Abstract
Both 5-methylcytosine (5mC) and its oxidized form 5-hydroxymethylcytosine (5hmC) have been proposed to be involved in tumorigenesis. Because the readout of the broadly used 5mC mapping method, bisulfite sequencing (BS-seq), is the sum of 5mC and 5hmC levels, the 5mC/5hmC patterns and relationship of these two modifications remain poorly understood. By profiling real 5mC (BS-seq corrected by Tet-assisted BS-seq, TAB-seq) and 5hmC (TAB-seq) levels simultaneously at single-nucleotide resolution, we here demonstrate that there is no global loss of 5mC in kidney tumors compared with matched normal tissues. Conversely, 5hmC was globally lost in virtually all kidney tumor tissues. The 5hmC level in tumor tissues is an independent prognostic marker for kidney cancer, with lower levels of 5hmC associated with shorter overall survival. Furthermore, we demonstrated that loss of 5hmC is linked to hypermethylation in tumors compared with matched normal tissues, particularly in gene body regions. Strikingly, gene body hypermethylation was significantly associated with silencing of the tumor-related genes. Downregulation of IDH1 was identified as a mechanism underlying 5hmC loss in kidney cancer. Restoring 5hmC levels attenuated the invasion capacity of tumor cells and suppressed tumor growth in a xenograft model. Collectively, our results demonstrate that loss of 5hmC is both a prognostic marker and an oncogenic event in kidney cancer by remodeling the DNA methylation pattern.
Keywords: 5mC, 5hmC, TET, kidney cancer
Introduction
Global loss and promoter-associated gain of DNA methylation have been considered as hallmarks of cancers and may contribute to tumorigenesis1,2,3. However, our knowledge regarding the functional role of the bimodal DNA methylation pattern remains limited because of the lack of single-base resolution DNA methylomes from tumors and matched normal tissues.
The genome-wide loss of DNA methylation in tumors was first identified by liquid chromatography in the 1980s, which demonstrated that the percentage of primary malignancies with hypomethylated DNA was intermediate between those of metastases and benign neoplasms4,5. Recently, with more accurate quantitative methods, DNA methylation levels have been observed to vary across diverse human cell and tissue types5,6. Thus, DNA methylation changes during tumorigenesis should be re-evaluated by comparing tumor samples with their matched normal tissues. In addition, it remains unknown whether global 5-methylcytosine (5mC) levels can distinguish tumors from their matched normal tissues.
As DNA hypermethylation is a potential therapeutic target, it is necessary to explore the mechanisms underlying the hypermethylation in tumors. Recently, TETs were discovered to convert 5mC to 5-hydroxymethylcytosine (5hmC), which may be linked to DNA demethylation7,8. Therefore, TET-mutated tumors are expected to accumulate 5mC compared with normal tissues. However, the reported effects of TET mutations on 5mC levels are conflicting9,10,11, and a low 5hmC level was observed in a subset of patients without TET mutations9. Thus, the relationship among TET mutations, 5hmC and 5mC levels and tumorigenesis remains obscure. Currently, most strategies for mapping genome-wide DNA methylome have limited genome coverage and resolution, such as the HELP (Hpall tiny fragment enrichment by ligation-mediated PCR) assay10 and the Illumina Infinium 27k array9. The broadly used genome-wide single-nucleotide resolution 5mC mapping method, bisulfite sequencing (BS-seq), does not distinguish between 5mC and 5hmC12,13. The readout of BS-seq is the sum of 5mC and 5hmC. Thus, measuring genome-wide single-nucleotide resolution patterns of 5mC and 5hmC separately is necessary to precisely define the roles of 5mC and 5hmC in carcinogenesis.
Here, we use renal cell carcinoma (RCC) as a model of solid tumor, which displays TET2 mutations in approximately 6% of patients14,15,16. By profiling 5hmC (Tet-assisted BS-seq (TAB-seq)17) and 5mC (BS-seq corrected by TAB-seq) levels simultaneously, we discovered that there was no significant difference in the global DNA methylation level between tumors and matched normal tissues. Loss of 5hmC occurs in virtually all the clear cell RCC (ccRCC, the major subtype of RCC) patients and is linked to hypermethylation especially in gene body regions.
Results
Loss of 5hmC but not 5mC is a hallmark of ccRCC
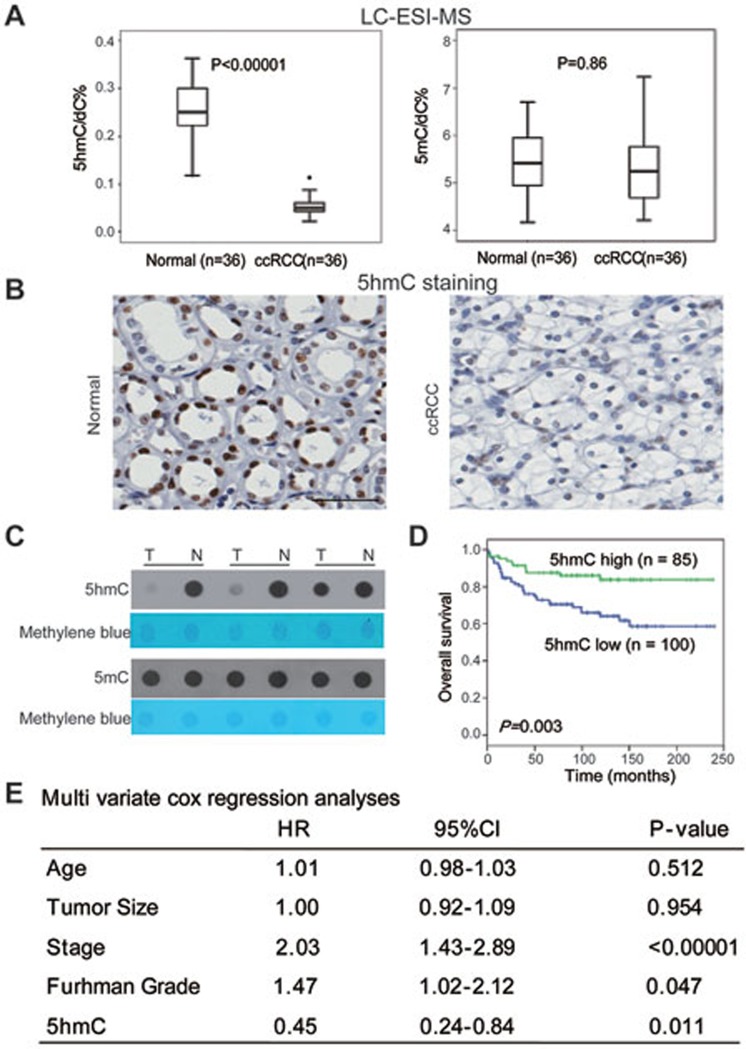
To explore the global changes of 5mC and 5hmC levels, we first performed the sensitive liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS) to measure global 5mC and 5hmC levels in 36 paired ccRCC and normal kidney samples. Consistent with recent findings in other types of cancers18,19, 5hmC levels decreased in all kidney tumor samples examined compared with those in matched normal tissues (Figure 1A and Supplementary information, Figure S1A). However, global 5mC levels did not change significantly (Figure 1A). Furthermore, immunohistochemical (IHC) staining and dot blot assays yielded similar results (Figure 1B, 1C and Supplementary information, Figure S1B). Similar results were also obtained in colorectal cancer (Supplementary information, Figure S1C) and hepatocellular carcinoma samples20. Thus, global 5hmC levels but not global 5mC levels can distinguish tumors from normal tissues in several types of cancer. These results suggest that loss of 5hmC could be a general feature of carcinogenesis.
Figure 1.
Loss of 5hmC but not 5mC is a hallmark of ccRCC. (A) 5hmC and 5mC levels were evaluated by the LC-ESI-MS method. P-values were calculated by the two-tailed student's t-test. (B) IHC staining of 5hmC in paired tumor and normal samples of one representative ccRCC patient. Scare bar, 100 μm. (C) Dot blot assays of 5hmC and 5mC. Three representative paired samples are shown. T, tumor; N, matched normal tissue. (D) Kaplan-Meier survival curves of ccRCC patients in the TMA. “5hmC high” and “5hmC low” represent cases in which > 10% and ≤ 10% cells are positive for IHC staining of 5hmC, respectively. P values were calculated by the log-rank test. n, the number of cases. (E) Multivariate Cox regression analyses of ccRCC cases in the TMA.
5hmC is an independent molecular marker of ccRCC progression
To explore whether the loss of 5hmC is involved in ccRCC progression, we correlated the 5hmC levels in ccRCC tissues with clinical characteristics using a tissue microarray (TMA) that contains more than 200 clinically annotated RCC cases including 185 ccRCC samples21. The TMA results confirmed significant 5hmC loss in ccRCC samples (Mann-Whitney U-test, P < 0.0001), but to a much lesser extent in other RCC subtypes (Supplementary information, Figure S1D). A univariate Kaplan-Meier assay revealed that patients with higher 5hmC levels (the IHC staining score > 10%) had significantly longer overall survival than patients with lower 5hmC levels (the IHC staining score ≤ 10%; Figure 1D and Supplementary information, Figure S1E). Further multivariate Cox proportional hazards regression analyses showed that the 5hmC levels in the tumor tissues independently provided predictive power, and lower 5hmC levels were correlated with shorter overall survival, as reflected by the hazard ratio of 0.45 (Figure 1E), suggesting that loss of 5hmC is critical for ccRCC progression.
Base-resolution analysis of 5hmC in paired tumor and adjacent normal tissues
To explore whether 5hmC loss during ccRCC tumorigenesis was genome wide or locus specific, we used TAB-seq to comprehensively profile the 5hmC patterns at single-nucleotide resolution for the tumor and matched normal kidney tissues of two ccRCC patients. A positive readout of 5hmC was gained from a single TAB-seq run. We generated sequences of 400 billion uniquely alignable base pairs (33× average genome coverage). Greater than 85% of all genomic CpG dinucleotides were covered by five or more uniquely mapped sequencing reads in both patients (Supplementary information, Table S1). Because a limited number of 5hmC sites were identified in non-CpG regions (data not shown), all following analyze focused on CpG sites only. The 5hmC status of individual CpG sites for the two normal kidney tissues was highly correlated between the two patients (Figure 2A). This result suggests that 5hmC modification is locus specific in normal kidney tissues and our data sets permitted accurate calling of genome-wide CpG 5hmC pattern. However, the 5hmC patterns in tumor samples were poorly correlated between these two patients, suggesting tumor heterogeneity. A substantially lower correlation was also identified between tumor and matched normal tissues (Figure 2A), indicating profound 5hmC reprogramming during tumorigenesis.
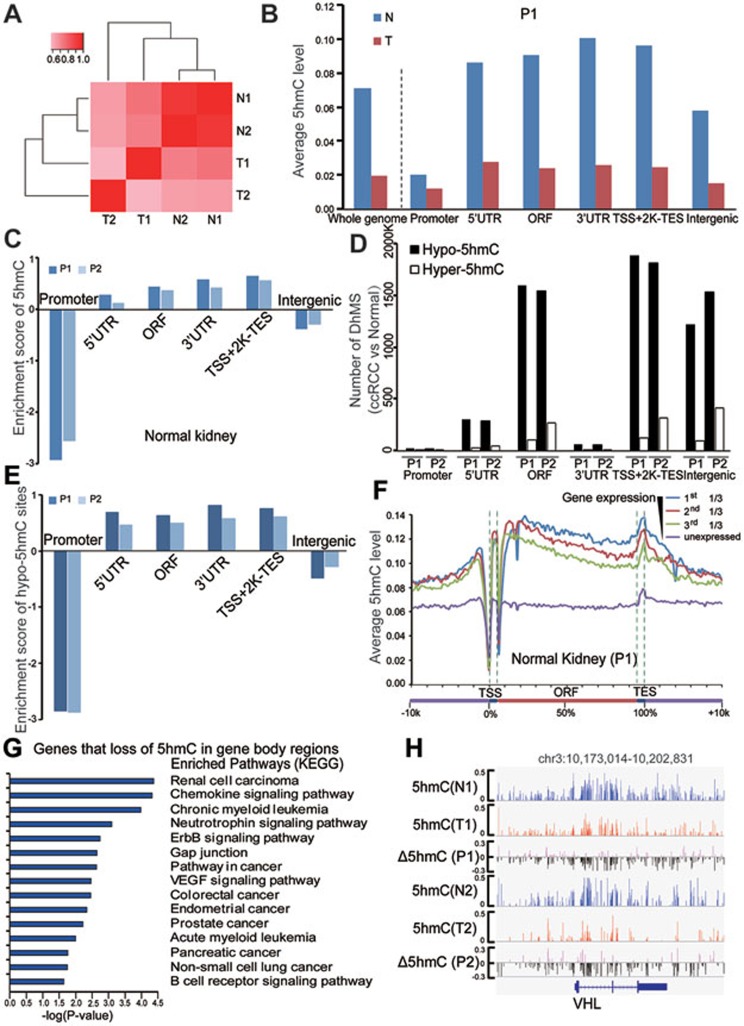
Figure 2.
Genome-wide single-nucleotide resolution mapping of 5hmC. (A) Correlations between 5hmC patterns with 5hmC level measured within 1 kb bins, and correlation coefficients are shown and colored from pink to red to indicate low to high. (B) Global changes of average 5hmC levels in different genomic elements determined by TAB-seq (promoter is defined as ± 500 bp of TSS). (C) The enrichment scores of called 5hmC sites in different genomic elements relative to expected. Score > 0 was defined as enriched. (D) The numbers of DhMS located in different gene-associated genetic elements. (E) The enrichment scores of hypo-5hmC sites in different genomic elements relative to expected. (F) The average 5hmC levels in normal kidney tissue across different gene-associated regions. Genes in the analyzed tissue were divided into four groups according to their gene expression levels (FPKM value). For each gene, a region from transcriptional start site (TSS) to transcriptional end site (TES) was divided into 100 bins and 5′-UTR and 3′-UTR contributed 5 bins each. Average 5hmC levels for each bin were calculated in the indicated samples. (G) The KEGG pathway analyses with the shared genes for both patients with loss of 5hmC in their gene body regions during tumorigenesis. The significance was evaluated by P-value. (H) Graphical representation of the dynamic 5hmC pattern during tumorigenesis at a 5hmC-enriched gene, VHL. T and N represent tumor and matched normal tissue, respectively. P1 and P2 represent patients 1 and 2, respectively.
Consistent with the LC-ESI-MS data, the average 5hmC level of tumor samples was lower than that of matched normal tissues in both patients (1.93% vs 7.07% for patient 1, 1.52% vs 6.96% for patient 2; Figure 2B and Supplementary information, Table S1). A total of 285 918 and 6 412 879 5hmC-modified sites were called in the tumor and matched normal tissues of the first patient, respectively (BH-adjusted P ≤ 0.05, coverage ≥ 5, Supplementary information, Table S1). Similarly, a total of 1 323 254 and 6 757 760 5hmC sites were called in the tumor and matched normal tissues of the second patient, respectively BH-adjusted P ≤ 0.05, coverage ≥ 5, Supplementary information, Table S1). As shown in Supplementary information, Figure S2A, the median 5hmC level in called hydroxymethylated CpG sites for both tumor and normal tissues is around 20%. In addition, profound global loss of 5hmC in tumor samples was identified across all genomic elements in both patients (Figure 2B and Supplementary information, Figure S2B). Meanwhile, compared with intergenic and promoter regions, 5hmC was enriched in gene body regions (from the transcription start site (TSS) to the transcription end site (TES))22 in normal and tumor tissues of both patients (Figure 2C and Supplementary information, Figure S2C). These results are consistent with the findings in nervous system23 and embryonic stem cells24.
We further determined 5hmC changes by comparing the 5hmC levels of tumor samples with those of matched normal tissues at genome-wide single-nucleotide resolution. Consistent with global 5hmC loss, we observed more hypo-5hmC sites (where the 5hmC level is lower in the tumor than in the paired normal tissue) than hyper-5hmC sites in both patients (where the 5hmC level is higher in the tumor; Figure 2D). Hypo-5hmC sites were also enriched in gene body regions compared with promoter and intergenic regions (Figure 2E). Thus, loss of 5hmC in ccRCC was also identified at single-nucleotide resolution across all genomic elements, particularly in gene body regions.
5hmC levels in gene bodies are positively correlated with gene expression
However, it remains unknown whether gene expression can be regulated by 5hmC modification in gene bodies. To address this, we divided genes analyzed into four groups according to their gene expression levels measured by RNA-seq (Supplementary information, Table S2). The data showed that 5hmC levels within gene bodies were positively correlated with expression levels of corresponding genes in normal kidney and tumor tissues for both patients (Figure 2F, Supplementary information, Figure S2D and data not shown). In addition, we explored whether genes displaying different 5hmC levels in tumor and paired normal tissues are also differentially expressed in these paired samples. Loss of 5hmC in gene body regions in tumors were identified in 1 706 and 2 388 genes for patients 1 and 2, respectively. Of the 1 706 genes identified for patient 1, 214 downregulated and 60 upregulated genes at gene expression levels in tumor samples compared with matched normal tissues were called. Of the 2 388 genes identified for patient 2, 273 downregulated and 204 upregulated genes were called. Similar analysis was applied to genes showing 5hmC loss in distal regulatory regions (2-12 kb upstream of TSS). Loss of 5hmC in promoter regions has been identified in 210 and 106 genes for patients 1 and 2, respectively. 20 downregulated and 10 upregulated genes out of the 210 genes were called for patient 1, and 12 downregulated and 11 upregulated genes out of the 106 genes were called for patient 2. The Fisher's exact test further demonstrated that loss of 5hmC in gene bodies and distal regulatory regions but not in promoter regions was significantly associated with gene silencing (Supplementary information, Figure S2E).
To explore the functional significance of 5hmC loss in gene bodies, we performed KEGG pathway enrichment analysis using 2 111 genes, which displayed significant loss of 5hmC in their gene bodies in tumors compared with normal tissues for both patients. The analysis showed that these genes were closely associated with various ccRCC-related pathways, such as renal cell carcinoma, the neurotrophin signaling pathway, the ErbB signaling pathway, and the VEGF signaling pathway (Figure 2G). Two of the most commonly mutated tumor suppressor genes, VHL and SETD2, displayed significant 5hmC loss in their gene bodies (Figure 2H and Supplementary information, Figure S2F; Fisher's exact test with Benjamini-Hochberg correction, P < 0.001). Consistent with 5hmC enrichment in gene body regions compared with promoter regions, only 49 genes (210 genes for patient 1 and 106 genes for patient 2) exhibited significant loss of 5hmC in promoter regions for both patients. KEGG pathway enrichment analysis did not identify any enriched functional clusters (data not shown). Collectively, these results suggest that loss of 5hmC in gene body regions is an epigenetic hallmark for ccRCC and may be associated with ccRCC progression.
Base-resolution analysis of 5mC in paired tumor and normal tissues
To accurately profile the genome-wide 5mC pattern, we combined the BS-seq and TAB-seq methods (Supplementary information, Table S3). Because limited numbers of nucleotide were identified as 5mC-modified sites in non-CpG regions (data not shown), all following analyses focused on CpG sites only. First, we observed that the average 5mC level of ccRCC tissues was marginally lower than that of matched normal tissues, as estimated by BS-seq in both patients (70.8% vs 75.3% for patient 1, 71.1% vs 72.9% for patient 2; Figure 3A and Supplementary information, Table S3). However, by subtracting the 5hmC contribution from the BS-seq data, the real 5mC levels in tumors of both ccRCC cases were slightly higher than that of matched normal tissues (Figure 3A). The average level of 5mC was increased in promoter and gene body regions but not in intergenic regions (Figure 3B). However, such DNA methylation pattern cannot be concluded from the BS-seq data (Supplementary information, Figure S3A). We found the high consistency between our BS-seq data and the published Human Methylation 450k array data15 which is also based on bisulfite treatment (Supplementary information, Figure S3B and Table S4). Moreover, 5hmC sites often overlapped with 5mC sites in both tumor and normal tissues (Figure 3C and Supplementary information, Figure S3C). Thus, the real 5mC aberrant pattern, particularly for 5hmC-modified sites, must be re-evaluated by BS-seq corrected with TAB-seq.
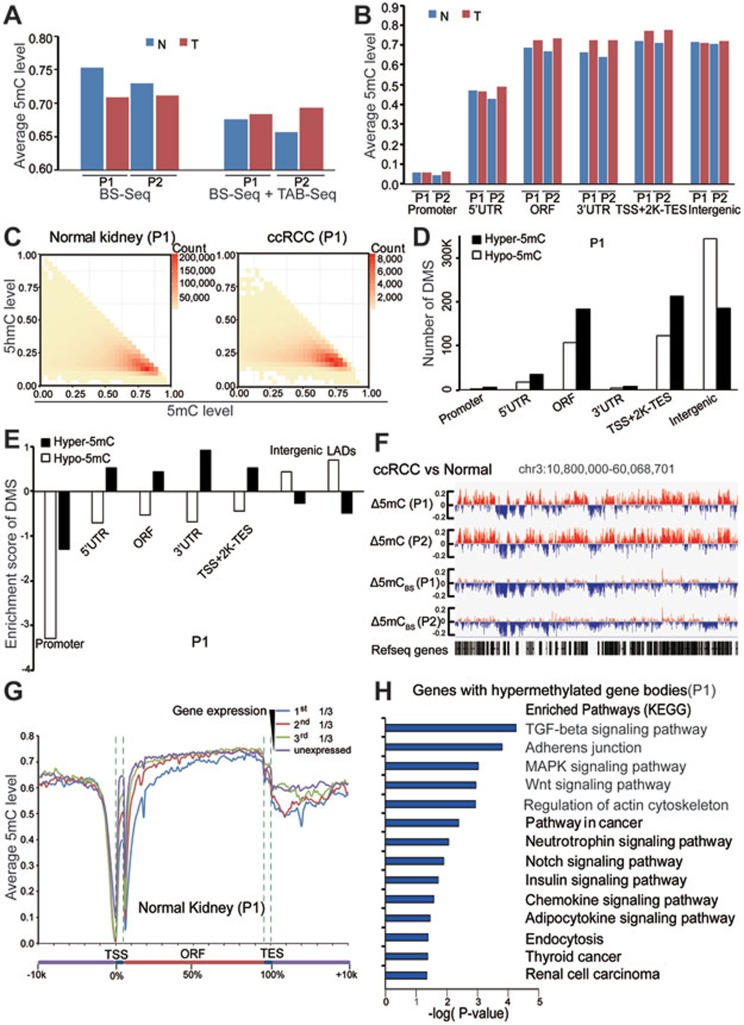
Figure 3.
Genome-wide single-nucleotide resolution mapping of 5mC. (A) Global changes of average 5mC levels in tumors and matched normal tissues. 5mC was calculated as the rate from BS-Seq (5hmC + 5mC) minus the 5hmC rate measured from TAB-seq. (B) Global changes of average 5mC levels in different genomic elements. (C) Heatmap of 5mC level at 5hmC sites in tumor and matched normal tissues. The scale bar from yellow to red represents the number of 5mC/5hmC modified sites from low to high. X and y axis showed the 5mC and 5hmC level of those modified sites, respectively. (D) The numbers of DMS in different genetic elements (promoter is defined as ± 500 bp of TSS). (E) The enrichment scores of DMS in different regulatory elements. LADs represent nuclear lamina-associated domains identified in TIG3 fibroblast cells. (F) Graphical representation of 5mC pattern in a region of chromosome 3. 5mCBS represents the methylation level from BS-seq only. (G) The average 5mC level throughout different gene-associated regions in normal kidney tissue from patient 1. Genes in the analyzed sample were divided into four groups according to their expression levels (FPKM value). (H) The KEGG pathway analyses of genes that gain methylation in gene body regions during tumorigenesis. The significance was illustrated by P-value. T and N represent tumor and matched normal tissue, respectively. P1 and P2 represent patients 1 and 2, respectively.
Hypermethylation is identified in both promoter and gene body regions in ccRCC
To assess the difference in 5mC levels at each CpG site between tumor and normal tissues, we first calculated differentially methylated sites (DMSs) based on the BS-Seq and TAB-Seq results (Supplementary information, Table S5). The data demonstrated that hypomethylated sites (hypo-5mC sites, where the 5mC level in tumor is lower than that in normal tissue) were enriched in intergenic regions (Figure 3D). Meanwhile, we observed a striking overlap between kidney tumor hypomethylated regions and nuclear lamina-associated domains (LADs) that were identified in TIG3 fibroblast cells25 (Figure 3E and Supplementary information, Figure S3E and S3F). Strikingly, hypermethylated sites (hyper-5mC sites, where the 5mC level in tumor is higher than that in normal tissue) were enriched in gene body regions (Figure 3D and 3E). A lower number of CpG sites were called as hyper-5mCBS sites by the BS-seq data alone especially in gene body regions (Supplementary information, Figure S3D). Both hypo-5mC and hyper-5mC sites were not enriched in promoter regions (Figure 3E and Supplementary information, Figure S3E). A representative locus at chromosome 3 is shown in Figure 3F. Consistent with findings in other types of tumors3,10,26, more genes with promoter hypermethylation (837 genes for patient 1 and 1 229 genes for patient 2) than those with promoter hypomethylation (580 genes for patient 1 and 457 genes for patient 2) were identified in ccRCC. In addition, 5mC levels in the promoters, especially in 5′-UTRs, were negatively associated with gene expression levels in normal kidney and tumor tissues in both patients (Figure 3G, Supplementary information, Figure S3G and data not shown). KEGG analysis for the genes with hypermethylated promoters showed that neuroactive ligand-receptor interaction and calcium signaling pathway are the two most enriched pathways (data not shown). Strikingly, profound gene body hypermethylation in tumors was observed in both patients (1 317 genes for patient 1 and 2 623 genes for patient 2). In contrast, much less genes with hypomethylated gene body regions in tumors were identified (272 and 237 genes for patients 1 and 2, respectively). KEGG analysis for the genes with hypermethylated gene bodies revealed several ccRCC-related functional pathways, such as adherens junction, pathway in cancer, neurotrophin signaling pathways, and RCC (Figure 3H and Supplementary information, Figure S3H). Collectively, these data suggest that besides promoter hypermethylation, gene body hypermethylation is a new epigenetic hallmark of ccRCC.
Loss of 5hmC in ccRCC is linked to methylation gain particularly in gene body regions
Since loss of 5hmC may lead to accumulation of 5mC, we determined whether loss of 5hmC is linked to hypermethylation in tumor. First, we observed that almost all CpG sites that contain 5hmC in normal tissues exhibited variable levels of methylation gains in tumor tissues in both promoter and gene body regions (Figure 4A and Supplementary information, Figure S4A). Consistent with previous data, methylation gains cannot be concluded from BS-seq data alone (Supplementary information, Figure S4C). Second, the hypo-5hmC sites significantly overlapped with hyper-5mC sites but not hypo-5mC sites in both patients (Figure 4B, Supplementary information, Figure S4B and Table S6; P < 0.001 for both patients; P-value for the number of the overlapped sites was calculated using a custom script based on BEDTools fisher27). Strikingly, analysis using 5mCBS data generated only by the BS-seq method failed to yield the significant association between hypo-5hmC and hyper-5mCBS sites (Figure 4B). Taken together, these results suggest that 5hmC loss is predominately associated with methylation gain in ccRCC.
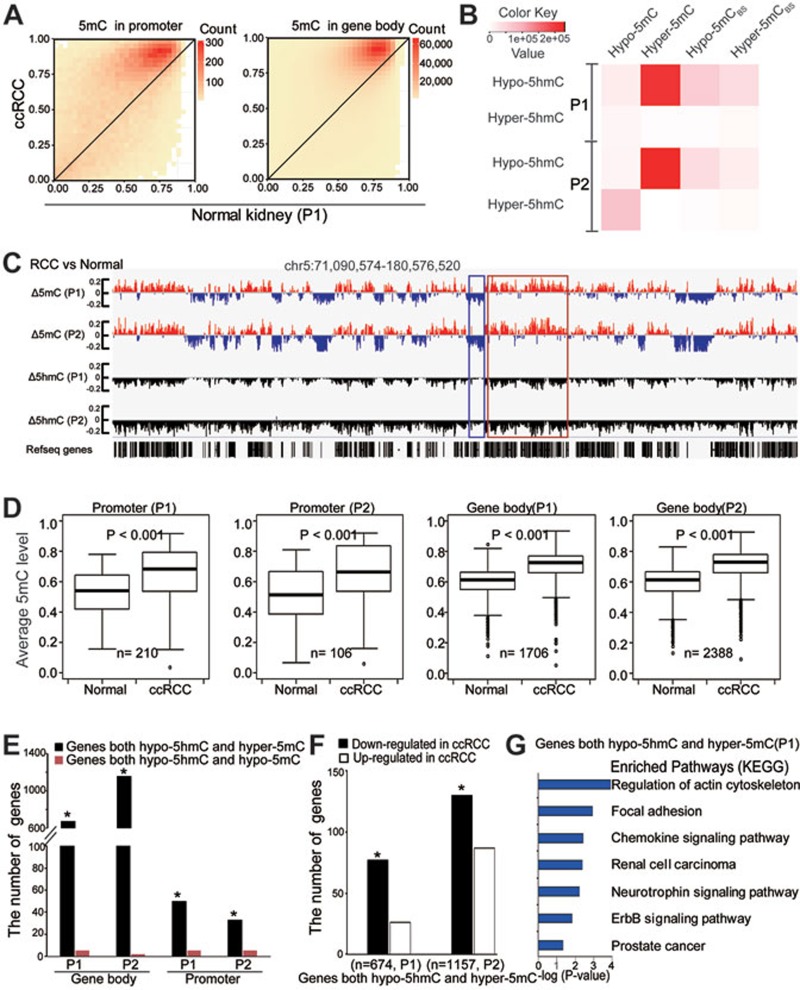
Figure 4.
Loss of 5hmC is linked to gene body hypermethylation in tumor tissues. (A) Heatmap of the estimated abundance of 5mC in tumor and matched normal tissues at 5hmC sites called in promoter or gene body regions. The scale bar from yellow to red represents the number of 5hmC sites from low to high. (B) Heatmap for the number of overlapped sites between DhMS and DMS. Hyper-5mCBS and Hypo-5mCBS sites were calculated by BS-seq data only. The color key from white to red represents the number of sites from low to high. (C) Wiggle tracks of a region from chromosome 5. Δ means the difference of 5mC or 5hmC level between ccRCC and normal samples. (D) Box-plot of average 5mC levels of the genes with hypo-5hmC promoters or gene bodies in tumor compared with those in normal tissues. (E) The number of overlapped genes among genes with hypo-5hmC, hyper-5mC and hypo-5mC promoter or gene body regions. The significance of the overlaps was tested using GeneOverlap (Fisher's exact test-based method; *P < 0.001). (F) The bar plots showing numbers of genes that harbor hypo-5hmC and hyper-5mC gene bodies and display changes in expression levels in ccRCC. n represents the number of genes with both hypo-5hmC and hyper-5mC signatures in gene bodies. *P < 0.001. (G) The KEGG pathway analyses of genes with both hypo-5hmC and hyper-5mC signatures in gene bodies. The significance was illustrated by P value. P1 and P2 represent patients 1 and 2, respectively.
To further examine whether the loss of 5hmC in tumors is involved in establishing the aberrant 5mC patterns observed in tumors, we partitioned the genome into 1 kb bins to quantify the difference in 5mC and 5hmC levels between tumors and matched normal tissues. A representative locus is shown in Figure 4C. Consistent with previous results at single-nucleotide resolution (Figure 4A and 4B), global hypermethylated and focal hypomethylated regions were enriched in gene-rich regions and intergenic regions, respectively (Figure 4C). Strikingly, the hypermethylated regions (red spikes) are located primarily within hypohydroxymethylated regions (black spikes). It is worth noting that the BS-seq method revealed long-range hypomethylation with focal hypermethylation in both patients (Figure 3F), similar to previous findings in other tumors, such as colorectal cancer28.
Moreover, the average 5mC levels of genes with hypo-5hmC promoter or gene body regions were increased in tumor tissues compared with matched normal tissues (Figure 4D). Fisher's exact test revealed that the hypo-5hmC genes significantly overlapped with hyper-5mC genes but not hypo-5mC genes in both promoter and gene body regions. Additionally, more genes showed the association (hypo-5hmC vs hyper-5mC) in the gene body regions compared to promoter regions (Figure 4E and Supplementary information, Figure S4E). Collectively, by combining the TAB-seq and BS-seq results, we identified the associate between 5hmC loss and hypermethylation, particularly in gene body regions.
Gene body hypermethylation in tumor tissues is associated with gene silencing
DNA methylation at promoters is related to gene expression29,30,31. Next, we explored the functional roles of gene body hypermethylation by integrating the 5hmC/5mC profiling with gene expression data. 674 and 1 157 genes were called as both hypo-5hmC and hyper-5mC in gene body regions in patients 1 and 2, respectively. Of the 674 genes for patient 1, 77 downregulated and 26 upregulated genes at gene expression levels in tumor tissue compared with matched normal tissue were called. Of the 1 157 genes for patient 2, 130 downregulated and 87 upregulated genes were called. Fisher's exact test revealed a significant association of gene expression downregulation with both hypo-5hmC and hyper-5mC gene bodies in the two patients (Figure 4F). It is worth noting that only a portion of genes with hypermethylation in gene bodies were downregulated, suggesting that changes of 5mC/5hmC in gene body regions may regulate the gene function beyond transcriptional regulation. Next, we explored the potential role of genes with both hypo-5hmC and hyper-5mC in gene body regions. KEGG pathway analyses demonstrated that these genes were closely associated with various ccRCC-related pathways in both patients, such as focal adhesion, renal cell carcinoma, neurotrophin signaling pathway, and ErbB signaling pathway (Figure 4G and Supplementary information, Figure S4F and Table S7). Taken together, these results suggest that loss of 5hmC is associated with hypermethylation in gene bodies and the latter is associated with silencing of part of tumor-related genes, indicating the oncogenic role of 5hmC loss in kidney cancer.
Downregulation of IDH1 contributes to global loss of 5hmC in ccRCC
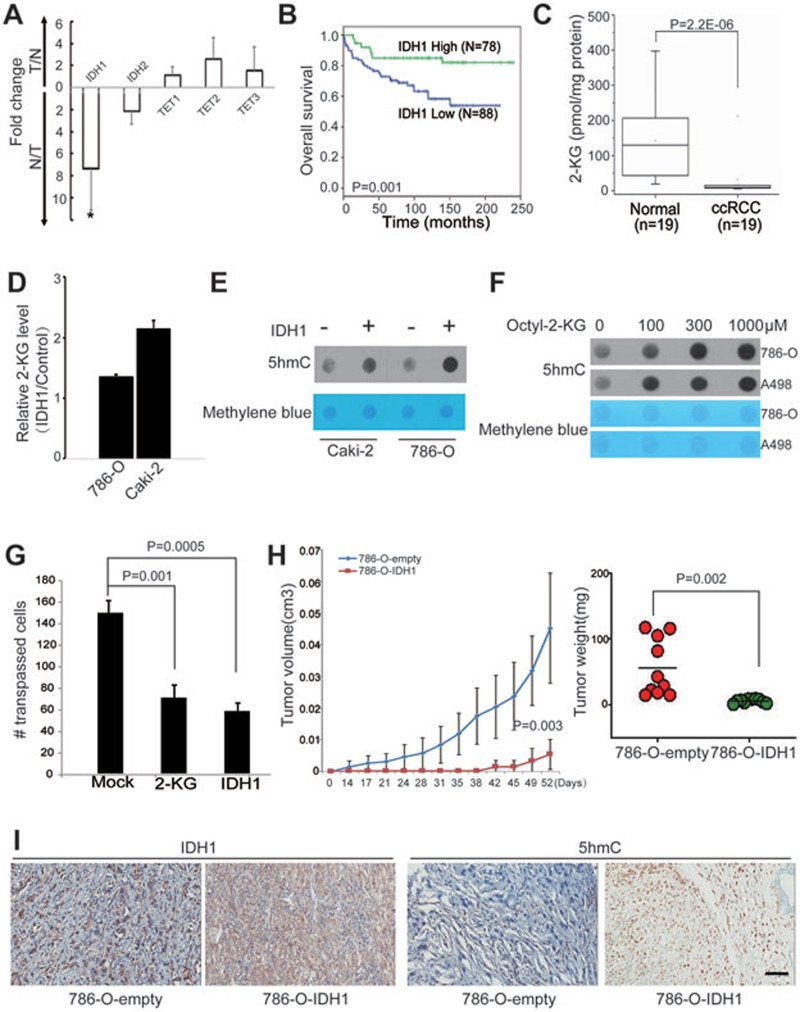
Next, we explored the molecular mechanisms underlying the 5hmC loss in ccRCC. Although 5hmC loss can be observed in all ccRCC samples examined, TET mutations are rare in ccRCC, suggesting that inhibition of TET-mediated 5mC oxidation might be responsible for the 5hmC loss in kidney cancer. TET-mediated 5mC oxidation requires cosubstrates, such as 2-ketoglutarate (2-KG), which is mainly generated by IDHs during the tricarboxylic acid cycle24. Thus, we examined the mRNA expression levels of IDHs and TETs in 6 ccRCC patients by RT-qPCR. We observed that only IDH1 was significantly downregulated in ccRCC compared with normal kidney cells (Figure 5A). According to IHC staining using the TMA, the protein level of IDH1 also decreased in tumor tissues compared with matched normal tissues (Supplementary information, Figure S5A-S5C). Moreover, similar to 5hmC, the lower IDH1 level in the tumor tissue predicts poor overall survival (Figure 5B). Thus, downregulation of IDH1 may represent one mechanism for loss of 5hmC in ccRCC through downregulation of 2-KG.
Figure 5.
IDH1 downregulation leads to loss of 5hmC, and restoring 5hmC levels attenuates tumor growth. (A) The relative expression levels of the indicated genes in ccRCC patient samples were measured by RT-qPCR. Data are shown as the mean ± SD (n = 6). P-values were calculated by the Wilcoxon test (*P = 0.002). (B) Kaplan–Meier survival curves of ccRCC patients in the TMA. “IDH1 high” and “IDH1 low” represent that > 10% and ≤ 10% cells are stained, respectively. P value was calculated by the log-rank test. (C) The 2-KG levels in ccRCC patient tissues were evaluated by the LC-ESI-MS method. P values were calculated by the Wilcoxon test (P = 2.2E−06). (D) The intracellular 2-KG level in ccRCC cell lines was measured by the LC-ESI-MS method. Y axis represents the intracellular 2-KG level in IDH1-transfected cells relative to that in control cells. (E) Dot blot assay of 5hmC levels in ccRCC cell lines with transient transfection of IDH1 or empty vector. Equal loading was validated by methylene blue staining. (F) 786-O and A498 ccRCC cells were treated with cell-permeable octyl-2-KG for 48 h. The 5hmC levels were measured by dot blot assay. (G) Cell invasion capacity was evaluated by transwell assay. 786-O ccRCC cells were treated with octyl-2-KG (300 μM) or transiently overexpressed IDH1 for 48 h. (H) Tumor growth curves of xenografts with 786-O-IDH1 or 786-O-empty vector cells. Tumor volume is shown as the mean ± SD (n = 10 mice). Tumor weights of the indicated xenografts at endpoint (52 days) are shown as the mean± SD (n = 10 mice). P-value was calculated by a two-tailed student's t-test. (I) A representative IHC staining of IDH1 and 5hmC in the indicated xenograft. Scare bar, 100 μm.
To further test this hypothesis, we examined the intracellular 2-KG levels of 19 paired tumor and normal kidney tissues by LC-ESI-MS. The results showed that intracellular 2-KG levels significantly decreased in tumor tissues (Figure 5C). Consistently, IDH1 overexpression increased intracellular 2-KG levels in two ccRCC cell lines (Figure 5D). A dot blot assay further demonstrated that ectopic expression of IDH1 can consistently restore 5hmC levels in these cell lines (Figure 5E). Taken together, these results suggest that downregulation of IDH1 is a mechanism underlying 5hmC loss in kidney cancer.
IDH1 may function as a tumor suppressor for ccRCC
To explore the oncogenic role of 5hmC depletion in ccRCC, we examined the biological consequences of manipulating 5hmC levels in ccRCC cell lines by both overexpressing IDH1 (Supplementary information, Figure S5E) and pharmacologically elevating intracellular 2-KG levels. Both treatments can increase intracellular 2-KG levels (Figure 5D and Supplementary information, Figure S5F) and reconstitute 5hmC levels in ccRCC cell lines (Figure 5E and 5F). Tumor invasion assay demonstrated that 5hmC reconstitution by these two treatments significantly blocked tumor cell invasion (Figure 5G). However, cell proliferation was largely unaffected in both conditions (Supplementary information, Figure S5G). Moreover, we generated stable polyclonal 786-O ccRCC cell lines transfected with IDH1 or empty vectors. Subcutaneous injection of the 786-O-IDH1 cells into nude mice induced slower growth and smaller tumor burden than injection of the 786-O-empty cells (Figure 5H). IHC staining indicated that both IDH1 and 5hmC levels were higher in 786-O-IDH1 tumors than in 786-O-empty vector xenografts (Figure 5I). In addition, H&E staining revealed more aggressive phenotype of the empty vector xenografts compared with the IDH1-overexpressing tumors (Supplementary information, Figure S5H). Taken together, manipulating intracellular 2-KG level can restore intracellular 5hmC levels in ccRCC, which can block tumor invasion and growth. Thus, IDH1 may function as a candidate tumor suppressor for ccRCC.
Discussion
Inactivation of the von Hippel-Lindau tumor suppressor (pVHL) is the most well-known oncogenic event in ccRCC32. However, VHL deletion in mice fails to elicit tumor formation, suggesting additional mechanisms are essential. Strikingly, a number of 2-KG-dependent enzymes that act on chromatin are themselves targets of mutation in ccRCC (e.g., KDM6A, KDM5C and TET2)14,15,16, suggesting that epigenetic pathways play pivotal roles in the pathogenesis of kidney cancer.
In this study, by exploring 5mC and 5hmC levels simultaneously in the same samples, we observed that global 5mC levels do not significantly change during kidney tumorigenesis. Consistent with previous findings in colorectal cancer28, kidney tumor hypomethylated sites are enriched in intergenic regions and LADs. Dynamic association and dissociation with nuclear lamina has been implicated as a key mechanism in developmental regulation of long-range gene silencing33, but further investigations are needed to elucidate the functional role of hypomethylated LADs in gene expression regulations. Inconsistent with previous findings, we observed that hyper-5mC sites are significantly enriched in gene body regions. Further analysis showed that gene body methylation is significantly associated with gene silencing for a few genes (Figure 4F). However, the mechanistic link between gene body hypermethylation and gene silencing is still lacking. It has been broadly accepted that methylation of a CpG island downstream of an active promoter in a mammalian gene clearly does not block the formation of a transcript34. Thus, the changes of 5mC/5hmC in gene body regions may regulate the gene function beyond regulating gene expression, such as mRNA splicing34. More comprehensive studies are needed to address these questions in the future. Collectively, these results support that currently approved hypomethylating agents (DNMT inhibitors) may facilitate the treatment of ccRCC through targeting the gene body methylation. However, a solid correlation between DNA methylation in gene body regions and therapeutic efficacy must be convincingly shown in the future before applying hypomethylating agents.
In addition, our results demonstrated that loss of 5hmC is linked to gene body DNA hypermethylation, suggesting that re-establishment of intracellular 5hmC levels may facilitate the treatment of ccRCC through specifically targeting gene body hypermethylation. We have demonstrated that loss of 5hmC is mediated in part by reduced expression of IDH1. This result is inconsistent with the gain-of-function mutations of IDH1 and IDH2 identified in several different tumors that result in accumulation of 2-hydroxyglutarate (2-HG), a 2-KG antagonist10,26,35. Recently, elevation of 2-HG, mediated in part by reduced expression of 2-HG dehydrogenase (L2-HGDH), has been shown to potentially represent a novel mechanism for the regulation of 5hmC in kidney cancer36. Consistent with these data, we also detect an increase of 2-HG levels in our samples (Supplementary information, Figure S5D). However, the molecular mechanism that drives the IDH1 and L2-HGDH downregulation in ccRCC remains unknown. The IDH1 and L2-HGDH gene loci are also methylated during tumorigenesis (data not shown), which may establish a positive feedback regulatory loop. Collectively, limited generation of 2-KG or elevated generation of the antagonist 2-HG contributes to the loss of 5hmC in kidney cancer. Thus, re-establishment of intracellular 5hmC levels may facilitate the treatment of ccRCC. Yen and colleagues demonstrated the efficacy of a selective IDH2-R140Q inhibitor at blocking 2-HG generation in primary AML cells37. However, mammalian cells express more than 60 dioxygenases that utilize 2-KG as a cosubstrates22,38, including TETs and histone demethylases. Thus, the synergistic role of 5hmC loss and other pathways regulated by 2-KG in ccRCC tumorigenesis needs to be explored further. Taken together, these results suggest that an enhanced understanding of epigenetic mechanisms that underlie ccRCC development might pave the way to new therapies that improve outcome for ccRCC patients. Further studies using similar strategies are required to elucidate whether similar epigenetic oncogenic pathways also work in other cancers, such as brain cancer, melanoma and colorectal cancer.
Materials and Methods
Surgical samples and genomic DNA and RNA extraction
The paired ccRCC samples without preoperative target therapy/chemotherapy were obtained from Department of Urology, Peking University First Hospital and Department of Urology, Peking University People's Hospital, respectively. Informed Consent Forms were obtained and approvals for the study were attained from the Ethical Committee of our institutes. The histopathologic results were reviewed by at least two independent pathologists. The part of surgical samples were stored in −80 °C and RNALater (Ambion, Cat#: AM7012) post-operation, respectively. The tumor and matched normal tissue from the same patient were used. Subsequently, genomic DNA (Qiagen, Cat#: 51306) and RNA (Life Tech, Cat#: 15596-018) were extracted according to the manufacturers' instructions.
Quantitative PCR
Total RNA was extracted from cells using Trizol reagent (Sigma) according to the manufacturer's instructions. After quantification, equivalent amounts of RNA were reverse transcribed using ImProm-IITM Reverse Transcription System (Promega, Cat#: A3800). Real-time quantitative PCR was performed using SYBRPremix (CWBio, Cat#: CW2391) and detected in the ABI7500 Real-time PCR system (Applied Biosystems, Foster City, CA, USA). Gene expression analysis was performed using the comparative ΔΔCT method with GAPDH for normalization. The primers used are as follows:
Table 1.
| GAPDH | F | 5′-GAAGGTGAAGGTCGGAGTC-3′ |
| R | 5′-GAAGATGGTGATGGGATTTC-3′ | |
| IDH1 | F | 5′-TCCGTCACTTGGTGTGTAGG-3′ |
| R | 5′-GGCTTGTGAGTGGATGGGTA-3′ | |
| IDH2 | F | 5′-TGAACTGCCAGATAATACGGG-3′ |
| R | 5′-CTGACAGCCCCCACCTC-3′ | |
| TET1 | F | 5′-GCTATACACAGAGCTCACAG-3′ |
| R | 5′-GCCAAAAGAGAATGAAGCTCC-3′ | |
| TET2 | F | 5′-CTTTCCTCCCTGGAGAACAGCTC-3′ |
| R | 5′-TGCTGGGACTGCTGCATGACT-3′ | |
| TET3 | F | 5′-GTTCCTGGAGCATGTACTTC-3′ |
| R | 5′-CTTCCTCTTTGGGATTGTCC-3′ |
Cell culture
The A498 ccRCC cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal bovine serum (FBS; Gibco). The ccRCC cell line Caki-2 was maintained in McCoy's 5a modified medium supplemented with 10% (vol/vol) FBS, and the 786-O RCC cell line was maintained in RPMI1640 medium supplemented with 10% (vol/vol) FBS. All cells were cultured in a 37 °C, humidified, 5% CO2-containing-atmosphere incubator (Thermo Scientific).
Dot blot
DNA samples were denatured in heating block at 95 °C for 10 min and chilled on ice for 5 min. Then samples were pointed on wet Hybrid Membrane and dried by suction. Briefly, the membrane was baked at 80 °C for 2 h and blocked with 5% non-fat milk in TBST (20 mM Tris-HCl, pH 7.4; 150 mM NaCl; 0.1% Tween-20) for 1 h at room temperature (RT). The membrane was further incubated with the primary antibody against 5hmC (dilution 1:10 000, Active motif) or 5mC (dilution 1:3 000, Active motif) for 1 h at RT. Then, the anti-rabbit or anti-mouse IgG-HRP antibody was added to the membrane for 30 min at RT. Subsequently the membrane was developed with ECL Kit (GE Amersham, Cat#: RPN2232).
LC-ESI-MS analysis
The isolated genomic DNA was enzymatically digested according to previously described method20. Briefly, DNA (750 ng) was first denatured by heating at 95 °C for 5 min and then chilling on ice for 2 min. After adding 1/10 volume (1 μl) of S1 nuclease buffer (30 mM CH3COONa, pH 4.6, 280 mM NaCl, 1 mM ZnSO4) and 50 units of S1 nuclease, the mixture (10 μl) was incubated at 37 °C for 12 h. To the solution was subsequently added 4 μl of alkaline phosphatase buffer (50 mM Tris-HCl, 10 mM MgCl2, pH 9.0), 0.001 units of venom phosphodiesterase I and five units of alkaline phosphatase. And then the mixture (40 μl) was incubated at 37 °C for an additional 4 h followed by extraction with equal volume of chloroform twice. The resulting aqueous layer was collected and lyophilized to dryness and then reconstituted in 70 μl water, which was subjected to LC-ESI-MS that contains an AB 3200 QTRAP mass spectrometer (Applied Biosystems) with an electrospray ionization source (Turbo Ionspray) and a Shimadzu LC-20AD HPLC (Tokyo, Japan) with two LC-20AD pumps. Data acquisition and processing were performed using AB SCIEX Analyst 1.5 Software (Applied Biosystems). The HPLC separation was performed on an ODS-N column (150 × 2.1 mm i.d., 5 μm, Weltech Co., Ltd., Wuhan, China) with a flow rate of 0.2 ml/min at 35 °C. Water (solvent A) and methanol (solvent B) were employed as mobile phase. The mass spectrometry detection was performed under positive ion mode. The target nucleosides were monitored by multiple reaction monitoring (MRM) mode using the mass transitions (precursor ions → product ions) of dC (228.4 → 112.2), T (243.3 → 127.2), dA (252.4 136.2), dG (268.4 → 152.4), 5-mdC (242.3 → 126.1), 5-hmdC (258.2 → 124.2). The MRM parameters of all nucleosides were optimized to achieve maximal detection sensitivity. Finally, the percentages of 5mC and 5hmC were calculated by the following formula, where M (cytosine), M (5mC) and M (5hmC) are the molar quantities of cytosine.
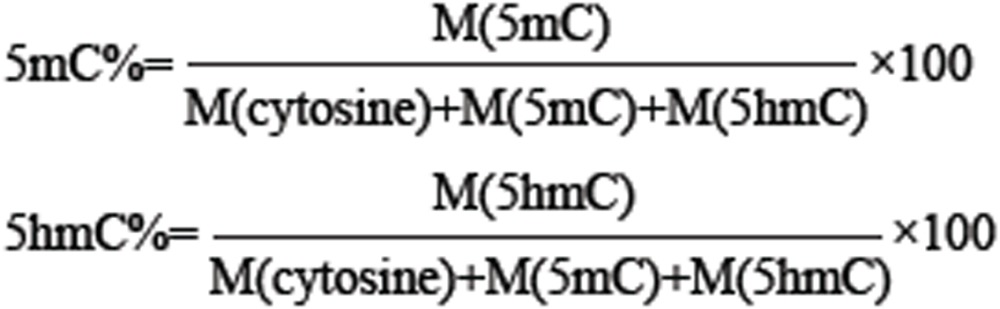 |
Western blot
Cells were harvested with lysis buffer (50 mM Tris-HCl (pH 7.4); 150 mM NaCl; 1% Triton X-100; 1 mM EDTA; 1 mM NaF; 1 mM Na3VO4 and 1× proteinase inhibitor cocktail (Sigma, P8340)). The protein concentration of the supernatant was quantified by a BCA assay (CWbiotech, China). Protein extracts, separated by SDS-PAGE and transferred onto PVDF membranes (Life Tech, Cat#: IB4010-02), were probed with antibodies against FLAG (1:5 000, Sigma-Aldrich, Cat#: F1804) overnight at 4 °C. Proteins of interest were detected with HRP-conjugated sheep anti-mouse IgG antibody (1: 5 000, GE Healthcare, Uppsala, Sweden) and visualized with the Pierce ECL Western blotting substrate (Thermo Scientific, Rockford, IL, USA), according to the manufacturer's instructions.
Cell invasion assay
A transwell system (Millipore, Cat#: PIEP12R48) was employed to assess cell migration according to the manufacturer's recommendation. 1 × 104 786-O ccRCC cells in 2% FBS Matrigel (BD, Cat#: 354234) were seeded into the upper chamber while the lower chamber Matrigel was filled with 10% FBS. After 24-h culture, cells which had not penetrated the filter were wiped out by cotton swabs, and cells which had migrated to the lower surface of the filter were fixed with 100% methanol and stained with 0.5% crystal violet and counted under microscope. Values for migration were calculated as the average number of migrated cells per microscopic field (×100) over nine fields. All the assays were repeated three times.
TMA, IHC and survival analysis
Patient tumors were formalin-fixed paraffin-embedded. TMA were constructed at 4 μm thickness, mounted on polylysine-coated slides, and succumbed to IHC analyses. Briefly, slide was subjected to de-paraffin, antigen retrieval, rinse, incubation with either rabbit anti-5hmC antibody (1:10 000, Active motif, Cat#: 39999) or mouse anti-IDH1 antibody (1:100, Protein Tech, Cat#: 12332-1-AP) overnight, and then incubation with the secondary antibody (EnVision Dual Link, Dako) for 30 min. After rinsing, DAB Enhancer (Dako) was used to toning for 1-2 min to enrich the brown color. The expression of IDH1 or 5hmC was evaluated by three experienced pathologists. The differences of 5hmC and IDH1 between controls and RCC patients were assessed using a standard nonparametric Mann-Whitney U-test. The relationship between 5hmC/IDH1 levels and clinical information was evaluated by univariate linear regression analysis. Multivariate Cox proportional hazards regression analysis with stepwise selection was used to evaluate independent prognostic factors associated with overall survival, and 5hmC level or IDH1 level, and other clinical information were used as covariates. For each covariate, a hazard ratio and an associated P-value were examined. The Kaplan-Meier method was used to estimate overall survival distribution. Differences in survival between distinctive survival groups were analyzed with the log-rank test. All P-values were two-tailed with 0.05 specified as statistically significant. Statistical analyses were performed using SPSS software version 13.
Animal care and xenograft
Twenty BALB/c nude mice (male 10, female 10, 6-8 weeks old) were purchased from VitalRiver (Beijing, China) and maintained under specific pathogen-free condition and handled according to animal care and use of Ethics Committee of Animal Experimentation in our institutes. Either 786-O-Vector cells or 786-O-IDH1 cells were inoculated (6 × 106 cells per 0.1 ml per mouse) s.c. at the right scapular of the mouse (n = 10/group, 5♂5♀). Tumor sizes were measured with a caliper at each time interval and the tumor volume was calculated using the formula: Volume = S × S × L/2, where S is the short length of the tumor in cm and L is the long length of the tumor in cm.
TAB-seq
The sequencing libraries were constructed as described17 with minor modification. Briefly, genomic DNA with spike-in controls was glucoslyated and oxidized using the kit from Wisegene (Cat#: K001). Then, the DNA was further directed to bisulfite conversion using the EZ DNA methylation Gold kit (Zymo Research) according to the instruction manual. The library was sequenced using Illumina HiSeq 2000. Paired reads were mapped uniquely to the reference genome (HG19, UCSC) by Bismark. Efficient conversion of unmodified cytosine to uracil and efficient conversion of 5mC to 5caU/U were calculated by spiked M. SssI-treated lambda DNA.
BS-Seq
5mC sequencing libraries were constructed as described with minor modifications39. Briefly, HG19 reference genome was downloaded from UCSC. The 48 502 bp Lambda genome was also included in the reference sequence for calculating bisulfite conversion rate. Paired-end BS-seq reads were mapped against the reference by Bismark_v0.6.440 with stringent parameters: -n 2 -l 60 -e 100 -X 600. A custom script was used to examine whether pair-end reads overlapped and the overlapped part is trimmed from one end to prevent counting twice from the same observation.
The consistency between our data and the published TCGA data
The published data were downloaded from the TCGA websites (https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/tumor/kirc/cgcc/jhu-usc.edu/humanmethylation450/methylation/). The level 3 data from 160 paired ccRCC and normal kidney tissue were used in the following analysis. Around 380 000 CpG sites covered by both all the samples of TCGA data and all the samples of our BS-seq cohort (depth ≥ 5×) were used in the following analysis. And the CpG sites on the sex chromosome were excluded from the analysis. The Pearson correlation coefficient was calculated between methylation levels from BS-seq data and β values from Human Methylation 450k array. Meanwhile, the association between methylation level of our sample and mean β values of the 160 TCGA samples was also shown.
Quantification of 5hmC levels of each CpG site
To quantify the 5hmC level, we first apply binomial distribution model to calculate the significance for the hydroxymethylation state of each CG site covered by at least one read. For each such base, we counted the number of “C” bases from TAB-Seq reads as hydroxymethylated (denoted as NC) and the number of “T” bases as not hydroxymethylated (denoted as NT). Then, we used the binomial distribution having parameters N as the sequencing depth (NC + NT) and p as the 5mC non-conversion rate (e.g., 3.48% for normal kidney tissue of P1, Supplementary information, Table S1), to assess the probability of observing NC greater than cytosines by chance. Then, Benjamini Hochberg method was used to correct the binomial P value. Only sites with Benjamini Hochberg corrected binomial P value ≤ 0.05 and reads coverage ≥ 5 were considered hydroxymethylated. The hydroxymethylation level is estimated as NC/(NC + NT). Only sites with 5mCBS level from BS-seq ≥ 5hmC level were considered in the following analyses.
Quantification of average 5hmC level for all CpG sites in each region
Successful detection of 5hmC is governed by two key parameters: (1) efficient protection of 5hmC; (2) efficient conversion of 5mC to 5caU/U (Tet oxidization rate). To reduce these effects, corrected average 5hmC level of each region was calculated by the following formula:
5hmC%corrected = 5hmC%measured/protection rate of 5hmC − 5mC%BS-seq × (1 − Tet oxidization rate5mC to 5caU/U). For 5hmC%measured, we counted the number of “C” bases from TAB-Seq reads as hydroxymethylated (denoted as NC) and the number of “T” bases as not hydroxymethylated (denoted as NT). Then, 5hmC%measured is estimated as NC/(NC + NT). Similarly, for 5mC%BS-seq, we counted the number of “C” bases from BS-Seq reads as methylated (denoted as MC) and the number of “T” bases as unmethylated (denoted as MT). Then, 5mC%BS-seq is estimated as MC/(MC + MT).
Quantification of differentially hydroxymethylated sites between normal and tumor tissues in each patient
To identify differentially hydroxymethylated sites between normal and tumor tissues of a patient, we applied the following criteria: (1) for the tissue-specific 5hmC-called sites, the 5hmC levels were ≥ 0.2; and (2) for the called 5hmC sites shared between normal and tumor tissues, the difference of 5hmC levels were ≥ 0.2 and Benjamini Hochberg corrected two-tailed Fisher's exact test P values were ≤ 0.05. The Fisher's exact test was performed using the following matrix: the number of “C” bases from TAB-Seq reads as hydroxymethylated (denoted as NC) and the number of “T” bases as not hydroxymethylated (denoted as NT; tumor vs matched normal tissue).
The 5hmC enrichment score in each genomic region
The 5hmC enrichment score was calculated by the following formula:
The enrichment scorein the genomic element = log2 (# called 5hmC sitesin the genomic element/# expected). # expected was computed as: # called 5hmC sitesin the genome × # CpG sitesin the genomic element/# total CpG sitesin the genome. # means the number of sites.
Quantification of differentially hydroxymethylated regions between tumor and normal tissue
The genomic regions containing at least 5 CG sites and each CG site covered by at least 5 reads were considered for further analysis. The corrected average 5hmC level of the region was calculated by the following formula:
5hmC%corrected = 5hmC%measured/protection rate of 5hmC − 5mC%BS-seq × (1 − Tet oxidization rate5mC to 5caU/U). The regions with difference of 5hmC level larger than 10% between normal and ccRCC tissues (two-tailed Fisher's exact test with Benjamini Hochberg adjusted P ≤ 0.05) were defined as differentially hydroxymethylated regions. The Fisher's exact test was performed using the following matrix: the number of called hydroxymethylated CpG sites (denoted as NC) and the number of covered CpG sites (denoted as N) within the region (tumor vs matched normal tissue).
Quantification of the methylation level for each CpG site by combining BS-seq and TAB-seq
For the CpG sites that were not called hydroxymethylated by TAB-seq data, the 5mC level was estimated as mi/(mi + ui), where mi was defined as the number of reads from BS-seq showing methylation over position i (both strands). ui was defined as the number of reads showing unmethylation over CpGi. Because CpG methylation is symmetric, mi and ui include observations associated with the cytosines on both strands for the i-th CpG. Only sites with read depth (mi + ui) ≥ 5 were considered in the downstream analysis. For CpG sites identified both methylated and hydroxymethylated, the 5mC level is calculated as 5mC level from BS-seq (5mCBS) − 5hmC level from TAB-seq data.
Identification of the DMSs
To identify DMSs between normal and tumor tissues of a patient, the permutation tests were used to compute statistical significance. The corrected 5mC levels of CpG sites covered by at least 5 reads were performed on 100 permutations and FDR was estimated by calculating the fraction of significant hits in the permuted data compared with the observed data at a specific P-value threshold. The sites were considered differentially methylated, if (1) the permuted P ≤ 0.1; and (2) the correct methylation level difference between normal and tumor tissues was ≥ 0.2.
Identification of the differentially methylated regions
The average 5mC level of the region was calculated by the following formula:
5mC%corrected = 5mC%BS-seq − 5hmC%TAB-seq. The methods to calculate the average 5mC level from BS-seq data and 5hmC level from TAB-seq data are shown previously. Genomic regions with difference of methylation level ≥ 10% between normal and ccRCC tissues (two-tailed Fisher's exact test with Benjamini Hochberg adjusted P ≤ 0.05) were defined as differentially methylated regions. The Fisher's exact test was performed using the following matrix: the number of “C” bases from BS-Seq reads as methylated (denoted as NC) and the number of “T” bases as unmethylated (denoted as NT; tumor vs matched normal tissue).
RNA-seq and statistics
The mRNA-Seq Sample Prep Kit (Illumina) was used to construct RNA-seq libraries, according to the manufacturer's instructions with little modifications. First, the raw FASTQ data files generated by the Illumina HiSeq 2000 platform were mapped to the human reference sequence (HG19) using Tophat program (tophat v2.0.1341) with the default parameters. Then the normalized FPKM levels of each sample with default parameters using the cufflinks program (cufflinks v2.2.142) were calculated. And the expressed genes (FPKM ≥ 1) and unexpressed genes (FPKM < 1) based on FPKM values were defined. Second, total read counts for each UCSC defined protein-coding gene were extracted using HTSeq (HTSeq version 0.6.1p143) with “intersection-nonempty” mode and read counts were loaded into R-package DEseq244 with the method GLM (a generalized linear model, for detail see DESeq2 package manual) to calculate fold changes for evaluating differentially expressed genes between normal and ccRCC tissues. Genes were considered differentially expressed with difference of normalized |log2 Fold Change (ccRCC to normal tissue)| ≥ 1.5.
Association between 5mC/5hmC and gene expression in each indicated sample
Genes were aligned from the TSS to the TES. For each gene, the region between TSS and TES was divided into 100 bins, and 5′-UTR and 3′-UTR represented five bins each. The 5mC and 5hmC level were calculated in each bin/region. Genes were stratified into four groups by gene expression in the indicated sample. FPKMs < 1 are identified as unexpressed genes and all the expressed genes are divided into three groups.
Association between 5mC/5hmC changes and gene expression changes between normal and tumor tissues
For a given whole set I of protein coding IDs and both subsets A (e.g., genes with hypo-5hmC in gene body regions) and B (e.g., genes downregulated at gene expression level) belong to I, and S was the common list of A and B. The significance of seeing S can be formulated as a hypergeometric distribution or a contingency table (which can be solved by Fisher's exact test; for detail see GeneOverlap documentation; URL: http://shenlab-sinai.github.io/shenlab-sinai/).
Gene ontology analysis
Gene ontology analysis was performed using DAVID (http://david.abcc.ncifcrf.gov/). KEGG terms with P < 0.05 were determined as statistically significant. The top 15 most enriched pathways were shown.
Accession number
The sequence data reported in this paper have been deposited in the genome sequence archive of Beijing Institute of Genomics, Chinese Academy of Sciences, gsa.big.ac.cn (accession no: PRJCA000102). And the GEO accession number is GSE63183.
Author Contributions
The project was conceived and the experiments were designed by WC, LJ and ZL. The TAB-seq and BS-seq library construction and genome analyses were performed by CK, ZJ and GZ. HC contributes TAB-seq. MQ, XZ, ZY, XZ, and KY were involved in statistical analysis and cellular experiments. The TMA data were evaluated by LZ, LY, YX and LX. YB performed the mass spectrometry experiments.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Dr Kevin P White for providing the TMA slides. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13040000 to WC and JL), the National Basic Research Program of China (973 Program; 2015CB856200 to JL), the 863 High-Tech Foundation (2014AA020608 to YK), the National Natural Science Foundation of China (91231112, 81422035 and 91519307 to WC, 81372746 to LZ, 81101940 to KC, and 31200958 to JZ), the Youth Innovation Promotion Association of Chinese Academy of Sciences (to KC and JZ), the KC Wong Education Foundation (to JZ), and the Chinese Academy of Sciences Funds for distinguished young scientists (to JZ).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Global dynamics of 5mC and 5hmC levels in kidney cancer.
Genome-wide dynamic patterns of 5hmC during kidney tumorigenesis.
Genome-wide single nucleotide resolution mapping of 5mC by combing BS-seq and TAB-seq method.
Loss of 5hmC facilitates the gene body hypermethylation in tumor tissue and promotes tumorigenesis.
Manipulating 5hmC generating pathway has potential therapeutical effect.
Summary of single-base 5hmC sequencing using TAB-seq.
Summary of RNA-seq.
Summary of shotgun bisulfite sequencing (BS-seq).
The consistency between TCGA KIRC 450K data and our BS-seq data.
Summary of DMS (differentially methylated sites).
The overlaps between aberrant 5hmC and 5mC changes at single nucleotide resolution during tumorigenesis.
The GO terms of genes with 5mC/5hmC changes in gene body regions.
References
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3:415–428. [DOI] [PubMed] [Google Scholar]
- Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 2003; 300:455. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003; 349:2042–2054. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res 1982; 10:2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res 1983; 11:6883–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013; 500:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 2011; 8:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 2010; 468(7325):839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18:553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki J, Taby R, Vasanthakumar A, et al. Effects of TET2 mutations on DNA methylation in chronic myelomonocytic leukemia. Epigenetics 2012; 7:201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 2010; 5:e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 2010; 38:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011; 469:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network TCGAR. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013; 499:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 2013; 45:860–867. [DOI] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012; 149:1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012; 150:1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Chaux A, Meeker AK, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2011; 2:627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, Shen F, Huang W, et al. Quantification of 5-methylcytosine and 5-hydroxymethylcytosine in genomic DNA from hepatocellular carcinoma tissues by capillary hydrophilic-interaction liquid chromatography/quadrupole TOF mass spectrometry. Clin Chem 2013; 59:824–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ghanim M, Xue L, et al. Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer. Science 2009; 323:1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol 2008; 4:152–156. [DOI] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 2012; 151:1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 2011; 42:451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008; 453:948–951. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012; 483(7390):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 2012; 44:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Weisenberger DJ, Aman JF, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 2010; 107:8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010; 328:916–919. [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer 2002; 2:673–682. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 2010; 38:603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. The DNA methylation paradox. Trends Genet 1999; 15:34–37. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010; 17:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EH, Livi CB, Rakheja D, et al. L-2-Hydroxyglutarate:an epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discov 2014; 4:1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Travins J, DeLaBarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 2013; 340:622–626. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle 2009; 8:1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zhang J, Wang JJ, et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 2013; 153:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28:511–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Global dynamics of 5mC and 5hmC levels in kidney cancer.
Genome-wide dynamic patterns of 5hmC during kidney tumorigenesis.
Genome-wide single nucleotide resolution mapping of 5mC by combing BS-seq and TAB-seq method.
Loss of 5hmC facilitates the gene body hypermethylation in tumor tissue and promotes tumorigenesis.
Manipulating 5hmC generating pathway has potential therapeutical effect.
Summary of single-base 5hmC sequencing using TAB-seq.
Summary of RNA-seq.
Summary of shotgun bisulfite sequencing (BS-seq).
The consistency between TCGA KIRC 450K data and our BS-seq data.
Summary of DMS (differentially methylated sites).
The overlaps between aberrant 5hmC and 5mC changes at single nucleotide resolution during tumorigenesis.
The GO terms of genes with 5mC/5hmC changes in gene body regions.