). During the intervention, both V̇o2 peak/kg and V̇o2 peak/kg fat-free mass increased by 14% and 13% (both P < 0.001), respectively, for the group as a whole. The participants lost 1.2 kg of weight and 3 cm of waist circumference (P = 0.025 and P < 0.001, respectively), and increased their skeletal muscle mtDNA content by 45% (P = 0.003, Table 2). Unexpectedly, there was an increase in fasting glucose at the end of the study. However, this was not associated with concomitant increase in glucose tolerance (2 h glucose during OGTT or HbA1c, Table 2). The intensity during the exercise was similar between the groups, having an average effect of 121 W and 130 W (P = 0.286, Fig. 1A) and a HRR of 62% and 63% (P = 0.418) in FH− and FH+, respectively. The FH+ group participated in 59% more exercise sessions and expended 61% more energy than the FH− group (Fig. 1B). This was due to both higher amount of exercise per month in the FH+ group (Fig. 1C) and a shorter period of active exercise in the FH− group (Fig. 1D). The changes in weight and waist circumference were more pronounced in the FH+ group, which could be related to the volume of exercise.
Table 2.
The changes during the 216-day intervention period
| All |
FH− |
FH+ |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Before |
After |
Before |
After |
Before |
After |
||||||||||
| Mean | SD | Mean | SD | Wilcoxon P value | Mean | SD | Mean | SD | Wilcoxon P value | Mean | SD | Mean | SD | Wilcoxon P value | |
| Weight, kg | 93.7 | 11.4 | 92.5 | 11.5 | 0.025 | 94.8 | 10.2 | 94.3 | 10.5 | 0.647 | 92.4 | 12.8 | 90.4 | 12.6 | 0.009 |
| BMI, kg/m2 | 28.7 | 2.9 | 28.3 | 3.0 | 0.025 | 29.0 | 2.9 | 28.8 | 3.0 | 0.616 | 28.4 | 2.8 | 27.8 | 2.9 | 0.009 |
| Waist circumference, cm | 99.6 | 8.0 | 96.9 | 7.9 | <0.001 | 101.0 | 7.8 | 99.2 | 7.8 | 0.036 | 98.1 | 8.2 | 94.3 | 7.3 | 0.001 |
| Hip circumference, cm | 105.0 | 5.6 | 104.1 | 5.8 | 0.010 | 106.1 | 4.9 | 105.5 | 5.4 | 0.208 | 103.8 | 6.3 | 102.6 | 6.1 | 0.015 |
| Waist/hip ratio | 0.95 | 0.04 | 0.93 | 0.05 | <0.001 | 0.95 | 0.05 | 0.94 | 0.05 | 0.142 | 0.95 | 0.04 | 0.92 | 0.05 | 0.001 |
| FFM, % | 76.7 | 5.3 | 76.6 | 5.8 | 0.806 | 75.7 | 4.6 | 75.2 | 5.5 | 0.546 | 78.0 | 5.8 | 78.4 | 5.7 | 0.535 |
| V̇o2peak/kg, ml·kg−1·min−1 | 31.4 | 4.7 | 35.7 | 5.6 | <0.001 | 30.5 | 4.7 | 34.6 | 5.5 | 0.003 | 32.5 | 4.6 | 36.9 | 5.7 | 0.003 |
| V̇o2peak/FFM, ml·kg FFM−1·min−1 | 41.0 | 5.0 | 46.5 | 5.8 | <0.001 | 40.4 | 5.3 | 45.7 | 5.7 | 0.001 | 41.7 | 4.6 | 47.3 | 6.0 | 0.003 |
| Fasting plasma glucose, mM | 4.3 | 0.5 | 4.7 | 0.6 | <0.001 | 4.3 | 0.6 | 4.6 | 0.4 | 0.029 | 4.3 | 0.4 | 4.9 | 0.7 | 0.001 |
| 2-h Glucose tolerance, mM | 5.8 | 1.3 | 5.5 | 1.5 | 0.245 | 5.8 | 1.4 | 5.7 | 1.7 | 0.717 | 5.8 | 1.2 | 5.4 | 1.3 | 0.175 |
| HbA1c, % | 4.3 | 0.3 | 4.3 | 0.3 | 0.196 | 4.3 | 0.3 | 4.3 | 0.3 | 0.178 | 4.3 | 0.4 | 4.3 | 0.4 | 0.685 |
| Fasting insulin, mU/ml | 7.5 | 3.2 | 8.8 | 5.7 | 0.097 | 7.4 | 3.0 | 7.5 | 2.7 | 0.586 | 7.7 | 3.6 | 10.3 | 7.7 | 0.066 |
| HOMA-IR ratio | 1.44 | 0.65 | 1.86 | 1.35 | 0.005 | 1.43 | 0.65 | 1.54 | 0.55 | 0.159 | 1.45 | 0.67 | 2.25 | 1.86 | 0.013 |
| HOMA-β, % | 309.8 | 376.2 | 170.7 | 123.7 | 0.005 | 354.4 | 483.6 | 166.4 | 123.9 | 0.035 | 242.9 | 175.5 | 175.9 | 127.4 | 0.053 |
| mtDNA/nDNA ratio | 2,509 | 727 | 3,636 | 1,598 | 0.003 | 2,443 | 835 | 3,323 | 991 | 0.158 | 2,587 | 590 | 3,904 | 1,977 | 0.053 |
The P value describes the significance of difference in each group before and after the intervention using Wilcoxon signed-rank test. Significant differences (P < 0.05) are in bold.
Fig. 1.
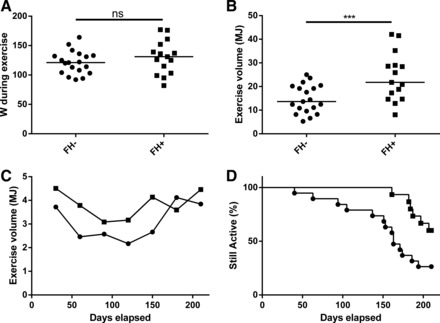
A: the average intensity of exercise during the training session measured in Watts (W) in participants without (FH−; ●) and with first-degree heredity of type 2 diabetes (FH+; ■). Each dot represents one individual, and the line indicates the median, P = 0.441. B: the exercise volume in the FH− and FH+ groups during the intervention. Each dot represents one individual, and the line indicates the median, P < 0.001. C: the mean exercise volume monthly by the active participants during the exercise intervention. Active is defined as not yet having recorded their last session. D: Kaplan-Meier graph of the percentage of participants actively taking part in the exercise intervention. Using the Mantel-Cox test, P = 0.012. ns, Nonsignificant. ***Significant difference: P < 0.001.
Due to the large difference in exercise volume between the groups, we used multiple regression to evaluate the effects of exercise in the FH− and FH+ groups separately. The exercise volume predicted decrease in weight and waist circumference and increase in V̇o2 peak/kg stronger in the FH− group (P = 0.018, P = 0.008, and P = 0.001, respectively), compared with the FH+ group (P = 0.249, P = 0.087, and P = 0.068, respectively). Exercise volume did not predict increase in mtDNA in FH− nor in FH+ in a significant way (P = 0.601 and P = 0.105, respectively, Table 3). When plotting change in weight, waist circumference, and V̇o2 peak/kg to exercise volume, the FH− group has a stronger relation compared with the FH+ (Fig. 2). The relative intensity in %HRR did not have any strong effects on these variables (Supplemental Table S1; Supplemental material for this article is available online at the journal website).
Table 3.
Univariate regression analysis of variables in their relation to exercise in megajoules
| FH− |
FH+ |
|||||
|---|---|---|---|---|---|---|
| β | SE | P value | β | SE | P value | |
| Age, yr | −0.255 | 0.097 | 0.018 | −0.076 | 0.063 | 0.249 |
| Weight, kg | −0.074 | 0.029 | 0.023 | −0.022 | 0.019 | 0.272 |
| BMI, kg/m2 | −0.313 | 0.103 | 0.008 | −0.127 | 0.068 | 0.087 |
| Waist circumference, cm | −0.011 | 0.078 | 0.889 | −0.081 | 0.039 | 0.061 |
| Hip circumference, cm | −0.003 | 0.001 | 0.007 | 0.000 | 0.001 | 0.674 |
| Waist/hip ratio | 0.443 | 0.140 | 0.006 | 0.069 | 0.067 | 0.322 |
| FFM, % | 0.492 | 0.111 | 0.001 | 0.187 | 0.093 | 0.068 |
| V̇o2peak/kg, ml·kg−1·min−1 | 0.446 | 0.167 | 0.018 | 0.118 | 0.134 | 0.397 |
| V̇o2peak/FFM, ml·kg FFM−1·min−1 | 0.019 | 0.014 | 0.184 | 0.018 | 0.016 | 0.279 |
| Fasting plasma glucose, mM | 0.049 | 0.056 | 0.393 | −0.031 | 0.031 | 0.341 |
| 2-h Glucose tolerance, mM | −0.008 | 0.007 | 0.295 | 0.007 | 0.005 | 0.181 |
| HbA1c, % | −0.113 | 0.095 | 0.250 | −0.082 | 0.150 | 0.593 |
| Fasting insulin, mU/ml | −0.017 | 0.018 | 0.371 | −0.01 | 0.037 | 0.787 |
| HOMA-IR ratio | −2.668 | 4.190 | 0.534 | −2.601 | 2.468 | 0.315 |
| HOMA-β, % | 34.4 | 63.6 | 0.601 | 108.0 | 60.0 | 0.105 |
Significant differences (P < 0.05) are in bold.
Fig. 2.
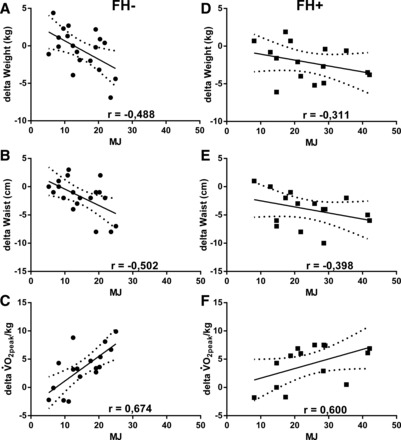
A–C: the relation between energy expenditure and change in weight (A), waist circumference (B), and peak oxygen consumption (V̇o2 peak)/kg (C) in the FH− group (●). D–F: relation between energy expenditure and weight (D), waist circumference (E), and V̇o2 peak/kg (F) in the FH+ group (■). Spearman's r is given for each correlation.
Gene expression in skeletal muscle in relation to exercise volume and FH status.
First, we investigated the effects of exercise on the change in mean centroid value (a representation of the expression of all genes included in a defined pathway) of a number of pathways that were identified in the baseline study (6). Genes involved in metabolism and glycolysis (GO: 0008152 and GO: 0006096) increased significantly (P = 0.045 and P = 0.007, respectively) due to exercise in the FH− group, but no such change could be observed in the FH+ group (P = 0.418 and P = 0.639, respectively) (Table 4). Relative intensity in %HRR did not affect the expression of these pathways (Supplemental Table S2).
Table 4.
Regression analysis of mean centroid of selected pathways influenced by exercise
| FH− |
FH+ |
|||||
|---|---|---|---|---|---|---|
| β | SE | P value | β | SE | P value | |
| Metabolism | 0.306 | 0.141 | 0.045 | −0.082 | 0.098 | 0.418 |
| Mitochondrion | −0.104 | 0.14 | 0.466 | 0.031 | 0.071 | 0.666 |
| TCA cycle | 0.613 | 0.382 | 0.128 | −0.059 | 0.412 | 0.889 |
| Glycolysis | 0.597 | 0.191 | 0.007 | 0.095 | 0.197 | 0.639 |
| Oxidative phosphorylation | 0.616 | 0.328 | 0.079 | −0.433 | 0.263 | 0.129 |
| Regulation of FA oxidation | −0.653 | 0.320 | 0.058 | 0.157 | 0.31 | 0.622 |
| FA metabolism | 0.744 | 0.373 | 0.063 | −0.257 | 0.279 | 0.377 |
| FA oxidation | 0.314 | 0.316 | 0.335 | −0.141 | 0.294 | 0.641 |
β and SE values are for gigajoules of exercise. FA, fatty acid.
Second, we used a regression model to test the impact of the intervention on gene expression where the expression level after the intervention of each gene was predicted using the exercise volume and the gene expression at the baseline level. In this model, the expression of 1,066 genes in the FH− group and 365 genes in the FH+ group changed in response to exercise with nominal significance. Two hundred genes in the FH− and 96 genes in the FH+ group increased with exercise (complete gene list can be found in the supplemental file). Next, the 200 (FH−) and 96 (FH+) nominally significant genes that increased with exercise were analyzed using Webgestalt to evaluate if the genes upregulated by exercise are overrepresented in certain pathways. For the FH− group, 30 GO pathways were associated with these genes: a majority represented metabolic pathways, including mitochondria, cellular respiration, oxidative phosphorylation, electron transport chain, and mitochondrial ATP synthesis. In the FH+ group, 22 GO pathways changed significantly in response to exercise (Table 5). These pathways are not clearly linked to those implicated for the FH− group, and the statistical association is lower. Using the KEGG database, expression of 10 pathways increased significantly in the FH− group, including oxidative phosphorylation, metabolic pathways, and TCA cycle. No KEGG pathways were significantly increased for the FH+ group (Table 5). The relative intensity of exercise was evaluated similarly, and 33 and 54 nominally significant genes increased with increased %HRR for the FH− and FH+ participants, but the response on pathway level had a low statistical association compared with exercise volume (Supplemental Table S3).
Table 5.
GO and KEGG Pathways overrepresented among genes showing increased expression with exercise in the FH− and the FH+ groups
| C | O | R | Adj P | Pathway ID | Description | |
|---|---|---|---|---|---|---|
| GO FH− | ||||||
| Biological process | 129 | 24 | 14.34 | 1.74e-18 | GO:0045333 | Cellular respiration |
| Biological process | 87 | 20 | 17.72 | 2.64e-17 | GO:0022904 | Respiratory electron transport chain |
| Biological process | 470 | 36 | 5.90 | 4.57e-16 | GO:0055114 | Oxidation-reduction process |
| Biological process | 117 | 21 | 13.84 | 4.57e-16 | GO:0022900 | Electron transport chain |
| Biological process | 277 | 28 | 7.79 | 2.89e-15 | GO:0015980 | Energy derivation by oxidation of organic compounds |
| Biological process | 380 | 31 | 6.29 | 1.75e-14 | GO:0006091 | Generation of precursor metabolites and energy |
| Biological process | 53 | 11 | 16.00 | 7.61e-09 | GO:0006119 | Oxidative phosphorylation |
| Biological process | 44 | 10 | 17.52 | 1.74e-08 | GO:0042773 | ATP synthesis coupled electron transport |
| Biological process | 44 | 10 | 17.52 | 1.74e-08 | GO:0042775 | Mitochondrial ATP synthesis coupled electron transport |
| Biological process | 35 | 9 | 19.82 | 3.99e-08 | GO:0006120 | Mitochondrial electron transport, NADH to ubiquinone |
| Molecular function | 552 | 31 | 4.44 | 3.28e-10 | GO:0016491 | Oxidoreductase activity |
| Molecular function | 48 | 11 | 18.12 | 6.11e-10 | GO:0016655 | Oxidoreductase activity, acting on NADH or NADPH, quinone or similar |
| Molecular function | 36 | 10 | 21.96 | 6.11e-10 | GO:0003954 | NADH dehydrogenase activity |
| Molecular function | 36 | 10 | 21.96 | 6.11e-10 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity |
| Molecular function | 36 | 10 | 21.96 | 6.11e-10 | GO:0050136 | NADH dehydrogenase (quinone) activity |
| Molecular function | 78 | 11 | 11.15 | 1.19e-07 | GO:0016651 | Oxidoreductase activity, acting on NADH or NADPH |
| Molecular function | 4,368 | 85 | 1.54 | 1.86e-05 | GO:0003824 | Catalytic activity |
| Molecular function | 4 | 3 | 59.30 | 0.0002 | GO:0004470 | Malic enzyme activity |
| Molecular function | 231 | 13 | 4.45 | 0.0002 | GO:0048037 | Cofactor binding |
| Molecular function | 132 | 10 | 5.99 | 0.0002 | GO:0009055 | Electron carrier activity |
| Cellular component | 1,240 | 76 | 4.98 | 1.99e-33 | GO:0005739 | Mitochondrion |
| Cellular component | 612 | 49 | 6.51 | 4.32e-25 | GO:0044429 | Mitochondrial part |
| Cellular component | 285 | 31 | 8.84 | 3.85e-19 | GO:0005743 | Mitochondrial inner membrane |
| Cellular component | 307 | 31 | 8.21 | 2.64e-18 | GO:0019866 | Organelle inner membrane |
| Cellular component | 430 | 33 | 6.24 | 5.41e-16 | GO:0005740 | Mitochondrial envelope |
| Cellular component | 62 | 16 | 20.98 | 6.04e-16 | GO:0070469 | Respiratory chain |
| Cellular component | 411 | 32 | 6.33 | 8.40e-16 | GO:0031966 | Mitochondrial membrane |
| Cellular component | 240 | 24 | 8.13 | 3.69e-14 | GO:0005759 | Mitochondrial matrix |
| Cellular component | 5,512 | 117 | 67.80 | 5.63e-14 | GO:0044444 | Cytoplasmic part |
| Cellular component | 116 | 18 | 12.62 | 5.63e-14 | GO:0044455 | Mitochondrial membrane part |
| GO FH+ | ||||||
| Biological process | 6 | 2 | 59.18 | 0.0458 | GO:0019368 | Fatty acid elongation, unsaturated fatty acid |
| Biological process | 255 | 7 | 4.87 | 0.0458 | GO:0046486 | Glycerolipid metabolic process |
| Biological process | 3 | 2 | 118.36 | 0.0458 | GO:0031642 | Negative regulation of myelination |
| Biological process | 7 | 2 | 50.73 | 0.0458 | GO:0030497 | Fatty acid elongation |
| Biological process | 63 | 4 | 11.27 | 0.0458 | GO:0008033 | tRNA processing |
| Biological process | 6 | 2 | 59.18 | 0.0458 | GO:0042761 | Very-long-chain fatty acid biosynthetic process |
| Biological process | 103 | 5 | 8.62 | 0.0458 | GO:0006399 | tRNA metabolic process |
| Biological process | 187 | 6 | 5.70 | 0.0458 | GO:0045017 | Glycerolipid biosynthetic process |
| Biological process | 4 | 2 | 88.77 | 0.0458 | GO:0034625 | Fatty acid elongation, monounsaturated fatty acid |
| Molecular function | 4 | 2 | 100.82 | 0.0106 | GO:0009922 | Fatty acid elongase activity |
| Molecular function | 1,403 | 17 | 2.44 | 0.0159 | GO:0016740 | Transferase activity |
| Molecular function | 7 | 2 | 57.61 | 0.0177 | GO:0004312 | Fatty acid synthase activity |
| Cellular component | 1,913 | 23 | 2.29 | 0.0064 | GO:0031090 | Organelle membrane |
| Cellular component | 614 | 11 | 3.42 | 0.0073 | GO:0042175 | Nuclear outer membrane-endoplasmic reticulum membrane network |
| Cellular component | 601 | 11 | 3.49 | 0.0073 | GO:0005789 | Endoplasmic reticulum membrane |
| Cellular component | 1,444 | 18 | 2.38 | 0.0073 | GO:0012505 | Endomembrane system |
| Cellular component | 5,404 | 42 | 1.48 | 0.0136 | GO:0044446 | Intracellular organelle part |
| Cellular component | 5,472 | 42 | 1.47 | 0.0146 | GO:0044422 | Organelle part |
| Cellular component | 11 | 2 | 34.71 | 0.0151 | GO:0031231 | Intrinsic to peroxisomal membrane |
| Cellular component | 11 | 2 | 34.71 | 0.0151 | GO:0005779 | Integral to peroxisomal membrane |
| Cellular component | 729 | 11 | 2.88 | 0.0151 | GO:0044432 | Endoplasmic reticulum part |
| Cellular component | 47 | 3 | 12.18 | 0.0184 | GO:0005884 | Actin filament |
| KEGG FH− | ||||||
| KEGG pathway | 103 | 19 | 13.87 | 2.59e-15 | 190 | Oxidative phosphorylation |
| KEGG pathway | 101 | 18 | 13.40 | 1.64e-14 | 5012 | Parkinson's disease |
| KEGG pathway | 142 | 19 | 10.06 | 3.07e-13 | 5010 | Alzheimer's disease |
| KEGG pathway | 160 | 20 | 9.40 | 3.07e-13 | 5016 | Huntington's disease |
| KEGG pathway | 969 | 39 | 3.03 | 2.17e-09 | 1100 | Metabolic pathways |
| KEGG pathway | 38 | 7 | 13.85 | 2.94e-06 | 280 | Valine, leucine and isoleucine degradation |
| KEGG pathway | 16 | 4 | 18.80 | 0.0002 | 630 | Glyoxylate and dicarboxylate metabolism |
| KEGG pathway | 6 | 3 | 37.59 | 0.0002 | 62 | Fatty acid elongation in mitochondria |
| KEGG pathway | 26 | 4 | 11.57 | 0.0013 | 20 | Citrate cycle (TCA cycle) |
| KEGG pathway | 37 | 4 | 8.13 | 0.0042 | 71 | Fatty acid metabolism |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; C, no. of genes in category, O, no. of observed genes in category, R, ratio of enrichment to expected genes, Adj P, P value adjusted for multiple comparisons.
DISCUSSION
In this study, we have evaluated the response to an exercise intervention in healthy, glucose-tolerant FH− and FH+ subjects with the two groups similar in regard to age, BMI, and V̇o2 peak. Notably, the FH+ individuals spent more time exercising than those in the FH− group, expending 61% more energy on exercise during the intervention period. It is possible that the exposure of FH+ participants to the consequences of diabetes in the family motivated them to increase their exercise to a greater extent during the study compared with the FH− participants. The observed difference in exercise volume led us to analyze the data in the FH− and FH+ groups separately, to avoid misinterpretation between the effects of exercise and FH background. In the regression analysis, the FH+ group responded less to exercise in terms of weight, waist circumference, and V̇o2 peak. Although this difference between the groups was not formally tested, it indicated that the FH+ background may limit the response to exercise. The individual intensity of the exercise was not a major determinant for exercise response in this study.
Several studies describe the exercise response in type 2 diabetic patients and have reported increased mitochondrial volume and increased activity of oxidative enzymes after exercise (21, 34, 35, 39, 41). In agreement, our analysis show increased mitochondrial density in both the FH− and FH+ groups (by 45%) after the intervention and also an increased expression of genes in the oxidative phosphorylation pathway in the FH− group. However, in this study, we also investigated the effect of exercise volume in relation to benefits gained. This analysis indicated that there is lesser gain per volume of exercise in the FH+ group vs. FH−, but no formal statistical test of this observation was done due to the difference in performed exercise between the groups. Further studies are clearly needed to determine the optimal, frequency, intensity, and type of exercise for this group at risk of metabolic disease.
Before the start of the exercise intervention, expression of genes in fatty acid oxidation and mitochondrial genes was decreased in FH+ compared with FH− individuals (6). The expression data, both from comparing mean centroid expression and when comparing overrepresented pathways in upregulated genes (GO and KEGG defined pathways), indicate that the FH− group has a stronger response to the exercise compared with FH+, suggesting that the FH+ background limits the beneficial effects of exercise on the gene expression level. The lower upregulation of those pathways in the FH+ group further emphasizes the reduced muscular adaptation to exercise and could possibly explain the lower basal expression of mitochondrial genes observed before the intervention, despite a similar habitual activity (6). Genes involved in oxidative phosphorylation were more strongly upregulated in the FH− compared with the FH+ group, indicating that lower expression of genes involved in oxidative phosphorylation in diabetic muscle (20, 24) can be attributed, at least partly, to the genetic background of FH+ rather than merely to activity levels (41). The adaptation to exercise is to some extent determined by genetic factors (38), and we speculate that the genetic variants responsible for low exercise response might be overrepresented in in the FH+ population and might be of importance for the development of type 2 diabetes.
The difference in exercise volume between the FH− and FH+ groups is a limitation of the study: it is, for example, not possible to know if FH− participants would gain less per megajoule of work performed at higher exercise volumes and thus have a more similar exercise response as the FH− group at these levels of exercise. However, our data support earlier findings with smaller increase in V̇o2 peak (23) and lower upregulation of muscle ATP production (13) and glucose metabolism (25) in FH+ participants and add a new level of observations in terms of a more detailed description of muscle expression.
In conclusion, even a relatively modest increase in physical activity during 7 mo improves physical fitness and thereby some cardiometabolic risk factors. Although no formal statistical test was done between the groups, this study indicates that individuals with a family history of diabetes seem to gain less by increasing the volume of exercise, in terms of gene expression, weight loss, waist circumference, and V̇o2 peak, indicating that the genetic background can influence the response in this group.
GRANTS
This study was supported by the Swedish Research Council Linnaeus grant: Lund University Diabetes Centre (Dnr 349-2006-237) and SFO Exodiab (Dnr 2009-1039); European Research Council Advanced Researcher Grant (GA 269045); NuGo; ALF; the Crafoord Foundation; SV Skånes Diabetes Förening; the Wallenberg Foundation; the Novo Nordisk Foundation; EXGENESIS; UMAS Fonder, Magn. Bergvalls Stiftelse; Syskonen Svenssons Fond; the Swedish Diabetes Research Foundation (2009-060); and the European Community's Seventh Framework Programme (FP7/2007–2013).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: C.E., P.A., H.P., M.D.N., T.R., F.M.K., A.B.T., P.W., K.-F.E., and O.H. analyzed data; C.E., K.S., P.A., C.L., A.B.T., K.-F.E., and O.H. interpreted results of experiments; C.E. and O.H. prepared figures; C.E., K.S., and O.H. drafted manuscript; C.E., T.E., K.S., H.P., M.D.N., A.B.T., P.W., K.-F.E., L.G., and O.H. edited and revised manuscript; C.E., T.E., K.S., P.A., H.P., M.D.N., T.R., F.M.K., C.L., A.B.T., P.W., K.-F.E., L.G., and O.H. approved final version of manuscript; T.E., A.B.T., P.W., K.-F.E., L.G., and O.H. conception and design of research; T.E., M.D.N., T.R., F.M.K., A.B.T., P.W., K.-F.E., and O.H. performed experiments.
Supplementary Material
REFERENCES
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biesenbach G, Bodlaj G, Sedlak M, Pieringer H, Kiesling G. Exercise program for older patients with insulin-treated type 2 diabetes: long-term effects on metabolic control and BMI. Z Gerontol Geriatr 42: 465–469, 2009. [DOI] [PubMed] [Google Scholar]
- 3.Blaak EE, Wolffenbuttel BH, Saris WH, Pelsers MM, Wagenmakers AJ. Weight reduction and the impaired plasma-derived free fatty acid oxidation in type 2 diabetic subjects. J Clin Endocrinol Metab 86: 1638–1644, 2001. [DOI] [PubMed] [Google Scholar]
- 4.DeFronzo RA, Jacot E, Jequier E, Maeder E, Wahren J, Felber JP. The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes 30: 1000–1007, 1981. [DOI] [PubMed] [Google Scholar]
- 5.Diabetes Prevention Program Research Group, Knowler WC, Fowler SE, Hamman RF, Christophi CA, Hoffman HJ, Brenneman AT, Brown-Friday JO, Goldberg R, Venditti E, and Nathan DM. 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. Lancet 374: 1677–1686, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elgzyri T, Parikh H, Zhou Y, Dekker Nitert M, Ronn T, Segerstrom AB, Ling C, Franks PW, Wollmer P, Eriksson KF, Groop L, Hansson O. First-degree relatives of type 2 diabetic patients have reduced expression of genes involved in fatty acid metabolism in skeletal muscle. J Clin Endocrinol Metab 97: E1332–E1337, 2012. [DOI] [PubMed] [Google Scholar]
- 7.Gan SK, Kriketos AD, Ellis BA, Thompson CH, Kraegen EW, Chisholm DJ. Changes in aerobic capacity and visceral fat but not myocyte lipid levels predict increased insulin action after exercise in overweight and obese men. Diabetes Care 26: 1706–1713, 2003. [DOI] [PubMed] [Google Scholar]
- 8.Groop L, Forsblom C, Lehtovirta M. Characterization of the prediabetic state. Am J Hypertens 10: 172S–180S, 1997. [DOI] [PubMed] [Google Scholar]
- 9.Groop L, Pociot F. Genetics of diabetes–Are we missing the genes or the disease? Mol Cell Endocrinol 382: 726–739, 2014. [DOI] [PubMed] [Google Scholar]
- 10.Heilbronn LK, Gregersen S, Shirkhedkar D, Hu D, Campbell LV. Impaired fat oxidation after a single high-fat meal in insulin-sensitive nondiabetic individuals with a family history of type 2 diabetes. Diabetes 56: 2046–2053, 2007. [DOI] [PubMed] [Google Scholar]
- 11.Hu FB, Sigal RJ, Rich-Edwards JW, Colditz GA, Solomon CG, Willett WC, Speizer FE, Manson JE. Walking compared with vigorous physical activity and risk of type 2 diabetes in women: a prospective study. JAMA 282: 1433–1439, 1999. [DOI] [PubMed] [Google Scholar]
- 12.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264, 2003. [DOI] [PubMed] [Google Scholar]
- 13.Kacerovsky-Bielesz G, Chmelik M, Ling C, Pokan R, Szendroedi J, Farukuoye M, Kacerovsky M, Schmid AI, Gruber S, Wolzt M, Moser E, Pacini G, Smekal G, Groop L, Roden M. Short-term exercise training does not stimulate skeletal muscle ATP synthesis in relatives of humans with type 2 diabetes. Diabetes 58: 1333–1341, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lattuada G, Costantino F, Caumo A, Scifo P, Ragogna F, De Cobelli F, Del Maschio A, Luzi L, Perseghin G. Reduced whole-body lipid oxidation is associated with insulin resistance, but not with intramyocellular lipid content in offspring of type 2 diabetic patients. Diabetologia 48: 741–747, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Leonard WR. Measuring human energy expenditure: what have we learned from the flex-heart rate method? Am J Hum Biol 15: 479–489, 2003. [DOI] [PubMed] [Google Scholar]
- 17.Li G, Zhang P, Wang J, Gregg EW, Yang W, Gong Q, Li H, Li H, Jiang Y, An Y, Shuai Y, Zhang B, Zhang J, Thompson TJ, Gerzoff RB, Roglic G, Hu Y, Bennett PH. The long-term effect of lifestyle interventions to prevent diabetes in the China Da Qing Diabetes Prevention Study: a 20-year follow-up study. Lancet 371: 1783–1789, 2008. [DOI] [PubMed] [Google Scholar]
- 18.Lindstrom J, Peltonen M, Eriksson JG, Ilanne-Parikka P, Aunola S, Keinanen-Kiukaanniemi S, Uusitupa M, Tuomilehto J; Finnish Diabetes Prevention Study. Improved lifestyle and decreased diabetes risk over 13 years: long-term follow-up of the randomised Finnish Diabetes Prevention Study (DPS). Diabetologia 56: 284–293, 2013. [DOI] [PubMed] [Google Scholar]
- 19.Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes 49: 2201–2207, 2000. [DOI] [PubMed] [Google Scholar]
- 20.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen J, Mogensen M, Vind BF, Sahlin K, Hojlund K, Schroder HD, Ortenblad N. Increased subsarcolemmal lipids in type 2 diabetes: effect of training on localization of lipids, mitochondria, and glycogen in sedentary human skeletal muscle. Am J Physiol Endocrinol Metab 298: E706–E713, 2010. [DOI] [PubMed] [Google Scholar]
- 22.Nyholm B, Qu Z, Kaal A, Pedersen SB, Gravholt CH, Andersen JL, Saltin B, Schmitz O. Evidence of an increased number of type IIb muscle fibers in insulin-resistant first-degree relatives of patients with NIDDM. Diabetes 46: 1822–1828, 1997. [DOI] [PubMed] [Google Scholar]
- 23.Ostergard T, Andersen JL, Nyholm B, Lund S, Nair KS, Saltin B, Schmitz O. Impact of exercise training on insulin sensitivity, physical fitness, and muscle oxidative capacity in first-degree relatives of type 2 diabetic patients. Am J Physiol Endocrinol Metab 290: E998–E1005, 2006. [DOI] [PubMed] [Google Scholar]
- 24.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A 100: 8466–8471, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perseghin G, Price TB, Petersen KF, Roden M, Cline GW, Gerow K, Rothman DL, Shulman GI. Increased glucose transport-phosphorylation and muscle glycogen synthesis after exercise training in insulin-resistant subjects. N Engl J Med 335: 1357–1362, 1996. [DOI] [PubMed] [Google Scholar]
- 26.Phielix E, Meex R, Moonen-Kornips E, Hesselink MK, Schrauwen P. Exercise training increases mitochondrial content and ex vivo mitochondrial function similarly in patients with type 2 diabetes and in control individuals. Diabetologia 53: 1714–1721, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Development Core Team. R: A Language and Environment for Statistical Computing. Boston, MA: R Foundation for Statistical Computing, 2008. [Google Scholar]
- 28.Redmon JB, Bertoni AG, Connelly S, Feeney PA, Glasser SP, Glick H, Greenway F, Hesson LA, Lawlor MS, Montez M, Montgomery B; Look AHEAD Research Group. Effect of the look AHEAD study intervention on medication use and related cost to treat cardiovascular disease risk factors in individuals with type 2 diabetes. Diabetes Care 33: 1153–1158, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter EA, Mikines KJ, Galbo H, Kiens B. Effect of exercise on insulin action in human skeletal muscle. J Appl Physiol 66: 876–885, 1989. [DOI] [PubMed] [Google Scholar]
- 30.Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH, Nilsson E, Tornberg A, Dekker Nitert M, Eriksson KF, Jones HA, Groop L, Ling C. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet 9: e1003572, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronn T, Volkov P, Tornberg A, Elgzyri T, Hansson O, Eriksson KF, Groop L, Ling C. Extensive changes in the transcriptional profile of human adipose tissue including genes involved in oxidative phosphorylation after a 6-month exercise intervention. Acta Physiol (Oxf) 211: 188–200, 2014. [DOI] [PubMed] [Google Scholar]
- 32.Russell RD, Kraemer RR, Nelson AG. Metabolic dysfunction in diabetic offspring: deviations in metabolic flexibility. Med Sci Sports Exerc 45: 8–15, 2013. [DOI] [PubMed] [Google Scholar]
- 33.Segerstrom AB, Holmback AM, Hansson O, Elgzyri T, Eriksson KF, Ringsberg K, Groop L, Wollmer P, Thorsson O. Relation between cycling exercise capacity, fiber-type composition, and lower extremity muscle strength and muscle endurance. J Strength Cond Res 25: 16–22, 2011. [DOI] [PubMed] [Google Scholar]
- 34.Shaw CS, Shepherd SO, Wagenmakers AJ, Hansen D, Dendale P, van Loon LJ. Prolonged exercise training increases intramuscular lipid content and perilipin 2 expression in type I muscle fibers of patients with type 2 diabetes. Am J Physiol Endocrinol Metab 303: E1158–E1165, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sparks LM, Johannsen NM, Church TS, Earnest CP, Moonen-Kornips E, Moro C, Hesselink MK, Smith SR, Schrauwen P. Nine months of combined training improves ex vivo skeletal muscle metabolism in individuals with type 2 diabetes. J Clin Endocrinol Metab 98: 1694–1702, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100: 9440–9445, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szendroedi J, Phielix E, Roden M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat Rev Endocrinol 8: 92–103, 2012. [DOI] [PubMed] [Google Scholar]
- 38.Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski M, Jensen T, Keller P, Scheele C, Vollaard NB, Nielsen S, Akerstrom T, MacDougald OA, Jansson E, Greenhaff PL, Tarnopolsky MA, van Loon LJ, Pedersen BK, Sundberg CJ, Wahlestedt C, Britton SL, Bouchard C. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol 108: 1487–1496, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toledo FG, Menshikova EV, Ritov VB, Azuma K, Radikova Z, DeLany J, Kelley DE. Effects of physical activity and weight loss on skeletal muscle mitochondria and relationship with glucose control in type 2 diabetes. Diabetes 56: 2142–2147, 2007. [DOI] [PubMed] [Google Scholar]
- 40.Ukropcova B, Sereda O, de Jonge L, Bogacka I, Nguyen T, Xie H, Bray GA, Smith SR. Family history of diabetes links impaired substrate switching and reduced mitochondrial content in skeletal muscle. Diabetes 56: 720–727, 2007. [DOI] [PubMed] [Google Scholar]
- 41.van Tienen FH, Praet SF, de Feyter HM, van den Broek NM, Lindsey PJ, Schoonderwoerd KG, de Coo IF, Nicolay K, Prompers JJ, Smeets HJ, van Loon LJ. Physical activity is the key determinant of skeletal muscle mitochondrial function in type 2 diabetes. J Clin Endocrinol Metab 97: 3261–3269, 2012. [DOI] [PubMed] [Google Scholar]
- 42.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33: W741–W748, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


