Abstract
Sepsis-induced skeletal muscle atrophy and weakness are due in part to decreased mTORC1-mediated protein synthesis and increased proteolysis via the autophagy-lysosomal system and ubiquitin-proteasome pathway. The REDD1 (regulated in development and DNA damage-1) protein is increased in sepsis and can negatively regulate mTORC1 activity. However, the contribution of REDD1 to the sepsis-induced change in muscle protein synthesis and degradation has not been determined. Sepsis was produced by cecal ligation and puncture in female REDD1−/− or wild-type (WT) mice, and end points were assessed 24 h later in gastrocnemius; time-matched, pair-fed controls of each genotype were included. Sepsis increased REDD1 protein 300% in WT mice, whereas REDD1 was absent in REDD1−/− muscle. Sepsis decreased protein synthesis and phosphorylation of downstream targets of mTORC1 (S6K1 Thr389, rpS6 Ser240/244, 4E-BP1 Ser65) in WT but not REDD1−/− mice. However, Akt and PRAS40 phosphorylation was suppressed in both sham and septic muscle from REDD1−/− mice despite unaltered PDK1, PP2A, or TSC2 expression. Sepsis increased autophagy as indicated by decreased ULK1 Ser757 phosphorylation and p62 abundance and increased LC3B-II/I in WT mice, whereas these changes were absent in septic REDD1−/− mice. Conversely, REDD1 deletion did not prevent the sepsis-induced decrease in IGF-I mRNA or the concomitant increase in IL-6, TNFα, MuRF1, and atrogin1 mRNA expression. Lastly, 5-day survival in a separate set of septic mice did not differ between WT and REDD1−/− mice. These data highlight the central role of REDD1 in regulating both protein synthesis and autophagy in skeletal muscle during sepsis.
Keywords: rgulated in development and DNA damage-1, mechanistic target of rapamycin complex 1, protein synthesis, autophagy, cecal ligation and puncture, proteolysis, critical illness
skeletal muscle atrophy is a debilitating consequence of sepsis and critical illness that increases the length of hospitalization and mortality (19). Loss of muscle mass and strength during sepsis results from the prolonged imbalance between rates of protein synthesis and degradation caused by several factors, including increased proinflammatory cytokine signaling, anabolic resistance, and excess glucocorticoids (1, 32, 48). Protein synthesis is controlled predominantly by the mechanistic (a.k.a. mammalian) target of rapamycin complex 1 (mTORC1), which phosphorylates 70-kDa ribosomal protein S6 kinase-1 (S6K1) and eukaryotic initiation factor (eIF) 4E-binding protein-1 (4E-BP1) to increase translation initiation and peptide chain elongation within muscle (13, 25). mTORC1 represents a central regulatory factor in the integration of various metabolic signals, including those induced by energy stress [AMP-activated protein kinase (AMPK), regulated in development and DNA damage-1 (REDD-1)] and growth factors (insulin/Akt). Upon activation of the insulin or insulin-like growth factor (IGF)-I receptor and the phosphoinositide 3-kinase signaling cascade, Akt is phosphorylated at Thr308 by 3-phosphoinositide-dependent protein kinase 1 (PDK1), and this in combination with mTORC2-mediated phosphorylation of Akt on Ser473 (23) results in its activation (2). Akt may stimulate protein synthesis via phosphorylation and inhibition of tuberous sclerosis complex (TSC)1/2, leading to Rheb-GTP loading and mTORC1 activation (26) and/or inactivation of glycogen synthase kinase-3β, thus releasing its inhibition of eIF2B (27). Impairment of mTORC1 kinase activity is a well-characterized phenomenon in skeletal muscle in response to sepsis and a causal mechanism in the loss of lean body mass (18, 32).
Upregulation of the two primary proteolytic pathways, autophagy and the ubiquitin-proteasome system (UPS), also contributes to the loss of muscle mass during sepsis (40, 49, 53). Appropriate autophagic balance is central to skeletal muscle mass and function, as it removes or recycles damaged cellular components, and deletion of autophagy genes (i.e., Atg7, Atg5) results in myopathy or is embryonically lethal (29, 36, 45). Autophagy is regulated predominantly by Akt/forkhead box O3 (FoxO3) signaling, whereas mTORC1-mediated phosphorylation of Unc-51-like autophagy-activating kinase 1 (ULK1) contributes to a lesser extent (54). Akt/FOXO signaling represents a link between protein synthesis, autophagy, and the UPS, the latter of which appears to be responsible for the breakdown of the majority of myofibrillar proteins (34, 54). Akt-mediated phosphorylation of FOXO prevents its nuclear translocation, thereby attenuating the transcription of muscle RING finger protein-1 (MuRF1) and muscle atrophy F-box 1/atrogin1, two genes tightly linked to UPS induction. Deletion of either of these atrogenes attenuates muscle loss following denervation, supporting the critical role of the UPS in atrophy (3).
As a negative regulator of mTORC1 activity, REDD1 is increased by cellular and catabolic stressors, including sepsis, glucocorticoids, starvation, AMPK activation, alcohol intoxication, DNA damage, hypoxia, and reactive oxygen species (7, 16, 31, 38). The mechanism by which REDD1 suppresses mTORC1 remains somewhat speculative, with recent in vitro work proposing that induction of PP2A-mediated dephosphorylation of Akt Thr308 leads to decreased TSC2 phosphorylation and enhanced Rheb-GTPase activity (i.e., increased Rheb-GDP loading) and, therefore, mTORC1 inhibition (10).
Although previously we have reported that sepsis increases REDD1 (48), the causal contribution of this increase to the reduction in mTORC1 signaling and protein synthesis is unknown. Therefore, mice with a global deletion of REDD1 were used to test the hypothesis that elimination of REDD1 would ameliorate the sepsis-induced inhibition of mTORC1 and thereby antagonize the impairment in protein synthesis and contribute to improved survival.
METHODS
Animals.
Female 15- to 16-wk-old wild-type (WT; B6/129F1) or REDD1 knockout (REDD1−/−) mice were used in all experiments. The REDD1−/− breeding pair was obtained from Dr. David Williamson (State University of New York at Buffalo), whereas Lexicon Genetics (The Woodlands, TX) generated the original REDD1−/− mice. The REDD1−/− mice (and WT controls) were bred in the Pennsylvania (Penn) State College of Medicine animal facility. Permission for their breeding and use was granted by Dr. Elena Feinstein (Quark Pharmaceuticals, Fremont, CA). To increase sample size, additional age-matched WT mice of an identical background (B6/129F1) were purchased from Charles River Laboratories (Wilmington, MA) and acclimated to the animal facility at the College of Medicine for >1 wk prior to experimental use. All mice were housed in shoebox cages with corn cob bedding under controlled environmental conditions (12:12-h light-dark cycle, 21–22°C, 30–70% humidity) and were provided Teklad Global no. 8604 diet (Harlan-Teklad, Boston, MA) and water ad libitum until the start of the experiment. The stage of estrous when surgery was performed was not controlled for in the female mice. Body composition (fat and lean mass) was assessed noninvasively by a single investigator in a blinded manner on conscious animals, using a 1H-NMR analyzer (Bruker LF90 Proton-NMR Minispec; Bruker Optics, The Woodlands, TX) prior to surgery. All experimental procedures were performed in accordance with the National Institutes of Health guidelines for the use of experimental animals and were approved by the Institutional Animal Care and Use Committee of the Penn State College of Medicine.
Induction of polymicrobial peritonitis.
Cecal ligation and puncture (CLP) was used to produce sepsis and was performed as described previously (48). Mice were anesthetized (isoflurane, 2–3% in O2 with 1.5% maintenance), and the abdomen was shaved and aseptically prepared. An abdominal incision was made, the distal two-thirds of the cecum was ligated, punctured through and through (i.e., 2 holes) with a 23-G needle, and a small amount of cecal material was expressed before the cecum was replaced, and the incision was closed with 5-0 surgical suture. The amount of cecal material expressed was not measured exactly but was approximately the same for all mice by visual inspection; CLP was performed by a single experienced surgeon blinded to animal genotype. Control mice underwent sham surgery, which included an identical incision and closure, but without ligation or puncture of the cecum. Following surgery, all animals received a subcutaneous injection of warm sterile 0.9% saline (1 ml). All mice were individually housed following surgery and given access to water until euthanization. Because sepsis acutely decreases food consumption, food was withheld from all mice in the acute sepsis study so that the nutritional status was identical across groups (46); hence, observed differences in protein synthesis and signal transduction are not due to differences in the nutritional state of the mice. The 24-h survival rate did not differ between genotypes, as survival was 100% in septic WT mice and 93% in septic REDD1 mice. Approximately 18–24 h after the induction of sepsis, mice were deeply anesthetized via isoflurane (4–5% in O2), and the gastrocnemius and plantaris muscle complex (referred to as muscle from here on) was excised and immediately frozen between aluminum blocks precooled to the temperature of liquid nitrogen. Blood was collected from the vena cava in heparinized syringes and centrifuged (10,000 g for 10 min) for isolation of plasma. Both frozen tissue and plasma were stored at −80°C until analysis.
Protein synthesis.
Global protein synthesis was assessed in the gastrocnemius and plantaris muscle complex in WT and REDD1−/− mice. All mice were injected intraperitoneally with l-[2,3,4,5,6-3H]phenylalanine [Phe; 150 mM, 30 μCi/ml, 0.5 ml] 15 min before tissue collection. High-performance liquid chromatography was used for the measurement of specific radioactivity of plasma Phe levels in the supernatant from trichloroacetic acid-treated plasma extracts. The global rate of [3H]Phe incorporation into skeletal muscle protein was assessed as described previously by our laboratory (51).
Survival study.
To determine whether global deletion of the REDD1 gene impacts survival, sepsis was induced in a second cohort of age-matched, WT (n = 15), and REDD1−/− (n = 15) female mice. Conditions were identical to those described above, except that food was provided ad libitum after surgery. All mice were monitored three to four times per day, and lethality was recorded every 12 h by the same investigator in a blinded manner. Any mouse that was determined to be moribund was immediately euthanized instead of waiting for spontaneous death (9, 41). At 5 days (120 h) after CLP, all surviving mice underwent measurement of body composition (1H-NMR) prior to anesthetization and removal of muscles for the determination of wet weight.
AICAR injection.
Based on data from our initial experiments, a separate study was performed to determine specifically whether the sepsis-induced decrease in muscle protein synthesis in REDD1−/− mice represented the maximal possible physiological decrement (e.g., a “floor effect”). To this end, three groups of fasted female mice (n = 6/group) were used: saline-injected WT as well as CLP-treated REDD1−/− mice injected with saline or 5-aminoimidazole-4-carboxamide-1-β-d-ribonucleoside (AICAR). Mice were injected subcutaneously with AICAR (0.5 mg/g body wt; Toronto Research Chemicals, Toronto, ON, Canada) to stimulate AMPK, and muscle was collected 60 min later; time-matched septic mice were injected with an equal volume of vehicle (sterile 0.9% saline). The dose of AICAR and the timing of the experiment were selected because previous studies indicated a significant reduction in muscle protein synthesis (42). In this series, in vivo protein synthesis was assessed using the SUnSET method and antibody against puromycin (Kerafast, Boston, MA), as described previously (20). Mice were injected intraperitoneally with 0.04 μmol/g body wt of puromycin dissolved in sterile saline 30 min prior to the removal of the plantaris muscle. Western blotting procedures were performed as descried below to visualize puromycin incorporation.
Western blotting.
Half of each muscle (50–90 mg) was homogenized using a mechanical homogenizer in 10 volumes of ice-cold buffer consisting of (in mmol/l) 20 HEPES, 2 EGTA, 0.2 EDTA, 100 potassium chloride, 50 β-glycerophosphate, 50 sodium fluoride, 0.5 sodium orthovanadate, 1 benzamidine, and 0.1 phenylmethanesulfonylfluoride. Protein concentration was quantified using the Bio-Rad Protein Assay Dye reagent (Hercules, CA), and SDS-PAGE was carried out using equal amounts of total protein per sample loaded onto 4–20% gradient gels (Bio-Rad, Hercules, CA). Following transfer to PVDF and prior to blocking, all membranes were treated with Ponceau-S stain and scanned so that sample loading could be verified and compared. Following washes in TBST, membranes were blocked in 5% nonfat milk, and primary antibody was added for overnight incubation at 4°C. Antibodies included (from Cell Signaling Technology, Beverly, MA, unless otherwise noted) total REDD1 RTP801/DDIT4 (ProteinTech, Chicago, IL) and REDD2 [RTP801L/DNA damage-inducible transcript 4-like (DDIT4L); Proteintech (12094-1-AP) or Fitzgerald, Acton, MA (70R-2699)], S6K1, S6K1 Thr389, ribosomal protein S6 (rpS6), rpS6 Ser240/244, 4E-BP1 and 4E-BP1 Ser65, AMPK and AMPK Thr172, acetyl-CoA carboxylase (ACC) and Ser79 ACC, eukaryotic elongation factor-2 (eEF2) and eEF2 Thr56, eEF2 kinase, eEF2 kinase Ser366 and Ser398 (Christopher Proud, South Australian Health and Medical Research Institute), proline-rich Akt substrate of 40 kDa (PRAS40) and PRAS40 Thr246, extracellular signal-regulated kinase (ERK) and ERK Thr202/Y204, p38 mitogen-activated protein kinase (p38) and p38 Thr180/Y182, eIF4E and eIF4 Ser209, eIF2Bε and eIF2Bε Ser536, eIF2α and eIF2α Ser51, PDK1 and PDK1 Ser241, protein phosphatase 2A (PP2A; c-subunit), tuberous sclerosis complex 2 (TSC2) and TSC2 Ser939, ULK1 and ULK1 Ser757, p62, light chain 3B (LC3B), Atg12, Atg7, Akt, Akt Ser473 and Thr308, and FOXO3 and FOXO3 Ser253. The FluorChem M Multifluor System (ProteinSimple, San Jose, CA) was used for visualization following exposure to enhanced chemiluminescence reagent (Thermo Scientific, Waltham, MA). Images were analyzed using AlphaView (ProteinSimple) and ImageJ software (National Institutes of Health).
mRNA content.
Total RNA was extracted using Tri-Reagent (Molecular Research Center, Cincinnati, OH) and an RNeasy mini kit (Qiagen, Valencia, CA), following the manufacturers' protocols. The remaining half of each muscle was homogenized in Tri-Reagent, followed by chloroform extraction, according to the manufacturers' instructions. An equal volume of 70% ethanol was added to the aqueous phase, and the mixture was loaded onto a Qiagen mini spin column. The Qiagen mini kit protocol was followed from this step onward, including the on-column DNase I treatment to remove residual DNA contamination. RNA was eluted from the column with RNase-free water, and an aliquot was used for quantitation (NanoDrop 2000; Thermo Fisher Scientific, Waltham, MA). The quality of the RNA was analyzed on a 1% agarose gel. Total RNA (1.5 μg) was reverse transcribed using superscript III RT (Invitrogen, Carlsbad, CA) in a total volume of 20 μl, following the manufacturer's instructions. Real-time quantitative PCR was performed with 0.5–1 μl of the reverse transcription reaction in a QuantStudio 12K Flex Real-Time PCR System using TaqMan gene expression assays (Applied Biosystems, Foster City, CA) for interleukin-6 (IL-6; Mm00446190_m1), tumor necrosis factor-α (TNFα; Mm00443258_m1), atrogin-1 (NM_026346.2), MuRF1 (NM_001039048.2), IGF-I (Mn01233690_m1), REDD1 (Mm00512504_g1), REDD2 (Mn0051f3313_m1), myostatin (Mm01254559_m1), and ribosomal protein L32 (Mm02528467_g1). The comparative quantification method 2−ΔΔCq was used in presenting gene expression of target genes in reference to the endogenous control (35).
Plasma and tissue analysis.
Measurement of glucose and lactate was performed on plasma samples collected at euthanization ∼24 h after sham or septic surgery. A rapid analyzer (Analox Instruments, Lunenburg, MA) was used for measuring each metabolite. Plasma concentrations of cytokines were determined by mouse-specific ELISA (Boster Biological, Fremont, CA). The protein concentration of TNFα and IL-6 in skeletal muscle was quantified by mouse-specific ELISA (R & D Systems, Minneapolis, MN), as described previously (33). The plasma insulin concentration was determined by ELISA (ALPCO, Salemn, NH). The plasma IGF-I concentration was also determined by ELISA (R & D Systems).
Statistical analysis.
Data were analyzed on commercial statistic software (SigmaPlot; Systat, San Jose, CA), using a two-way ANOVA (genotype × sepsis) with Student-Newman-Keuls post hoc test when appropriate. Survival data were analyzed using Kaplan-Meier survival curve analysis (GraphPad, San Diego, CA). Data are presented as means ± SE and considered significant when P < 0.05.
RESULTS
Body weight, both initially at the start of the study and ∼24 h after surgery and/or induction of sepsis, did not differ between groups (Table 1). Likewise, the percentage of fat and lean mass normalized to individual body weight did not differ across the groups prior to surgery (Table 1). Body composition was not determined after sepsis, as the insult is too acute to see reproducible changes in either fat or lean mass. However, the gastrocnemius and quadriceps were isolated intact and weighed (Table 1). There were no statistical differences for either of these muscles when expressed in terms of absolute mass (Table 1) or normalized to body weight (data not shown). The plasma glucose concentration was decreased in both septic-WT and septic-REDD1−/− mice compared with their respective nonseptic control values; however, the sepsis-induced decrement in glucose was less in mice lacking REDD1 (Table 1). Conversely, sepsis increased the plasma lactate concentration in both genotypes; however, the magnitude of the hyperlactacidemia was smaller in the septic REDD1−/− mice (Table 1).
Table 1.
Effect of sepsis in WT and REDD1−/− mice on body weight and composition, plasma glucose, lactate, and insulin concentrations
| Sham-WT | Sham-REDD1−/− | Septic-WT | Septic-REDD1−/− | |
|---|---|---|---|---|
| Body weight, g | ||||
| Presurgery (initial) | 22.9 ± 0.7 | 23.9 ± 0.6 | 23.1 ± 0.4 | 23.7 ± 0.4 |
| Postsurgery/sepsis | 20.9 ± 0.7 | 21.3 ± 0.5 | 21.5 ± 0.3 | 22.0 ± 0.4 |
| Body composition presurgery, % | ||||
| Fat mass | 7.6 ± 0.5 | 6.7 ± 0.5 | 7.3 ± 0.4 | 7.3 ± 0.4 |
| Lean mass | 73.6 ± 0.8 | 74.2 ± 0.6 | 73.4 ± 0.7 | 74.8 ± 0.3 |
| Muscle weight postsurgery/CLP, mg | ||||
| Gastrocnemius | 108 ± 3 | 107 ± 3 | 108 ± 3 | 106 ± 2 |
| Quadriceps | 136 ± 6 | 144 ± 5 | 138 ± 3 | 139 ± 5 |
| Plasma concentrations | ||||
| Glucose, mg/dl | 162 ± 4a | 168 ± 8a | 84 ± 5b | 106 ± 5c |
| Lactate, mg/dl | 53 ± 4a | 54 ± 3a | 87 ± 3b | 68 ± 2a,b |
| Insulin, ng/ml | 0.52 ± 0.07a | 0.80 ± 0.10a | 0.59 ± 0.13a | 1.46 ± 0.24b |
Values are expressed as means ± SE; n = 10, 10, 15, and 13 for Sham-WT, Sham-REDD1−/−, Septic-WT, and Septic-REDD1−/−, respectively. WT, wild type; CLP, cecal ligation and puncture. Prior to surgery and CLP, body weight and body composition (1H-NMR) were determined. Sepsis was then induced by CLP, and body weight was determined 24 h later at the time of euthanization. At this time point, muscle weight (right gastrocnemius and quadriceps) and blood were collected for the determination of plasma glucose, lactate, and insulin concentrations. Statistical differences between groups (P < 0.05) are indicated by different letters.
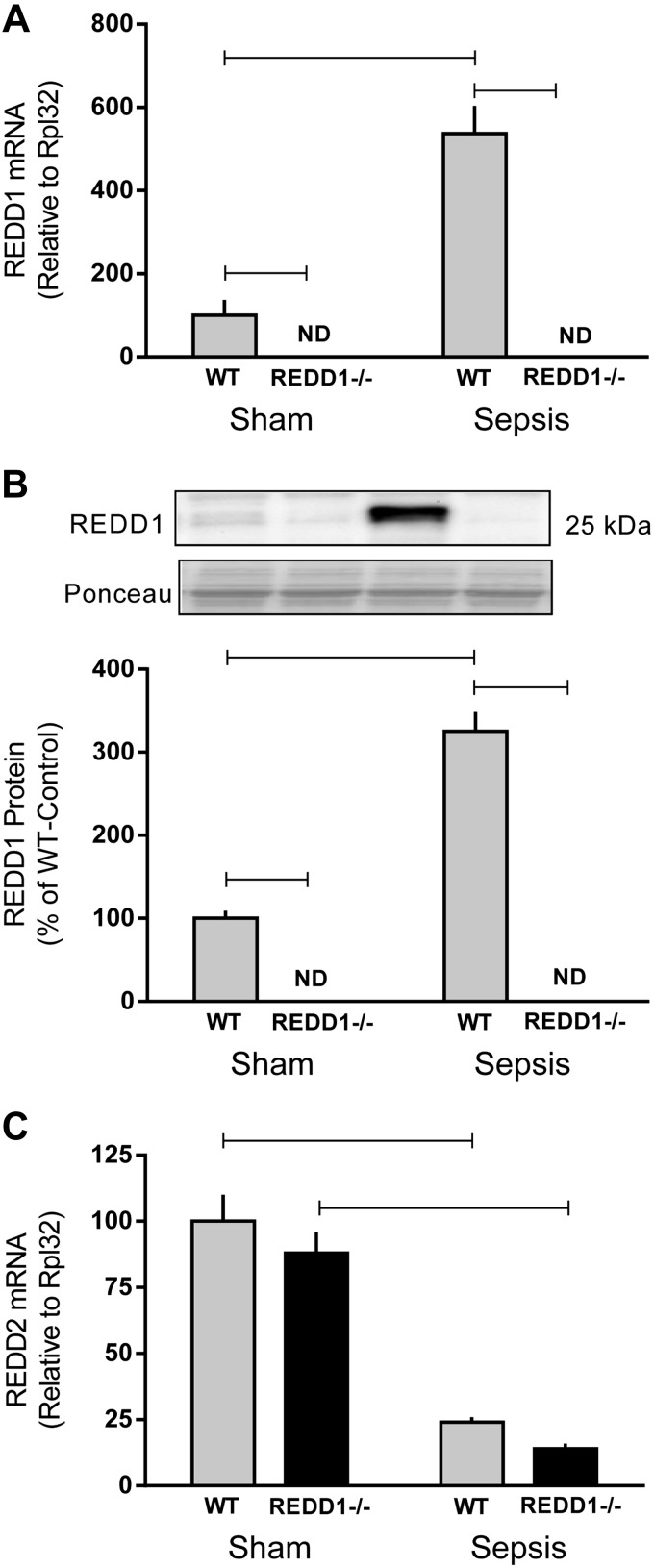
Sepsis increased mRNA content (500%) and protein expression (300%) of REDD1 in skeletal muscle of WT mice, whereas REDD1 was essentially undetectable in sham and septic knockout mice (Fig. 1, A and B). An increase in REDD2, which shares sequence homology with REDD1, can also inhibit mTOR (39). REDD2 mRNA did not differ between WT and REDD1−/− mice under nonseptic control conditions, and sepsis decreased REDD2 mRNA by ∼80% regardless of genotype (Fig. 1C). REDD2 protein content is not reported because of uncertainty related to Western blot data using several commercially available antibodies (more in discussion).
Fig. 1.
Sepsis-induced alternations in regulated in development and DNA damage (REDD)1 and REDD2 mRNA and protein from wild-type (WT) and REDD1−/− mice. REDD1 mRNA content (A) and protein expression (B) as well as REDD2 mRNA content (C) were determined by quantitative real-time PCR and Western blot analysis of gastrocnemius. REDD1 mRNA was nondetectable (ND) in REDD1−/− mice. Gray bars represent WT mice (n = 10 sham and 15 septic), and black bars correspond to REDD1−/− mice (n = 10 sham and 13 septic). Horizontal bars indicate statistical differences between groups (P < 0.05). Values are expressed as means ± SE.
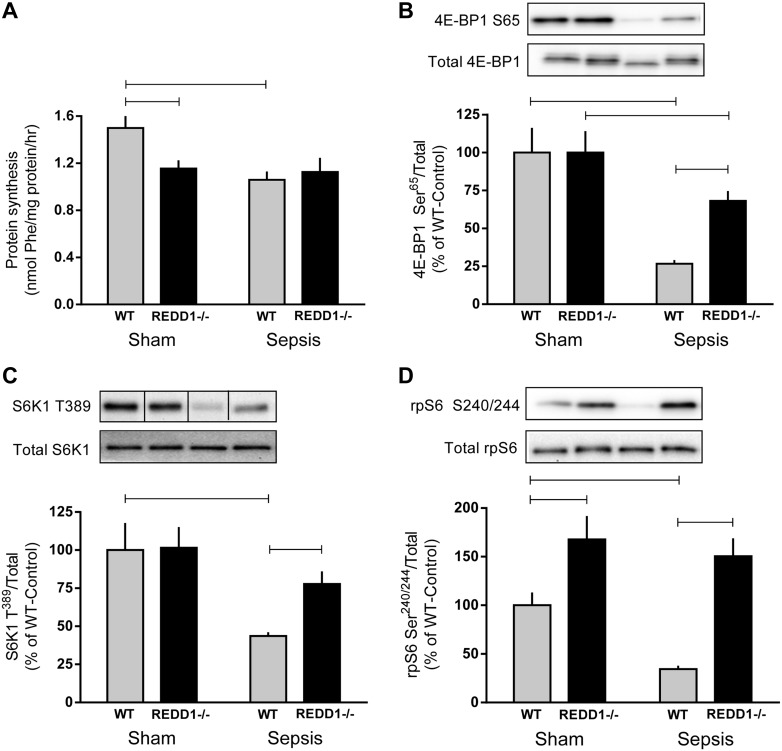
In accord with the role of REDD1 as a negative regulator of mTORC1 (7), sepsis decreased the rate of protein synthesis 30% and suppressed the phosphorylation of 4E-BP1 Ser65 (73%), S6K1 Thr389 (57%), and rpS6 Ser240/244 (65%) in WT mice (Fig. 2, A–D). In contrast, no sepsis-induced decrease in protein synthesis was observed in REDD1−/− mice compared with sham REDD1−/− mice. Furthermore, the phosphorylation of 4E-BP1 Ser65, S6K1 Thr389, and rpS6 Ser240/244 was greater in septic REDD1−/− than septic WT mice. Sepsis reduced 4E-BP1 Ser65 phosphorylation in REDD1−/− muscle; however, the phosphorylation of S6K1 Thr389 and rpS6 Ser240/244 did not differ between the sham and septic REDD1−/− mice.
Fig. 2.
Sepsis decreases protein synthesis and mechanistic target of rapamycin complex 1 (mTORC1) signaling in WT but not REDD1−/− mice. Protein synthesis (A), eukaryotic initiation factor (eIF) 4E-binding protein-1 (4E-BP1) Ser65 (B), ribosomal protein (rp)S6 kinase 1 (S6K1) Thr389 (C), and rpS6 Ser240/244 (D) phosphorylation was assessed in the gastrocnemius of sham control and septic mice from each genotype. Bar graphs (B–D) represent quantification of Western blot images relative to the total amount of the respective protein relative to the sham-WT value, which was set to 100%. Gray bars represent WT mice (n = 10 sham and 15 septic), and black bars correspond to REDD1−/− mice (n = 10 sham and 13 septic). Black lines between bands indicate where images were spliced to adjust sample order on the membrane for presentation. Horizontal bars indicate statistical differences between groups (P < 0.05). Values are expressed as means ± SE.
To determine whether the rate of muscle protein synthesis in the septic REDD1−/− mice had been maximally decreased (i.e., a “floor effect”), a separate study was performed in which REDD1−/− mice were injected with the pharmacological agent AICAR, which stimulates AMPK and decreases muscle protein synthesis (4, 42). The relative rate of protein synthesis was 100 ± 11% in WT control mice, 73 ± 5% in REDD1−/− septic mice, and 44 ± 6% for REDD1−/− septic mice injected with AICAR. One-way ANOVA with Student-Newman-Keuls followup indicated a significant difference (P < 0.05) between all groups.
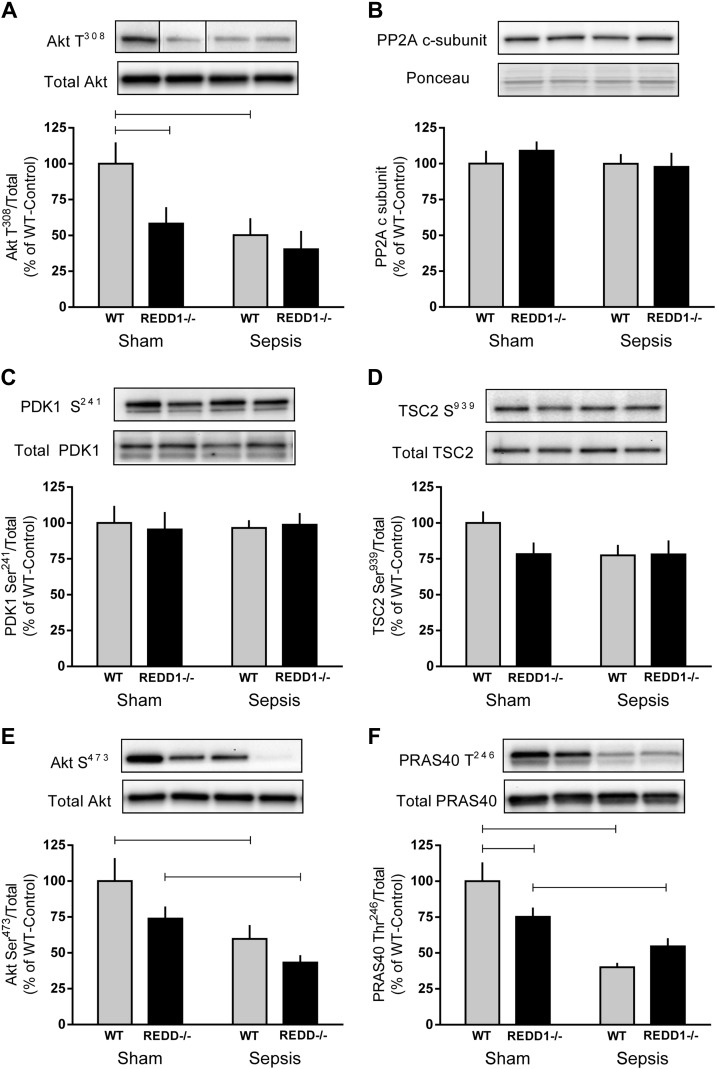
The current model of REDD1 signaling based on in vitro work proposes that REDD1 induces PP2A-mediated dephosphorylation of Akt Thr308 to inhibit mTORC1 (10). Therefore, REDD1−/− mice would be expected to display elevated levels of Akt Thr308, leading to enhanced mTORC1 activity. In contrast, decreased phosphorylation of Akt Thr308 (40%) was observed in sham-REDD1−/− mice relative to sham-WT mice (Fig. 3A). Furthermore, there was no difference in Akt Thr308 phosphorylation between sham-REDD1−/− mice and either group of septic mice, with all three groups exhibiting a 50–60% reduction in Akt phosphorylation compared with sham-WT values. Decreased Akt Thr308 does not appear to be due to a change in the protein content of PP2A (c-subunit), which dephosphorylates Akt Thr308, or to a change in PDK1 Ser241, which phosphorylates Akt (Fig. 3, B and C), as these proteins were unaltered by sepsis or REDD1 deletion. Additionally, the phosphorylation of TSC2 Ser939, an Akt substrate, did not differ among the four groups (Fig. 3D). Similarly to the decrease in Akt Thr308 phosphorylation, sepsis decreased the phosphorylation of Akt Ser473 in WT and REDD1−/− mice, consistent with suppression of mTORC2 (Fig. 3E). Changes in PRAS40 Thr246 phosphorylation also mimicked that of Akt; under sham conditions PRAS40 phosphorylation was decreased in REDD1−/− mice, whereas sepsis decreased levels even further in both genotypes (Fig. 3F).
Fig. 3.
Deletion of REDD1 and sepsis modulates Akt phosphorylation in skeletal muscle. Bar graphs represent the quantification of Western blot images relative to the total amount of the respective protein or Ponceau S stain for Akt Thr308 (A), protein phosphatase 2A (PP2A; c-subunit; B), phosphoinositide-dependent protein kinase 1 (PDK1) Ser241 (C), tuberous sclerosis complex 2 (TSC2) Ser939 (D), Akt Ser473 (E), and proline-rich Akt substrate of 40 kDa (PRAS40) Thr246 (F) phosphorylation. Gray bars represent WT mice (n = 10 sham and 15 septic), and black bars correspond to REDD1−/− mice (n = 10 sham and 13 septic). Black lines between bands indicate where images were spliced to adjust sample order on the membrane for presentation. All values are expressed relative to sham-WT value, which was set to 100%. Horizontal bars indicate statistical differences between groups (P < 0.05). Values are expressed as means ± SE.
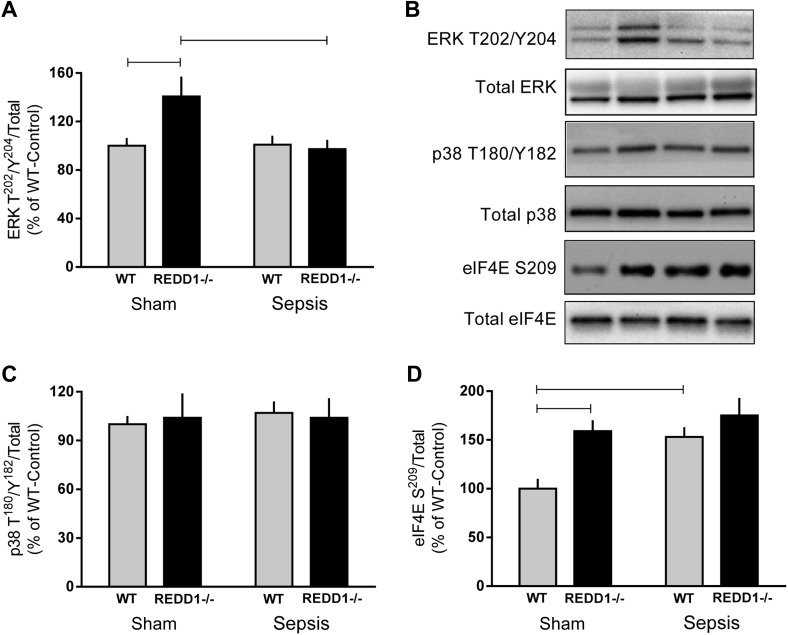
Protein synthesis can also be regulated by several mTORC1-independent pathways that have been investigated specifically herein. First, ERK and p38 phosphorylate MNK (MAPK signal-integrating kinase)-1/2, which in turn phosphorylates eIF4E, thereby potentially enhancing translation (50). Phosphorylation of ERK (but not p38) was increased in nonseptic control REDD1−/− mice compared with WT mice (Fig. 4A). While there was no sepsis-induced increase in ERK phosphorylation in WT septic mice, ERK phosphorylation was lower in REDD1−/− septic mice compared with control REDD1−/− values. There was neither a genotype nor a sepsis effect on the phosphorylation state of p38 (Fig. 4C) or JNK (data not shown). Phosphorylation of eIF4E on Ser209 was increased by REDD1 deletion under control conditions and in WT mice by sepsis (Fig. 4D). There was no interaction of REDD1 deletion and sepsis.
Fig. 4.
Alterations in ERK, p38, and eIF4E phosphorylation in skeletal muscle in response to REDD1 deletion and sepsis. Bar graphs represent the quantification of Western blot images relative to the total amount of the respective protein ERK Thr202/Y204 (A), p38 Thr180/Y182 (C), and eIF4E Ser209 (D) phosphorylation. Gray bars represent WT mice (n = 10 sham and 15 septic), and black bars correspond to REDD1−/− mice (n = 10 sham and 13 septic). All values are expressed relative to the sham-WT value, which was set to 100%. There was no significant genotype or sepsis effect on the total amount of any of the 3 proteins (data not shown). Horizontal bars indicate statistical differences between groups (P < 0.05). Values are expressed as means ± SE. B: representative Western blot images for the 4 treatment groups.
eIF2B activity can also regulate the global rate of protein synthesis, and such activity is controlled in part by phosphorylation of eIF2Bε and eIF2α (28). However, there was no significant genotype or sepsis effect on the phosphorylation of eIF2Bε Ser536 or eIF2α Ser51 or on the total amount of either protein in skeletal muscle (data not shown).
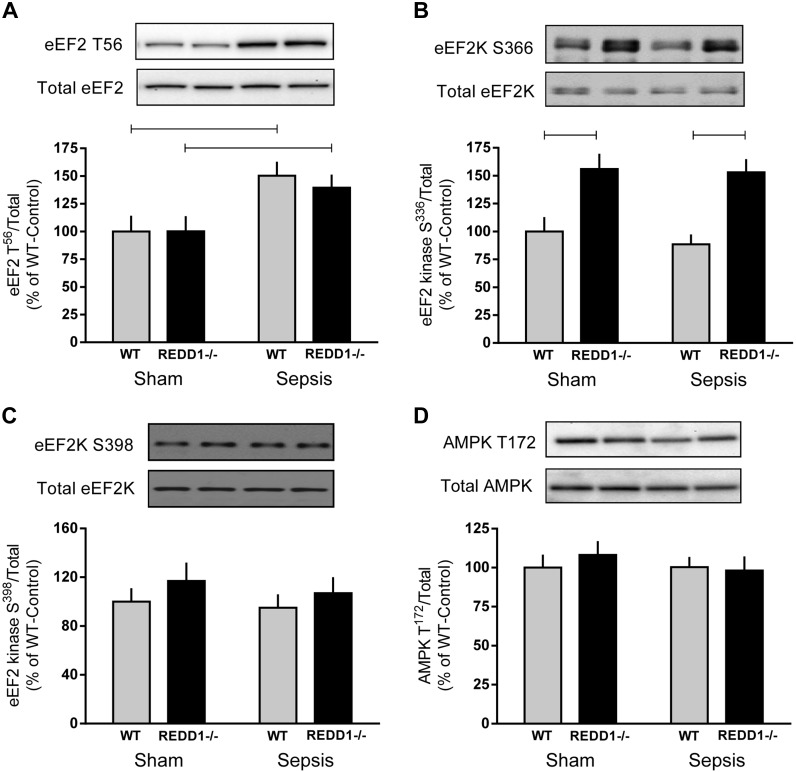
Finally, changes in the rate of translation elongation can also influence protein synthesis (6). Phosphorylation of eEF2 on Thr56 did not differ between WT and REDD1−/− mice under either control or septic conditions, and sepsis increased eEF2 phosphorylation by 40–50% in both genotypes (Fig. 5A). To determine the potential mechanism for this change in phosphorylation state, two distinct phosphorylation sites for eEF2 kinase were assessed. S6K1-mediated phosphorylation of eEF2 kinase on Ser366 was increased similarly in REDD1−/− mice under both control and septic conditions, and there was no sepsis-induced change in phosphorylation (Fig. 5B). There was no consistent change among the four groups in the phosphorylation of eEF2 kinase on Ser398, which is mediated by AMPK, and this was consistent with the lack of genotype and/or sepsis effect on AMPK phosphorylation (Fig. 5, C and D, respectively). We confirmed that AMPK was not activated by sepsis and/or REDD1 deletion, as Ser79 phosphorylation of ACC in muscle did not differ between any of the four experimental groups (data not shown).
Fig. 5.
Alterations in the phosphorylation of eukaryotic elongation factor (eEF)-2, eEF2 kinase (eEF2K), and AMP kinase (AMPK) in skeletal muscle in response to REDD1 deletion and sepsis. Bar graphs represent the quantification of Western blot images relative to the total amount of the respective protein eEF2 Thr56 (A), eEF2K Ser366 (B), eEF2K Ser398 (C), and AMPK Thr172 (D) phosphorylation. Gray bars represent WT mice (n = 10 sham and 15 septic), and black bars correspond to REDD1−/− mice (n = 10 sham and 13 septic). All values are expressed relative to sham-WT value, which was set to 100%. There was no significant genotype or sepsis effect on the total amount of any of the 4 proteins (data not shown). Horizontal bars indicate statistical differences between groups (P < 0.05). Values are expressed as means ± SE. Representative Western blots for the 4 treatment groups are shown.
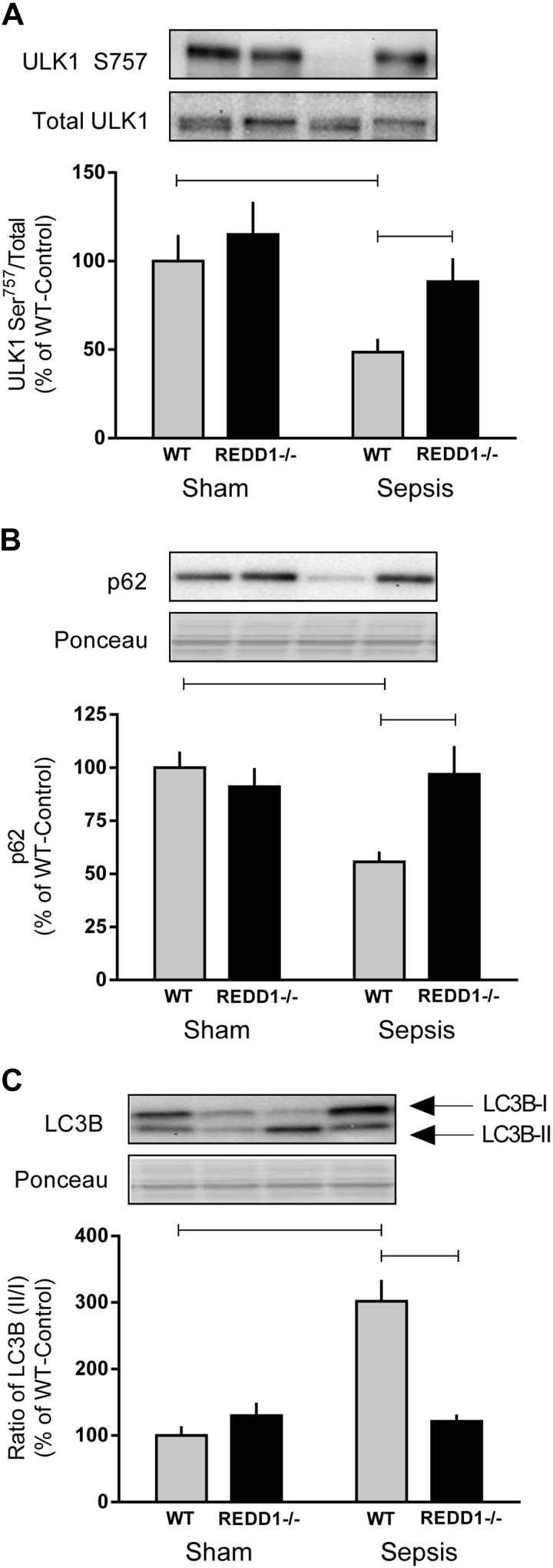
Because muscle mass depends on the balance between rates of synthesis and degradation, surrogate markers of each of the two primary proteolytic pathways (autophagy and UPS) were assessed. Sepsis decreased the phosphorylation of ULK1 Ser757 and p62 expression, whereas it increased the abundance of LC3B-II vs. LC3-I (Fig. 6, A–C). Collectively, these data imply that sepsis increases autophagy in WT mice. Deletion of REDD1 prevented the sepsis-induced increase in autophagy, as these markers did not differ from nonseptic sham mice (Fig. 6, A–C). However, protein expression of the autophagy-related genes Atg7 and Atg12 was not altered in response to REDD1 knockout or sepsis (data not shown).
Fig. 6.
Markers of autophagy were induced in septic muscle of WT but not REDD1−/− mice. Indicators of autophagy, including Unc-like autophagy-activating kinase 1 (ULK1) Ser757 phosphorylation (A), p62 (B), and light chain 3B (LC3B; C), were quantified from Western blot images and expressed relative to the total amount of the respective protein. Where the corresponding total protein was not measured, Ponceau S stain was used to verify loading. Gray bars represent WT mice (n = 10 sham and 15 septic), and black bars correspond to REDD1−/− mice (n = 10 sham and 13 septic). All values are expressed relative to sham-WT, which was set to 100%. Horizontal bars indicate statistical differences between groups (P < 0.05). Values are expressed as means ± SE.
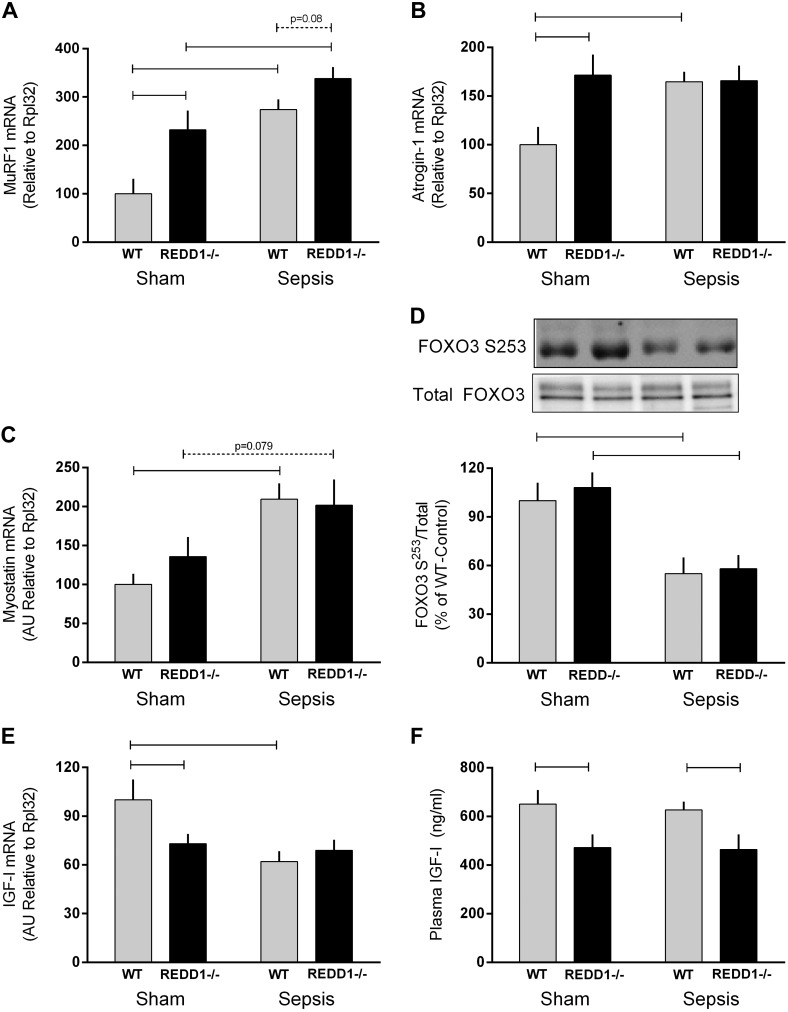
Activation of the UPS is often inferred from the mRNA content of the atrogenes MuRF1 and atrogin-1, as deletion of these genes can partially or completely prevent muscle atrophy (3). MuRF1 and atrogin-1 mRNA were increased in muscle from REDD1−/− mice under sham conditions (Fig. 7, A and B). Sepsis increased both MuRF1 and atrogin-1 mRNA in WT mice, whereas only a sepsis-induced increase (P = 0.08) in MuRF1 was detected in REDD1−/− mice (Fig. 7, A and B). Myostatin can also negatively regulate muscle mass at least in part by stimulating protein degradation (37). Although there was no genotype effect on myostatin mRNA content, sepsis increased myostatin in WT mice, and a similar trend was detected in REDD1−/− mice (P = 0.07; Fig. 7C). Myostatin also regulates muscle atrophy via FOXO-dependent induction of MuRF1 and atrogin-1 (44). Accordingly, sepsis decreased phosphorylation of FOXO3 Ser253 in both WT and REDD1−/− mice to the same extent (Fig. 7D).
Fig. 7.
Sepsis-induced changes in the mRNA content for muscle RING finger 1 (MuRF1), atrogin-1, myostatin, and IGF-I as well as forkhead box O3 (FoxO3) Ser253 phosphorylation and plasma IGF-I concentration in WT and REDD1−/− mice. The various mRNAs in muscle were determined via RT-PCR and normalized to ribosomal protein L32, which did not change significantly between groups. Western blot images were quantitated and expressed relative to the total amount of the respective protein. Gray bars represent WT mice (n = 10 sham and 15 septic), and black bars correspond to REDD1−/− mice (n = 10 sham and 13 septic), with all values expressed relative to sham-WT value, which was set to 100%. Horizontal bars indicate statistical differences between groups (P < 0.05). Values are expressed as means ± SE.
IGF-I via both its autocrine/paracrine and its classical endocrine effects regulates muscle protein balance by stimulating protein synthesis and inhibiting degradation (17). In this regard, IGF-I mRNA was decreased in sham-REDD1−/− compared with sham-WT mice (Fig. 7E). IGF-I mRNA was decreased by sepsis in WT mice, although no further decrease (vs. sham-REDD1−/−) was observed in septic-REDD1−/− mice. In contrast to the muscle mRNA content, the circulating concentration of IGF-I did not differ under nonseptic control conditions between the WT and REDD1−/− mice, and sepsis decreased the plasma IGF-I concentration similarly (30–40%) regardless of genotype (Fig. 7F).
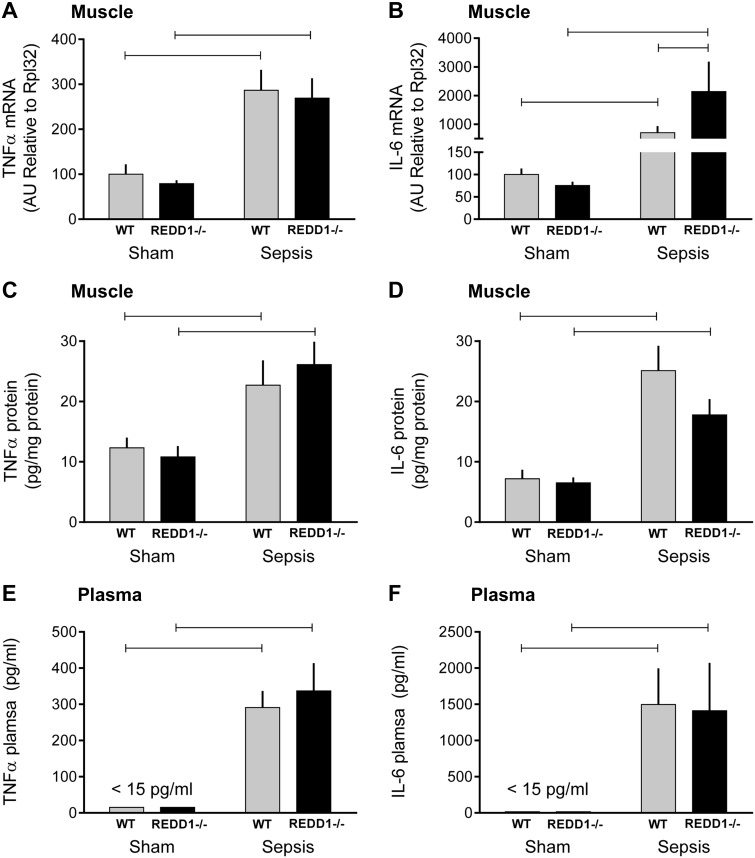
Finally, we assessed both muscle mRNA and protein as well as the plasma concentration for two key inflammatory cytokines, TNFα and IL-6, that can contribute to muscle wasting (8, 24, 30). There was no genotype effect on TNFα mRNA/protein in muscle or TNFα in plasma under control conditions, and sepsis increased TNFα to the same extent in WT and REDD1−/− mice (Fig. 8, A, C, and E). Similarly, there was no difference in IL-6 mRNA/protein in muscle or plasma between WT and REDD1−/− mice (Fig. 8, B, D, and F). Whereas sepsis increased IL-6 protein in muscle and plasma in both groups similarly, the sepsis-induced increase in IL-6 mRNA was exaggerated in the REDD1−/− mice compared with septic WT mice.
Fig. 8.
Effect of REDD1 deletion on sepsis-induced changes in muscle and plasma cytokines. TNFα (A) and IL-6 (B) mRNA were determined, as were TNFα (C) and IL-6 (D) protein in gastrocnemius from WT and REDD1−/− mice under sham control and septic conditions. Also, the impact of REDD1 deletion on sepsis-induced changes in plasma TNFα (E) and IL-6 (F) was determined. Gray bars represent WT mice (n = 10 sham and 15 septic), and black bars correspond to REDD1−/− mice (n = 10 sham and 13 septic), with all values expressed relative to sham-WT, which was set to 100%. Horizontal bars indicate statistical differences between groups (P < 0.05). Values are expressed as means ± SE.
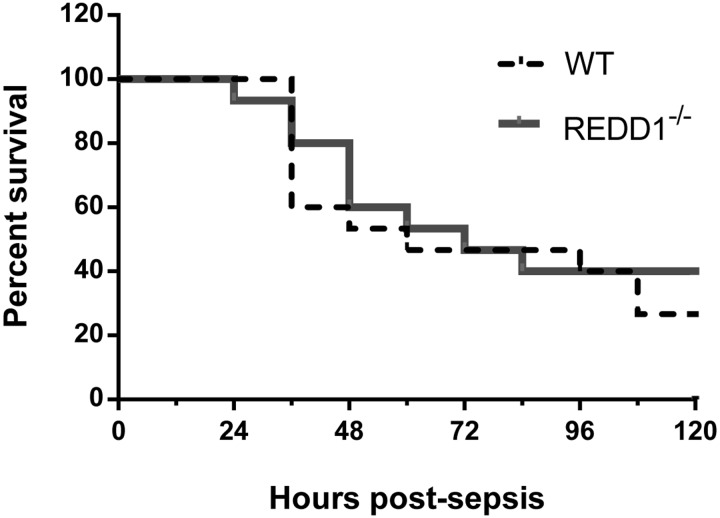
To assess whether global REDD1 disruption influences survival during sepsis, an additional group of WT and REDD1−/− female mice underwent CLP surgery and were monitored for the subsequent 5-day period. As displayed by the Kaplan-Meier survival curve in Fig. 9, survival did not differ between the two genotypes at 5 days postinfection. For the mice euthanized at the 5-day time point, body composition, including fat and lean mass, did not differ between WT and REDD1−/− mice (data not shown). Furthermore, gastrocnemius and quadriceps weight did not differ after 5 days of sepsis in WT or REDD1−/− mice (data not shown).
Fig. 9.
REDD1 disruption does not alter survival during a prolonged septic insult. An additional group of WT (n = 15) and REDD1−/− (n = 15) mice underwent cecal ligation and puncture, and survival was monitored every 12 h for 5 days (120 h). The dashed line corresponds to %WT mice alive at the given time point, and the solid line represents %REDD1−/− mice alive at each time point. No differences between groups were detected.
DISCUSSION
REDD1 is elevated in muscle during sepsis and as a repressor of mTORC1 was posited to be causally related to the sepsis-induced decrease in protein synthesis. The current findings, at least in part, support this hypothesis, as sepsis did not decrease mTORC1 signaling or protein synthesis in muscle from REDD1−/− mice as observed in septic-WT female mice. Additionally, deletion of REDD1 prevented the sepsis-induced increase in autophagy, as assessed by the phosphorylation state of ULK1 as well as the increased LC3B and decreased content for p62. In contrast, atrogene (MuRF1 and atrogin1) mRNA content was elevated in sham- REDD1−/− mice compared with sham WT mice. Whereas sepsis increased expression of these genes in WT mice, MuRF1 was further increased by sepsis in REDD1−/− mice. Likewise, sepsis increased the TNFα and IL-6 mRNA and protein in muscle as well as the plasma concentration of both cytokines regardless of genotype. Although the sepsis-induced increase in IL-6 mRNA was greater in REDD1−/− mice than in WT mice, this difference was not manifested at the level of IL-6 protein in muscle or by a genotype difference in circulating IL-6. Neither the basal nor the sepsis-induced changes in muscle protein metabolism could be explained by compensatory changes in REDD2 mRNA. Although deletion of REDD1 clearly modulates a cohort of the changes in protein synthesis and degradation acutely (∼24 h) produced by sepsis, REDD1 null mice did not have a long-term (5 day) survival advantage.
Regulation of muscle protein balance and signal transduction by REDD1 deletion under basal nonseptic conditions.
REDD1−/− mice do not exhibit overt phenotypic differences from WT mice, as muscle weights, the proportion of type I and II myofibers, and contractility have been reported to be similar (5). Our data confirmed these findings related to muscle weight and extended them by demonstrating no difference in whole body fat or lean body mass between WT and REDD1-null mice. Previously, increased protein synthesis and mTORC1 signaling (e.g., S6K1 and 4E-BP1 phosphorylation) have been observed in muscle of overnight-fasted REDD1−/− male mice compared with WT mice (21, 22). In contrast, we detected a decreased rate of synthesis in 24-h-fasted female REDD1−/− mice compared with fasted WT mice. This difference was associated with a concomitant reduction in Akt Thr308 phosphorylation, although this genotypic difference was not transmitted down the mTORC1 signal transduction pathway, as the phosphorylation state of 4E-BP1 and S6K1 did not differ between WT and REDD1−/− mice, suggesting no change in mTORC1 activity. Moreover, despite the similar phosphorylation of S6K1, phosphorylation of rpS6 was increased in REDD1−/− mice compared with WT mice under control conditions. The reason for these discordant signaling events and the difference in protein synthesis is not known but does not appear to be due to differences in the prevailing plasma concentration of insulin or IGF-I. Differences in results between the current study and previous work may also represent a sexual dimorphic response, but this area will require additional systemic studies to elucidate. Finally, while unexpected, reduced protein synthesis in REDD1−/− mice could theoretically provide a survival advantage during nutrient insufficiency, as it would decrease the use of nutrients, thereby partitioning them to other tissues.
In the absence of a genotypic change in mTOR kinase activity, this raises the question as to the mechanism for the observed decrease in protein synthesis. Of those alternatives investigated, none of the potential mTORC1-independent regulatory mechanisms appeared operational in the current experimental context. For example, increased ERK activity (i.e., increased Thr202/Y204 phosphorylation) and corresponding increase in Ser209 phosphorylation of eIF4E would be expected to enhance, not suppress, cap-dependent translation (47). Furthermore, there was no significant difference in the total amount or the phosphorylation state of eIF2α (Ser51) or eIF2Bε (Ser536) between WT and REDD1−/− mice under control conditions, implying that the decreased protein synthesis is likely independent of a change in eIF2B activity. Finally, despite the increased Ser366 phosphorylation of eEF2K, which would be expected to decrease its kinase activity (6), there was also no detectable difference for total and Thr56-phosphorylated eEF2 between WT and REDD1−/− mice. Collectively, these latter data suggest that the genotype difference in protein synthesis was not mediated by a differential effect on mRNA elongation.
Akt has been implicated in the regulation of mTORC1 by REDD1 (10), and in the present study we observed a decrease in Akt Thr308 phosphorylation in sham-REDD1−/− mice. Moreover, Thr246 phosphorylation of PRAS40, the authentic downstream target of Akt, was coordinately decreased in REDD1−/− mice compared with WT controls. The differential phosphorylation of Thr308-phosphorylated Akt could not be explained by the activity of the upstream kinase PDK1 (Ser241 phosphorylation) or the phosphatase PP2A. In contrast, there was no genotypic difference in mTORC2-regulated Ser473-phosphorylated Akt. Accordingly, although there is evidence of dysregulated insulin signaling in REDD1−/− mice (5, 14, 52), this is the first evidence of decreased Akt phosphorylation under basal fasted conditions. Although PRAS40 phosphorylation is decreased, the ramification of this decreased Akt activity remains equivocal, as TSC2 Ser939 phosphorylation and mTOR kinase activity appear to be generally well maintained.
Deletion of REDD1 increased atrogene mRNA content in muscle from fasted sham-REDD1−/− mice compared with sham-WT mice, suggesting enhanced UPS-mediated muscle proteolysis. These results differ from those reported by Britto et al. (5), where there was no effect of REDD1 deletion on muscle atrogene expression, ubiquitin content, or in vitro-determined tyrosine release. We do not have an explanation for the inconsistency, as both studies used a comparable methodological approach in fasted female WT and REDD1−/− mice. In contrast, both studies failed to detect changes in indices of autophagy, such as ULK phosphorylation, p62 depletion, or an increased LC3B-II/I ratio between REDD1−/− and WT mice under basal conditions.
All of the above-mentioned genotypic differences in muscle protein balance appeared to be independent of increases in selected catabolic stimuli such as TNFα and IL-6 in muscle and blood as well as decreases in anabolic stimuli such as plasma insulin and IGF-I. Others have reported that the excretion of corticosterone, a major stress hormone, did not differ between WT and REDD1−/− mice (5). However, we did detect a reduction in IGF-I mRNA in muscle from REDD1−/− mice, and a decreased autocrine action of this growth factor might explain at least in part the concomitant reduction in Akt Thr308 phosphorylation in these animals.
An increase in REDD2 can also inhibit mTORC1 (39), and compensatory changes may occur in REDD1−/− mice. Muscle extracts from all four experimental groups were analyzed for REDD2 protein levels using two different DDIT4L antibodies. These included a rabbit polyclonal antibody (cat. no. 12094-1-AP; Proteintech) and a rabbit antibody raised to a synthetic peptide that corresponds to the middle region of REDD2 (cat. no. 70R-2699; Fitzgerald). Immunoblots, using either of the above two antibodies, did not detect a band(s) that would correspond to the migration of a protein of 22 kDa, the predicted molecular weight of REDD2. Using an ectopically expressed epitope tagged REDD2 in human embryonic kidney-293FT cells, we were able to ascertain that REDD2 migrates on SDS-PAGE with a mobility expected for its size and that the commercial antibodies are able to detect REDD2 on immunoblot. These data suggest that REDD2 is modified in vivo such that the antibodies used do not recognize the protein or that REDD2 is of low abundance in skeletal muscle in vivo. In this regard, it is noteworthy that neither DDIT4 nor DDIT4L was listed among the 8,309 protein identifications from a recent deep proteome study of mouse skeletal muscle (12) and may be consistent with proteins characterized by very low abundance or short-lived proteins whose expression may be transient in nature, depending on cellular status, and are consequently difficult to detect.
Impact of REDD1 deletion on mTORC1 and autophagy in sepsis.
The current data corroborate past reports, as sepsis increased REDD1, decreased Akt/mTOR signal transduction and protein synthesis, and appeared to increase proteolysis via upregulation of the UPS and enhanced autophagy (18, 32, 40). Our data demonstrate that this sepsis-induced increase in REDD1 regulates muscle mTOR kinase activity, as the decreased phosphorylation of 4E-BP1 and S6K1 as well as the decreased phosphorylation of its downstream substrate rpS6 was partially or completely prevented in septic REDD1−/− mice. As a result of this maintained mTORC1 signaling in REDD1−/− mice, there was no sepsis-induced decrease in muscle protein synthesis. One potential limitation of the current study was that REDD1−/− mice had a lower basal level of muscle protein synthesis, and it was possible that a further decrement in synthetic rate could not be achieved in these mice (i.e., a floor effect). To address this issue, a separate study was performed in which REDD1−/− mice were injected acutely with AICAR to effectively activate AMPK (42). Using this experimental model, we convincingly demonstrated a further decrease in muscle protein synthesis in REDD1−/− mice. Collectively, these data suggest that although basal muscle protein synthesis is reduced, deletion of REDD1 completely prevents the sepsis-induced decrease in muscle protein synthesis. These conclusions are consistent with data showing that deletion of REDD1 also prevents dexamethasone-induced muscle atrophy and suppression of mTORC1 signaling and protein synthesis (5).
In a recent study (10), REDD1 was shown to interact with both Akt and the catalytic subunit of PP2A to promote the dephosphorylation of Akt Thr308 independent of a change in Akt Ser473 phosphorylation. Furthermore, mutations in REDD1 that prevented its interaction with either protein blocked the REDD1-mediated decrease in Akt Thr308 phosphorylation, providing support for a model in which REDD1 acts to target the catalytic subunit of PP2A to Akt to promote selective dephosphorylation of Thr308. The finding in the present study that Akt Thr308 phosphorylation was lower in septic WT mice in which REDD1 expression was significantly increased (compared to sham WT mice), but not in septic REDD1−/− mice in which REDD1 was not increased by sepsis, supports the previously proposed model (10). However, in the present study, phosphorylation of Akt Thr308 was also decreased in sham-REDD1−/− mice compared with sham-WT mice despite no further attenuation with sepsis. Although this decrease may appear to be counterintuitive, REDD1−/− mice have dysregulated insulin signaling, which may have contributed to this finding (5, 14, 52). Regardless of the exact mechanism, the sepsis-induced decrease in Akt phosphorylation at both Thr308 and Ser473 as well as the decreased phosphorylation of PRAS40 (Akt substrate) were not prevented in REDD1−/− mice. Hence, the maintenance of mTORC1 and muscle protein synthesis in septic-REDD1−/− mice appears to be independent of a corresponding change in Akt.
REDD1 also appears to mediate the sepsis-induced increase in muscle autophagy, as deletion of REDD1 largely prevented the decrease in ULK1 Ser757 phosphorylation and p62 protein expression as well as the increased LC3B-II/I ratio. The sepsis-induced decrement in ULK Ser757 phosphorylation in WT mice and the prevention of this response in REDD1−/− mice were associated with coordinate changes in mTORC1 activity and were consistent with the recognized role of mTOR in regulating autophagy (11, 55). These results are also consistent with previous reports in which MEFs lacking REDD1 were protected from the serum deprivation (10 h)-induced decrease in mTORC1 activity, ULK1 Ser757 phosphorylation, p62, and induction of autophagy (11, 43). However, our data differ from those showing that the deletion of REDD1 does not prevent increased autophagy following acute dexamethasone administration (5). AMPK can also promote autophagy by directly activating ULK1 via other phosphorylation sites (15); however, this mechanism does not appear to be operational in the current setting, as sepsis did not increase AMPK activity, as evidenced by lack of genotype or treatment effect on AMPK Thr172 phosphorylation or ACC Ser79 phosphorylation.
In contrast to the regulatory role of REDD1 outlined above for the sepsis-induced increase in autophagy, deletion of REDD1 did not antagonize the increase in MuRF1 and atrogin-1, suggesting the sepsis-induced increase in the UPS remains elevated in REDD1−/− mice. Furthermore, increases in the proinflammatory milieu of muscle and blood can regulate muscle protein balance (17). However, sepsis-induced elevation of TNFα and IL-6 protein in muscle per se and in the systemic circulation were comparably increased in WT and REDD1−/− mice, suggesting that differences in these cytokines are unlikely to mediate genotypic differences in rates of protein synthesis or autophagy between WT and REDD1−/− mice.
Overall our current data suggest that REDD1 mediates the impairment in mTORC1 signaling and autophagy observed in muscle during sepsis. They also highlight a significant role for REDD1 in the regulation of IGF-I/Akt signaling, protein synthesis, and protein breakdown in both fasted and septic conditions. Finally, although the maintenance of mTORC1 signaling and protein synthesis in REDD1−/− mice did not significantly enhance survival, such changes might improve recovery and rehabilitation of those patients surviving critical illness.
GRANTS
This work was supported in part by Grants R01-GM-38032 (C. H. Lang), F32-AA023422 (J. L. Steiner), F32-GM-112401 (K. T. Crowell), and DK-15658 (S. R. Kimball).
DISCLOSURES
The authors declare they have no competing interests that might be perceived to influence the results and discussion reported in this paper.
AUTHOR CONTRIBUTIONS
J.L.S., S.R.K., and C.H.L. conception and design of research; J.L.S. and K.T.C. performed experiments; J.L.S. and C.H.L. analyzed data; J.L.S., S.R.K., and C.H.L. interpreted results of experiments; J.L.S. prepared figures; J.L.S. drafted manuscript; J.L.S., K.T.C., S.R.K., and C.H.L. edited and revised manuscript; J.L.S., K.T.C., S.R.K., and C.H.L. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Chris Proud for the generous gift of the antibody that recognizes Thr56-phosphorylated eEF2 and Ser398-phosphorylated eEF2K, Chen Yang for breeding the REDD1−/− mice, Dr. David Williamson for supplying the REDD1−/− breeding animals, Quark Pharmaceuticals for permission to use the REDD1−/− mice, and Maithili Navaratnarajah, Anne Pruznak, and Gina Deiter for their excellent technical assistance.
REFERENCES
- 1.Alamdari N, Toraldo G, Aversa Z, Smith I, Castillero E, Renaud G, Qaisar R, Larsson L, Jasuja R, Hasselgren PO. Loss of muscle strength during sepsis is in part regulated by glucocorticoids and is associated with reduced muscle fiber stiffness. Am J Physiol Regul Integr Comp Physiol 303: R1090–R1099, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J 15: 6541–6551, 1996. [PMC free article] [PubMed] [Google Scholar]
- 3.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3: 1014–1019, 2001. [DOI] [PubMed] [Google Scholar]
- 4.Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem 277: 23977–23980, 2002. [DOI] [PubMed] [Google Scholar]
- 5.Britto FA, Begue G, Rossano B, Docquier A, Vernus B, Sar C, Ferry A, Bonnieu A, Ollendorff V, Favier FB. REDD1 deletion prevents dexamethasone-induced skeletal muscle atrophy. Am J Physiol Endocrinol Metab 307: E983–E993, 2014. [DOI] [PubMed] [Google Scholar]
- 6.Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem 279: 12220–12231, 2004. [DOI] [PubMed] [Google Scholar]
- 7.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 18: 2893–2904, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooney R, Kimball SR, Eckman R, Maish G, Shumate M, Vary TC. TNF-binding protein ameliorates inhibition of skeletal muscle protein synthesis during sepsis. Am J Physiol Endocrinol Metab 276: E611–E619, 1999. [DOI] [PubMed] [Google Scholar]
- 9.Demers G, Griffin G, De Vroey G, Haywood JR, Zurlo J, Bédard M. Animal research. Harmonization of animal care and use guidance. Science 312: 700–701, 2006. [DOI] [PubMed] [Google Scholar]
- 10.Dennis MD, Coleman CS, Berg A, Jefferson LS, Kimball SR. REDD1 enhances protein phosphatase 2A-mediated dephosphorylation of Akt to repress mTORC1 signaling. Sci Signal 7: ra68, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis MD, McGhee NK, Jefferson LS, Kimball SR. Regulated in DNA damage and development 1 (REDD1) promotes cell survival during serum deprivation by sustaining repression of signaling through the mechanistic target of rapamycin in complex 1 (mTORC1). Cell Signal 25: 2709–2716, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshmukh AS, Murgia M, Nagaraj N, Treebak JT, Cox J, Mann M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol Cell Proteomics 14: 841–853, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dufner A, Thomas G. Ribosomal S6 kinase signaling and the control of translation. Exp Cell Res 253: 100–109, 1999. [DOI] [PubMed] [Google Scholar]
- 14.Dungan CM, Wright DC, Williamson DL. Lack of REDD1 reduces whole body glucose and insulin tolerance, and impairs skeletal muscle insulin signaling. Biochem Biophys Res Commun 453: 778–783, 2014. [DOI] [PubMed] [Google Scholar]
- 15.Egan D, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy 7: 643–644, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell 10: 995–1005, 2002. [DOI] [PubMed] [Google Scholar]
- 17.Frost RA, Lang CH. Alteration of somatotropic function by proinflammatory cytokines. J Anim Sci 82 E-Suppl: E100–E109, 2004. [DOI] [PubMed] [Google Scholar]
- 18.Frost RA, Lang CH. mTor signaling in skeletal muscle during sepsis and inflammation: where does it all go wrong? Physiology 26: 83–96, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnacho-Montero J, Madrazo-Osuna J, García-Garmendia JL, Ortiz-Leyba C, Jiménez-Jiménez FJ, Barrero-Almodóvar A, Garnacho-Montero MC, Moyano-Del-Estad MR. Critical illness polyneuropathy: risk factors and clinical consequences. A cohort study in septic patients. Intensive Care Med 27: 1288–1296, 2001. [DOI] [PubMed] [Google Scholar]
- 20.Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J 25: 1028–1039, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon BS, Steiner JL, Lang CH, Jefferson LS, Kimball SR. Reduced REDD1 expression contributes to activation of mTORC1 following electrically induced muscle contraction. Am J Physiol Endocrinol Metab 307: E703–E711, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon BS, Williamson DL, Lang CH, Jefferson LS, Kimball SR. Nutrient-induced stimulation of protein synthesis in mouse skeletal muscle is limited by the mTORC1 repressor REDD1. J Nutr 145: 708–713, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 11: 859–871, 2006. [DOI] [PubMed] [Google Scholar]
- 24.Haddad F, Zaldivar F, Cooper DM, Adams GR. IL-6-induced skeletal muscle atrophy. J Appl Physiol (1985) 98: 911–917, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem 272: 26457–26463, 1997. [DOI] [PubMed] [Google Scholar]
- 26.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4: 648–657, 2002. [DOI] [PubMed] [Google Scholar]
- 27.Jefferson LS, Fabian JR, Kimball SR. Glycogen synthase kinase-3 is the predominant insulin-regulated eukaryotic initiation factor 2B kinase in skeletal muscle. Int J Biochem Cell Biol 31: 191–200, 1999. [DOI] [PubMed] [Google Scholar]
- 28.Kimball SR, Horetsky RL, Jefferson LS. Implication of eIF2B rather than eIF4E in the regulation of global protein synthesis by amino acids in L6 myoblasts. J Biol Chem 273: 30945–30953, 1998. [DOI] [PubMed] [Google Scholar]
- 29.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang CH, Frost RA. Sepsis-induced suppression of skeletal muscle translation initiation mediated by tumor necrosis factor alpha. Metabolism 56: 49–57, 2007. [DOI] [PubMed] [Google Scholar]
- 31.Lang CH, Frost RA, Vary TC. Acute alcohol intoxication increases REDD1 in skeletal muscle. Alcohol Clin Exp Res 32: 796–805, 2008. [DOI] [PubMed] [Google Scholar]
- 32.Lang CH, Frost RA, Vary TC. Regulation of muscle protein synthesis during sepsis and inflammation. Am J Physiol Endocrinol Metab 293: E453–E459, 2007. [DOI] [PubMed] [Google Scholar]
- 33.Lang CH, Silvis C, Deshpande N, Nystrom G, Frost RA. Endotoxin stimulates in vivo expression of inflammatory cytokines tumor necrosis factor alpha, interleukin-1beta, -6, and high-mobility-group protein-1 in skeletal muscle. Shock 19: 538–546, 2003. [DOI] [PubMed] [Google Scholar]
- 34.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol 17: 1807–1819, 2006. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 36.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab 10: 507–515, 2009. [DOI] [PubMed] [Google Scholar]
- 37.McFarlane C, Plummer E, Thomas M, Hennebry A, Ashby M, Ling N, Smith H, Sharma M, Kambadur R. Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF-kappaB-independent, FoxO1-dependent mechanism. J Cell Physiol 209: 501–514, 2006. [DOI] [PubMed] [Google Scholar]
- 38.McGhee NK, Jefferson LS, Kimball SR. Elevated corticosterone associated with food deprivation upregulates expression in rat skeletal muscle of the mTORC1 repressor, REDD1. J Nutr 139: 828–834, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyazaki M, Esser KA. REDD2 is enriched in skeletal muscle and inhibits mTOR signaling in response to leucine and stretch. Am J Physiol Cell Physiol 296: C583–C592, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mofarrahi M, Sigala I, Guo Y, Godin R, Davis EC, Petrof B, Sandri M, Burelle Y, Hussain SN. Autophagy and skeletal muscles in sepsis. PLoS One 7: e47265, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nystrom G, Pruznak A, Huber D, Frost RA, Lang CH. Local insulin-like growth factor I prevents sepsis-induced muscle atrophy. Metabolism 58: 787–797, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruznak AM, Kazi AA, Frost RA, Vary TC, Lang CH. Activation of AMP-activated protein kinase by 5-aminoimidazole-4-carboxamide-1-beta-d-ribonucleoside prevents leucine-stimulated protein synthesis in rat skeletal muscle. J Nutr 138: 1887–1894, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao S, Dennis M, Song X, Vadysirisack DD, Salunke D, Nash Z, Yang Z, Liesa M, Yoshioka J, Matsuzawa S, Shirihai OS, Lee RT, Reed JC, Ellisen LW. A REDD1/TXNIP pro-oxidant complex regulates ATG4B activity to control stress-induced autophagy and sustain exercise capacity. Nat Commun 6: 7014, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez J, Vernus B, Chelh I, Cassar-Malek I, Gabillard JC, Hadj Sassi A, Seiliez I, Picard B, Bonnieu A. Myostatin and the skeletal muscle atrophy and hypertrophy signaling pathways. Cell Mol Life Sci 71: 4361–4371, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandri M, Coletto L, Grumati P, Bonaldo P. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J Cell Sci 126: 5325–5333, 2013. [DOI] [PubMed] [Google Scholar]
- 46.Schmitz D, Wilsenack K, Lendemanns S, Schedlowski M, Oberbeck R. beta-Adrenergic blockade during systemic inflammation: impact on cellular immune functions and survival in a murine model of sepsis. Resuscitation 72: 286–294, 2007. [DOI] [PubMed] [Google Scholar]
- 47.Shenberger JS, Zhang L, Hughlock MK, Ueda T, Watanabe-Fukunaga R, Fukunaga R. Roles of mitogen-activated protein kinase signal-integrating kinases 1 and 2 in oxidant-mediated eIF4E phosphorylation. Int J Biochem Cell Biol 39: 1828–1842, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steiner JL, Lang CH. Sepsis attenuates the anabolic response to skeletal muscle contraction. Shock 43: 344–351, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tiao G, Fagan JM, Samuels N, James JH, Hudson K, Lieberman M, Fischer JE, Hasselgren PO. Sepsis stimulates nonlysosomal, energy-dependent proteolysis and increases ubiquitin mRNA levels in rat skeletal muscle. J Clin Invest 94: 2255–2264, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol 24: 6539–6549, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vary TC, Jefferson LS, Kimball SR. Amino acid-induced stimulation of translation initiation in rat skeletal muscle. Am J Physiol Endocrinol Metab 277: E1077–E1086, 1999. [DOI] [PubMed] [Google Scholar]
- 52.Williamson DL, Li Z, Tuder RM, Feinstein E, Kimball SR, Dungan CM. Altered nutrient response of mTORC1 as a result of changes in REDD1 expression: effect of obesity vs. REDD1 deficiency. J Appl Physiol 117: 246–256, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wray CJ, Mammen JM, Hershko DD, Hasselgren PO. Sepsis upregulates the gene expression of multiple ubiquitin ligases in skeletal muscle. Int J Biochem Cell Biol 35: 698–705, 2003. [DOI] [PubMed] [Google Scholar]
- 54.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 6: 472–483, 2007. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis 3: e386, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]