Abstract
Dysregulated cell cycle progression has a critical role in tumorigenesis. Cell division cycle 27 (CDC27) is a core subunit of the anaphase-promoting complex/cyclosome, although the specific role of CDC27 in cancer remains unknown. In our study, we explored the biological and clinical significance of CDC27 in colorectal cancer (CRC) growth and progression and investigated the underlying molecular mechanisms. Results showed that CDC27 expression is significantly correlated with tumor progression and poor patient survival. Functional assays demonstrated that overexpression of CDC27 promoted proliferation in DLD1 cells, whereas knockdown of CDC27 in HCT116 cells inhibited proliferation both in vitro and in vivo. Further mechanistic investigation showed that CDC27 downregulation resulted in G1/S phase transition arrest via the significant accumulation of p21 in HCT116 cells, and the upregulation of CDC27 promoted G1/S phase transition via the attenuation of p21 in DLD1 cells. Furthermore, we also demonstrated that CDC27 regulated inhibitor of DNA binding 1 (ID1) protein expression in DLD1 and HCT116 cells, and rescue assays revealed that CDC27 regulated p21 expression through modulating ID1 expression. Taken together, our results indicate that CDC27 contributes to CRC cell proliferation via the modulation of ID1-mediated p21 regulation, which offers a novel approach to the inhibition of tumor growth. Indeed, these findings provide new perspectives for the future study of CDC27 as a target for CRC treatment.
Despite the increased understanding of its pathogenic risk and the development of progressive therapeutic strategies, colorectal cancer (CRC) remains a major cause of cancer morbidity and ranks as the second leading cause of cancer death in China.1, 2, 3, 4 CRC development is a multistep process that involves complex cascades of molecular events,5 therefore understanding the molecular mechanisms that drive malignant tumor formation and progression will contribute to the identification of novel molecular targets and provide more effective methods for CRC prevention and therapy.
The anaphase-promoting complex/cyclosome (APC/C) is a major regulator of protein degradation during mitosis,6, 7, 8 consists of at least 14 subunits9 and are involved in various critical cellular events, such as mitotic progression, and controlling differentiation.10, 11 APC/C degrades its substrates through binding to two co-activators: CDH1 and cell division cycle 20 (CDC20). CDC20 is active in early mitosis, whereas CDH1 activity is restricted to late mitosis and the G1 phase.12 Cell division cycle 27 (CDC27) is a core subunit of APC/C that is responsible for binding CDH1 and CDC20, thereby activating APC/C to recognize and degrade target substrates.13 Emerging evidence has revealed that CDC20 and CDH1 have opposing functions in tumorigenesis. CDH1 has been shown to function largely as a tumor suppressor, whereas CDC20 exhibits an oncogenic function, which was observed in a variety of human tumors.14, 15, 16, 17, 18 However, despite being a key binding partner of CDC20 and CDH1, there is no research on whether CDC27 has a role in cancer. However, recent sequencing studies have reported that CDC27 mutation is detected in various cancers.19, 20, 21, 22 In particular, for CRC, the CDC27 mutation rate was more than 5%, which attracted our interest in exploring the potential function of CDC27 in CRC formation and progression.23
Inhibitor of DNA binding 1 (ID1) is a negative regulator of basic helix–loop–helix transcription factors and is involved in regulating a variety of cellular processes, including growth, senescence, and differentiation.24, 25, 26, 27, 28 The overexpression of ID1 has been observed in a variety of cancers where it contributes to tumor formation and invasion.26 Previous studies have demonstrated that ID1 transcriptionally inhibits the expression of the cyclin-dependent kinase inhibitors p21 in prostate cancer.29, 30 Nevertheless, the relationship between CDC27 and ID1 in cancer cells remains unknown.
In the present study, we aimed to explore whether CDC27 has a key role in CRC, and our findings provide new insights into the mechanisms of CRC tumorigenesis and support the potential of CDC27 as a therapeutic target in CRC treatment.
Results
CDC27 is frequently upregulated in CRC cell lines and tumors
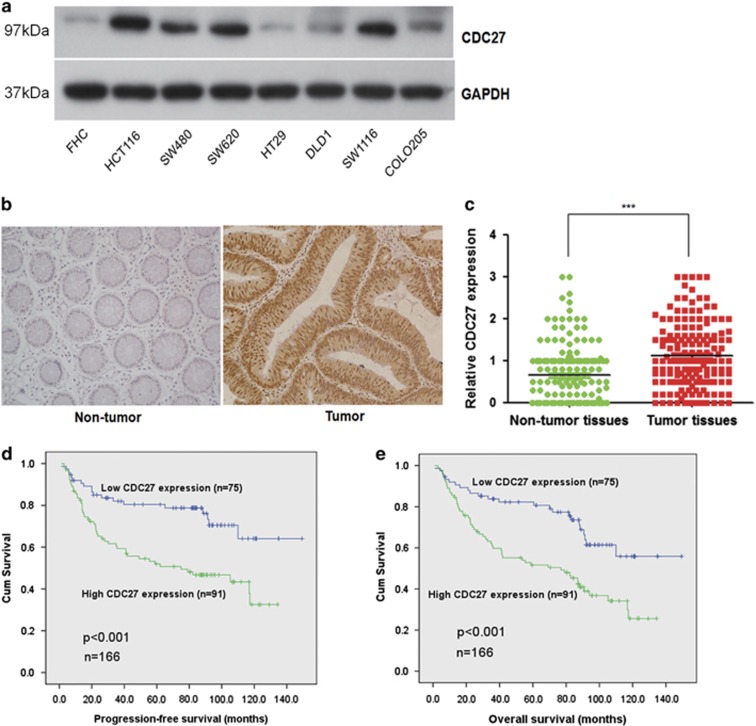
To investigate the role of CDC27 in human CRC development, we first examined CDC27 expression at the protein level in eight CRC cell lines using western blotting. Western blotting analysis revealed that CDC27 protein expression was upregulated in CRC cell lines compared to the normal colon epithelial cell line FHC (Figure 1a). Furthermore, we evaluated endogenous CDC27 expression by immunohistochemistry using paraffin-embedded tissue sections (n=166) of histopathologically confirmed CRC. This analysis revealed that CDC27 was predominantly expressed in the nucleus, and tumor tissues showed a stronger staining intensity compared with paired adjacent non-tumor tissues (Figures 1b and c).
Figure 1.
CDC27 is overexpressed in CRC cell lines and patient tissues and is associated with decreased survival in CRC. (a) Upregulation of CDC27 protein expression was detected by western blotting analysis in a normal colon epithelial cell line (FHC) and seven CRC cell lines. GAPDH was used as a reference control. (b) Representative images of CDC27 expression in CRC tissues and paired adjacent normal mucosal tissues by immunohistochemistry (× 200 magnification). (c) Relative immunohistochemstry analysis for CDC27 expression in CRC tissues and adjacent normal mucosal tissues; each point in the graph represents the CDC27 expression H-score of an individual patient tumor (n=166, ***P<0.001, paired Student's t-test). (d and e) Kaplan–Meier survival analysis of the association between CDC27 expression and PFS or OS (log-rank test)
CDC27 expression is positively correlated with cancer progression and proliferation of CRC
To understand the functional significance of CDC27 in CRC, we analyzed the relationship between CDC27 expression and the clinicopathological features of CRC patients. The clinical characteristics of the patients are summarized in Table 1. High positive CDC27 expression was detected in 91 (54.8%) of the tumor tissue samples. Moreover, the results revealed that CDC27 expression was significantly associated with tumor size (P=0.035), TNM stage (P=0.013), and distant metastasis (P=0.03). Collectively, our results provide evidence that CDC27 is associated with CRC proliferation and progression.
Table 1. Clinicopathological findings and correlation with CDC27 expression.
| Variables | N (%) | CDC27-low (%) | CDC27-high (%) | P value |
|---|---|---|---|---|
| Total cases | 166 | 75 (45.2) | 91 (54.8) | |
| Age (years) | ||||
| <60 | 116 (69.9) | 53 (31.9) | 63 (38.0) | 0.867 |
| ≥60 | 50 (30.1) | 22 (13.4) | 28 (16.7) | |
| Gender | ||||
| Male | 89 (53.6) | 37 (22.3) | 52 (31.3) | 0.35 |
| Female | 77 (46.4) | 38 (22.9) | 39 (23.5) | |
| Tumor location | ||||
| Colon | 88 (53.0) | 38 (22.9) | 50 (30.1) | 0.64 |
| Rectum | 78 (47.0) | 37 (22.3) | 41 (24.7) | |
| Tumor size (cm) | ||||
| <5 | 60 (36.1) | 34 (20.5) | 26 (15.7) | 0.035a |
| ≥5 | 106 (63.9) | 41 (24.7) | 65 (39.2) | |
| Tumor invasive depthb | ||||
| T1–T2 | 39 (23.5) | 19 (11.4) | 20 (12.0) | 0.713 |
| T3–T4 | 127 (76.5) | 56 (33.7) | 71 (42.8) | |
| AJCC/TNM stage | ||||
| I-II | 75 (45.2) | 42 (25.3) | 33 (19.9) | 0.013a |
| III-IV | 91 (54.8) | 33 (19.9) | 58 (34.9) | |
| Lymph node status | ||||
| <1 | 91 (54.8) | 47 (28.3) | 44 (26.5) | 0.085 |
| ≥1 | 75 (45.2) | 28 (16.9) | 47 (28.3) | |
| Distant metastasis | ||||
| No metastasis | 126 (75.9) | 63 (37.95) | 63 (37.95) | 0.03a |
| Metastasis | 40 (24.1) | 12 (7.2) | 28 (16.9) | |
| Preoperative CEA (ng/ml) | ||||
| <5 | 81 (48.8) | 38 (22.9) | 43 (25.9) | 0.755 |
| ≥5 | 85 (51.2) | 37 (22.3) | 48 (28.9) | |
Abbreviation: CEA, carcino-embryonic antigen.
Note: The numbers in parentheses indicate the percentages of tumors with a specific clinical or pathologic feature for a given CDC27 subtype.
Statistically significant.
According to the 7th Edition of the AJCC Cancer Staging Manual.
CDC27 overexpression is associated with decreased survival in CRC
Next, we analyzed the relationship between CDC27 expression and patient survival. The survival time of the 166 CRC patients ranged from 1.1 to 149.4 months. As shown in Figures 1d and e, the results showed that CDC27 expression in CRC was negatively correlated with patient progression-free survival (PFS) and overall survival (OS). Kaplan–Meier survival analysis and log-rank tests revealed that CDC27 expression was negatively associated with PFS (P<0.001, HR=2.518, 95% CI=1.481–4.281) and OS (P<0.001, HR=2.348, 95% CI=1.454–3.791). The rates of PFS (46.2% versus 74.7%) and OS (38.5% versus 68.0%) were significantly lower in the CDC27 high-expression group than that in the low-expression group. Multivariate Cox proportional hazards regression analysis indicated that CDC27 expression served as an independent prognostic factor for PFS (P=0.04, HR=1.78, 95% CI=1.03–3.07) and OS (P=0.02, HR=1.79, 95% CI=1.09–2.94; Table 2). Collectively, this analysis strongly indicates that CDC27 expression may serve as a potential independent prognostic factor for PFS and OS in CRC patients.
Table 2. Multivariate analysis for PFS and OS.
| Variable | PFS, HR (95% CI) | P value | OS, HR (95% CI) | P value |
|---|---|---|---|---|
| Age (years, <60 versus ≥60) | 1.11 (0.63–1.97) | 0.70 | 1.21 (0.72–2.03) | 0.48 |
| Gender (male versus female) | 1.13 (0.68–1.88) | 0.64 | 1.25 (0.78–2.00) | 0.35 |
| Tumor location (colon versus rectum) | 1.18 (0.70–1.99) | 0.53 | 1.49 (0.93–2.41) | 0.10 |
| Tumor size (cm, <5 versus ≥5) | 1.07 (0.56–2.05) | 0.84 | 1.17 (0.66–2.08) | 0.59 |
| Tumor invasive depth (T1–2 versus T3–4) | 0.84 (0.32–2.2) | 0.72 | 0.88 (0.37–2.06) | 0.76 |
| TNM stage (I-II versus III-IV) | 10.65 (5.2–21.7) | <0.001a | 8.46 (4.69–15.2) | <0.001a |
| Lymph node status (<1 versus ≥1) | 2.21 (1.17–4.19) | 0.02a | 2.55 (1.41–4.64) | 0.002a |
| Distant metastasis (no versus yes) | 7.4 (4.02–13.65) | <0.001a | 6.53 (3.74–11.48) | <0.001a |
| CDC27 (low versus high) | 1.78 (1.03–3.07) | 0.04a | 1.79 (1.09–2.94) | 0.02a |
Abbreviations: PFS, progression-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval.
Statistically significant.
Downregulation of CDC27 inhibits cell growth in colon cancer cells
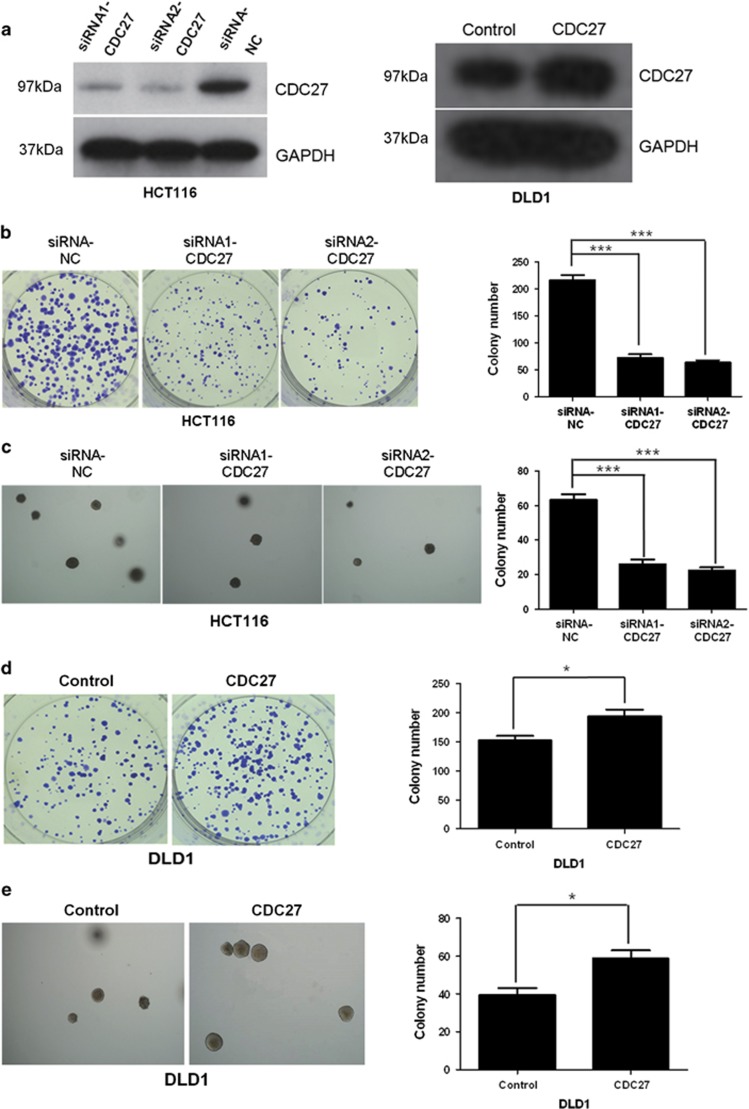
We next sought to investigate whether downregulation of CDC27 could affect the proliferation of CRC cells. HCT116 cells were transiently transfected with small interfering RNAs (siRNAs) targeting CDC27 (siRNA1-CDC27, siRNA2-CDC27) or negative control siRNA (siRNA-NC), and DLD1 cells were transiently transfected with CDC27-carrying plasmids or empty vector, respectively. The expression level of CDC27 in these cells was confirmed by western blotting (Figure 2a). The results of the colony formation assays indicated that the cells transfected with siRNA exhibited a weakened capacity for colony formation compared with the control group in HCT116 (Figure 2b). Similar results were obtained in another CRC cell line, SW480 (Supplementary Figure 1). We also performed soft agar colony formation assays, and the colony number of the CDC27 siRNA group was distinctly less than that of the control group in HCT116 cell line (Figure 2c). Moreover, exogenous CDC27 expression in DLD1 can promote cell proliferation (Figures 2d and e). Therefore, our results provide evidence that CDC27 can promote proliferation in CRC cell lines.
Figure 2.
CDC27 promotes proliferation of CRC cells. (a) The efficient overexpression or suppression of CDC27 was identified by western blotting. (b) Representative images show the colony formation ability of HCT116 cells with CDC27 transiently suppressed. The number of the colonies was quantified. ***P<0.001 using Student's t-test. (c) Soft agar colony formation assays. Representative images are shown; the numbers of colonies containing more than 50 cells were scored. ***P<0.001 using Student's t-test. (d and e) CDC27 overexpression promoted cell growth in both colony formation assays and soft agar colony formation assays. Representative images are presented. *P<0.05 using Student's t-test. These experiments were repeated at least three times. Error bars, mean±S.D.
CDC27 influences the G1/S phase transition of the cell cycle through regulating p21 expression
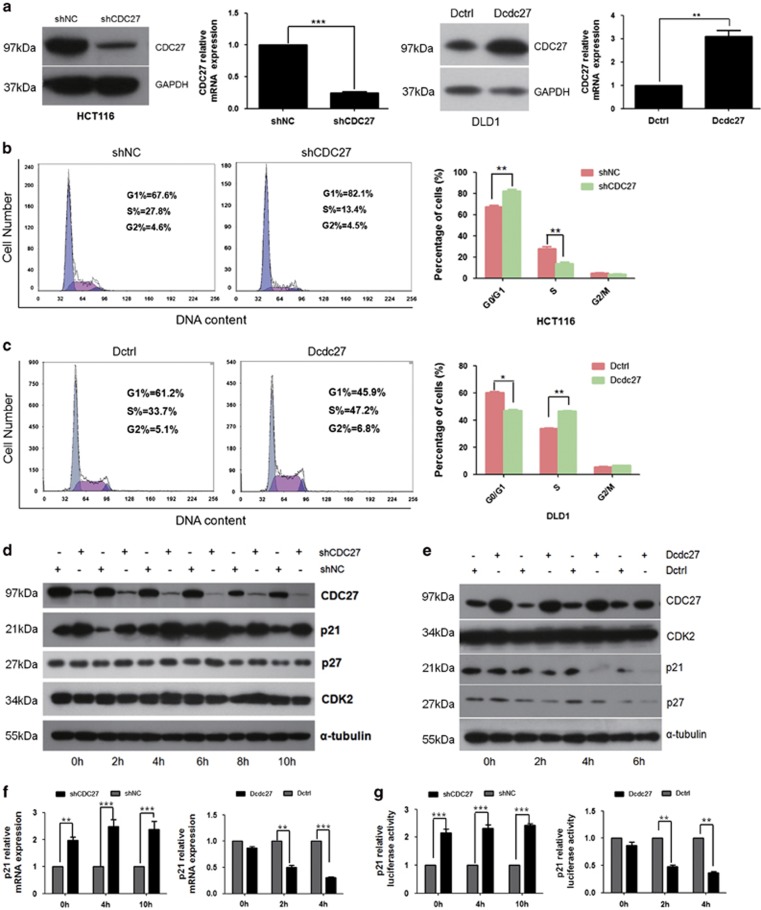
Given our findings thus far, we conducted a cell cycle analysis to identify the mechanisms by which CDC27 downregulation can inhibit proliferation. To persistently suppress CDC27 expression, we constructed two HCT116 stable cell lines: shCDC27 and shNC. In addition, we constructed a stable DLD1 cell line overexpressing CDC27 (Dcdc27) and a control DLD1 cell line (Dctrl) to detect alterations in cell cycle distribution. The overexpression or knockdown efficiency was confirmed through real-time PCR and western blotting (Figure 3a). As shown in Figure 3b, HCT116-shCDC27 remarkably increased the number of cells in G1 phase compared with the control group. Correspondingly, overexpression of CDC27 promoted the G1/S transition in DLD1 stable cell lines (Figure 3c). To clarify the specific role of CDC27 in the G1/S phase transition, cells were synchronized at the G0/G1 phase and harvested every 2 h after recovery of the serum supply. Key regulators of the G1 phase were evaluated by western blotting. Intriguingly, we found that knockdown of CDC27 led to a dramatic accumulation of p21 protein at each time point examined, whereas p27 expression was only weakly increased at certain time points (Figure 3d). In addition, the expression of p21 remarkably decreased in the Dcdc27 cell lines at 2, 4, and 6 h after serum restoration compared with that in the Dctrl stable cell lines, whereas p27 expression decreased in a marginal manner (Figure 3e). Therefore, we concluded that CDC27 influences the cell cycle transition by mainly regulating the expression of p21. These results were confirmed using real-time PCR, and the data showed that p21 was negatively regulated by CDC27 at the mRNA level (Figure 3f). Moreover, the indicated stable cell lines were transfected with luciferase reporter plasmids driven by the p21 promoter. The results demonstrated that CDC27 inhibited p21 promoter activity (Figure 3g). Therefore, we concluded that the alteration of p21 expression can be at least in part attributed to the regulation of CDC27 at transcription level.
Figure 3.
CDC27 inhibits p21-dependent arrest at the G1/S phase transition. (a) CDC27 overexpression in DLD1 stable cell lines and persistent suppression in HCT116 stable cell lines identified using real-time PCR and western blotting. Mean±S.D. of triplicate samples are shown. **P<0.01, ***P<0.001, Student's t-test. (b and c) The indicated stable cell lines were subjected to cell cycle distribution analysis by flow cytometry. Images and qualification of the cell cycle distribution in three independent experiments are shown; *P<0.05, **P<0.01, Student's t-test. (d and e) The HCT116 stable cell lines or DLD1 stable cell lines were synchronized by serum starvation for 24 h, and protein was harvested every 2 h after serum restoration. Western blotting analysis was used to detect changes in p21, p27 expression in the indicated cell lines. α-Tubulin was used as a reference control. (f) Cells were harvested at indicated time points after synchronization. Expression of p21 mRNA was analyzed by real-time PCR. Mean±S.D. of triplicate samples are shown. **P<0.01, ***P<0.001, Student's t-test. (g) pGL3-p21 was transfected into both HCT116 and DLD1 stable cell lines, and cells were harvested at the indicated time after synchronization for luciferase reporter assays. Mean±S.D. of triplicate samples are shown. **P<0.01, ***P<0.001, Student's t-test
CDC27 mediates p21-dependent cell cycle arrest by modulating ID1 expression
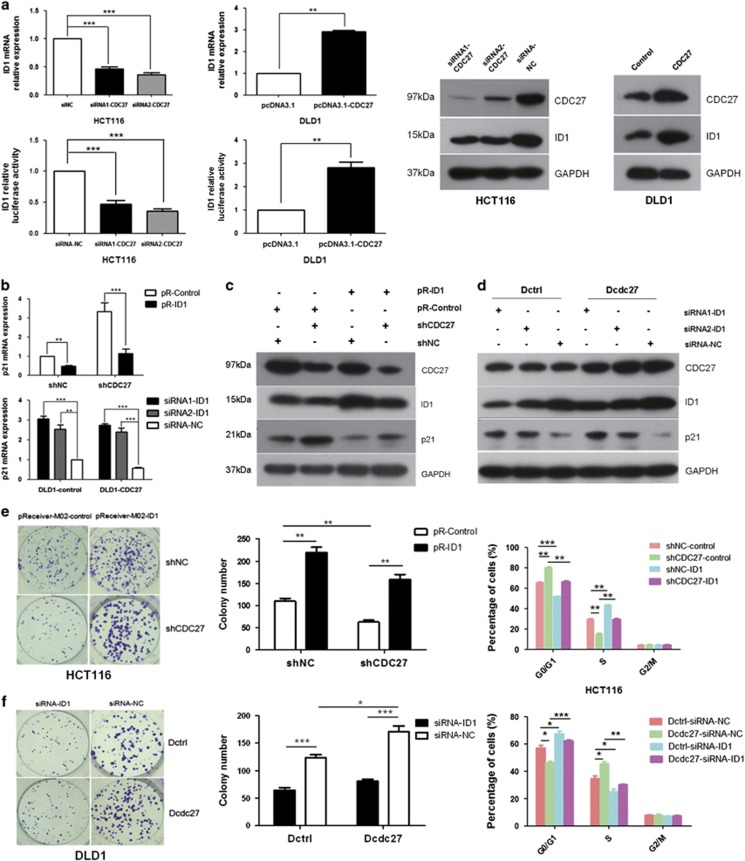
ID1 is a key molecular regulator upstream of p21, and we wondered if ID1 participate in the regulation of p21 expression by CDC27. To test the hypothesis, we evaluated whether the expression of ID1 can be influenced by CDC27 using real-time PCR, luciferase reporter assays, and western blotting. Results indicated that ID1 expression was dramatically decreased upon transiently interference of CDC27 in HCT116 cells, whereas increased upon CDC27 overexpression in DLD1 cells (Figure 4a). Next, we transfected exogenous ID1 expression vectors (pR-ID1) or empty vector (pR-Control) into shCDC27 or shNC stable HCT116 cell lines, and simultaneously siRNAs of ID1 (siRNA1-ID1, siRNA2-ID1) or negative control sequence (siRNA-NC) were transfected into the Dcdc27 or Dctrl stable DLD1 cell lines. Alterations of p21 expression were detected by real-time PCR and western blotting. The results showed that overexpression of exogenous ID1 could significantly reduce p21 accumulation in HCT116-shCDC27 cell lines. Similarly, the p21 expression levels in Dcdc27 cells were increased after transient interference of ID1 (Figures 4b–d). Therefore, we concluded that CDC27 affects p21 expression in an ID1-mediated manner.
Figure 4.
CDC27 regulates p21 expression through modulating ID1 expression. (a) Real-time PCR, luciferase reporter assays (left) and western blotting (right) were performed to detect ID1 expression in the indicated cells with CDC27 was transiently overexpressed or suppressed. Mean±S.D. of triplicate samples are shown. **P<0.01, ***P<0.001 using Student's t-test. (b–d) Cells were harvested at 4 h after synchronization by serum starvation for p21 mRNA and protein expression level detection. Mean±S.D. of triplicate samples are shown. **P<0.01, ***P<0.001, Student's t-test. (e and f) ID1 was transiently overexpressed or suppressed in the indicated stable cell lines. Representative images of colony formation rescue assays and quantification were presented. Cells were subjected to cell cycle distribution analysis by flow cytometry. Mean±S.D. of triplicate samples are shown. *P<0.05, **P<0.01, ***P<0.001, Student's t-test
Exogenous ID1 expression can reverse the inhibition of proliferation induced by CDC27 downregulation
Given the results obtained thus far, we next sought to determine whether ID1 affects the proliferation of CRC cells induced by CDC27. As shown in Figure 4e, the exogenous expression of ID1 reversed the inhibition of proliferation and suppressed the G1/S transition arrest caused by CDC27 downregulation in HCT116 stable cell lines. Moreover, transient transfection of ID1 siRNA suppressed cell proliferation and inhibited the G1/S transition caused by CDC27 in DLD1 stable cell lines (Figure 4f). Therefore, ID1 has a crucial role in CDC27-regulated CRC cell proliferation.
CDC27 promotes tumor growth in a xenograft mouse model
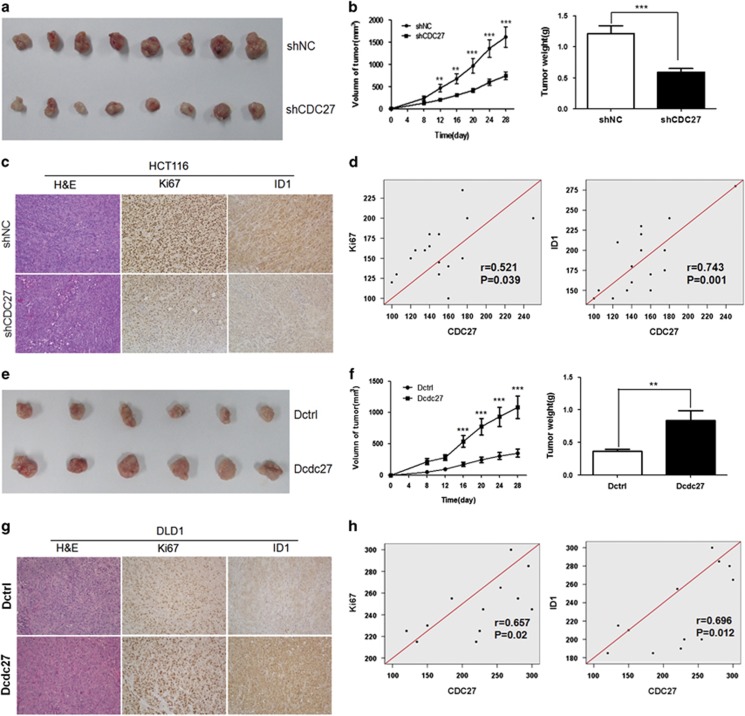
Next, we established a BALB/c nude mouse xenograft model using the stable HCT116 cell lines and DLD1 cell lines. Cells were injected subcutaneously into the flanks of male nude mice. Tumor size was measured every 4 days, and the tumor volume was calculated. Four weeks later, the mice were killed, and the tumors were collected. Results indicated that knockdown of CDC27 in HCT116 stable cell lines led to a significant decrease in tumor weight and volume compared with the control group (n=8, Figures 5a and b), whereas mice injected with Dcdc27 cells exhibited greater tumor growth capacity compared with the control group (n=6, Figures 5e and f). Furthermore, we used immunohistochemistry to evaluate the expression of ID1 and the nuclear cell proliferation marker Ki67 in the excised mouse tumors. As shown in Figure 5c, ID1 was stained in both the nuclear and cytoplasm. The tumors from the HCT116-shCDC27 group exhibited weaker Ki67 and ID1 staining intensity, whereas the tumors from the Dcdc27 group exhibited stronger Ki67 and ID1 staining intensity compared with the control group (Figure 5g). Moreover, the expression of both ID1 and Ki67 was positively correlated with CDC27 expression (Figures 5d and h). These results suggest that CDC27 contributes to tumor proliferation in CRC.
Figure 5.
CDC27 promotes tumor growth in vivo. (a and e) The xenografts tumors were collected at 28 days after injection. (b and f) Data points are presented as the mean±S.D. tumor volume and the weight of the tumors were analyzed. **P<0.01, ***P<0.001. (c and g) The tumor sections were under H&E staining and immunohistochemstry staining using antibody against Ki67 and ID1 (× 200 magnification). (d and h) Correlations between selected molecular expression were analyzed by Pearson's test in mice tumor tissues
Discussion
In the present study, our results showed that CDC27 has a key role in CRC proliferation through regulating p21 expression and consequently control G1/S phase transition. CDC27 may serve as an independent prognostic factor for CRC patients.
A major driver of cellular transformation is the loss of proper control of the mammalian cell cycle.31 Disturbance of this balance by disrupting the program that regulates cell cycle entry can result in the transformation of normal cells into tumor cells.32, 33, 34 As a key regulator of cell cycle progression at G1/S transition, p21 has a critical role in tumorigenesis.34, 35, 36, 37 Our study demonstrated that downregulation of CDC27 can lead to G1/S transition arrest via the accumulation of p21 (Figures 3b and d). We also performed apoptosis assays, although we did not find evidence that CDC27 has a key role in apoptosis (data not shown). In CRC, ID1 is also an important marker for tumor progression.38 Intriguingly, we observed CDC27 positively regulated ID1 expression in DLD1 and HCT116 cell lines (Figure 4a). To further investigate the relationship between CDC27 and ID1 in CRC, we detect the expression of CDC27 and ID1 both in CRC cell lines and 12 pairs of patient tissues. The results indicated the expression of CDC27 and ID1 is significantly correlated in the 12 pairs of patient tissues (Supplementary Figure 2A), despite that there was no correlation in CRC cell lines (Supplementary Figure 2B). Given that ID1 acts as a classical upstream regulator of p21,30 we next explored whether ID1 took part in p21 accumulation caused by CDC27 downregulation. Consistent with our assumption, exogenous overexpression of ID1 rendered cells' refractory to p21 accumulation (Figure 4c), and abolished the inhibition of proliferation caused by CDC27 downregulation (Figure 4e).
Dysregulation of APC/C has been shown to contribute to tumorigenesis.39, 40 APC/C is not only a complicated, large-size complex involving in various signaling pathways and cell biological process, but also works dependently on its subunits interaction and numerous substrates degradation in different phases of cell cycle, thereby any subunit or co-activator alteration may affect or even abolish its function and lead to cascade of response with a wide range of biological effects.9, 10 Several studies have shown that the two co-activators CDC20 and CDH1 act, respectively, as an oncogene and a tumor suppressor in cancer. However, few studies paid attention to the independent roles of its subunits in cancer. CDC27 is a core subunit of APC/C, but the function of CDC27 in cancer remains unclear. Regardless of APC function alteration, our study emphasized the role of CDC27 in CRC. To the best of our knowledge, our study is the first to explore the function of CDC27 in cancer, and our findings provide a novel perspective for further study of the functions of APC/C subunits.
Furthermore, it is worth mentioning that increasing evidence indicated that cancer stem-like cells (CSCs) are a rare subpopulation of cancer cells capable of propagating the disease and causing cancer recurrence, thus searching for the potential targeting of CSCs is a challenging task.41, 42 Previous studies have reported that p21 is linked to maintenance of self-renewal capacity in stem cells,43, 44 for that low level or absence of p21 expression leads to more efficient and faster reprogramming in mouse and human cells.45, 46 In addition, ID1 can also participates in self-renewal capacity in murine cortical neural stem cells, and studies in a murine model of hematopoiesis revealed that Id1−/− whole-bone marrow displayed impaired self-renewal capacity relative to wild-type controls.47 Moreover, previous studies have demonstrated that ID1 participates in maintaining self-renewal through repressive effects on expression of p21 in hematopoietic stem cells and in bone marrow for the generation of endothelial progenitor cells.48, 49 In our study, we demonstrated that CDC27 regulated p21 expression through modulating ID1 expression. Therefore, we are curious about if CDC27 was involved in CSC. We have examined the expression of two classical colorectal epithelial stemness markers CD44 and CD133 using the HCT116 (shCDC27, shNC) and DLD1 (Dcdc27, Dctrl) stable cell lines. Interestingly, the expression of CD44 dramatically decreased in the HCT116-shCDC27 cell lines, and increased in the Dcdc27 cell lines compared with the control group (Supplementary Figure 3), whereas there was no difference in the expression of CD133. The findings above indicate there is a possibility that CDC27 is involved in CSC, and becomes a potential target of CSCs for monitoring the progress of cancer therapy and for evaluating new therapeutic approaches.
However, there are still many topics that remain to be addressed. First, recent studies have demonstrated that CDC27 mutations are involved in various cancers; thus, we sought to investigate the presence of important disease-associated alleles, although we have not yet identified key mutations that can significantly affect tumor proliferation or progression. Second, ID1 is overexpressed in a variety of solid tumors,50, 51, 52, 53 and ID1 upregulation correlates with both poor prognosis and chemoresistance.54, 55, 56, 57 Therefore, we strongly suspect that CDC27 may also have a role in other cancers. Considering p21 is well positioned to function as both a sensor and an effector of multiple anti-proliferative signals.58 ID1 is not the unique mediator in CDC27-p21 regulation, and additional mechanisms may involve in p21 regulation caused by CDC27. In addition, for the reason that CDC27 expression is significantly associated with distant metastasis in CRC (Table 1), we have carried out experiments to explore the role of CDC27 in metastasis and invasiveness in CRC. Therefore, whether CDC27 can promote metastasis via modulating certain potential downstream molecular in CRC, and whether there is a correlation between CDC27 and its downstream molecular in clinical samples, remain to be determined in future studies.
In conclusion, our study is the first to demonstrate that CDC27 is associated with tumor proliferation and progression in CRC. These results suggest that CDC27 may serve as a prognostic biomarker and therapeutic target for CRC treatment.
Materials and Methods
Cell lines and cell culture
All cell lines used were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA), authentication by short tandem repeat profiling/karyotyping/isoenzyme analysis. Cell culture methods were listed in Supplementary Methods and Materials.
Patient tissue specimens and clinicopathological characteristics
Patient tissues used in our study were collected from 166 patients histologically and clinically diagnosed with CRC between 1999 and 2005 at the Sun Yat-sen University Cancer Center (Guangzhou, China). The detailed selection criterion was described in the Supplementary Methods and Materials.
Immunohistochemistry
The detailed method has been described in previous study.59 Primary antibodies used were as follow: CDC27 (Santa Cruz, CA, USA, sc-9972, 1 : 50), Ki67 (Santa, sc-15402, 1 : 100), and ID1 (Santa, sc-488, 1 : 100). The expression of CDC27 was evaluated using H-scores. The final H score was obtained according to the intensity and proportion of the area stained. CDC27 expression intensity was scored as negative=0, weak=1, moderate=2, or strong=3. The final H score was calculated by multiplying the intensity score by the percentage of the staining area. Receiver operating characteristic curve analysis was used to determine a cutoff value for CDC27 high expression or low expression. The sensitivity and specificity for the outcome under study was plotted, thus generating a receiver operating characteristic. The score that was closest to the point with both maximum sensitivity and specificity were selected as the cutoff value.
RNA extraction and real-time PCR
Total RNA from colorectal cells was extracted using Trizol (Invitrogen, Carlsbad, CA, USA) reagent according to the manufacturer's instructions, and then cDNA was synthesized using the GoScript Reverse Transcription System (Promega, Madison, WI, USA). Real-time PCR was performed using a SYBR Green PCR Kit (Invitrogen). GAPDH was used as an endogenous control to normalize the amount of target gene mRNA. The primer sequences were shown in Supplementary Table 1.
Stable cell line construction
CDC27-overexpressing or control vectors were transfected into DLD1 cells using Lipo2000. Cells were harvested at 48 h after transfection and were incubated with G418 for 12 days, with the medium refreshed every 3 days. For persistent suppression of CDC27 expression, short hairpin RNA oligonucleotides were cloned into the pSuper-retro-puro vector to knockdown CDC27 endogenous expression. Retrovirus vector pSuper-retro-puro-short hairpin RNA or pSuper-retro-puro empty vector and PIK packaging plasmids were cotransfected into 293T cells, and virus supernatant was harvested 48 h after transfection. Retrovirus supernatant was used to infect HCT116 cells, and puromycin was added 48 h after infection to select stable cell lines for 7 days, with the medium refreshed every 3 days.
Western blotting
Total protein was extracted from cells using cell lysis buffer containing protease inhibitor (Sigma-Aldrich, St. Louis, MO, USA, P8340). Western blotting was performed as previously described.59 Primary antibodies used were summarized in Supplementary Methods and Materials.
Colony formation and soft agar assays
For colony formation assays, HCT116 and SW480 cells were transfected with siRNAs of CDC27 or negative control siRNA, and DLD1 cells were transfected with CDC27-carring plasmids or empty vector. Cells were plated into six-well plates at 48 h after transfection. After incubating at 37 °C for 14 days, visible colonies were fixed and stained with 0.5% crystal violet in methanol. For the soft agar assay, the cells in single-cell suspension were plated in 0.3% agarose over a 0.6% agarose bottom layer in six-well plates and incubated for 14 days. The number of colonies containing more than 50 cells was counted.
Cell cycle analysis
Cells were harvested, washed, and fixed in pre-cooled 75% ethanol mixed with PBS at −20 °C overnight. The next day, the cells were washed and resuspended in 500 μl of pre-cooled PBS including RNase (Thermo Scientific, Lafayette, CO, USA) at a concentration of 10 μg/ml, and the cells were incubated at 37 °C for 30 min before staining. A 30-μl solution containing propidium iodide (Sigma) at a concentration of 1 mg/ml was added to the cells and incubated at 4 °C in the dark for at least 1 h. All cell samples were harvested and analyzed on a flow cytometer (Beckman Gallios, Beckman Gallios, Inc., Fullerton, CA, USA).
Luciferase reporter assays
The luciferase reporter assays were performed as previously described.60
Tumor formation assays in nude mice
Animal experiments were approved by the Sun Yat-Sen University Cancer Center Institutional Animal Care and Usage Committee. Male BALB/c nude mice (4–5 weeks old, 15–18 g) were purchased from the Slaccas Experimental Animal Center (Shanghai, China). Mice were injected with HCT116 or DLD1 stable cell lines (n=8 and n=6, respectively). 2 × 106 cells were injected subcutaneously into each nude mouse. The tumor volume was calculated using the following formula: V=(width2 × length)/2. After 28 days, the mice were euthanized, and the primary tumors were collected and weighed.
Statistical analysis
All statistical analyses were completed using the SPSS version 16.0 statistical software package (SPSS Inc., Chicago, IL, USA). Distributed data are expressed as mean±S.D. Student's t-test and two-tailed χ2 test were used. Pearson's test was applied for correlation analysis. A P value of less than 0.05 was considered statistically significant.
Acknowledgments
We thank Jiemin Chen and Ling Zhou of the laboratory for their helpful assistance on the manuscript. This work was supported by National High Technology Research and Development Program of China (863 Program, No. 2012AA02A204), and the National Natural Science Foundation of China (No. 81272513, No. 81272638).
Glossary
- CRC
colorectal cancer
- CDC27
cell division cycle 27
- APC/C
anaphase-promoting complex/cyclosome
- ID1
inhibitor of DNA binding 1
- CDC20
cell division cycle 20
- PFS
progression-free survival
- OS
overall survival
- siRNA
small interfering RNA
- CSC
cancer stem-like cell
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014; 64: 104–117. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer2010; 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg 2009; 22: 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Huang S, Lei Y, Zhang T, Wang K, Liu B et al. FGF8 promotes colorectal cancer growth and metastasis by activating YAP1. Oncotarget 2015; 6: 935–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Chapelle A. Genetic predisposition to colorectal cancer Nat Rev Cancer 2004; 4: 769–780. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Regulation of the APC and the exit from mitosis. Nat Cell Biol 1999; 1: E47–E53. [DOI] [PubMed] [Google Scholar]
- Page AM, Hieter P. The anaphase-promoting complex: new subunits and regulators Annu Rev Biochem 1999; 68: 583–609. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol 2007; 8: 894–903. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang J, Wan L, Zhou X, Wang Z, Wei W. Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol Ther 2015; 151: 141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 2006; 7: 644–656. [DOI] [PubMed] [Google Scholar]
- Pines J. The APC/C: a smorgasbord for proteolysis. Mol Cell 2009; 34: 135–136. [DOI] [PubMed] [Google Scholar]
- Song MS, Carracedo A, Salmena L, Song SJ, Egia A, Malumbres M et al. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell 2011; 144: 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 2011; 20: 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DZ, Ma Y, Ji B, Liu Y, Hwu P, Abbruzzese JL et al. Increased CDC20 expression is associated with pancreatic ductal adenocarcinoma differentiation and progression. J Hematol Oncol 2012; 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon DS et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res 2006; 12: 405–410. [DOI] [PubMed] [Google Scholar]
- Kwan PS, Lau CC, Chiu YT, Man C, Liu J, Tang KD et al. Daxx regulates mitotic progression and prostate cancer predisposition. Carcinogenesis 2013; 34: 750–759. [DOI] [PubMed] [Google Scholar]
- Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kaji M. Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. J Surg Oncol 2012; 106: 423–430. [DOI] [PubMed] [Google Scholar]
- Wu WJ, Hu KS, Wang DS, Zeng ZL, Zhang DS, Chen DL et al. CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J Transl Med 2013; 11: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann E, Koks S, Ho X, Maasalu K, Martson A. Whole exome sequencing of a single osteosarcoma case inverted question markintegrative analysis with whole transcriptome RNA-seq data. Hum Genomics 2014; 8: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhlin CC, Goh G, Healy JM, Fonseca AL, Scholl UI, Stenman A et al. Whole-exome sequencing characterizes the landscape of somatic mutations and copy number alterations in adrenocortical carcinoma. J Clin Endocrinol Metab 2015; 100: E493–E502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield K, Summersgill B, Yost S, Sultana R, Labreche K, Dudakia D et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat Commun 2015; 6: 5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg J, Mills IG, Klevebring D, Liu W, Neiman M, Xu J et al. The mitochondrial and autosomal mutation landscapes of prostate cancer. Eur Urol 2013; 63: 702–708. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014; 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Rademacher BL, Pittsenbarger J, Huang CY, Harvey CT, Lafortune MC et al. ID1 enhances docetaxel cytotoxicity in prostate cancer cells through inhibition of p21. Cancer Res 2010; 70: 3239–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci 2000; 113: 3897–3905. [DOI] [PubMed] [Google Scholar]
- Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell 2003; 3: 525–530. [DOI] [PubMed] [Google Scholar]
- Benezra R. Role of Id proteins in embryonic and tumor angiogenesis. Trends Cardiovasc Med 2001; 11: 237–241. [DOI] [PubMed] [Google Scholar]
- Benezra R, Rafii S, Lyden D. The Id proteins and angiogenesis. Oncogene 2001; 20: 8334–8341. [DOI] [PubMed] [Google Scholar]
- Asirvatham AJ, Carey JP, Chaudhary J. ID1-, ID2-, and ID3-regulated gene expression in E2A positive or negative prostate cancer cells. Prostate 2007; 67: 1411–1420. [DOI] [PubMed] [Google Scholar]
- Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol 1997; 17: 5888–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasch R, Robbins JA, Cross FR. The emerging role of APC/CCdh1 in controlling differentiation, genomic stability and tumor suppression. Oncogene 2010; 29: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. The role of aneuploidy in promoting and suppressing tumors. J Cell Biol 2009; 185: 935–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol 2001; 2: 21–32. [DOI] [PubMed] [Google Scholar]
- Pines J, Rieder CL. Re-staging mitosis: a contemporary view of mitotic progression. Nat Cell Biol 2001; 3: E3–E6. [DOI] [PubMed] [Google Scholar]
- Calvisi DF, Donninger H, Vos MD, Birrer MJ, Gordon L, Leaner V et al. NORE1A tumor suppressor candidate modulates p21CIP1 via p53. Cancer Res 2009; 69: 4629–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler S, Hahnel PS, Schad A, Dammann R, Schuler M. RASSF1A mediates p21Cip1/Waf1-dependent cell cycle arrest and senescence through modulation of the Raf-MEK-ERK pathway and inhibition of Akt. Cancer Res 2009; 69: 1748–1757. [DOI] [PubMed] [Google Scholar]
- Viatour P, Sage J. Newly identified aspects of tumor suppression by RB. Dis Model Mech 2011; 4: 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZR, Zhang ZY, Zhang H, Jiang L, Wang MW, Sun XF. Overexpression of Id-1 protein is a marker in colorectal cancer progression. Oncol Rep 2008; 19: 419–424. [PubMed] [Google Scholar]
- Jackson PK. Linking tumor suppression, DNA damage and the anaphase-promoting complex. Trends Cell Biol 2004; 14: 331–334. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 2009; 10: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K et al. Cancer stem cell markers in common cancers—therapeutic implications. Trends Mol Med 2008; 14: 450–460. [DOI] [PubMed] [Google Scholar]
- Jain MV, Jangamreddy JR, Grabarek J, Schweizer F, Klonisch T, Cieslar-Pobuda A et al. Nuclear localized Akt enhances breast cancer stem-like cells through counter-regulation of p21(Waf1/Cip1) and p27(kip1). Cell Cycle 2015; 14: 2109–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 2000; 287: 1804–1808. [DOI] [PubMed] [Google Scholar]
- Wasik AM, Grabarek J, Pantovic A, Cieslar-Pobuda A, Asgari HR, Bundgaard-Nielsen C et al. Reprogramming and carcinogenesis—parallels and distinctions. Int Rev Cell Mol Biol 2014; 308: 167–203. [DOI] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 2009; 23: 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 2009; 460: 1136–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SS, Zhao Y, Nie L, Cochrane SW, Huang Z, Sun XH. Id1, but not Id3, directs long-term repopulating hematopoietic stem-cell maintenance. Blood 2007; 110: 2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarrocchi A, Jankovic V, Shaked Y, Nolan DJ, Mittal V, Kerbel RS et al. Id1 restrains p21 expression to control endothelial progenitor cell formation. PLoS ONE 2007; 2: e1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic V, Ciarrocchi A, Boccuni P, DeBlasio T, Benezra R, Nimer SD. Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc Natl Acad Sci U S A 2007; 104: 1260–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindl M, Oberhuber G, Obermair A, Schoppmann SF, Karner B, Birner P. Overexpression of Id-1 protein is a marker for unfavorable prognosis in early-stage cervical cancer. Cancer Res 2001; 61: 5703–5706. [PubMed] [Google Scholar]
- Schindl M, Schoppmann SF, Strobel T, Heinzl H, Leisser C, Horvat R et al. Level of Id-1 protein expression correlates with poor differentiation, enhanced malignant potential, and more aggressive clinical behavior of epithelial ovarian tumors. Clin Cancer Res 2003; 9: 779–785. [PubMed] [Google Scholar]
- Lin CQ, Singh J, Murata K, Itahana Y, Parrinello S, Liang SH et al. A role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res 2000; 60: 1332–1340. [PubMed] [Google Scholar]
- Meteoglu I, Meydan N, Erkus M. Id-1: regulator of EGFR and VEGF and potential target for colorectal cancer therapy. J Exp Clin Cancer Res 2008; 27: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien CA, Kreso A, Ryan P, Hermans KG, Gibson L, Wang Y et al. ID1 and ID3 regulate the self-renewal capacity of human colon cancer-initiating cells through p21. Cancer Cell 2012; 21: 777–792. [DOI] [PubMed] [Google Scholar]
- Cheung HW, Ling MT, Tsao SW, Wong YC, Wang X. Id-1-induced Raf/MEK pathway activation is essential for its protective role against taxol-induced apoptosis in nasopharyngeal carcinoma cells. Carcinogenesis 2004; 25: 881–887. [DOI] [PubMed] [Google Scholar]
- Hu H, Han HY, Wang YL, Zhang XP, Chua CW, Wong YC et al. The role of Id-1 in chemosensitivity and epirubicin-induced apoptosis in bladder cancer cells. Oncol Rep 2009; 21: 1053–1059. [DOI] [PubMed] [Google Scholar]
- Li B, Cheung PY, Wang X, Tsao SW, Ling MT, Wong YC et al. Id-1 activation of PI3K/Akt/NFkappaB signaling pathway and its significance in promoting survival of esophageal cancer cells. Carcinogenesis 2007; 28: 2313–2320. [DOI] [PubMed] [Google Scholar]
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009; 9: 400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ye W, Wu J, Meng X, Liu RY, Ying X et al. Overexpression of sirt7 exhibits oncogenic property and serves as a prognostic factor in colorectal cancer. Clin Cancer Res 2014; 20: 3434–3445. [DOI] [PubMed] [Google Scholar]
- Meng X, Wu J, Pan C, Wang H, Ying X, Zhou Y et al. Genetic and epigenetic down-regulation of microRNA-212 promotes colorectal tumor metastasis via dysregulation of MnSOD. Gastroenterology 2013; 145: 426–436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.