Abstract
A significant effort is made by the cell to maintain certain phospholipids at specific sites. It is well described that proteins involved in intracellular signaling can be targeted to the plasma membrane and organelles through phospholipid-binding domains. Thus, the accumulation of a specific combination of phospholipids, denoted here as the ‘phospholipid code', is key in initiating cellular processes. Interestingly, a variety of extracellular proteins and pathogen-derived proteins can also recognize or modify phospholipids to facilitate the recognition of dying cells, tumorigenesis and host–microbe interactions. In this article, we discuss the importance of the phospholipid code in a range of physiological and pathological processes.
Facts
We have defined the ‘phospholipid code' as the pattern of phospholipids being exposed intracellularly and extracellularly by host cells and microbes that can regulate a diverse array of cellular processes.
The biological consequences of cell death can be determined by the phospholipid code being displayed to the immune system.
An altered phospholipid code in the tumor microenvironment can favor tumor progression and serve as a therapeutic target.
A wide variety of pathogens exploit the host phospholipid code to facilitate infection and survival inside the host. Likewise, the phospholipid code of pathogens may be targeted by peptides of innate immunity to protect host cells from infection.
Open Questions
What is the evidence to support the existence of a phospholipid code?
What is the function of a phospholipid code?
How can the phospholipid code be targeted to treat human diseases?
The display of specific phospholipids at various intracellular locations is essential in regulating a range of important cellular processes. It has been well characterized that the recruitment of proteins to the plasma membrane is fundamental for initiating and regulating signal transduction events. Besides protein–protein interactions, signaling proteins are often targeted directly to various membrane sites by interacting with specific phospholipids. Details of these processes as well as other key intracellular functions of phospholipids are outlined in Box 1. In addition to the intracellular role of phospholipids, membrane-derived extracellular vesicles (for example, apoptotic bodies, microparticles, exosomes, matrix vesicles), healthy and dying/dead cells, as well as enveloped pathogens can also display a distinct pattern of phospholipids extracellularly to regulate processes including skeletal development, immunity and coagulation.1, 2, 3 Interestingly, pathogens have also evolved to express a variety of molecules that can either interact with or modify their host's phospholipids intracellularly and extracellularly to aid the infection process. Similarly, host organisms are also armed innately with numerous phospholipid-recognition proteins that can interact with pathogens and protect the host from microbial assault. These observations, which will be discussed further herein, strongly indicate the existence of a complex intracellular and extracellular ‘phospholipid code' being displayed on host cells and microbes.
Phospholipids as key regulators of intracellular processes.
The localization of specific phospholipids at various intracellular sites can have an important role in regulating signaling events. For example, SHP-1 phosphatase can be localized to the plasma membrane through recognition of phosphatidic acid (PA)122 and subsequently regulate signal transduction by dephosphorylating proteins such as B cell linker protein.123 Similarly, the recruitment of phospholipase Cδ1 to phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) located at the inner leaflet of the plasma membrane through its pleckstrin homology domain can mediate hydrolysis of PI(4,5)P2, and subsequently generate second messengers diacylglycerol and inositol 1,4,5-trisphosphate.124
In addition to cell signaling, the recruitment of proteins to specific membrane compartments via phospholipid recognition can modulate a variety of other processes including endocytosis, phagocytosis, membrane/organelle fusion, membrane–cytoskeleton adhesion and ion channel function. For example, adaptor protein 180, a key component of clathrin-mediated endocytosis, can interact with both PI(4,5)P2 and clathrin simultaneously to facilitate the formation of clathrin lattice at the plasma membrane.125 Likewise, the Ca2+-sensing protein, synaptotagmin, mediates neurotransmitter release via interacting with PI(4,5)P2 and t-SNARE at the pre-synaptic axon terminal of neurons.126 Similarly, assembly of factors required for early endosome fusion involves the recruitment of early endosome antigen 1 protein to phosphatidylinositol 3-phosphate (PI(3)P) located at the cytosolic leaflet of endosome via its FYVE domain.127
As the availability of certain phospholipids is key in orchestrating the cellular localization of various phospholipid-binding proteins, a number of enzymatic- and non-enzymatic-based mechanisms are present to tightly regulate this process. The levels of phosphatidylinositol mono-/bis-/tri-phosphate are controlled by the activities of lipid kinases and phosphatases. For example, synthesis of PI(4,5)P2 from phosphatidylinositol 4-phosphate (PI(4)P) at the plasma membrane is mediated by type I PI(4)P 5-kinases.128 Likewise, the phosphoinositide phosphatase Sac1 has an important role in regulating the turnover of PI(4)P at the endoplasmic reticulum and golgi complex.129, 130 Besides phosphatidylinositols, levels of the mitochondrial glycerophospholipid, cardiolipin, are also tightly controlled. Cardiolipin is abundant in the inner mitochondrial membrane where it acts as a key component of the electron transport chain, as well as having an essential role in the initiation of apoptosis through association with cytochrome C.131 To coordinate these essential mitochondrial processes, cardiolipin synthase as well as a range of phospholipases exquisitely maintain cardiolipin levels within the mitochondria.132 Another key mechanism in regulating the availability of phospholipids at different locations, particularly on which surface of the phospholipid bilayer, is mediated by nonspecific and specific lipid transporters. The presence of nonspecific lipid transporters at the endoplasmic reticulum is thought to maintain equal distribution of phospholipids on both the luminal and cytosolic surface.133 In contrast, phospholipid asymmetry at the plasma membrane, with phosphatidylserine (PS) and phosphatidylethanolamine (PE) found mainly at the inner leaflet, is regulated by a number of lipid transporters including P-type ATPases and ABC transporters134 (for further discussion, see below). In addition to controlling the absolute levels and distribution of phospholipids, the availability of certain phospholipids can simply be masked by the binding of phospholipid-binding proteins. For example, profilin, a PI(4,5)P2-binding protein involved in cytoskeletal restructuring and dynamic actin turnover, can competitively inhibit phospholipase C-mediated hydrolysis of PI(4,5)P2 when bound.104, 135
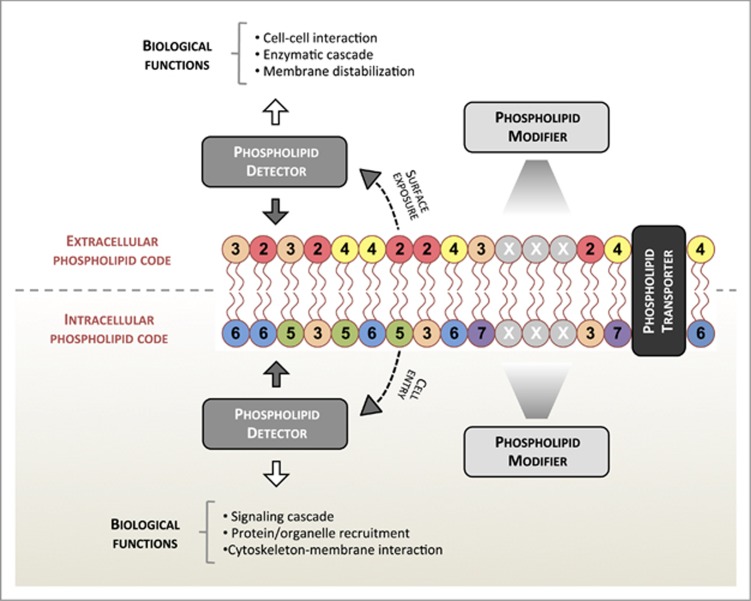
We would like to define the ‘phospholipid code' as the pattern of phospholipids being exposed intracellularly and extracellularly by host cells and microbes that could influence the fate of altered-self (dying/dead cells) recognition, tumor progression and host–microbes interactions. How the phospholipid code may function in these settings and be regulated by molecules derived from host and microbes is shown schematically in Figure 1. These well-described processes are chosen as examples to demonstrate clearly how the phospholipid code could operate both intracellularly and extracellularly in health and disease. However, it should be noted that in addition to the roles of phospholipids in these key processes and those outlined in Box 1, the phospholipid code could also have an important role in other processes such as mineral deposition, myotube formation and spermatogenesis. Furthermore, although this review will focus mainly on protein–lipid interactions, it is also important to note that protein–protein as well as protein–carbohydrate interactions may also work in concert with the phospholipid code.
Figure 1.
The phospholipid code. The phospholipid code may become exposed either extracellularly or intracellularly. Different colors and numbers represent different types of phospholipids present on the membrane. The phospholipid code can be recognized by phospholipid detectors and subsequently orchestrate a variety of biological functions. Certain phospholipid detectors may also pass through the membrane to exert their activities. The phospholipid code can be maintained or changed by enzymes that directly modify phospholipid head groups (phospholipid modifiers) or by phospholipid transporters that regulate the arrangement of lipids present on each side of the phospholipid bilayer. It is important to note that the intracellular phospholipid code can also be displayed on other organelles
Sensing Dying/Dead Cells via the Phospholipid Code
The immune system has an important role in the detection and elimination of foreign pathogens (non-self) from healthy cells (self). Likewise, the ability of the immune system to distinguish healthy cells from dying/dead cells (altered self) under physiological and pathological conditions (for example, tissue remodeling and tissue injury) is also a key process to ensure efficient clearance of unwanted or damaged cells.4 It is well described that cells can undergo cell death via a number of distinct pathways, generating different types of dying/dead cells including apoptotic and necrotic cells.4 Importantly, dying/dead cells can expose a variety of molecules, particularly phospholipids, to trigger their recognition and removal by the immune system.3
In healthy cells, the specific distribution of different phospholipid species across the cytosolic and outer leaflet of the plasma membrane is mediated by a group of membrane-bound enzymes broadly termed flippases and floppases.5 These enzymes are responsible for ATP-dependent inward and outward transport of lipids, respectively, to maintain phospholipid asymmetry at the membrane. For example, the anionic phospholipids PS and PE are maintained predominantly at the cytosolic leaflet of the plasma membrane through aminophospholipid translocase flippases, while phosphatidylcholine (PC), sphingolipid and cholesterol are transported to the outer leaflet via floppases.5 In addition to these ATP-dependent enzymes, scramblases, which can mediate ATP-independent bi-directional movement of lipids across the membrane, are also involved in maintaining phospholipid asymmetry.6, 7 During the course of apoptosis (a form of programmed cell death), the lipid distribution at the plasma membrane is modified by a variety of mechanisms, resulting in the exposure of anionic lipids such as PS to the outer membrane,3 and thus changing the ‘extracellular phospholipid code'. Recently, Xk-related protein 8 and the nematode homolog CED-8 were identified to be involved in promoting PS exposure on apoptotic cells following caspase-mediated cleavage and subsequently enhancing phospholipid scrambling activity at the plasma membrane.8 Furthermore, the flippase ATP11C was also identified as a key regulator of PS exposure on apoptotic cells, in which caspase-mediated inactivation of ATP11C promoted the distribution of PS on the outer leaflet of the plasma membrane during apoptosis9 (Figure 2a). It is also worth noting that PS can be modified by reactive oxygen species (ROS; a form of ‘phospholipid modifier') to generate oxidized PS in certain cell types, a process that could aid PS externalization10, 11 and recognition by phagocytes.12
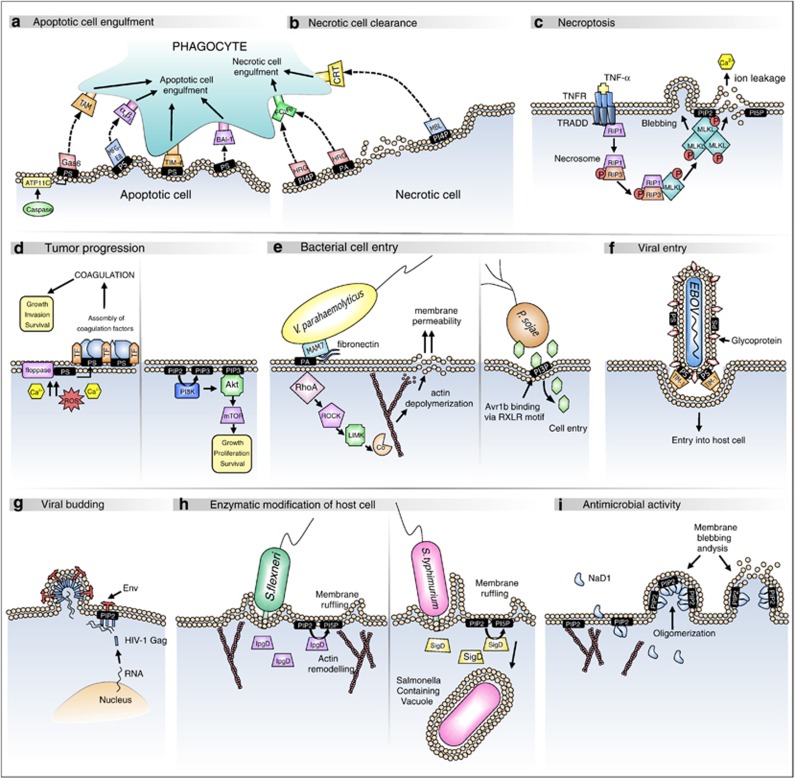
Figure 2.
Controlling a variety of extracellular and intracellular processes via the phospholipid code. (a) During apoptosis, loss of phospholipid asymmetry due to inactivation of ATP11C flippase induces PS exposure. PS acts as an ‘eat me' signal and can be detected by receptors on neighboring phagocytes (directly) and serum opsonins (indirectly) to trigger engulfment. (b) Similarly, cells undergoing necrosis exposes additional phospholipids such as PI(4)P and PA, which can be recognized by serum opsonins such as histidine-rich glycoprotein and mannose-binding lectin to trigger phagocytic engulfment. (c) The caspase-independent programmed cell death pathway of necroptosis involves formation of the ‘necrosome' that activates MLKL, leading to its translocation to the plasma membrane. MLKL oligomerization upon contact with PIP2 (PI(4,5)P2) and PI(5)P then mediates membrane permeabilization. (d) In the tumor microenvironment, elevated cytosolic Ca2+ and ROS production can trigger a loss in phospholipid asymmetry and expose anionic phospholipids on tumor vascular endothelium. PS exposure and TF overexpression can then promote assembly of circulating coagulation factors such as FIXa and FVIIIa on the tumor cell membrane to initiate blood clotting, which can drive tumor progression through elevated growth factor signaling (left). Abberant regulation of PI3K can lead to elevated levels of the plasma membrane second messenger PIP3 (PI(3,4,5)P3), a process that could mediate recruitment and activation of Akt and subsequently mTOR to promote tumor proliferation and survival (right). (e) The Gram-negative bacteria, V. parahaemolyticus uses MAM7, an adhesion molecule that interacts with both PA and fibronectin on intestinal epithelial cells, to mediate host cell entry (left). The oomycetes P. sojae translocates an effector molecule, Avr1b into the host cell membrane to repress host defenses, via binding of its RXLR motif to outer membrane PI(3)P (right). (f) Viruses such as the ebola virus use ‘apoptotic mimicry' to enter target cells. Ebola virus exposes PS on its viral envelope that binds to the TIM-1 receptor on host cells to facilitate viral uptake. (g) In HIV-1 infected cells, the HIV-1 Gag protein is trafficked to the plasma membrane via its matrix (MA) domain where it interacts with PIP2 to initiate viral budding. (h) Enzymatic modification of host cell phospholipids by microbial virulence factors can aid infection. S. flexneri secretes IpgD into intestinal host cells that acts as phosphoinositol 4-phosphatase to deplete plasma membrane PIP2, leading to actin remodeling and membrane ruffling (left). SigD from S. typhimurium displays similar phosphatase activity to induce membrane ruffling and facilitate formation of Salmonella-containing vacuoles, thereby enabling bacterial replication (right). (i) The plant defensin NaD1 induces cytolysis of both fungal and mammalian tumor cells through binding to plasma membrane PIP2, with which it forms an oligomeric complex, leading to membrane permeabilization. TAM, Tyro-3, Axl, Mer; Gas6, growth arrest-specific 6; MGF-E8, milk fat globule-EGF factor 8; TIM-4, T cell/transmembrane, Ig, mucin-4; BAI-1, brain-specific angiogenesis inhibitor 1; HRG, histidine-rich glycoprotein; CRT, calreticulin; MBL, mannose-binding lectin; RhoA, ras homolog gene family, member A; ROCK, rho-associated protein kinase; LIMK, LIM motif-containing protein kinase; Co, cofilin; EBOV, Ebola virus; TIM-1, T cell immunoglobulin and mucin domain 1; Env, envelope glycoprotein
The exposure of PS on the surface of apoptotic cells can function as a key ‘eat-me' signal to facilitate apoptotic cell recognition by phagocytes via a variety of ‘phospholipid detectors' (Table 1). For example, PS can be recognized by cell surface receptors including TIM-4 (Kobayashi et al.13 and Miyanishi et al.14) and BAI-1.15 PS can also be recognized by soluble opsonins such as MFG-E8 and Gas6, that are in turn detected by cell surface molecules αVβ3 and TAM receptors, respectively16, 17, 18 (Figure 2a). The recognition of PS by phagocytes through various phospholipid detectors is an important molecular step to trigger the engulfment of apoptotic cells,3 and exemplifies how modification of the phospholipid code on the surface of apoptotic cells can promote their clearance. Besides apoptotic cells, the exposure of PS on pyrenocytes (that is, the erythroid nucleus extruded from the erythroblast during the final stage of erythrocyte differentiation) can also facilitate their removal by macrophages.19, 20 Furthermore, it is important to note that the phospholipid code on dying cells can undergo further changes when the plasma membrane is permeabilized to generate necrotic cells.4 This is particularly evident when healthy cells are exposed to extreme chemical or physical assault to generate primary necrotic cells, or when apoptotic cells are not promptly removed and resulted in the formation of late apoptotic cells (also known as secondary necrotic cells).4 Under these necrotic conditions, phospholipids that are normally localized at the inner leaflet of the plasma membrane can become exposed to the extracellular milieu.4 This represents a unique situation in which the partitioning of the extracellular and intracellular phospholipid code is abolished. Interestingly, there is growing evidence supporting the notion that serum opsonins such as histidine-rich glycoprotein and mannose-binding lectin can detect other types of phospholipids such as PI(4)P and PA on necrotic cells to facilitate their removal by phagocytes3, 21, 22, 23 (Figure 2b).
Table 1. Host- and pathogen-derived phospholipid detectors.
| Phospholipid detector | Lipid binding | Comments | Reference |
|---|---|---|---|
| Intracellular proteins | |||
| SH2 domain-containing phosphatase 1 (SHP-1) | PA, PI(3,4,5)P3, PI(4,5)P2, PS, dipalmitoylphosphatidylglycerol | Protein tyrosine phosphatase that can negatively regulate signaling pathways by dephosphorylating signaling proteins such as BLNK and SLP-76 | Frank et al.122 |
| Phospholipase C delta 1 (PLCδ1) | PI(4,5)P2 | Phospholipase that hydrolyzes PtdIns(4,5)P2 to generate second messengers diacylglycerol and IP3 | Ochocka et al.124 |
| Adaptor protein 180 (AP180) | PI(4,5)P2 | Membrane-bound protein involved in the formation of clathrin lattice during clathrin-mediated endocytosis | Ford et al.125 |
| Early endosome antigen 1 protein (EEA1) | PI(3)P | Endosomal membrane protein required for early endosome fusion | Kutateladze et al.127 |
| Profilin | PI(4,5)P2 | Abundant actin-binding protein that regulates actin polymerization and membrane trafficking | Goldschmidt-Clermont et al.104 Krishnan et al.135 |
| Mixed lineage kinase-like domain (MLKL) | PI(4,5)P2, PI(5)P | Soluble protein involved in the necroptosis pathway, phosphorylated by RIPK3 and translocated to the plasma membrane to induce membrane permeabilization | Dondelinger et al,27 |
| Cytochrome C | Cardiolipin | Inner mitochondrial membrane-bound protein that associates with cardiolipin and acts as a peroxidase to facilitate the release of Cytochrome C into the cytosol upon apoptotic stimuli | Paradies et al.131 |
| Akt | PI(3,4,5)P3 | Soluble protein involved in a wide range of cellular processes including activation of pro-survival pathways | Wong et al.56 Markman et al.57 |
| Extracellular host-derived proteins | |||
| Milk fat globule-EGF factor 8 (MFG-E8) | PS | Mammalian soluble protein secreted by macrophage that enhances apoptotic cell clearance | Hanayama et al.16 |
| Growth arrest-specific protein 6 (Gas6) | PS | Mammalian serum protein that facilitates apoptotic cell engulfment | Ishimoto et al.17 |
| Histidine-rich glycoprotein (HRG) | PA, cardiolipin, sulfitide, PI(4)P, LPS | Mammalian multifunctional plasma protein involved in coagulation, pathogen/necrotic cell clearance | Poon et al.4 |
| T-cell immunoglobulin mucin-4 (TIM-4) | PS | Mammalian cell surface receptor involved in apoptotic cell uptake | Kobayashi et al.13 Miyanishi et al.14 |
| Brain-specific angiogenesis inhibitor 1 (BAI-1) | PS, cardiolipin, sulfitide, PI(4)P | Mammalian cell surface receptor involved in angiogenesis, pathogen/apoptotic cell clearance | Park et al.15 |
| FIXa:FVIIIa (intrinsic tenase complex; Xase) | PS | Complex formed by Factors IXa and VIIIa during activation of the coagulation cascade that binds to exposed PS on suface of platelets to facilitate platelet aggregation | Adams et al.36 |
| Nicotiana alata Defensin 1 (NaD1) | A range of phosphoinositol mono-/bis-/tri-phosphate, cardiolipin, sulfitide | Plant cationic antimicrobial peptide of Nicotiana alata that exhibits antifungal and anticancer activities | Poon et al.3 |
| Tomato pistil-predominant 3 (TPP3) | PI(4,5)P2 | Plant cationic antimicrobial peptide of Solanum lycopersicum that exhibits antifungal and anticancer activities | Baxter et al.70 |
| Meidcago trunatula defensin 4 (MtDef4) | PA | Plant cationic antimicrobial peptide of Medicago trunatula that exhibits antifungal activities | Sagaram et al.48 |
| Pathogen-derived proteins | |||
| Multivalent adhesion molecule 7 (MAM7) | PA | Gram-negative bacterial adhesion protein that binds PA and fibronectin to mediate infection of intestinal epithelial cells | Krachler et al.74 |
| CagA | PS | Multifunctional effector protein of Helicobacter pylori that modifies host cell signaling | Murata-Kamiya et al.83 |
| LTcyt | PS | Lethal toxin of Clostridium sordellii that functions as a glucosyltransferase in the cytoplasm of host cells | Mesmin et al.98 |
| HIV-1 Gag | PI(4,5)P2, PS | HIV-1 structural protein that mediates viral assembly at the plasma membrane of host cells | Chukkapalli et al.99 |
| EIAV Gag | PI(4,5)P2,, PI(3)P, PI(3,5)P2 | Equine inffective anemia virus (EIAV) protein that mediates viral assembly at the plasma and intracellular membranes of host cells | Fernandes et al.101 |
| Vaccinia profilin | PI(4)P, PI(4,5)P2 | Vaccinia viral protein that binds strongly to phosphoinositides | Machesky et al.102 |
| Avr1b | PI(3)P, PI(4)P | Effector protein produced by the oomycetes Phytophthora sojae that enters plant and animal cells | Kale et al.81 |
| Exoenzyme S | PE | Virulence factor secreted by Helicobacter pylori and other bacteria to facilitate host cell adhesion | Lingwood et al.76 |
In addition to apoptosis and primary necrosis, the caspase-independent programmed cell death pathway of necroptosis has also been shown to depend on recognition of specific phospholipids at the inner leaflet of the plasma membrane (the ‘intracellular phospholipid code') to trigger cell lysis. When apoptosis is blocked, for example, using the pan caspase inhibitor zVAD-fmk, assembly of the ‘necrosome' is triggered within the cytosol, a complex formed by receptor-interacting serine/threonine-protein kinase-1 and -3 (RIPK1 and RIPK3, respectively),24 followed by recruitment and activation of mixed lineage kinase domain-like (MLKL) protein.25, 26 Recently, it has been proposed that during necroptosis, MLKL is translocated to the plasma membrane where it oligomerizes upon interaction with PI(4,5)P2 and PI(5)P via its N-terminal four helical bundle domain to induce membrane permeabilization27 (Figure 2c). Interestingly, whether necroptotic, primary necrotic and secondary necrotic cells are recognized by the same phospholipid detector(s) to mediate their removal is unclear.
Alteration of the Phospholipid Code During Tumor Progression
Modification of the extracellular phospholipid code
Besides dying/dead cells, cells associated with tumor progression (a form of altered self) can also undergo modification of their lipid composition. The loss of plasma membrane phospholipid asymmetry is reported in a number of malignant cell types as well as in the vasculature of solid tumors.28 In particular, environmental stressors within the tumor microenvironment can alter the activity of ‘phospholipid transporters' and thereby the ability of cells to maintain plasma membrane phospholipid asymmetry, leading to a change in the phospholipid code. For example, elevated production of ROS is a common feature of solid tumors. Along with hypoxic conditions, ROS can elevate cytosolic Ca2+ levels and lead to the activation of Ca2+-dependent scramblases and floppases to promote the exposure of PS and, to a lesser extent, PE on the lumenal surface of tumor vascular endothelium28, 29, 30 (Figure 2d).
In addition to dysregulation of phospholipid transporters, altered gene expression of multidrug-resistant proteins (ABC transporters), which can also function to outwardly transport phospholipids across the membrane, has been shown to increase PS exposure on the cell surface of certain tumor cell types.31, 32 In EPG85-257 gastric carcinoma cells overexpressing multidrug-resistant 1 P-glycoprotein (MDR1 Pgp), the elevated exposure of cell surface PS was suggested to result from MDR1 Pgp-mediated floppase activity.31 Furthermore, EPG85-257 cells and MCF7 human breast cancer cells overexpressing the breast cancer resistance protein, BCRP, were also shown to outwardly transport PS, leading to increased cell surface exposure. However, the specific role of BCRP in the observed outward phospholipid transfer was not determined.32
Despite PS exposure on dying/dead cells being a key modification of the extracellular phospholipid code to trigger recognition and clearance by phagocytes (see above), it is interesting to note that in the context of cancer, PS exposure can act favorably towards tumor cell survival and metastasis by initiating the coagulation cascade. It is well documented that activation of the blood coagulation cascade is a common feature of cancer and is associated with increased malignancy and poor outcomes.33, 34, 35 Under non-pathological conditions, activation of the blood coagulation cascade is triggered in response to tissue injury or trauma and occurs through the activation of a suite of coagulation factors and complexes.36 It is initiated by exposure of the subendothelial transmembrane glycoprotein, tissue factor (TF), to blood, where it forms a complex with circulating factor VIIa (TF:FVIIa). This complex then activates zymogens, factor IX and factor X to generate thrombin and several other factors and complexes to initiate clotting. Elevated cytosolic calcium under these conditions leads to increased PS exposure on platelets, with PS acting as a negatively charged platform upon which downstream complexes FIXa:FVIIIa (intrinsic tenase complex) and FXa:FVa (prothrombinase complex) bind to facilitate platelet aggregation36 (Figure 2d). In the tumor microenvironment, TF activity is enhanced by elevated cell surface PS exposure, which can trigger the recruitment of circulating factors to initiate coagulation.37 Activation of the coagulation cascade by tumor cells can subsequently stimulate the release of a range of pro-survival factors associated with cancer progression. For example, the clot-forming protein, fibrin, can sequester growth factors such as VEGF, bVEGF and IGF-1 to tumor cells as well as act as a scaffold for the formation of new blood vessels, while thrombin can interact with cell surface receptors to upregulate growth factors as well as matrix metalloproteinases to further promote angiogenesis and metastasis.38 Thus, the extracellular phospholipid code on tumor cells and tumor vascular endothelium can have an important role in cancer progression.
Therapeutic potential of targeting the extracellular phospholipid code
A change in the extracellular phospholipid code following exposure of anionic phospholipids on cell surfaces (as described above) has become a valuable biomarker of malignancy. To this end, a number of tools have been developed to monitor the exposure of phospholipids during tumor progression. The rat mAb, 9D2, detects a range of anionic phospholipids including PS. 9D2 and annexin V (a PS-binding protein) were both shown to target PS at the vasculature of several tumor types in mice, with PS exposure shown to be transient and likely to be dependent on oxidative conditions.39 Similarly, a PE-specific probe, biotinylated duramycin, was targeted to tumor blood vessels in a range of mouse tumors in vivo, a process that can be further enhanced by oxidative stress.30 3G4, a mouse mAb that binds anionic phospholipids was targeted specifically to the surface of cultured endothelial cells and breast cancer cells under oxidative conditions, as well as tumor vasculature.40 Here, 3G4-mediated phospholipid binding was dependent on the presence of serum protein beta-2 glycoprotein I, which was suggested to either form a conformational epitope or to crosslink with the mAb, to facilitate binding. Importantly, 3G4 was shown to reduce the growth of tumors in vivo, possibly via Fc receptor-mediated antibody-dependent cell clearance by monocytes and macrophages.40 The effects of 3G4 on tumor growth were subsequently found to be enhanced by further increasing anionic phospholipid exposure on tumor vasculature, either using the chemotherapy drug, docetaxel or via treatment with radiation therapy in mouse models of cancer.41, 42 Similar effects were observed for the PS-targeted mouse mAb mch1N11, whereby a combination of cisplatin (a chemotherapeutic) and mch1N11 showed greater therapeutic efficacy in treating murine lung tumors than with cisplatin or mch1N11 alone.43 Progressing from in vitro studies and animal models, a commercially available chimeric version of 3G4 (known as bavituximab) has been used in combination therapy in human clinical trials. In a 2014 phase II clinical trial among 49 locally advanced or metastatic non-small-cell lung cancer patients, bavituximab combined with carboplatin and paclitaxel treatment resulted in a 41% response rate and was considered to show tolerable safety and efficacy.44 More recently, a phase I clinical trial with bavituximab in combination with paclitaxel has shown more promising results among 14 HER2-negative metastatic breast cancer patients, with an 85% response rate.45 In addition, an ongoing phase III trial involving the treatment of advanced non-small-cell lung cancer with bavituximab and docetaxel is currently in progress.46 Collectively, these studies highlight the potential of targeting and manipulating the extracellular phospholipid code therapeutically in cancer settings.
Besides phospholipid-binding antibodies, innate defense molecules can also target the altered cell surface phospholipid profile of tumor cells. Cationic antimicrobial peptides (CAPs) are a class of small (<5 kDa) positively charged innate defense molecules that act primarily to destabilize or penetrate the membrane of microbial cells, in some cases via interactions with specific phospholipids47, 48 (functioning as phospholipid detectors). Interestingly, CAPs originated from different species including plants, insects, amphibians, mammals have been shown to possess antiproliferative activity towards mammalian tumor cells both in vitro and in vivo (reviewed in detail by Riedl et al.49). It has been suggested that increased expression of sialic acid-rich mucins50, 51 and heparan sulfate glycosaminoglycans,52 as well as the exposure of anionic phospholipids such as PS32, 53 on the surface of tumor cells, are likely to contribute to the selectivity of these CAPs towards tumor cells. For example, NK-2, a synthetic cationic peptide derived from the porcine antimicrobial peptide NK-lysin, was tested on a range of human tumor cell lines and displayed significantly greater cytotoxic activity towards cell types that were exposing elevated levels of outer membrane PS.54 Similarly, treatment of a range of mammalian tumor cell lines with four enantiomeric 9-mer CAPs derived from beetle defensins revealed a strong positive correlation between the sensitivity of cell types to CAP-mediated cytotoxicity and PS exposure.55
Modification of the intracellular phospholipid code
Besides changes in the extracellular phospholipid code, the intracellular phospholipid code can also be modified in tumor cells and regulate cancer progression. In particular, modulating the levels of either phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3) or PA at the inner leaflet of the plasma membrane of tumor cells has been shown to promote tumorigenesis by altering signaling events. For PI(3,4,5)P3, its level at the plasma membrane is markedly augmented by the dysregulation of the PI3K/Akt/mTOR pathway in many cancers.56 Generated by PI3K in response to growth factors, PI(3,4,5)P3 functions as a phospholipid second messenger that mediates the recruitment and activation of Akt (a pro-survival protein) by acting as a docking site at the plasma membrane for Akt. Subsequently, downstream activation of mTOR by Akt can regulate a variety of cellular processes including cell proliferation and survival57 (Figure 2d). Mutations commonly seen in the α-catalytic subunit of PI3K58 as well as mutations or deletions in PI3K pathway's main regulatory gene, PTEN,59, 60 have been reported in many malignant phenotypes, resulting in elevated levels of PI(3,4,5)P3. Likewise, PA has also been implicated in the progression of cancer through its role as an upstream mediator of mTOR signaling. PA is generated by phospholipase D-mediated cleavage of the choline head group of PC and activates mTOR-mediated mitogenic signaling via binding to the rapamycin binding domain of mTOR.61 Phospholipase D overexpression has been reported for many cancer types including breast cancer cell lines and tissue samples, as well as renal and gastric cancers and is associated with mTOR-mediated pro-survival phenotypes.62, 63, 64, 65 Thus, modification of the intracellular phospholipid code by changes in the expression and/or activity of various phospholipid modifiers can ultimately regulate tumor progression.
Therapeutic potential of targeting the intracellular phospholipid code
Cancer treatments involving chemotherapeutics that target phospholipid modifiers have the potential to ‘correct' the altered intracellular phospholipid code at the tumor cell membrane. For example, inhibition of PI3K, of which there are four human isoforms, can effectively modify levels of PI(3,4,5)P3 at the inner plasma membrane leaflet to limit aberrant downstream pro-survival processes. PI3K inhibitors that are used in the clinic can be classified into three categories, including pan-PI3K inhibitors, isoform-specific inhibitors and dual PI3K-mTOR inhibitors (reviewed in detail by Scott et al. 2009).66 To a lesser extent, the potential of using phospholipase D inhibitors as anticancer therapeutics has also been explored. Phospholipase D inhibitors have been shown to block invasive migration of human breast cancer lines and enhance the sensitivity of tumor cells towards radiotherapy.67, 68 Besides modulating the activity of phospholipid modifiers, targeting phospholipids directly at the inner leaflet of the tumor cell membrane has also been examined as a therapeutic approach. As mentioned above, certain CAPs can kill tumor cells by permeabilizing the cell membrane.49 Recently, it was demonstrated that the plant defensin NaD1, a CAP isolated from the ornamental tobacco Nicotiana alata, can selectively induce membrane lysis (primary necrosis) of mammalian tumor cells through binding PI(4,5)P2 at the inner leaflet of the plasma membrane at low micromolar concentrations.69 Although how NaD1 enters the tumor cell and gains access to the intracellular phospholipid code is unclear, the ability of NaD1 to form a multimeric complex with the phosphatidylinositol head group of PI(4,5)P2 suggests the possibility of NaD1 acting as a phospholipid detector that can mask the cellular function of PI(4,5)P2, discussed further below.69 More recently, the closely related tomato defensin, TPP3, was also shown to induce PI(4,5)P2-mediated tumor cell permeabilization via a similar mechanism.70
Exploiting the phospholipid code during host–pathogen interaction
Pathogens commonly hijack and take advantage of host molecules to facilitate their invasion and replication. An obvious example is the ability of viruses to bind certain receptors expressed by specific cell types to facilitate cell entry, and subsequently utilize intracellular machineries for nucleotide and protein production.71, 72 In addition, some viruses encode proteins that share structural and functional similarities with host molecules, such as herpes and myxoma viruses, that mimic anti-apoptotic protein BCL-2 to prolong viral replication before the initiation of host-programmed cell death.73 Not surprisingly, pathogens including bacteria, viruses, fungi and oomycetes also exploit the host's phospholipid code for a variety of functions during an infection (Table 1).
Pathogenic Entry via the Host Extracellular Phospholipid Code
Phospholipid-associated adhesion proteins
The efficient and targeted adhesion of pathogens to a specific type of host cell is a critical process during an infection and the extracellular phospholipid code on host cells is frequently exploited by a range of pathogens to mediate cell attachment. A variety of Gram-negative bacteria including Vibrio parahaemolyticus, Vibrio cholerae and Yersinia pseudotuberculosis encode MAM7 (seven mammalian cell entry domain-containing multivalent adhesion molecule) that can bind to both PA as well as extracellular matrix protein fibronectin, forming a complex to mediate adhesion of MAM7-expressing bacteria to fibroblasts74 (Figure 2e). Recently, it was shown that during adhesion of the marine bacterium Vibrio parahaemolyticus to intestinal epithelial cells, the interaction between MAM7 and cell surface PA activates the GTPase signaling protein, RhoA. It was hypothesized that RhoA signaling could facilitate actin depolymerization through the activation of ROCK/LIMK/cofilin downstream signaling, resulting in impaired epithelial barrier function and allowing bacterial movement into the cell.75 Likewise, Helicobacter pylori secretes a virulence factor, exoenzyme S, that binds to exposed PE on the host gastric epithelial cell surface to facilitate attachment.76 Cell surface PE has also been reported as the lipid ligand for adhesion proteins of several other bacterial species, including enteropathogenic and enterohemorragic Escherichia coli strains,77 Chlamydia trachomatis and Chlamydia pneumoniae.78 In addition to utilizing PE to facilitate host cell adhesion, enteropathogenic Escherichia coli has also been reported to induce transient accumulation of both PI(4,5)P2 and PI(3,4,5)P3 at the inner leaflet of host cells at the site of interaction to promote bacterial attachment, possibly by stimulating local actin accumulation to facilitate the formation of an actin-rich pedestal.79 As the adhesion of pathogens to host cells is key for initiating the infection process, targeting pathogen-derived adhesion molecules by inhibiting their ability to interact with phospholipids on host cells is an attractive therapeutic approach to limit infection. Indeed, targeting the pathogen-derived MAM7 (an adhesion molecule that can function as a phospholipid detector) using a peptide-based inhibitor was found to prevent the adhesion of methicillin-resistant Staphylococcus aureus to fibroblasts,80 possibly by interfering with the interaction between MAM7 and PA on host cells.
Phospholipid-associated effector proteins
In addition to cell adhesion proteins, the ability of pathogen-derived effector molecules to bind and enter into host cells can also be mediated via the recognition of specific phospholipids. For example, the oomycete Phytophthora sojae contains an effector molecule, Avr1b, that translocates into the cytoplasm of host cells to repress host defenses via a host-targeting signal. This signal contains an N-terminal RXLR motif which recognizes PI(3)P on the outer plasma membrane of both plant and human cells.81 Besides the well-known intracellular functions of PI(3)P, PI(3)P was detected on the cell surface of soybean roots and human lung epithelial cells, with the exposure of PI(3)P found to be necessary for Avr1b binding and entry in both cell types81 (Figure 2e). Furthermore, another effector protein from the same species, Avh5, was found to enter soybean root and human lung epithelial cells via PI(3)P binding, with both the RXLR motif as well as a lysine-rich region shown to be required for cell entry.82 Likewise, fungal-derived effectors AvrL567 (Melampsora lini), Avr2 (Fusarium oxysporum f. sp. lycopersici) and AvrLm6 (Leptosphaeria maculans) containing the RXLR motif were also shown to bind PI(3)P and facilitate protein entry into soybean root cells.81 Interestingly, Helicobacter pylori can modify the host phospholipid code by inducing PS exposure on epithelial cells via its bacterial Type IV secretion system upon initial membrane contact. Exposure of PS then facilitates translocation of CagA, a secreted virulence factor that interacts directly with PS via its N-terminal region, into host cells. This enables CagA to subsequently modify host cell signaling to induce the formation of gastric carcinoma.83 Thus, pathogen-derived effector molecules that exhibit phospholipid detector properties can exploit the extracellular phospholipid code on host cells to gain cellular entry.
Impersonating and hijacking the host extracellular phospholipid code to aid infection
As described above, apoptotic cells can expose PS as an ‘eat-me' signal on the cell surface to facilitate their recognition and uptake by phagocytes. Strikingly, a large number of viruses as well as other microbes can acquire an outer membrane that resembles an apoptotic cell by displaying PS, a process known as apoptotic mimicry, to facilitate uptake/infection into immune cells. A key advantage of this mechanism of infection is that, by hijacking the apoptotic cell clearance machinery, these pathogens may also avoid eliciting host immunity.84 For example, it has recently been suggested that the Ebola virus can infect epithelial cells through the binding of PS exposed on the viral envelope to the host cell surface PS recognition receptor TIM-1. Although there was no direct evidence for PS-mediated viral internalization, the interaction between PS and TIM-1 was demonstrated to be critical for viral transduction85, 86 (Figure 2f). TIM family receptors and other host-derived phospholipid detectors have also been implicated in PS-mediated uptake of several other viruses including hepatitis A,87 Lake Victoria marburgvirus,88 dengue virus89 and a number of lentiviruses.90 However, it is worth noting that not all PS recognition receptors are involved in this apoptotic mimicry process. In particular, PS-binding receptors such as BAI-1, RAGE and stablins-1 and -2 have been shown not to support viral infection.85, 89, 90 In addition to viruses that demonstrate apoptotic cell mimicry, PS exposure on the viral envelope by vaccinia virus was suggested to allow the virus to mimic an apoptotic body (an intact fragment of an apoptotic cell following cell disassembly), thereby triggering macropinocytosis into the host cell.91 Interestingly, herpes virus can hijack the host extracellular phospholipid code by hiding inside apoptotic bodies to facilitate their entry into macrophages.92 Thus, controlling the disassembly of infected apoptotic cells may limit the spread of certain infections.93, 94 Furthermore, the protozoan Leishmania amazonesis in the ‘amastigote' stage of development was shown to expose PS on its surface to enable macropinocytotic internalization by macrophages, a process that could aid infection.95 It is worth noting that apoptotic mimicry may also be achieved by viruses through modification of the host's extracellular phospholipid code. Viral infection of host cells could result in an increase in cytosolic Ca2+, leading to a change in phospholipid translocase activity and altering the phospholipid asymmetry at the plasma membrane that favors the exposure of PS at the outer leaflet.96 Under these conditions, the exposure of PS on infected host cells could facilitate their uptake by phagocytes to perpetuate the infection process. As PS is the key phospholipid that mediates the apoptotic mimicry process by pathogens, targeting PS via blocking antibodies is a potential approach to treat a variety of infections. In particular, Bavituximab/3G4 (previously discussed for its use in cancer treatment), was demonstrated by Thorpe and colleagues (2008) to exhibit in vivo antiviral properties towards Pichinde virus in guinea pigs as well as mouse cytomegalovirus in mice.96 In 2011, Bavituximab reached phase II clinical trial status for the treatment of hepatitis C virus, although no current developments involving its use for hepatitis C virus treatment are reported.97
Hijacking the Host Intracellular Phospholipid Code to Promote Infection
Direct phospholipid binding by pathogen-derived proteins
Once pathogens or pathogen-derived molecules gain access into different intracellular compartments of the host cell, they could utilize the existing host intracellular phospholipid code to target effector molecules to various locations. For example, the glucosyltransferase domain of lethal toxin released by Clostridium sordellii can bind preferentially to PS compared with PA, PG, PI and PI(4,5)P2 containing liposomes.98 Functionally, targeting the glucosyltransferase domain of lethal toxin to PS has been suggested to facilitate its localization at the plasma membrane to mediate glucosylation of Rac-GDP, a process that could alter the GDP/GTP cycle of G proteins and result in a change in cell morphology.98 Likewise, targeting viral proteins to the plasma membrane is also a key process in mediating viral assembly and budding. In particular, Gag proteins of certain retroviruses have been shown to traffic to the plasma membrane via their N-terminal matrix (MA) domain, which then binds PI(4,5)P2 to initiate budding99 (Figure 2g). The MA domain of HIV-1 Gag has been proposed to interact with the plasma membrane via PI(4,5)P2 binding, with the subsequent exposure of its N-terminal myristoyl moiety inserting into the membrane necessary to stabilize the interaction, as well as induce multimerization of HIV-1 Gag.99 The Gag protein of equine infective anemia virus can also interact with PI(4,5)P2 during viral budding, with PI(4,5)P2 suggested to recruit the MA domain of equine infective anemia virus Gag to the plasma membrane to facilitate Gag trimerization.100 Recently, it was demonstrated that the equine infective anemia virus Gag MA protein can also localize with both PI(3)P and phosphatidylinositol 3,5-bisphosphate at membrane compartments consistent with early endosomes, suggesting that this protein is trafficked not only to the plasma membrane but to intracellular vesicles as well.101 Besides simply using the phospholipid code to target molecules to various intracellular locations, the notion that pathogen-derived proteins could mask the ability of other cellular proteins to bind to or enzymatically modify various phospholipids has also been suggested. In support of this concept, the vaccinia virus has been shown to express a profilin homolog that binds strongly to PI(4,5)P2 and PI(4)P.102 Although the vaccinia virus profilin is not required for infectivity,103 it has been postulated that it may interfere with host cell lipid signaling by binding to PI(4,5)P2, thereby inhibiting the ability of phospholipase C to hydrolyze PI(4,5)P2 and possibly preventing an inflammatory response by the host cell.104
Enzymatic modification of the host phospholipids
It is important to note that a variety of microbes can also directly manipulate the host phospholipid code through pathogen-derived phosphatases and kinases. For example, a number of bacterial-derived phosphatases can be delivered into host cells via the Type III secretion system to reduce PI(4,5)P2 levels at the inner leaflet of the plasma membrane, causing a range of effects on the host cell. Vibrio parahaemolyticus can secrete the effector protein VPA0450, a phosphoinositide 5-phosphatase that dephosphorylates PI(4,5)P2 to PI(4)P, into host cells to induce membrane blebbing and rapid cell lysis.105 Shigella flexneri can introduce the virulence factor IpgD, a phosphoinositide 4-phosphatase that dephosphorylates PI(4,5)P2 to PI(5)P, into mammalian cells to aid infection by modulating a variety of host cellular processes, including actin filament remodeling to assist invasion106 (Figure 2h) as well as PI3K/Akt/mTOR signaling to inhibit apoptosis and enable bacterial colonization within host cells.107 Similarly, the Salmonella typhimurium effector molecule SigD, a homolog of Shigella flexneri IpgD, also displays phosphoinositide 4-phosphatase activity towards PI(4,5)P2 and causes membrane blebbing and cell rounding in infected cells. The effect of SigD on host cells has been suggested to decrease cellular fluid absorption, thus contributing to host diarrhea and facilitating transmission of Salmonella typhimurium.108 More recently, it has been reported that the phosphatase activity of Salmonella typhimurium-derived effector molecules can induce membrane ruffling and generation of the macropinosome, resulting in the formation of Salmonella-containing vacuoles to aid bacterial replication following internalization109, 110 (Figure 2h).
In addition to the modification of the host phospholipid code by pathogen-derived phosphatases, certain pathogens can exploit host cell-derived phospholipid modifiers (phosphatases and kinases) to alter the intracellular phospholipid code. During Yersinia pseudotuberculosis infection, endocytosis of the bacterium into PI(4,5)P2-rich prevacuoles can induce dephosphorylation of PI(4,5)P2 through rab5-dependent recruitment of endogenous phosphatidylinositide 5-phosphatases. This process can subsequently promote vacuole maturation and aid bacterial internalization into epithelial cells.111 In hepatitis C virus-infected U2OS human osteosarcoma-derived cells, endogenous phosphatidylinositol 4-kinase-α was shown to act as a co-factor for hepatitis C virus RNA replication through generating PI(4)P at the endoplasmic reticulum of infected cells, which could recruit PI(4)P-binding proteins involved in viral replication.112 Likewise, the coxsackievirus B3 was shown to utilize endogenous phosphatidylinositol 4-kinase-β in HeLa cells to generate an approximately five-fold increase in PI(4)P at enteroviral replication organelles. It was reported that the association of viral RNA polymerase with PI(4)P at the sites of phosphatidylinositol 4-kinase-β activity could facilitate viral replication.113 Thus, the host intracellular phospholipid code can be modified and exploited by a range of infectious agents.
Targeting the pathogen phospholipid code by the host
As mentioned earlier, host-derived innate defense molecules, in particular CAPs, must effectively damage or penetrate the membranes of invading microbes to overcome infection. Importantly, CAPs often exploit an increase in negative charge on the microbial cell membrane to facilitate entry and/or lysis.114 Therefore, the membrane lipid composition of infectious pathogens can determine the efficacy and selectivity of CAPs, and in this way the host can target the ‘pathogen phospholipid code' as a defense mechanism. For example, the membranes of many Gram-negative bacteria contain a high level of PE, compared with Gram-positive species.115 It has been proposed that the presence of PE in the Gram-negative bacterial membrane can promote the congregation of CAPs and CAP-mediated membrane disruption through a process known as ‘anionic lipid clustering', in which the presence of CAPs can drive the accumulation of anionic lipids and CAPs in discrete regions of the membrane.116 Hence, Gram-negative bacteria with higher degrees of PE in their membrane are likely to promote anionic lipid clustering and are more sensitive to CAP-mediated membrane disruption. Indeed, the ability of natural and synthetic CAPs to induce anionic lipid clustering in PE-containing membranes has been reported.117 Likewise, the antimicrobial activity of the synthetic lysine-rich peptide, OAK C12K-7α8, was found to correlate positively with the level of membrane PE across a range of bacterial species.116
Besides the bacterial outer membrane, phospholipids in the membrane of fungal pathogens are also targeted by various CAPs, in particular plant defensins (Table 1). The Medicago truncatula legume defensin, MtDef4, has been shown to induce membrane lysis of the filamentous fungal pathogen Fusarium graminareum via phospholipid binding. The antifungal activity of MtDef4 was proposed to be mediated through an interaction between a cationic loop region of the defensin and PA on the fungal hyphal membrane.48 Likewise, the aforementioned ornamental tobacco defensin, NaD1, which binds PI(4,5)P2 as well as a range of other phosphorylated phosphatidylinositol species, also exhibits antifungal activity against filamentous fungi, in part mediated by phospholipid binding.69 NaD1 was solved by X-ray crystallography in complex with PI(4,5)P2, revealing the formation of a dimeric ‘cationic grip' PI(4,5)P2 binding pocket, repeated to form an extended oligomeric arch. This interaction was proposed to induce membrane destabilization of target cells, with site-directed mutagenesis of key lipid-binding residues confirming the requirement of PI(4,5)P2 for efficient membrane lysis of both Fusarium oxysporum as well as mammalian tumor cells69, 118 (Figure 2i). Similar PI(4,5)P2-binding mechanism was also described for the tomato defensin TPP3, which binds specifically to PI(4,5)P2 and shares the conserved cationic grip binding pocket as NaD1 to mediate cell lysis.70 It should be noted that the cationic loop region identified as the PA binding site in MtDef4, is conserved among many plant defensins119, 120 and interestingly, the equivalent region (between β2 and β3 strands) was also found to be the PI(4,5)P2 binding site for both NaD1 and TPP3,69, 70 indicating that this region is functionally conserved to bind phospholipid. Intriguingly, the ability of these plant innate defense molecules to induce membrane permeabilization via binding to inner membrane PI(4,5)P2 and forming oligomers resembles the aforementioned PI(4,5)P2-dependent targeting and oligomerization by MLKL in mammalian cells undergoing necroptosis.27 It is tempting to speculate that the targeting of PI(4,5)P2 by both plant defensins and the mammalian MLKL to facilitate membrane disruption may reflect convergent evolutionary pathways in which the primary downstream function is to induce cell lysis through interactions with this key phospholipid. Collectively, the availability of specific phospholipids at the microbial membrane can also operate as a form of phospholipid code and act as a major target of innate antimicrobial peptides.
Concluding Remarks
By definition, ‘code' (as a noun) is ‘a system of words, letters, figures or symbols used to represent others'.121 Consistent with this description, the phospholipid code as discussed in this article comprises a system of phospholipids (PS, PC, PE, phosphatidylglycerol, phosphatidylinositol, etc.) that functions as an information platform to mediate a variety of cellular processes intracellularly and extracellularly. Importantly, a code is only meaningful if the appropriate decoding mechanism is available. In the case for the phospholipid code, the code is deciphered by the availability of specific phospholipid detectors (phospholipid-binding proteins), and these phospholipid detectors can originate from both the host and the invading pathogen. As the phospholipid code is a key component of dying cell recognition, tumor progression and host–microbes interactions, understanding and therapeutically targeting the phospholipid code could lead to novel treatment of diseases.
Glossary
- PC
phosphatidylcholine
- PS
phosphatidylserine
- PA
phosphatidic acid
- PE
phosphatidylethanolamine
- PI(3)P
phosphatidylinositol 3-phosphate
- PI(4)P
phosphatidylinositol 4-phosphate
- PI(5)P
phosphatidylinositol 5-phosphate
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PI(3,5)P2
phosphatidylinositol 3,5-bisphosphate
- PI(3,4,5)P2
phosphatidylinositol 3,4,5-trisphosphate
- ROS
reactive oxygen species
- MLKL
mixed lineage kinase domain-like protein
- TF
tissue factor
- CAP
cationic antimicrobial peptide
- NaD1
Nicotiana alata defensin 1
- MAM7
seven mammalian cell entry domain-containing multivalent adhesion molecule
- MA
matrix
The authors declare no conflict of interest.
Footnotes
Edited by D Vaux
References
- Lentz BR. Exposure of platelet membrane phosphatidylserine regulates blood coagulation. Prog Lipid Res 2003; 42: 423–438. [DOI] [PubMed] [Google Scholar]
- Wuthier RE, Lipscomb GF. Matrix vesicles: structure, composition, formation and function in calcification. Front Biosci 2011; 16: 2812–2902. [DOI] [PubMed] [Google Scholar]
- Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol 2014. [DOI] [PMC free article] [PubMed]
- Poon IKH, Hulett MD, Parish CR. Molecular mechanisms of late apoptotic/necrotic cell clearance. Cell Death Differ 2010; 17: 381–397. [DOI] [PubMed] [Google Scholar]
- Clark MR. Flippin' lipids. Nat Immunol 2011; 12: 373–375. [DOI] [PubMed] [Google Scholar]
- van Meer G. Dynamic transbilayer lipid asymmetry. Cold Spring Harb Persp Biol 2011; 3: a004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys 2010; 39: 407–427. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 2013; 341: 403–406. [DOI] [PubMed] [Google Scholar]
- Segawa K, Kurata S, Yanagihashi Y, Brummelkamp TR, Matsuda F, Nagata S. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 2014; 344: 1164–1168. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Gleiss B, Tyurina YY, Tyurin VA, Elenström-Magnusson C, Liu S-X et al. A role for oxidative stress in apoptosis: oxidation and externalization of phosphatidylserine is required for macrophage clearance of cells undergoing Fas-mediated apoptosis. J Immunol 2002; 169: 487–499. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Serinkan FB, Tyurin VA, Kini V, Yalowich JC, Schroit AJ et al. Lipid antioxidant, etoposide, inhibits phosphatidylserine externalization and macrophage clearance of apoptotic cells by preventing phosphatidylserine oxidation. J Biol Chem 2004; 279: 6056–6064. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Sun M, Zhang R, Febbraio M, Silverstein R, Hazen SL. Oxidized phosphatidylserine–CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J Exp Med 2006; 203: 2613–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Karisola P, Peña-Cruz V, Dorfman DM, Jinushi M, Umetsu SE et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 2007; 27: 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature 2007; 450: 435–439. [DOI] [PubMed] [Google Scholar]
- Park D, Tosello-Trampont A-C, Elliott MR, Lu M, Haney LB, Ma Z et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 2007; 450: 430–434. [DOI] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature 2002; 417: 182–187. [DOI] [PubMed] [Google Scholar]
- Ishimoto Y, Ohashi K, Mizuno K, Nakano T. Promotion of the uptake of PS liposomes and apoptotic cells by a product of growth arrest-specific gene, gas6. J Biochem 2000; 127: 411–417. [DOI] [PubMed] [Google Scholar]
- Scott RS, McMahon EJ, Pop SM, Reap EA, Caricchio R, Cohen PL et al. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 2001; 411: 207–211. [DOI] [PubMed] [Google Scholar]
- Toda S, Segawa K, Nagata S. MerTK-mediated engulfment of pyrenocytes by central macrophages in erythroblastic islands. Blood 2014; 123: 3963–3971. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kawane K, Koike M, Mori Y, Uchiyama Y, Nagata S. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature 2005; 437: 754–758. [DOI] [PubMed] [Google Scholar]
- Poon IK, Hulett MD, Parish CR. Histidine-rich glycoprotein is a novel plasma pattern recognition molecule that recruits IgG to facilitate necrotic cell clearance via FcγRI on phagocytes. Blood 2010; 115: 2473–2482. [DOI] [PubMed] [Google Scholar]
- Patel KK, Poon IK, Talbo GH, Perugini MA, Taylor NL, Ralph TJ et al. New method for purifying histidine-rich glycoprotein from human plasma redefines its functional properties. IUBMB Life 2013; 65: 550–563. [DOI] [PubMed] [Google Scholar]
- Kuroki Y, Honma T, Chiba H, Sano H, Saitoh M, Ogasawara Y et al. A novel type of binding specificity to phospholipids for rat mannose-binding proteins isolated from serum and liver. FEBS Lett 1997; 414: 387–392. [DOI] [PubMed] [Google Scholar]
- Declercq W, Berghe TV, Vandenabeele P. RIP kinases at the crossroads of cell death and survival. Cell 2009; 138: 229–232. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang H, Wang Z, He S, Chen S, Liao D et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012; 148: 213–227. [DOI] [PubMed] [Google Scholar]
- Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci USA 2012; 109: 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondelinger Y, Declercq W, Montessuit S, Roelandt R, Goncalves A, Bruggeman I et al. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep 2014; 7: 971–981. [DOI] [PubMed] [Google Scholar]
- Balasubramanian K, Schroit AJ. Aminophospholipid asymmetry: a matter of life and death. Annu Rev Physiol 2003; 65: 701–734. [DOI] [PubMed] [Google Scholar]
- Ran S, Thorpe PE. Phosphatidylserine is a marker of tumor vasculature and a potential target for cancer imaging and therapy. Int J Radiat Oncol Biol Phys 2002; 54: 1479–1484. [DOI] [PubMed] [Google Scholar]
- Stafford JH, Thorpe PE. Increased exposure of phosphatidylethanolamine on the surface of tumor vascular endothelium. Neoplasia 2011; 13: 299–IN292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl A, Lage H, Muller P, Pomorski T, Herrmann A. Transport of phosphatidylserine via MDR1 (multidrug resistance 1) P-glycoprotein in a human gastric carcinoma cell line. Biochem J 2002; 365: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlecke H, Pohl A, Alder-Baerens N, Lage H, Herrmann A. Enhanced exposure of phosphatidylserine in human gastric carcinoma cells overexpressing the half-size ABC transporter BCRP (ABCG2). Biochem J 2003; 376: 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Rickles F, Moritz TE, Henderson W, Zacharski L, Forman W et al. Abnormalities of blood coagulation tests in patients with cancer. Am J Clin Pathol 1987; 88: 596–602. [DOI] [PubMed] [Google Scholar]
- Gouin-Thibault I, Achkar A, Samama M. The thrombophilic state in cancer patients. Acta Haematol 2000; 106: 33–42. [DOI] [PubMed] [Google Scholar]
- Lima LG, Monteiro RQ. Activation of blood coagulation in cancer: implications for tumour progression. Biosci Rep 2013; 33: 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams RL, Bird RJ. Review article: Coagulation cascade and therapeutics update: relevance to nephrology. Part 1: Overview of coagulation, thrombophilias and history of anticoagulants. Nephrology 2009; 14: 462–470. [DOI] [PubMed] [Google Scholar]
- Utsugi T, Schroit AJ, Connor J, Bucana CD, Fidler IJ. Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res 1991; 51: 3062–3066. [PubMed] [Google Scholar]
- Falanga A, Marchetti M, Vignoli A. Coagulation and cancer: biological and clinical aspects. J Thromb Haemost 2013; 11: 223–233. [DOI] [PubMed] [Google Scholar]
- Ran S, Downes A, Thorpe PE. Increased exposure of anionic phospholipids on the surface of tumor blood vessels. Cancer Res 2002; 62: 6132–6140. [PubMed] [Google Scholar]
- Ran S, He J, Huang X, Soares M, Scothorn D, Thorpe PE. Antitumor effects of a monoclonal antibody that binds anionic phospholipids on the surface of tumor blood vessels in mice. Clin Cancer Res 2005; 11: 1551–1562. [DOI] [PubMed] [Google Scholar]
- Huang X, Bennett M, Thorpe PE. A monoclonal antibody that binds anionic phospholipids on tumor blood vessels enhances the antitumor effect of docetaxel on human breast tumors in mice. Cancer Res 2005; 65: 4408–4416. [DOI] [PubMed] [Google Scholar]
- He J, Luster TA, Thorpe PE. Radiation-enhanced vascular targeting of human lung cancers in mice with a monoclonal antibody that binds anionic phospholipids. Clin Cancer Res 2007; 13: 5211–5218. [DOI] [PubMed] [Google Scholar]
- Judy BF, Aliperti LA, Predina JD, Levine D, Kapoor V, Thorpe PE et al. Vascular endothelial-targeted therapy combined with cytotoxic chemotherapy induces inflammatory intratumoral infiltrates and inhibits tumor relapses after surgery. Neoplasia 2012; 14: 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digumarti R, Bapsy P, Suresh AV, Bhattacharyya G, Dasappa L, Shan JS et al. Bavituximab plus paclitaxel and carboplatin for the treatment of advanced non-small-cell lung cancer. Lung Cancer 2014; 86: 231–236. [DOI] [PubMed] [Google Scholar]
- Chalasani P, Marron M, Roe D, Clarke K, Iannone M, Livingston RB et al. A phase I clinical trial of bavituximab and paclitaxel in patients with HER2 negative metastatic breast cancer. Cancer Med 2015; 4: 1051–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peregrine. Phase III Trial: SUNRISE (Stimulating ImmUne RespoNse thRough BavItuximab in a PhaSE III Lung Cancer Study) 2015. Available from http://www.peregrineinc.com/clinical-trials/bavituximab-trials.html.
- Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002; 66: 236–248. [DOI] [PubMed] [Google Scholar]
- Sagaram US, El-Mounadi K, Buchko GW, Berg HR, Kaur J, Pandurangi RS et al. Structural and functional studies of a phosphatidic acid-binding antifungal plant defensin MtDef4: identification of an RGFRRR motif governing fungal cell entry. PLoS One 2013; 8: e82485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl S, Zweytick D, Lohner K. Membrane-active host defense peptides–challenges and perspectives for the development of novel anticancer drugs. Chem Phys Lipids 2011; 164: 766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon W-H, Park H-D, Lim K, Hwang B-D. Effect of O-glycosylated mucin on invasion and metastasis of HM7 human colon cancer cells. Biochem Biophys Res Commun 1996; 222: 694–699. [DOI] [PubMed] [Google Scholar]
- Bafna S, Kaur S, Batra S. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene 2010; 29: 2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson RD, Yang Y, Suva LJ, Kelly T. Heparan sulfate proteoglycans and heparanase—partners in osteolytic tumor growth and metastasis. Matrix Biol 2004; 23: 341–352. [DOI] [PubMed] [Google Scholar]
- Kirszberg C, Lima LG, de Oliveira ADS, Pickering W, Gray E, Barrowcliffe TW et al. Simultaneous tissue factor expression and phosphatidylserine exposure account for the highly procoagulant pattern of melanoma cell lines. Melanoma Res 2009; 19: 301–308. [DOI] [PubMed] [Google Scholar]
- Schröder-Borm H, Bakalova R, Andrä J. The NK-lysin derived peptide NK-2 preferentially kills cancer cells with increased surface levels of negatively charged phosphatidylserine. FEBS Lett 2005; 579: 6128–6134. [DOI] [PubMed] [Google Scholar]
- Iwasaki T, Ishibashi J, Tanaka H, Sato M, Asaoka A, Taylor D et al. Selective cancer cell cytotoxicity of enantiomeric 9-mer peptides derived from beetle defensins depends on negatively charged phosphatidylserine on the cell surface. Peptides 2009; 30: 660–668. [DOI] [PubMed] [Google Scholar]
- Wong K-K, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev 2010; 20: 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markman B, Dienstmann R, Tabernero J. Targeting the PI3K/Akt/mTOR pathway–beyond rapalogs. Oncotarget 2010; 1: 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004; 304: 554–554. [DOI] [PubMed] [Google Scholar]
- Bartholomeusz C, Gonzalez-Angulo AM. Targeting the PI3K signaling pathway in cancer therapy. Expert Opin Ther Targets 2012; 16: 121–130. [DOI] [PubMed] [Google Scholar]
- Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer 2011; 11: 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 2001; 294: 1942–1945. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zheng Y, Foster DA. Phospholipase D confers rapamycin resistance in human breast cancer cells. Oncogene 2003; 22: 3937–3942. [DOI] [PubMed] [Google Scholar]
- Noh D-Y, Ahn S-J, Lee R-A, Park I-A, Kim J-H, Suh P-G et al. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett 2000; 161: 207–214. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ehara H, Akao Y, Shamoto M, Nakagawa Y, Banno Y et al. Increased activity and intranuclear expression of phospholipase D2 in human renal cancer. Biochem Biophys Res Commun 2000; 278: 140–143. [DOI] [PubMed] [Google Scholar]
- Uchida N, Okamura S, Kuwano H. Phospholipase D activity in human gastric carcinoma. Anticancer Res 1998; 19: 671–675. [PubMed] [Google Scholar]
- Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol 2013; 10: 143–153. [DOI] [PubMed] [Google Scholar]
- Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL et al. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol 2009; 5: 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son JC, Kang DW, Yang KM, Choi K-Y, Son TG. Phospholipase D inhibitor enhances radiosensitivity of breast cancer cells. Exp Mol Med 2013; 45: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IK, Baxter AA, Lay FT, Mills GD, Adda CG, Payne JA et al. Phosphoinositide-mediated oligomerization of a defensin induces cell lysis. Elife 2014; 3: e01808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter AA, Richter V, Lay FT, Poon IK, Adda CG, Veneer PK et al. The tomato defensin TPP3 binds phosphatidylinositol (4, 5)-bisphosphate via a conserved dimeric cationic grip conformation to mediate cell lysis. Mol Cell Biol 2015; 35: 1964–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 1996; 381: 667–673. [DOI] [PubMed] [Google Scholar]
- Agnello V, Ábel G, Elfahal M, Knight GB, Zhang Q-X. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA 1999; 96: 12766–12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvansakul M, van Delft MF, Lee EF, Gulbis JM, Fairlie WD, Huang D et al. A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak. Mol Cell 2007; 25: 933–942. [DOI] [PubMed] [Google Scholar]
- Krachler AM, Ham H, Orth K. Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens. Proc Natl Acad Sci USA 2011; 108: 11614–11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Stones DH, Hawley CA, Watson CA, Krachler AM. Multivalent adhesion molecule 7 clusters act as signaling platform for host cellular GTPase activation and facilitate epithelial barrier dysfunction. PLoS Pathog 2014; 10: e1004421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood C, Huesca M, Kuksis A. The glycerolipid receptor for Helicobacter pylori (and exoenzyme S) is phosphatidylethanolamine. Infect Immun 1992; 60: 2470–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DB, Philpott D, Abul-Milh M, Huesca M, Sherman PM, Lingwood CA. Phosphatidylethanolamine recognition promotes enteropathogenic E. coli and enterohemorrhagic E. coli host cell attachment. Microb Pathog 1999; 27: 289–301. [DOI] [PubMed] [Google Scholar]
- Krivan HC, Nilsson B, Lingwood C, Ryu H. Chlamydia trachomatis and Chlamydia pneumoniae bind specifically to phosphatidylethanolamine in HeLa cells and to GalNAcβ1-4Galβ1-4Glc sequences found in asialo-GM 1 and asialo-GM 2. Biochem Biophys Res Commun 1991; 175: 1082–1089. [DOI] [PubMed] [Google Scholar]
- Sason H, Milgrom M, Weiss AM, Melamed-Book N, Balla T, Grinstein S et al. Enteropathogenic Escherichia coli subverts phosphatidylinositol 4, 5-bisphosphate and phosphatidylinositol 3, 4, 5-trisphosphate upon epithelial cell infection. Mol Biol Cell 2009; 20: 544–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley CA, Watson CA, Orth K, Krachler AM. A MAM7 peptide-based inhibitor of Staphylococcus aureus adhesion does not interfere with in vitro host cell function. PLoS One 2013; 8: e81216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale SD, Gu B, Capelluto DG, Dou D, Feldman E, Rumore A et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 2010; 142: 284–295. [DOI] [PubMed] [Google Scholar]
- Sun F, Kale SD, Azurmendi HF, Li D, Tyler BM, Capelluto DG. Structural basis for interactions of the Phytophthora sojae RxLR effector Avh5 with phosphatidylinositol 3-phosphate and for host cell entry. Mol Plant Microbe Interact 2013; 26: 330–344. [DOI] [PubMed] [Google Scholar]
- Murata-Kamiya N, Kikuchi K, Hayashi T, Higashi H, Hatakeyama M. Helicobacter pylori exploits host membrane phosphatidylserine for delivery, localization, and pathophysiological action of the CagA oncoprotein. Cell Host Microbe 2010; 7: 399–411. [DOI] [PubMed] [Google Scholar]
- Alcami A, Koszinowski UH. Viral mechanisms of immune evasion. Immunol Today 2000; 21: 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Tank S, Kondratowicz AS, Davey RA, Rennert PD, Maury W. Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J Virol 2013; 87: 8327–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Tank S, Maury W. Phosphatidylserine receptors: enhancers of enveloped virus entry and infection. Virology 2014; 468: 565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire JJ, Umetsu SE, Macaubas C, Hoyte EG, Cinnioglu C, Cavalli-Sforza LL et al. Immunology: hepatitis A virus link to atopic disease. Nature 2003; 425: 576–576. [DOI] [PubMed] [Google Scholar]
- Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci USA 2011; 108: 8426–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E et al. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 2012; 12: 544–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizono K, Chen IS. Role of phosphatidylserine receptors in enveloped virus infection. J Virol 2014; 88: 4275–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 2008; 320: 531–535. [DOI] [PubMed] [Google Scholar]
- De Mesquita R, Biberfeld P. Demonstration of herpesvirus particles in apoptotic bodies of a brain lymphoma in an SIV-immunodeficient monkey. Int J Cancer 1995; 63: 472–473. [DOI] [PubMed] [Google Scholar]
- Atkin-Smith GK, Tixeira R, Paone S, Mathivanan S, Collins C, Liem M et al. A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nat Commun 2015; 6: 7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IK, Chiu Y-H, Armstrong AJ, Kinchen JM, Juncadella IJ, Bayliss DA et al. Unexpected link between an antibiotic, pannexin channels and apoptosis. Nature 2014; 507: 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanderley JL, Moreira ME, Benjamin A, Bonomo AC, Barcinski MA. Mimicry of apoptotic cells by exposing phosphatidylserine participates in the establishment of amastigotes of Leishmania (L) amazonensis in mammalian hosts. J Immunol 2006; 176: 1834–1839. [DOI] [PubMed] [Google Scholar]
- Soares MM, King SW, Thorpe PE. Targeting inside-out phosphatidylserine as a therapeutic strategy for viral diseases. Nat Med 2008; 14: 1357–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peregrine. Bavituximab antiviral 2015. Available from: http://www.peregrineinc.com/pipeline/cot.html.
- Mesmin B, Robbe K, Geny B, Luton F, Brandolin G, Popoff MR et al. A phosphatidylserine-binding site in the cytosolic fragment of Clostridium sordellii lethal toxin facilitates glucosylation of membrane-bound Rac and is required for cytotoxicity. J Biol Chem 2004; 279: 49876–49882. [DOI] [PubMed] [Google Scholar]
- Chukkapalli V, Hogue IB, Boyko V, Hu W-S, Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4, 5)-bisphosphate is essential for efficient gag membrane binding. J Virol 2008; 82: 2405–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Bachtiar I, Piszczek G, Bouamr F, Carter C, Tjandra N. Solution NMR characterizations of oligomerization and dynamics of equine infectious anemia virus matrix protein and its interaction with PIP2. Biochemistry 2008; 47: 1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes F, Chen K, Ehrlich LS, Jin J, Chen MH, Medina GN et al. Phosphoinositides direct equine infectious anemia virus gag trafficking and release. Traffic 2011; 12: 438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Cole NB, Moss B, Pollard TD. Vaccinia virus expresses a novel profilin with a higher affinity for polyphosphoinositides than actin. Biochemistry 1994; 33: 10815–10824. [DOI] [PubMed] [Google Scholar]
- Blasco R, Cole NB, Moss B. Sequence analysis, expression, and deletion of a vaccinia virus gene encoding a homolog of profilin, a eukaryotic actin-binding protein. J Virol 1991; 65: 4598–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Machesky LM, Baldassare JJ, Pollard TD. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science 1990; 247: 1575–1578. [DOI] [PubMed] [Google Scholar]
- Broberg CA, Zhang L, Gonzalez H, Laskowski-Arce MA, Orth K. A Vibrio effector protein is an inositol phosphatase and disrupts host cell membrane integrity. Science 2010; 329: 1660–1662. [DOI] [PubMed] [Google Scholar]
- Niebuhr K, Giuriato S, Pedron T, Philpott DJ, Gaits F, Sable J et al. Conversion of PtdIns (4, 5) P2 into PtdIns (5) P by the S. flexneri effector IpgD reorganizes host cell morphology. EMBO J 2002; 21: 5069–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendaries C, Tronchère H, Arbibe L, Mounier J, Gozani O, Cantley L et al. PtdIns (5) P activates the host cell PI3-kinase/Akt pathway during Shigella flexneri infection. EMBO J 2006; 25: 1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D, Mallo GV, Terebiznik MR, Payrastre B, Finlay BB, Brumell JH et al. Alteration of epithelial structure and function associated with PtdIns (4, 5) P2 degradation by a bacterial phosphatase. J Gen Physiol 2007; 129: 267–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowski MA, Braun V, Brumell JH. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic 2008; 9: 2022–2031. [DOI] [PubMed] [Google Scholar]
- Kerr M, Castro N, Teasdale R, Karunaratne S. The Phosphoinositides: Key Regulators of Salmonella Containing Vacuole (SCV) Trafficking and Identity 2012. INTECH Open Access Publisher. [Google Scholar]
- Sarantis H, Balkin DM, De Camilli P, Isberg RR, Brumell JH, Grinstein S. Yersinia entry into host cells requires Rab5-dependent dephosphorylation of PI (4, 5) P2 and membrane scission. Cell Host Microbe 2012; 11: 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol 2011; 85: 8870–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu N-Y, Ilnytska O, Belov G, Santiana M, Chen Y-H, Takvorian PM et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 2010; 141: 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 2005; 3: 238–250. [DOI] [PubMed] [Google Scholar]
- Wang G. Role of bacterial membrane lipid composition in antimicrobial sensitivity. In. Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. CAB International: Oxford, UK, 2010, pp 118–119. [Google Scholar]
- Epand RM, Epand RF. Bacterial membrane lipids in the action of antimicrobial agents. J Pept Sci 2011; 17: 298–305. [DOI] [PubMed] [Google Scholar]
- Wadhwani P, Epand R, Heidenreich N, Bürck J, Ulrich A, Epand R. Membrane-active peptides and the clustering of anionic lipids. Biophys J 2012; 103: 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay FT, Mills GD, Poon IK, Cowieson NP, Kirby N, Baxter AA et al. Dimerization of plant defensin NaD1 enhances its antifungal activity. J Biol Chem 2012; 287: 19961–19972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Weerden NL, Anderson MA. Plant defensins: common fold, multiple functions. Fungal Biol Rev 2013; 26: 121–131. [Google Scholar]
- Vriens K, Cammue B, Thevissen K. Antifungal plant defensins: mechanisms of action and production. Molecules 2014; 19: 12280–12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson A. Oxford Dictionary of English 2010. OUP: Oxford, UK. [Google Scholar]
- Frank C, Keilhack H, Opitz F, Zschornig O, Bohmer FD. Binding of phosphatidic acid to the protein-tyrosine phosphatase SHP-1 as a basis for activity modulation. Biochemistry 1999; 38: 11993–12002. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Tagawa Y, Mitomo K, Arimura Y, Hatano N, Katagiri T et al. Src homology region 2 (SH2) domain-containing phosphatase-1 dephosphorylates B cell linker protein/SH2 domain leukocyte protein of 65 kDa and selectively regulates c-Jun NH2-terminal kinase activation in B cells. J Immunol 2000; 165: 1344–1351. [DOI] [PubMed] [Google Scholar]
- Ochocka AM, Pawelczyk T. Isozymes delta of phosphoinositide-specific phospholipase C and their role in signal transduction in the cell. Acta Biochim Pol 2003; 50: 1097–1110. [PubMed] [Google Scholar]
- Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A et al. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 2001; 291: 1051–1055. [DOI] [PubMed] [Google Scholar]
- Kim JY, Choi BK, Choi MG, Kim SA, Lai Y, Shin YK et al. Solution single-vesicle assay reveals PIP2-mediated sequential actions of synaptotagmin-1 on SNAREs. EMBO J 2012; 31: 2144–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze T, Overduin M. Structural mechanism of endosome docking by the FYVE domain. Science 2001; 291: 1793–1796. [DOI] [PubMed] [Google Scholar]
- Krauss M, Haucke V. Phosphoinositide-metabolizing enzymes at the interface between membrane traffic and cell signalling. EMBO Rep 2007; 8: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell 2001; 12: 2396–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S, Schorr M, Mayinger P. Regulation of intracellular phosphatidylinositol-4-phosphate by the Sac1 lipid phosphatase. Traffic 2005; 6: 116–130. [DOI] [PubMed] [Google Scholar]
- Paradies G, Paradies V, De Benedictis V, Ruggiero FM, Petrosillo G. Functional role of cardiolipin in mitochondrial bioenergetics. Biochim Biophys Acta 2014; 1837: 408–417. [DOI] [PubMed] [Google Scholar]
- Mejia EM, Dolinsky VW, Hatch GM. Role of phospholipases in regulation of cardiolipin biosynthesis and remodeling in the heart and mammalian cells. In: Phospholipases in Health and Disease. Springer: New York, NY, USA, 2014, pp 39–53. [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 2008; 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadeel B, Xue D. The ins and outs of phospholipid asymmetry in the plasma membrane: roles in health and disease. Crit Rev Biochem Mol Biol 2009; 44: 264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan K, Moens PD. Structure and functions of profilins. Biophys Rev 2009; 1: 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]