Abstract
It has been shown that p53 has a critical role in the differentiation and functionality of various multipotent progenitor cells. P53 mutations can lead to genome instability and subsequent functional alterations and aberrant transformation of mesenchymal stem cells (MSCs). The significance of p53 in safeguarding our body from developing osteosarcoma (OS) is well recognized. During bone remodeling, p53 has a key role in negatively regulating key factors orchestrating the early stages of osteogenic differentiation of MSCs. Interestingly, changes in the p53 status can compromise bone homeostasis and affect the tumor microenvironment. This review aims to provide a unique opportunity to study the p53 function in MSCs and OS. In the context of loss of function of p53, we provide a model for two sources of OS: MSCs as progenitor cells of osteoblasts and bone tumor microenvironment components. Standing at the bone remodeling point of view, in this review we will first explain the determinant function of p53 in OS development. We will then summarize the role of p53 in monitoring MSC fidelity and in regulating MSC differentiation programs during osteogenesis. Finally, we will discuss the importance of loss of p53 function in tissue microenvironment. We expect that the information provided herein could lead to better understanding and treatment of OS.
Facts
P53 is a guardian of cell differentiation.
P53 regulates genomic stability, growth, proliferation, and immunoproperties of mesenchymal stem cells (MSCs).
P53 is a negative regulator of osteogenic differentiation of MSCs.
Loss of function of p53 in MSCs compromises their osteogenic differentiation and affects the properties of bone tumor microenvironment (BME) components, therefore it dictates the conditions for osteosarcoma (OS) development.
Open Questions
To identify in vivo and in vitro key molecules involved in the process of bone remodeling, in the context of loss of function of p53.
Are there any molecules produced by p53-null MSCs that could affect osteoclast properties and compromise bone homeostasis?
How do they relate to the diagnosis and prognosis of OS?
TP53 belongs to the so-called ‘p53 gene family' of transcription factors, which includes also the proteins p63, p73, and p53 itself.1, 2, 3 Having been discovered since 1979, p53 is the most studied member of the family with over 60 000 papers so far published. This large mass of scientific data evidentiate a huge complexity for p53 functional program, ranging from the regulation of metabolism4, 5, 6 and mitochondria/oxygen radicals7, 8 to the deeply analyzed DNA damage repair system,9, 10, 11, 12, 13, 14 autophagy,15, 16 and, last but not the least, its role in cell stem maintenance and lineage determination.17, 18 Despite all these investigations, efforts, and advances in knowledge, many crucial intriguing points still remain unanswered to fully understand the physiological and pathological role of p53. These wide range of effects raise from several angles, including, for example, its regulation at the transcriptional level, at the level of micro-RNA,19, 20, 21, 22 and splicing isoforms23, 24 to its translational regulation and its stability/degradation at the protein level.25, 26, 27, 28, 29 In parallel to so much effort in understanding the function of p53, significant efforts are also underway on its potential clinical exploitation.30, 31, 32, 33, 34, 35, 36, 37 Although being identified after ~20 years, already now, p63 and p73 show a similar complexity, and also the ability to interact with p53 at the structural and functional level,34, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49 where the p63 function is highly relevant in skin formation and homeostasis,50, 51 as well as in cancer46, 52, 53 and stem cell regulation.54, 55, 56, 57
P53 and OS in clinical settings
P53 and tumor
The p53 family of transcription factors have several members including p53, p63, and p73. Each member of this family expresses unique mRNA variants resulting from alternative splicing, promoters, and transcription initiation sites.58 Thus, a single gene can exist in multiple isoforms with distinct biological functions.59, 60 P53 protein, encoded by the TP53 gene in humans and the Trp53 gene in mice, is well known for its role as the ‘guardian of the genome' and exerts a pivotal role in maintaining the genetic stability.61, 62, 63 It can prevent tumor formation by regulating cell cycle,64 apoptosis,65 senescence,66 and metabolism67 by binding to responsive elements on DNA (p53RE).64, 68 Abnormal regulation of the p53 family has a critical role in tumorigenesis; indeed, TP53 mutations have been detected in over 50% of all human cancers.69, 70
Silent mutations in the tumor suppressor gene TP53 and/or the retinoblastoma gene RB1 have been reported to be the main causes of the development of sporadic OS.71 In vitro experiments comparing MSCs with malignant OS cells, as well as in vivo studies using transgenic mice with targeting p53 and Rb (retinoblastoma gene RB1; retinoblastoma protein) silencing in MSCs, have elegantly demonstrated that when p53 alone was deleted, the incidence of OS could reach 60%.72 Another function of p53 in suppressing tumor is to act as ‘a guardian of differentiation'.59
Notably, p53 guards osteogenic, myogenic, adipogenic, hematopoietic, and neural differentiation of adult stem cells.73, 74
P53 in OS
OS is a bone tumor affecting long bones in childhood and adolescence.75 Seven subtypes of OS have been characterized according to histological analysis of the osteoid matrix produced by aberrant osteoblasts: osteoblastic, fibroblastic, chondroblastic, telangiectatic, epithelial, small cell, and giant-rich cell.76 The abundant deposition of osteoid matrix and osteoblast-like features of the malignant cells are the dominant characteristics of the osteoblastic phenotype. This subtype manifests the highest incidence representing 75% of screened OS.76, 77, 78 Within OS of the osteoblastic subtype, aberrant preosteoblasts and osteoblasts produce their own osteoid mineralized matrix close to the area of growth plate (GP). Although chromosomal abnormalities have a decisive role in the development of OS,79 the karyotype is not essential for the subtype classification. OS frequently occurs in human patients with Li-Fraumeni syndrome and with hereditary retinoblastoma. Li-Fraumeni patients carry a germline p53 mutation in one allele compromising the function of p53.80, 81, 82 Different studies have reported that preosteoblasts and osteoblasts represent the cells of origin of OS.78, 83 Importantly, cellular microenvironment is also decisive in determining the fate of stem cells and in promoting tumor formation.84 The osteogenic differentiation of p53-deficient or mutant MSCs can be affected by signals from BME and promote eventually OS. Intrabone inoculation of undifferentiated p53−/− and p53−/−Rb−/− MSCs generated osteoblastic OS and developed metastasis characterized by osteoid areas in the lung, spleen, and heart.33 These data suggest that, along with specific bone microenvironment conditions, undifferentiated MSCs with compromised p53 function can represent the cells of origin of OS (Figure 1). The initiation of the tumor could, in part, be affected by a failure of MSCs in maintaining a balance with other differentiation gene programs, such as adipogenesis and chondrogenesis.78 Notably, p53−/−Rb−/− MSCs reflect the phenotype for the development of sarcoma.79 Interestingly, the feature of the osteoblast subtype of OS is tightly associated with the impaired activity of p53 in mouse MSCs and osteoblasts.85
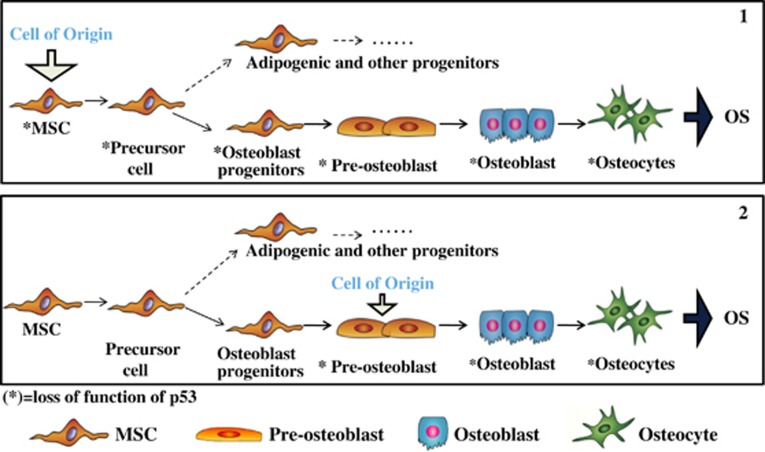
Figure 1.
Loss of function of p53 in undifferentiated MSCs and origin of OS. Preosteoblasts and osteoblasts can be considered as cells of origin for OS development (2) as well as undifferentiated MSCs (1). When mesenchymal progenitor cells (1) or preosteoblast cells (2) become aberrant following mutational events of p53 tumor suppressor gene (*p53), they show compromised growth, proliferation and terminal differentiation. The arrow (*) indicates MSCs with loss of function of p53: *MSCs
Formation of OS
Aberrant proliferation of preosteoblasts and osteoblasts
The bone is an alive and active tissue, crossed by blood vessels that form a complex sinusoidal vascular network. Its basic structure is composed of both trabecular and cortical bone, with trabecules of trabecular bone interspersed in the bone marrow and in direct contact with the bone marrow microenvironment. The surface of the trabecules includes both active and quiescent osteoblasts, which originate the endosteal niche.86 During bone remodeling, the osteoblasts, cells of mesenchymal origin,87 and the osteoclasts, cells of hematopoietic origin, cooperate and work in proximity of the endosteum niche, respectively, generating and resorbing the bone. Osteoblasts and osteoclasts can communicate, regulate, and activate each other through the secretion of specific key molecules (coupling growth factors) released during bone resorption. Insulin-like growth factors and transforming growth factor-beta (TGF-β) are examples of two coupling growth factors secreted during bone degradation, which have been proven to stimulate the osteoblast activity.88 However, MSCs and osteoblasts can also secrete molecules that can influence the osteoclast activity, and affect bone remodeling in the same cases.89 Indeed, throughout the lifetime bone remodeling is kept at a constant rate to balance bone formation and bone degradation, and to guarantee bone homeostasis. However, along with aging, this exquisite equilibrium is subjected to alterations mainly as a consequence of hormonal dysfunctions. Indeed, steroid hormone deficit enhances the resorbing activity of osteoclasts, which could terminate in an osteoporotic condition.90 Conversely, an increase in bone mass and bone density is representative of osteosclerosis and osteopetrosis conditions. Higher intake of bone in osteosclerosis is exclusively promoted by osteoblasts, whereas osteopetrosis is due to aberrant activity of osteoclast-mediated bone resorption.91 Notably, during the process of endochondral bone formation, which occurs until adolescence, the mesenchymal progenitor cells first differentiate into chondrocytes, which generate new cartilage on the GP. The chondrocytes will be slowly replaced by osteogenic progenitor cells and osteoblasts to produce the bone.84 Interestingly, under normal conditions p53 acts as a negative regulator of osteoblastogenesis by repressing the promoter activity of transcription factors required in the early phase of osteogenic commitment, such as Osterix,92 Cbfα-1, and Runx2 in osteoprogenitor cells73, 93 (Figure 2). According to these evidences, p53-null mice are considered as a model for increased bone remodeling and osteosclerosis.94 During MSC osteogenic differentiation, p53 can inhibit osteogenesis95, 96 along with the downregulating expression of critical osteogenic transcription factors including Osterix and Runx2.92 Higher bone density and formation rate have been reported in p53-deficient mice along with increased bone resorption, which is not directly regulated by p53.92
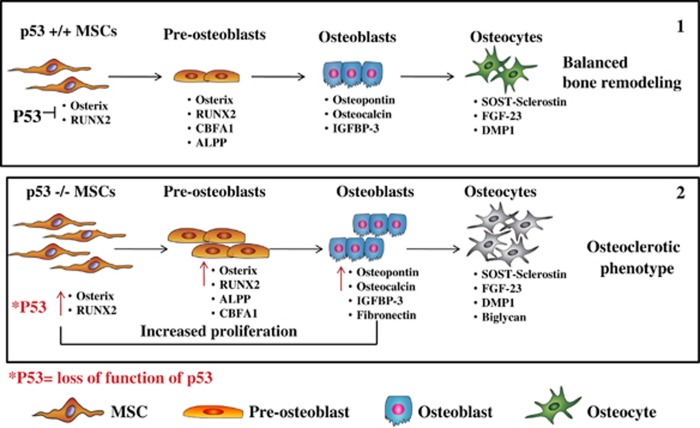
Figure 2.
P53-null MSCs show abnormal osteogenesis compared with the wild-type MSCs. In non-aberrant conditions, the expression of Osterix and Runx2 is upregulated during osteogenic differentiation of osteogenic committed cells to promote their differentiation and maturation towards osteoblasts and osteocytes, and to ensure a balanced bone remodeling (1). P53-null MSCs express before the commitment towards upregulated levels of both Osterix and Runx2. This compromises their differentiation towards mature osteoblasts and osteocytes, culminating in impaired bone remodeling and in the osteosclerotic phenotype observed in p53-deficient mice (2)
MSCs are a source of osteoblasts
MSCs are a subset of adult progenitor cells that exist in almost all adult tissues (bone marrow, adipose tissue, skin, and liver). Adult MSCs have proven to be cells of mesodermal origin, which can give rise to skeleton, muscle, heart, spleen, and other internal organs.97 These cells exert a key function in the maintenance of tissue homeostasis, tissue regeneration, and wound repair.98 MSCs show immunoregulatory properties,99 self-renewing, and differentiation ability into mesenchymal lineages (i.e. chondrocytes, osteoblasts, adipocytes, endothelial cells, pericytes).100, 101 P53 can regulate key transcription factor genes involved in exclusive programs of differentiation and dedifferentiation of somatic cells, with an impact on stemness and development. Given that p53 is a tumor suppressor gene and gate keeper of cell differentiation, in this review we want bring to light the connection of p53 status in MSCs, BME, and OS development. Notably, p53-null MSCs exhibit accelerated growth rate and aberrant osteogenic differentiation compared with wild-type MSCs, which contributes to tumor bone formation. Indeed, distinct studies highlight that p53-deficient bone marrow-derived MSCs can proliferate faster, and appear to differentiate earlier during in vitro osteogenic differentiation compared with the wild-type MSCs.93, 95, 102 However, this ‘tricky' appearance to differentiate earlier into osteoblasts reflects a more complex scenario; indeed, p53-null MSCs are impaired in achieving terminal differentiation towards mature osteocytes.92 MSCs represent a source of precursor for osteogenic progenitor cells and osteoblasts. P53 mutations that lead to defects in the control of cell growth of osteogenic progenitor cells represent the main source of sporadic OS.
The in vitro knockdown of p53 in mouse embryonic fibroblasts (MEFs), which are cells representative of an embryonic stage of development, induced higher expression level of Osterix and Runx273 but not of terminal differentiation markers such as Osteocalcin and Sost-Sclerostin.103 Conversely, p53 knockdown in multipotent bone marrow stromal cells (MBA-15), which resembles adult progenitor cells,73, 104 promoted terminal osteogenic differentiation.73 Consistently, also after reintroduction of wild-type p53 in OS cell line, apoptosis and terminal differentiation were promoted.104 We can emphasize that p53 can regulate bone formation and the differentiation of early osteogenic precursors as demonstrated by the knockdown of p53 in MEF, and, furthermore, it can also promote terminal differentiation in MBA-15.
P53 and MSCs
P53 and MSC proliferation
Isolated MSCs can be maintained in culture in vitro for several passages without being severely compromised in their properties.105 The induction of p21 or cyclin-dependent kinase (CDK) inhibitor p21Cip1/Waf1 mainly promoted by p53 is associated with cell-cycle arrest (Figure 3). This suggests that alterations in cell-cycle regulators represent one of the main causes inducing aberrations in MSCs.106 Transformation of MSCs is highly correlated with simultaneous abnormalities of p53 and p21, and this could represent the event that could lead to the origin of mesodermal tumors.106 Nevertheless, it has been proven that loss of p53 in MSCs promote higher growth rate, chromosomal instability, resistance to apoptosis, and senescence107, 108 (Figure 3). Interestingly, p53 has a key role in regulating both differentiation of mesenchymal precursors and quiescence of hematopoietic stem cells in the bone marrow environment.109
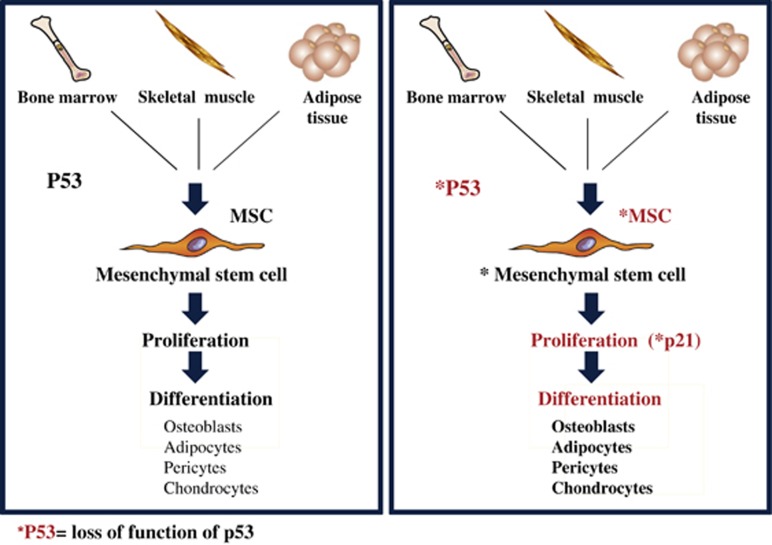
Figure 3.
Loss of function of p53 compromises proliferation and differentiation of MSCs. MSCs can be isolated from adult organs, such as bone marrow, skeletal muscle, adipose tissue, and others, with a higher prevalence from the bone marrow (BM). MSCs can be identified in vitro for their surface markers and their multipotential differentiation properties. P53 has a role in regulating growth and proliferation of MSCs. Mutational events of p53 or p53 deficiency compromise the proliferation rate of MSCs mainly through p21 or CDK inhibitor p21Cip1/Waf1
Notably, bone marrow is one of the important sites for hematopoiesis in adults where hematopoietic stem cells are kept in a stemness condition.100, 110 Bone and bone marrow are functionally and anatomically correlated87 composing a unique compartment, which has a role in hematopoiesis and in bone homeostasis.100, 111 Transplanted nestin-marked human MSCs into the bone marrow cavity of nonobese diabetic/severe-combined immunodeficiency mice persisted 10 weeks after transplantation. Interestingly, these transplanted cells were able to differentiate into all the cells of the hematopoietic environment.112 Indeed, nestin-positive cells in the bone marrow have been recognized to have all the properties of MSCs, and were closely located with hematopoietic stem cells to support their quiescence.113 In vivo depletion of nestin-positive cells reduced the percentage of hematopoietic progenitor cells hosted in the bone marrow.113
P53 and MSCs differentiation
In vitro gene knockdown of TP53 and Trp53 have revealed the importance of p53 in mesenchymal differentiation of multiprogenitor cells. However, controversial role of p53 during differentiation of MSCs have been reported.92, 95 Cell cycle and differentiation represent two interconnected processes in which p53 can exert distinct functions depending on the cell type. Absence of p53 can block the terminal cell differentiation, resulting in the accumulation of early and intermediate progenitors, which can lead to alterations of that specific differentiation pathway such as osteogenesis, adipogenesis, or myogenesis (Figure 4). For example, p53 negatively regulates adipogenesis by repressing the key adipogenic transcription factors PPAR-γ (proliferator-activated receptor-γ) and C/EBP-α (CCAAT/enhancer-binding protein-α).74, 114 Adipocytes and osteoblasts are both cells derived from multipotent progenitor cells. The undifferentiated status of the cells is kept by the repression of transcription factors that repress each other to preserve multipotency. However, upon appropriate stimulations, MSCs make sequential cell fate choices.115 The commitment of MSCs towards a specific mesenchymal differentiation program is coordinated also by p53, which regulates their multipotential state. In in vivo studies, p53 is upregulated in adipocytes from genetically obese mice in a fed state. Transgenic mice overexpressing functional p53 gain less body mass and adipose tissue when compared with wild-type mice; this has suggested an inhibitory role for p53, which may be exerted by changes in metabolism.116 Indeed, in the absence of functioning p53, a shift from oxidative phosphorylation towards glycolysis was observed.117 Furthermore, even myogenic differentiation can be monitored by p53. Distinct studies have shown an increase in p53 mRNA levels during myogenic differentiation in vitro.118, 119, 120 P53 might be involved in this process by regulating the retinoblastoma protein, Rb, which has a pivotal role in the differentiation of muscle through cell-cycle arrest and also by specific genes involved in the myogenic differentiation program.121, 122, 123 The association of p53 expression and cell-cycle regulators, which are target genes of p53, such as p21, was observed during the development of the mouse nervous system.124, 125 Indeed, p53 monitors the differentiation of neural stem cells via its regulating pathways including cooperation with phosphatase and tensin homolog (PTEN).125 Notably, accumulation of mutant p53 in neural stem cells in the subventricular zone of the brain could generate aberrant neural progenitor cells and promote glioma formation.126 However, it is still unclear how p53 functions in the specific signaling context to regulate neural stem cells differentiation.127 In hematopoiesis, p53 induces hematopoietic stem cells to differentiate into proper mature blood cells and function in maintaining their quiescence.73 So far, the development of skeletal muscles and blood system has not yet been found abnormal in p53-null mice. Several in vitro studies have confirmed the role of p53 as a negative regulator in cell differentiation pathways, which reflects the complexity of the underlying mechanisms (Figure 4).
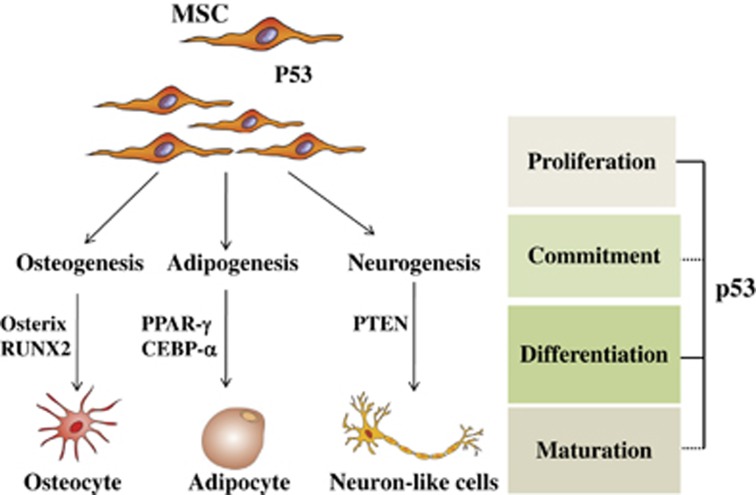
Figure 4.
P53 is a negative regulator of differentiation pathways of MSCs. P53 can negatively regulate differentiation of mesenchymal progenitor cells such as osteogenesis, myogenesis, adipogenesis, and neurogenesis pathways by downregulating the expression levels of key transcription factor genes. In undifferentiated MSCs, p53 maintains lower expression levels of key transcription factor genes involved in the early phases of differentiation, such as Osterix and Runx2 for osteogenesis, PPAR-γ, CEBP-α for adipogenesis, Rb for myogenesis, and PTEN for neurogenesis. P53 status exerts a decisive role on proliferation, commitment, differentiation, and maturation of MSCs
P53 and MSCs dictating tumor microenvironment
P53 and tumor microenvironment
Interestingly, our previous work has proved that MSCs that lack p53 exhibit tumor-promoting characteristic through high secretion of nitric oxide (NO) and higher vigorous immunomodulation when compared with wild-type MSCs.128 We found that the higher secretion of NO from p53-deficient MSCs have an inhibitory effect on T cells, and promote tumor growth.128 Thus, loss of p53 function in MSCs can promote their transformation by regulating their immunoproperties, growth, and proliferation. Importantly, p53 can dictate tumor microenvironment in an MSC-related manner. Indeed, except for tumor cells, also non-tumoral cells in tumor stroma were reported to gain p53 mutations, which were associated with regional lymph-node metastases.129 When p53 is inactivated, tumor stroma shows less response to anticancer drugs, such as cisplatinum, etoposide, and vincristine as a consequence of failure in upregulating p53-inducible genes and inducing apoptosis of tumor cells.130 Alternatively, ablation of p53 in tumor stroma has promoted tumor growth by upregulating the expression of stromal cell-derived factor 1/C–X–C motif chemokine 12.131, 132 Notably, tumor stroma with dysfunctional p53 can enhance differentiation of myeloid-derived suppressor cell, exacerbate immunosuppression, and promote tumor progression.133 Furthermore, the role of p53 in the tumor microenvironment under hypoxic conditions has also been reported. Indeed, p53 targets the subunit HIF-1-α of hypoxia-inducible factor (HIF), necessary for metabolism adaptation, avoiding its proteasomic degradation through murine double minute 2 protein.134 Ablation of p53 increases the expression of HIF-1-α in a hypoxia condition, which in turn induces the expression of vascular endothelial growth factor (VEGF) in tumor cells and promotes angiogenesis, neovascularization, tumor growth, and invasion. VEGF and HIF-1-α are, furthermore, overexpressed in several types of human cancers, especially with HIF-1-α in metastatic OS.135
P53 and MSC in OS tumor microenvironment
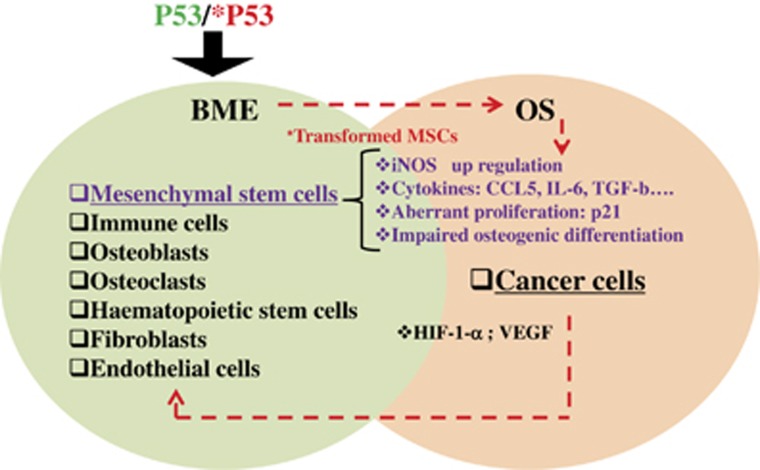
During bone growth, several factors and extracellular matrix components secreted by mesenchymal progenitor cells and chondrocytes in the GP will recruit BME components to guarantee a balanced bone remodeling. The BME hosts different types of cells, including osteoblasts, osteoclasts, mesenchymal cell precursors, hematopoietic stem cells, chondrocytes, and endothelial cells, as well as fibroblasts stromal cells and immune cells. In this special scenario, p53 status has a determinant role (Figure 5). In the context of bone remodeling, along with the coexistence of aberrant conditions arising from a p53 mutational landscape, the BME components could contribute to altered bone homeostasis compromising the cross-talk between MSCs, osteoblasts, osteoclasts, and hematopoietic cells, and therefore it dictates the environment for tumor initiation.84 Given the plasticity of MSCs to generate and differentiate into several cell types, including osteoblasts, it is not surprising that MSCs with p53 aberrations have been suggested to be the cells of origin for bone tumor, including OS, chondrosarcoma, Ewing's sarcoma, and sarcoma.75 We previously have discussed about the impaired osteogenic differentiation of p53-deficient MSCs; however, we here want to emphasize that p53-null MSCs also represent an important cellular component of tumor microenvironment. Interestingly, it has been demonstrated that in tumor BME, MSCs can support OS growth through the expression of CCL5 (chemokine ligand 5).136 Interestingly, CCL5/CCR5 (C–C chemokine receptor type 5) axis can promote OS migration through the extracellular signal-regulated kinase pathway, which induces the nuclear factor κ-light-chain enhancer of activated B cells (NF-κb).136 Conversely, growth factors secreted from cancer cells and during bone resorption, such as tumor necrosis factor-α, TGF-β, bone morphogenetic protein 2 (BMP2), and interleukin-6 (IL-6), can promote osteoclast maturation by directing the expression of RANK (receptor activator of the receptor activator of nuclear factor-κb) on the surface of osteoclast precursor cells increasing bone erosion. The increased bone degradation culminates in the release of BMP2 and TGF-β, which severely contribute to tumor growth and stimulate the osteoclast activity.137 TGF-β can evoke MSC-secreted IL-6, which acts by promoting OS metastasis via STAT-3 (signal transducer and activator of signal-3).138 These data emphasize how the loss of function of p53 is a determinant in dictating the conditions that contribute to initiate OS: on the one hand, it can affect proliferation, immunoproperties, and compromise osteogenesis of undifferentiated MSCs, but on the other hand, it can affect the properties of BME components compromising the talk between BME and cancer cells, a further condition that supports OS initiation and development.
Figure 5.
P53 status and the vicious cycle between BME and OS. In a p53 mutational landscape, BME components represent a soil for tumor promotion. In this scenario, p53-null MSCs exhibit higher proliferation rate, impaired osteogenesis, and altered immunoregulatory properties such as higher expression levels of iNOS (inducible nitric oxide synthase), CCL5, IL-6, and TGF-β. These molecules can, in turn, stimulate bone tumor-forming cells to produce cytokines to support their growth and affect BME. This generates a vicious cycle of cross-talking between BME and OS, which is dictated from p53 status
Conclusion
In this review, we aimed to bring to light that p53 has a pivotal role in keeping the balance between bone formation and bone degradation. Indeed, p53 not only regulates the genomic stability of MSCs but also their osteogenic differentiation functioning as ‘bone remodeling surveillant' to prevent bone tumor initiation. Wang et al.92 and other groups have proposed a negative role of p53 in regulating osteogenesis and other mesenchymal differentiation programs. We set the evidence that p53 mutational events occurring in undifferentiated MSCs or in osteoblasts at different stages of commitment can promote OS initiation72 as a consequence of alterations of osteogenic differentiation, bone remodeling, and bone homeostasis.139 Indeed, OS is a heterogeneous tumor that includes cells at different stages of commitment during osteogenesis.140 Interestingly, the osteosclerotic condition observed in p53-null mice imposes the phenotype of OS development.106 We summarize that p53 can affect osteogenic differentiation of MSCs and largely contribute to OS initiation: (1) it can promote or abrogate differentiation of multipotent progenitor cells acting as a negative mediator of transcription factors of early osteogenic differentiation; (2) it can regulate the genomic stability, growth, and proliferation of MSCs; (3) it can affect the immunoproperties of MSCs through growth factors and chemokine secretion; (4) it can affect the BME-regulating immune properties, growth, proliferation, and differentiation of microenvironment components. Further investigations on the molecular mechanisms through which loss of function of p53 can affect properties of MSCs and osteoprogenitor cells should be considered. This will ameliorate the knowledge of p53 function in the context of bone biology, and also will be helpful in identifying new strategies for targeting key molecules necessary for OS formation and survival.
Glossary
- OS
osteosarcoma
- MSC
mesenchymal stem cells
- BME
bone tumor microenvironment
- Rb
retinoblastoma gene RB1; retinoblastoma protein
- TGF-β
transforming growth factor-β
- GP
growth plate
- Cbfα -1
core-binding factor α-1
- MEF
mouse embryonic fibroblast
- MBA-15
multipotent bone marrow stromal cells
- CDK
cyclin-dependent kinase
- PPAR-γ
proliferator activated receptor-γ
- C/EBP-α
CCAAT/ enhancer binding protein-α
- PTEN
phosphatase and tensin homolog
- NO
nitric oxide
- MDSC
myeloid-derived suppressor cell
- HIF
hypoxia-inducible factor
- VEGF
vascular endothelial growth factor
- CCL5
chemokine ligand 5
- BMP2
bone morphogenetic protein 2
- IL-6
interleukin-6
The authors declare no conflict of interest.
Footnotes
Edited by G Raschellà
References
- Zambetti GP. Expanding the reach of the p53 tumor suppressor network. Cell Death Differ 2014; 21: 505–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon H, Sharon M, Rotter V. Modulation of alternative splicing contributes to cancer development: focusing on p53 isoforms, p53beta and p53gamma. Cell Death Differ 2014; 21: 1347–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T, Wiman KG. TP53: an oncogene in disguise. Cell Death Differ 2015; 22: 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene 2013; 32: 5129–5143. [DOI] [PubMed] [Google Scholar]
- Xu J, Wang J, Hu Y, Qian J, Xu B, Chen H et al. Unequal prognostic potentials of p53 gain-of-function mutations in human cancers associate with drug-metabolizing activity. Cell Death Dis 2014; 5: e1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Melino G. The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression. Trends Biochem Sci 2015; 40: 425–434. [DOI] [PubMed] [Google Scholar]
- Ci Y, Shi K, An J, Yang Y, Hui K, Wu P et al. ROS inhibit autophagy by downregulating ULK1 mediated by the phosphorylation of p53 in selenite-treated NB4 cells. Cell Death Dis 2014; 5: e1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evstafieva AG, Garaeva AA, Khutornenko AA, Klepikova AV, Logacheva MD, Penin AA et al. A sustained deficiency of mitochondrial respiratory complex III induces an apoptotic cell death through the p53-mediated inhibition of pro-survival activities of the activating transcription factor 4. Cell Death Dis 2014; 5: e1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Fava LL, Krumschnabel G, Peintner L, Tanzer MC, Soratroi C et al. Death of p53-defective cells triggered by forced mitotic entry in the presence of DNA damage is not uniquely dependent on caspase-2 or the PIDDosome. Cell Death Dis 2013; 4: e942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phesse TJ, Myant KB, Cole AM, Ridgway RA, Pearson H, Muncan V et al. Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ 2014; 21: 956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezina L, Aksenova V, Fedorova O, Malikova D, Shuvalov O, Antonov AV et al. KMT Set7/9 affects genotoxic stress response via the Mdm2 axis. Oncotarget 2015; 6: 25843–25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezina L, Aksenova V, Ivanova T, Purmessur N, Antonov AV, Tentler D et al. KMTase Set7/9 is a critical regulator of E2F1 activity upon genotoxic stress. Cell Death Differ 2014; 21: 1889–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair BC, Krishnan SR, Sareddy GR, Mann M, Xu B, Natarajan M et al. Proline, glutamic acid and leucine-rich protein-1 is essential for optimal p53-mediated DNA damage response. Cell Death Differ 2014; 21: 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashzeveg N, Taira N, Lu ZG, Kimura J, Yoshida K. Palmdelphin, a novel target of p53 with Ser46 phosphorylation, controls cell death in response to DNA damage. Cell Death Dis 2014; 5: e1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon HU, Yousefi S, Schmid I, Friis R. ATG5 can regulate p53 expression and activation. Cell Death Dis 2014; 5: e1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garufi A, Pucci D, D'Orazi V, Cirone M, Bossi G, Avantaggiati ML et al. Degradation of mutant p53H175 protein by Zn(II) through autophagy. Cell Death Dis 2014; 5: e1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Nakasaki M, Todorova D, Lake B, Yuan CY, Jamora C et al. p53 Induces skin aging by depleting Blimp1+ sebaceous gland cells. Cell Death Dis 2014; 5: e1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ng DS, Mah WC, Almeida FF, Rahmat SA, Rao VK et al. A unique role for p53 in the regulation of M2 macrophage polarization. Cell Death Differ 2015; 22: 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori ME, Barbini C, Haas TL, Marroncelli N, Patrizii M, Biffoni M et al. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ 2014; 21: 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman Y, Bublik DR, Pilpel Y, Oren M. miR-661 downregulates both Mdm2 and Mdm4 to activate p53. Cell Death Differ 2014; 21: 302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato O, Boeri M, Moro M, Verri C, Mensah M, Conte D et al. Mir-660 is downregulated in lung cancer patients and its replacement inhibits lung tumorigenesis by targeting MDM2-p53 interaction. Cell Death Dis 2014; 5: e1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZJ, Nong XY, Lv YR, Sun HH, An PP, Wang F et al. Mir-509-5p joins the Mdm2/p53 feedback loop and regulates cancer cell growth. Cell Death Dis 2014; 5: e1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Fernandes K, Terrier O, Lane DP, Bourdon JC. Modulation of p53beta and p53gamma expression by regulating the alternative splicing of TP53 gene modifies cellular response. Cell Death Differ 2014; 21: 1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatter TL, Hung N, Bowie S, Campbell H, Rubio C, Speidel D et al. Delta122p53, a mouse model of Delta133p53alpha, enhances the tumor-suppressor activities of an attenuated p53 mutant. Cell Death Dis 2015; 6: e1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sane S, Abdullah A, Boudreau DA, Autenried RK, Gupta BK, Wang X et al. Ubiquitin-like (UBX)-domain-containing protein, UBXN2A, promotes cell death by interfering with the p53-Mortalin interactions in colon cancer cells. Cell Death Dis 2014; 5: e1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang C, Wang XL, Ly P, Belyi V, Xu-Monette ZY et al. E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ 2014; 21: 1792–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Li SZ, Zhang ZY, Hu XM, Hou PN, Gao L et al. Nemo-like kinase is critical for p53 stabilization and function in response to DNA damage. Cell Death Differ 2014; 21: 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuget S, Bonacci T, Soubeyran P, Iovanna J, Dusetti NJ. Oxidative stress-induced p53 activity is enhanced by a redox-sensitive TP53INP1 SUMOylation. Cell Death Differ 2014; 21: 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoreva TA, Tribulovich VG, Garabadzhiu AV, Melino G, Barlev NA. The 26S proteasome is a multifaceted target for anti-cancer therapies. Oncotarget 2015; 6: 24733–24749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MS, Schmezer P, Breuer R, Haas SF, Essers MA, Krammer PH et al. The traditional Chinese medical compound Rocaglamide protects nonmalignant primary cells from DNA damage-induced toxicity by inhibition of p53 expression. Cell Death Dis 2014; 5: e1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongiorno-Borbone L, Giacobbe A, Compagnone M, Eramo A, De Maria R, Peschiaroli A et al. Anti-tumoral effect of desmethylclomipramine in lung cancer stem cells. Oncotarget 2015; 6: 16926–16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbacher A, Gutekunst M, Oren M, Aulitzky WE, van der Kuip H. RITA can induce cell death in p53-defective cells independently of p53 function via activation of JNK/SAPK and p38. Cell Death Dis 2014; 5: e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Fan YH, Xu X, Zhang H, Dou J, Tang Y et al. A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways. Cell Death Dis 2014; 5: e1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Rotblat B, Ansell K, Amelio I, Caraglia M, Misso G et al. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis 2014; 5: e1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Landre V, Knight RA, Lisitsa A, Melino G, Antonov AV. Polypharmacology of small molecules targeting the ubiquitin-proteasome and ubiquitin-like systems. Oncotarget 2015; 6: 9646–9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Bravo-San Pedro JM, Vitale I, Aaronson SA, Abrams JM, Adam D et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ 2015; 22: 58–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Melino G. CRISPR: a new method for genetic engineering - a prokaryotic immune component may potentially open a new era of gene silencing. Cell Death Differ 2015; 22: 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostecka A, Sznarkowska A, Meller K, Acedo P, Shi Y, Sakil HAM et al. JNK-NQO1 axis drives TAp73-mediated tumor suppression upon oxidative and proteasomal stress. Cell Death Dis 2014; 5: e1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova V, Mancini M, Agostini M, Knight RA, Annicchiarico-Petruzzelli M, Barlev NA et al. TAp73 transcriptionally represses BNIP3 expression. Cell Cycle 2015; 14: 2484–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Tomasini R, Rufini A, Elia AJ, Agostini M, Amelio I et al. TAp73 is required for spermatogenesis and the maintenance of male fertility. Proc Natl Acad Sci USA 2014; 111: 1843–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Abramo M, Besker N, Desideri A, Levine AJ, Melino G, Chillemi G. The p53 tetramer shows an induced-fit interaction of the C-terminal domain with the DNA-binding domain. Oncogene 2015; doi:10.1038/onc.2015.388. [DOI] [PMC free article] [PubMed]
- Adamovich Y, Adler J, Meltser V, Reuven N, Shaul Y. AMPK couples p73 with p53 in cell fate decision. Cell Death Differ 2014; 21: 1451–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatt MP, Cancino GI, Miller FD, Kaplan DR. P63 and p73 coordinate p53 function to determine the balance between survival, cell death, and senescence in adult neural precursor cells. Cell Death Differ 2014; 21: 1546–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Inoue S, Markert EK, Levine AJ, Knight RA, Mak TW et al. TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1 alpha degradation. Proc Natl Acad Sci USA 2015; 112: 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Markert EK, Rufini A, Antonov AV, Sayan BS, Tucci P et al. P73 regulates serine biosynthesis in cancer. Oncogene 2014; 33: 5039–5046. [DOI] [PubMed] [Google Scholar]
- Yallowitz AR, Alexandrova EM, Talos F, Xu S, Marchenko ND, Moll UM. P63 is a prosurvival factor in the adult mammary gland during post-lactational involution, affecting PI-MECs and ErbB2 tumorigenesis. Cell Death Differ 2014; 21: 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunjobpo W, Dulloo I, Igarashi K, Concin N, Matsuo K, Sabapathy K. Suppression of acetylpolyamine oxidase by selected AP-1 members regulates DNp73 abundance: mechanistic insights for overcoming DNp73-mediated resistance to chemotherapeutic drugs. Cell Death Differ 2014; 21: 1240–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklison-Chirou MV, Killick R, Knight RA, Nicotera P, Melino G, Agostini M. How does p73 cause neuronal defects? Mol Neurobiol 2015; doi:10.1007/s12035-015-9381-1. [DOI] [PubMed]
- Niklison-Chirou MV, Steinert JR, Agostini M, Knight RA, Dinsdale D, Cattaneo A et al. TAp73 knockout mice show morphological and functional nervous system defects associated with loss of p75 neurotrophin receptor. Proc Natl Acad Sci USA 2013; 110: 18952–18957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viticchie G, Agostini M, Lena AM, Mancini M, Zhou HQ, Zolla L et al. p63 supports aerobic respiration through hexokinase II. Proc Natl Acad Sci USA 2015; 112: 11577–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnley P, Rahman M, Wang H, Zhang Z, Sun X, Zhuge Q et al. Role of the p63-FoxN1 regulatory axis in thymic epithelial cell homeostasis during aging. Cell Death Dis 2013; 4: e932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Liang S, Bergholz J, He H, Walsh EM, Zhang Y et al. DeltaNp63alpha activates CD82 metastasis suppressor to inhibit cancer cell invasion. Cell Death Dis 2014; 5: e1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah Z, Bar-Mag T, Kohn Y, Pichiorri F, Palumbo T, Melino G et al. Tumor suppressor WWOX binds to Delta DNp63 alpha and sensitizes cancer cells to chemotherapy. Cell Death Dis 2013; 4: e480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memmi EM, Sanarico AG, Giacobbe A, Peschiaroli A, Frezza V, Cicalese A et al. P63 sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Natl Acad Sci USA 2015; 112: 3499–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G, Memmi EM, Pelicci PG, Bernassola F. Maintaining epithelial stemness with p63. Sci Signal 2015; 8: re9. [DOI] [PubMed] [Google Scholar]
- Vasileva EA, Shuvalov OU, Garabadgiu AV, Melino G, Barlev NA. Genome-editing tools for stem cell biology. Cell Death Dis 2015; 6: e1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Amelio I, Agostini M, Melino G. MicroRNAs and p63 in epithelial stemness. Cell Death Differ 2015; 22: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaman JM, Waridel F, Estreicher A, Vannier A, Limacher JM, Gilbert D et al. The human tumour suppressor gene p53 is alternatively spliced in normal cells. Oncogene 1996; 12: 813–818. [PubMed] [Google Scholar]
- Murray-Zmijewski F, Lane DP, Bourdon JC. P53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ 2006; 13: 962–972. [DOI] [PubMed] [Google Scholar]
- Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S et al. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ 2011; 18: 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature 1991; 351: 453–456. [DOI] [PubMed] [Google Scholar]
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature 1979; 278: 261–263. [DOI] [PubMed] [Google Scholar]
- Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 1979; 17: 43–52. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet 1992; 1: 45–49. [DOI] [PubMed] [Google Scholar]
- Shaw P, Bovey R, Tardy S, Sahli R, Sordat B, Costa J. Induction of apoptosis by wild-type p53 in a human colon tumor-derived cell line. Proc Natl Acad Sci USA 1992; 89: 4495–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature 1991; 352: 345–347. [DOI] [PubMed] [Google Scholar]
- Berkers Celia R, Maddocks Oliver DK, Cheung Eric C, Mor I, Vousden Karen H. Metabolic regulation by p53 family members. Cell Metab 18: 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC, Deguin-Chambon V, Lelong JC, Dessen P, May P, Debuire B et al. Further characterisation of the p53 responsive element – identification of new candidate genes for trans-activation by p53. Oncogene 1997; 14: 85–94. [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010; 2: a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol 2010; 2: a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch JM, Blair V, Kelsey AM, Evans DG, Harris M, Tricker KJ et al. Cancer phenotype correlates with constitutional TP53 genotype in families with the Li-Fraumeni syndrome. Oncogene 1998; 17: 1061–1068. [DOI] [PubMed] [Google Scholar]
- Lin PP, Pandey MK, Jin F, Raymond AK, Akiyama H, Lozano G. Targeted mutation of p53 and Rb in mesenchymal cells of the limb bud produces sarcomas in mice. Carcinogenesis 2009; 30: 1789–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molchadsky A, Shats I, Goldfinger N, Pevsner-Fischer M, Olson M, Rinon A et al. P53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS One 2008; 3: e3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. p53 is balancing development, differentiation and de-differentiation to assure cancer prevention. Carcinogenesis 2010; 31: 1501–1508. [DOI] [PubMed] [Google Scholar]
- Heare T, Hensley MA, Dell'Orfano S. Bone tumors: osteosarcoma and Ewing's sarcoma. Curr Opin Pediatr 2009; 21: 365–372. [DOI] [PubMed] [Google Scholar]
- Fletcher CDM, Unni KK, Mertens F (eds). World Health Organization Classification ofTumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. IARC Press: Lyon, France, 2002. [Google Scholar]
- Bacci G, Longhi A, Fagioli F, Briccoli A, Versari M, Picci P. Adjuvant and neoadjuvant chemotherapy for osteosarcoma of the extremities: 27 year experience at Rizzoli Institute, Italy. Eur J Cancer 2005; 41: 2836–2845. [DOI] [PubMed] [Google Scholar]
- Mutsaers AJ, Ng AJ, Baker EK, Russell MR, Chalk AM, Wall M et al. Modeling distinct osteosarcoma subtypes in vivo using Cre:lox and lineage-restricted transgenic shRNA. Bone 2013; 55: 166–178. [DOI] [PubMed] [Google Scholar]
- Ottaviani G, Jaffe N. The etiology of osteosarcoma. Cancer Treat Res 2009; 152: 15–32. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature 1990; 348: 747–749. [DOI] [PubMed] [Google Scholar]
- Varley JM. Germline TP53 mutations and Li-Fraumeni syndrome. Hum Mutat 2003; 21: 313–320. [DOI] [PubMed] [Google Scholar]
- Hansen MF, Koufos A, Gallie BL, Phillips RA, Fodstad O, Brogger A et al. Osteosarcoma and retinoblastoma: a shared chromosomal mechanism revealing recessive predisposition. Proc Natl Acad Sci USA 1985; 82: 6216–6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 2006; 133: 3231–3244. [DOI] [PubMed] [Google Scholar]
- Alfranca A, Martinez-Cruzado L, Tornin J, Abarrategi A, Amaral T, de Alava E et al. Bone microenvironment signals in osteosarcoma development. Cell Mol Life Sci 2015; 72: 3097–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev 2008; 22: 1662–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke B. Normal bone anatomy and physiology. Clin J Am Soc Nephrol 2008; 3: S131–S139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 2003; 425: 841–846. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature 2003; 423: 337–342. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang P, Liu L, Zou L. Hierarchical organization and regulation of the hematopoietic stem cell osteoblastic niche. Crit Rev Oncol Hematol 2013; 85: 1–8. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Deavall DG, Orphanides G. Sex steroids, ANGELS and osteoporosis. BioEssays 2003; 25: 195–199. [DOI] [PubMed] [Google Scholar]
- Filvaroff E, Derynck R. Bone remodelling: a signalling system for osteoclast regulation. Curr Biol 1998; 8: R679–R682. [DOI] [PubMed] [Google Scholar]
- Wang X, Kua HY, Hu Y, Guo K, Zeng Q, Wu Q et al. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J Cell Biol 2006; 172: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Steinman HA, Gagnon J, Smith TW, Henderson JE, Kream BE et al. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol. 2006; 172: 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li B. Genetic studies of bone diseases: evidence for involvement of DNA damage response proteins in bone remodeling. Int J Biomed Sci 2007; 3: 217–228. [PMC free article] [PubMed] [Google Scholar]
- Tataria M, Quarto N, Longaker MT, Sylvester KG. Absence of the p53 tumor suppressor gene promotes osteogenesis in mesenchymal stem cells. J Pediatr Surg 2006; 41: 624–632; discussion 32. [DOI] [PubMed] [Google Scholar]
- Armesilla-Diaz A, Elvira G, Silva A. P53 regulates the proliferation, differentiation and spontaneous transformation of mesenchymal stem cells. Exp Cell Res 2009; 315: 3598–3610. [DOI] [PubMed] [Google Scholar]
- Aloni-Grinstein R, Shetzer Y, Kaufman T, Rotter V. P53: the barrier to cancer stem cell formation. FEBS Lett 2014; 588: 2580–2589. [DOI] [PubMed] [Google Scholar]
- Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 1997; 276: 71–74. [DOI] [PubMed] [Google Scholar]
- Shi Y, Hu G, Su J, Li W, Chen Q, Shou P et al. Mesenchymal stem cells: a new strategy for immunosuppression and tissue repair. Cell Res 2010; 20: 510–518. [DOI] [PubMed] [Google Scholar]
- Bianco P. Stem cells and bone: a historical perspective. Bone 2015; 70: 2–9. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD et al. Multilineage potential of adult human mesenchymal stem cells. Science 1999; 284: 143–147. [DOI] [PubMed] [Google Scholar]
- He Y, de Castro LF, Shin MH, Dubois W, Yang HH, Jiang S et al. P53 loss increases the osteogenic differentiation of Bmscs. Stem Cells 2015; 33: 1304–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz KA, Lanciloti NJ, Moore MK, Campione AL, Chandar N. P53 transactivity during in vitro osteoblast differentiation in a rat osteosarcoma cell line. Mol Carcinogen 1999; 25: 132–138. [DOI] [PubMed] [Google Scholar]
- Radinsky R, Fidler IJ, Price JE, Esumi N, Tsan R, Petty CM et al. Terminal differentiation and apoptosis in experimental lung metastases of human osteogenic sarcoma cells by wild type p53. Oncogene 1994; 9: 1877–1883. [PubMed] [Google Scholar]
- Friedenstein AJ, Chailakhyan RK, Latsinik NV, Panasyuk AF, Keiliss-Borok IV. Stromal cells responsible for transferring the microenvironment of the hemopoietic tissues. Cloning in vitro and retransplantation in vivo. Transplantation 1974; 17: 331–340. [DOI] [PubMed] [Google Scholar]
- Rodriguez R, Rubio R, Masip M, Catalina P, Nieto A, de la Cueva T et al. Loss of p53 induces tumorigenesis in p21-deficient mesenchymal stem cells. Neoplasia 2009; 11: 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Nauta AJ, Osborn MJ, Panoskaltsis Mortari A, McElmurry RT, Bell S et al. Sarcoma derived from cultured mesenchymal stem cells. Stem Cells. 2007; 25: 371–379. [DOI] [PubMed] [Google Scholar]
- Mueller LP, Luetzkendorf J, Mueller T, Reichelt K, Simon H, Schmoll HJ. Presence of mesenchymal stem cells in human bone marrow after exposure to chemotherapy: evidence of resistance to apoptosis induction. Stem Cells 2006; 24: 2753–2765. [DOI] [PubMed] [Google Scholar]
- Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G et al. P53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 2009; 4: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 1978; 4: 7–25. [PubMed] [Google Scholar]
- Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 2008; 3: 301–313. [DOI] [PubMed] [Google Scholar]
- Muguruma Y, Yahata T, Miyatake H, Sato T, Uno T, Itoh J et al. Reconstitution of the functional human hematopoietic microenvironment derived from human mesenchymal stem cells in the murine bone marrow compartment. Blood 2006; 107: 1878–1887. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010; 466: 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell 2007; 131: 242–256. [DOI] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006; 7: 885–896. [DOI] [PubMed] [Google Scholar]
- Garber K. Energy deregulation: licensing tumors to grow. Science 2006; 312: 1158–1159. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O et al. P53 regulates mitochondrial respiration. Science 2006; 312: 1650–1653. [DOI] [PubMed] [Google Scholar]
- Mazzaro G, Bossi G, Coen S, Sacchi A, Soddu S. The role of wild-type p53 in the differentiation of primary hemopoietic and muscle cells. Oncogene 1999; 18: 5831–5835. [DOI] [PubMed] [Google Scholar]
- Soddu S, Blandino G, Scardigli R, Coen S, Marchetti A, Rizzo MG et al. Interference with p53 protein inhibits hematopoietic and muscle differentiation. J Cell Biol 1996; 134: 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy O. P53 gene is up-regulated during skeletal muscle cell differentiation. Biochem Biophys Res Commun 1993; 192: 714–719. [DOI] [PubMed] [Google Scholar]
- Tamir Y, Bengal E. P53 protein is activated during muscle differentiation and participates with MyoD in the transcription of muscle creatine kinase gene. Oncogene 1998; 17: 347–356. [DOI] [PubMed] [Google Scholar]
- Porrello A, Cerone MA, Coen S, Gurtner A, Fontemaggi G, Cimino L et al. P53 regulates myogenesis by triggering the differentiation activity of pRb. J Cell Biol 2000; 151: 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H, Griesmann H, Beitzinger M, Hofmann L, Beinoraviciute-Kellner R, Sauer M et al. p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell 2006; 10: 281–293. [DOI] [PubMed] [Google Scholar]
- van Lookeren Campagne M, Gill R. Tumor-suppressor p53 is expressed in proliferating and newly formed neurons of the embryonic and postnatal rat brain: comparison with expression of the cell cycle regulators p21Waf1/Cip1, p27Kip1, p57Kip2, p16Ink4a, cyclin G1, and the proto-oncogene Bax. J Comp Neurol 1998; 397: 181–198. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Di Giovanni S. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep 2009; 10: 576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE et al. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell 2009; 15: 514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Matsuyama K, Mori F, Nakajima K. Supraspinal sites that induce locomotion in the vertebrate central nervous system. Adv Neurol 2001; 87: 25–40. [PubMed] [Google Scholar]
- Huang Y, Yu P, Li W, Ren G, Roberts AI, Cao W et al. p53 regulates mesenchymal stem cell-mediated tumor suppression in a tumor microenvironment through immune modulation. Oncogene 2014; 33: 3830–3838. [DOI] [PubMed] [Google Scholar]
- Patocs A, Zhang L, Xu Y, Weber F, Caldes T, Mutter GL et al. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N Engl J Med 2007; 357: 2543–2551. [DOI] [PubMed] [Google Scholar]
- Dudley AC, Shih SC, Cliffe AR, Hida K, Klagsbrun M. Attenuated p53 activation in tumour-associated stromal cells accompanies decreased sensitivity to etoposide and vincristine. Br J Cancer 2008; 99: 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addadi Y, Moskovits N, Granot D, Lozano G, Carmi Y, Apte RN et al. P53 status in stromal fibroblasts modulates tumor growth in an SDF1-dependent manner. Cancer Res 2010; 70: 9650–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovits N, Kalinkovich A, Bar J, Lapidot T, Oren M. P53 attenuates cancer cell migration and invasion through repression of SDF-1/CXCL12 expression in stromal fibroblasts. Cancer Res 2006; 66: 10671–10676. [DOI] [PubMed] [Google Scholar]
- Guo G, Marrero L, Rodriguez P, Del Valle L, Ochoa A, Cui Y. Trp53 inactivation in the tumor microenvironment promotes tumor progression by expanding the immunosuppressive lymphoid-like stromal network. Cancer Res 2013; 73: 1668–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q et al. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev 2000; 14: 34–44. [PMC free article] [PubMed] [Google Scholar]
- El Naggar A, Clarkson P, Zhang F, Mathers J, Tognon C, Sorensen PH. Expression and stability of hypoxia inducible factor 1alpha in osteosarcoma. Pediatr Blood Cancer 2012; 59: 1215–1222. [DOI] [PubMed] [Google Scholar]
- Wang SW, Wu HH, Liu SC, Wang PC, Ou WC, Chou WY et al. CCL5 and CCR5 interaction promotes cell motility in human osteosarcoma. PLoS One 2012; 7: e35101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchimaru T, Hoshino T, Aikawa T, Yasuda H, Kobayashi T, Kadonosono T et al. Bone resorption facilitates osteoblastic bone metastatic colonization by cooperation of insulin-like growth factor and hypoxia. Cancer Sc. 2014; 105: 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WT, Bian ZY, Fan QM, Li G, Tang TT. Human mesenchymal stem cells (hMSCs) target osteosarcoma and promote its growth and pulmonary metastasis. Cancer Lett 2009; 281: 32–41. [DOI] [PubMed] [Google Scholar]
- Rivlin N, Koifman G, Rotter V. P53 orchestrates between normal differentiation and cancer. Semin Cancer Biol 2014. [DOI] [PubMed]
- Basu-Roy U, Basilico C, Mansukhani A. Perspectives on cancer stem cells in osteosarcoma. Cancer Lett 2013; 338: 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]