Abstract
Chronic lymphocytic leukemia (CLL) is a B-cell malignancy with a mature phenotype. In spite of its relatively indolent nature, no radical cure is as yet available. CLL is not associated with either a unique cytogenetic or a molecular defect, which might have been a potential therapeutic target. Instead, several factors are involved in disease development, such as environmental signals which interact with genetic abnormalities to promote survival, proliferation and an immune surveillance escape. Among these, PI3-Kinase signal pathway alterations are nowadays considered to be clearly important. The TCL1 gene, an AKT co-activator, is the cause of a mature T-cell leukemia, as well as being highly expressed in all B-CLL. A TCL1 transgenic mouse which reproduces leukemia with a distinct immunophenotype and similar to the course of the human B-CLL was developed several years ago and is widely used by many groups. This is a review of the CLL biology arising from work of many independent investigators who have used TCL1 transgenic mouse model focusing on pathogenetic, microenviroment and therapeutic targets.
Facts
Aggressive form chronic lymphocytic leukemia (CLL) is still incurable.
The TCL1-tg mouse model is most similar to aggressive human CLL.
The TCL1-tg model has fundamentally contributed to the elucidation of CLL pathogenic mechanisms.
Many novel therapeutic strategies have been tested using TCL1-tg mice.
Open Questions
Can new combination therapies be investigated using the TCL1-tg model?
Can microenvironment contribution and tumor immune suppression be more easily studied through animal models than in human patients?
Can microRNAs targeting TCL1 or TCL1-specific inhibitors be used as therapies against CLL?
CLL is the most common B-cell malignancy in Western countries. CLL lymphocytes are similar to memory B-cells bearing a mature immunophenotype and showing different activation and maturation states.1 CLL patients manifest distinct disease courses2, 3 and prognostic molecular markers identify patients at different risk: leukemic clones with few IgHV-gene mutations (U-CLL) but with many CD38+ or ZAP70+ B-cells, lead to an aggressive disease, chemotherapy resistance and is usually fatal; clones with mutated IgHV (M-CLL), few CD38+ or ZAP70+ B-cells, exhibit an indolent asymptomatic course which generally responds to therapy.4 The monoclonal nature of leukemic cells suggests the existence of genetic lesions in the CLL. Recurrent cytogenetic aberrations include: deletion at 13q14.3 (55% of cases) is associated with an indolent form and loss of miR-15a and miR-16-1 genes;5 deletions at 17p13 (7%) or 11q22-23 (18%) with consequent loss of TP53 at 17p, ATM and miR-34b/miR-34c at 11q are associated with a more aggressive form;6, 7 trisomy 12 (16%) is associated with an intermediate form of CLL. Nucleotide sequencing has discovered recurrent mutations in a number of genes such as TP53, NOTCH1, SF3B1, BIRC3 and ATM, which are indicative of derailed multiple pathways in CLL cells. It is also known that p53 mutations result in selective resistance to alkylating agents, such as fludarabine. In addition, treatment with DNA-damaging agents is correlated with an occurrence of p53 mutations in a clinical setting.8 Besides genetic lesions, pathogenic mechanisms may also represent survival signals arising from the microenvironment, through the B-cell receptor (BCR), integrins, chemokines and cytokine receptors, which allow CLL cells to actively proliferate and accumulate.9, 10
In addition, the T-cell leukemia-1 oncogene (TCL1) is expressed in almost all CLL patients and high-TCL1 protein levels correlate with the aggressive prognostic markers such as unmutated VH status, ZAP70 expression and chromosome 11q22-23 deletions.11 Accordingly, lower TCL1 levels are associated with a higher probability of positive response to chemoimmunotherapy.12 Animal models help to decipher pathogenic mechanisms of a disease and to evaluate the efficacy and mechanisms of novel therapies. A number of CLL mouse models have recently been reviewed by Simonetti et al.13 The authors compared different models and identified the Eμ-TCL1 transgenic mouse (TCL1-tg) as the most similar to aggressive type human CLL, in terms of immunophenotype, BCR repertoire and disease course. Importantly, TCL1 overexpression exhibits a 100% disease penetrance.
TCL1: Functions, Roles in CLL and Animal Models
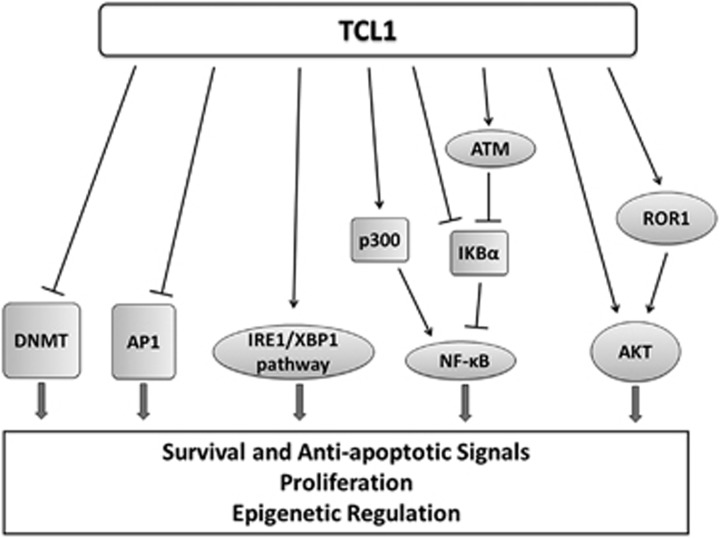
The TCL1 gene was discovered as the causative oncogene of T-prolymphocytic leukemia (T-PLL), where it is overexpressed in almost 100% of cases by a chromosomal translocation.14 TCL1 is also expressed in human seminomas,15 and in CD4+/CD56+ skin blastic tumors16 and in other B-cell lymphomas.17 TCL1 is a low-molecular weight protein and its first recognized function was the activation of phosphoinositide 3-kinase (PI3K) pathway, implicated in cell proliferation and survival (Figure 1), through direct binding with the AKT1/2 kinases.18 TCL1 binds to several other proteins and among these interacting proteins, the most relevant in CLL are: the receptor tyrosine kinase-like orphan receptor-1 (ROR1),19 the p300 transcription factor and the AP1 components FOS and JUN,20 the NFkB inhibitor alpha (IkBα),21 the XBP1 transcription factor22 and the DNA methyltransferases (DNMTs)23, 24 (Figure 2).
Figure 1.
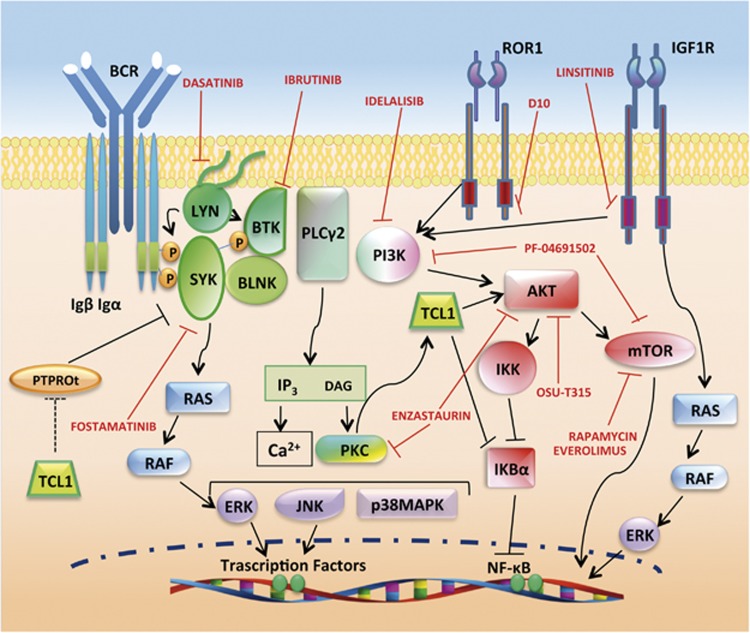
Targeting of BCR and PI3K/AKT signaling as therapeutic strategy in CLL. BCR signaling has a major role in the development of CLL. After antigen ligation on BCR, three main protein tyrosine kinases, LYN, SYK and BTK, are activated. PLC2 and PI3K are important downstream effectors of BCR signaling. PI3K activates downstream kinases such as AKT, which in turn induces NFkB and mTOR routes. Activation of PLC2 leads to the release of intracellular Ca2+ and activation of PKC, both of which are crucial for the activation of mitogen-activated protein kinases (MAPKs), such as ERK, c-JUN NH2-terminal kinase (JNK) and p38 MAPK and transcription factors, including NFκB. PI3K/AKT pathway can be induced also by tyrosine kinase receptors such as IGFR1 and ROR1. TCL1 enhances AKT signal and can sustain activation of the BCR downstream factors SYK and LYN by indirect inhibition of the phosphatase PTPROt. Red symbols and letters indicate new therapeutic targets as discussed in the text
Figure 2.
TCL1 function in CLL pathogenesis. TCL1 binds and regulates molecular factors implicated in proliferation, survival, inhibition of apoptosis and epigenetic regulation, thus contributing to CLL transformation
Physiologic functions of TCL1 protein mainly concern B-cell maturation, early embryonic development and stem cells regulation.15, 17, 25, 26, 27 Physiological roles of TCL1 was elucidated also by modifying TCL1 expression in mice. The knocking out (KO) of Tcl1 shows light impairment in B- and T-cell differentiation,28 while KO has stronger phenotypes in the embryonic stem cell proliferation/differentiation balance,29 embryo development15 and skin, especially in the hair follicle regeneration.27 This last KO phenotype is rescued when the strain is crossed to a TCL1 transgenic mouse specific for epidermal basal layer, under the Keratin14-promoter.27
The overexpression of TCL1 in transgenic animal models recapitulates faithfully leukemia of T-cell or B-cell origin according to the promoter used: the overexpression of TCL1 in T cells under Lck-promoter, recapitulates human T-PLL30 and the overexpression of TCL1 in B cells under the VH-promoter-IgH-Eμ-enhancer (TCL1-tg), recapitulates CLL.31, 32 As in humans, leukemia developed in the TCL1-tg model is characterized by clonal expansion of B cells with B220+/IgM+/CD5+ immunophenotype, unmutated IGHV, increased proliferation and enhanced AKT phosphorylation, which represent an aggressive form of CLL.31, 32, 33 Leukemic cells are firstly detected in the peritoneal cavity, at 2 months age; then tumor cells become detectable in peripheral blood (PB) and expand to the spleen (at 4 months) and bone marrow (BM; at 8 months).31 Tumor cells in TCL1 mice have wild-type (WT) p53 and initially respond to fludarabine treatment, after which drug resistance develops.34 Notably, the leukemic cells from a TCL1-tg donor can be transplanted by intra-peritoneum or intra-venous injection into syngeneic WT or immunodeficient mice (e.g., SCID) to accelerate the disease course and to generate a genetically homogeneous population of leukemic mice, which allows for the systematic study of novel therapies, without waiting for its natural course in non-transplanted animals. This technique can further be emphasized by serially adoptive cell transfer of leukemic cells into SCID mice. The repeated transplantations result in a clonal selection of B cells, which can be used for the analysis of particular conditions, for instance, BCR specificity.
Therapeutic Targets in TCL1-tg Mouse Models
Nowadays, many studies have used these TCL1-tg mouse models (Table 1), and then contribute to our present knowledge of CLL biology and generate fundamental data for the development of new therapeutic approaches aimed at the overcome of drug resistance and the curative treatment of CLL. The following is a review of the data obtained from these mice models.
Table 1. TCL1-tg animal models in B-CLL investigation.
| Function | Mouse model | Findings | Relevance | Ref. |
|---|---|---|---|---|
| TCL1 transgenic mouse models | TCL1-tg | TCL1 overexpression is causative for CLL | Mouse without UTRs of human TCL1 | Bichi et al.31 |
| TCL1FL-tg | MicroRNAs regulation | Mouse with UTRs of human TCL1 | Efanov et al.32 | |
| PI3K/AKT | TCL1-tg cells transplanted into C57bl/6 (i.v.) | AKT targeted therapy (OSU-T315) | Preclinical in vitro and in vivo | Liu et al.37 |
| TCL1-tg cells transplanted into B6/C3H (i.p.) | TCL1/AKT/mTOR pathway; mTOR targeted (rapamycin) | CLL pathogenesis; preclinical in vivo | Zanesi et al.38 | |
| TCL1-tg cells transplanted into C57bl/6 (i.p.) | Dual PI3K/mTOR inhibitor (PF-04691502) | Preclinical in vitro and in vivo | Blunt et al.39 | |
| TCL1-tg | Anti-IGFR1-targeted therapy (linsitinib) | Preclinical in vitro and in vivo; CT | Yaktapour et al.40 | |
| TCL1-tg crossed with ROR-Tg | ROR1/TCL1 complex; anti-ROR1 Ab therapy (D10) | Preclinical in vitro and in vivo | Widhopf et al.19 | |
| TCL1-tg crossed with Pkc null | PKC/TCL1/AKT route; PCK targeted (enzastaurin) | CLL pathogenesis; CT | Holler et al.41 | |
| NFkB | TCL1-tg | IkBα/TCL1 interaction and NFkB activation | Basic research; CLL pathogenesis | Guadio et al.21 |
| TCL1-tg cells transplanted into SCID (i.v.) | Anti-HSP90-targeted therapy (17-DMAG alvespimycin) | Preclinical in vivo; CT | Hertlein et al.42 | |
| TCL1-tg | XBP1/TCL1 interaction, BCR signaling and IRE1/XBP1 targeted therapy (A-106) | Basic research; CLL pathogenesis; preclinical in vitro and in vivo | Kriss et al.22 | |
| TCL1-tg | CLL therapy by ROS induction (Auranofin) | Preclinical in vivo; CT | Fiskus et al.43 | |
| Epigenetic regulation | TCL1-tg | TCL1/p50/HDAC1 complex and DNA methylation | Basic research; CLL pathogenesis | Chen et al.23 |
| TCL1-tg crossed with Id4+/− | ID4 repression and CLL progression | CLL etiology | Chen et al.44 | |
| TCL1-tg | TCL1/DNMT3A-B interaction and DNA methylation | Basic research; CLL pathogenesis | Palamarchuk et al.24 | |
| TCL1-tg | DNMT3A-B expression and leukemogenesis | CLL pathogenesis | Chen et al.45 | |
| TCL1-tg cells transplanted into SCID (i.v.) | HDAC inhibition (AR-42) | Preclinical in vivo; CT | Lucas et al.47 | |
| TCL1-tg crossed with p53 null | p53/miR15-16/Mcl1 axis | CLL pathogenesis | Liu et al.51 | |
| TCL1-tg mice | MDM2/p53/miR34a axis | CLL pathogenesis | Asslaber et al.96 | |
| TCL1FL-tg cells transplanted into FVB (i.p.) | miR-181b anti-leukemic activity | Preclinical in vitro and in vivo | Bresin et al.58 | |
| B-cell receptor | TCL1-tg crossed with dnRag1-Tg | TCL1 enhancement of BCR auto-reactivity | CLL pathogenesis | Nganga et al.65 |
| TCL1-tg | TCL1-induced PTPROt inhibition and BCR signal support | CLL pathogenesis | Motiwala et al.66 | |
| TCL1-tg crossed with PTPROt-Tg | PTPROt overexpression and CLL phenotype rescue | CLL pathogenesis | Motiwala et al.67 | |
| TCL1-tg | BCR resemblance to U-CLLs | CLL pathogenesis | Yan et al.33 | |
| TCL1-tg cells serially transferred into SCID | Clonal selection and antigen drive | CLL pathogenesis | Chen et al.69 | |
| TCL1-tg | Autonomous BCR signaling | CLL pathogenesis | Duhren-von minden et al.70 | |
| TCL1-tg | Intrinsic/extrinsic BCR activation | CLL pathogenesis | Lacovelli et al.71 | |
| Inhibitors of BCR signalosome | TCL1-tg | SYK targeted therapy (fostamatinib R788) | CLL pathogenesis; preclinical in vivo; CT | Suljagic et al.72 |
| TCL1-tg crossed with XID | Btk inactivation and CLL pathogenesis | CLL pathogenesis | Woyach et al.74 | |
| TCL1-tg | BTK targeted therapy (ibrutinib PCI-32765) | Preclinical in vivo; CT | Woyach et al.74 | |
| TCL1-tg cells serially transferred into SCID | Ibrutinib mechanism of action | Preclinical in vivo; CT | Ponader et al.76 | |
| TCL1-tg crossed with Hs1 null | Hs1 inactivation and CLL progression | CLL pathogenesis | Scielzo et al.78 | |
| TCL1-tg cells transplanted into C57bl/6 (i.p.) | Tyrosine kinase inhibitor mechanism of action (dasatinib) | Preclinical in vitro and in vivo; CT | ten Hacken et al.79 | |
| Leukemia-environment interplay | TCL1-tg cells serially transferred into SCID | CLL cells proliferation in LNs | CLL pathogenesis | Chen et al.69 |
| TCL1-tg | BCR signaling activation in LNs | CLL pathogenesis | Mittal et al.81 | |
| TCL1-tg crossed with Cxcr5 null | CXCL13/CXCR5 axis and CLL cells proliferation into LNs | CLL pathogenesis | Heinig et al.82 | |
| TCL1-tg | Stroma remodeling by CLL cells; LTβR targeted (LTβR-Ig) | CLL pathogenesis; preclinical in vivo | Heinig et al.82 | |
| TCL1-tg crossed with BAFF-Tg | BAFF/NFkB activation and CLL progression | CLL pathogenesis | Enzler et al.83 | |
| TCL1-tg crossed with APRIL-Tg | APRIL/TNFR activation and CLL progression | CLL pathogenesis | Lascano et al.84 | |
| TCL1-tg crossed with Mif null | MIF and activation of CLL cells and M2 macrophages | CLL pathogenesis | Reinart et al.85 | |
| TCL1-tg crossed with Cd44 null | MIF/CD44 interaction and BCR signaling | CLL pathogenesis | Fedorchenko et al.86 | |
| TCL1-tg | CD44 antibody-targeting (IM7) | Preclinical in vitro and in vivo | Fedorchenko et al.86 | |
| TCL1-tg | IL-10-mediated immunosuppression by CLL cells | CLL pathogenesis | Di Lillo et al.87 | |
| TCL1-tg and transplants into young TCL1-tg (i.v.) | T-cells expression profiling and tumor immune response | CLL pathogenesis | Gorgun et al.88 | |
| TCL1-tg and TCL1-tg cells transferred into C57bl/6 (i.p.) | T-cells skewing by CLL cells | CLL pathogenesis | Hofbauer et al.89 | |
| TCL1-tg | T-cells synapse and immunomodulation (lenalidomide) | CLL pathogenesis; preclinical in vivo; CT | Ramsay et al.90 | |
| TCL1-tg and transplants into young TCL1-tg (i.v.) | T-cells defects and PD-1/PD-L1 expression | CLL pathogenesis | McClanahan et al.93 | |
| TCL1-tg and TCL1-tg cells transferred into C57bl/6 (i.p.) | T-cells exhaustion and PD-1/PD-L1 blockade | CLL pathogenesis | Gassner et al.92 | |
| TCL1-tg cells transplanted into C57bl/6 (i.p.) | T-cell immune response rescue by PD-1/PD-L1 blockade | CLL pathogenesis; preclinical in vivo | McClanahan et al.91 | |
| TCL1-tg | Macrophages activation (αCD40 and CpG) | CLL pathogenesis; preclinical in vivo | Wu et al.94 | |
| TCL1-tg crossed with Tir8 null | Tir8 inactivation and CLL progression | CLL pathogenesis | Bertilaccio et al.95 |
CT, clinical trials; LNs, lymph nodes; UTRs, untranslated regions.
PI3K/AKT pathway
As mentioned above, TCL1 directly binds to AKT (Figure 1) which enhances its phosphorylation and nuclear translocation.18, 35 AKT, originally isolated from leukemia and lymphoma-prone mice cells, is over expressed in many tumors and is a key factor in CLL, integrating survival and proliferative signals from the environment through BCR, growth factors, integrins, chemokines and TNF receptors.36 Oral administration of the AKT-inhibitor OSU-T315 in mice transplanted with TCL1-tg cells was shown to prolong survival37 and allowed for the elucidation of the drug mechanism acitivity: OSU-T315 displaces AKT from lipid rafts, thus impairing AKT activation regardless of activating pathways.
One of the AKT downstream factors is the protein kinase mammalian target of rapamycin (mTOR), which controls cell growth, proliferation and survival. Zanesi et al. established a syngenic transplantation model, where leukemic cells isolated from a TCL1-tg donor spleen can be indefinitely maintained in vivo. Treatment of transplanted mice with the mTOR inhibitor rapamycin slowed leukemia and prolonged survival.38
Recently, exciting results have been published using the dual PI3K/mTOR inhibitor PF-04691502 on the TCL1-tg.39 PF-04691502 is a potent antitumor agent able to inhibit all the PI3K isoforms and both the mTOR complexes 1 and 2 (mTORC1/2). This is necessary to overcome redundancy between PI3K isoforms and mTORC2 positive feedback on AKT phosphorylation, which were observed with the Food and Drug Administration (FDA)-approved specific inhibitors of PI3Kδ (idelalisib) and mTORC1 (everolimus). The study revealed the pro-apoptotic activity of PF-04691502 through caspase activation in both human and mouse CLL cells. Moreover, in vivo treatment of TCL1-tg mice allowed for further insight into the clinical effects of PF-04691502: inhibition of CXCL12-mediated migration toward spleen and lymph nodes (LNs) induced redistribution of the tumor cells from lymphoid organs to the blood, followed by a marked reduction of tumor burden due to the cytotoxic activity of the drug. The splenic architecture was maintained in treated mice, although tumor cells were not completely eradicated, reflecting some resistant subpopulation.
Alternatively, the AKT pathway can be affected through inhibition of upstream signals. For example, the insulin-like growth factor-1 receptor (IGF1R) is overexpressed in CLL and mediates IGF1-induced activation of PI3K/AKT and mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) pathways. Inhibition of IGF1R by oral administration of linsitinib in TCL1-tg mice produced a significant decrease in malignant cells.40
Also, AKT phosphorylation can be enhanced through overexpression of ROR1. TCL1/ROR1 double-tg mice revealed the formation of complexes between the two factors and more aggressive leukemia due to increased proliferation and decreased apoptosis. In vivo administration of anti-ROR1 specific antibody, D10 revokes the potentiating effect of ROR1 on TCL1-tg cells, suggesting that this may be a novel therapeutic target in ROR1-expressing cancers.19
Finally, the overexpression of a member of the protein kinase C family, PKCβ, involved in signal transduction of growth factors and BCR and known to be a PI3K-independent AKT activator, correlates with poor-prognosis in CLL patients. The cross talk between TCL1/AKT and PKCβ was demonstrated in TCL1-tg mouse.41 In fact, genetic removal of PKCβ prevents CLL development in crossed TCL1-tg/Pkcβ−/− mice. However, TCL1 overexpression restores AKT signaling and B-cells production, which were abrogated in parental Pkcβ−/− mice, suggesting hierarchical order with TCL1/AKT acting downstream to PKCβ. Besides the examination of the PKC/TCL1/AKT route in CLL, these results led to the in vitro evaluation of enzastaurin, which inhibits both PKCβ and AKT, for therapeutic activity in CLL cells and subsequently to clinical trials (NCT00452257; Figure 1).
Nuclear factor kappa-light-chain-enhancer of activated B cells (NFkB)
Anti-apoptotic activity of NFkB, mainly activated through BCR signaling, is an important factor in CLL etiology and several pieces of evidence indicate that TCL1 is involved in NFkB activation (Figure 2). TCL1 interacts with the p300 transcription factor, enhancing its ability to activate NFkB in human B cells.20 Also, TCL1 can directly interact with ATM and the NFkB inhibitor IkBα, thus enhancing the phosphorylation and degradation of IkBα, with consequent activation of NFkB both in human and mouse CLL.21 In vivo treatment with the inhibitor of chaperone protein HSP90 (17-DMAG or alvespimycin), depletes IkB kinase complex subunits (IKK) and inhibits NFkB transcriptional activity, resulting in reduced expression of anti-apoptotic proteins BCL2 and MCL1 and caspase-dependent apoptosis.42 The 17-DMAG is being tested in phase I clinical trials in CLL patients (NCT01126502).
Endoplasmic reticulum (ER) stress response
ER stress response and IRE1/XBP1 pathway are aberrantly activated in human CLL and in TCL1-tg mice.22 TCL1 is directly involved in ER response by physical interaction with XBP1 and alteration of its transcriptional activity, resulting in constitutive activation of BCR signaling and influencing cross talk with other factors such as IRF4, BLIMP1 and AID. The importance of the IRE1/XBP1 pathway in leukemia maintenance is demonstrated by the observation that in vivo treatments with the specific inhibitor A-106 selectively induces apoptosis in TCL1-tg leukemic cells.22
On the other hand, prolonged activation of ER stress response by reactive oxygen species (ROS) may be exploited to induce cell death in CLL cells.43 Auranofin is a gold-containing drug, currently used in the rheumatoid arthritis treatment, which induces ROS levels. Oral administration of this FDA-approved compound to TCL1-tg mice markedly reduced leukemia expansion.43 Auranofin is currently investigated in phase II clinical trial for CLL therapy (NCT01419691).
Epigenetic regulation
DNA methylation and histone modifications shape gene expression without changes in DNA sequences. Abnormalities affecting these epigenetic events are implicated in pathological conditions, including cancer and leukemia.
DNA methyltransferases
Methylation at cytosine residues of DNA is realized by DNMTs and causes repression of transcription. TCL1 appears to be directly implicated in such epigenetic regulation, although, diverse reports describe opposite roles. Chen et al. found increased methylation levels in human as well as mouse CLL cells23 and identified a repressor complex, constituted by TCL1, p50 subunit of NFkB and histone deacetylase 1 (HDAC1), which induces transcriptional silencing before DNA methylation. For example, transcriptional repression of the inhibitor of DNA-binding protein-4 (ID4) in TCL1 mice results in acceleration of leukemia development.44 Conversely, Palamarchuk et al. described decreased methylation levels in TCL1-tg mice and in CLL patients.24 The authors found a strong interaction between TCL1 and de novo DNMT3A and 3B, with a drastic inhibition of enzymatic activity, suggesting a leukemogenesis mechanism involving inhibition of de novo methylation. However, Chen et al. observed the lack of DNMT3A and 3B in the genesis of the transformation of splenocytes from TCL1-tg mice, but increasing protein levels at later stages.45 Discrepancies in these data may reflect the complex timing of epigenetic changes with early event required for transformation and secondary events accumulating as a consequence of leukemogenesis. The analysis of the methylation status and transcriptional activation of these genes in Tcl1−/− mice might provide additional clues on this issue.
Deacetylases
Deacetylases (DACs) are a family of enzymes, subdivided into classes I and II, which remove acetyl groups from a broad range of proteins. Histone proteins are the most studied target of DACs, but transcription factors, chaperones and signaling components are as much as important. Owing to the regulatory effects on cell growth and differentiation, inhibitors of DACs possess antitumor activity and are currently used in diverse solid cancers therapy. For example, HDAC inhibitors facilitate the formation of an active death-inducing signaling complex, leading to the rapid activation of caspase-8.46 Preclinical studies have also demonstrated the pro-apoptotic efficacy for class I-specific DACs inhibitors (romidepsin) in CLL cells and are currently in clinical trials. A novel class I/II DACs inhibitor, AR-42 (OSU-HDAC42) has been tested in a transplanted model of TCL1-tg mouse47 and demonstrated to be more potent than vorinostat, a pan-DAC inhibitor approved for cutaneous T-cell lymphoma therapy. AR-42 treatment in these mice resulted in diminished tumor growth and prolonged survival and this compound is now in phase I clinical trial (NCT01129193).
MicroRNAs
MicroRNAs (miRNAs) are highly conserved small noncoding RNAs that function in post-transcriptional regulation. As miRNAs exhibit a variety of crucial functions related to cell growth, development and differentiation, dysregulation of miRNAs has been associated with diseases, including cancer. The first evidence for miRNAs involvement in human cancer was indeed from a study on CLL: 13q14.3 deletion, frequently observed in the patients, was shown to remove two microRNA genes, miR-15 and miR-16; their loss causes deregulation of BCL2.5, 48, 49, 50 Subsequent studies, with the support of the TCL1-tg mouse model, disclosed an involvement of p53-miR15/16-MCL1 axis in the regulation of resistance.51 Loss of p53 in TCL1-tg/p53−/− crossed mice causes a decrease of miR15/16 together with an upregulation of MCL1 and the consequent development of a more severe CLL. This association is fully consistent with the poor prognosis observed in CLL patients with the 17p deletion.
Extensive miRNA expression profiling on CLL cells from well-annotated cohorts of patients, identified miRNA signatures associated with prognosis and progression in B-CLL or with cytogenetic subgroups.52, 53, 54 Thus, inhibition of ‘oncomiRs' by anti-miRs or replacement of tumor suppressor miRNAs by mature sequence oligos (mimics) may represent novel therapeutic strategies. As CLL pathogenesis depends on several pathways and a single miRNA may have hundreds of mRNA targets, miRNA-based therapy might constitute a multibranched approach with higher potentialities than specific inhibitors.55
We have recently explored the therapeutic potential of miR-181b on the TCL1FL-tg mouse model.32 In addition, to its known function to modulate TCL1, MCL1 and BCL2,11, 56, 57 we observed the ability of this miRNA to downregulate important leukemia-promoting pathways such as AKT and ERK.58 Concordant with these findings, the treatment with miR-181b mimics reduces leukemic expansion and prolongs overall survival in TCL1FL-tg mice. This result represents the first in vivo demonstration for therapeutic efficacy of miRNA replacement strategy in CLL. Studies on additional miRNAs as well as combination with other conventional or innovative compounds might open new therapeutic possibilities, as suggested by the overcoming of resistance to the BCL2 inhibitors, through down-modulation of MCL1 and PI3K/AKT/mTOR pathway with siRNA or specific drugs.59
B-cell Receptor
CLL cells proliferate only in lymphoid organs, where accessory cells (i.e., nurse-like cells (NLC), T cells and stromal cells) provide the correct environment (mainly chemokines and cytokines) to sustain proliferation and survival of malignant cells9 (Figure 3). In this scenario, BCR signaling has a key role. A large amount of data, either from patients' studies or experimental models, supports the hypothesis of sustained antigen-dependent stimulation of BCR as a promoting event for clonal amplification of CLL cells.1, 10, 60 The sustained engagement of the BCR activates downstream targets such as NFkB, AKT and ERK,61, 62 which promote the expression of anti-apoptotic proteins, mainly BCL2 and MCL1.36, 63
Figure 3.
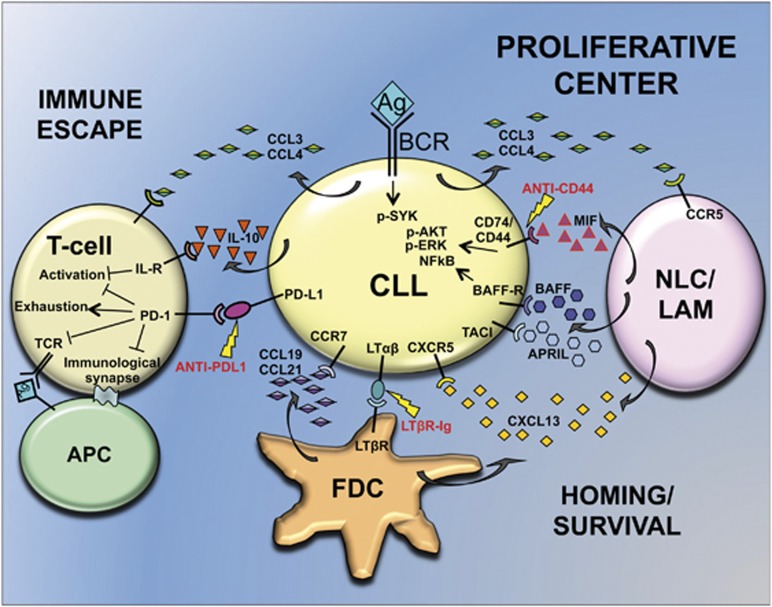
Leukemia-environment interplay within secondary lymphoid organs. A growth-promoting niche named proliferative center is the result of cross talk between tumor and accessory cells. CLL cells are attracted within LNs through chemokines (i.e., CXCL13 and CCL19/21) secreted by FDC and NLC, that are thought to correspond to leukemia-associated macrophages (LAM). Here, accessory cells provide a proliferative/survival milieu by secreting cytokines like MIF, BAFF and APRIL. CLL cells in turn, stimulate cytokine production by accessory cells, induce stromal cell differentiation through LTαβ/LTβR interaction and recruit NLCs and T cells through CCL3/4 secretion. An exhausted phenotype on T cells is also accomplished by CLL cells, through PD-L1/PD-1 interaction and IL-10 secretion to realize immune escape. Lightning symbols and red letters indicate new therapeutic targets as discussed in the text
BCR response is highly correlated with TCL1 levels in the CLL cells and with the formation of activation complexes at the BCR, by TCL1, AKT and ZAP70 kinases.64 Interplay between TCL1 and BCR activity was also suggested by studies on dnRag1/TCL1 double-tg.65 Enforced BCR auto-reactivity, due to RAG1 impairment, induces an indolent accumulation of CD5+ B cells, similar to monoclonal B-cell lymphocytosis; the TCL1 overexpression provides an additional lesion on this background, promoting progression to CLL. Moreover, TCL1 can sustain activation of the BCR downstream factors spleen tyrosine kinase (SYK) and LYN, by inhibiting AP1-dependent transcription of the phosphatase, PTPROt.20, 66 PTPROt ability to regulate BCR signaling components have been established using the TCL1/PTPROt double-tg mouse,67 which exhibits decreased splenic cells growth and increased lifespan. For example, the chemokine CCL3, which is upregulated by BCR signaling, is repressed in double-tg respect to TCL1-tg mice.
CLL patients exhibit stereotyped BCRs, with unique HCDR3 features and recurrent VH-DH-JH rearrangements, particularly in the IGHV unmutated cases.68 This observation led to the hypothesis that a subset of B-cells presenting stereotyped BCRs is selected by specific antigens such as auto antigens or microbial antigens and eventually these subsets get transformed by additional genetic abnormalities.1 TCL1-tg BCRs show HCDR3 characteristics and V(D)J rearrangements similar to U-CLLs.33 When B-lymphocytes reactive to auto antigen such as phosphatidylcholine (PtC) are serially transferred from TCL1-tg into SCID mice, a more aggressive leukemic clone is selected, showing increased reactivity with PtC over time. This finding is in agreement with an antigen selection and drive theory for leukemogenesis.69 An alternative hypothesis for BCR-induced CLL pathogenesis is proposed by Duhren-von Minden et al,70 who found an autonomous, ligand-independent BCR signaling in CLL samples from both human patients and TCL1-tg mice. This represents a new intriguing point of view, which does not exclude the hypothesis of extrinsic antigen involvement in CLL pathogenesis. Recently, the intrinsic/extrinsic types of BCR activation were examined in TCL1 mice.71 CLL is supported by an aberrant auto antigen-driven response and BCR interactions are positively selected by low-affinity auto antigens during leukemia development. Thus, the two BCR responses might have independent roles with autonomous BCR activation being essential for disease initiation and the low-affinity interaction with external auto antigens providing powerful co-stimulatory signals.
Altogether these data sustain the importance of therapeutic approaches based on targeting BCR cascade. The close resemblance between BCRs from U-CLL patients and TCL1-tg mice validates the use of this model for preclinical studies to test the efficacy of inhibitors that block specific components of BCR pathways.
Inhibitors of BCR Signalosome
Activation of BCR, either by extrinsic or intrinsic stimuli, transmits to membrane-associated signalosome proteins, composed of ‘proximal' kinases such as LYN, SYK, Bruton's tyrosine kinase (BTK), BLNK and PI3K. These, in turn, activate ‘distal' kinases, primarily ERK and AKT.10 Theoretically, each of the signalosome components might be a good candidate for targeted therapy, and increasing number of promising results have been achieved in this direction, some of which with the support of mouse models (Figure 1).
Spleen tyrosine kinase
An extensive study on BCR signaling in TCL1-tg mice was performed by Suljagic et al. using SYK inhibitors.72 The authors found that SYK and its direct substrate BLNK are constitutively phosphorylated in some mice. In any case, BCR engagement led to the activation of downstream signals such as ERK, AKT, GSK3 and FOXO and this activation was reversed by the SYK inhibitor R406. Leukemic cells from TCL1-tg mice treated with R406 prodrug fostamatinib (R788) showed reduced phosphorylation at SYK, BLNK and ERK, decreased proliferation and increased apoptosis. Survival of treated mice was extended, leading in some cases to the eradication of malignant clones.72 Fostamatinib is currently tested in phase II clinical trial for B-cell lymphomas and CLL (NCT00446095).
Bruton's tyrosine kinase
BTK inhibitor ibrutinib (PCI-32765) is one of the most recent FDA-approved drugs for refractory and aggressive 17p deleted form of CLL.73 XID mice bear a point mutation in Btk gene, which prevents its kinase activity.74 XID/TCL1 crossed mice have lower tumor burden in PB and superior survival than TCL1-tg. Similarly to Btk genetic inactivation, ibrutinib treatment induced an increased overall survival in treated mice, which was shown to be dependent from the inhibition of BCR-induced ERK phosphorylation.74 TCL1-tg mouse also allowed for better understanding ibrutinib mechanism of action and observations from patients. For example, during a phase I–II clinical trial on CLL patients (NCT01105247) it was noted a transient increase in lymphocytosis, followed by rapid decline during the days of treatment;75 this effect is probably caused by ibrutinib-mediated block of chemokine signaling and secretion, that inhibits CLL cells homing in lymphoid organs, and it was also observed in Eμ-TCL1 mice.76 Nevertheless, ibrutinib prolongs overall survival in these mice and induces significant decrease of CLL cells survival and proliferation demonstrating dual activity.76
LYN kinase and hematopoietic cell-specific LYN substrate-1 (HS1)
HS1 is phosphorylated by SYK and LYN kinases on BCR engagement and its hyperphosphorylation correlates with a worst outcome in CLL patients.77 TCL1-tg/Hs1−/− mice show accumulation of leukemic cells in all lymphoid tissues and shorter survival than TCL1-tg mice.78 As the genetic inactivation of Hs1 produces the same effects of hyperphosphorylation, Scielzo et al. conclude that phosphorylation has an inhibitory effect on HS1. The therapeutic potential for targeting LYN and HS1 in CLL has been assessed in TCL1-tg transplantable mouse model.79 In vitro treatment with the tyrosine kinase inhibitor, dasatinib, prevents HS1 and ERK phosphorylation, induces apoptosis and blocks CXCL12 chemotaxis and the interaction with stromal cells. In vivo administration reduces LYN activity and the percentages of leukemic cells and delays CLL progression. Thus, dasatinib mechanism of action involves both cell survival and migration to specific tissues. Furthermore, the study highlighted the importance to analyze LYN/HS1 axis in CLL patients; in fact, dasatinib variable results in clinical trials80 may depend on LYN/HS1 activation status.79 Many clinical trials on dasatinib treatment in CLL are currently ongoing or have been completed (clinicaltrials.gov).
Leukemia-Environment Interplay
Tumor microenvironment contributes to drug resistance and is crucial for the establishment of CLL proliferative centers, where malignant cells find optimal conditions to proliferate and survive. While environmental factors sustain tumor cells, tumor cells actively strive to establish a microenvironment in their favor, secreting chemokines that attract supportive cells and realizing complex strategies to escape immune surveillance.9 Thus, CLL-microenvironment interplay represents an interesting therapeutic target (Figure 3). The importance of microenvironment is also apparent in TCL1-tg as leukemic cells, obtained from a TCL1-tg donor and selected by serial transfer in SCID mice, display different response to BCR signaling based on cell residence:69 these cells actively proliferate in spleen and LN, but not in BM, peritoneal cavity and blood, despite their common origin (i.e., the spleen of the donor) and their clonal nature. Historically, BM has been considered the most important CLL microenvironment, where the stromal BM cells (SBMCs) secrete CXCL12 chemokine, and attract CXCR4-expressing CLL cells. However, increasing evidence support the relevance of LN. Gene expression analyses of leukemic cells in CLL patients, revealed a specific pattern of activation according to the tissue/organ source, such as PB, BM and LN.81 In particular, genes associated with signaling of BCR, BAFF/APRIL and some chemokines are overexpressed in LNs. High expression of phospho-SYK (BCR signaling) and phospho-p65 (NFkB pathway) are also found in LN from TCL1-tg mice.
Recently, Heinig et al. provided a clarifying dissection of the biological and molecular mechanisms underlying CLL cells homing to the LN.82 By crossing TCL1-tg with Cxcr5−/− mice, these authors demonstrated this receptor to be indispensable for recruitment and proliferation of CLL cells into the germinal center mediated by CXCL13-expressing follicular dendritic cells (FDCs). In addition, TCL1-tg leukemic cells themselves are able to induce stromal cell differentiation through cell-bound lymphotoxin (lymphotoxin-αβ (LTαβ)). The proposed model is a recruitment of CLL cells by FDCs, via CXCL13/CXCR5 interaction, into a growth-promoting stromal niche, which provides BCR stimulation and paracrine cytokines (mainly BAFF). Reciprocally, leukemic cells induce stroma remodeling and CXCL13 secretion in a loop controlled by LTαβ. This mechanism offers at least two therapeutic targets: the CXCL13-CXCR5 axis and the LTαβ interaction with its receptor (LTβR), as demonstrated by in vivo treatment with an LTβR-Ig fusion protein, which abrogates the paracrine feedback loop between leukemic and stromal cells and retards leukemia growth.82
Survival cytokines
Accessory cells in the leukemia microenvironment produce survival factors that inhibit spontaneous apoptosis. For example, BAFF and APRIL cytokines are highly expressed by NLCs. Overexpression of BAFF, by crossing TCL1-tg with BAFF-tg mice, induces faster development and more aggressive leukemia due to increased expression of anti-apoptotic proteins. The protective signal of BAFF is partly mediated by the NFkB pathway, also activated by TCL1.83 Similar results come from TCL1/APRIL double-tg.84 APRIL effects on CLL cells mainly rely on CD267/TACI TNF-receptor member and this is of therapeutic relevance as selective targeting of APRIL-TACI interaction may inhibit leukemic cells survival without affecting normal B-cells carrying another TNF-receptor member, namely CD269/BCMA.84
The macrophage migration inhibitory factor (MIF) is a proinflammatory cytokine acting on B cells, through the CD74/CD44 receptor complex. MIF loss in TCL1-tg/Mif−/− mice delays CLL development and reduces leukemic cells survival.85 In addition, MIF can also act as a chemokine, recruiting M2 macrophages in leukemic organs, which, differently to cytotoxic type M1, promote tumor progression and suppress the immune response. These lymphoma-associated macrophages might correspond to NLCs.85 Further studies revealed a fundamental role for CD44 coreceptor. The TCL1-tg/Cd44−/− crossed mice86 recapitulate the same phenotype of the TCL1-tg/Mif−/− mouse model,85 exhibiting a reduced phosphorylation of major BCR downstream kinases (SYK, AKT and ERK) and reduced MCL1, no longer inducible by BCR engagement. As CD44 genetic deletion is mimicked by antibody-based targeting,86 MIF/CD44 signal pathway represents a potential target for therapy, which might be investigated in the TCL1-tg mouse model.
Tumor immune suppression
Besides upregulation of survival and proliferative pathways by exploiting accessory cells, CLL cells induce also profound alteration of T-cell functions to realize tumor immune escape. Indeed, the rescue of immune surveillance by T cells is one of the greatest challenges for definitive CLL therapy. Immunosuppressive activity of CLL cells in the TCL1-tg mouse model has been well characterized and proved to be highly similar to human patients. Regulatory functions for CLL cells are supported by studies of Di Lillo et al.87 who showed that molecular mechanism underlying CLL-induced T-cell immunosuppression may be mediated by the cytokine IL-10. The competence of leukemic cells to express IL-10 either in human or mouse is normally found in B10 regulatory B cells, known to negatively regulate the immune response. In fact, mouse CLL cells suppress T-cell and monocyte/macrophage activation through IL-10-dependent pathways both in vitro and in vivo.
Gene expression profiling and protein expression analyses of CLL T-cells compared with WT or young TCL1-tg mice88 revealed that main changes involve proliferation, differentiation and cytokine/chemokine-response pathways, leading to functional impairment of antigen recognition, immune response, T-helper differentiation and cytotoxicity. Of note, using the transplantation technique a causal relationship between CLL, B-cells and T-cells changes has been found.88 Further rigorous transplantation experiments definitively demonstrated that CLL cells rapidly induce T-cell differentiation into memory compartment, probably through a tumor antigen-driven selection.89 Hence, T cells from human and TCL1-tg CLLs present a number of dysfunctions such as an impaired ability to form immunological synapse on conjugation with antigen-presenting cells, due to defects in cytoskeleton remodeling and in the recruitment of the T-cell receptor (TCR).90 The immunomodulatory drug lenalidomide is able to restore such defect and is extensively investigated in CLL clinical trials, especially in combination with other drugs (clinicaltrials.gov). Additional impairments in T-cell differentiation concern: the skewing from a naïve to an antigen-experienced memory compartment particularly in LNs,89 the reduction of CD4/CD8 ratios with the loss of central memory toward CD8+ effector memory pool,91 the exhausted phenotype characterized by poor effector function, the reduced cytokine production, the replicative senescence and finally the continued expression of inhibitory receptors.92 The latter is mainly represented by the receptors lymphocyte-activation gene 3 and programmed cell death 1 (PD-1) with PD-L1/2 ligands, which mediate dephosphorylation of signaling molecules downstream of the TCR. Actually, aberrant PD-1/PD-L1 signaling is involved in all the above mentioned T-cells dysfunctions.91, 92 In fact, normalization of the CD4/CD8 ratio, activation of T cells with restoring of effector cells cytotoxicity and immunological synapses formation, resolution of systemic inflammation and reversal of myeloid skewing are observed in the TCL1-tg mouse after anti-PD-1 antibody systemic treatment.93 Although a direct cytotoxic effect of the antibody on CLL cells was excluded, the treated mice showed a significant reduction of tumor load in disease-affected tissues, suggesting that PD-1/PD-L1 blockade is effective in tumor control restoration through immune effector functions.93 These remarkable results may represent an innovative strategy to decisively strengthen targeted therapy toward PI3K/mTOR or BCR signalosome inhibition.
Alternatively, to overcome the defective T-cell antitumor response, immunotherapy may be addressed to other cytotoxic effectors like macrophages.94 Treatment with CpG-containing oligodeoxynucleotides (CpG) synergizes with anti-CD40 mAb (αCD40) to activate macrophage antitumor response against human and mouse CLL cells, resulting in little or no tumor growth. In vitro analysis suggests that killing ability is partly mediated by nitric oxide synthesis. However, both human and mouse CLL cells express CD40 and toll-like receptor 9 (TLR9) and respond to treatment with αCD40 and CpG themselves, showing increased proliferation and modest protection from apoptosis. Thus, antitumor effects of activated macrophages must overcome the proliferative and anti-apoptotic effects of these stimuli on tumor cells.94 In agreement with this observation, the lack of TIR8 receptor which normally inhibits the signaling of TLRs, results in earlier and more aggressive CLL in TCL1-tg/Tir8−/− crossed mice.95 Nevertheless, ligands for TLRs other than TLR9 might provide better and more specific activation of macrophages, resulting in improvement of CD40-based immunotherapy.
Conclusions
TCL1-tg mouse model has been extensively investigated and the similarity with human U-CLL is striking. Thus, the TCL1-tg mouse represents a very useful model for a wide variety of studies, from basic molecular and cellular mechanisms, to fine dissection of the pathophysiology of CLL cells, including their interplay with tumor microenvironment and the preclinical evaluation of novel therapies. Further, a combination of two or more inhibitors, targeting pathways that cooperate to the insurgency, progression and relapse of CLL, is a topic that has never been tested in this mouse model and would be of considerable relevance for CLL therapy. TCL1 is not ubiquitously expressed being its expression, so far, limited to lymphoid, myeloid, cutaneous and embryonic cells.15, 17, 25, 26, 27 For this reason, therapies targeting TCL1 such as miRNAs or specific inhibitors of TCL1 protein in combination with pharmacological compounds currently used in the treatment of human B-CLL might represent also an interesting alternative to be tested on this animal model.
Acknowledgments
This study was supported by funds of the Ricerca Corrente of the Ministero della Salute and by the Italian Association for Cancer Research (AIRC) grant IG 15828 to GR; by the AIRC grant 5xmille n. 9980 and FAR grant 2012–2014 from the University of Ferrara to MN. We apologize for any missing references, which could be due to the complexity of the work and of the disease. We also thank Mrs. Ann Anthony for editorial assistance.
Glossary
- CLL
chronic lymphocytic leukemia
- TCL1
T-cell leukemia/lymphoma-1
- T-PLL
T-prolymphocytic leukemia
- KO
Knock out
- WT
wild type
- FDA
Food and Drug Administration
- BCR
B-cell receptor
- PI3K
phosphoinositide 3-kinase
- mTOR
mammalian target of rapamycin
- ER
Endoplasmic reticulum
- NFkB
nuclear factor kappa-light-chain-enhancer of activated B cells
- miR
miRNA microRNA
- BTK
Bruton's tyrosine kinase
- SYK
spleen tyrosine kinase
- LN
lymph node
- BM
bone marrow
- PB
peripheral blood
- FDCs
follicular dendritic cells
- LTαβ
lymphotoxin-αβ
- PD-1
programmed cell death 1
- ERK
extracellular signal-regulated kinase
- MIF
macrophage migration inhibitory factor
- PtC
phosphatidylcholine
- ROR1
receptor tyrosine kinase-like orphan receptor-1
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med 2005; 352: 804–815. [DOI] [PubMed] [Google Scholar]
- Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood 1975; 46: 219–234. [PubMed] [Google Scholar]
- Binet JL, Lepoprier M, Dighiero G, Charron D, D'Athis P, Vaugier G et al. A clinical staging system for chronic lymphocytic leukemia: prognostic significance. Cancer 1977; 40: 855–864. [DOI] [PubMed] [Google Scholar]
- Kay NE, O'Brien SM, Pettitt AR, Stilgenbauer S. The role of prognostic factors in assessing 'high-risk' subgroups of patients with chronic lymphocytic leukemia. Leukemia 2007; 21: 1885–1891. [DOI] [PubMed] [Google Scholar]
- Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA 2008; 105: 5166–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Raa GD, Moerland PD, Leeksma AC, Derks IA, Yigittop H, Laddach N et al. Assessment of p53 and ATM functionality in chronic lymphocytic leukemia by multiplex ligation-dependent probe amplification. Cell Death Dis 2015; 6: e1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1910–1916. [DOI] [PubMed] [Google Scholar]
- Sturm I, Bosanquet AG, Hermann S, Guner D, Dorken B, Daniel PT. Mutation of p53 and consecutive selective drug resistance in B-CLL occurs as a consequence of prior DNA-damaging chemotherapy. Cell Death Differ 2003; 10: 477–484. [DOI] [PubMed] [Google Scholar]
- Burger JA, Ghia P, Rosenwald A, Caligaris-Cappio F. The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood 2009; 114: 3367–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood 2011; 118: 4313–4320. [DOI] [PubMed] [Google Scholar]
- Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res 2006; 66: 11590–11593. [DOI] [PubMed] [Google Scholar]
- Browning RL, Geyer SM, Johnson AJ, Jelinek DF, Tschumper RC, Call TG et al. Expression of TCL-1 as a potential prognostic factor for treatment outcome in B-cell chronic lymphocytic leukemia. Leuk Res 2007; 31: 1737–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti G, Bertilaccio MT, Ghia P, Klein U. Mouse models in the study of chronic lymphocytic leukemia pathogenesis and therapy. Blood 2014; 124: 1010–1019. [DOI] [PubMed] [Google Scholar]
- Virgilio L, Narducci MG, Isobe M, Billips LG, Cooper MD, Croce CM et al. Identification of the TCL1 gene involved in T-cell malignancies. Proc Natl Acad Sci USA 1994; 91: 12530–12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narducci MG, Fiorenza MT, Kang SM, Bevilacqua A, Di Giacomo M, Remotti D et al. TCL1 participates in early embryonic development and is over expressed in human seminomas. Proc Natl Acad Sci USA 2002; 99: 11712–11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herling M, Teitell MA, Shen RR, Medeiros LJ, Jones D. TCL1 expression in plasmacytoid dendritic cells (DC2s) and the related CD4+ CD56+ blastic tumors of skin. Blood 2003; 101: 5007–5009. [DOI] [PubMed] [Google Scholar]
- Narducci MG, Pescarmona E, Lazzeri C, Signoretti S, Lavinia AM, Remotti D et al. Regulation of TCL1 expression in B- and T-cell lymphomas and reactive lymphoid tissues. Cancer Res 2000; 60: 2095–2100. [PubMed] [Google Scholar]
- Pekarsky Y, Koval A, Hallas C, Bichi R, Tresini M, Malstrom S et al. Tcl1 enhances Akt kinase activity and mediates its nuclear translocation. Proc Natl Acad Sci USA 2000; 97: 3028–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widhopf GF 2nd, Cui B, Ghia EM, Chen L, Messer K, Shen Z et al. ROR1 can interact with TCL1 and enhance leukemogenesis in Emu-TCL1 transgenic mice. Proc Natl Acad Sci USA 2014; 111: 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Palamarchuk A, Maximov V, Efanov A, Nazaryan N, Santanam U et al. Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proc Natl Acad Sci USA 2008; 105: 19643–19648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudio E, Spizzo R, Paduano F, Luo Z, Efanov A, Palamarchuk A et al. Tcl1 interacts with Atm and enhances NF-kappaB activation in hematologic malignancies. Blood 2012; 119: 180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriss CL, Pinilla-Ibarz JA, Mailloux AW, Powers JJ, Tang CH, Kang CW et al. Over expression of TCL1 activates the endoplasmic reticulum stress response: a novel mechanism of leukemic progression in mice. Blood 2012; 120: 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Raval A, Johnson AJ, Hertlein E, Liu TH, Jin VX et al. Epigenetic changes during disease progression in a murine model of human chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2009; 106: 13433–13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamarchuk A, Yan PS, Zanesi N, Wang L, Rodrigues B, Murphy M et al. Tcl1 protein functions as an inhibitor of de novo DNA methylation in B-cell chronic lymphocytic leukemia (CLL). Proc Natl Acad Sci USA 2012; 109: 2555–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorenza MT, Torcia S, Canterini S, Bevilacqua A, Narducci MG, Ragone G et al. TCL1 promotes blastomere proliferation through nuclear transfer, but not direct phosphorylation, of AKT/PKB in early mouse embryos. Cell Death Differ 2008; 15: 420–422. [DOI] [PubMed] [Google Scholar]
- Matoba R, Niwa H, Masui S, Ohtsuka S, Carter MG, Sharov AA et al. Dissecting Oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS One 2006; 1: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragone G, Bresin A, Piermarini F, Lazzeri C, Picchio MC, Remotti D et al. The Tcl1 oncogene defines secondary hair germ cells differentiation at catagen-telogen transition and affects stem-cell marker CD34 expression. Oncogene 2009; 28: 1329–1338. [DOI] [PubMed] [Google Scholar]
- Kang SM, Narducci MG, Lazzeri C, Mongiovi AM, Caprini E, Bresin A et al. Impaired T- and B-cell development in Tcl1-deficient mice. Blood 2005; 105: 1288–1294. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Miyazaki S, Ashida M, Tanaka T, Tashiro F, Miyazaki J. Functional analysis of Tcl1 using Tcl1-deficient mouse embryonic stem cells. PLoS One 2013; 8: e71645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgilio L, Lazzeri C, Bichi R, Nibu K, Narducci MG, Russo G et al. Deregulated expression of TCL1 causes T cell leukemia in mice. Proc Natl Acad Sci USA 1998; 95: 3885–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA 2002; 99: 6955–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efanov A, Zanesi N, Nazaryan N, Santanam U, Palamarchuk A, Croce CM et al. CD5+CD23+ leukemic cell populations in TCL1 transgenic mice show significantly increased proliferation and Akt phosphorylation. Leukemia 2010; 24: 970–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan XJ, Albesiano E, Zanesi N, Yancopoulos S, Sawyer A, Romano E et al. B cell receptors in TCL1 transgenic mice resemble those of aggressive, treatment-resistant human chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2006; 103: 11713–11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AJ, Lucas DM, Muthusamy N, Smith LL, Edwards RB, De Lay MD et al. Characterization of the TCL-1 transgenic mouse as a preclinical drug development tool for human chronic lymphocytic leukemia. Blood 2006; 108: 1334–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine J, Kunstle G, Obata T, Sha M, Noguchi M. The protooncogene TCL1 is an Akt kinase coactivator. Mol Cell 2000; 6: 395–407. [DOI] [PubMed] [Google Scholar]
- Longo PG, Laurenti L, Gobessi S, Sica S, Leone G, Efremov DG. The Akt/Mcl-1 pathway plays a prominent role in mediating antiapoptotic signals downstream of the B-cell receptor in chronic lymphocytic leukemia B cells. Blood 2008; 111: 846–855. [DOI] [PubMed] [Google Scholar]
- Liu TM, Ling Y, Woyach JA, Beckwith K, Yeh YY, Hertlein E et al. OSU-T315: a novel targeted therapeutic that antagonizes AKT membrane localization and activation of chronic lymphocytic leukemia cells. Blood 2015; 125: 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanesi N, Aqeilan R, Drusco A, Kaou M, Sevignani C, Costinean S et al. Effect of rapamycin on mouse chronic lymphocytic leukemia and the development of nonhematopoietic malignancies in Emu-TCL1 transgenic mice. Cancer Res 2006; 66: 915–920. [DOI] [PubMed] [Google Scholar]
- Blunt MD, Carter MJ, Larrayoz M, Smith LD, Aguilar-Hernandez M, Cox KL et al. The PI3K/mTOR inhibitor PF-04691502 induces apoptosis and inhibits microenvironmental signaling in CLL and the Emicro-TCL1 mouse model. Blood 2015; 125: 4032–4041. [DOI] [PubMed] [Google Scholar]
- Yaktapour N, Ubelhart R, Schuler J, Aumann K, Dierks C, Burger M et al. Insulin-like growth factor-1 receptor (IGF1R) as a novel target in chronic lymphocytic leukemia. Blood 2013; 122: 1621–1633. [DOI] [PubMed] [Google Scholar]
- Holler C, Pinon JD, Denk U, Heyder C, Hofbauer S, Greil R et al. PKCbeta is essential for the development of chronic lymphocytic leukemia in the TCL1 transgenic mouse model: validation of PKCbeta as a therapeutic target in chronic lymphocytic leukemia. Blood 2009; 113: 2791–2794. [DOI] [PubMed] [Google Scholar]
- Hertlein E, Wagner AJ, Jones J, Lin TS, Maddocks KJ, Towns WH 3rd et al. 17-DMAG targets the nuclear factor-kappaB family of proteins to induce apoptosis in chronic lymphocytic leukemia: clinical implications of HSP90 inhibition. Blood 2010; 116: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskus W, Saba N, Shen M, Ghias M, Liu J, Gupta SD et al. Auranofin induces lethal oxidative and endoplasmic reticulum stress and exerts potent preclinical activity against chronic lymphocytic leukemia. Cancer Res 2014; 74: 2520–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Claus R, Lucas DM, Yu L, Qian J, Ruppert AS et al. Silencing of the inhibitor of DNA binding protein 4 (ID4) contributes to the pathogenesis of mouse and human CLL. Blood 2011; 117: 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Sherman MH, Hertlein E, Johnson AJ, Teitell MA, Byrd JC et al. Epigenetic alterations in a murine model for chronic lymphocytic leukemia. Cell Cycle 2009; 8: 3663–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, MacFarlane M, Harper N, Wheat LM, Dyer MJ, Cohen GM. Histone deacetylase inhibitors potentiate TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in lymphoid malignancies. Cell Death Differ 2004; 11(Suppl 2): S193–S206. [DOI] [PubMed] [Google Scholar]
- Lucas DM, Alinari L, West DA, Davis ME, Edwards RB, Johnson AJ et al. The novel deacetylase inhibitor AR-42 demonstrates pre-clinical activity in B-cell malignancies in vitro and in vivo. PLoS One 2010; 5: e10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2002; 99: 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Croce CM. Role of miR-15/16 in CLL. Cell Death Differ 2015; 22: 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ 2010; 17: 215–220. [DOI] [PubMed] [Google Scholar]
- Liu J, Chen G, Feng L, Zhang W, Pelicano H, Wang F et al. Loss of p53 and altered miR15-a/16-1short right arrowMCL-1 pathway in CLL: insights from TCL1-Tg:p53(−/−) mouse model and primary human leukemia cells. Leukemia 2014; 28: 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005; 353: 1793–1801. [DOI] [PubMed] [Google Scholar]
- Visone R, Rassenti LZ, Veronese A, Taccioli C, Costinean S, Aguda BD et al. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood 2009; 114: 3872–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini M, Cutrona G, Bassi C, Fabris S, Zagatti B, Colombo M et al. MicroRNAome expression in chronic lymphocytic leukemia: comparison with normal B-cell subsets and correlations with prognostic and clinical parameters. Clin Cancer Res 2014; 20: 4141–4153. [DOI] [PubMed] [Google Scholar]
- Nana-Sinkam SP, Croce CM. MicroRNA in chronic lymphocytic leukemia: transitioning from laboratory-based investigation to clinical application. Cancer Genet Cytogenet 2010; 203: 127–133. [DOI] [PubMed] [Google Scholar]
- Ouyang YB, Lu Y, Yue S, Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion 2012; 12: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Veronese A, Rassenti LZ, Balatti V, Pearl DK, Acunzo M et al. miR-181b is a biomarker of disease progression in chronic lymphocytic leukemia. Blood 2011; 118: 3072–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresin A, Callegari E, D'Abundo L, Cattani C, Bassi C, Zagatti B et al. miR-181b as a therapeutic agent for chronic lymphocytic leukemia in the Emicro-TCL1 mouse model. Oncotarget 2015; 6: 19807–19818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary GS, Al-Harbi S, Mazumder S, Hill BT, Smith MR, Bodo J et al. MCL-1 and BCL-xL-dependent resistance to the BCL-2 inhibitor ABT-199 can be overcome by preventing PI3K/AKT/mTOR activation in lymphoid malignancies. Cell Death Dis 2015; 6: e1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger JA, Chiorazzi N. B cell receptor signaling in chronic lymphocytic leukemia. Trends Immunol 2013; 34: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal A, Pastore RD, Asgary Z, Keller SA, Cesarman E, Liou HC et al. Survival of leukemic B cells promoted by engagement of the antigen receptor. Blood 2001; 98: 3050–3057. [DOI] [PubMed] [Google Scholar]
- Barragan M, Bellosillo B, Campas C, Colomer D, Pons G, Gil J. Involvement of protein kinase C and phosphatidylinositol 3-kinase pathways in the survival of B-cell chronic lymphocytic leukemia cells. Blood 2002; 99: 2969–2976. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 2005; 102: 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herling M, Patel KA, Weit N, Lilienthal N, Hallek M, Keating MJ et al. High TCL1 levels are a marker of B-cell receptor pathway responsiveness and adverse outcome in chronic lymphocytic leukemia. Blood 2009; 114: 4675–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nganga VK, Palmer VL, Naushad H, Kassmeier MD, Anderson DK, Perry GA et al. Accelerated progression of chronic lymphocytic leukemia in Emu-TCL1 mice expressing catalytically inactive RAG1. Blood 2013; 121: 3855–3866, S3851-S3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala T, Zanesi N, Datta J, Roy S, Kutay H, Checovich AM et al. AP-1 elements and TCL1 protein regulate expression of the gene encoding protein tyrosine phosphatase PTPROt in leukemia. Blood 2011; 118: 6132–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala T, Kutay H, Zanesi N, Frissora FW, Mo X, Muthusamy N et al. PTPROt-mediated regulation of p53/Foxm1 suppresses leukemic phenotype in a CLL mouse model. Leukemia 2015; 29: 1350–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest 1998; 102: 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Batliwalla F, Holodick NE, Yan XJ, Yancopoulos S, Croce CM et al. Autoantigen can promote progression to a more aggressive TCL1 leukemia by selecting variants with enhanced B-cell receptor signaling. Proc Natl Acad Sci USA 2013; 110: E1500–E1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhren-von Minden M, Ubelhart R, Schneider D, Wossning T, Bach MP, Buchner M et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 2012; 489: 309–312. [DOI] [PubMed] [Google Scholar]
- Iacovelli S, Hug E, Bennardo S, Duehren-von Minden M, Gobessi S, Rinaldi A et al. Two types of BCR interactions are positively selected during leukemia development in the Emu-TCL1 transgenic mouse model of CLL. Blood 2015; 125: 1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suljagic M, Longo PG, Bennardo S, Perlas E, Leone G, Laurenti L et al. The Syk inhibitor fostamatinib disodium (R788) inhibits tumor growth in the Emu- TCL1 transgenic mouse model of CLL by blocking antigen-dependent B-cell receptor signaling. Blood 2010; 116: 4894–4905. [DOI] [PubMed] [Google Scholar]
- Byrd JC, Brown JR, O'Brien S, Barrientos JC, Kay NE, Reddy NM et al. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N Engl J Med 2014; 371: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyach JA, Bojnik E, Ruppert AS, Stefanovski MR, Goettl VM, Smucker KA et al. Bruton's tyrosine kinase (BTK) function is important to the development and expansion of chronic lymphocytic leukemia (CLL). Blood 2014; 123: 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med 2013; 369: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponader S, Chen SS, Buggy JJ, Balakrishnan K, Gandhi V, Wierda WG et al. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood 2012; 119: 1182–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scielzo C, Ghia P, Conti A, Bachi A, Guida G, Geuna M et al. HS1 protein is differentially expressed in chronic lymphocytic leukemia patient subsets with good or poor prognoses. J Clin Invest 2005; 115: 1644–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scielzo C, Bertilaccio MT, Simonetti G, Dagklis A, ten Hacken E, Fazi C et al. HS1 has a central role in the trafficking and homing of leukemic B cells. Blood 2010; 116: 3537–3546. [DOI] [PubMed] [Google Scholar]
- ten Hacken E, Scielzo C, Bertilaccio MT, Scarfo L, Apollonio B, Barbaglio F et al. Targeting the LYN/HS1 signaling axis in chronic lymphocytic leukemia. Blood 2013; 121: 2264–2273. [DOI] [PubMed] [Google Scholar]
- Amrein PC, Attar EC, Takvorian T, Hochberg EP, Ballen KK, Leahy KM et al. Phase II study of dasatinib in relapsed or refractory chronic lymphocytic leukemia. Clin Cancer Res 2011; 17: 2977–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal AK, Chaturvedi NK, Rai KJ, Gilling-Cutucache CE, Nordgren TM, Moragues M et al. Chronic lymphocytic leukemia cells in a lymph node microenvironment depict molecular signature associated with an aggressive disease. Mol Med 2014; 20: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinig K, Gatjen M, Grau M, Stache V, Anagnostopoulos I, Gerlach K et al. Access to follicular dendritic cells is a pivotal step in murine chronic lymphocytic leukemia B-cell activation and proliferation. Cancer Discov 2014; 4: 1448–1465. [DOI] [PubMed] [Google Scholar]
- Enzler T, Kater AP, Zhang W, Widhopf GF 2nd, Chuang HY, Lee J et al. Chronic lymphocytic leukemia of Emu-TCL1 transgenic mice undergoes rapid cell turnover that can be offset by extrinsic CD257 to accelerate disease progression. Blood 2009; 114: 4469–4476. [DOI] [PubMed] [Google Scholar]
- Lascano V, Guadagnoli M, Schot JG, Luijks DM, Guikema JE, Cameron K et al. Chronic lymphocytic leukemia disease progression is accelerated by APRIL-TACI interaction in the TCL1 transgenic mouse model. Blood 2013; 122: 3960–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinart N, Nguyen PH, Boucas J, Rosen N, Kvasnicka HM, Heukamp L et al. Delayed development of chronic lymphocytic leukemia in the absence of macrophage migration inhibitory factor. Blood 2013; 121: 812–821. [DOI] [PubMed] [Google Scholar]
- Fedorchenko O, Stiefelhagen M, Peer-Zada AA, Barthel R, Mayer P, Eckei L et al. CD44 regulates the apoptotic response and promotes disease development in chronic lymphocytic leukemia. Blood 2013; 121: 4126–4136. [DOI] [PubMed] [Google Scholar]
- DiLillo DJ, Weinberg JB, Yoshizaki A, Horikawa M, Bryant JM, Iwata Y et al. Chronic lymphocytic leukemia and regulatory B cells share IL-10 competence and immunosuppressive function. Leukemia 2013; 27: 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgun G, Ramsay AG, Holderried TA, Zahrieh D, Le Dieu R, Liu F et al. E(mu)-TCL1 mice represent a model for immunotherapeutic reversal of chronic lymphocytic leukemia-induced T-cell dysfunction. Proc Natl Acad Sci USA 2009; 106: 6250–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer JP, Heyder C, Denk U, Kocher T, Holler C, Trapin D et al. Development of CLL in the TCL1 transgenic mouse model is associated with severe skewing of the T-cell compartment homologous to human CLL. Leukemia 2011; 25: 1452–1458. [DOI] [PubMed] [Google Scholar]
- Ramsay AG, Johnson AJ, Lee AM, Gorgun G, Le Dieu R, Blum W et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest 2008; 118: 2427–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan F, Riches JC, Miller S, Day WP, Kotsiou E, Neuberg D et al. Mechanisms of PD-L1/PD-1 mediated CD8 T-cell dysfunction in the context of aging-related immune defects in the Emu-TCL1 CLL mouse model. Blood 2015; 126: 212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassner FJ, Zaborsky N, Catakovic K, Rebhandl S, Huemer M, Egle A et al. Chronic lymphocytic leukaemia induces an exhausted T cell phenotype in the TCL1 transgenic mouse model. Br J Haematol 2015; 170: 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan F, Hanna B, Miller S, Clear AJ, Lichter P, Gribben JG et al. PD-L1 Checkpoint Blockade Prevents Immune Dysfunction and Leukemia Development in a Mouse Model of Chronic Lymphocytic Leukemia. Blood 2015; 126: 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu QL, Buhtoiarov IN, Sondel PM, Rakhmilevich AL, Ranheim EA. Tumoricidal effects of activated macrophages in a mouse model of chronic lymphocytic leukemia. J Immunol 2009; 182: 6771–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertilaccio MT, Simonetti G, Dagklis A, Rocchi M, Rodriguez TV, Apollonio B et al. Lack of TIR8/SIGIRR triggers progression of chronic lymphocytic leukemia in mouse models. Blood 2011; 118: 660–669. [DOI] [PubMed] [Google Scholar]
- Asslaber D, Pinon JD, Seyfried I, Desch P, Stocher M, Tinhofer I et al. microRNA-34a expression correlates with MDM2 SNP309 polymorphism and treatment-free survival in chronic lymphocytic leukemia. Blood 2010; 115: 4191–4197. [DOI] [PubMed] [Google Scholar]